Abstract
Breast cancer is the second leading cause of cancer-related mortality in women. Successful development of sensitive nanoprobes for breast cancer cell detection is of great importance for breast cancer diagnosis and symptomatic treatment. Herein, inspired by the intrinsic peroxidase property of gold nanoclusters, high loading, and targeting ability of ErbB2/Her2 antibody functionalized liposomes, we report that gold nanoclusters-loaded, target-directed, functionalized liposomes can serve as a robust sensing platform for amplified colorimetric detection of HER2-positive breast cancer cells. This approach allows HER2-positive breast cancer cell identification at high sensitivity with high selectivity. In addition, the colorimetric “readout” offers extra advantages in terms of low-cost, portability, and easy-to-use applications. The practicality of this platform was further proved by successful detection of HER2-positive breast cancer cells in human serum samples and in breast cancer tissue, which indicated our proposed method has potential for application in cancer theranostics.
Keywords: Colorimetric sensor, signal amplification, liposomes, gold nanoclusters, artificial enzymes, breast cancer cells
Introduction
With one in twenty-nine women dying from the disease 1, 2, breast cancer is the second leading cause of cancer-related mortality among females worldwide 3-6. Early detection and diagnosis is of great importance for the improvement of treatability and curability of breast cancer 7. Human epidermal growth factor receptor 2 (HER2), which belongs to a family of the tyrosine kinase 8, is a 185 kDa transmembrane protein encoded by oncogene HER2/neu or c-erbB-2, and its overexpression is present in approximately 20%-30% of human breast cancers 9. As compared to other types of breast cancer, HER2-positive breast cancer, mainly due to HER2 gene amplification 10, is associated with increased malignancy, enhanced aggressive behavior, higher recurrence, poor prognosis and reduced overall survival 11-16. Therefore, developing a sensitive and accurate tool for detection of HER2 positive overexpressed breast cancer cells would be beneficial to cancer diagnosis, prognosis, monitoring of tumor sensitivity to anticancer drugs, and further symptomatic treatment 17-19.
Gold nanoclusters (AuNCs), as prominent fluorescence labels for sensitive detection of cancer cells, have attracted considerable interest because of their unique optical properties 20-28. For example, Wang et al. prepared the gold nanoclusters-reduced graphene oxide nanocomposites. It was observed that gold nanoclusters anchored on reduced graphene oxide retained their near-infrared fluorescent property for cancer cell imaging while reduced graphene oxide could carry anticancer agents such as doxorubicin for chemotherapy 23. In addition, Qi et al. developed the novel nanoconjugates containing of ultrasmall water-soluble gold nanoclusters as the fluorescent part and folic acid as the targeting ligand. The receptor specific cancer targeting ability and near-infrared fluorescence of the nanoconjugates make them as the promising candidates for optical-imaging-based cancer detection 24. These fluorescence imaging methods based on AuNCs have become an advanced and versatile tool for cancer research 29. However, they require fluorescence imaging equipment for readout, which make the fluorescence detection not adequate for the development of convenient household devices that can be practically suited for point-of-care applications.
Colorimetric detection is a simple and efficient method for determining targets by monitoring the color change 30-32. This colorimetric “readout” avoids the usage of any advanced or complicated instrumentation and thus may provide portable, low cost opportunities 33-35. A variety of colorimetric reagents, such as organic chromophoric probes 36-38 and plasmonic nanoparticle 39-41 have been used for visual detection of specific targets. For example, by the chemical dissolution of silver nanoparticles into silver ions, together with the subsequent induction of aggregation of alkyne-functionalized gold nanoparticles, Tian and Long et al. demonstrated a metal-linked immunosorbent assay (MeLISA) for the colorimetric detection of disease biomarkers in serum 42. Yang et al. also developed a novel method for the rapid, sensitive and selective colorimetric detection of copper ions as copper ions could decrease L-cysteine-induced gold nanoparticles aggregation. This platform could be efficiently used for colorimetric immunoassays 43. Specifically, due to the high sensitive and obvious color reaction, enzyme-based colorimetric detection systems are emerging as alternatives to conventional chromogenic sensors 44-46. As compared to natural enzymes, the artificial enzymes are attractive as colorimetric probes because of their easy preparation, low cost, high stability against stringent conditions, tunability in catalytic activities and long-term storage 47-49. Thus far, artificial enzyme-based biosensors are extremely appealing for a wide range of applications in environmental monitoring, disease diagnostics, as well as food and water safety 50-56. Among investigated host nanomaterials of artificial enzymes, gold nanoclusters (AuNCs) have drawn particular attention 57-60. Compared with other reported nanomaterial-based artificial enzymes, the AuNCs are more prominent for bioanalysis due to their easy modification, ultra-small size and excellent biocompatibility 47, 57. In addition, unlike nature enzymes, the BSA-AuNCs have strong robustness and can be used over a wide range of pH and temperature 57, which hold enormous potential for breast cancer cell detection.
Liposomes are nanoscale lipid vesicles consisting of single or multiple bilayered membrane structures with amphiphilic lipid molecules 61, 62. Since the first description in 1965 63-65, liposomes have attracted great interest as effective drug delivery systems, as well as versatile diagnostic and therapeutic tools for a lot of diseases. Liposomes exhibit good biodegradability and biocompatibility, outstanding size controllability, excellent surface tuning property, as well as high drug loading capacity 66-69. Up to now, a number of liposome formulations have been approved by the U.S. Food and Drug Administration (FDA) for clinical therapeutic purposes 69-71. As a platform to detect cancer cells, nanomaterial-based signal amplification systems have received increasing attention. These include the usage of nanomaterial-based artificial enzymes as amplifiers.
In light of the advantages of the liposome and artificial enzyme-based signal amplification techniques, we sought to develop a robust sensing platform for amplified colorimetric detection of HER2-positive breast cancer cells by utilizing artificial enzyme AuNCs-loaded liposomes. As illustrated in Scheme 1, we first obtain the AuNCs-loaded liposomes (AuNCs-LPs) by the traditional extrusion method. Subsequently, the HER2 antibodies (anti-HER2) can be anchored to the surface of AuNCs-LPs (BSA-AuNCs-LPs-anti-HER2) via EDC/NHS chemistry. Then the BSA-AuNCs-LPs-anti-HER2 can efficiently target the HER2-positive breast cancer cells via the cell-surface HER2 receptor recognition. After centrifugation and washes, the peroxidase-like activity of BSA-AuNCs-LPs-anti-HER2 adsorbed on HER2-positive breast cancer cells can be effectively utilized for quantitative colorimetric detection of HER2-positive breast cancer cells. Here, we provide the first evidence that the BSA-AuNCs-LPs-anti-HER2 can be applied as a novel signal amplification system for HER2-positive breast cancer cell detection. The combination of enzymatically catalyzed reactions with liposome-enrichment events resulted in a further improvement in cancer cell detection 72, highlighting the potential of BSA-AuNCs-LPs-anti-HER2 to serve as a promising diagnostic tool in oncology.
Scheme 1.
Schematic representation of (a) preparation of anti-HER2 conjugated liposome-AuNCs hybrid (BSA-AuNCs-LPs-anti-HER2) and (b) HER2-positive breast cancer cell detection by using BSA-AuNCs-LPs-anti-HER2.
Materials and Methods
Materials and measurements
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-dodecanoyl (N-dod-PE) and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) were purchased from Avanti Polar Lipids (Alabaster, AL). Human ErbB2/Her2 antibody (anti-HER2) was purchased from R&D Systems (Minneapolis, MN). 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) was purchased from Alfa Aesar. Sulfo-NHS (N-hydroxysulfosuccinimide) was purchased from Thermo Fisher Scientific (Pittsburgh, PA). Dulbecco's phosphate buffered saline (PBS) was purchased from Invitrogen (Carlsbad, CA). Aptamer was purchased from Elim Biopharmaceuticals, Inc. (Hayward, CA). The sequence of MUC1 aptamer was 5'-NH2-GCA GTT GAT CCT TTG GAT ACC CTG G-3'. Human serum was purchased from Sigma-Aldrich. Breast cancer tissue array with HER2 from 0, 1+, 2+ and 3+ were purchased from US Biomax. All other reagents were all of analytical reagent grade and used as received. Nanopure water (18.2 MΩ; Millpore Co., USA) was used throughout the experiment. Transmission electron microscopy (TEM) images were recorded using a JEOL 2100 transmission electron microscope operating with an accelerating voltage of 200 kV. Scanning electron microscopic (SEM) images were recorded using a Hitachi S-4700 scanning electron microscope. Zeta potential and size measurements were performed with a ZetaPALS zeta potential analyzer. Absorbance measurements were carried out by using a spectramax plus 384 microplate reader.
Synthesis of BSA-templated gold nanoclusters (BSA-AuNCs)
The BSA-templated gold nanoclusters (BSA-AuNCs) were synthesized by a previously one-pot green method 73. In a typical experiment, aqueous HAuCl4 solution (5 mL, 10 mM, 37 °C) was added to BSA solution (5 mL, 50 mg/mL, 37 °C) under vigorous stirring. NaOH solution (0.5 mL, 1 M) was introduced 2 min later, and the reaction was allowed to proceed under vigorous stirring at 37 °C for 12 h. The solution was then dialyzed in double distilled water to remove unreacted HAuCl4 and NaOH. The final solution was stored at 4 °C until use.
BSA-AuNCs-LPs-anti-HER2 preparation
The gold nanoclusters-loaded liposomes (AuNCs-LPs) were prepared by the extrusion method 74-79. Briefly, a mixture of DOPC:N-dod-PE (95:5, mol:mol) was solubilized in chloroform and dried in a rotary evaporator under reduced pressure. Then the lipid film was dissolved in the solution of gold nanoclusters (5 mg/mL, the concentration of BSA was adopted to confirm the concentration of BSA-AuNCs) while being agitated at 650 rpm with a stir bar to yield a 5 mM lipid solution. After that, liposomes were extruded via a NorthernLipids Extruder with a 200 nm polycarbonate nanoporous membrane. After extrusion, the solution of AuNCs-LPs was dialyzed in H2O using a Slide-A-Lyzer dialysis cassette (MWCO 300 kDa) overnight at room temperature (RT). Liposomes were conjugated to anti-HER2 or aptamer via the N-dod-PE anchor. EDC (2 mg) and NHS (3 mg) were mixed with 1 mmol of liposomes in PBS (pH 7.4) and incubated for 6 h at room temperature. A Slide-A-Lyzer dialysis cassette (MWCO 10 kDa) was used to remove unreacted EDC and NHS. Then anti-HER2 or aptamer was added to EDC-modified liposomes at a molar ratio of 1:1000 (antibody (aptamer):phospholipid) and incubated overnight at room temperature. Unreacted antibodies (aptamers) were removed using a Float-A-Lyzer G2 (MWCO 300 kDa).
To determine the antibody density of anti-HER2 on liposomes, 2 μm borosilicate beads (Duke Scientific, Palo Alto, CA), large enough to be detected by flow cytometry, were coated with a layer of lipids from liposomes by agitating small unilamellar liposomes with microbeads in PBS for 6 h. Then, microbeads were washed 3 times with PBS through centrifugation-suspension cycles to separate free liposomes. Conjugation of PE-anti-HER2 to microbead encapsulating liposomes was performed using EDC/NHS chemistry. The surface density of anti-HER2 conjugated to each microbead was determined with reference to Quantum Simply Cellular microbeads, which have defined numbers of antibody binding sites per bead 75, 78, 79.
Cell culture
MCF7, SKBR3, MDA-MB-231, MDA-MB-436, MDA-MB-468, MDA-MB-157, HCC1806, Hs578T and MCF10A cells were all obtained from American Type Culture Collection (ATCC, Manassas, VA). MCF7, MDA-MB-231 and MDA-MB-436 cells were cultured in DMEM (Corning, Corning, NY), supplemented with 10% FBS (Life Technologies, Carlsbad, CA) + 1% Pen/Strep (Life Technologies, Carlsbad, CA). SKBR3 cells were cultured in McCoy5a (Life Technologies, Carlsbad, CA) + 10% FBS. MDA-MB-468 and HCC1806 cells were cultured in RPMI-1640 (Corning) + 10% FBS. All cell lines were grown in a humidified atmosphere (5% CO2) at 37 °C. MDA-MB-157 cells were cultured in L-15 (ATCC) + 10% FBS. Hs578T cells were cultured in DMEM, supplemented with 10% FBS + 10 mg/ml of Insulin. MCF10A cells were cultured in DMEM/F12 (Life Technologies, Carlsbad, CA) + 5% Horse Serum (Life Technologies, Carlsbad, CA) + 20 ng/ml of Epidermal Growth Factor (EGF) (Peprotech, NJ) + 0.5 mg/ml of Hydrocortisone (Sigma-Aldrich, St. Louis, MO) + 100 ng/ml Cholera Toxin (Sigma-Aldrich, St. Louis, MO) + 10 mg/ml of Insulin (Sigma-Aldrich, St. Louis, MO) + 1% Pen/Strep (Life Technologies, Carlsbad, CA).
Cytotoxicity assays
MTT assays were used to probe cellular viability. SKBR3 Cells were seeded at a density of 5000 cells well-1 (100 μL total volume well-1) in 96-well assay plates. After 24 h incubation, the as-prepared liposome, BSA-AuNCs and BSA-AuNCs-LPs-anti-HER2, at the indicated concentrations, were added for further incubation of 24 h. To determine toxicity, 10 μL of MTT solution was added to each well of the microtiter plate and the plate was incubated in the CO2 incubator for an additional 4 h. Then the cells were lysed by the addition of 100 μL of DMSO. Absorbance values of formazan were determined with microplate reader at 490 nm (corrected for background absorbance at 630 nm). Three replicates were done for each treatment group.
Bioassay
In a typical test, chemicals were added into 100 μL buffer solution (PBST, 25 mM Na2HPO4, pH 4.0, with 0.05% Tween-20) in an order of certain amounts of the BSA-AuNCs-LPs-anti-HER2, 2 μL TMB (final concentration 800 μM), and 6.2 μL H2O2 (final concentration 50 mM) at 25 °C. After 30 min reaction, the peroxidase activities of the mixtures were carried out by using a spectramax plus 384 microplate reader.
Breast cancer cell detection
Cells were first harvested by centrifugation at 1000 rpm for 5 min, and washed three times with phosphate buffered saline (PBS, pH 7.4). For the colorimetric detection, BSA-AuNCs-LPs-anti-HER2 were incubated with different cells lines at 37 °C for 1.5 h, then harvested by centrifugation at 1000 rpm, and washed three times with phosphate buffered saline (PBS, pH 7.4). After that, cells were dispersed in detection buffer (PBST, 25 mM Na2HPO4, pH 4.0, with 0.05% Tween-20). Then, TMB (final concentration 800 μM) and H2O2 (final concentration 50 mM) were added at 25 °C. After 30 min reaction, the peroxidase activities of the mixtures were determined.
Cancer cell detection in human serum samples
Human serum was diluted with PBS buffer with ratio of 1:1 before being spiked with SKBR3 cells at different concentrations. The as-prepared BSA-AuNCs-LPs-anti-HER2 was then used to detect SKBR3 cells in diluted serum following the same procedure as that for cancer cell detection.
Detection of HER2-positive breast cancer tissues
For disaggregation, breast cancer tissues with HER2 from 0, 1+, 2+ and 3+ were first digested with 2 mg/ml of collagenase, type IV, 2 mg/mL hyaluronidase and 1 mg/mL DNase for 30 minutes at 37 °C, with intermittent shaking 80-84. After filtering, the resulting cell suspensions were washed three times with phosphate buffered saline (PBS, pH 7.4). For the colorimetric detection, BSA-AuNCs-LPs-anti-HER2 were incubated with different cancer cells (20000 cells) at 37 °C for 1.5 h, then harvested by centrifugation at 1000 rpm, and washed three times with phosphate buffered saline (PBS, pH 7.4). After that, cancer cells were dispersed in detection buffer (PBST, 25 mM Na2HPO4, pH 4.0, with 0.05% Tween-20). Then, TMB (final concentration 800 μM) and H2O2 (final concentration 50 mM) were added at 25 °C. After 30 min reaction, the peroxidase activities of the mixtures were determined.
Results and discussion
Synthesis and characterization of AuNCs-LPs
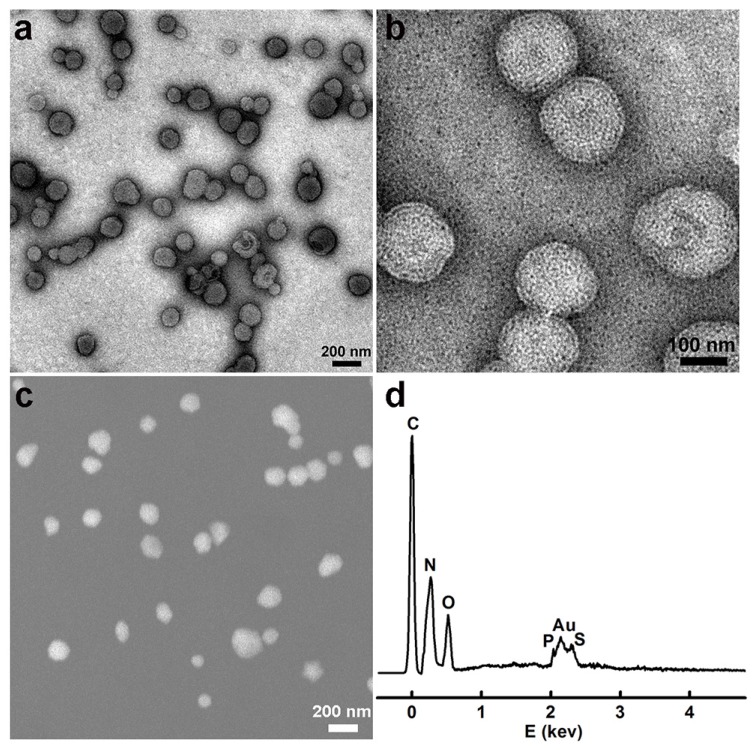
We first synthesized the gold nanoclusters-loaded liposomes (AuNCs-LPs) using the lipid film hydration and membrane extrusion method 85, 86. AuNCs-LPs assembled from a mixture of DOPC:N-dod-PE (95:5, mol:mol), encapsulating AuNCs (Figure S1) in the liposome formulations with the ratio of about 1925 : 1 (AuNCs : liposome) (Figure S2) 87-89. The formation of AuNCs-LPs was verified with transmission electron microscopy (TEM) and scanning electron microscopic (SEM) measurements (Figure S3 and S4). AuNCs-LPs had spherical structures with an average diameter around 150 nm. Dynamic light scattering (DLS) indicated that AuNCs-LPs were narrowly dispersed and the diameters of unconjugated AuNCs-LPs were 175.04 ± 2.45 nm. The smaller size from TEM/SEM observations as compared with the DLS measurement may be due to dehydration during the TEM/SEM sample preparation process 90-92. Zeta potential measurements revealed that the AuNCs-LPs were highly negatively charged (-37.67 ± 0.32 mV). Afterwards, anti-HER2 was conjugated to the AuNCs-LPs through EDC/NHS chemistry (BSA-AuNCs-LPs-anti-HER2). TEM and SEM images (Figure 1a-c) indicated the BSA-AuNCs-LPs-anti-HER2 remained spherical and monodispersed. The loading of AuNCs in the liposome was confirmed by energy dispersive X-ray spectroscopy (EDS). As shown in Figure 1d, the EDS measurement of the as-prepared nanohybrid showed primarily Au signals, suggesting that the vast majority of AuNCs incorporated in the liposome. The diameters of BSA-AuNCs-LPs-anti-HER2 were 173.80 ± 1.80 nm as indicated by DLS analysis, which were similar to the AuNCs-LPs. And the surface charge of BSA-AuNCs-LPs-anti-HER2 was altered from -37.67 mV for AuNCs-LPs to -36.17 mV. The antibody density was quantified using standardized microbeads of known density (Table 1). The average antibody density was 1680 ± 20 molecules/μm2 or 159 ± 2 molecules/liposome.
Figure 1.
(a, b) TEM images of the BSA-AuNCs-LPs-anti-HER2 under different magnifications; (c) SEM images of the BSA-AuNCs-LPs-anti-HER2; (d) EDS spectra of the as-synthesized BSA-AuNCs-LPs-anti-HER2.
Table 1.
Characterization of liposome, AuNCs and liposome-AuNCs hybrids.
| Size (DLS, nm) | Polydispersity Index (PDI) | Zeta potential (mV) | HER2 antibody density (molecules/μm2) | |
|---|---|---|---|---|
| liposome | 162.44 ± 1.49 | 0.082 | -38.04 ± 0.29 | |
| BSA-AuNCs | 5.82 ± 0.92 | 0.174 | -19.71 ± 2.08 | |
| BSA-AuNCs dispersed in PBST | 5.76 ± 0.87 | 0.168 | ||
| AuNCs-LPs | 175.04 ± 2.45 | 0.112 | -37.67 ± 0.32 | |
| BSA-AuNCs-LPs-anti-HER2 | 173.80 ± 1.80 | 0.084 | -36.17 ± 0.85 | 1680 ± 20 |
| BSA-AuNCs-LPs-AptMUC1 | 174.12 ± 2.26 | 0.106 | -37.32 ± 0.48 |
Peroxidase-like catalytic activity of BSA-AuNCs-LPs-anti-HER2
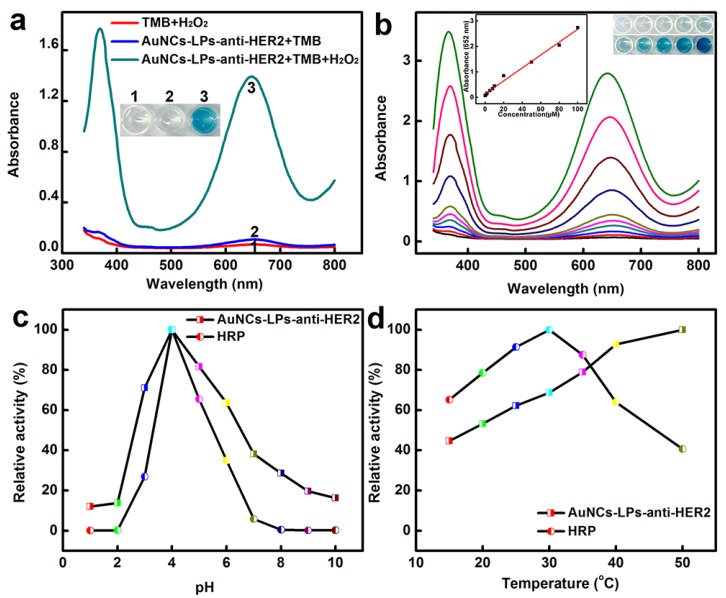
The peroxidase-like activity of BSA-AuNCs-LPs-anti-HER2 was evaluated in the catalysis of peroxidase substrates 3,3′,5,5′-tetramethylbenzidine (TMB). BSA-AuNCs-LPs-anti-HER2 in PBST could catalyze the oxidation of TMB in the presence of H2O2, and produce a deep blue color, with maximum absorbance at 652 nm 93. Tween-20 in PBST could effectively cause the breakdown of the liposomes and the release of AuNCs (Figure S5), leading to the enzymatic colorimetric reactions. TEM in Figure S6 confirmed the breakdown of the liposomes in the presence of PBST. In contrast, BSA-AuNCs-LPs-anti-HER2 or H2O2 alone did not produce the significant color change (Figure 2a). These results confirmed that the peroxidase-like activity toward TMB came from the intrinsic catalytic property of AuNCs in BSA-AuNCs-LPs-anti-HER2. Figure 2b shows the absorbance changes against different concentrations of BSA-AuNCs-LPs-anti-HER2. Dramatic improvement in catalytic activity was observed with the steady increase of the concentration of the BSA-AuNCs-LPs-anti-HER2. To optimize the enzymatic reaction of the BSA-AuNCs-LPs-anti-HER2, we investigated the time-dependent absorbance evolution at 652 nm of the as-prepared BSA-AuNCs-LPs-anti-HER2 incubated with TMB and H2O2 solutions. The characteristic absorbance at 652 nm increased gradually with time and reached a plateau at ≈ 30 min (Figure S7). Thus, 30 min was selected as the incubation time. Furthermore, like other nanomaterial-based peroxidase mimics, the activity of BSA-AuNCs-LPs-anti-HER2 was also dependent on pH (Figure 2c), temperature (Figure 2d) and H2O2 concentrations (Figure S8).
Figure 2.
(a) The absorbance spectra and visual color changes of TMB in different reaction systems: (1) TMB + H2O2, (2) BSA-AuNCs-LPs-anti-HER2 + TMB, and (3) BSA-AuNCs-LPs-anti-HER2 + TMB + H2O2 in PBST (25 mM Na2HPO4, pH 4.0, with 0.05% Tween-20) at 25 °C after 30 min incubation. (b) The absorbance spectra and visual color changes of TMB in presence of different concentrations of BSA-AuNCs-LPs-anti-HER2 after 30 min incubation. Inset: the absorption values at 652 nm depend on the concentrations of BSA-AuNCs-LPs-anti-HER2. (c, d) The peroxidase-like activity of the BSA-AuNCs-LPs-anti-HER2 is dependent on pH (c) and temperature (d).
Breast cancer cell detection using AuNCs-LPs
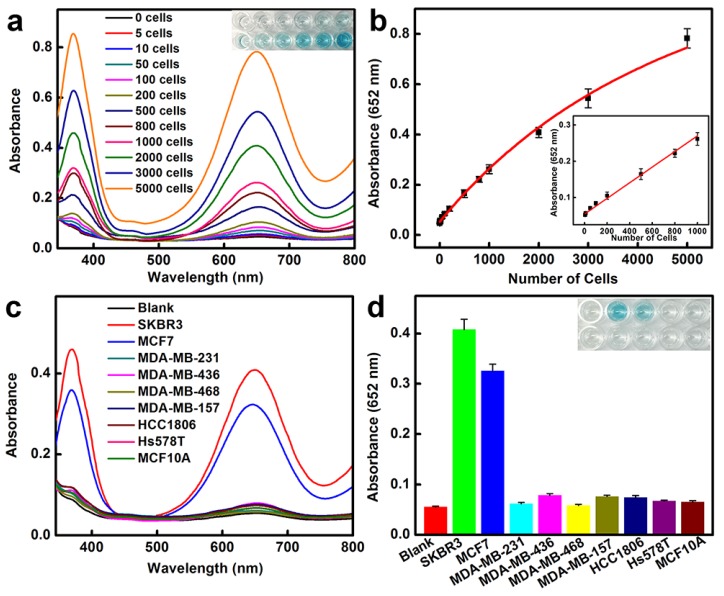
We then applied the peroxidase-like activity of BSA-AuNCs-LPs-anti-HER2 for quantitative colorimetric detection of cancer cells (Figure 3a). Upon conjugation of anti-HER2 to the AuNCs-LPs, the BSA-AuNCs-LPs-anti-HER2 could serve as a novel probe for selective, quantitative, and fast colorimetric detection of HER2-positive breast cancer cells. The BSA-AuNCs-LPs-anti-HER2 showed negligible cytotoxicity to the cells as shown by MTT results (Figure S9). To calculate the sensitivity, different amounts of HER2-positive SKBR3 cells 94 were first incubated with BSA-AuNCs-LPs-anti-HER2 in PBS for 1.5 hours and subsequently centrifuged. The precipitate was collected and rinsed with PBS three times to remove the unattached BSA-AuNCs-LPs-anti-HER2. The control experiments indicated that free BSA-AuNCs-LPs-anti-HER2 could be completely removed from the HER2-positive breast cancer cells by centrifugation (Figure S10). In the presence of TMB and H2O2, the cell conjugated BSA-AuNCs-LPs-anti-HER2 in PBST would catalyze a color reaction that could be judged by the naked eye and easily be quantitatively monitored by UV-Vis absorbance spectrometry. Figure 3a showed absorbance changes as a function of cell number. Results indicated an increase in the absorbance with increasing number of SKBR3 cells, which suggested that more BSA-AuNCs-LPs-anti-HER2 bound to HER2 receptor on the surface of SKBR3 cells. The absorbance of TMB at 652 nm linearly depends on the number of target cells ranging from 5 cells to 1000 cells, with a detect limit as low as 5 cells (Figure 3b). As compared with BSA-AuNCs-anti-HER2 without the lipid component, the detection sensitivity of BSA-AuNCs-LPs-anti-HER2 is significantly enhanced because of the liposome-enrichment method (Figure S11). Similarly, the ability of BSA-AuNCs-LPs-anti-HER2 to detect the other type of HER2-positive breast cancer cell line MCF7 95 was also proved (Figure S12). This colorimetric detection method also demonstrated the specificity of BSA-AuNCs-LPs-anti-HER2 for HER2-positive breast cancer cells (Figure 3c, d). In our experiments, nine different human breast cell lines were used, including one nonneoplastic, fibrocystic breast cell line (MCF10A), two HER2-positive breast cancer cell lines (SKBR3 and MCF7) and six triple negative breast cancer cell lines (MDA-MB-231, MDA-MB-436, MDA-MB-468, MDA-MB-157, HCC1806, Hs578T) 6. Following 1.5 h in culture, BSA-AuNCs-LPs-anti-HER2 showed much stronger binding to HER2-positive breast cancer cells than other cells. The absorbance changes for SKBR3 and MCF7 cells were more than 4 times higher than those for other breast cells. The differences were due to the high levels of HER2 receptor expression on SKBR3 and MCF7 cells.
Figure 3.
HER2-positive breast cancer cell detection by using BSA-AuNCs-LPs-anti-HER2. (a) The absorbance spectra and visual color changes upon analyzing different number of SKBR3 cells. (b) The absorption values at 652 nm depend on the number of SKBR3 cells. Inset: the linear plot. The error bars represent the standard deviation of three measurements. (c-d) Selectivity analysis for HER2-positive breast cancer cell detection using BSA-AuNCs-LPs-anti-HER2 by monitoring the absorbance spectra (c) and color changes (d). The error bars represent the standard deviation of three measurements.
In order to further broaden the utility of AuNCs-LPs-based colorimetric detection system, we used an aptamer instead of antibody as the recognition agent for cancer cell detection. Aptamers, an emerging class of targeting ligands selected by an in vitro iterative process called systematic evolution of ligands by exponential enrichment (SELEX) 96, are well recognized as an alternative to antibodies 97. Aptamers consist of single-stranded oligonucleotides; unique sequences may exhibit high affinity to a broad range of targets including metal ions, organic molecules, amino acids, peptides, proteins, and even whole cells and bacteria. Aptamers have certain advantages as target ligands, such as ease of synthesis and modification 98, relatively small size, excellent affinity, outstanding specificity, and lack of immunogenicity 99, which may serve as excellent targeting ligands in the construction of biosensors 100-104, biosystem imaging 105-107 and targeted therapeutic systems 108-111. MCF7 cells were selected as model cells to prove the viability of the developed analytical approach as MCF7 cells represent one of the most widely used experimental models on breast cancer studies 112. The tumor biomarker MUC1 is a transmembrane glycoprotein, whose expression increases at least 10-fold on the surface of MCF7 cells when compared to normal breast cells 7, 108, 113. The 25-base oligonucleotide aptamers of MUC1 (AptMUC1) presented high affinity to MUC1 proteins 7, 108, 114 and were conjugated to the AuNCs-LPs (BSA-AuNCs-LPs-AptMUC1) (Table 1, Figure S13) to serve as the recognition agent for selective MCF7 detection. The sensitivity of our provided strategy was investigated by monitoring the absorbance change of TMB in the presence of different numbers of MCF7 cells (Figure S14). In the absence of the target cancer cell (MCF7 cell), the BSA-AuNCs-LPs-AptMUC1 would be removed from the detection system by centrifugation and wash steps. In the presence of MCF7 cell, the BSA-AuNCs-LPs-AptMUC1 would efficiently adsorb onto the target cells for following colorimetric analysis. A target-concentration-dependent signal intensity was observed in the MCF7 cells. With our proposed strategy, a high detection sensitivity of 20 cells can be achieved on the basis of the synergistic action of high catalytic activity of AuNCs and high recognition ability of AptMUC1 to MUC1 on the MCF7 cells 115. The detection sensitivity of BSA-AuNCs-LPs-AptMUC1 is much higher as compared with AuNCs-AptMUC1 without the lipid component (Figure S15). These results demonstrate that the proposed assay presents a sensitive sensing platform for breast cancer cell detection.
Determination of HER2-positive breast cancer cells in biological fluids and HER2-positive breast cancer tissues
The high sensitivity combined with excellent specificity of BSA-AuNCs-LPs-anti-HER2 to HER2-positive breast cancer cells indicated that our strategy might be directly applied for detecting HER2-positive breast cancer cells in real samples. Therefore, we examined the practicality of the assay for detection of SKBR3 cells in human serum samples. As can be seen in Figure 4a-b, the absorbance of TMB at 652 nm linearly depends on the number of target cells ranging from 5 to 1000 cells and the slope of this curve is very close to that of SKBR3 cell detection in buffer solution from Figure 3b. To further explore the feasibility of the developed method for clinical analysis, the as-prepared BSA-AuNCs-LPs-anti-HER2 was used to detect HER2-positive breast cancer tissues (Figure 4c-h). Immunohistochemistry (IHC) images shown in Figure 4c-f indicated the breast cancer tissues with HER2 from 0, 1+, 2+ and 3+. Samples scored as 0 or 1+ were considered negative for HER2 overexpression, 2+ was weak positive and 3+ was strong positive, with complete membrane staining in ≥10% of tumor cells 8, 116. As compared to samples of HER2-negative breast tissue, the samples of HER2-positive breast cancer tissue showed much higher absorbance signal at 652 nm owing to the BSA-AuNCs-LPs-anti-HER2 oxidation of TMB (Figure 4g-h). The response enhanced with increase in HER2 IHC score indicating that the developed biosensor is able to effectively detect HER2-positive breast cancer tissue, hence valuable in the clinical diagnosis. All these results reveal the practicality of using BSA-AuNCs-LPs-anti-HER2 for the detection of HER2-positive breast cancer cell and tissue in real samples. Compared with other probes for the detection of HER2-positive breast cancer cells (Table 2) 117-124, our probe has the advantages of easy preparation, excellent compatibility, high sensitivity and low cost, which holds great promise in cancer theranostics.
Figure 4.
(a-b) HER2-positive breast cancer cell detection by using BSA-AuNCs-LPs-anti-HER2 in human serum (50%). (a) The absorbance spectra upon analyzing different number of SKBR3 cells. (b) The absorption values at 652 nm depend on the number of SKBR3 cells. Inset: the linear plot. The error bars represent the standard deviation of three measurements. (c-f) Immunohistochemistry (IHC) staining of breast cancer tissues with HER2 from 0 (c), 1+ (d), 2+ (e) and 3+ (f). Scale bar = 100 μm. (g-h) HER2-positive breast cancer tissue detection by using BSA-AuNCs-LPs-anti-HER2. The absorbance spectra response (g) and absorption values at 652 nm (h) enhanced with increase in HER2 IHC score (0, 1+, 2+, 3+).
Table 2.
Comparison of methods for detection of HER2-positive breast cancer cells.
| Probe | Detection method | Cell type and detection limit | Assay time | Application | Ref |
|---|---|---|---|---|---|
| Herceptin-conjugated liquid crystal microdroplets | Polarized optical microscopic images | SKBR3 | 3 h | Detection of SKBR3 cells in biological fluids | [117] |
| Anti-HER2 IgY antibody-functionalized single-walled carbon nanotubes | Raman signal | SKBR3 | 24 h | Detection and destruction of breast cancer cells | [118] |
| Anti-Her2 antibody conjugated superparamagnetic iron oxide nanoparticles | Superconducting quantum interference device-detected magnetic relaxometry | MCF7/Her2-18 (940,000 cells at a depth of 4.5 cm) | 16 min | In vivo breast tumor cell detection | [119] |
| Herceptin-conjugated super paramagnetic iron oxide nanoparticles | Iron staining with Prussian blue staining method |
SKBR3 and T47D | 1 h | Detection tumor cells | [120] |
| Polyhedral oligomeric silsesquioxanes-containing conjugated polymer loaded PLGA nanoparticles with trastuzumab (herceptin) functionalization | Fluorescence imaging | SKBR3 | 2 h | Targeted biological imaging and detection | [121] |
| EP1045Y conjugated CdSeTe/CdS/ZnS quantum dots | Fluorescence imaging | MCF7 | 2 h | Identify the HER2 overexpressed breast tumor subtype in tumor-bearing mouse | [122] |
| HER2-specific monoclonal antibodies conjugated quantum dots | Fluorescence imaging | SKBR3 | 2 h | Detection of HER2 expression in breast cancer cells and tissues | [123] |
| Multifunctional (monoclonal anti-HER2/c-erb-2 antibody and S6 RNA aptamer-conjugated) oval-shaped gold-nanoparticle-based nanoconjugate | A simple colorimetric and highly sensitive two-photon scattering assay | SKBR3 (100 cells/mL) | 30 min | Diagnosis of cancer cell lines | [124] |
| HER2 antibodies anchored AuNCs-loaded liposomes | Colorimetric detection | SKBR3 (5 cells) | 2 h | Detection of HER2-positive breast cancer cells in human serum samples and in breast cancer tissue | Our work |
Conclusion
In summary, we have demonstrated a naked-eye detection of HER2-positive breast cancer cells using peroxidase mimetic-based colorimetric method combined with signal amplification via gold nanoclusters-loaded liposomes (AuNCs-LPs). With the conjugation of ErbB2/Her2 antibody (anti-HER2) to the AuNCs-LPs, the nanostructures can be applied for the highly sensitive detection of the HER2-positive breast cancer cell lines SKBR3 and MCF7, as well as discrimination of SKBR3 and MCF7 cells from other different breast cell lines. In addition, aptamer instead of antibody was also utilized as the recognition probe for breast cancer cell detection, which proved university of our platform. This strategy is simple and economic in operation, and no sophisticated experimental techniques or equipment are required. Furthermore, the usage of the target-specific antibody or aptamer imparts good selectivity, and the application of the amplified colorimetric method also brings excellent sensitivity. More importantly, the proposed method can be successfully used to detect HER2-positive breast cancer cells in human serum samples and in breast cancer tissue. Therefore, we expect that this approach may offer a new approach for developing low cost, sensitive and simple sensor for detecting of HER2-positive breast cancer cells, and will be extraordinarily beneficial in a wide spectrum of applications including clinical diagnostics, bioanalysis, and bionanotechnology.
Supplementary Material
Supplementary figures.
Acknowledgments
The authors would like to acknowledge funding from the NIH (NCI DP2 CA174495-01).
Abbreviations
- HER2
human epidermal growth factor receptor 2
- AuNCs
gold nanoclusters
- FDA
food and drug administration
- AuNCs-LPs
AuNCs-loaded liposomes
- anti-HER2
HER2 antibodies
- BSA-AuNCs-LPs-anti-HER2
anti-HER2 conjugated liposome-AuNCs hybrid
- N-dod-PE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-dodecanoyl
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- EDC
1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride
- Sulfo-NHS
N-hydroxysulfosuccinimide
- PBS
phosphate buffered saline
- TEM
transmission electron microscopy
- SEM
scanning electron microscopic
- BSA-AuNCs
BSA-templated gold nanoclusters
- EGF
epidermal growth factor
- DLS
dynamic light scattering
- TMB
3,3′,5,5′-tetramethylbenzidine
- SELEX
systematic evolution of ligands by exponential enrichment
- AptMUC1
aptamer of MUC1
- BSA-AuNCs-LPs-AptMUC1
AptMUC1 conjugated to the AuNCs-LPs
- IHC
immunohistochemistry.
References
- 1.Lainé A-L, Adriaenssens E, Vessières A, Jaouen G, Corbet C, Desruelles E. et al. The in vivo performance of ferrocenyl tamoxifen lipid nanocapsules in xenografted triple negative breast cancer. Biomaterials. 2013;34:6949–56. doi: 10.1016/j.biomaterials.2013.05.065. [DOI] [PubMed] [Google Scholar]
- 2.Seib FP, Kaplan DL. Doxorubicin-loaded silk films: Drug-silk interactions and in vivo performance in human orthotopic breast cancer. Biomaterials. 2012;33:8442–50. doi: 10.1016/j.biomaterials.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckhardt BL, Francis PA, Parker BS, Anderson RL. Strategies for the discovery and development of therapies for metastatic breast cancer. Nat Rev Drug Discov. 2012;11:479–97. doi: 10.1038/nrd2372. [DOI] [PubMed] [Google Scholar]
- 4.He X, Bao X, Cao H, Zhang Z, Yin Q, Gu W. et al. Tumor-penetrating nanotherapeutics loading a near-infrared probe inhibit growth and metastasis of breast cancer. Adv Funct Mater. 2015;25:2831–9. [Google Scholar]
- 5.You J-O, Guo P, Auguste DT. A drug-delivery vehicle combining the targeting and thermal ablation of HER2+ breast-cancer cells with triggered drug release. Angew Chem Int Ed. 2013;52:4141–6. doi: 10.1002/anie.201209804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao Y, Auguste DT. Array-based identification of triple-negative breast cancer cells using fluorescent nanodot-graphene oxide complexes. Biosens Bioelectron. 2016;81:431–7. doi: 10.1016/j.bios.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Wu P, Gao Y, Zhang H, Cai C. Aptamer-guided silver-gold bimetallic nanostructures with highly active surface-enhanced Raman scattering for specific detection and near-infrared photothermal therapy of human breast cancer cells. Anal Chem. 2012;84:7692–9. doi: 10.1021/ac3015164. [DOI] [PubMed] [Google Scholar]
- 8.Joshi BP, Zhou J, Pant A, Duan X, Zhou Q, Kuick R. et al. Design and synthesis of near-infrared peptide for in vivo molecular imaging of HER2. Bioconjug Chem. 2016;27:481–94. doi: 10.1021/acs.bioconjchem.5b00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakha EA, Pinder SE, Bartlett JMS, Ibrahim M, Starczynski J, Carder PJ. et al. Updated UK recommendations for HER2 assessment in breast cancer. J Clin Pathol. 2015;68:93–9. doi: 10.1136/jclinpath-2014-202571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkorian K, Ellis A, Twyman LJ. Catalytic hyperbranched polymers as enzyme mimics; exploiting the principles of encapsulation and supramolecular chemistry. Chem Soc Rev. 2012;41:6138–59. doi: 10.1039/c2cs35238a. [DOI] [PubMed] [Google Scholar]
- 11.Garrett JT, Arteaga CL. Resistance to HER2-directed antibodies and tyrosine kinase inhibitors: Mechanisms and clinical implications. Cancer Biol Ther. 2011;11:793–800. doi: 10.4161/cbt.11.9.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke T, Reeves J, Lanigan A, Stanton P. HER2 as a prognostic and predictive marker for breast cancer. Ann Oncol. 2001;12:S23–S8. doi: 10.1093/annonc/12.suppl_1.s23. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Duan J-H, Song Y-M, Ma J, Wang F-D, Lu X. et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Transl Med. 2012;10:148. doi: 10.1186/1479-5876-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Xia H-S, Gong Y-P, Peng J, Peng C-W, Hu M-B. et al. The quantitative detection of total HER2 load by quantum dots and the identification of a new subtype of breast cancer with different 5-year prognosis. Biomaterials. 2010;31:8818–25. doi: 10.1016/j.biomaterials.2010.07.091. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Sun S-R, Gong Y-P, Qi C-B, Peng C-W, Yang X-Q. et al. Quantum dots-based molecular classification of breast cancer by quantitative spectroanalysis of hormone receptors and HER2. Biomaterials. 2011;32:7592–9. doi: 10.1016/j.biomaterials.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 16.Slamon D, Godolphin W, Jones L, Holt J, Wong S, Keith D. et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–12. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 17.Etayash H, Jiang K, Azmi S, Thundat T, Kaur K. Real-time detection of breast cancer cells using peptide-functionalized microcantilever arrays. Sci Rep. 2015;5:13967. doi: 10.1038/srep13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129–44. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 19.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Hu Y, Cen Y, Chu X, Lu Y. A dual-emission fluorescent nanocomplex of gold-cluster-decorated silica particles for live cell imaging of highly reactive oxygen species. J Am Chem Soc. 2013;135:11595–602. doi: 10.1021/ja4035939. [DOI] [PubMed] [Google Scholar]
- 21.Su X, Jiang H, Wang X. Thiols-induced rapid photoluminescent enhancement of glutathione-capped gold nanoparticles for intracellular thiols imaging applications. Anal Chem. 2015;87:10230–6. doi: 10.1021/acs.analchem.5b02559. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Cui Y, Zhao Y, Liu R, Sun Z, Li W. et al. Bifunctional peptides that precisely biomineralize Au clusters and specifically stain cell nuclei. Chem Commun. 2012;48:871–3. doi: 10.1039/c1cc15926g. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Li J, Amatore C, Chen Y, Jiang H, Wang X-M. Gold nanoclusters and graphene nanocomposites for drug delivery and imaging of cancer cells. Angew Chem Int Ed. 2011;50:11644–8. doi: 10.1002/anie.201105573. [DOI] [PubMed] [Google Scholar]
- 24.Qiao J, Mu X, Qi L, Deng J, Mao L. Folic acid-functionalized fluorescent gold nanoclusters with polymer as linker for cancer cells imaging. Chem Commun. 2013;49:8030–2. doi: 10.1039/c3cc44256j. [DOI] [PubMed] [Google Scholar]
- 25.Liu C-L, Wu H-T, Hsiao Y-H, Lai C-W, Shih C-W, Peng Y-K. et al. Insulin-directed synthesis of fluorescent gold nanoclusters: Preservation of insulin bioactivity and versatility in cell imaging. Angew Chem Int Ed. 2011;50:7056–60. doi: 10.1002/anie.201100299. [DOI] [PubMed] [Google Scholar]
- 26.He H, Xie C, Ren J. Nonbleaching fluorescence of gold nanoparticles and its applications in cancer cell imaging. Anal Chem. 2008;80:5951–7. doi: 10.1021/ac8005796. [DOI] [PubMed] [Google Scholar]
- 27.Shang L, Dorlich RM, Brandholt S, Schneider R, Trouillet V, Bruns M. et al. Facile preparation of water-soluble fluorescent gold nanoclusters for cellular imaging applications. Nanoscale. 2011;3:2009–14. doi: 10.1039/c0nr00947d. [DOI] [PubMed] [Google Scholar]
- 28.Wang H-H, Lin C-AJ, Lee C-H, Lin Y-C, Tseng Y-M, Hsieh C-L. et al. Fluorescent gold nanoclusters as a biocompatible marker for in vitro and in vivo tracking of endothelial cells. ACS Nano. 2011;5:4337–44. doi: 10.1021/nn102752a. [DOI] [PubMed] [Google Scholar]
- 29.Bi S, Ji B, Zhang Z, Zhang S. A chemiluminescence imaging array for the detection of cancer cells by dual-aptamer recognition and bio-bar-code nanoprobe-based rolling circle amplification. Chem Commun. 2013;49:3452–4. doi: 10.1039/c3cc39296a. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Xu Z, Yoon J. Fluorescent and colorimetric chemosensors for detection of nucleotides, FAD and NADH: highlighted research during 2004-2010. Chem Soc Rev. 2011;40:2222–35. doi: 10.1039/c0cs00169d. [DOI] [PubMed] [Google Scholar]
- 31.Lodeiro C, Capelo JL, Mejuto JC, Oliveira E, Santos HM, Pedras B. et al. Light and colour as analytical detection tools: A journey into the periodic table using polyamines to bio-inspired systems as chemosensors. Chem Soc Rev. 2010;39:2948–76. doi: 10.1039/b819787n. [DOI] [PubMed] [Google Scholar]
- 32.Du J, Zhu B, Chen X. Urine for Plasmonic nanoparticle-based colorimetric detection of mercury ion. Small. 2013;9:4104–11. doi: 10.1002/smll.201300593. [DOI] [PubMed] [Google Scholar]
- 33.Xia F, Zuo XL, Yang RQ, Xiao Y, Kang D, Vallee-Belisle A. et al. Colorimetric detection of DNA, small molecules, proteins, and ions using unmodified gold nanoparticles and conjugated polyelectrolytes. P Natl Acad Sci USA. 2010;107:10837–41. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109:1948–98. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali MM, Li Y. Colorimetric sensing by using allosteric-DNAzyme-coupled rolling circle amplification and a peptide nucleic acid-organic dye probe. Angew Chem Int Ed. 2009;48:3512–5. doi: 10.1002/anie.200805966. [DOI] [PubMed] [Google Scholar]
- 36.Coll C, Martínez-Máñez R, Marcos MD, Sancenón F, Soto J. A simple approach for the selective and sensitive colorimetric detection of anionic surfactants in water. Angew Chem Int Ed. 2007;46:1675–8. doi: 10.1002/anie.200603800. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JF, Zhou Y, Yoon J, Kim JS. Recent progress in fluorescent and colorimetric chemosensors for detection of precious metal ions (silver, gold and platinum ions) Chem Soc Rev. 2011;40:3416–29. doi: 10.1039/c1cs15028f. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Yoon J. Recent progress in fluorescent and colorimetric chemosensors for detection of amino acids. Chem Soc Rev. 2012;41:52–67. doi: 10.1039/c1cs15159b. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Z, Wu C, Liu H, Zou Y, Zhang X, Kang H. et al. An aptamer cross-linked hydrogel as a colorimetric platform for visual detection. Angew Chem Int Ed. 2010;49:1052–6. doi: 10.1002/anie.200905570. [DOI] [PubMed] [Google Scholar]
- 40.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–40. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 41.Kato D, Oishi M. Ultrasensitive detection of DNA and RNA based on enzyme-free click chemical ligation chain reaction on dispersed gold nanoparticles. ACS Nano. 2014;8:9988–97. doi: 10.1021/nn503150w. [DOI] [PubMed] [Google Scholar]
- 42.Yu R-J, Ma W, Liu X-Y, Jin H-Y, Han H-X, Wang H-Y. et al. Metal-linked immunosorbent assay (MeLISA): the enzyme-free alternative to ELISA for biomarker detection in serum. Theranostics. 2016;6:1732–9. doi: 10.7150/thno.16129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu C-H, Wang Y-W, Ye S-L, Chen G-N, Yang H-H. Ultrasensitive detection of Cu2+ with the naked eye and application in immunoassays. NPG Asia Mater. 2012;4:e10. [Google Scholar]
- 44.Lin Y, Ren J, Qu X. Catalytically active nanomaterials: A promising candidate for artificial enzymes. Acc Chem Res. 2014;47:1097–105. doi: 10.1021/ar400250z. [DOI] [PubMed] [Google Scholar]
- 45.Lin Y, Ren J, Qu X. Nano-gold as artificial enzymes: Hidden talents. Adv Mater. 2014;26:4200–17. doi: 10.1002/adma.201400238. [DOI] [PubMed] [Google Scholar]
- 46.Murakami Y, Kikuchi J, Hisaeda Y, Hayashida O. Artificial enzymes. Chem Rev. 1996;96:721–58. doi: 10.1021/cr9403704. [DOI] [PubMed] [Google Scholar]
- 47.Tao Y, Lin Y, Huang Z, Ren J, Qu X. Incorporating graphene oxide and gold nanoclusters: A synergistic catalyst with surprisingly high peroxidase-like activity over a broad pH range and its application for cancer cell detection. Adv Mater. 2013;25:2594–9. doi: 10.1002/adma.201204419. [DOI] [PubMed] [Google Scholar]
- 48.Rebilly J-N, Colasson B, Bistri O, Over D, Reinaud O. Biomimetic cavity-based metal complexes. Chem Soc Rev. 2015;44:467–89. doi: 10.1039/c4cs00211c. [DOI] [PubMed] [Google Scholar]
- 49.Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42:6060–93. doi: 10.1039/c3cs35486e. [DOI] [PubMed] [Google Scholar]
- 50.Orozco J, García-Gradilla V, D'Agostino M, Gao W, Cortés A, Wang J. Artificial enzyme-powered microfish for water-quality testing. ACS Nano. 2013;7:818–24. doi: 10.1021/nn305372n. [DOI] [PubMed] [Google Scholar]
- 51.Cai R, Yang D, Peng S, Chen X, Huang Y, Liu Y. et al. Single nanoparticle to 3D supercage: Framing for an artificial enzyme system. J Am Chem Soc. 2015;137:13957–63. doi: 10.1021/jacs.5b09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y, Deng L, Li J, Guo S, Wang E, Dong S. Hemin-graphene hybrid nanosheets with intrinsic peroxidase-like activity for label-free colorimetric detection of single-nucleotide polymorphism. ACS Nano. 2011;5:1282–90. doi: 10.1021/nn1029586. [DOI] [PubMed] [Google Scholar]
- 53.Song Y, Qu K, Zhao C, Ren J, Qu X. Graphene oxide: Intrinsic peroxidase catalytic activity and its application to glucose detection. Adv Mater. 2010;22:2206–10. doi: 10.1002/adma.200903783. [DOI] [PubMed] [Google Scholar]
- 54.Zheng X, Liu Q, Jing C, Li Y, Li D, Luo W. et al. Catalytic gold nanoparticles for nanoplasmonic detection of DNA hybridization. Angew Chem Int Ed. 2011;50:11994–8. doi: 10.1002/anie.201105121. [DOI] [PubMed] [Google Scholar]
- 55.de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012;7:821–4. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 56.Gao L, Zhuang J, Nie L, Zhang J, Zhang Y, Gu N. et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat Nanotechnol. 2007;2:577–83. doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- 57.Wang X-X, Wu Q, Shan Z, Huang Q-M. BSA-stabilized Au clusters as peroxidase mimetics for use in xanthine detection. Biosens Bioelectron. 2011;26:3614–9. doi: 10.1016/j.bios.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 58.Zheng C, Zheng A-X, Liu B, Zhang X-L, He Y, Li J. et al. One-pot synthesized DNA-templated Ag/Pt bimetallic nanoclusters as peroxidase mimics for colorimetric detection of thrombin. Chem Commun. 2014;50:13103–6. doi: 10.1039/c4cc05339g. [DOI] [PubMed] [Google Scholar]
- 59.Yu C-J, Chen T-H, Jiang J-Y, Tseng W-L. Lysozyme-directed synthesis of platinum nanoclusters as a mimic oxidase. Nanoscale. 2014;6:9618–24. doi: 10.1039/c3nr06896j. [DOI] [PubMed] [Google Scholar]
- 60.Das S, Goswami A, Hesari M, Al-Sharab JF, Mikmeková E, Maran F. et al. Reductive deprotection of monolayer protected nanoclusters: An efficient route to supported ultrasmall Au nanocatalysts for selective oxidation. Small. 2014;10:1473–8. doi: 10.1002/smll.201302854. [DOI] [PubMed] [Google Scholar]
- 61.Gao W, Vecchio D, Li J, Zhu J, Zhang Q, Fu V. et al. Hydrogel containing nanoparticle-stabilized liposomes for topical antimicrobial delivery. ACS Nano. 2014;8:2900–7. doi: 10.1021/nn500110a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bigdeli A, Palchetti S, Pozzi D, Hormozi-Nezhad MR, Baldelli Bombelli F, Caracciolo G. et al. Exploring cellular interactions of liposomes using protein corona fingerprints and physicochemical properties. ACS Nano. 2016;10:3723–37. doi: 10.1021/acsnano.6b00261. [DOI] [PubMed] [Google Scholar]
- 63.Allen TM, Cullis PR. Liposomal drug delivery systems: From concept to clinical applications. Adv Drug Deliv Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 64.Chen K-J, Liang H-F, Chen H-L, Wang Y, Cheng P-Y, Liu H-L. et al. A thermoresponsive bubble-generating liposomal system for triggering localized extracellular drug delivery. ACS Nano. 2013;7:438–46. doi: 10.1021/nn304474j. [DOI] [PubMed] [Google Scholar]
- 65.Feng L, Gao M, Tao D, Chen Q, Wang H, Dong Z. et al. Cisplatin-prodrug-constructed liposomes as a versatile theranostic nanoplatform for bimodal imaging guided combination cancer therapy. Adv Funct Mater. 2016;26:2207–17. [Google Scholar]
- 66.Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discov. 2005;4:145–60. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Situ A, Kang Y, Villabroza KR, Liao Y, Chang CH. et al. Irinotecan delivery by lipid-coated mesoporous silica nanoparticles shows improved efficacy and safety over liposomes for pancreatic cancer. ACS Nano. 2016;10:2702–15. doi: 10.1021/acsnano.5b07781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dave N, Liu J. Protection and promotion of UV radiation-induced liposome leakage via DNA-directed assembly with gold nanoparticles. Adv Mater. 2011;23:3182–6. doi: 10.1002/adma.201101086. [DOI] [PubMed] [Google Scholar]
- 69.Pornpattananangkul D, Zhang L, Olson S, Aryal S, Obonyo M, Vecchio K. et al. Bacterial toxin-triggered drug release from gold nanoparticle-stabilized liposomes for the treatment of bacterial infection. J Am Chem Soc. 2011;133:4132–9. doi: 10.1021/ja111110e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner A, Platzgummer M, Kreismayr G, Quendler H, Stiegler G, Ferko B. et al. GMP production of liposomes—A new industrial approach. J Liposome Res. 2006;16:311–9. doi: 10.1080/08982100600851086. [DOI] [PubMed] [Google Scholar]
- 71.Cao Z, Tong R, Mishra A, Xu W, Wong GCL, Cheng J. et al. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew Chem Int Ed. 2009;48:6494–8. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 72.Sheng Q, Cheng N, Bai W, Zheng J. Ultrasensitive electrochemical detection of breast cancer cells based on DNA-rolling-circle-amplification-directed enzyme-catalyzed polymerization. Chem Commun. 2015;51:2114–7. doi: 10.1039/c4cc08954e. [DOI] [PubMed] [Google Scholar]
- 73.Xie JP, Zheng YG, Ying JY. Protein-directed synthesis of highly fluorescent gold nanoclusters. J Am Chem Soc. 2009;131:888–9. doi: 10.1021/ja806804u. [DOI] [PubMed] [Google Scholar]
- 74.Dave N, Liu JW. Programmable assembly of DNA-functionalized liposomes by DNA. ACS Nano. 2011;5:1304–12. doi: 10.1021/nn1030093. [DOI] [PubMed] [Google Scholar]
- 75.Guo P, You J-O, Yang J, Moses MA, Auguste DT. Using breast cancer cell CXCR4 surface expression to predict liposome binding and cytotoxicity. Biomaterials. 2012;33:8104–10. doi: 10.1016/j.biomaterials.2012.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Wang J, Shigematsu H, Xu W, Shih WM, Rothman JE. et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat Chem. 2016;8:476–83. doi: 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andrews CD, Huh M-S, Patton K, Higgins D, Van Nest G, Ott G. et al. Encapsulating immunostimulatory CpG oligonucleotides in listeriolysin O-liposomes promotes a Th1-type response and CTL activity. Mol Pharm. 2012;9:1118–25. doi: 10.1021/mp2003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo P, You J-O, Yang J, Jia D, Moses MA, Auguste DT. Inhibiting metastatic breast cancer cell migration via the synergy of targeted, pH-triggered siRNA delivery and chemokine axis blockade. Mol Pharm. 2014;11:755–65. doi: 10.1021/mp4004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gunawan RC, Almeda D, Auguste DT. Complementary targeting of liposomes to IL-1α and TNF-α activated endothelial cells via the transient expression of VCAM1 and E-selectin. Biomaterials. 2011;32:9848–53. doi: 10.1016/j.biomaterials.2011.08.093. [DOI] [PubMed] [Google Scholar]
- 80.Patterson SD, Rossi JM, Paweletz KL, Fitzpatrick VD, Begley CG, Busse L. et al. Functional EpoR pathway utilization is not detected in primary tumor cells isolated from human breast, non-small cell lung, colorectal, and ovarian tumor tissues. PLOS ONE. 2015;10:e0122149. doi: 10.1371/journal.pone.0122149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Makdissi FBA, Machado LVST, Oliveira AGC, Benvenuti TT, Katayama MLH, Brentani MM. et al. Expression of E-cadherin, Snail and Hakai in epithelial cells isolated from the primary tumor and from peritumoral tissue of invasive ductal breast carcinomas. Braz J Med Biol Res. 2009;42:1128–37. doi: 10.1590/s0100-879x2009001200002. [DOI] [PubMed] [Google Scholar]
- 82.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M. et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 83.Ray S, Langan RC, Mullinax JE, Koizumi T, Xin H-W, Wiegand GW. et al. Establishment of human ultra-low passage colorectal cancer cell lines using spheroids from fresh surgical specimens suitable for in vitro and in vivo studies. J Cancer. 2012;3:196–206. doi: 10.7150/jca.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 85.Gao L, Fei J, Zhao J, Li H, Cui Y, Li J. Hypocrellin-loaded gold nanocages with high two-photon efficiency for photothermal/photodynamic cancer therapy in vitro. ACS Nano. 2012;6:8030–40. doi: 10.1021/nn302634m. [DOI] [PubMed] [Google Scholar]
- 86.Wang F, Liu B, Ip ACF, Liu J. Orthogonal adsorption onto nano-graphene oxide using different intermolecular forces for multiplexed delivery. Adv Mater. 2013;25:4087–92. doi: 10.1002/adma.201301183. [DOI] [PubMed] [Google Scholar]
- 87.Epstein H, Afergan E, Moise T, Richter Y, Rudich Y, Golomb G. Number-concentration of nanoparticles in liposomal and polymeric multiparticulate preparations: Empirical and calculation methods. Biomaterials. 2006;27:651–9. doi: 10.1016/j.biomaterials.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 88.Sardan M, Kilinc M, Genc R, Tekinay AB, Guler MO. Cell penetrating peptide amphiphile integrated liposomal systems for enhanced delivery of anticancer drugs to tumor cells. Farad Discuss. 2013;166:269–83. doi: 10.1039/c3fd00058c. [DOI] [PubMed] [Google Scholar]
- 89.Hwang S, Maitani Y, Qi X-R, Takayama K, Nagai T. Remote loading of diclofenac, insulin and fluorescein isothiocyanate labeled insulin into liposomes by pH and acetate gradient methods. Int J Pharm. 1999;179:85–95. doi: 10.1016/s0378-5173(98)00392-5. [DOI] [PubMed] [Google Scholar]
- 90.Gong M-Q, Wu J-L, Chen B, Zhuo R-X, Cheng S-X. Self-assembled polymer/inorganic hybrid nanovesicles for multiple drug delivery to overcome drug resistance in cancer chemotherapy. Langmuir. 2015;31:5115–22. doi: 10.1021/acs.langmuir.5b00542. [DOI] [PubMed] [Google Scholar]
- 91.Li M, Tang Z, Lv S, Song W, Hong H, Jing X. et al. Cisplatin crosslinked pH-sensitive nanoparticles for efficient delivery of doxorubicin. Biomaterials. 2014;35:3851–64. doi: 10.1016/j.biomaterials.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 92.Khlebtsov BN, Khlebtsov NG. On the measurement of gold nanoparticle sizes by the dynamic light scattering method. Colloid J. 2011;73:118–27. [Google Scholar]
- 93.Marquez LA, Dunford HB. Mechanism of the oxidation of 3,5,3',5'-tetramethylbenzidine by myeloperoxidase determined by transient- and steady-state kinetics. Biochemistry. 1997;36:9349–55. doi: 10.1021/bi970595j. [DOI] [PubMed] [Google Scholar]
- 94.Guo P, Huang J, Wang L, Jia D, Yang J, Dillon DA. et al. ICAM-1 as a molecular target for triple negative breast cancer. P Natl Acad Sci USA. 2014;111:14710–5. doi: 10.1073/pnas.1408556111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fiandra L, Mazzucchelli S, De Palma C, Colombo M, Allevi R, Sommaruga S. et al. Assessing the in vivo targeting efficiency of multifunctional nanoconstructs bearing antibody-derived ligands. ACS Nano. 2013;7:6092–102. doi: 10.1021/nn4018922. [DOI] [PubMed] [Google Scholar]
- 96.Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. P Natl Acad Sci USA. 2013;110:8170–5. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–6. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 98.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–43. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 99.Melancon MP, Zhou M, Zhang R, Xiong C, Allen P, Wen X. et al. Selective uptake and imaging of aptamer- and antibody-conjugated hollow nanospheres targeted to epidermal growth factor receptors overexpressed in head and neck cancer. ACS Nano. 2014;8:4530–8. doi: 10.1021/nn406632u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Zhu J, Han L, Jin L, Zhu C, Wang E. et al. Graphene-based aptamer logic gates and their application to multiplex detection. ACS Nano. 2012;6:6659–66. doi: 10.1021/nn300997f. [DOI] [PubMed] [Google Scholar]
- 101.Lee JS, Kim SG, Jun J, Shin DH, Jang J. Aptamer-functionalized multidimensional conducting-polymer nanoparticles for an ultrasensitive and selective field-effect-transistor endocrine-disruptor sensors. Adv Funct Mater. 2014;24:6145–53. [Google Scholar]
- 102.Zheng F, Cheng Y, Wang J, Lu J, Zhang B, Zhao Y. et al. Aptamer-functionalized barcode particles for the capture and detection of multiple types of circulating tumor cells. Adv Mater. 2014;26:7333–8. doi: 10.1002/adma.201403530. [DOI] [PubMed] [Google Scholar]
- 103.Liu H, Xiang Y, Lu Y, Crooks RM. Aptamer-based origami paper analytical device for electrochemical detection of adenosine. Angew Chem Int Ed. 2012;51:6925–8. doi: 10.1002/anie.201202929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tan W, Donovan MJ, Jiang J. Aptamers from cell-based selection for bioanalytical applications. Chem Rev. 2013;113:2842–62. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu C, Chen T, Han D, You M, Peng L, Cansiz S. et al. Engineering of switchable aptamer micelle flares for molecular imaging in living cells. ACS Nano. 2013;7:5724–31. doi: 10.1021/nn402517v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakatsuka MA, Mattrey RF, Esener SC, Cha JN, Goodwin AP. Aptamer-crosslinked microbubbles: Smart contrast agents for thrombin-activated ultrasound imaging. Adv Mater. 2012;24:6010–6. doi: 10.1002/adma.201201484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sunbul M, Jäschke A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew Chem Int Ed. 2013;52:13401–4. doi: 10.1002/anie.201306622. [DOI] [PubMed] [Google Scholar]
- 108.Chang M, Yang CS, Huang DM. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5:6156–63. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]
- 109.Han D, Zhu G, Wu C, Zhu Z, Chen T, Zhang X. et al. Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano. 2013;7:2312–9. doi: 10.1021/nn305484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yuan Q, Wu Y, Wang J, Lu D, Zhao Z, Liu T. et al. Targeted bioimaging and photodynamic therapy nanoplatform using an aptamer-guided G-quadruplex DNA carrier and near-infrared light. Angew Chem Int Ed. 2013;52:13965–9. doi: 10.1002/anie.201305707. [DOI] [PubMed] [Google Scholar]
- 111.Oh SS, Lee BF, Leibfarth FA, Eisenstein M, Robb MJ, Lynd NA. et al. Synthetic aptamer-polymer hybrid constructs for programmed drug delivery into specific target cells. J Am Chem Soc. 2014;136:15010–5. doi: 10.1021/ja5079464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xie X, Xu W, Liu X. Improving colorimetric assays through protein enzyme-assisted gold nanoparticle amplification. Acc Chem Res. 2012;45:1511–20. doi: 10.1021/ar300044j. [DOI] [PubMed] [Google Scholar]
- 113.Yu C, Hu Y, Duan J, Yuan W, Wang C, Xu H. et al. Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitro. PLoS ONE. 2011;6:e24077. doi: 10.1371/journal.pone.0024077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li C, Meng Y, Wang S, Qian M, Wang J, Lu W. et al. Mesoporous carbon nanospheres featured fluorescent aptasensor for multiple diagnosis of cancer in vitro and in vivo. ACS Nano. 2015;9:12096–103. doi: 10.1021/acsnano.5b05137. [DOI] [PubMed] [Google Scholar]
- 115.Wang K, Fan D, Liu Y, Wang E. Highly sensitive and specific colorimetric detection of cancer cells via dual-aptamer target binding strategy. Biosens Bioelectron. 2015;73:1–6. doi: 10.1016/j.bios.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 116.Cayre A, Mishellany F, Lagarde N, Penault-Llorca F. Comparison of different commercial kits for HER2 testing in breast cancer: looking for the accurate cutoff for amplification. Breast Cancer Res. 2007;9:R64. doi: 10.1186/bcr1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ding W, Gupta KC, Park S-Y, Kim Y-K, Kang I-K. In vitro detection of human breast cancer cells (SK-BR3) using herceptin-conjugated liquid crystal microdroplets as a sensing platform. Biomater Sci. 2016;4:1473–84. doi: 10.1039/c6bm00404k. [DOI] [PubMed] [Google Scholar]
- 118.Xiao Y, Gao X, Taratula O, Treado S, Urbas A, Holbrook RD. et al. Anti-HER2 IgY antibody-functionalized single-walled carbon nanotubes for detection and selective destruction of breast cancer cells. BMC Cancer. 2009;9:351. doi: 10.1186/1471-2407-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hathaway HJ, Butler KS, Adolphi NL, Lovato DM, Belfon R, Fegan D. et al. Detection of breast cancer cells using targeted magnetic nanoparticles and ultra-sensitive magnetic field sensors. Breast Cancer Res. 2011;13:R108. doi: 10.1186/bcr3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shamsipour F, Zarnani AH, Ghods R, Chamankhah M, Forouzesh F, Vafaei S. et al. Conjugation of monoclonal antibodies to super paramagnetic iron oxide nanoparticles for detection of her2/neu antigen on breast cancer cell lines. Avicenna J Med Biotechnol. 2009;1:27–31. [PMC free article] [PubMed] [Google Scholar]
- 121.Li K, Liu Y, Pu K-Y, Feng S-S, Zhan R, Liu B. Polyhedral oligomeric silsesquioxanes-containing conjugated polymer loaded PLGA nanoparticles with trastuzumab (herceptin) functionalization for HER2-positive cancer cell detection. Adv Funct Mater. 2011;21:287–94. [Google Scholar]
- 122.Ding J, Zhou Y, Li J, Jiang L, He Z, Zhu J-J. Screening of HER2 overexpressed breast cancer subtype in vivo by the validation of high-performance, long-term, and noninvasive fluorescence tracer. Anal Chem. 2015;87:12290–7. doi: 10.1021/acs.analchem.5b03580. [DOI] [PubMed] [Google Scholar]
- 123.Tabatabaei-Panah A-S, Jeddi-Tehrani M, Ghods R, Akhondi M-M, Mojtabavi N, Mahmoudi A-R. et al. Accurate sensitivity of quantum dots for detection of HER2 expression in breast cancer cells and tissues. J Fluoresc. 2013;23:293–302. doi: 10.1007/s10895-012-1147-9. [DOI] [PubMed] [Google Scholar]
- 124.Lu W, Arumugam SR, Senapati D, Singh AK, Arbneshi T, Khan SA. et al. Multifunctional oval-shaped gold-nanoparticle-based selective detection of breast cancer cells using simple colorimetric and highly sensitive two-photon scattering assay. ACS Nano. 2010;4:1739–49. doi: 10.1021/nn901742q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.