Abstract
The piggyBac transposable element was originally isolated from the cabbage looper moth, Trichoplusia ni, in the 1980s. Despite its early discovery and specificity compared to the other Class II elements, the diversity and evolution of this superfamily have been only partially analyzed. Two main types of elements can be distinguished: the piggyBac-like elements (PBLE) with terminal inverted repeats, untranslated region, and an open reading frame encoding a transposase, and the piggyBac-derived sequences (PGBD), containing a sequence derived from a piggyBac transposase, and which correspond to domesticated elements. To define the distribution, their structural diversity and phylogenetic relationships, analyses were conducted using known PBLE and PGBD sequences to scan databases. From this data mining, numerous new sequences were characterized (50 for PBLE and 396 for PGBD). Structural analyses suggest that four groups of PBLE can be defined according to the presence/absence of sub-terminal repeats. The transposase is characterized by highly variable catalytic domain and C-terminal region. There is no relationship between the structural groups and the phylogeny of these PBLE elements. The PGBD are clearly structured into nine main groups. A new group of domesticated elements is suspected in Neopterygii and the remaining eight previously described elements have been investigated in more detail. In all cases, these sequences are no longer transposable elements, the catalytic domain of the ancestral transposase is not always conserved, but they are under strong purifying selection. The phylogeny of both PBLE and PGBD suggests multiple and independent domestication events of PGBD from different PBLE ancestors.
Keywords: transposable element, piggyBac, molecular evolution, domestication
Introduction
Transposable elements (TEs) are mobile and repetitive genetic elements, abundant in all eukaryotic genomes investigated so far. Two classes of elements are distinguished according to their respective transposition mechanisms (Wicker et al. 2007). With few exceptions (like SINEs, MITES, LARD elements), TEs encode their own transposition machinery. They use a reverse transcriptase and integrase, leading to a “copy and paste” mechanism (retrotransposons or Class I) or a transposase in a “cut and paste” mechanism (DNA transposons or Class II). Excluding dead copies (i.e. with no coding capacity and not mobilizable), both classes exist as autonomous active copies, which encode all the factors required for their mobility and as nonautonomous but trans-mobilizable copies depending on the transposition machinery of their autonomous relatives (Feschotte et al. 2002).
While TEs are generally considered as selfish sequences or genomic parasites, they are also important evolutionary factors in both structural and functional dynamics of the genomes. Indeed, one of the most direct contributions of TEs to host genome evolution is their potential role in the emergence of new genes and functions through an exaptation process also known as a “molecular domestication” where the use of TE sequences for a new function is usually associated to the loss of their mobility capacities (Britten 1996; Kidwell and Lisch 1997; Miller et al. 1999; Kidwell 2002; Volff 2006; Sinzelle et al. 2009; Joly-Lopez et al. 2016). In these cases, while “classical” TEs are present in multi-copies and inserted at different positions in the genome, within and between species, domesticated elements are generally found as single orthologous copies in different species (if the domestication is old enough). In addition, a low ratio (Ka/Ks < 1) of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitution rates suggests that the sequence is under strong purifying selection if the new function is already acquired, while a high ratio (Ka/Ks > 1) indicates that the sequences are under positive selection, that is, when the adaptive peak is not yet reached (Hurst 2002).
This process can be illustrated by RAG1 in V(D)J recombination (Kapitonov and Jurka 2005; Hencken et al. 2012) and the CENPB centromere protein (Casola et al. 2008), which derive from the Transib and the pogo transposases, respectively. More recently, in the primate lineage, a mariner-like transposase was fused to an SET histone methyltransferase domain by de novo exonization. The fusion protein retains the ancestral DNA binding activity of the transposase and acts as a transcriptional regulator of dispersed mariner-like repeat elements (Cordaux et al. 2006; Liu et al. 2007).
The piggyBac element (formerly IFP2) is a typical Class II element, originally isolated from a mutant baculovirus in a cell culture of the Lepidopteran cabbage looper moth Trichoplusia ni (Fraser et al. 1983; Cary et al. 1989). More precisely, this element jumped from T. ni to the baculovirus. This 2,475 bp autonomous mobile element inserts in TTAA target sites and is bounded by 13 bp terminal inverted repeats (TIRs) and 19 bp sub-terminal asymmetric inverted repeats (STIR) located 3 and 31 bp from the 5′ and 3′ TIRs, respectively (Cary et al. 1989; Fraser et al. 1996; Lobo et al. 1999). The single open reading frame is 1,782 bp long, coding for a protein of 594 amino acids with a molecular weight of 64 kDa. Interestingly, piggyBac is also functional in other organisms, including yeasts, protozoa, vertebrates and plants (Yusa 2015). Due to its high transposition activity in several species of insects, it has become one of the most widely used systems for the germline transformations, as well as a genetic tool for gene tagging or trapping, and “an insertional mutagen” (Yusa 2015).
Since its discovery in 1983, piggyBac has for a long time remained the only member of the currently known piggyBac superfamily. The taxonomic distribution, initially believed to be restricted to the insect orders (Coleoptera, Diptera, Hymenoptera, Lepidoptera, and Orthoptera; see Wang et al. 2008 and references therein), has been significantly expanded to cover several eukaryotic groups. Indeed, a number of piggyBac-like elements (PBLE) from other species or from baculovirus have been identified (Penton et al. 2002; Arkhipova and Meselson 2005; Pritham et al. 2005; Wang et al. 2006; Xu et al. 2006; Hikosaka et al. 2007; Ray et al. 2008; Sun et al. 2008; Wang et al. 2008; Wu et al. 2008; Carpes et al. 2009; Wang et al. 2009; Daimon et al. 2010; Pagan et al. 2010; Luo et al. 2011; Wu et al. 2011; Luo et al. 2014; Wu and Wang 2014).
Most mammalian genomes also contain decayed piggyBac transposons. They ceased their activity due to several mutations or rearrangements (Pace and Feschotte, 2007; Pagan et al. 2010). The picture emerging from the initial analyses of the human, mouse, rat, and dog genomes shows that there is no evidence for piggyBac activity during the past 40 Ma (Lander et al. 2001; Gibbs et al. 2004; Lindblad-Toh et al. 2005; Pace and Feschotte 2007). However, piggyBac evolution can be different from one lineage to another. For instance, recent data suggest a continuous colonization of the vesper bat genomes. Several waves of amplification of piggyBac have succeeded over the past 40 Ma and the invasion seems to be ongoing, while a new member of PBLE, piggyBat, has been identified in the little brown bat Myotis lucifugus. This element is active in its native form and in transposition assays in bat and human cultured cells, as well as in Saccharomyces cerevisiae (Ray et al. 2008; Mitra et al. 2013).
Using computational analysis of genomic data, several species have also revealed a number of genes, derived from various TEs, including piggyBac transposases (Sarkar et al. 2003). Indeed, the human genome contains at least five domesticated piggyBac, designated from PGBD1 to PGBD5 (for piggyBac-derived genes). On the one hand, PGBD1 and PGBD2 were probably present in the common ancestor of mammals, while PGBD3 and PGBD4 are restricted to primates. On the other hand, PGBD5, the only sequences interrupted by multiple introns, are not only orthologous in mammals and fish (Sarkar et al. 2003) but also in lamprey and lancelet suggesting an ancient domestication event about 525 Ma before cephalocordates and vertebrates split from urochordates (Pavelitz et al. 2013).
Otherwise, domestication of piggyBac transposases can be observed in several evolutionary lineages. In the ciliates Paramecium tetraurelia (Baudry et al. 2009) and Tetrahymena thermophila (Cheng et al. 2010), the genome undergoes massive DNA amplification during macronucleus development, and extensive programmed genome rearrangement, including elimination of TEs and internal eliminated sequences (IES). These eliminations, essential to reconstruct functional genes, are due to domesticated piggyBac transposases, named piggyMac (PGM) in P. tetraurelia and TPB2 in T. thermophila (Baudry et al. 2009; Cheng et al. 2010). In Xenopus, the KOBUTA-domesticated transposase has been conserved for 100 Ma and seems to be involved in DNA-binding or DNA-recombination activity. Moreover, it can inactivate the Uribo autonomous transposase through heterodimerization (Hikosaka et al. 2007).
The general features shared by the members of the piggyBac superfamily are the TTAA integration target sites and the precise excision of the element leading to the restoration of the pre-integration site. In addition, highly conserved blocks can be detected in the core region, including several aspartic acid (D) and glutamic acid (E) residues (Sarkar et al. 2003; Keith et al. 2008). Although this region does not readily show similarity to the widespread DDE catalytic domains of many Class II transposases and Class I integrases, a weak similarity to the IS4 family protein was identified (Sarkar et al. 2003), leading to the prediction that D268 and D346 in the T. ni transposase might be the conserved aspartic acid of a DDE/D catalytic domain. Mutational analyses of these positions, as well as another highly conserved D447, revealed that these residues are absolutely required for all steps of transposition. While not conserved in piggyMac and TPB2, a fourth aspartic acid D450 could also be involved for the excision of the element in cell cultures, while a glutamate substitution can be tolerated (Keith et al. 2008). Another peculiar feature of all piggyBac transposases is the conserved Cysteine residues, forming a putative zinc-binding homeodomain (PHD) finger in the C-terminal region (Sarkar et al. 2003; Keith et al. 2008).
The availability of numerous almost complete eukaryotic genome sequences has considerably enriched the repertoire of annotated piggyBac elements, providing an opportunity to better characterize the origin, distribution, diversity, structure and evolution of this superfamily as well as those of the piggyBac-derived genes. Until now, in many cases the evolutionary scenarios leading to the presumed domestications have not been fully reconstructed particularly because the ancestral copies were difficult to identify unambiguously.
The objective of this work is to identify and characterize piggyBac-related elements, including domesticated sequences, in a large spectrum of organisms. Structural and sequence comparisons suggest that PBLE (bona fide transposons) can be grouped in four different structures due to the presence or absence of subterminal repeats with highly divergent catalytic domains and C-terminal regions. Concerning domesticated piggyBac, we identified a new group of PGBD sequences, besides the eight groups already described. Evolutionary scenarios based on the structural features, phylogenetic relationships and fate of these elements are discussed.
Materials and Methods
Data Mining
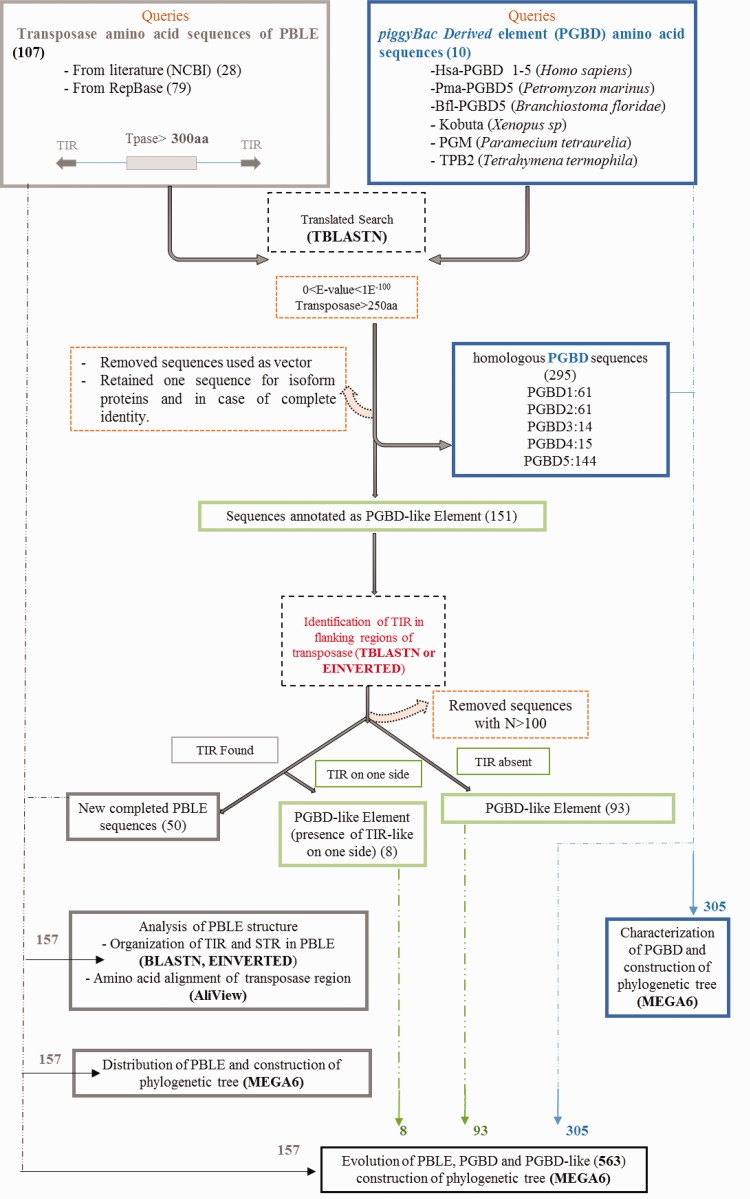
One hundred and seventeen piggyBac transposases were extracted from databases (NCBI, Repbase, and genomes available). Among those, 107 sequences are related to PBLE including 28 sequences from literature and 79 consensus sequences from Repbase with a transposase longer than 300 aa. Sequences containing long truncations, insertions or deletions were not retained. The other ten sequences are related to piggyBac-derived elements (PGBD) including five sequences from Homo sapiens, two sequences orthologous to PGBD5 of Humans, namely, Pma from the agnathe Petromyzon marinus and Bfl from the cephalochordate Branchiostoma floridae, the KOBUTA element from Xenopus sp and two piggyMac sequences, namely PGM from P. tetraurelia and TPB2 from T. termophila (fig. 1 and supplementary material S1, Supplementary Material online).
Fig. 1.—
Data mining of piggyBac element. Search of elements belonging to the piggyBac superfamily was done using known copies (literature or database) as BLAST queries. Ten PGBD and 107 PBLE copies were used as queries. After a first run, 295 sequences annotated as PGBD (PGBD1, 2, 3, 4, and 5) and 151 new sequences, annotated as PGBD-like element, belonging to 58 species were retrieved. TIR were looked for in the 5′ and 3′ flanking regions (5 kb in both directions) of these new sequences. Fifty of them were found with two TIR and 101 with one (5′ or 3′) or no TIR. In fine, a total of 563 sequences were available. The sequences with more than 100 N were eliminated.
Each of these copies was used as query to look for homologous sequences in the NCBI nr database and genomes available by TBlastN. The new sequences identified were in turn used as query in order to identify more PBLE and PGBD. The stringency of the mining steps was between 0 and 1E− 100 (TBlastN) and only transposases of at least 250 amino acids were retained. Sequences used as vector were removed and in case of isoform proteins or identical sequences only one sequence was selected.
Structural Analysis of Copies
The annotation of piggyBac sequences in databases can sometime be confusing. For instance, the term PGBD-like can be used indifferently for PBLE, PGBD or for degenerated copies (with internal deletion, truncation…). In the present work, PBLE will refer to sequences (active or not) with two TIRs, two untranslated regions (UTRs) and a transposase. PGBD will correspond to domesticated sequences (i.e., sequences in single copy in orthologous position in different species and a Ka/Ks indicating the existence of positive or purifying selection). Finally, the PGBD-like term will be restricted to the remaining sequence i.e. with one or no TIRs or UTRs and a transposase that can be partially deleted or truncated.
The BLAST searches were done using the transposase sequence as query. Therefore, to discriminate between PBLE sequences from PGBD-like elements, TBlastN was used to align each sequence on the host genome. Then, 5 kb of both flanking regions were added to detect potential TIR. Flanking sequences containing more than 100 N were removed. When TIRs were found on both sides, the sequence was considered as new complete PBLE, while sequences flanked by a single TIR or no TIR were considered as PGBD-like element (supplementary material S1, Supplementary Material online). Then, in PBLE, direct repeats (DR) and sub-terminal repeats (STIR) were searched by alignment of flanked sequences of the transposase using BlastN (supplementary material S2, Supplementary Material online). At the end of this screening and filtering steps, a total of 157 sequences of PBLE (107 from Repbase and literature plus 50 new sequences) and 406 PGBD or PGBD-like sequences were retrieved (fig. 1 and supplementary material S1, Supplementary Material online).
PBLE vs. PGBD
PBLE transposases were aligned with AliView (version 1.17.1; Larsson 2014) using Muscle with default parameters. Blocks with a conservation level higher than 30% (relative to the consensus given by AliView) with no long indels (>20aa) were retained for phylogenetic analyses (supplementary material S3, Supplementary Material online). The phylogenies (Maximum likelihood, but different methods led to similar topologies), based on the previous blocks, were inferred with MEGA6 (Tamura et al. 2013) after a search for the best evolutionary scenario with ProtTest 2.4 server (AIC, matrix LG + F+G). For the phylogeny of PGBD and all copies (PBLE, PGBD, and PGBD like), the same procedure was followed (supplementary materials S4 and S5, Supplementary Material online).
Results
Structures, characteristics and phylogenies of PBLE and PGBD elements were analyzed separately because they correspond to different types of sequences with different evolutionary trajectories. The former are putatively active transposons, while the latter are domesticated. Then, to infer the phylogenetic relationships between all the piggyBac-like sequences, the PGBD-like elements were added to these two groups. Beyond the analysis of the molecular evolution of piggyBac, our objective was to determine which type of piggyBac elements gave rise to the domesticated sequences using the phylogenetic proximities.
PBLE Landscape
Among the 107 sequences used here as queries, 64 (59%) correspond to potentially functional transposases, while the others are defective due to the presence of multiple stop codons and/or frameshifts. The TBlastN allowed us to identify 50 new sequences from different genomes.
Characterization of PBLE: Structures and Distribution among Species
Members of the piggyBac superfamily have a TTAA sequence as target site duplication (TSD) and have TIR of 12-19 bp long (Fraser et al. 1983). From the analysis of the 157 PBLE, we show that rarely other insertion sites, including TTTT, ATAG, TTAT, ATAT, and ATAA, can be detected for a unique sequence (supplementary material S2, Supplementary Material online), and that the average length of the TIR is 14 ± 2 bp (without the TIR of 50 bp from Paracoccidioides brasiliensis). Moreover, while the first six nucleotides are relatively well conserved with the following motif C[C/A/T][C/A/T][T/G/A][T/A/G][T/G/A], the remaining part of the TIR can be highly divergent from one copy to another (supplementary material S2, Supplementary Material online). The element size varies from 1,721 to 8,451 bp with an average value of 2,813 ± 84 bp.
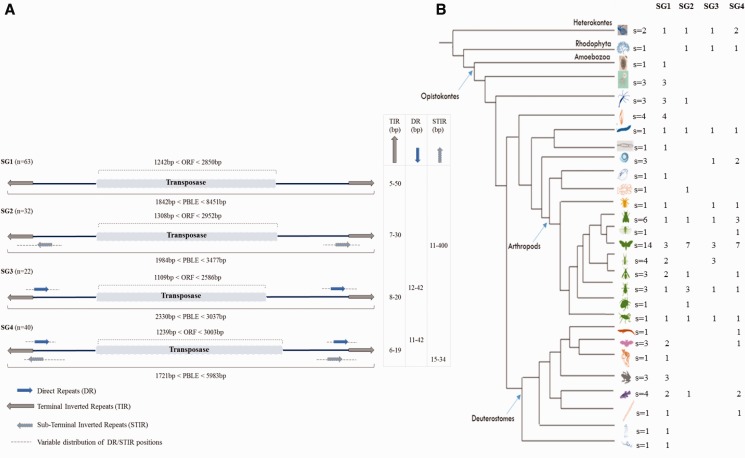
A detailed analysis of the 5′ and 3′ ends revealed that, besides the presence of TIR, 94 PBLE contain DR and/or sub-TIR (STIR) at their ends. Based on the presence/absence of these repeats, four structural groups (SG) of PBLEs can be defined (fig. 2 and supplementary material S2, Supplementary Material online). The first group (SG1), comprises 63 sequences characterized by TIR ranging between 5 and 19 bp with the exception of a long TIR of 50 bp in the fungi P. brasiliensis. The second group (SG2) contains TIR (7–30 bp) and imperfect STIR, varying from 11 to 400 bp; 16 out of 32 sequences showed an overlapping region between TIR and STIR. The third one (SG3) is characterized by TIR (8–20 bp) and DR (12–42 bp); 5 sequences out of 22 show an overlapping region between TIR and DR. The last group (SG4), containing 40 sequences, is more complex since all sequences contains not only TIRs (6-19 bp), but also the DR (11–42 bp) and STIRs (15-34 bp). We note that 36 out of 40 transposons belonging to this group have STIR and DR that are similar. For example, the 5' and 3' DR of piggyBac-5_Ccri are identical (same sequence and orientation) to the 5' and 3' STIR respectively. Such a phenomenon is possible because the common sequence is palindromic (supplementary material S2, Supplementary Material online). For the other members of this group, and as already mentioned for the SG2 and SG3, overlaps between DR/STIR and TIR can be observed. Finally, no consensus sequence has been found for DRs or STIRs.
Fig. 2.—
Structure of PBLEs and their distribution among species. (A) According to the presence/absence of DR and STIRs, four groups of PBLE were proposed (from SG1 to SG4). The number of sequences (n) in each group is given in brackets. (B) Specific distribution of the four SGs. The number of species (s) is given for each branch of the phylogenetic tree and according to each structure. Involved species (Heterokontes, Rhodophyta, Amoebozoa, and Opistokontes) are detailed, from the top down, as follows: [Phytophthora infestans, P. ramorum], [Chondrus crispus], [Entamoeba invadens], [Paracoccidioides brasiliensis, Mucor circinelloides, Nosema apis], [Acropora millepora, Hydra magnipapillata, Nematostella vectensis], [Aplysia californica, Biomphalaria glabrata, Crassostrea gigas, Lingula anatina], [Schmidtea mediterranea], [Adineta vaga], [Ancylostoma ceylanicum, Haemonchus contortus, Trichinella spiralis], [Branchipoda crustacean], [Lepeophtheirus salmonis], [Stegodyphus mimosarum], [Aedes aegypti, Anopheles gambiae, Drosophila ananassae, D. biarmipes, D. bipectinata, D. eugracilis], [Mengenilla moldrzyki], [Anticarsia gemmatalis, Agrotis ipsilon, Amyelois transitella, Bombyx mori, Ctenoplusia agnata, Chilo suppressalis, Helicoverpa armigera, Heliconius melpomene, Heliothis virescens, Macdunnoughia crassisigna, Pectinophora gossypiella, Papilio xuthus, Spodoptera frugiperda, Trichoplusia ni], [Cerapachys biroi, Orussus abietinus, Solenopsis invicta, Vollenhovia emeryi], [Athalia rosae, Megachile rotundata, Nasonia vitripennis], [Aphis gossypii, Acyrthosiphon pisum, Diaphorina citri], [Tribolium castaneum], [Locusta migratoria], [Alligator mississippiensis], [Myotis davidii, Myotis lucifugus, Pteropus vampyrus], [Microcebus murinus], [Xenopus borealis, X. laevis, X. tropicalis], [Fundulus heteroclitus, Latimeria chalumnae, Oreochromis niloticus, Salmo salar], [Branchiostoma floridae], [Ciona intestinalis], [Saccoglossus kowalevskii].
Another interesting feature is length variability of the four previous groups. Indeed, when the different parts of the element are considered [5′ and 3′ TIR, UTRs and open reading frame (ORF)], the coefficients of variation of UTRs (5′ and 3′) are much higher (from 2 to 8 times) than those of TIR and ORF for all SGs considered, suggesting more selective pressures on the latter (supplementary material S6, Supplementary Material online). Moreover, the SG1 group characterized by the absence of subterminal DR and STIRs, seems to be more variable than the other SG, except for the ORF region.
The specific distribution of the four SGs of PBLE (fig. 2), shows that some species can contain a single SG and others from two to four SG. In particular, the SG1 group is present in many species including protozoa, red algae, fungi and metazoan, while the others SG are less widely distributed. Therefore, the predominance of SG1 over the other groups (63 SG1, 32 SG2, 22 SG3 and 40 SG4, χ2 P < 10− 5), its large specific distribution and its higher variability could suggest that the piggyBac sequences containing only TIR are the ancestral structure. The underlying hypothesis to this proposition is that the presence of sequences belonging to other SG could be due to evolutionary convergence and/or to horizontal transfers. An argument in favor of the convergence hypothesis is the absence of consensus sequences in the UTR. However, the alternative hypothesis suggesting the presence of the four SGs in the common ancestor of all species here considered, followed by independent loss in several lineages, leading to a patchy distribution and a rapid evolution of DR and STIR sequences cannot be excluded.
The piggyBac-like Transposase Protein Family
In spite of a strong variability of the element lengths, the alignment of amino-acid sequences of the putative transposases encoded by the 157 PBLE shows several highly conserved motifs (supplementary material S3, Supplementary Material online). However, as already mentioned by Sarkar et al. (2003), the N-terminal region (positions 1–130, using T. ni transposase as reference), suspected to be a DNA-binding-domain interacting with TIR, is not well conserved and no rational alignment can be provided.
The central part of transposase (positions 130–522, T. ni as reference) contains several conserved and clearly delimited blocks (supplementary material S3, Supplementary Material online). Among them are those surrounding the residues of the catalytic domain DDD-D/G in positions 268, 346, 447, and 450. The four first Aspartates are strictly conserved in the putatively active elements and only one sequence with a G is found for the last position in the nematode Trichinella spiralis. However, according to Keith et al. (2008), a Glutamate can be tolerated at this position. Therefore, this last residue can be less constrained than the others and may be essential but not involved in the catalytic site itself.
The C-terminal region (positions 559–594) overlapping the piggyBac nuclear localization signal (NLS = PVMKKRTYCTYCPSKIRRKAN from position 551 to 571, Keith et al. 2008) is relatively well conserved. However, as mentioned by these authors, if this motif is a functional NLS, the definition of the complete NLS seems to be more difficult. The ZnF motif of the C-terminal region starts in the NLS motif, which includes the first two Cysteines (bold/underlined in the previous motif). Comparison of the different sequences shows that this region may present five to seven conserved Cysteines and sometimes a Histidine. For instance, there are seven cysteines in the piggyBac transposase of T. ni but five in that of Aplysia californica (sequence named AcaPBLE). Moreover, spacing between these residues are relatively well conserved. Keith et al. (2008) suggested that the ZnF region might not be involved in the DNA binding process but in other processes, including protein-protein interaction as required for a putative dimerization of the transposase.
Phylogenetic Analysis
Despite their high divergence in sequence, most PBLE transposases were found to contain conserved domains for a total of about 237 residues. Thus, it is possible to infer a phylogenetic tree based on these conserved regions (supplementary material S3, Supplementary Material online). The PBLE tree (supplementary material S7, Supplementary Material online) presents short terminal branches reflecting the high similarity between these elements. The simplified phylogeny (fig. 3) shows that there is no congruence between the phylogeny of the transposases and the general structure of the elements. Therefore, no clear evolutionary scenario can be proposed to explain the distribution of the different SG. In addition, the flexibility of the UTR length and sequences, compared to other parts of the elements, and the absence of consensus sequence between DRs or between STIRs, suggest that the structures of elements belonging to the same SG might be the result of evolutionary convergences.
Fig. 3.—
Phylogenetic tree of PBLE. This phylogeny is based on amino-acid sequences covering about 237 residues (the alignment is given in supplementary material S3, Supplementary Material online). For simplicity, only 29 sequences are represented, but the complete tree is available in supplementary material S7, Supplementary Material online. After a search of the best evolutionary scenario (ProTest 2.4), this unrooted tree was generated in MEGA6 with the maximum likelihood (ML) method, using LG + F + G matrix. Only bootstrapping values (100 replications) higher than 70% are mentioned on the branch. Red dots, green squares, yellow lozenges and blue triangles refer to the different SGs, that is, SG1, SG2, SG3, and SG4, respectively. Sequence names: the term PBLE (piggyBac-Like Element) is used for sequences extracted from literature, or copies newly characterized in this study, while the term piggyBac is restricted to sequences extracted from Repbase (real name of these sequences in this database).
Analysis of the PGBD
During evolution, functional features of transposases can be exapted to create new genes with specific cellular functions (see for instance the V(D)J system of mammalian derived from the transib transposase; Kapitonov and Jurka 2005). In this respect, several copies of piggyBac have been described as domesticated sequences (table 1). More precisely, in the human genome, five genes (PGBD1–PGBD5) derived from piggyBac transposases (Sarkar et al. 2003). Otherwise, the KOBUTA gene of Xenopus (Hikosaka et al. 2007), the PGM (Baudry et al. 2009) and TPB2 (Cheng et al. 2010) genes of the ciliates P. tetraurelia and T. thermophila, respectively, and the Pma and Bfl sequences of agnathe and cephalochordate (Pavelitz et al. 2013), are also originated from piggyBac transposases. The human PGBD5 and the last two sequences (Pma and Bfl) are orthologous, suggesting an ancient domestication (Pavelitz et al. 2013).
Table 1.
Structural and Putative Functions of Eight Domesticated piggyBac Elements
| Gene ID | Name | References | Organism | Length of Coding Region (aa) | Presence of Introns | Catalytic Motif D D D D 1 2 3 4 | CRD | Additional Domains or Genes | Functions |
|---|---|---|---|---|---|---|---|---|---|
| PGBD1 | piggyBac-derived 1 | Sarkar et al. 2003 | Homo sapiens/ Mammals (61) | 809 a , b 312 < aa < 826 | – | D D _ D G E | − | Zn_SCAN (290aa) | Unknown |
| PGBD2 | piggyBac-derived 2 | Sarkar et al. 2003 | Homo sapiens/ Mammals (61) | 592 a 586 < aa < 759 | – | G D G D _ | + | – | Unknown |
| PGBD3 | piggyBac-derived 3 | Sarkar et al. 2003 Newman et al. 2008 | Homo sapiens/ Primates (14) | 593 a ,c | – | D N D D G | + | CSB gene | Involved in Cockayne syndrome |
| PGBD4 | piggyBac-derived 4 | Sarkar et al. 2003 | Homo sapiens/ Primates (15) | 585 a 585 < aa < 603 | – | D D D D | + | – | Unknown |
| KOBUTA | KOBUTA | Hikosaka et al. 2007 | Xenopus tropicalis, laevis, borealis | 610 | – | D D N D N | + | – | Unknown |
| PGM | piggyMac | Baudry et al. 2009 | Paramecium tetraurelia | 1,065 | 2 | D D D N | + | – | Required for programmed genome rearrangements |
| TPB2 | Cheng et al. 2010 | Tetrahymena thermophila | 1,220 | 12 | |||||
| PGBD5 | piggyBac-derived 5 | Sarkar et al. 2003 Pavelitz et al. 2013 | Homo sapiens/ Myomerozoa (146) | 554 a 343 < aa < 732 | 5–7 | _ _ _ D N | − | – | Neural specific |
Note.—This table summarizes the information from several references including the organism (the number of sequences is in brackets), the length of the coding region, the presence and number of introns, the fate of the piggyBac catalytic motif using the four aspartate of PBLE as reference, presence/absence of Cysteine Rich Domain (CRD) in C-terminal region, existence of additional domains or part of genes, and putative functions.
Refers to Homo sapiens.
PGBD1 is composed by two parts. The first part, localized in 5′, includes five exons encoding a sequence of 290 aa corresponding to a Zn_Scan domain. The second part, derived from the piggyBac transposase, encodes a sequence of 519 aa. Therefore, the total PGBD1 is a sequence of 809 aa of unknown function.
PGBD3 is located in the fifth intron of the CSB gene, The CSB-PGBD3 fusion encodes a protein of 1,061 aa including 468 residues of CSB and the entire transposase of PGBD3 (593 aa).
These eight genes were used as query against general database to extract new PGBD sequences. The objective was to get a better idea of the specific distribution of these sequences. This allowed us to retrieve 295 PGBD sequences. All these sequences can be clustered as eight groups. The orthology was verified from a detailed analysis of the 5′ and 3′ flanking regions (as shown in supplementary material S8, Supplementary Material online). All these sequences are found in single copies. They present a coding region in which all or a part of the piggyBac transposase is identified. Within each group, the similarity level between the sequences is higher than 85% (see below), and between groups no alignment can be made except for the parts corresponding to the piggyBac transposase. Therefore, sequences of the different groups probably correspond to different domestication events.
Based on the five genes described in the human genome, five groups of PGBD sequences (from PGBD1 to PGBD5) can be defined according to the similarity level of their amino-acid sequences (table 1, supplementary materials S4 and S9, Supplementary Material online). PGBD1 members (n = 62) are the result of an ancestral fusion between five exons containing LER or SCAN domains (leucine-rich regions) in the N-terminal regions and the transposase of a piggyBac element (Sarkar et al. 2003). The size of PGBD1 members varies from 312 (Rno-PGBD1) to 826 (Bbi-PGBD1) amino acids, suggesting one or several indels in the different parts of the transposase during evolution. Nevertheless, they present a high level of similarity (average similarity =85%). In the PGBD2 group (n = 62), a single uninterrupted exon corresponding to the piggyBac transposase is found. As mentioned by Sarkar et al. (2003) and Pavelitz et al. (2013), we observed two exons in 5′ of the transposase sequence. Based on all PGBD2 sequences extracted from the database, the length of the encoded proteins (uninterrupted exon) varies from 586 (Pal-PGBD2) to 759 (Lve-PGBD2) amino acids. The average similarity between the members of this group is 88%. The PGBD3 (n = 15) transposase is inserted into the fifth intron of the Cockayne Syndrome group B gene (CSB). Indeed, unlike other PGBD, the PGBD3 transposase is flanked by a potential 3′ splicing site in the 5′ region and a polyadenylation signal in the 3′ region. Thus, an alternative splicing of this region leads to a regular CSB product or to the CSB-PGBD3 fusion protein which has been conserved since the common ancestor of human and marmoset lineages, that is, 43 Ma (Newman et al. 2008). Moreover, it has been demonstrated that the CSB-PGBD3 protein regulates gene expression from AP1, TEAD, and CTCF sites but not from MER85 sites (Gray et al. 2012). The average similarity of the PGBD3 sequences retrieved in various primates is very high (98%).
PGBD4 sequences (n = 16) present a single ORF encoding for a protein of 585 aa. Only the product of the Ppa-PGBD4 from Pan paniscus has a longer size (603 aa). The average percentage of similarity between the members of this group is also very high (98%).
PGBD5 is the largest group (n = 147). It contains several introns (six in the Bfl gene of cephalochordates, seven in Pma of agnathes and six in all other vertebrates). According to Pavelitz et al. (2013) and assuming that all PGBD5 behave similarly, there is no alternative splicing and all introns are spliced. Furthermore, a potential polyadenylation signal is observed in 3′ region of gnathostomates. The length of the PGBD5 encoded protein varies from 343 aa (Eca-PGBD5 in Equus caballus) to 732 aa (Eed-PGBD5 in Elephantulus edwardii). This size variation seems to be due to a weak conservation of the N-terminal part of the protein (supplementary material S4, Supplementary Material online). Nonetheless, the similarity between these sequences with the N-terminal sequence is close to 76% and 87% without this region.
Two other domesticated elements present introns within their transposase-derived sequence. Indeed, PGM contains two introns with a coding region about 1,065 aa while TPB2 contains 12 introns with an ORF coding for 1,220 aa. Finally, the KOBUTA element of Xenopus presents a single ORF with no introns encoding a protein of 610 aa.
The analysis of the Ka/Ks ratio provides arguments in favor of the domestication of these sequences. Indeed, this ratio calculated within each PGBD group varies from 0.123 ± 0.003 in PGBD2 to 0.432 ± 0.004 in PGBD1 (PGBD3 = 0.143 ± 0.005, PGBD4 = 0.150 ± 0.009, PGBD5 = 0.397 ± 0.002). This suggests the existence of a purifying or stabilizing selection. In this context, the DDD catalytic domain of the piggyBac transposase is not conserved among the domesticated sequences, including those retrieved from databases and those described in literature (Sarkar et al. 2003; Newman et al. 2008; Pavelitz et al. 2013), suggesting that this characteristic was not selected for during all exaptation processes. This unconserved catalytic domain is particularly observed for the members of the PGBD5, which is probably the oldest domesticated sequence (table 1). In addition, in the PGBD1 and PGBD5 the C-terminal region is truncated, removing the ZnF motif.
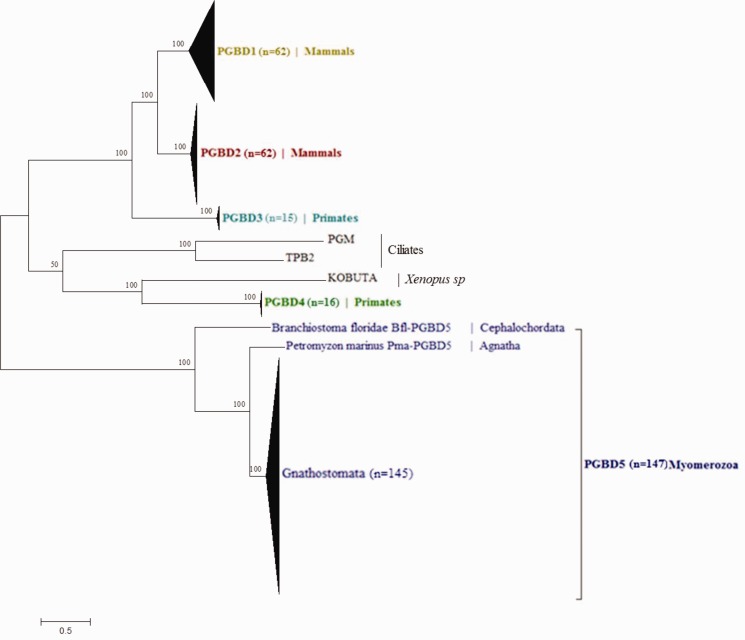
The unrooted tree inferred from the 305 PGBDs (fig. 4) showed that the eight clades of PGBD are clearly identified and well supported (all bootstrap values are equal to 100). PGBD1 and PGBD2, exclusively found in mammals, are related to PGBD3 only present in primates, while PGBD4, also detected exclusively in primates, is closely related to KOBUTA. PGM and TPB2 of ciliates are grouped together. PGBD5, found in a large spectrum of species, is quite distinct from the other PGBDs. As previously described (Pavelitz et al. 2013), this reflects an early domestication event. In this respect, its absence in echinoderms, hemichordates and urochordates (fig. 5), suggests a domestication event at least in the ancestor of the Myomerozoa lineage.
Fig. 4.—
Phylogenetic tree of ‘domesticated’ elements. This phylogenetic tree comprises 10 PGBDs used as queries, including Human PGBD (Hsa-PGBD1, 2, 3, 4, 5), two orthologous to PGBD5 of Humans, namely Pma from the agnathe Petromyzon marinus and Bfl from the cephalochordate Branchiostoma floridae, Tetrahymena thermophila (TPB2) and Paramecium tetraurelia (PGM) piggyMac, KOBUTA from Xenopus sp, and 295 homologous PGBD sequences including 61 PGBD1, 61 PGBD2, 14 PGBD3, 15 PGBD4 and 144 PGBD5 sequences. Conserved transposase regions including 240 residues (see supplementary material S4, Supplementary Material online) were used to generate a maximum likelihood (ML) tree with LG + F + G matrix (best evolutionary scenario proposed by Protest 2.4). The number of sequences in the PGBDs groups is given in brackets.
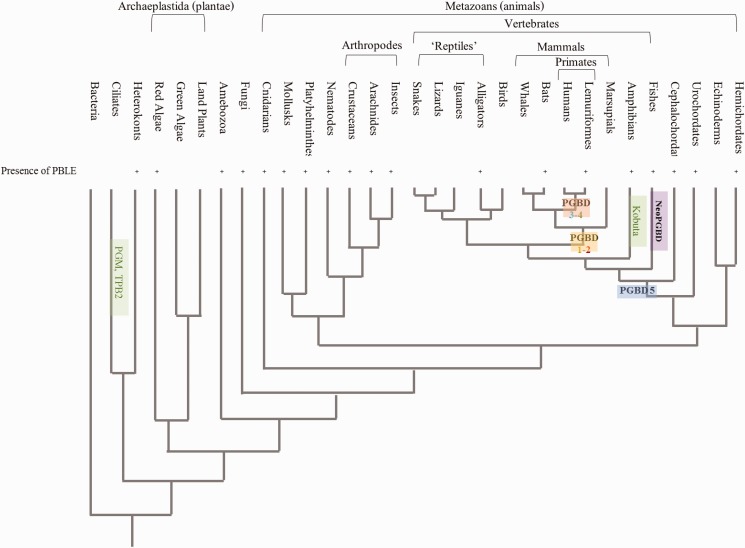
Fig. 5.—
Distribution of PGBD sequences in eukaryotes. Each domesticated group of elements is highlighted by specific colors. PGBD1 and PGBD2 are found in mammals, PGBD3 and PGBD4 in primates, PGBD5 present a large spectrum of species belonging to Myomerozoa (including cephalochordates and all vertebrates). NeoPGBD is only specific to teleost fishes (Actinopterygii). TPB2 and PGM are found in ciliates and KOBUTA in Xenopus sp. The presence of PBLE elements is mentioned by “+”. The general phylogeny used here is redrawn from ref. http://www.talkorigins.org/faqs/comdesc/phylo.html#fig1 and has been modified to add some organisms.
Relationship between PGBD/PGBD-like and PBLE
In addition to the 157 PBLE (elements with both ends) and 305 PGBD identified previously, 101 PGBD-like-elements including eight sequences containing a single TIR (5′ or 3′) and 93 sequences with no TIR were detected (supplementary material S1, Supplementary Material online). In order to infer the relationship between all piggyBac-related elements, a maximum likelihood tree was built from the most conserved blocks. These blocks roughly correspond to the region surrounding the catalytic domain DDD. This provides an alignment of a total of 170 aa after concatenation (supplementary material S5, Supplementary Material online).
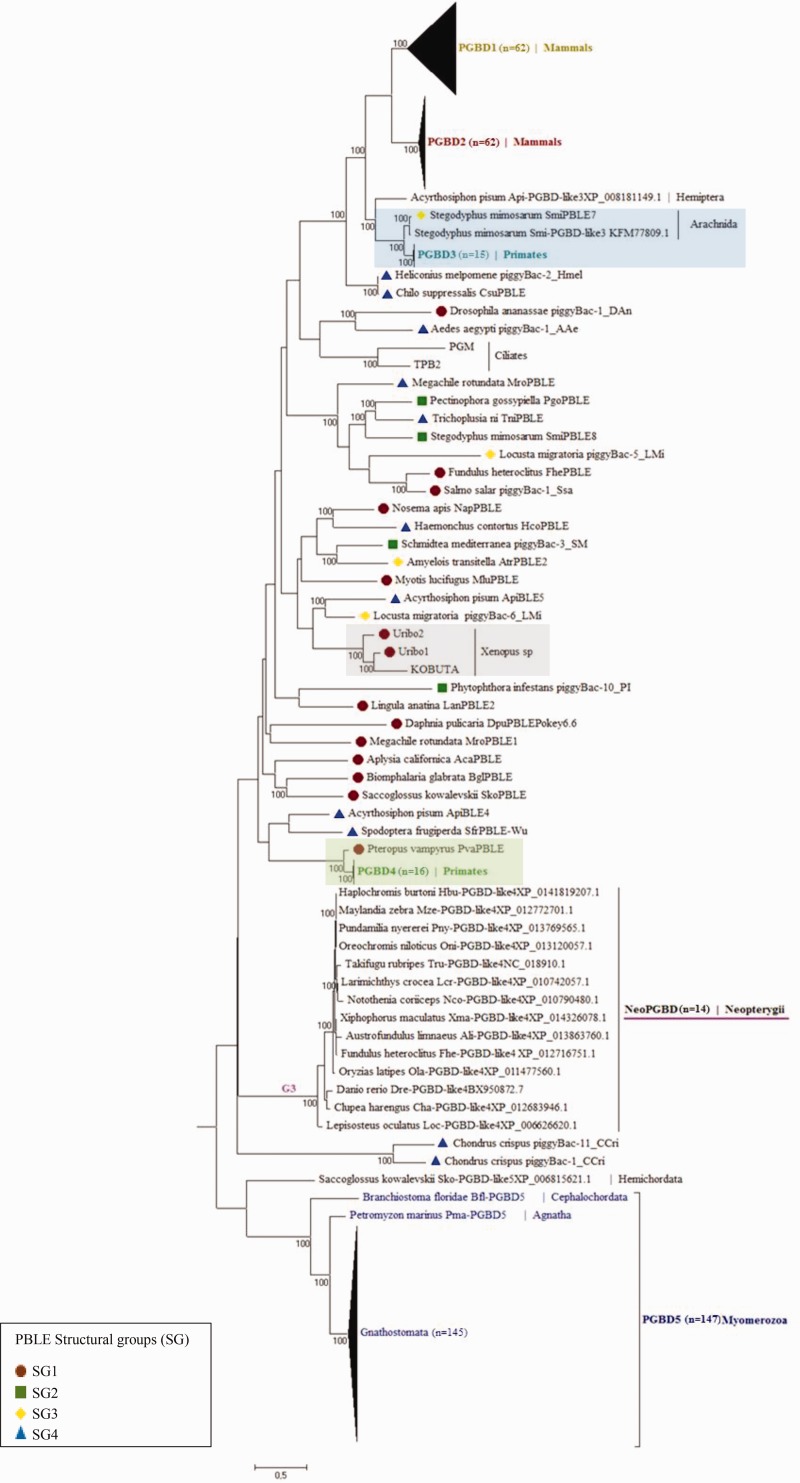
In this tree (fig. 6 and supplementary material S10, Supplementary Material online), PGBD1, PGBD2 and PGBD3 remain clustered compared to the tree of figure 4. Interestingly, three sequences—one from the aphid Acyrthosiphum pisum (Api-PGBD-like3) and two from the spider Stegodyphus mimosarum (SmiPBLE7 and Smi-PGBD-like3)—appear closely related to those of PGBD3; nevertheless, Api-PGBD-like3 and Smi-PGBD-like3, found in multiple copies with indels, appear not to be domesticated. The analysis of flanking regions of these ORFs (5 kb on both side) reveals in 5′ the presence of a potential 3′ splicing site (TTTTCTCTTCATATTTTTTTAG in SmiPBLE7 and Smi-PGBD-like3, TTTTTACTAGTTTTAG in Api-PGBD-like3 and CCTTTTTTCCGTTTTAG in PGBD3) and in 3′, a potential polyadenylation signal (AATAAA). These observations suggest that all these sequences share a common ancestor. Moreover, the 3′ splicing site (TTTTTTCTGTGTTAATATCTAG) and polyadenylation signal are also found in two other lepidopteran sequences (piggyBac-2_Hmel from Heliconius melpomene and CsuPBLE from Chilo suppressalis), closely related to the three groups of domesticated elements PGBD1, 2 and 3. According to the phylogenetic analysis (fig. 6), these two sequences diverged before the emergence of the three groups. The most parsimonious explanation is that the splicing site and the polyadenylation signal were present in the common ancestor of all these sequences and were then lost along the branches leading to PGBD1 and PGBD2, while being retained in the clade containing PGBD3. Outside of this clade, these motifs can be present but with a patchy distribution.
Fig. 6.—
Phylogenetic tree of piggyBac superfamily. This phylogeny is based on amino acid sequences covering about 170 residues (supplementary material S5, Supplementary Material online). For simplicity, only 355 sequences are represented, including 305 PGBD sequences, 17 PGBD-like sequences and 33 PBLE sequences. Maximum likelihood (ML) method, with LG + F + G matrix, is used to construct this tree. Only bootstrap values (100 replications) higher than 70% are labeled. Groups including PGBD elements and PBLE and/or PGBD-like, are framed in colors. Red dots, green squares, yellow lozenges, and blue triangles refer to the different PBLE SGs, that is, SG1, SG2, SG3, and SG4, respectively. The putative domesticated sequences (NeoPGBD), found only in Actinopterygii, are underlined in purple. Sequence names: the term PBLE is used for sequences extracted from literature, or copies newly characterized in this study, while the term piggyBac is restricted to sequences extracted from Repbase (real name of these sequences in this database). TPB2 is from Tetrahymena thermophila, PGM from Paramecium tetraurelia and KOBUTA from Xenopus sp.
The KOBUTA element forms a robust cluster with two other elements of Xenopus (Uribo1 and Uribo2). The PGBD4 and the PvaPBLE of the chiropteran Pteropus vampyrus group together (similarity = 96% and Ka/Ks = 0.23±0.01). PGBD5 is always distinct as previously described. Nonetheless, it must be stressed that a PGBD-like sequence of a hemichordata (Sko-PGBD-like5) seems closely related to this group (supplementary material S10, Supplementary Material online). However, this sequence is intronless and presents several deletions and substitutions. While degenerated, this sequence was kept in our analysis since this is the only one close to the PGBD5 group. But, it must be stressed that this sequence is probably no longer functional as PBLE nor domesticated as PGBD5 members.
In the complete tree inferred from PBLE, PGBD like, and PGBD, three distinct groups (G1, G2, and G3) are specific to Actinopterygii species (supplementary material S10, Supplementary Material online). On the one hand, the first two contain complete or partial transposase of PBLE or PGBD-like copies that can be present in several copies in each species. On the other hand, the third group corresponds to sequences found as single copy in 14 teleost fish species (supplementary material S1, Supplementary Material online). All these sequences are annotated PGBD-like4 in database. No ortholog is detected in other vertebrates. This group, here named NeoPGBD, presents a conserved putative ORF with few substitutions or gaps but with no frameshift or non-sense mutation (supplementary material S8, Supplementary Material online). The only exception is for Ali-PGBD-like4 showing a stop codon. The ORF length varies from 649 to 682 aa with an average similarity of 86% and Ka/Ks value of 0.094 ± 0.004. This is consistent with strong purifying selection acting on this insertion. On one hand, no promoter, no splicing signal and no binding site of transcription factor is found in the 5′ region, but a highly conserved sequence of 365 bp (identity > 90%) of unknown function is located immediately upstream the ORF. On the other hand, a potential polyadenylation signal AATAAA seems to be conserved in the 3′ region except in Lpe-PGBD-like4. When the analysis of the flanking regions is extended (5 kb on each side), the identity level remains relatively high (65% in 5' and 70% in 3'), and no gene or associated domain can be detected.
Discussion
A large diversity of transposons belonging to the piggyBac superfamily has been documented in eukaryotes, except in plants, from data mining in databanks. This study provides a detailed characterization of these elements, their specific distribution and their possible evolution.
A total of 157 PBLE, including 50 new sequences, were analyzed. The target site is TTAA. Rarely, for only five elements (unique copy), other TSD can be observed (supplementary material S2, Supplementary Material online). This is probably due to mutations, since the 5′ TSD and 3′ TSD sequences are not the same. In only two cases (Yabusame1, AcePBLE), the two TSD are identical, but Yabusame1 does not seem active in spite of a putative intact ORF (Daimon et al. 2010).
TIRs of PBLE appear to be divergent both in length and sequence except for the first highly conserved cytosine. TIRs play an essential role in transposase recognition and cleavage from the target site (Elick et al. 1997; Li et al. 2001). Mutagenesis experiments with the piggyBac element confirmed that the 3′ terminal G plays an important role in the selection of the excision site and that the deletion of a single 3′G nucleotide from one of the termini is sufficient to abolish excision (Elick et al. 1997). For other transposons, similar examples also indicate that mutations of the first two base pairs of TIR lead to defective excision processes (Haniford and Kleckner 1994). In this study, we show that the first three residues of TIR were not usually CCC/GGG but C[C/A/T] [C/A/T]. Nevertheless, these copies are functional with an excision activity (Xu et al. 2006; Hikosaka et al. 2007; Luo et al. 2011; Mitra et al. 2013). Therefore, the functional impact of the second and third residues of TIR is not so clear since they are not always responsible for defects in the excision process.
Structural architecture of PBLE is also variable and the occurrence of DRs close to TIR was previously reported (Wu and Wang 2014). In the present work, four SGs were identified according to the presence/absence of DR and/or STIR. This suggests that these elements are structurally highly flexible. Two hypotheses can be proposed to explain the specific distribution of the different structures. On one hand, the SG1 (with only two TIRs) is the ancestral structure, while the other ones derive from the ancestral sequence by independent acquisitions of DR and STIR (convergence) and/or by horizontal transfers. On the other hand, the four SGs were already present in the common ancestor of all species in which PBLE copies have been found, and their patchy distribution is due to independent loss in several lineages. However, the absence of consensus sequences between these regions and the incongruence with the tree derived from transposases are in favor of independent acquisitions (convergences) and rapid evolution of these regions. The inconsistencies observed between the transposase trees, the element structures, and with the species phylogeny also suggest that piggyBac might be frequently and successfully horizontally transferred. This may be due to a “host factors independent” activity, and could explain why this element is a powerful vector for genome engineering.
Furthermore, the presence of internal repeats (DR/STIR), sometimes with palindromic motifs, raises several questions. Are these regions involved in the element activity via the transposase binding and stabilization of the transposase-TIR/DR/STIR complex? This probably reflects a rapid coevolution between the transposase and the different terminal repeated sequences, but this remains to be functionally demonstrated. For the mariner-like elements (MLE), it was shown that conserved palindromic and mirror motifs within TIR, are important features of the transposase-TIR interaction (Bigot et al. 2005).
PBLE transposase includes a D268D346D447D450 catalytic domain and a Cysteine Rich Domain (CRD) in C-terminal region (Keith et al. 2008). An additional Histidine can be observed in some sequences between the fifth and the sixth Cysteines (Fraser et al. 1983; Arkhipova and Meselson 2005). However, while Cysteines are found in almost all sequences and are relatively well conserved, the presence and position of the Histidine is highly variable. Thus, Histidine impact on the transposase structure and functionality is not clear.
Like all TEs, piggyBac are prone to indels, recombinations and mutations that inactivate many copies, which can be rapidly lost in absence of trans-mobilization (Robertson 1993; Le Rouzic et al. 2007; Hua-Van et al. 2011). Nevertheless, molecular domestication events may occur, in which TE-insertions are advantageous, maintained by natural selection and turned into regular genes (Le Rouzic et al. 2007; Sinzelle et al. 2009; Hua-Van et al. 2011). Such sequences can be distinguished from defective copies from a low ratio Ka/Ks, indicating that they evolve under purifying selection and do not correspond to pseudogenes (Sarkar et al. 2003; Newman et al. 2008; Pavelitz et al. 2013). Otherwise, in the present study, some evidence suggests a new domestication event of a piggyBac element. This new group is exclusively found in Neopterygii (NeoPGBD). This sequence is present in single copy per genome and its ORF is well conserved in several species orders, suggesting an ancient domestication in Neopterygii, that is, at least 250 Ma (Betancur et al. 2013). Similarly, to the previous PGBD groups already described, the Ka/Ks ratio is low. Moreover, the high conservation of the region upstream the sequence of these PGBD suggests the existence of selective pressures to maintain a putative function, which remains unknown.
The presence of these exaptations raises the question of their emergence: what is the origin of the PGBD sequences? Answering this question remains difficult, but several hypotheses can be proposed. First, several PBLEs seems to be potentially active, or at least they were within the recent past. Second, the general phylogeny, including PBLE, PGDB and PGDB-like, shows that a close relationship can be observed between PGBD and PBLE or PGBD-like. For instance, PvaPBLE of Pteropus vampyrus is closely related to the members of PGBD4. The PBLE (Uribo1 and Uribo2) and KOBUTA also form a robust clade. Similarly, with PGBD3 members, SmiPBLE7 and Smi-PGBD-like3 of Stegodyphus mimosarum and Api-PGBD-like3 of Acyrthosiphon pisum, are grouped together.
For PGBD5, the domestication probably occurred along the branch leading to the Myomerozoa, as described by Pavelitz et al. (2013), based on transposase sequence, intron location, and microsynteny. However, an earlier domestication, followed by a loss of this element in all branches except one leading to Myomerozoa, cannot be excluded. To check this hypothesis, a BlastN search, using PGBD5 and its flanking sequences as query, in urochordates (Ciona intestinalis, Oikopleura dioica), echinoderms (Strongylocentrotus purpuratus, Acanthaster planci) and hemichordate (Saccoglossus kowalevskii), do not allow us to detect PGBD5-like sequences with introns. Indeed, only a few fragments of intronless piggyBac–like ORF can be found. Moreover, no sequence similar to those of the flanking region of PGBD5 can be found in hemichordate, echinoderms, and urochordates. From these observations, and assuming that the genome assemblies of these species are correct, an evolutionary scenario can be proposed. The insertion at the origin of PGBD5 occurred in the ancestor of the Myomerozoa. This initial insertion rapidly acquired a new function and was probably under a high selective pressure leading to a selective sweep on the entire region including the flanking sequences.
Therefore, based on the phylogenetic proximities between PBLE and PGBD sequences, and because domesticated sequences are distinct monophyletic groups, we can hypothesize that each PGBD group (including NeoPGBD), as well as PGM, TPB2 and KOBUTA genes, derive from a single and specific ancestral PBLE sequence, suggesting that the domestication event at the origin of each group occurs once.
The evolutionary trajectory of PGBD is not systematically accompanied by modifications of transposase activity (see, for instance, Sarkar et al. 2003; Pavelitz et al. 2013). In this respect, piggyMac, TPB2 and KOBUTA have conserved the DDD catalytic domain and the C-terminal region of the PBLE and are involved in excision mechanisms. This DDD motif is also found in the members of the PGBD4 group, but there is no proof that this is associated to a transposase activity (Mitra et al. 2008). For PGBD3, while the DDD motif is not strictly conserved, the C-terminal transposase domain of the human CSB-PGBD3 fusion protein is able to mobilize the MER85 (Gray et al. 2012). Moreover, the protein encoded by human PGBD5 seems to be involved in stereotypical cut-and-paste DNA transposition in human cells, but in this case, the genomic integration required distinct aspartic acid residues, and specific DNA sequences (including TIR) compared to those of Uribo2, piggyBac, piggyMac and piggyBat (Henssen et al. 2015). For the remaining PGBD (1 and 2), the DDD motif is not conserved and no mobilization activity can be suspected. Therefore, a switch to new unknown host functions, since these sequences are under purifying selection, probably occurred.
Several cases of true or putative exaptation of TEs have been reported in all domains of the tree of life (see, for instance, Hoen and Bureau, 2012 for exaptation in plants). However, it seems that all families of TEs are not “equal” in terms of their fate, since some of them tend to be more prone to be domesticated than others. Is it a relevant observation or a sampling effect? Such an observation has been recently made for the members of the Mutator-like superfamily (Joly-Lopez et al. 2016). Are the members of the piggyBac superfamily prone to such fate? If so, what could be the reason for this? Is it due to specific internal features, a transpositional mechanism and/or particular genomic locations of these elements?
While several features of piggyBac can be listed, it remains difficult to know which one(s) are responsible for their domestication success. For instance, full-length copies of piggyBac can be found in a large spectrum of species (this work), so this element can potentially move in a large set of organisms and excise precisely without host damage (see the references in Mitra et al. 2008). In addition, according to Newman et al. (2008), piggyBac could be a natural “exon trap” as shown for PGBD3 since a potential 3′ splicing site and a polyadenylation signal can be detected. These allow its insertion into CSB intron 5, generating an N-terminal fusion protein. Moreover, the analysis of the 5′ flanking regions PGBD1 and PGBD2 reveals that five and two exons, respectively, derived from the host gene (supplementary material S9, Supplementary Material online). Again, TniPBLE transposase is able to tolerate N-terminal fusion and to retain a significant transposition activity. In this respect, this element seems more flexible than Sleeping Beauty, Tol2 and Mos1 transposases (Wu et al. 2006). Based on the “3′ exon trap” hypothesis, Newman et al. (2008) also suggest that piggyBac could benefit from the efficient host promoter (see also Gray et al. 2012). All these characteristics may be the reason for its colonizing success and its capacity to be domesticated.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
MB, MM, and PC conceived and designed research. MB performed research. MB, AHV, MM, and PC contributed analysis tools. JDR participated to the sequences analysis. MB, AHV, MM, and PC wrote the article.
Supplementary Material
Acknowledgments
This work was financially supported by the Centre National de la Recherche Scientifique [UMR 9191], the University Paris-Sud, the Tunisian Ministry of Higher Education and Scientific Research and the University of Tunis El Manar. Authors thank Mireille Bétermier and Julien Bischerour for their helpful comments and Malcolm Eden for the English review of the manuscript.
Literature Cited
- Arkhipova IR, Meselson M. 2005. Diverse DNA transposons in rotifers of the class Bdelloidea. Proc Natl Acad Sci U S A. 102 (33):11781–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry C, et al. 2009. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia . Genes Dev. 23:2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur R.R, et al. 2013. The tree of life and a new classification of bony fishes. PLOS Curr Tree Life. Edition 1.5. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed]
- Bigot Y, Brillet B, Augé-Gouillou C. 2005. Conservation of palindromic and mirror motifs within inverted terminal repeats of mariner-like elements. J Mol Biol. 351:108–116. [DOI] [PubMed] [Google Scholar]
- Britten RJ. 1996. DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci U S A. 93:9374–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpes MP, et al. 2009. Molecular analysis of a mutant Anticarsia gemmatalis multiple nucleopolyhedrovirus (AgMNPV) shows an interruption of an inhibitor of apoptosis gene (iap-3) by a new class-II piggyBac-related insect transposon. Insect Mol Biol. 18:747–757. [DOI] [PubMed] [Google Scholar]
- Cary LC, et al. 1989. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172:156–169. [DOI] [PubMed] [Google Scholar]
- Casola C, Hucks D, Feschotte C. 2008. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol. 25:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Vogt A, Mochizuki K, Yao MC. 2010. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila . Mol Biol Cell. 21:1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Udit S, Batzer MA, Feschotte C. 2006. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 103:8101–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon T, et al. 2010. Recent transposition of yabusame, a novel piggyBac-like transposable element in the genome of the silkworm, Bombyx mori . Genome 53:585–593. [DOI] [PubMed] [Google Scholar]
- Elick TA, Lobo N, Fraser MJ. 1997. Analysis of cis-acting DNA elements required for piggyBac transposable element excision. Mol Gen Genet. 255:605–610. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler SR. 2002. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, D.C: : American Society of Microbiology Press; p. 1147–1158. [Google Scholar]
- Fraser MJ, Smith GE, Summers MD. 1983. Acquisition of host cell DNA sequences by baculoviruses: Relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J Virol. 47:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MJ, Ciszczon T, Elick T, Bauser C. 1996. Precise excision of TTAA specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 5:141–151. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521. [DOI] [PubMed] [Google Scholar]
- Gray LT, Fong KK, Pavelitz T, Weiner AM. 2012. Tethering of the conserved piggyBac transposase fusion protein CSB-PGBD3 to chromosomal AP-1 proteins regulates expression of nearby genes in humans. PLoS Genet. 8(9):e1002972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D, Kleckner N. 1994. Tn10 transposition in vivo: temporal separation of cleavages at the two transposon ends and roles of terminal basepairs subsequent to interaction of ends. EMBO J. 13:3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hencken CG, Li X, Craig NL. 2012. Functional characterization of an active Rag-like transposase. Nat Struct Mol Biol. 19:834–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen AG, et al. 2015. Genomic DNA transposition induced by human PGBD5. Botchan MR, ed. eLife, 4:e10565. doi:10.7554/eLife.10565. [DOI] [PMC free article] [PubMed]
- Hikosaka A, Kobayashi T, Saito Y, Kawahara A. 2007. Evolution of the Xenopus piggyBac transposon family TxpB: domesticated and untamed strategies of transposon subfamilies. Mol Biol Evol. 24:2648–2656. [DOI] [PubMed] [Google Scholar]
- Hoen DR, Bureau T. 2012. Transposable element exaptation in plant In Grandbastien MG, Casacuberta JM, editors. Plant transposable elements: impact on genome, structure and function. Topics in Current Genetics; p. 219-251. [Google Scholar]
- Hua-Van A, Le Rouzic A, Boutin TS, Filée J, Capy P. 2011. The struggle for life of the genome’s selfish architects. Biol Direct. 6(1):19–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18(9):486–487. [DOI] [PubMed] [Google Scholar]
- Joly-Lopez Z, Hoen DR, Blanchette M, Bureau TE. 2016. Phylogenetic and genomic analyses resolve the origin of important plant genes derived from transposable elements. Mol Biol Evol. doi:10.1093/molbev/msw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2005. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith JH, Schaeper CA, Fraser TS, Fraser MJ., Jr. 2008. Mutational analysis of highly conserved aspartate residues essential to the catalytic core of the piggyBac transposase. BMC Mol Biol. 9:73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. 1997. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A. 94:7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30 (22):3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic A, Boutin TS, Capy P. 2007. Long-term evolution of transposable elements. Proc Natl Acad Sci U S A. 104:19375–19380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lobo N, Bauser CA, Fraser MJ. 2001. The minimum internal and external sequence requirements for transposition of the eukaryotic transformation vector piggyBac. Mol Genet Genom. 266:190–198. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819. [DOI] [PubMed] [Google Scholar]
- Liu, et al. 2007. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 27(3):1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo N, Li X, Fraser MJ. 1999. Transposition of the piggyBac element in embryos of Drosophila melanogaster, Aedes aegypti and Trichoplusia ni . Mol Gen Genet. 261:803–810. [DOI] [PubMed] [Google Scholar]
- Luo GH, Wu M, Wang XF, Zhang W, Han ZJ. 2011. A new active piggyBac-like element in Aphis gossypii . Insect Sci. 18(6):652–662. [Google Scholar]
- Luo GH, et al. 2014. Molecular characterization of the piggyBac-like element, a candidate marker for phylogenetic research of Chilo suppressalis (Walker) in China. BMC Mol Biol. 15:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, McDonald JF, Nouaud D, Anxolabéhère D. 1999. Molecular domestication – More than a sporadic episode in evolution. Genetica 107:197–207. [PubMed] [Google Scholar]
- Mitra R, Fain-Thornton J, Craig NL. 2008. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 27:1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, et al. 2013. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc Natl Acad Sci U S A. 110:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. 2008. An abundant evolutionarily conserved CSB-piggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 4(3):e1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK, Feschotte C. 2007. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 17:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan HJ, Smith JD, Hubley RM, Ray DA. 2010. PiggyBac-ing on a primate genome: novel elements, recent activity and horizontal transfer. Genome Biol Evol. 4:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelitz T, Gray LT, Padilla SL, Bailey AD, Weiner AM. 2013. PGBD5: a neural-specific intron containing piggyBac transposase domesticated over 500 million years ago and conserved from cephalochordates to humans. Mobile DNA 4:23–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton EH, Sullender BW, Crease TJ. 2002. Pokey, a new DNA transposon in Daphnia (cladocera: crustacea). J Mol Evol. 55:664–673. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C, Wessler SR. 2005. Unexpected diversity and differential success of DNA transposons in four species of Entamoeba protozoans. Mol Biol Evol. 22(9):1751–1763. [DOI] [PubMed] [Google Scholar]
- Ray DA, et al. 2008. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus . Genome 18:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM. 1993. The mariner transposable element is widespread in insects. Nature 362:241–245. [DOI] [PubMed] [Google Scholar]
- Sarkar A, et al. 2003. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol Genet Genom. 270(2):173–180. [DOI] [PubMed] [Google Scholar]
- Sinzelle L, Izsvák Z, Ivics Z. 2009. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci. 66:1073–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZC, Wu M, Miller TA, Han ZJ. 2008. piggyBac-like elements in cotton bollworm, Helicoverpa armigera (Hubner). Insect Mol Biol. 17:9–18. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN. 2006. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays 28:913–922. [DOI] [PubMed] [Google Scholar]
- Wang J, Ren X, Miller TA, Park Y. 2006. piggyBac-like elements PLE in the tobacco budworm, Heliothis virescens (Fabricius). Insect Mol Biol. 15:435–443. [DOI] [PubMed] [Google Scholar]
- Wang JJ, Du YZ, Wang SZ, Brown SJ, Park Y. 2008. Large diversity of the piggyBac-like elements in the genome of Tribolium castaneum . Insect Biochem Mol Biol. 38(4):490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2009. piggyBac-like elements in the pink bollworm, Pectinophora gossypiella . Insect Mol Biol. 19:177–184. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
- Wu C, Wang S. 2014. PLE-wu, a new member of piggyBac transposon family from insect, is active in mammalian cells. J Biosci Bioeng. 118(4):359–366. [DOI] [PubMed] [Google Scholar]
- Wu M, Sun ZC, Hu CL, Zhang GF, Han ZJ. 2008. An active piggyBac-like element in Macdunnoughia crassisigna . Insect Sci. 15:521–528. [Google Scholar]
- Wu M, et al. 2006. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 103(41):15008–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, et al. 2011. Cloning and characterization of piggyBac-like elements in lepidopteran insects. Genetica 139(1):149–154. [DOI] [PubMed] [Google Scholar]
- Xu HF, et al. 2006. Identification and characterization of piggyBac-like elements in the genome of domesticated silkworm, Bombyx mori . Mol Genet Genom. 276:31–40. [DOI] [PubMed] [Google Scholar]
- Yusa K. 2015. piggyBac transposon. Microbiol Spectr. 3(2): MDNA3-0028-2014. doi:10.1128/microbiolspec.MDNA3-0028-2014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.