Abstract
Development is generally regarded as a unidirectional process that results in the acquisition of specialized cell fates. During this process, cellular identity is precisely defined by signaling cues that tailor the chromatin landscape for cell-specific gene expression programs. Once established, these pathways and cell states are typically resistant to disruption. However, loss of cell identity occurs during tumor initiation and upon injury response. Moreover, terminally differentiated cells can be experimentally provoked to become pluripotent. Chromatin reorganization is key to the establishment of new gene expression signatures and thus new cell identity. Here we explore an emerging concept that lysine acetyltransferase enzymes drive cellular plasticity in the context of somatic cell reprogramming and tumorigenesis.
Keywords: acetylation, histone, embryonic stem cells, reprogramming, plasticity
Graphical Abstract
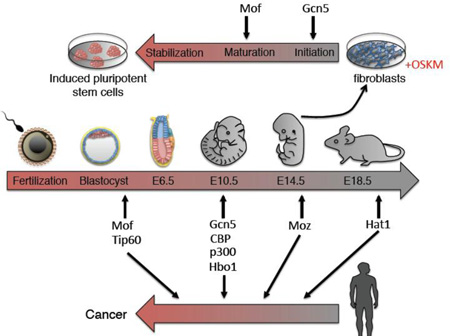
The role of KAT enzymes in embryonic development, pluripotency induction and tumorigenesis is depicted.
Introduction
Cell fate is established by signaling pathways that drive requisite gene expression programs. Central to modulation of these pathways are reversible protein post-translational modifications (PTMs) that permit dynamic cellular regulation and flexibility in response to signaling cues and stimuli. To date, over two hundred different PTMs have been identified that influence countless aspects of signaling regulation1. Importantly, PTMs also play a prominent role in altering chromatin structure and function.
Chromatin is composed of repeating nucleosome units of approximately 146bp of DNA wrapped around a histone core octamer, containing two copies of each histone protein H2A, H2B, H3 and H42. Numerous PTMs occur on histones, including acetylation, phosphorylation, methylation, ubiquitylation, sumoylation, glycosylation, citrullination, ADP-ribosylation, and other types of acylations3; 4; 5; 6; 7. Collectively, these modifications translate cellular signals within the nucleus, functioning to negotiate chromatin accessibility as well as recruitment of additional proteins and enzymatic complexes for different biological processes8. Hence, histone modifications greatly impact folding of the 3D genome and play fundamental roles in establishing appropriate gene expression networks that dictate cell fate throughout development.
Lysine acetylation was first detected on histones over fifty years ago and has long been associated with gene activation9; 10. Reversible acetylation is balanced by the activities of lysine acetyltransferases (KATs), which are writer enzymes that catalyze acetyl group transfer from acetyl coenzyme A (acetyl-CoA) to the epsilon amino side chain of lysine resides, and histone deacetylases (HDACs), which are eraser enzymes that remove these marks. In this manner, specific lysines are acetylated within the globular domains of histones H3 and H4, as well as within the unstructured amino-terminal tails of H2A, H2B, H3 and H411; 12 (Figure 1). Acetylation of histones neutralizes the positive charge on lysine residues and relieves inter- and intra-nucleosomal interactions to create a less compact chromatin environment that promotes transcriptional activation13; 14; 15; 16; 17, as well as DNA repair, replication, and native centromere assembly18; 19; 20. Furthermore, acetylated residues cooperate with other PTMs to boost these pathways and block other lysine modifications, including methylation and ubiquitylation, which can elicit opposite regulatory outcomes21. Accordingly, depletion of individual KATs also vastly alters the pattern of other local histone modifications22.
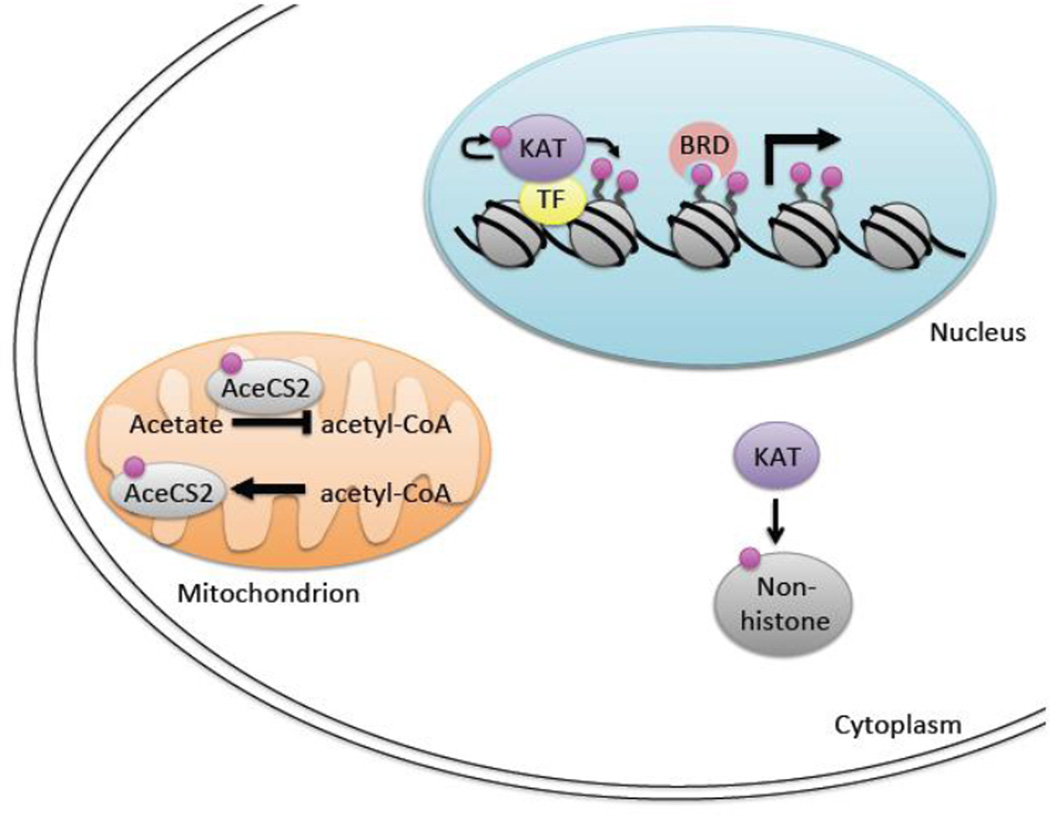
Figure 1.
Cellular lysine acetylation. Representative lysine acetylation events are displayed in the nucleus, cytoplasm, and mitochondria. Within the nucleus, KATs are recruited by transcription factors (TFs) to chromatin, where they acetylate surrounding histones and function to reduce chromatin compaction. Following acetylation, bromodomain-containing proteins (BRD) may interact with acetylated histones to further promote gene activation. In the cytoplasm, it is shown that KATs acetylate non-histone proteins. KAT-mediated non-histone acetylation further occurs in the nucleus and mitochondria. As an example, lysine acetylation of the Acetyl-CoA synthetase 2 enzyme (AceCS2) blocks acetyl-CoA production in mitochondria. Pink circle: acetylation.
Beyond physically altering chromatin structure, deposition of acetyl-moieties on histones additionally creates docking sites for bromodomain23, YEATS (named from the family members Yaf9, ENL, AF9, Taf14 and Sas5) domain24, and select PHD (Plant homeodomain) finger containing reader proteins25; 26; 27. These domains are present within many transcription-related proteins and chromatin modifying enzymes, including KATs28, and they act as adapter motifs that link acetylated histones to other regulatory factors and complexes. Therefore, histone acetylation is fundamental to promoting chromatin accessibility for the transcriptional machinery.
Reversible lysine acetylation also occurs on non-histone proteins, outside of chromatin. In mammals, over 8000 acetyl-lysine sites are present on proteins that reside primarily in nuclear, cytoplasmic and mitochondrial subcellular compartments29; 30; 31; 32, and many of these modification sites are conserved across different species, implying their significance33; 34. Protein acetylation imparts a variety of functional outcomes, ranging from altering enzymatic activity and protein-protein interactions to influencing nucleic acid binding, protein stability and subcellular localization35. This includes auto-acetylation of a number of KATs, which promotes catalytic activity and/or substrate binding36; 37; 38; 39; 40. Acetylated sites also crosstalk with other PTMs in non-histone proteins. For example, acetylation blocks lysine ubiquitylation and ensuing degradation of proteins such as the p53 transcription factor41 and the Smad7 negative regulator from the transforming growth factor beta (TGFβ) pathway42. Furthermore, many key mitochondrial proteins are subject to acetylation, including enzymes involved in glycolysis, gluconeogenesis, the tricarboxylic acid (TCA) cycle, the urea cycle, fatty acid metabolism and glycogen metabolism43. Although the contribution of KATs to mitochondrial protein acetylation remains unclear, as high acetyl-CoA levels within this organelle may drive non-enzymatic acetylation for the bulk of acetylation events, acetyl lysine deposition is tightly linked to cellular metabolism on multiple levels. For instance, acetyl-CoA supply across the cell is restricted by acetylation-mediated inactivation of both cytosolic Acetyl-CoA synthetase 1 (AceCS1) and mitochondrial Acetyl-CoA synthetase 2 (AceCS2) enzymes44; 45. Interconnection of acetylation states and carbon source utilization are particularly important in stem cells, as the glycolytic state of embryonic stem cells (ESCs) supports acetyl-CoA production to promote histone acetylation and maintain stemness46. In contrast, differentiating ESCs experience a rapid drop in glycolysis that leads to a reduction in both acetyl-CoA and histone acetylation.
Clearly lysine acetylation events on both histone and non-histone proteins control numerous biological processes. Thus, it is not surprising that multiple KATs and HDACs are required for embryonic viability and are de-regulated in cancer. These enzymes play important roles in delineating cell-specific gene expression pathways and therefore function as key determinants of cell fate. Recent work indicates that specific KATs also promote cellular plasticity by creating a more open chromatin configuration amenable to activation of new gene expression networks and act as powerful drivers of chromatin reorganization during nuclear reprogramming processes, such as somatic cell reprogramming, trans-differentiation and oncogenesis. Here, we focus specifically on the multi-faceted ability of KATs to direct embryonic development, confer cellular plasticity, and contribute to nuclear reprogramming.
Lysine acetyltransferase (KAT) families in development
KAT enzymes are functionally diverse and commonly exist within large multi-subunit complexes that collectively facilitate specific acetylation events. The human genome encodes 17 KATs47, that are traditionally divided into two classes depending on their subcellular localization. A-type KATs reside in the nuclear compartment and accordingly facilitate changes in chromatin compaction and transcription, while B-type KATs are present in cytosol and acetylate newly synthesized histone H3 and H4 subunits prior to de novo nucleosome assembly48; 49. However, this classification is likely far too simplistic, as additional KATs have recently been uncovered in mitochondria50; 51, golgi apparatus52 and endoplasmic reticulum53. Based on sequence and structural similarities in the catalytic domain, KATs are more appropriately grouped into five major families: GNAT (Gcn5-related N-acetyltransferase), CBP/p300 (CREB-binding protein / E1A-associated protein of 300 kDa), MYST (Moz, Ybf2/Sas3, Sas2, Tip60), nuclear receptor co-activators, and basal transcription factors11. Many KATs play fundamental roles throughout development and more specifically within ESCs, which have the unique ability to both self-renew and differentiate into all cell types of the three primary germ layers. Comparatively, ESCs generally display higher histone acetylation levels, enhanced chromatin accessibility and transcriptional hyperactivity than their differentiated counterparts54, implying that histone acetylation is an important driver of cellular plasticity. In the following section, the different KAT families and representative members that coordinate protein acetylation in the context of embryonic development are discussed (Table 1) (Figure 2).
Table 1.
Summary of KAT enzymes. The family, subcellular localization, multi-subunit complex name, substrates, and reported mouse phenotypes are listed for select lysine acetyltransferases.
| Family | Name | Subcellular localization |
Multi- subunit complex |
Primary histone substrates and non-histone substrate examples |
Mouse phenotypes | References |
|---|---|---|---|---|---|---|
| GNAT | Kat1 / Hat1 | Nucleus / Cytoplasm |
HAT-B / NuB4 |
free H4K5/K12 | Hat1−/−: die at birth |
48,
104, 105, 108, 109 |
| Kat2a / Gcn5 | Nucleus | SAGA / ATAC |
H3K9/K14, Myc, C/EBPα | Gcn5−/−: embryonic lethal (E8.5) |
64–69, 71, 91, 92, 97 |
|
| Kat2b / Pcaf | Nucleus | SAGA / ATAC |
H3K14, E2f1, Myc, Ezh2, Akt1, Acly, β-Catenin |
Pcaf−/−: viable with no
reported embryonic phenotypes |
64–69, 91, 92, 97 |
|
| Hat4 / Naa60 | Golgi | free H4K20/K79/K91 | Not determined | 52 | ||
| CBP/ p300 |
Kat3a / CBP | Nucleus / Cytoplasm |
H3K18 / K27 |
Cbp−/−: embryonic
lethal E9.5- E10.5 |
97, 127, 128 | |
| Kat3b / p300 | Nucleus / Cytoplasm |
H3K18 / K27 |
p300−/−: embryonic
lethal E9- E11.5 |
97, 127, 128 | ||
| MYS T | Kat5 / Tip60 | Nucleus / Cytoplasm |
TIP60 | H4K5/K8/K12, H2AK5, H2A.Z, H2A.X, p53, Atm |
Tip60−/−: embryonic
lethal between E3.5-E7.5 |
135–137, 144 |
| Kat6a / Moz / Myst3 |
Nucleus | MOZ | H3K14, p53, Runx2 |
Moz−/−: embryonic
lethal around E15 |
155, 158, 159 | |
| Kat6b / Morf / Myst4 |
Nucleus | MORF | H3K14, Runx2 |
Morfgt/gt:
viable / brain development defect |
155, 161, 162 | |
| Kat7 / Hbo1 / Myst2 |
Nucleus | HBO1 | H3K14, H4K5/K8/K12 | Hbo1−/−: embryonic lethal E10.5 | 155–157, 163 | |
| Kat8 / Mof / Myst1 |
Nucleus | NSL / MSL | H4K16, p53, Atm, Nrf2 | Mof−/−: embryonic lethal E4.5 | 148–150 | |
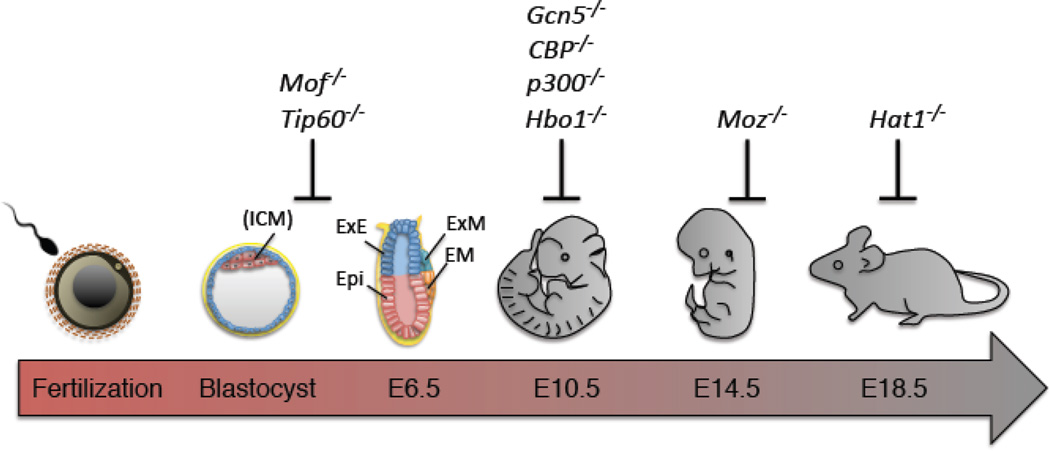
Figure 2.
The involvement of KATs in development. A timeline of mouse development is shown highlighting the points of embryonic lethality in different KAT null mouse lines. ICM: inner cell mass, Epi: Epiblast, ExE: extra-embryonic ectoderm, EM: embryonic mesoderm, ExM: extra-embryonic mesoderm.
GNAT Superfamily
The GNAT superfamily is evolutionarily conserved from bacteria to mammals. This family is composed of A-type (Kat2a/Gcn5, Kat2b/Pcaf, Kat9/Elp3) and B-type KATs (Kat1/Hat1), as well as KATs that reside in other subcellular compartments (Gcn5l1, Atat1, Hat4, Nat8 and Nat8b). Altogether, these KATs control a variety of cellular process, such as transcriptional activation55; 56, transcript elongation57, histone deposition48, DNA repair52; 58; 59 and microtubule stability60. GNAT family members characteristically contain three to four common motifs in the catalytic core61, including a highly conserved Arg/Gln-X-X-Gly-X-Gly/Ala sequence that recognizes and binds acetyl-CoA62. Moreover, Kat2a/Gcn5 and Kat2b/Pcaf contain a carboxy-terminal bromodomain that associates with acetylated lysine residues23.
Gcn5 and Pcaf
Kat2a, hereafter referred to as Gcn5 (general control non-repressible 5), was the first transcription-related KAT identified in eukaryotes and is arguably the best-studied member of the GNAT family. Originally discovered in yeast as a transcriptional co-activator63, the Gcn5 ortholog in Tetrahymena thermophila (p55) was later discovered to have acetyltransferease activity17; 55. Together, these observations established a molecular link between histone acetylation and the regulation of gene expression, and also provided a foundation for the discovery of additional KATs, including the closely related family member Kat2b, which will henceforth be discussed by its initial name Pcaf (p300/CBP-associated factor)64.
Recombinant Gcn5 and Pcaf preferentially acetylate specific sites in free histones (e.g. lysine 14 in histone H3 (H3K14) for Gcn5), but fail to effectively acetylate nucleosomes in vitro, indicating that neither KAT is independently sufficient to modify chromatin64; 65; 66. In vivo, Gcn5 or Pcaf reside within two major native multi-subunit complexes, SAGA (Spt-Ada-Gcn5 acetyltransferase)66; 67 and ATAC (ADA Two-A containing)68; 69. Incorporation into these complexes enhances KAT activity and has long been suggested to expand lysine target specificity70 as well as the functionality of Gcn5 and Pcaf. However, recent work indicates that while the SAGA and ATAC increase catalytic efficiency of human Gcn5, by approximately 10- and 6-fold respectively, the specificity of Gcn5-mediated acetylation on histone octamers does not change when Gcn5 is alone or integrated into these complexes71. H3K14 still remains the primary acetylation site in vitro, while H3K9, H3K23, H3K27, H3K36, H4K5 and H4K8 residues are acetylated to a lesser extent by Gcn571. Beyond promoting catalytic activity, the SAGA and ATAC complexes regulate different functions due to their structural modularity. The SAGA complex contains four functional units that elegantly control transcriptional activation, telomere maintenance72, mRNA export73 and DNA repair74. One module consists of the acetyltransferase unit (Gcn5, Ada2b, Ada3, Sgf29) that harbors KAT activity and facilitates SAGA recruitment to H3K4me2/3 sites via the tandem Tudor domains of Sgf2975; 76 and/or to acetyl lysine residues through the bromodomain of Gcn577. A second enzymatic module, called the deubiquitination (DUB) module (Usp22, Atxn7, Atxn7L3 and Eny2) promotes H2BK120 deubiquitination during transcription78. The Spt module (Trrap, Spt3, Spt20, Staf42, Staf65γ) mediates interactions with TBP (TATA-binding protein)79 and various transcription factors, including Myc and E2f180; 81. The Taf (TBP-associated factor) module (Taf5l, Taf6l, Taf9, Taf10, Taf12) further connects SAGA to the general transcription machinery82.
In comparison to the SAGA complex, ATAC similarly contains a slightly modified KAT module (Gcn5, Ada2a, Ada3, Sgf29), but lacks many of the general transcription factors present in SAGA. Furthermore, ATAC houses a second KAT activity, which is mediated by Atac2, and contains Yeats2, which is a reader of both acetyl-lysine and crotonyl-lysine modifications 83; 84. The different subunit compositions of SAGA and ATAC reflect their disparate functions, as SAGA primarily associates with gene promoters, while ATAC is found at both promoters and tissue-specific enhancers85. The SAGA and ATAC complexes also respond to different stimuli and activate distinct subsets of inducible genes86; 87. Additionally, SAGA has been described as a general transcriptional co-activator complex88, although it is clearly recruited to specific gene promoters in ESCs89. SAGA and ATAC as well control distinct biological processes via non-histone acetylation, such as ATAC-specific acetylation of Cyclin A, which promotes mitotic progression90.
In agreement with the wide functionality of both the SAGA and ATAC complexes, Gcn5 is required for normal embryonic development in mice91; 92. Gcn5 null (Gcn5−/−) mice fail to progress beyond 10.5 days of embryonic development (E10.5), largely due to increased apoptosis in mesoderm lineages91. Furthermore, Gcn5hat/hat mice lacking catalytically active Gcn5 die by E16.5 and display both cranial neural tube closure defects and exencephaly93. The delay in lethality of these mice compared to Gcn5 null counterparts indicates that Gcn5 regulates embryogenesis through KAT-dependent as well as KAT-independent mechanisms. Gcn5 also mediates later stages of development, including neural stem cell (NSC) proliferation94 and normal skeletal patterning95, but is not required to maintain self-renewal or pluripotency of mouse embryonic stem cells (mESCs)89; 96. Gcn5 specifically co-activates Myc and E2f1 gene expression networks in mESCs, although loss of Gcn5 does not affect self-renewal or pluripotency, possibly due to redundancy with Pcaf or other KATs89. Contrary to Gcn5, Pcaf null mice develop normally91; 92. However, homozygous double null (Gcn5−/−;Pcaf−/−) embryos are severely developmentally delayed and die by E7.591, implying that Pcaf and Gcn5 share redundant functions during early embryogenesis. Compatible with this hypothesis, Gcn5 expression is increased in certain Pcaf−/− tissues that normally express Pcaf, such as liver and lung92, partially explaining why Pcaf null mice develop normally. Additionally, Pcaf and Gcn5 act redundantly to regulate H3K9 acetylation (H3K9ac) in mouse embryonic fibroblasts (MEFs)97 and control mouse adipocyte differentiation98. Nevertheless, Gcn5 does not universally compensate for Pcaf, as loss of Pcaf alone also produces notable phenotypes, including defective neurite outgrowth following spinal cord injury99. Interestingly, components of the SAGA and ATAC complex have also been linked to development. Trrap, the accessory subunit within SAGA and also the TIP60 complex that facilitates transcription factor interactions, is essential for early embryonic development and ESC self-renewal100; 101; 102, while Atac2 is required for embryo viability103.
Hat1
Kat1, more commonly referred to as Hat1, is a cytoplasmic KAT enzyme that acetylates newly synthesized free histones during chromatin assembly48. Together, Hat1 and histone binding protein RbAp46 form the HAT-B complex104. Following new histone synthesis, HAT-B associates with sNasp (somatic Nuclear autoantigenic sperm protein)-bound H3-H4 dimers to acetylate H4K5 and H4K12105. These modified histones are then transported to the nucleus for de novo chromatin assembly and subsequently deacetylated during chromatin maturation106. In addition to functioning within the cytosol, Hat1 is also present in the nuclear compartment107. While the nuclear function of Hat1 is still poorly understood, studies performed in yeast suggest that Hat1 functions as part of the NuB4 (Nuclear type B HAT specific for H4) complex, containing Hat1, RbAp46, sNasp and H3-H4, dimers to control histone deposition and/or DNA repair-based chromatin reassembly108. Interestingly, the Hat4 GNAT family member also regulates many of these processes, but instead acetylates free histone H4 on K20, K79 and K91 residues52.
Hat1 plays an essential role during embryonic development, as Hat1−/− mice die at birth or shortly after birth from lung maturation defects resulting from cellular hyperproliferation109. These mice are smaller in size than their wild-type counterparts and display craniofacial abnormalities, likely due to enhanced bone growth and reduced cartilage production109. In addition, Hat1−/− mouse embryonic fibroblasts are strikingly sensitive to DNA damaging agents, and exhibit high genome instability109.
CBP/p300 Family
The CBP/p300 family is composed of these two homologous enzymes110, also known as Kat3a and Kat3b, respectively47. Both contain a well-conserved acetyltransferase domain111, and a number of protein interaction domains that facilitate binding with over 400 proteins and promote many non-histone acetylation events112. Given their high sequence and structural similarity, CBP and p300 generally function in an analogous manner, yet still modulate distinct processes. Both proteins act as transcriptional co-activators, incapable of directly binding DNA and are hence recruited via interactions with sequence-specific transcription factors113; 114; 115. In vitro, recombinant CBP and p300 readily acetylate each of the four core histones in nucleosomes without the assistance of accessory factors110; 116, and they appear to preferentially acetylate H3K18 and H3K27 residues in vivo97. CBP and p300 also contribute to H3K56ac, a modification that is elevated in both embryonic stem cells and multiple forms of cancer117; 118. Furthermore, CBP and p300 dually function as crotonyltransferase enzymes that deposit crotonyl moieties on histones to activate transcription119. At the genomic level, CBP and p300 binding is enriched at both promoters and enhancers120; 121. However, CBP/p300 and H3K27ac most notably mark active enhancers during early development that drive transcription programs associated with cell and tissue specification122; 123; 124. Similarly, p300 has also been linked to super-enhancers in mESCs125, defined as enlarged enhancer regions densely co-bound by the Mediator transcriptional co-activator complex and pluripotency regulators Oct4, Sox2 and Nanog126.
CBP and p300 are largely co-expressed during mouse embryogenesis and therefore share certain developmental functions. Both are required for normal embryonic development127; 128; 129. Knockout of p300 (p300−/−) in mice leads to embryonic lethality between E9 and E11.5, and is accompanied by defects in cell proliferation, heart development and neural tube closure127. Similarly, CBP−/− mice die between E9.5 and E10.5 of embryonic development with severe neurulation defects, and display abnormalities in hematopoietic differentiation. Interestingly, a subset of p300 heterozygous (p300+/−) mice are also embryonic lethal and compound heterozygote (CBP+/−;p300+/−) mice die in utero with open neural tubes, suggesting the combined level of CBP and p300 expression is important for normal development127. Beyond this, CBP heterozygous mice exhibit skeletal defects, consistent with CBP mutations observed in patients with the haploinsufficiency disorder, Rubinstein-Taybi syndrome130. In mESCs, loss of p300 leads to premature differentiation131, while combined knockdown of CBP and p300 further enhance this differentiation defect and abrogate normal self-renewal capacity132. In line with these observations, binding of CBP and p300 in mESCs overlaps with the pluripotency master regulators Oct4, Sox2 and particularly Nanog132; 133. Moreover, direct binding of CBP/p300 to Nanog establishes long-range chromatin interactions necessary for mESC maintenance.
Interestingly, both Gcn5 and Pcaf interact with p300 and CBP. These two KAT families share both distinct and overlapping functions during development, as illustrated by the finding that about 25% of embryos that carry one null allele of both Gcn5 and p300 die, even though embryos that are heterozygous for either null allele alone are viable134.
MYST Family
The MYST family has five mammalian members: Tip60 (Kat5/Htatip), Mof (Kat8/Myst1), Moz (Kat6a/Myst3), Morf (Kat6b/Myst4), and Hbo1 (Kat7/Myst2). Each member contains a well-conserved MYST domain that includes a C2HC zinc finger as well as an acetyl-CoA binding motif homologous to that found in GNAT family members135. Furthermore, individual members harbor specialized domains that bind modified histones, including PHD and chromodomains136. Similar to the GNAT family, the MYST KATs also function in macro- molecular complexes and regulate a wide variety of biological and developmental processes.
Tip60
In mammals, the transcriptional co-regulator Tip60 (HIV Tat-interacting protein of 60 kDa) assembles into the multi-subunit TIP60 complex. This complex contains at least 16 proteins and has two enzymatic platforms, including Tip60 acetyltransferase activity that drives H2A and H4 acetylation, and p400 ATP-dependent chromatin remodeling activity that deposits the H2A.Z histone variant into chromatin137. Functionally, the TIP60 complex primarily associates with active promoters, via binding to proximal promoter R-loops and various transcription factors, including Myc, E2f1, and β-catenin102; 138; 139; 140; 141; 142. As well, TIP60 acetyltransferse activity functions in homologous recombination (HR)-based repair of DNA double strand breaks (DSB)137; 143. In agreement with these observations, multiple members of the TIP60 complex are vital regulators of normal embryogenesis and/or ESC regulation in mice. In developing mice, inactivation of either the TIP60 complex genes, Tip60 (Tip60−/−), Trrap (Trrap−/−) or Dmap1 (Dmap1−/−) leads to early peri-implantation lethality101; 144; 145. The severe phenotype of the Trrap−/− embryos likely reflects disruption of TIP60 and SAGA complexes. Individual knockdown in the expression levels of seven TIP60 members (Tip60, Trrap, p400, Dmap1, Ruvbl1, Ruvbl2 and Yeats4) also abrogates normal mESC identity102. Furthermore, the TIP60 complex regulates the Myc mESC network142; 146 and maintains the primed state of developmental genes in mESCs102.
Mof
Mof (males absent on the first) was originally identified in Drosophlia as a regulator of dosage compensation, leading to H4K16ac and hyperactivation of the single male X chromosome147. However, it is still unclear whether Mof manages similar dosage compensation effects in mammals. In humans, Mof assembles into two primary KAT complexes: the highly conserved MSL (male-specific lethal) multi-protein complex that specifically acetylates H4K16148, and the nine subunit NSL (non-specific lethal) complex that targets H4K5, K8 and K16 acetylation149. In vivo, Mof is required for mouse embryonic development beyond the blastocyst stage150. Mof−/− mice die at E4.5, display increased chromatin compaction and a striking loss of H4K16ac preceding apoptosis150. Accordingly, Mof is also needed to maintain pluripotency and self-renewal of mESCs151. Loss of Mof drives massive transcriptional changes, including down-regulation of the pluripotency genes encoding Oct4, Sox2 and Nanog and up-regulation of lineage specific genes151. The MSL complex is largely responsible for maintaining H4K16ac levels in mESCs and primarily localizes within gene bodies of ESC-specific targets, whereas NSL associates with promoter regions of housekeeping genes152. However, Mof occupancy has also been linked to active enhancers153; 154. Moreover, the MSL complex protects mESC identity, as it directly enhances Tsix transcription to ensure X chromosome activation154.
Moz, Morf and Hbo1
The MYST acetyltransferases, Moz (monocytic leukemia zinc-finger protein) and Morf (MOZ-related factor) have similar structural organization and independently form MOZ/MORF tetrameric complexes, containing Ing5 (inhibitor of growth 5), Eaf6 (homolog yeast of Esa1-associated factor 6), and either Brpf1/2/or 3 (bromodomain-and PHD finger-containing protein) paralogs155. Likewise, the HBO1 complex includes the Hbo1 acetyltransferase, Ing4/5, Eaf6 and either Brpf1/2/3 or Jade1/2/3 (gene for apoptosis and differentiation in epithelia)155; 156. Both MOZ and MORF complexes primarily acetylate H3K14 and function as co-activators for Runx and p53 transcription factors155, while the Brpf-containing HBO1 complex targets H3K14/ K23 acetylation, and the Jade-containing HBO1 complex mediates H4K5/K8/K12 acetylation157.
Although, each of these KATs participates within similar multi-subunit complexes, they exhibit very different developmental roles as reflected by different phenotypes of the null mice. Mice lacking Moz (Moz−/−) die around E15 due to defective hematopoietic stem cell (HSC) development and maintenance158; 159. This fate is partially dependent on Moz KAT activity, as mice expressing catalytically inactive Moz (Kat−/− Moz) survive until birth, yet die sooner than their wild-type counterparts and generally display reduced body weight as well as decreased spleen and thymus size, corresponding to impaired HSC proliferation160. In comparison, a 90% reduction in Morf expression, within Morf gene trap mice (Morf gt/gt), results in low birth weight, craniofacial abnormalities, and defective brain development161, as well as reduced adult neural stem cell production162. Hbo1 null mice die at E10.5, due to increased apoptosis in mesenchymal tissues and display near complete loss in H3K14ac163.
Nuclear Reprogramming
Throughout development, pluripotent cells differentiate in a spatial and temporal manner to adopt stable somatic cell identities. Once established, specified cells are typically resistant to changes in cell fate. However, in rare contexts, such as during tumor initiation, gene networks are rewired resulting in cell identity changes164. Furthermore, cell specification is readily reversed and/or altered using experimental techniques, such as transcription factor directed somatic cell reprogramming165 and transdifferentiation166. Simply put, the switch in cell fate that occurs when a committed cell is triggered to dedifferentiate or to adopt a new cell identity, based on transcriptional changes, is referred to as nuclear reprogramming167; 168; 169; 170. Predictably, massive reorganization and resetting of the original chromatin landscape is key to nuclear reprogramming, and both histone acetylation changes and KATs have been implicated as potential drivers of these processes. Next, we specifically discuss the ability of histone acetylation and KATs to modulate chromatin plasticity in different nuclear reprogramming contexts including somatic cell reprogramming and tumor initiation.
Somatic cell reprogramming
In 2006, Takahashi and Yamanaka reported that ectopic expression of the stem cell-related transcription factors Oct4, Sox2 Klf4 and Myc (OSKM) was capable of reversing mature somatic cells back to a pluripotent state165. This process, known as somatic cell reprogramming, results in the generation of induced pluripotent stem cells (iPSCs), that closely resemble ESCs 171; 172; 173. Reprogramming thus offers unparalleled potential to create undifferentiated patient-specific cells for regenerative medicine applications, disease modeling and drug discovery.
Mechanistically, somatic cells undergo reprogramming through three phases termed initiation, maturation and stabilization174; 175 (Figure 3). The initiation phase is triggered by the induction of OSKM and is characterized by increased cellular proliferation and a requisite mesenchymal-to-epithelial transition (MET) driven by bone morphogenetic protein (BMP) signaling, which results in loss of the somatic cell profile (Snai1/2, Zeb1/2) and gain of epithelial-associated expression (Cdh1, Epcam, Ocln)174; 176; 177. Furthermore, additional transcriptional changes affect cytoskeletal organization, RNA processing and metabolism178. In comparison, the maturation phase is linked to activation of the first set of pluripotent genes (endogenous Oct4, Nanog, Esrrb), whereas the stabilization phase coincides with acquisition of a self-sustaining pluripotency network that is independent of ectopic OSKM transgene expression, marked by the full complement of pluripotent genes (endogenous Sox2, Dppa4, Pecam) and reactivation of the somatically silenced X chromosome174; 179; 180; 181. Wnt signaling is also necessary for late reprogramming events182. Moreover, changes in cellular metabolism are tightly linked to reprogramming, as dedifferentiating somatic cells suppress oxidative phosphorylation and acquire enhanced glycolytic potential to ramp up acetate supply, as seen in mESCs183.
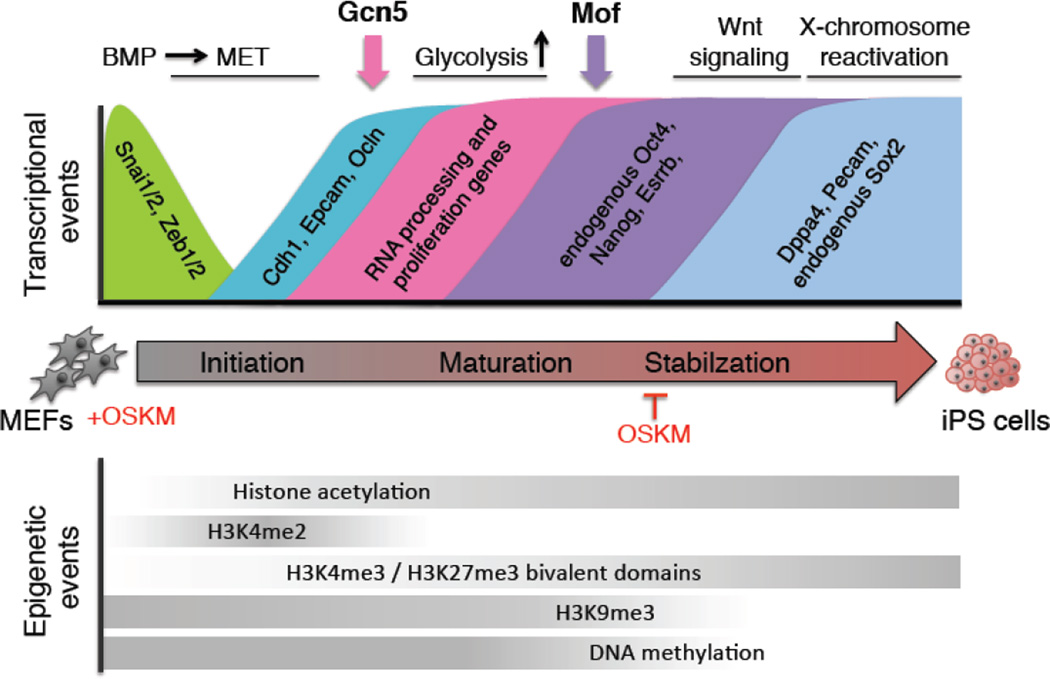
Figure 3.
Somatic cell reprogramming. The key transcriptional (top) and epigenetic events (bottom) are detailed across the initiation, maturation and stabilization phases of reprogramming. The involvement of Gcn5 and Mof is highlighted. MEF: mouse embryonic fibroblast, iPS: induced pluripotent stem cells, MET: mesenchymal-to-epithelial transition, OSKM: Oct4, Sox2, Klf4, Myc.
Throughout the reprogramming process, alterations in gene expression are coupled to reorganization of the chromatin landscape and DNA methylation patterns184. During the initiation phase, chromatin compaction is reduced due to a loss of repressive H3K27me3 and deposition of active H3K4me2 and H3K4me3 marks178; 185; 186; 187. Bivalent domains harboring both H3K27me3 and H3K4me3 are gradually established to poise developmental genes178. Furthermore, suppressive H3K9me3 modifications are erased and DNA demethylation occurs at pluripotency genes, such as Nanog178; 188. Beyond these differences, acetylation of nearly all H3 and H4 lysine residues are elevated in iPS cells compared to MEFs188; 189. Furthermore, high H3K9ac levels augment reprogramming190 and the expression levels of distinct KATs increase as somatic cells dedifferentiate89; 189, implying that histone acetylation is important for attaining pluripotency. Indeed, HDAC inhibitors enhance reprogramming efficiency191; 192; 193, however, the mechanistic details of this enhancement remain unclear, as valproic acid promotes reprogramming in the absence of Myc193, while butyrate drives initiation events in a Myc-dependent manner in mice192, and only partially compensates for Myc loss during human reprogramming191. Nonetheless, these observations are particularly intriguing given that Myc facilitates recruitment of KAT co-activator complexes (SAGA and TIP60)80; 81; 138, and that Myc is required to maintain histone acetylation and gene activation in neural progenitor cells194.
Accordingly, multiple components of the SAGA complex act as essential regulators of reprogramming, including Gcn5, Trrap, Sgf29 and Taf1289. Early in reprogramming, Myc directly up-regulates both Gcn5 and Sgf29 expression to facilitate a positive feedforward mechanism, whereby Myc subsequently interacts with Gcn5 to co-activate gene expression89. During this time, Myc plays fundamental roles in accessing open chromatin within promoter regions to drive early gene activation, suppressing the fibroblast expression signature, and boosting OSK engagement at enhancers 186; 195. Although, Myc has been proposed to work as a general amplifier of transcription, based on its role in cancer196; 197, Gcn5 is not a universal Myc co-activator during early reprogramming. Instead, Gcn5 cooperates with Myc at specific target genes, including a subset of genes that encode RNA processing factors linked to alternative splicing.89 Similar to transcription and epigenetic changes that occur as somatic cells become pluripotent, alternative splicing networks are also altered198. Interestingly, elevated expression of the RNA processing factors, mediated by Myc and Gcn5, corresponds with downstream changes in alternative splicing patterns linked to reprogramming and pluripotency89; 198; 199. Furthermore, these splicing events are compromised by depletion of Gcn5, Trrap, Sgf29 and Taf12 subunits of the SAGA complex89. Thus, Myc and the SAGA complex establish a key alternative splicing pathway by promoting expression of RNA processing factors within the initiation phase of reprogramming. Since this discovery, Myc-mediated alternative splicing has also been implicated in tumorigenesis, but the role of Gcn5 in this context remains unclear200; 201.
The impact of KATs on the maturation and the stabilization phases of reprogramming has yet to be systematically investigated, but Mof appears to be important for progression through these phases. As mentioned above, Mof is needed to ensure proper chromatin organization and mESC-specific expression patterns through acetylation of H4K16151. Ectopic Mof expression also improves OSKM-mediated reprogramming of human fibroblasts, while knockdown of Mof abrogates the process202. Interestingly, both Mof mRNA and protein levels increase throughout reprogramming and peak in iPSCs, in a manner similar to the Nanog maturation marker202. Notably, this spike in Mof expression also correlates with global enhancement in H4K16ac, a modification linked to chromatin decompaction203. During the maturation phase, Mof directly binds both the Oct4 and Nanog promoters to deposit H4K16ac and promote Wdr5-mediated H3K4me3202. Thus, Mof plays a fundamental role in establishing the pluripotency network of iPSCs. However, the role of Mof in reprogramming may extend beyond these observations. Given that the MSL complex controls X chromosome activation in mESCs154, and the necessity for X chromosome reactivation during iPSC generation181, it will be of interest to see if Mof also directs reactivation of the X chromosome during reprogramming.
It is also likely that additional KATs are necessary for iPSC generation. In particular, members of the CBP/p300 family may function late in reprogramming. Recently, Tex10 was identified as a Sox2-interacting protein that recruits p300 to deposit H3K27ac at super-enhancers in order to maintain self-renewal and pluripotency of mESCs125. During reprogramming, Tex10 expression levels are elevated with similar kinetics to that of the late reprogramming marker, endogenous Sox2. Also, Tex10 is required for both mouse and human reprogramming125, suggesting that p300 also contributes to defining active super-enhancers within iPSCs. Based on their roles in mESCs, CBP and p300 may facilitate long-range chromatin interactions in iPSCs132. Moreover, CBP/p300 may either positively or negatively effect reprogramming based on selective interactions with distinct transcription factors. For instance, Wnt signaling drives late-stage iPSC formation182, and CBP is a well-known transcriptional co-activator of β-catenin182, implying that CBP modulates Wnt signaling in this context. In agreement, reprogramming performed in the presence of conditioned Wnt3a media enhances reprogramming efficiency, but is completely abrogated upon treatment with ICG-001, an inhibitor that specifically blocks CBP binding to β-catentin204; 205. Therefore, CBP and β-catenin appear to facilitate reprogramming, yet the underlying mechanism remains to be explored.
Tip60 may also be important for reprogramming, given that seven TIP60 complex components, including Tip60 itself, are required to maintain mESC identity102 and Myc interacts with the TIP60 complex in mESCs146. However, it is currently unknown if TIP60 regulates conversion of somatic cells to iPSCs. Furthermore, the expression of other MYST family members, including Hat1, Morf and Hbo1, is elevated in iPSCs (unpublished observation)189. Of these, Morf has also been implicated in neural stem cell (NSC) maintenance162; 206, while Hat1 is highly expressed in mESCs, NSCs, and hematopoietic stem cells (HSCs)207; 208; 209, further suggesting a role for these enzymes in promoting cellular plasticity during reprogramming.
Tumor Initiation and progression
Pluripotent cells and tumor initiating cells share many common features, including acquired self-renewal capacity, restricted differentiation, enhanced proliferation and glycolytic energy preference210. Cancer cells also establish ESC-like gene expression patterns211, and loss of tumor suppressor function promotes somatic cell reprogramming212; 213; 214; 215; 216. Furthermore, the OSKM reprogramming factors each play roles in tumorigenesis217; 218; 219; 220; 221, suggesting that common epigenetic and transcription pathways drive pluripotent properties during reprogramming and tumor development. Accordingly, several KATs have been linked to tumorigenesis. Here we discuss the impact of select KATs on cancer development and progression (Figure 4).
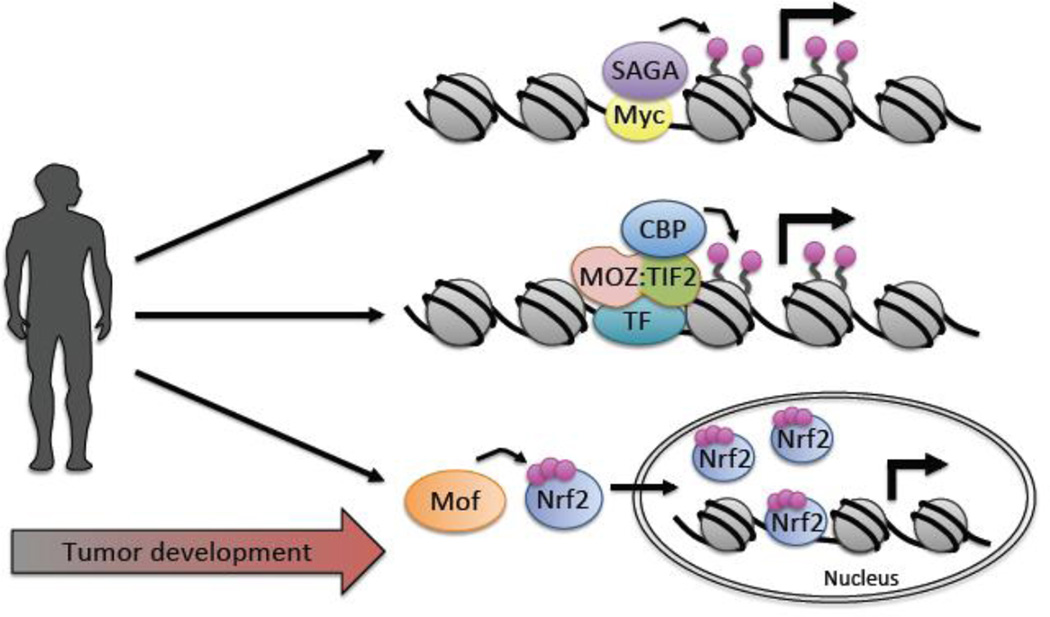
Figure 4.
The impact of KATs on tumorigenesis. Representative themes and examples of KAT involvement in tumor development are highlighted. Top. Functioning as co-activators, KATs promote transcriptional up-regulation of oncogene targets. Here, Myc recruits KAT co-activator complexes including SAGA (shown) and TIP60 (not shown) that drive histone acetylation and subsequent gene activation. Middle. KAT fusion proteins result in mistargeting of KAT activity. Here, the MOZ:TIF2 fusion protein triggers CBP recruitment and gene activation of Moz target genes. Bottom. KATs mediate non-histone acetylation. Here, Mof acetylates Nrf2 resulting in nuclear retention and activation of Nrf2 gene targets. Pink circle: acetylation.
Myc is a commonly amplified proto-oncogene that promotes cell growth in human cancers221 and self-renewal in ESCs211. Thus, precise Myc regulation is crucial for normal cellular behavior. In cancer, Myc has been implicated as both a general amplifier of active genes, as well as a gene-specific transcriptional regulator196; 197; 222. As noted above, both TIP60 and Gcn5/SAGA act as co-activator complexes for Myc in mESCs. Tip60 interacts with Myc at approximately 50% of target genes102; 142; 146, and Gcn5-bound loci strongly associate with sites bound by Myc, E2f1 and H3K9/14ac89. N-Myc and Gcn5 also regulate common transcription programs in NSCs94. Therefore, given these precedents, it seems likely that TIP60 and/or SAGA may similarly contribute to the activation of Myc transcriptional networks in cancer146. Gcn5, Pcaf, and Tip60 also directly acetylate Myc to increase protein stability 223, implying that these enzymes facilitate Myc-driven tumorigenesis on multiple levels. Moreover, recent work indicates that Myc actively promotes these mechanisms, as Myc up-regulates Gcn5 expression in NSCs194, reprogramming cells89 and human colon cancer224. Beyond these observations, Gcn5 is overexpressed in a variety of carcinomas including advanced stage gastric cancer225, colon cancer224, and non-small cell lung cancer (NSCLC), where Gcn5 specifically promotes cell growth by functioning as an E2f1 co-activator at Cyclin D1, Cyclin E1 and E2f1 genes226.
Like Gcn5, Pcaf functions as a co-activator in tumor maintenance. For example, in medulloblastoma and glioblastoma cells, Pcaf interacts with Gli1 to promote hyperactive Hedgehog-Gli1 signaling and high cellular proliferation227. Notably, Pcaf mediates many of its oncogenic functions via acetylation of non-histone targets including Ezh2228, Akt1229, Acly (ATP-citrate lyase)230, and β-catenin231. Pcaf-mediated acetylation of Ezh2, Acly and β-catenin leads to improved protein stability and tumor-promoting characteristics in lung228; 230 and colon cancer224, respectively. Additionally, the GNAT family member, Hat1, has also been reported to promote cell growth in esophageal squamous cell carcinomas228.
CBP/p300 and Moz/Morf KATs undergo recurrent chromosomal translocations in acute myeloid leukemia (AML), resulting in MOZ-CBP, MOZ-p300, and MORF-CBP fusion proteins232. These KATs also rearrange with other genes, such as MLL (mixed lineage leukemia) to generate MLL-CBP and MLL-p300 fusion proteins that retain the bromodomain and KAT domain. MLL-CBP models of therapy-related myeloproliferative disease in mice demonstrate that MLL-CBP not only promotes proliferation but also alters gene expression patterns233. Furthermore, MOZ fuses with the nuclear receptor co-activator, TIF2 (transcription intermediary factor 2), to drive the conversion of committed myeloid progenitors into self-renewing leukemic stem cells that promote AML in mice234; 235. Chromosomal translocations also occur between MOZ and LEUTX (leucine twenty homeobox) genes in therapy-related AML236, and MORF fuses with KANSL1 in retroperitoneal leiomyoma237. While a detailed functional understanding of many of these fusion proteins is lacking, it is predicted that translocations involving KATs have multiple potential effects on tumor development, including altered co-factor recruitment and enzymatic activity, resulting in KAT mistargeting and changes in the acetylation landscape of histone and non-histone proteins. For example, the MOZ-TIF2 fusion protein contains both the MYST domain of MOZ and the CBP interaction domain (CID) of TIF2. However, the KAT domain of MOZ is not required, but rather the C2HC nucleosome recognition domain of MOZ and the CID of TIF2 are essential for transformation, suggesting that CBP constitutively co-activates MOZ target genes235.
Numerous reports indicate KATs also function in the progression of solid tumors. In particular, p300 overexpression correlates with poor patient prognosis in prostate cancer, hepatocellular carcinoma and nasopharyngeal carcinoma, while increased cytoplasmic CBP and p300 levels are linked to melanoma progression and tumor size112. Given that CBP and p300 interact with a multitude of proteins, it is predicted that overexpression of these KATs impacts their protein binding selectivity. Hence, oncogenes may exploit this phenomenon to reorganize chromatin, and direct tumorigenic pathways. Moz and Morf have also recently been found in a pan-cancer analysis as being recurrently amplified across 11 cancer types221. Furthermore, Moz is required to drive Eµ-Myc lymphoma in mice238, implying its involvement in tumor initiation. Hbo1 expression is elevated in testicular, ovarian, bladder, stomach / esophageal as well as breast carcinomas239; 240, and Hbo1 expression often anti-correlates with ERα (estrogen receptor α) levels due to Hbo1 KAT-specific destabilization of ERα in human breast tumors241. Moreover, Mof is commonly overexpressed in NSCLC242; 243, where Mof both acetylates Nrf2 (nuclear factor erythroid-2-related factor 2) to promote nuclear retention and downstream transcription of Nrf2 target genes242, as well as drives S-phase progression by directly activating Skp2 (S-phase kinase-associated protein 2) transcription243.
Conclusions
It is clear that KATs play a central role in ESC maintenance and development. Comparatively, these enzymes differ in substrate specificity and complex formation, and hence control a variety of biological processes. Interestingly, these enzymes are also important regulators of reversing the more rigid states of differentiated cells, during both somatic cell reprogramming and tumor development. Distinct KATs directly control the expression of ESC-specific genes, which are commonly elevated in cancer. They acetylate non-histone proteins to drive pluripotency and tumorigenesis. Furthermore, KATs act as co-activators for multiple transcription factors, such as Myc, to drive changes in chromatin organization and ultimately cell state. Understanding these mechanisms offers new potential for anti-neoplastic interventions. For instance, cancer cells often become addicted to high levels of Myc, implying they may also be addicted to KAT activity. Given that KAT complexes harbor druggable domains, such as the KAT domain and bromodomain, targeting these complexes presents a novel therapeutic approach.
Based on the fact that KATs effectively promote cell plasticity, it is also conceivable that these enzymes mediate directed transdifferentiaiton pathways. Furthermore, KATs, may act in normal tissue homeostasis and injury response networks, where dedifferentiation drives mammalian progenitor cell production within the intestine, skin, lung and nervous system164. In the future, it will be interesting to see if KATs function in these pathways, and whether distinct KATs can be exploited to improve cellular transdifferentiaiton and tissue regeneration.
Research Highlights.
KATs play major roles in ESC maintenance and normal embryonic development
KATs coordinate chromatin reorganization during reprogramming and tumor development
Gcn5 and Myc activate an alternative splicing network essential for reprogramming
Mof mediates activation of the first wave of pluripotency genes during reprogramming
Multiple KATs are misregulated in various forms of cancer
Acknowledgments
We thank Dr. Laurent David (Université de Nantes) for graphical contributions to Figure 3. This work is supported by a NIH R01067718 grant to S.Y.R.D., and J.L.W is a recipient of a Foundation Award from the CIHR. C.L.H. is supported by a Venture Sinai Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khoury GA, Baliban RC, Floudas CA. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep. 2011:1. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, Sabari BR, Deng Z, Allis CD, Ren B, Khochbin S, Zhao Y. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015;16:1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 12.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 13.Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 14.Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Cerbo V, Mohn F, Ryan DP, Montellier E, Kacem S, Tropberger P, Kallis E, Holzner M, Hoerner L, Feldmann A, Richter FM, Bannister AJ, Mittler G, Michaelis J, Khochbin S, Feil R, Schuebeler D, Owen-Hughes T, Daujat S, Schneider R. Acetylation of histone H3 at lysine 64 regulates nucleosome dynamics and facilitates transcription. Elife. 2014;3:e01632. doi: 10.7554/eLife.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Buhler M, Margueron R, Schneider R. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859–872. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 17.Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci U S A. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 19.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 20.Ohzeki J, Shono N, Otake K, Martins NM, Kugou K, Kimura H, Nagase T, Larionov V, Earnshaw WC, Masumoto H. KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation. Dev Cell. 2016;37:413–427. doi: 10.1016/j.devcel.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 22.Feller C, Forne I, Imhof A, Becker PB. Global and specific responses of the histone acetylome to systematic perturbation. Mol Cell. 2015;57:559–571. doi: 10.1016/j.molcel.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, Li H, Shi X. AF9 YEATS domain links histone acetylation to DOT1L–mediated H3K79 methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali M, Yan K, Lalonde ME, Degerny C, Rothbart SB, Strahl BD, Cote J, Yang XJ, Kutateladze TG. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J Mol Biol. 2012;424:328–338. doi: 10.1016/j.jmb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling S. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 29.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 30.Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, Lain S, Cox J, Matthias P, Mann M, Bradner JE, Choudhary C. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 31.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CF, Zerweck J, Schutkowski M, Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun. 2013;4:2327. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- 32.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 33.Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WR, Olsen JV, Jensen LJ, Choudhary C. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal. 2011;4:ra48. doi: 10.1126/scisignal.2001902. [DOI] [PubMed] [Google Scholar]
- 34.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 36.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 37.Yuan H, Rossetto D, Mellert H, Dang W, Srinivasan M, Johnson J, Hodawadekar S, Ding EC, Speicher K, Abshiru N, Perry R, Wu J, Yang C, Zheng YG, Speicher DW, Thibault P, Verreault A, Johnson FB, Berger SL, Sternglanz R, McMahon SB, Cote J, Marmorstein R. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012;31:58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albaugh BN, Arnold KM, Lee S, Denu JM. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286:24694–24701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci U S A. 2008;105:12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang C, Wu J, Zheng YG. Function of the active site lysine autoacetylation in Tip60 catalysis. PLoS One. 2012;7:e32886. doi: 10.1371/journal.pone.0032886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 43.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, Bomze D, Elena-Herrmann B, Scherf T, Nissim-Rafinia M, Kempa S, Itskovitz-Eldor J, Meshorer E, Aberdam D, Nahmias Y. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 48.Sobel RE, Cook RG, Allis CD. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 49.Ruiz-Carrillo A, Wangh LJ, Allfrey VG. Processing of newly synthesized histone molecules. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 50.Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton LM, Garcia BA, Aleckovic M, Kang Y, Kaluz S, Devi N, Van Meir EG, Hitosugi T, Seo JH, Lonial S, Gaddh M, Arellano M, Khoury HJ, Khuri FR, Boggon TJ, Kang S, Chen J. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell. 2014;53:534–548. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X, Yu W, Shi L, Sun L, Liang J, Yi X, Li Q, Zhang Y, Yang F, Han X, Zhang D, Yang J, Yao Z, Shang Y. HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol Cell. 2011;44:39–50. doi: 10.1016/j.molcel.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 53.Mak AB, Pehar M, Nixon AM, Williams RA, Uetrecht AC, Puglielli L, Moffat J. Post-translational regulation of CD133 by ATase1/ATase2-mediated lysine acetylation. J Mol Biol. 2014;426:2175–2182. doi: 10.1016/j.jmb.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 56.Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis CD, Berger SL. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 58.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39:1390–1397. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalebic N, Sorrentino S, Perlas E, Bolasco G, Martinez C, Heppenstall PA. alphaTAT1 is the major alpha-tubulin acetyltransferase in mice. Nat Commun. 2013;4:1962. doi: 10.1038/ncomms2962. [DOI] [PubMed] [Google Scholar]
- 61.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–15. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 62.Wolf E, Vassilev A, Makino Y, Sali A, Nakatani Y, Burley SK. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94:439–449. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 63.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 65.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 66.Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 67.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 68.Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, 3rd, Washburn MP, Abmayr SM, Workman JL. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 71.Riss A, Scheer E, Joint M, Trowitzsch S, Berger I, Tora L. Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. J Biol Chem. 2015;290:28997–29009. doi: 10.1074/jbc.M115.668533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 74.Ramachandran S, Haddad D, Li C, Le MX, Ling AK, So CC, Nepal RM, Gommerman JL, Yu K, Ketela T, Moffat J, Martin A. The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated gammaH2AX Formation. Cell Rep. 2016;15:1554–1565. doi: 10.1016/j.celrep.2016.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, Fleharty BE, Paulson A, Allali-Hassani A, Zhou JQ, Mer G, Grant PA, Workman JL, Zang J, Min J. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ringel AE, Cieniewicz AM, Taverna SD, Wolberger C. Nucleosome competition reveals processive acetylation by the SAGA HAT module. Proc Natl Acad Sci U S A. 2015;112:E5461–E5470. doi: 10.1073/pnas.1508449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 78.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 81.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 83.Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, Zhao Y, Allis CD, Shi X, Li H. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016;26:629–632. doi: 10.1038/cr.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 85.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci. 2010;67:611–628. doi: 10.1007/s00018-009-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol. 2009;29:1649–1660. doi: 10.1128/MCB.01076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, Xia Z, Salinger AP, Wilson M, Vizeacoumar F, Datti A, Li W, Cooney AJ, Barton MC, Blencowe BJ, Wrana JL, Dent SY. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 2015;29:803–816. doi: 10.1101/gad.255109.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orpinell M, Fournier M, Riss A, Nagy Z, Krebs AR, Frontini M, Tora L. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010;29:2381–2394. doi: 10.1038/emboj.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 92.Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci U S A. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinez-Cerdeno V, Lemen JM, Chan V, Wey A, Lin W, Dent SR, Knoepfler PS. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS One. 2012;7:e39456. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin W, Zhang Z, Chen CH, Behringer RR, Dent SY. Proper Gcn5 histone acetyltransferase expression is required for normal anteroposterior patterning of the mouse skeleton. Dev Growth Differ. 2008;50:321–330. doi: 10.1111/j.1440-169X.2008.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY. Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev Dyn. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- 97.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY. Gcn5 and PCAF Regulate PPARgamma and Prdm16 Expression To Facilitate Brown Adipogenesis. Mol Cell Biol. 2014;34:3746–3753. doi: 10.1128/MCB.00622-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Puttagunta R, Tedeschi A, Soria MG, Hervera A, Lindner R, Rathore KI, Gaub P, Joshi Y, Nguyen T, Schmandke A, Laskowski CJ, Boutillier AL, Bradke F, Di Giovanni S. PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat Commun. 2014;5:3527. doi: 10.1038/ncomms4527. [DOI] [PubMed] [Google Scholar]
- 100.Sawan C, Hernandez-Vargas H, Murr R, Lopez F, Vaissiere T, Ghantous AY, Cuenin C, Imbert J, Wang ZQ, Ren B, Herceg Z. Histone acetyltransferase cofactor Trrap maintains self-renewal and restricts differentiation of embryonic stem cells. Stem Cells. 2013;31:979–991. doi: 10.1002/stem.1341. [DOI] [PubMed] [Google Scholar]
- 101.Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet. 2001;29:206–211. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- 102.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, Zha J. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol. 2009;29:1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 105.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Annunziato AT, Seale RL. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 107.Parthun MR. Histone acetyltransferase 1: More than just an enzyme? Biochim Biophys Acta. 2012;1819:256–263. doi: 10.1016/j.bbagrm.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ge Z, Wang H, Parthun MR. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J Biol Chem. 2011;286:16790–16799. doi: 10.1074/jbc.M110.216846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagarajan P, Ge Z, Sirbu B, Doughty C, Agudelo Garcia PA, Schlederer M, Annunziato AT, Cortez D, Kenner L, Parthun MR. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 111.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 112.Dancy BM, Cole PA. Protein lysine acetylation by p300/CBP. Chem Rev. 2015;115:2419–2452. doi: 10.1021/cr500452k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 114.Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A–associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 115.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A–associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 116.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 117.Costantini C, Ko MH, Jonas MC, Puglielli L. A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochem J. 2007;407:383–395. doi: 10.1042/BJ20070040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, Grunstein M. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, Roeder RG, Allis CD. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell. 2015;58:203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 121.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ding J, Huang X, Shao N, Zhou H, Lee DF, Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, Wang H, Waghray A, Papatsenko D, Sanchez-Priego C, Li D, Yuan Y, Lemischka IR, Shen L, Kelley K, Deng H, Shen X, Wang J. Tex10 Coordinates Epigenetic Control of Super-Enhancer Activity in Pluripotency and Reprogramming. Cell Stem Cell. 2015;16:653–668. doi: 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 128.Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 129.Partanen A, Motoyama J, Hui CC. Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int J Dev Biol. 1999;43:487–494. [PubMed] [Google Scholar]
- 130.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci U S A. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhong X, Jin Y. Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J Biol Chem. 2009;284:9168–9175. doi: 10.1074/jbc.M805562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fang F, Xu Y, Chew KK, Chen X, Ng HH, Matsudaira P. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells. 2014;32:1805–1816. doi: 10.1002/stem.1705. [DOI] [PubMed] [Google Scholar]
- 133.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 134.Phan HM, Xu AW, Coco C, Srajer G, Wyszomierski S, Evrard YA, Eckner R, Dent SY. GCN5 and p300 share essential functions during early embryogenesis. Dev Dyn. 2005;233:1337–1347. doi: 10.1002/dvdy.20445. [DOI] [PubMed] [Google Scholar]
- 135.Sapountzi V, Cote J. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci. 2011;68:1147–1156. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 138.Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F–dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol. 2015;22:999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ravens S, Yu C, Ye T, Stierle M, Tora L. Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells. Epigenetics Chromatin. 2015;8:45. doi: 10.1186/s13072-015-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, Legube G, Doyon Y, Durocher D, Cote J. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol Cell. 2016;62:409–421. doi: 10.1016/j.molcel.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mohan KN, Ding F, Chaillet JR. Distinct roles of DMAP1 in mouse development. Mol Cell Biol. 2011;31:1861–1869. doi: 10.1128/MCB.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Su J, Wang F, Cai Y, Jin J. The Functional Analysis of Histone Acetyltransferase MOF in Tumorigenesis. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol. 2008;28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ravens S, Fournier M, Ye T, Stierle M, Dembele D, Chavant V, Tora L. Mof-associated complexes have overlapping and unique roles in regulating pluripotency in embryonic stem cells and during differentiation. Elife. 2014:3. doi: 10.7554/eLife.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Taylor GC, Eskeland R, Hekimoglu-Balkan B, Pradeepa MM, Bickmore WA. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 2013;23:2053–2065. doi: 10.1101/gr.155028.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chelmicki T, Dundar F, Turley MJ, Khanam T, Aktas T, Ramirez F, Gendrel AV, Wright PR, Videm P, Backofen R, Heard E, Manke T, Akhtar A. MOF-associated complexes ensure stem cell identity and Xist repression. Elife. 2014;3:e02024. doi: 10.7554/eLife.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 156.Mishima Y, Miyagi S, Saraya A, Negishi M, Endoh M, Endo TA, Toyoda T, Shinga J, Katsumoto T, Chiba T, Yamaguchi N, Kitabayashi I, Koseki H, Iwama A. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011;118:2443–2453. doi: 10.1182/blood-2011-01-331892. [DOI] [PubMed] [Google Scholar]
- 157.Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, Tan S, Yang XJ, Kutateladze TG, Cote J. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27:2009–2024. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, Ochiya T, Kitabayashi I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20:1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thomas T, Corcoran LM, Gugasyan R, Dixon MP, Brodnicki T, Nutt SL, Metcalf D, Voss AK. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20:1175–1186. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Perez-Campo FM, Borrow J, Kouskoff V, Lacaud G. The histone acetyl transferase activity of monocytic leukemia zinc finger is critical for the proliferation of hematopoietic precursors. Blood. 2009;113:4866–4874. doi: 10.1182/blood-2008-04-152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thomas T, Voss AK, Chowdhury K, Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–2548. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- 162.Merson TD, Dixon MP, Collin C, Rietze RL, Bartlett PF, Thomas T, Voss AK. The transcriptional coactivator Querkopf controls adult neurogenesis. J Neurosci. 2006;26:11359–11370. doi: 10.1523/JNEUROSCI.2247-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31:845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol. 2016;17:413–425. doi: 10.1038/nrm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 166.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 167.Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- 168.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 169.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 173.Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, Lee G, Rinn JL, Meissner A, Park PJ, Hochedlinger K. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 175.David L, Polo JM. Phases of reprogramming. Stem Cell Res. 2014;12:754–761. doi: 10.1016/j.scr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 176.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 177.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, Cheloufi S, Stadtfeld M, Figueroa ME, Robinton D, Natesan S, Melnick A, Zhu J, Ramaswamy S, Hochedlinger K. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 180.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pasque V, Tchieu J, Karnik R, Uyeda M, Sadhu Dimashkie A, Case D, Papp B, Bonora G, Patel S, Ho R, Schmidt R, McKee R, Sado T, Tada T, Meissner A, Plath K. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ho R, Papp B, Hoffman JA, Merrill BJ, Plath K. Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep. 2013;3:2113–2126. doi: 10.1016/j.celrep.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hussein SM, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, Mosbergen R, Li M, Lee DS, Cloonan N, Wood DL, Munoz J, Middleton R, Korn O, Patel HR, White CA, Shin JY, Gauthier ME, Le Cao KA, Kim JI, Mar JC, Shakiba N, Ritchie W, Rasko JE, Grimmond SM, Zandstra PW, Wells CA, Preiss T, Seo JS, Heck AJ, Rogers IM, Nagy A. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- 188.Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Benevento M, Tonge PD, Puri MC, Nagy A, Heck AJ, Munoz J. Fluctuations in histone H4 isoforms during cellular reprogramming monitored by middle-down proteomics. Proteomics. 2015;15:3219–3231. doi: 10.1002/pmic.201500031. [DOI] [PubMed] [Google Scholar]
- 190.Hezroni H, Tzchori I, Davidi A, Mattout A, Biran A, Nissim-Rafinia M, Westphal H, Meshorer E. H3K9 histone acetylation predicts pluripotency and reprogramming capacity of ES cells. Nucleus. 2011;2:300–309. doi: 10.4161/nucl.2.4.16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, Yusa K, Bradley A, Meyers DJ, Mukherjee C, Cole PA, Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285:25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep. 2013;5:357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 199.Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, Wang E, Trcka D, Thompson T, O’Hanlon D, Slobodeniuc V, Barbosa-Morais NL, Burge CB, Moffat J, Frey BJ, Nagy A, Ellis J, Wrana JL, Blencowe BJ. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabo A, Amati B, Wee KB, Guccione E. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 201.Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, Karlin KL, Dominguez-Vidana R, Hartman JD, Renwick A, Scorsone K, Bernardi RJ, Skinner SO, Jain A, Orellana M, Lagisetti C, Golding I, Jung SY, Neilson JR, Zhang XH, Cooper TA, Webb TR, Neel BG, Shaw CA, Westbrook TF. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mu X, Yan S, Fu C, Wei A. The Histone Acetyltransferase MOF Promotes Induces Generation of Pluripotent Stem Cells. Cell Reprogram. 2015;17:259–267. doi: 10.1089/cell.2014.0102. [DOI] [PubMed] [Google Scholar]
- 203.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 204.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Kim HY, Moon SH, Ha JR, Kahn M. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sheikh BN, Dixon MP, Thomas T, Voss AK. Querkopf is a key marker of self-renewal and multipotency of adult neural stem cells. J Cell Sci. 2012;125:295–309. doi: 10.1242/jcs.077271. [DOI] [PubMed] [Google Scholar]
- 207.Park IK, He Y, Lin F, Laerum OD, Tian Q, Bumgarner R, Klug CA, Li K, Kuhr C, Doyle MJ, Xie T, Schummer M, Sun Y, Goldsmith A, Clarke MF, Weissman IL, Hood L, Li L. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood. 2002;99:488–498. doi: 10.1182/blood.v99.2.488. [DOI] [PubMed] [Google Scholar]
- 208.Lim DA, Suarez-Farinas M, Naef F, Hacker CR, Menn B, Takebayashi H, Magnasco M, Patil N, Alvarez-Buylla A. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 209.Wagh V, Doss MX, Sabour D, Niemann R, Meganathan K, Jagtap S, Gaspar JA, Ardestani MA, Papadopoulos S, Gajewski M, Winkler J, Hescheler J, Sachinidis A. Fam40b is required for lineage commitment of murine embryonic stem cells. Cell Death Dis. 2014;5:e1320. doi: 10.1038/cddis.2014.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Semi K, Matsuda Y, Ohnishi K, Yamada Y. Cellular reprogramming and cancer development. Int J Cancer. 2013;132:1240–1248. doi: 10.1002/ijc.27963. [DOI] [PubMed] [Google Scholar]
- 211.Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 2011;3:75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 218.Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 219.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, Brohee S, Salmon I, Dubois C, del Marmol V, Fuks F, Beck B, Blanpain C. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 220.Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson AJ, Kudlow JE, Lobo-Ruppert SM, Ruppert JM. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, Sougnez C, Gabriel SB, Hernandez B, Shen H, Laird PW, Getz G, Meyerson M, Beroukhim R. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, Tonelli C, Faga G, Bianchi V, Ronchi A, Low D, Muller H, Guccione E, Campaner S, Amati B. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yin YW, Jin HJ, Zhao W, Gao B, Fang J, Wei J, Zhang DD, Zhang J, Fang D. The Histone Acetyltransferase GCN5 Expression Is Elevated and Regulated by c-Myc and E2F1 Transcription Factors in Human Colon Cancer. Gene Expr. 2015;16:187–196. doi: 10.3727/105221615X14399878166230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wisnieski F, Calcagno DQ, Leal MF, Chen ES, Gigek CO, Santos LC, Pontes TB, Rasmussen LT, Payao SL, Assumpcao PP, Lourenco LG, Demachki S, Artigiani R, Burbano RR, Smith MC. Differential expression of histone deacetylase and acetyltransferase genes in gastric cancer and their modulation by trichostatin A. Tumour Biol. 2014;35:6373–6381. doi: 10.1007/s13277-014-1841-0. [DOI] [PubMed] [Google Scholar]
- 226.Chen L, Wei T, Si X, Wang Q, Li Y, Leng Y, Deng A, Chen J, Wang G, Zhu S, Kang J. Lysine acetyltransferase GCN5 potentiates the growth of non-small cell lung cancer via promotion of E2F1, cyclin D1, and cyclin E1 expression. J Biol Chem. 2013;288:14510–14521. doi: 10.1074/jbc.M113.458737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squatrito M, Helin K. Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 2013;73:6323–6333. doi: 10.1158/0008-5472.CAN-12-4660. [DOI] [PubMed] [Google Scholar]
- 228.Wan J, Zhan J, Li S, Ma J, Xu W, Liu C, Xue X, Xie Y, Fang W, Chin YE, Zhang H. PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res. 2015;43:3591–3604. doi: 10.1093/nar/gkv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhang S, Sun G, Wang Z, Wan Y, Guo J, Shi L. PCAF-mediated Akt1 acetylation enhances the proliferation of human glioblastoma cells. Tumour Biol. 2015;36:1455–1462. doi: 10.1007/s13277-014-2522-8. [DOI] [PubMed] [Google Scholar]
- 230.Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, Xiong Y, Lei QY. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell. 2013;51:506–518. doi: 10.1016/j.molcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ge X, Jin Q, Zhang F, Yan T, Zhai Q. PCAF acetylates {beta}-catenin and improves its stability. Mol Biol Cell. 2009;20:419–427. doi: 10.1091/mbc.E08-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 233.Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, Akashi K, Gilliland DG. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 235.Deguchi K, Ayton PM, Carapeti M, Kutok JL, Snyder CS, Williams IR, Cross NC, Glass CK, Cleary ML, Gilliland DG. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 236.Chinen Y, Taki T, Tsutsumi Y, Kobayashi S, Matsumoto Y, Sakamoto N, Kuroda J, Horiike S, Nishida K, Ohno H, Uike N, Taniwaki M. The leucine twenty homeobox (LEUTX) gene, which lacks a histone acetyltransferase domain, is fused to KAT6A in therapy-related acute myeloid leukemia with t(8;19)(p11;q13) Genes Chromosomes Cancer. 2014;53:299–308. doi: 10.1002/gcc.22140. [DOI] [PubMed] [Google Scholar]
- 237.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. Novel KAT6B–KANSL1 fusion gene identified by RNA sequencing in retroperitoneal leiomyoma with t(10;17)(q22;q21) PLoS One. 2015;10:e0117010. doi: 10.1371/journal.pone.0117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sheikh BN, Lee SC, El-Saafin F, Vanyai HK, Hu Y, Pang SH, Grabow S, Strasser A, Nutt SL, Alexander WS, Smyth GK, Voss AK, Thomas T. MOZ regulates B-cell progenitors and, consequently, Moz haploinsufficiency dramatically retards MYC-induced lymphoma development. Blood. 2015;125:1910–1921. doi: 10.1182/blood-2014-08-594655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF, Jr, Fukusato T, Smith MM. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436:108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hu X, Stern HM, Ge L, O’Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, Chant J, Stokoe D, Lackner MR, Cavet G. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 241.Iizuka M, Susa T, Takahashi Y, Tamamori-Adachi M, Kajitani T, Okinaga H, Fukusato T, Okazaki T. Histone acetyltransferase Hbo1 destabilizes estrogen receptor alpha by ubiquitination and modulates proliferation of breast cancers. Cancer Sci. 2013;104:1647–1655. doi: 10.1111/cas.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chen Z, Ye X, Tang N, Shen S, Li Z, Niu X, Lu S, Xu L. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. Br J Pharmacol. 2014;171:3196–3211. doi: 10.1111/bph.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhao L, Wang DL, Liu Y, Chen S, Sun FL. Histone acetyltransferase hMOF promotes S phase entry and tumorigenesis in lung cancer. Cell Signal. 2013;25:1689–1698. doi: 10.1016/j.cellsig.2013.04.006. [DOI] [PubMed] [Google Scholar]