Abstract
Western blotting is a ubiquitous tool used extensively in the clinical and research settings to identify proteins and characterize their levels. It has rapidly become a mainstay in research laboratories due to its specificity, low cost, and ease of use. The specificity arises from the orthogonal processes used to identify proteins. Samples are first separated based on size and then probed with antibodies specific for the protein of interest. This confirmatory approach helps avoid pitfalls associated with antibody cross-reactivity and specificity issues. While the technique has evolved since its inception, the last decade has witnessed a paradigm shift in Western blotting technology. The introduction of capillary and microfluidic platforms has significantly decreased time and sample requirements while enabling high-throughput capabilities. These advances have enabled Western analysis down to the single cell level in highly parallel formats, opening vast new opportunities for studying cellular heterogeneity. Recent innovations in microscale Western blotting are surveyed, and the potential for enhancing detection using advances in label-free biosensing is briefly discussed.
Introduction
Western blotting (WB) is a semi-quantitative technique used extensively in research to specifically identify proteins and characterize their levels.1–3 Introduced in 1979, it quickly established itself as a robust, powerful, and cost effective approach for protein analysis.1,3 The tremendous impact of WB comes from its specificity, which arises from the orthogonal mechanisms used to identify proteins. Proteins are first separated based on size using gel electrophoresis and then detected with antibodies specific for the protein of interest; combining two independent mechanisms for identification.1–4 The advantage of this orthogonal approach over sensing using antibodies alone is illustrated by the ongoing discussion of antibody specificity.
Antibodies that specifically bind proteins have had an enormous impact in basic research, medical diagnostics, and biotechnology. They are of central importance in WB analysis and for widely used research methods such as immunoprecipitation and immunofluorescence imaging. Antibodies have also played a pivotal role in the development of medical diagnostics. The enzyme-linked immunosorbent assay (ELISA) represents the gold standard for detecting and quantifying levels of diagnostic biomarkers in serum and other body fluids. Immunohistochemistry is extensively employed in the clinical setting to confirm and identify the presence of cancerous tissues and other hallmarks of disease. The ubiquitous use of antibodies in the medical and life sciences has resulted in a significant market, with global demand surpassing US$80 billion a year.5
There is, however, mounting concern over the reliability of antibodies obtained from different sources or even the same source, which has led to issues in repeating measurements and confirming results between labs.6–11 Antibody variability can arise from many sources, which has sharpened attention on antibody validation.12,13 The potential magnitude of this problem was exemplified in a recent study testing 1,124 antibodies in HEK293 cell lysates. Using consistent protocols spread across five independent labs, this study concluded that only 452 of the 1,124 antibodies tested even recognized their intended targets.13 This can be further aggravated by cross-reactivity and studies showing that even when antibodies recognize their intended target, binding can be influenced by the environment or sample preparation methods.14–16 Clearly, methods relying solely on single protein recognition events require extensive control and validation protocols to confirm specificity.
Multidimensional approaches such as WB, on the other hand, help ameliorate these complications by comparing orthogonal information to identify proteins.2,4 In conventional WB analysis, proteins are first separated based on size using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The anionic surfactant SDS linearizes the proteins and uniformly decorates them with negative charge, leading to an overall charge proportional to the size of the protein. In an electric field, proteins separate based on size as they pass through the porous polyacrylamide (PA) sieving gel. PA porosity can be tuned by changing the concentration of polymer and amount of crosslinker, providing a flexible and easily optimized platform for protein separation based on size. Once separated, the protein bands are transferred to a membrane such as PVDF (polyvinylidene difluoride) or nitrocellulose by electroblotting, forming a replica on the more porous support. The porous membrane enables access of probe antibodies to the protein bands for immunodetection.1–3
The electroblotting step represented the key experimental breakthrough in the development of WB and closely followed principles developed earlier by Edwin Southern for DNA blotting.4,17 The latter became known as Southern blotting, and the other “geographical” assays, including WB, were named accordingly.1,3 The efficiency of protein transfer from the gel to the membrane, retention during processing, and subsequent detection/amplification, largely determine the detection performance in WB.4
While WB is a robust, powerful, and easily implemented approach for protein identification, it does have key limitations.2,18,19 As generally applied, the approach is slow, semi-quantitative, and labor intensive, with most steps done manually. Gel preparation and separation, transfer, blocking, multiple incubation and washing steps, and finally imaging all contribute to a significant investment in time (hours to overnight). Since most platforms have not been miniaturized, the process is also sample and antibody intensive. Conventional WB, for example, typically requires micrograms of sample and antibody. 2,18,19 The former can be an issue in sample limited applications and the latter can be expensive. Finally, the detection of multiple targets often requires stripping the membrane (removing the antibody) and re-probing, which is time consuming, has limited re-probing cycles, and inevitably results in antigen loss.4
To overcome performance limitations and expand its capabilities, the last decade has seen a transition from method refinement to a complete repackaging of the WB hardware.20 Many of these advances have leveraged the considerable progress made in microfluidics and on-chip microanalytical systems made during this same period.21–24 The inherently small amount of consumables needed in these platforms, their scalability, enhanced signal-to-noise, and rapid analysis times are well-suited to address many of the current limitations in WB analysis and advance the method for new applications. Here we highlight a few of these exciting developments, which help illustrate the potential of these new platforms.
Multiplexed Microfluidic Probing Following Western Blotting
As mentioned, multiplexed detection in conventional WB is time consuming, requires harsh chemical treatments, and leads to loss of protein from the transfer membrane. An approach for integrating multiplexed microfluidic immunodetection with conventional WB was introduced by Jiang and coworkers to overcome these drawbacks.25 In this report, conventional protocols were used to separate lysates of NIH-3T3 cells using SDS-PAGE and transfer the separated protein replica onto a PVDF membrane. To detect the immobilized proteins, a simple microfluidic chip was fabricated in PDMS consisting of seven co-linear channels (150 μm wide × 100 μm deep and 3.5 cm in length) equipped with inlet and outlet ports at the ends.
After staining the protein bands with Ponceau S, the microfluidic chip was sealed against the PVDF membrane. The microchannels were aligned parallel to the separation axis, with the channels overlaying the protein bands. Each channel in the microfluidic chip was used to deliver a primary antibody probe solution to those regions of the protein bands overlaid by the microchannel. This enables multiplexed labeling across the width of the protein features on the PVDF membrane, similar in concept to the micromosaic immunoassay.26 Following incubation, the PDMS chip was carefully peeled away from the PVDF membrane, which was incubated with fluorescently labeled secondary antibody for detection. The simultaneous detection of seven proteins was demonstrated with similar sensitivity and detection limits as conventional WB. A similar format was used to multiplex detection of inflammatory markers in cell lysates.27
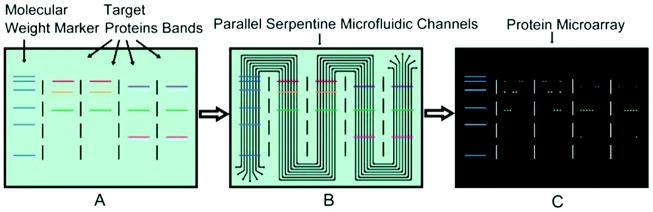
A slightly more complicated serpentine microfluidic design was later introduced for multiplexed probing of multiple separation lanes.28 As shown in Fig. 1, the PDMS chip incorporated seven parallel channels designed to weave along multiple lanes in a PVDF membrane, sharing one set of inlet and outlet ports. This enables each microchannel (probe antibody) to interact along each lane in a multilane separation, thus increasing throughput. Beyond the multiplexed capabilities, this general approach requires significantly less antibody (1% of conventional WB), has some advantages in optimizing labeling conditions, and is easily adapted with existing WB platforms.25
Figure 1.
(A) A PVDF membrane blotted with electrophoretically separated proteins and ladder markers. (B) A microfluidic network of seven parallel serpentine channels was sealed against the PVDF membrane, overlaying the protein bands. Probe antibodies were introduced in each microfluidic lane, enabling multiplexed detection (C) across each protein band. (Reprinted with permission from ref. 28. Copyright 2015 Royal Society of Chemistry.)
Microarray Spotting of Samples for Western Blotting
Another approach that can be adapted with conventional WB uses advances in microarraying technology to design highly parallel assays. In one report, microarray printing enabled efficient spotting of cell lysates onto a PA gel in a pattern matching a conventional 96 well-plate.29 The spotting was used to print 96 identical blocks, with each block containing one size standard and six sample spots. During printing, the relative humidity was held at 80% to keep the gel and samples hydrated. Once printed, semidry electrophoresis was completed in 12 minutes and the separated proteins were electroblotted onto a nitrocellulose membrane (1 hour to overnight). Following transfer, a 96 well isolation mask was aligned onto the membrane, enabling separate antibody probing of each block. In theory, this geometry enables six different lysates to be probed with 96 different antibodies, one for each block. Higher content, moreover, can be introduced using multiple probing or multiplexed detection steps. Incubation with dye-labeled secondary antibodies and infrared fluorescence detection was used to measure the developed membrane.
In this study, the time evolution of 91 phosphorylation sites in 67 proteins was tracked following epidermal growth factor stimulation of A432 human carcinoma cells.29 Moreover, each spot in the printed array required only 250 ng of protein (approximately 1000 cells) and 16 ng of detection antibody.29 While the overall process is similar in length to conventional WB, significant advantages in multiplexing and throughput are realized using this microarraying approach.
Capillary Gel Electrophoresis Coupled with Direct Blotting
While the previous methods largely adapted conventional WB approaches to increase performance, recent trends have sought to completely change the ways in which the separation step, blotting step, or both are carried out. For example, capillary electrophoresis (CE) encompasses a versatile and an effective suite of analytical tools for separations.30–34 In particular, capillary gel electrophoresis (CGE) uses small capillaries loaded with a polymer sieving matrix to separate SDS treated proteins based on size35, making SDS-CGE a natural replacement for SDS-PAGE in WB.20,36 Advantages of SDS-CGE include dramatically reduced sample volumes, increased separation efficiency, and more accurate protein sizing capabilities.31,35 CE techniques also lend themselves to automation making them attractive for method development.37,38
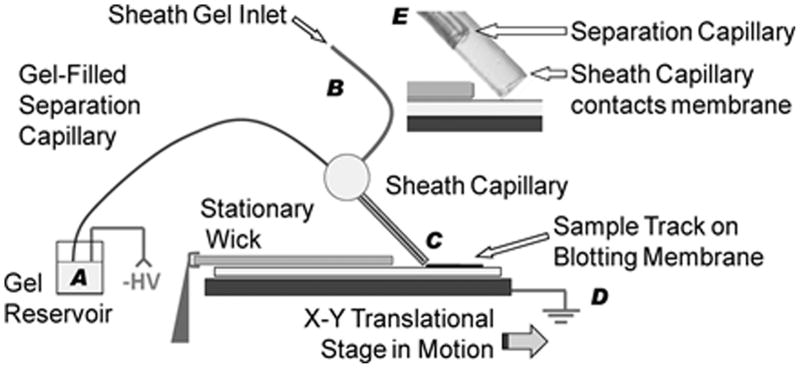
As shown in Fig. 2, Kennedy and coworkers36 integrated SDS-CGE into a WB format using a direct delivery arrangement for blotting the separated proteins.19,39–41 The separation capillary was loaded with an entangled polymer gel and fitted with a gel-filled sheath capillary surrounding the outlet.36 A flow of gel through the sheath was needed to stabilize the electrophoresis current and prevent bubble formation in the separation capillary. The sheathed outlet was brought into light contact with an underlying PVDF membrane, and negative high voltage was applied at the capillary inlet reservoir and grounded at the stage supporting the membrane. Separated protein bands exiting the capillary were directly deposited onto the PVDF membrane, which was slowly translated beneath the sheathed outlet to spatially disperse the bands for subsequent detection. An equal volume mixture of methanol and running buffer kept the membrane moist and was found necessary for efficient protein deposition. The methanol increases capture efficiency at the hydrophobic membrane by dissociating the SDS-protein complex. Following band deposition onto the PVDF membrane, conventional immunoassays can be used to probe the membrane. For these experiments, rapid immunoassays were performed using the SNAP i.d. (Millipore) system that actively drives reagents through the membrane with vacuum.
Figure 2.
Protein separation using capillary gel electrophoresis (CGE) coupled with direct blotting onto a PVDF membrane. Proteins were separated in a sieving matrix filled capillary using a negative high voltage at the inlet (A) and ground (D). Sheath flow was introduced at (B) to stabilize the separation current and the emerging protein bands were directly blotted (C) onto a PVDF membrane that was slowly translated under the sheath capillary. Inset (E) shows the end of the separation capillary with respect to the surrounding sheath capillary. (Reprinted with permission from ref. 36. Copyright 2011 American Chemical Society.)
To demonstrate protein-sizing capabilities, two fluorescently labeled ladder markers, FITC-insulin and FITC-BSA, were mixed directly with a test sample consisting of carbonic anhydrase.36 Following GCE separation and direct blotting, an initial fluorescence image of the membrane mapped the locations of the ladder markers. Subsequent probing with antibodies specific for carbonic anhydrase and imaging revealed an intermediate band as expected.
Using the SDS-CGE direct blotting approach, WB analysis of the small protein lysozyme (MW=14 kDa) was completed in less than an hour with limits of detection of 5 μg/ml (50 pg injected). While larger proteins like BSA take slightly longer (1.5 hr.) due to their slower migration times, this approach results in a significant savings in time over conventional WB. Increased broadening in the delivered protein bands was observed compared to conventional on-column detection, which was attributed mainly to parabolic flow within the sheath at the end of the capillary.36
Sieving Microchip Electrophoresis Coupled with Direct Blotting
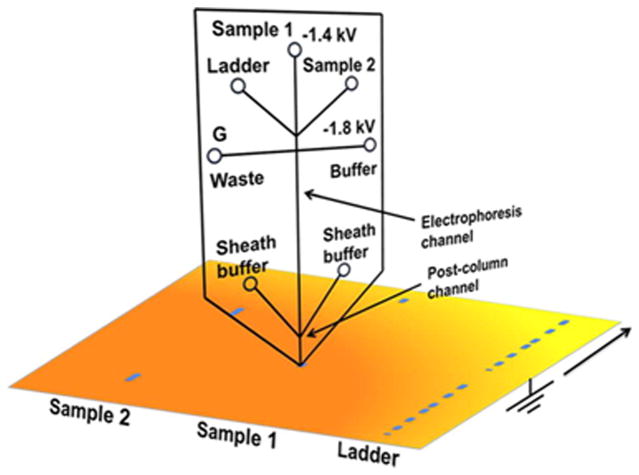
A subsequent report addressed the sheath broadening and further reduced assay times by replacing the CGE separation with a microchip approach.42 Microfluidic separations offer many advantages; they are fast, efficient, easily scaled, and consume minimal amounts of reagents.31,43–46 The microchip shown in Fig. 3 was fabricated in glass with three sample reservoirs connected in a tee-format with buffer and waste reservoirs. The separation channel itself was 2 cm long (50 μm wide, and 15 μm deep) and filled with an entangled polymer sieving matrix. As in the capillary design, a postcolumn sheath flow of sieving material was required to maintain stable currents. This postcolumn sheath channel was slightly wider (90 μm) than the separation channel and extended 300 μm beyond its end. A syringe pump was used to feed the two inlets supplying the sheath flow. As in the original report, direct blotting was used to load the separated proteins onto a PVDF membrane using similar protocols.36,42
Figure 3.
Protein separation using microchip gel electrophoresis coupled with direct blotting onto a PVDF membrane. The microchip was fabricated with a 2 cm long, 15 μm deep, and 50 μm wide electrophoresis channel and reservoirs for sample, size ladder, buffer, waste, and sheath buffer. Sample and ladder standards were injected into the electrophoresis channel by floating the buffer and waste reservoirs and applying the desired voltage between the sample and grounded membrane support. As in the capillary design, a sheath flow was required to stabilize current. The post-column sheath channel was slightly wider (90 μm) than the separation channel and extends 300 μm. As before, emerging proteins were directly blotted onto a PVDF membrane translated below the sheath channel. (Reprinted with permission from ref. 42. Copyright 2013 American Chemical Society.)
As expected, a dramatic reduction in protein separation times was realized in the microchip configuration.43 A protein size ladder consisting of seven protein-SDS complexes ranging from 11 kDa to 155 kDa was separated with the sieving electrophoresis microchip in 5 minutes, with separation efficiencies ranging from 5×104 to 1.8×105 theoretical plates (detected at the exit of the separation channel).42 Moreover, separation times were reduced to 2 minutes at higher voltages by sacrificing some of the separation efficiency. This can be compared with the 20-minute separation times typically realized with capillaries.35,36 The microchip design also enabled better control over sheath geometry thus reducing broadening effects once flow rate of the sheath buffer was optimized. While the blotted protein electropherogram retained comparable theoretical plates as on-column detection, the overall separation had slightly lower resolution on the membrane. Sensitivity and limits of detection were evaluated for actin, carbonic anhydrase II, and lysozyme following development of the blotted proteins, with limits of detection ranging from 0.03 μg/ml to 0.06 μg/ml. Of particular interest for comparison is lysozyme, which led to a noticeably improved LOD of 0.06 μg/ml compared to 5 μg/ml in the previous work.36,42
This promising approach was further optimized to improve separations and multiplex the detection using a slightly modified microchip and blotting protocol.47 The new microchip design extended the separation channel length from a few centimeters to 8.6 cm, thus enabling better resolution of similarly sized proteins at the expense of slightly longer separation times. To multiplex the immunodetection, individual separation tracks are transferred to the PVDF membrane, one for each probe antibody to be used. These membrane regions are physically separated, exposed to the probe antibody, and developed using standard protocols. The key advantages here are the small amounts of sample required, the elimination of stripping procedures used to re-probe membranes in conventional WB, and the significant multiplexing capabilities afforded by this approach. In this initial demonstration, 11 proteins were individually detected using only 400 ng of sample.47
With the faster protein separation step afforded by the microchip, total analysis time was reduced from 1–1.5 hours using CGE to 22–32 minutes.36,42,47 For high throughput, it was noted that 60 injections could be separated on the same microchip with minimal degradation in performance. Performance, moreover, is completely restored upon gel replacement, enabling chips to be reused.42 While more rapid than the SDS-GCE Western approach and considerably faster than conventional WB, the microchip WB approach is still throttled by the immunoassay step (20–30 minutes). For higher throughput, this can be somewhat mitigated by laying down sequential separations on the same membrane for subsequent immunodetection.42,47 Migration time repeatability, however, limits the performance using this approach and was found to vary from 3 to 6% for the proteins studied.
Fully Integrated Microchip for Western Blotting
A separate microchip approach has been explored that integrates the separation and blotting steps onto a single microfluidic platform. Microfluidic analysis platforms can integrate a large number of processes onto a single chip, which offers many advantages.48,49 Compared to their macroscopic counterparts, these processes are often faster and more efficient while leading to substantial reductions in sample and reagent requirements. Herr and coworkers have made significant progress in miniaturizing protein analysis onto microchip formats. These creative approaches help illustrate the potential of these platforms for microscale WB analysis.50–68
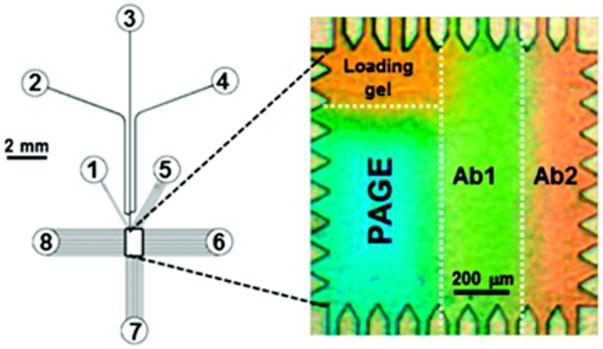
Initial work focused on developing microchips that integrated separation and blotting steps.50,52,55,58,60 As shown in Fig. 4, a glass microfluidic chip was fabricated with a central 1 mm × 1.6 mm rectangular chamber connected to various reservoirs to introduce sample, buffer, and apply sequential voltage programs.52,55,58,60 The central chamber housed a thin PA gel that was spatially modified to create zones for sample loading/stacking, separation, and blotting. The regional properties of the PA gel were controlled through a multistep photopolymerization protocol to modify pore size and functionality of the gel. Initial work immobilized the probe antibody for the desired protein target in the blotting region.50,52,55
Figure 4.
Fully integrated microchip for multiplexed native Westerns. Reservoirs 1–8 provide access for sample, buffer and control voltages, which connect to a central assay chamber through a network of microfluidic channels. An expanded view of the central 1 mm × 1.5 mm microchamber is shown to the right. The patterned PA gel contains a sample loading zone, PAGE separation area, and two antibody-immobilized regions (Ab1 and Ab2) for immunoblotting. Proteins separated based on charge-to-mass in the PAGE region were electrotransferred to the antibody blotting area, where retention indicates binding to the immobilized probe. (Reprinted with permission from ref. 55. Copyright 2011 American Chemical Society.)
Native protein samples (not treated with SDS) introduced and stacked at the loading zone were separated based on their charge-to-mass ratio in the neighboring separation PA gel using electrophoresis.52,55 Once separated, the proteins were laterally transferred to the blotting zone using a voltage applied orthogonal to the previous separation step. The blotting region with immobilized probe antibody captured the protein of interest while bands not recognized by the antibody were swept away, out of the blotting zone.
The microchip design enabled full-field imaging so that protein mobility in the separation area could be directly linked with antibody recognition in the blotting region. To demonstrate the capabilities of this approach, free prostate antigen (fPSA) extracted from seminal fluid was detected.52 Native PAGE in the microfluidic chamber required only 36 seconds with an average separation resolution of 1.59. An additional 30–60 s was required to transfer the separated bands into the blotting region, where electric field shaping and gel properties resulted in 90% transfer efficiency with only 6% loss in separation resolution. Only the band associated with fPSA was retained in the blotting region by the immobilized anti-fPSA antibody. Most impressively, the entire native fPSA analysis required less than 5 minutes, required approximately 1 μg of immobilized detection antibody, and yielded a detection S/N of 40 at 500 nM fPSA.52 This represents a significant savings in time and illustrates the distinct advantages inherent in miniaturized designs. This unified microchip design was later extended to incorporate several blotting regions for multiplexed protein detection as shown in Fig. 4.55
A limitation in the previously described approach is that proteins are not separated by size as they are in conventional SDS-PAGE. The reduced affinity between surfactant-coated proteins and the immobilized antibody made peak capture difficult using SDS treated samples.58 To circumvent this, a new approach was introduced that used electrostatic interactions to trap separated proteins in the blotting zone for subsequent probing. As before, the microfluidic PA chamber was photopatterned to create a loading/stacking zone, a separation gel, and a new electrostatic immobilization region where anionic charged moieties were copolymerized in the gel. Protein samples were treated with the cationic surfactant cetyltrimethylammonium bromide (CTAB), commonly used in Eastern blotting, to coat proteins with positive charge.2,58,69 Following sample loading and CTAB-PAGE separation to size the protein mixture, the protein bands were electrotransferred laterally into the electrostatic immobilization gel. Electrostatic interactions between the CTAB decorated proteins and the negatively charged gel immobilized the protein bands for subsequent blocking and probing with antibodies.58 This general scheme was later expanded to enable SDS-PAGE separation using an electrostatic capture zone incorporating positively charged poly-L-lysine.60,70 Unlike the original designs, both approaches enable direct correlations between protein size and immunodetection, albeit at the expense of assay speed (hours).60
On-Capillary Protein Immobilization Approaches
In conventional WB, separated proteins in the PA gel are generally difficult to access directly with antibody probes, thus necessitating the replica transfer step to a more porous membrane.1–3 The last decade, however, has seen a rapid expansion of approaches using a protein immobilization step following separation that enables subsequent antibody probing. These approaches eliminate the transfer step, which reduces the complexity of the assay, increases throughput, and facilitates automation. This has evolved into a suite of commercial instruments for automated, high throughput WB in parallel formats.59,71–74
Capillary electrophoretic protein separation followed by an immobilization step for immunoprobing was first introduced by O’Neill et. al. in 2006.75 Isoelectric focusing was used to separate proteins in a short capillary (5 cm long with 100 μm i.d.). Separated proteins were then photochemically captured onto the walls using photoactive moieties lining the capillary. Capturing the proteins to the capillary wall preserves the separation information and enables subsequent probing with antibodies. This provides the confirmatory information that gives WB its specificity for targeted proteins.
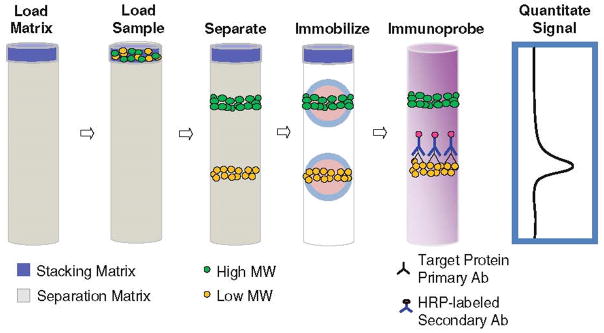
As shown in Fig. 5, this basic approach has been extended and commercialized to separate proteins based on molecular weight followed by UV immobilization to the capillary wall using proprietary chemistry (SimpleWestern, ProteinSimple).59,71–74,76 Following immobilization, the column is flushed with a matrix removing buffer and blocking buffer before incubated sequentially with primary and secondary antibodies. To detect the immobilized bands, chemiluminescence signals from HRP linked secondary antibodies are detected using a CCD camera.
Figure 5.
Sequence of steps for WB analysis using the automated SimpleWestern method. The sample is loaded, stacked, and separated in a short capillary filled with sieving matrix. Photoactivation immobilizes the separated proteins to the capillary wall, which are then probed with primary antibodies. HRP-labeled secondary antibodies label the primary, leading to detection of the targeted protein. (Reprinted with permission from ref. 76. Copyright 2015 Humana Press.)
With their introduction, SimpleWesterns have had a substantial impact since they automate the WB process, can analyze proteins ranging from 12 to 440 kDa in size, require only nanograms of sample loading, and can run 25 analyses in parallel in about 3–5 hours.59,71–74 Comparisons between SimpleWesterns and manual WB have confirmed significant advantages in throughput, repeatability, and quantification.59,71,73,74 Both intra- and inter-assay reproducibility are improved in the automated systems and are less dependent on operator skill or technique.18,74 There are, however, low capture efficiencies in the immobilization step and less flexibility in gel options, which can limit high-resolution measurements at the extremes of molecular weight.57,74
In-Gel Protein Immobilization Approaches
Recently, another approach for eliminating the transfer step and carrying out in-gel sieving, immobilization, and probing has appeared that has enabled Western analysis down to the single cell level.56,57,64–66,68 The general approach uses a bifunctional gel that serves as a sieving matrix for protein separation and immobilization scaffold, following brief UV exposure, capturing separated proteins for subsequent probing with antibodies. This method eliminates the transfer step and lends itself to scalable, microfabricated platforms.
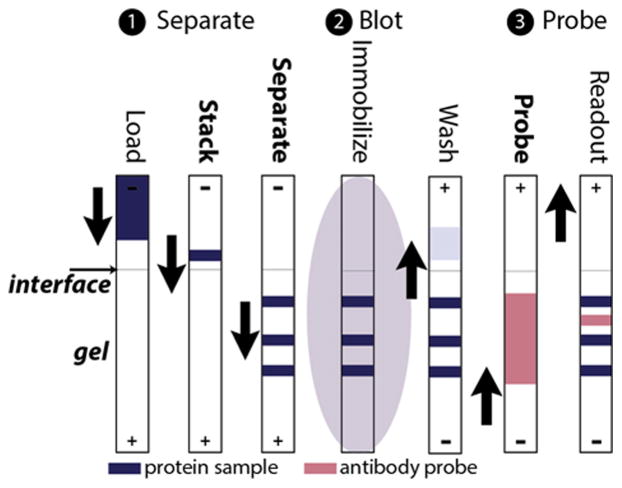
Initial reports used microfabricated chips with 1 cm long separation channels (70 μm wide and 10 μm deep) wet etched into borosilicate glass slides.56,64 The enclosed channels were loaded with photopatterned PA gel incorporating a benzophenone methacrylamide monomer that can covalently bind separated proteins upon UV exposure. Following protein separation in the gel, 30 second exposure to UV light immobilized the separated protein bands which were then probed with antibodies, electrophoretically passed through the same channel.56,64 This sequence of steps is shown schematically in Fig. 6.
Figure 6.
Microchannel Western blotting using protein immobilization following separation. (1) Proteins were loaded, stacked, and separated in an enclosed separation channel, 1 cm long × 10 μm deep, 70 μm wide, filled with photoactive PA gel. (2) Separated proteins were immobilized in the gel with UV exposure. (3) Antibody was introduced via electrophoresis to probe the immobilized protein bands. (Reprinted with permission from ref. 64. Copyright 2014 American Chemical Society.)
The potential for high throughput was demonstrated by fabricating 48 separate microchannel assays on a standard glass microscope slide.56 A photoactive gel was designed that had a tunable porosity (PACTgel). The sieving gel is photopatterned to reproducibly create the loading interface and incorporates benzophenone moieties for photoimmobilization of proteins following separation. The parallel format was tested by measuring the transcription factor NFκB in lysates of NFκB-transfected 293T cells, purified HIV proteins, and HIV antibodies in human sera. Analysis was completed in less than an hour with competitive detection metrics. Notably, the efficiency of protein capture in the photoactive 3-D gel was greater than 75%, which can be compared with approximately 0.01% efficiency in the SimpleWestern approach.56
A discontinuity in pore size at the interface between the sample loading and separation segments of the gel was found important for optimizing separation efficiency and antibody probing.64 A step in pore size from large to small at the interface transitioned the assay from transient isotachophoresis, used to stack the sample, to protein sizing in the PAGE separation. A discontinuous tris tricine buffer system was also used to improve separation efficiency and more effectively separate smaller proteins, which can remain stacked in other buffers.64
Controlling interfacial pore size was also found important for optimizing antibody loading and transport to the immobilized protein bands.64 Interestingly, a discrete plug of antibody swept down the channel was found to provide better detection than loading the entire channel. Not only do discrete plugs reduce antibody consumption, they also decreased background signal and assay time. The reduced background enhanced the signal-to-noise by a factor of two and improved the correlation of variation from 18.8% in whole channel loading to 11.4% in the swept plug approach.64
This same in-gel separation and probing approach has been extended to the single cell level, opening exciting possibilities for studying cellular heterogeneity.65,66,68 As shown in Fig. 7, an array of open microwells, optimized to capture single cells (30 μm deep × 32 μm diameter), were fabricated in a thin, 30 μm thin film of photoactive PA gel. As before, the bifunctional gel serves as the sieving matrix for protein separation and immobilization scaffold following brief UV exposure. The separation length is limited by the spacing between microwell rows, which was optimized at a 1 mm gap to maximize assay density while providing sufficient length to separate proteins ranging from 15 to 90 kDa.65,66,68 More recently, a greyscale photolithography approach has enabled a gradient of pore sizes to be fabricated over the limited length of the separation axis, expanding the mass range of the separation.68
Figure 7.
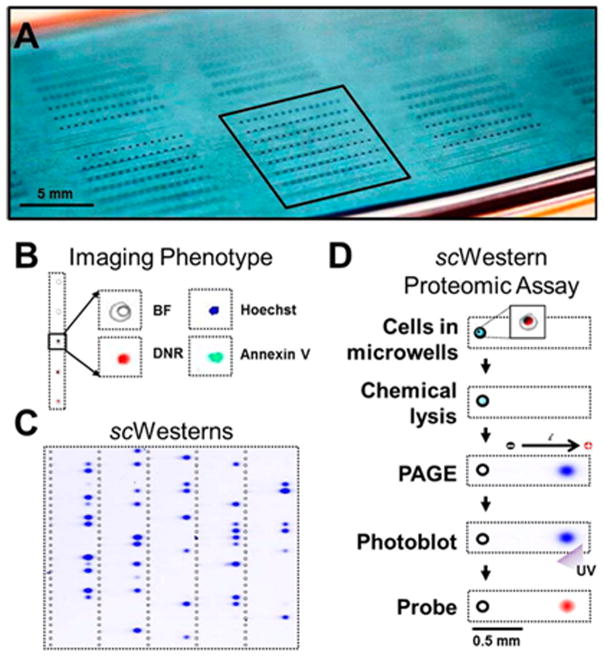
(A) An array of microwells fabricated into a thin layer (30 μm) of photoactive PA gel for single cell analysis. (B) Cells trapped in the wells can be imaged for phenotyping and (C) lysed for Western analysis. (D) Separated proteins were immobilized in the photoactive gel using UV light and probed with antibody. (Reprinted with permission from ref. 66. Copyright 2014 American Chemical Society.)
A suspension of cells was distributed across the microchip and allowed to settle.65,66 Repeated rinse cycles removed unwanted cells on the surface of the microchip while leaving a statistical population of occupied microwells. The trapped cells were lysed, the lysate was separated using PAGE, and the proteins were immobilized in the gel matrix using photoactivation of benzophenone. The slide was incubated with probe and secondary antibodies and finally analyzed using a fluorescence microarray scanner.66
This general approach was applied to characterize the single cell response of U373 MG human glioblastoma cells to the chemotherapeutic, daunomycin, and to map cell variability in stem cell signaling and differentiation.65,66 Detection limits below 30,000 protein copies were reported with total analysis times under 4 hours. Thousands of simultaneous assays can potentially be carried out on a chip the size of a conventional microscope slide, enabling significant throughput capabilities. As shown in Fig. 7, the thin, transparent gel also enables imaging of the trapped cells prior to lysis, providing additional phenotype mapping prior to single cell Western analysis.66
Future Prospects: Label-Free Biosensing for Multiplexed Detection
Miniaturized platforms have led to transformative improvements in WB automation and throughput. Given the activity in developing new microfluidic approaches and micro-total analysis systems, these directions are likely to further expand the capabilities and applications of microscale WB. For the most part, the preferred detection method in WB remains conventional antibody probing followed by incubation with HRP-linked secondary antibodies for detection. Significant advances, however, are being made in label-free biosensing that may offer interesting new capabilities for specifically detecting separated proteins in real-time.77–80 These approaches are generally rapid, quantitative, require minimal antibody, and are easily multiplexed; thus offering intriguing metrics worth exploring.81,82
Surface plasmon resonance (SPR), for example, has been developed extensively for sensing at surfaces and provides rapid, quantitative measurements of antigen binding to immobilized antibodies. SPR has also been integrated with CE, illustrating the feasibility of combining separation and sensing steps. Whelan and Zare, for instance, integrated SPR detection following CE to specifically detect IgG. Protein-A was immobilized on the SPR sensor surface which was positioned at the outlet of the CE.83 IgG exiting the separation capillary was specifically detected at the SPR sensor with a detection limit of 2 fmol and reported dynamic range of three orders of magnitude. Other reports have outlined improvements in interfacing SPR with CE and coupling it with microfluidic separations.84,85
Small optical microresonators have also been extensively developed for label-free sensing and recently integrated with separations.79,80,86–90 Whispering gallery mode (WGM) resonators are small, axially symmetric dielectrics that can resonantly trap and recirculate light via total internal reflection. Their spectral resonances are extremely narrow and shift in response to changes in local refractive index, providing a mechanism for label-free biosensing.79,80,86 Antibodies immobilized at the microresonator surface, for example, can recognize and bind antigens, which alter the local refractive index and shifts the WGM resonance. The small size of WGM resonators (tens of microns), ease of multiplexed detection, and favorable figures of merit may enable new capabilities for microscale WB when combined with separations.
WGM detection has been integrated with both liquid chromatography and CE.88–90 For integration with CE, Fan and coworkers developed an on-column approach using thinned capillaries that support WGM resonances. The thinned wall of the capillary enabled the evanescent field of the circulating WGM to sense the interior environment of the capillary. To demonstrate detection capabilities, refractive index changes associated with neutral glycerol sample plugs driven through the capillary with electrophoresis, were characterized. These measurements reported detection limits of 10−7 RIU (refractive index units) and were the first to illustrate the feasibility of integrating WGM sensing with CE. While impressive, this particular approach results in fragile and difficult to reproduce capillaries that are not easily modified for immunosensing.
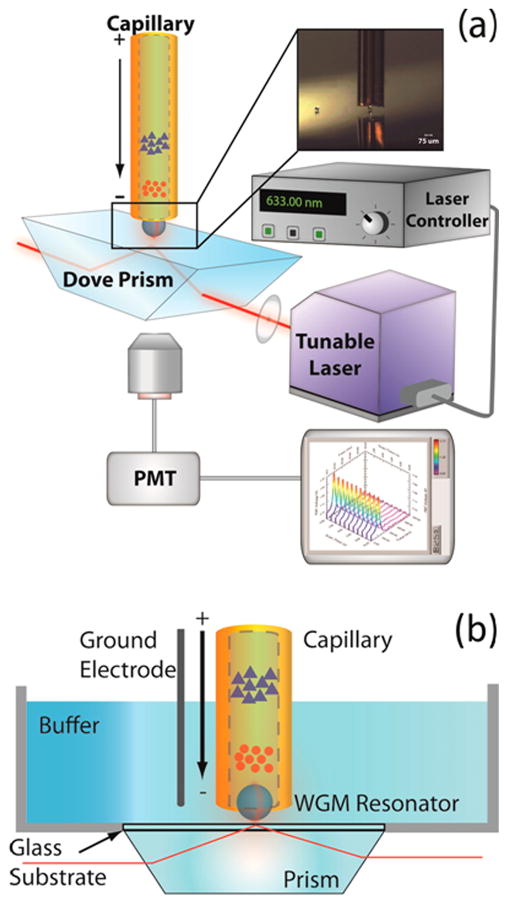
A more recent approach integrated WGM refractive index sensing using a more flexible end-column geometry.88 As shown in Fig. 8, a Dove prism is used to couple excitation light from a tunable diode laser into a 53 μm diameter spherical microresonator positioned in the lumen of a 75 μm i.d. separation capillary. Spectral shifts in the WGM resonance were monitored to measure refractive index changes associated with the separation of three ions (Na+, Li+, and K+). The microsphere did not adversely affect the separation efficiency and 10−7 RIU detection limits were reported using a signal modulation scheme.88
Figure 8.
(A) Schematic of the approach used to integrate end-column WGM refractive index sensing with capillary electrophoresis. The evanescent field created from total internal reflection in the Dove prism excites resonances in a 53 μm barium titanate microsphere positioned in the lumen of the separation capillary. Scattered light from the microsphere was collected from below and detected with a photomultiplier tube (PMT) to track shifts in its resonance as the tunable diode laser is scanned. (B) Magnified view showing the grounded buffer reservoir and detection geometry. Ions separated in the capillary change the refractive index around the WGM resonator as they pass by, leading to shifts in the WGM resonance. (Reprinted with permission from ref. 88. Copyright 2016 American Chemical Society.)
Integrating specific biosensing with functionalized WGM resonators is conceptually similar to the on-chip antibody capture method and offers unique advantages. WGM sensing is compatible with both capillary and microchip based separations, each resonator (assay) requires trace amounts of antibody, and quantitative assays can be completed in minutes, significantly improving analysis times.81,91,92 However, much work is needed to explore the practical limits of such an approach. Biosensing performance is strongly dependent on analyte transport to the sensing surface, which depends on a number of factors.93–96 The size and shape of the sensing element, presence of flow, channel geometry, and development of depletion zones around the sensing element can all influence the observed response. These factors, moreover, become limiting at low concentrations where the unreasonably long timescales associated with analyte transport to the sensing surface prevents practical applications.93,94 For spherical WGM resonators, however, their size and shape lead to favorable mass transport metrics compared to smaller, two-dimensional sensing elements. Moreover, many practical applications do not require pushing the envelope of detection limits. In such applications, the potential for real-time, multiplexed detection of proteins following a separation step offers exciting opportunities.81,88,97
Summary
Expanding WB applications requires a departure from conventional slab gels and electroblotting to incorporate capabilities better suited to handle small sample sizes and high-throughput demands. Exciting new WB approaches incorporating capillary separations and integrated microfluidic chips have already decreased analysis times, increased throughput, and decreased sample requirements down to the single cell level. The examples highlighted here help illustrate some of these breakthroughs and point to an exciting path forward for developing rapid, miniaturized WB platforms. The development of improved detection methods, moreover, that can expand multiplexing capabilities, push detection limits, and increase quantitation is an area ripe for exploration. Progress in these areas will likely continue at a rapid pace as improved methods for microfluidics, microscale analysis systems, and label-free biosensing continue to advance the field.
Acknowledgments
It is an honor to be included in this special issue dedicated to the memory of our colleague Professor Craig E. Lunte. Craig was a dear friend whose generous spirit and humor is truly missed. An expert in CE, Craig helped us initiate projects coupling sensing with separations and was pleased we were finally trying to do something useful (his words). We gratefully acknowledge support from NSF (CBET 1133814), the Madison and Lila Self Foundation (B.J.S.), and NIH T32-GM008359 (D.C.K.).
References
- 1.Burnette WN. Western Blotting - Electrophoretic Transfer of Proteins from Sodium Dodecyl Sulfate-Polyacrylamide Gels to Unmodified Nitrocellulose and Radiographic Detection with Antibody and Radioiodinated Protein-A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 2.Kurien BT, Scofield RH. Western blotting. Methods. 2006;38:283–293. doi: 10.1016/j.ymeth.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Towbin H, Staehelin T, Gordon J. Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets - Procedure and Some Applications. P Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurien BT, Scofield RH, editors. Protein Blotting and Detection: Methods and Protocols. Vol. 536 Humana Press; 2009. [Google Scholar]
- 5.http://www.biocompare.com/Editorial-Articles/177815-2015-Antibody-Market-Report/
- 6.Marx V. Finding the right antibody for the job. Nat Methods. 2013;10:703–707. doi: 10.1038/nmeth.2570. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury A, Pluckthun A. Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 8.Baker M. Antibody Anarchy: A Call to Order. Nature. 2015;527:545–551. doi: 10.1038/527545a. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht E, Norheim F, Thiede B, Holen T, Ohashi T, Schering L, Lee S, Brenmoehl J, Thomas S, Drevon CA, Erickson HP, Maak S. Irisin - a myth rather than an exercise-inducible myokine. Sci Rep-Uk. 2015;5 doi: 10.1038/srep08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel MC, Wieland T, Tsujimoto G. How reliable are G-protein-coupled receptor antibodies? N-S Arch Pharmacol. 2009;379:385–388. doi: 10.1007/s00210-009-0395-y. [DOI] [PubMed] [Google Scholar]
- 11.Jensen BC, Swigart PM, Simpson PC. Ten commercial antibodies for alpha-1-adrenergic receptor subtypes are nonspecific. N-S Arch Pharmacol. 2009;379:409–412. doi: 10.1007/s00210-008-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordeaux J, Welsh AW, Agarwal S, Killiam E, Baquero MT, Hanna JA, Anagnostou VK, Rimm DL. Antibody validation. Biotechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcon E, Jain H, Bhattacharya A, Guo HB, Phanse S, Pu SY, Byram G, Collins BC, Dowdell E, Fenner M, Guo XH, Hutchinson A, Kennedy JJ, Krastins B, Larsen B, Lin ZY, Lopez MF, Loppnau P, Miersch S, Nguyen T, Olsen JB, Paduch M, Ravichandran M, Seitova A, Vadali G, Vogelsang MS, Whiteaker JR, Zhong GQ, Zhong N, Zhao L, Aebersold R, Arrowsmith CH, Emili A, Frappier L, Gingras AC, Gstaiger M, Paulovich AG, Koide S, Kossiakoff AA, Sidhu SS, Wodak SJ, Graslund S, Greenblatt JF, Edwards AM. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat Methods. 2015;12:725–U747. doi: 10.1038/nmeth.3472. [DOI] [PubMed] [Google Scholar]
- 14.Willingham MC. Conditional epitopes: Is your antibody always specific? J Histochem Cytochem. 1999;47:1233–1235. doi: 10.1177/002215549904701002. [DOI] [PubMed] [Google Scholar]
- 15.Saper CB, Sawchenko PE. Magic peptides, magic antibodies: Guidelines for appropriate controls for immunohistochemistry. J Comp Neurol. 2003;465:161–163. doi: 10.1002/cne.10858. [DOI] [PubMed] [Google Scholar]
- 16.Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, Gu TT, Kharchenko PV, Kuan S, Latorre I, Linder-Basso D, Luu Y, Ngo Q, Perry M, Rechtsteiner A, Riddle NC, Schwartz YB, Shanower GA, Vielle A, Ahringer J, Elgin SCR, Kuroda MI, Pirrotta V, Ren B, Strome S, Park PJ, Karpen GH, Hawkins RD, Lieb JD. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southern EM. Detection of Specific Sequences among DNA Fragments Separated by Gel-Electrophoresis. J Mol Biol. 1975;98:503. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh R, Gilda JE, Gomes AV. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev Proteomic. 2014;11:549–560. doi: 10.1586/14789450.2014.939635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck S. Protein Blotting with Direct Blotting Electrophoresis. Anal Biochem. 1988;170:361–366. doi: 10.1016/0003-2697(88)90643-4. [DOI] [PubMed] [Google Scholar]
- 20.Jin S, Kennedy RT. New developments in Western blot technology. Chinese Chem Lett. 2015;26:416–418. [Google Scholar]
- 21.Nuchtavorn N, Suntornsuk W, Lunte SM, Suntornsuk L. Recent applications of microchip electrophoresis to biomedical analysis. J Pharmaceut Biomed. 2015;113:72–96. doi: 10.1016/j.jpba.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Delamarche E, Juncker D, Schmid H. Microfluidics for processing surfaces and miniaturizing biological assays. Adv Mater. 2005;17:2911–2933. [Google Scholar]
- 23.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 24.Janasek D, Franzke J, Manz A. Scaling and the design of miniaturized chemical-analysis systems. Nature. 2006;442:374–380. doi: 10.1038/nature05059. [DOI] [PubMed] [Google Scholar]
- 25.Pan WY, Chen W, Jiang XY. Microfluidic Western Blot. Anal Chem. 2010;82:3974–3976. doi: 10.1021/ac1000493. [DOI] [PubMed] [Google Scholar]
- 26.Bernard A, Michel B, Delamarche E. Micromosaic immunoassays. Anal Chem. 2001;73:8–12. doi: 10.1021/ac0008845. [DOI] [PubMed] [Google Scholar]
- 27.Chang HN, Leroueil PR, Selwa K, Gasper CJ, Tsuchida RE, Wang JJ, McHugh WM, Cornell TT, Baker JR, Goonewardena SN. Profiling Inflammatory Responses with Microfluidic Immunoblotting. Plos One. 2013;8 doi: 10.1371/journal.pone.0081889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He S, Zhang Y, Wang P, Xu XZ, Zhu K, Pan WY, Liu WW, Cai KY, Sun JS, Zhang W, Jiang XY. Multiplexed microfluidic blotting of proteins and nucleic acids by parallel, serpentine microchannels. Lab Chip. 2015;15:105–112. doi: 10.1039/c4lc00901k. [DOI] [PubMed] [Google Scholar]
- 29.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA, Jones RB. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods. 2010;7:148–U195. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harstad RK, Johnson AC, Weisenberger MM, Bowser MT. Capillary Electrophoresis. Anal Chem. 2016;88:299–319. doi: 10.1021/acs.analchem.5b04125. [DOI] [PubMed] [Google Scholar]
- 31.Creamer JS, Oborny NJ, Lunte SM. Recent advances in the analysis of therapeutic proteins by capillary and microchip electrophoresis. Anal Methods-Uk. 2014;6:5427–5449. doi: 10.1039/C4AY00447G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osbourn DM, Weiss DJ, Lunte CE. On-line preconcentration methods for capillary electrophoresis. Electrophoresis. 2000;21:2768–2779. doi: 10.1002/1522-2683(20000801)21:14<2768::AID-ELPS2768>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaul S, Faiman MD, Lunte CE. Determination of GABA, glutamate and carbamathione in brain microdialysis samples by capillary electrophoresis with fluorescence detection. Electrophoresis. 2011;32:284–291. doi: 10.1002/elps.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnett SD, Lunte CE. Investigation of the mechanism of pH-mediated stacking of anions for the analysis of physiological samples by capillary electrophoresis. Electrophoresis. 2003;24:1745–1752. doi: 10.1002/elps.200305399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu ZF, Lu JJ, Liu SR. Protein separation by capillary gel electrophoresis: A review. Anal Chim Acta. 2012;709:21–31. doi: 10.1016/j.aca.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson GJ, Cipolla CM, Kennedy RT. Western Blotting Using Capillary Electrophoresis. Anal Chem. 2011;83:1350–1355. doi: 10.1021/ac102671n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen LA, Christiansen M, Vuust J, Andersen PS. High throughput mutation screening by automated capillary electrophoresis. Comb Chem High T Scr. 2000;3:393–409. doi: 10.2174/1386207003331508. [DOI] [PubMed] [Google Scholar]
- 38.Bossuyt X. Separation of serum proteins by automated capillary zone electrophoresis. Clin Chem Lab Med. 2003;41:762–772. doi: 10.1515/CCLM.2003.116. [DOI] [PubMed] [Google Scholar]
- 39.Beck S, Pohl FM. DNA Sequencing with Direct Blotting Electrophoresis. Embo J. 1984;3:2905–2909. doi: 10.1002/j.1460-2075.1984.tb02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen YK, Jiang YW, Chang SC, Wang AB. Western Blotting by Thin-Film Direct Coating. Anal Chem. 2014;86:5164–5170. doi: 10.1021/ac5010162. [DOI] [PubMed] [Google Scholar]
- 41.Goto S, Savory N, Abe K, Kinoshita H, Ikebukuro K. Development of an automated direct blotting electrophoresis system for bioanalytical applications. Anal Methods-Uk. 2015;7:4881–4884. [Google Scholar]
- 42.Jin S, Anderson GJ, Kennedy RT. Western Blotting Using Microchip Electrophoresis Interfaced to a Protein Capture Membrane. Anal Chem. 2013;85:6073–6079. doi: 10.1021/ac400940x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu DP, Qin JH, Lin BC. Electrophoretic separations on microfluidic chips. J Chromatogr A. 2008;1184:542–559. doi: 10.1016/j.chroma.2007.11.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bousse L, Mouradian S, Minalla A, Yee H, Williams K, Dubrow R. Protein sizing on a microchip. Anal Chem. 2001;73:1207–1212. doi: 10.1021/ac0012492. [DOI] [PubMed] [Google Scholar]
- 45.Backofen U, Matysik FM, Lunte CE. A chip-based electrophoresis system with electrochemical detection and hydrodynamic injection. Anal Chem. 2002;74:4054–4059. doi: 10.1021/ac020110j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro ER, Manz A. Present state of microchip electrophoresis: State of the art and routine applications. J Chromatogr A. 2015;1382:66–85. doi: 10.1016/j.chroma.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 47.Jin S, Furtaw MD, Chen H, Lamb DT, Ferguson SA, Arvin NE, Dawod M, Kennedy RT. Multiplexed Western Blotting Using Microchip Electrophoresis. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rios A, Zougagh M, Avila M. Miniaturization through lab-on-a-chip: Utopia or reality for routine laboratories? A review. Anal Chim Acta. 2012;740:1–11. doi: 10.1016/j.aca.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 49.Patabadige DEW, Jia S, Sibbitts J, Sadeghi J, Sellens K, Culbertson CT. Micro Total Analysis Systems: Fundamental Advances and Applications. Anal Chem. 2016;88:320–338. doi: 10.1021/acs.analchem.5b04310. [DOI] [PubMed] [Google Scholar]
- 50.He M, Herr AE. Microfluidic Polyacrylamide Gel Electrophoresis with in Situ Immunoblotting for Native Protein Analysis. Anal Chem. 2009;81:8177–8184. doi: 10.1021/ac901392u. [DOI] [PubMed] [Google Scholar]
- 51.He M, Herr AE. Automated microfluidic protein immunoblotting. Nat Protoc. 2010;5:1844–1856. doi: 10.1038/nprot.2010.142. [DOI] [PubMed] [Google Scholar]
- 52.He M, Herr AE. Polyacrylamide Gel Photopatterning Enables Automated Protein Immunoblotting in a Two-Dimensional Microdevice. J Am Chem Soc. 2010;132:2512. doi: 10.1021/ja910164d. [DOI] [PubMed] [Google Scholar]
- 53.Chen XF, Kapil MA, Hughes AJ, Herr AE. Single-Microchannel, Multistep Assay Reports Protein Size and Immunoaffinity. Anal Chem. 2011;83:6573–6579. doi: 10.1021/ac200982j. [DOI] [PubMed] [Google Scholar]
- 54.He M, Novak J, Julian BA, Herr AE. Membrane-Assisted Online Renaturation for Automated Microfluidic Lectin Blotting. J Am Chem Soc. 2011;133:19610–19613. doi: 10.1021/ja207963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tia SQ, He M, Kim D, Herr AE. Multianalyte On-Chip Native Western Blotting. Anal Chem. 2011;83:3581–3588. doi: 10.1021/ac200322z. [DOI] [PubMed] [Google Scholar]
- 56.Hughes AJ, Herr AE. Microfluidic Western blotting. P Natl Acad Sci USA. 2012;109:21450–21455. doi: 10.1073/pnas.1207754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hughes AJ, Lin RKC, Peehl DM, Herr AE. Microfluidic integration for automated targeted proteomic assays. P Natl Acad Sci USA. 2012;109:5972–5977. doi: 10.1073/pnas.1108617109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Karns K, Tia SQ, He M, Herr AE. Electrostatic Protein Immobilization Using Charged Polyacrylamide Gels and Cationic Detergent Microfluidic Western Blotting. Anal Chem. 2012;84:2533–2540. doi: 10.1021/ac3000013. [DOI] [PubMed] [Google Scholar]
- 59.Chen JQ, Heldman MR, Herrmann MA, Kedei N, Woo W, Blumberg PM, Goldsmith PK. Absolute quantitation of endogenous proteins with precision and accuracy using a capillary Western system. Anal Biochem. 2013;442:97–103. doi: 10.1016/j.ab.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung M, Kim D, Herr AE. Microchamber Western Blotting Using Poly-L-Lysine Conjugated Polyacrylamide Gel for Blotting of Sodium Dodecyl Sulfate Coated Proteins. Anal Chem. 2013;85:7753–7761. doi: 10.1021/ac401012j. [DOI] [PubMed] [Google Scholar]
- 61.Duncombe TA, Herr AE. Photopatterned free-standing polyacrylamide gels for microfluidic protein electrophoresis. Lab Chip. 2013;13:2115–2123. doi: 10.1039/c3lc50269d. [DOI] [PubMed] [Google Scholar]
- 62.Herr AE. Disruptive by Design: A Perspective on Engineering in Analytical Chemistry. Anal Chem. 2013;85:7622–7628. doi: 10.1021/ac4010887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tentori AM, Hughes AJ, Herr AE. Microchamber Integration Unifies Distinct Separation Modes for Two-Dimensional Electrophoresis. Anal Chem. 2013;85:4538–4545. doi: 10.1021/ac4001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerver RE, Herr AE. Microfluidic Western Blotting of Low-Molecular-Mass Proteins. Anal Chem. 2014;86:10625–10632. doi: 10.1021/ac5024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hughes AJ, Spelke DP, Xu ZC, Kang CC, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11:749–U794. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang CC, Lin JMG, Xu ZC, Kumar S, Herr AE. Single-Cell Western Blotting after Whole-Cell Imaging to Assess Cancer Chemotherapeutic Response. Anal Chem. 2014;86:10429–10436. doi: 10.1021/ac502932t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duncombe TA, Tentori AM, Herr AE. Microfluidics: reframing biological enquiry. Nat Rev Mol Cell Bio. 2015;16:554–567. doi: 10.1038/nrm4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duncombe TA, Kang CC, Maity S, Ward TM, Pegram MD, Murthy N, Herr AE. Hydrogel Pore-Size Modulation for Enhanced Single-Cell Western Blotting. Adv Mater. 2016;28:327–334. doi: 10.1002/adma.201503939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buxbaum E. Cationic electrophoresis and electrotransfer of membrane glycoproteins. Anal Biochem. 2003;314:70–76. doi: 10.1016/s0003-2697(02)00639-5. [DOI] [PubMed] [Google Scholar]
- 70.Hou CL, Herr AE. Microfluidic integration of Western blotting is enabled by electrotransfer-assisted sodium dodecyl sulfate dilution. Analyst. 2013;138:158–163. doi: 10.1039/c2an36033k. [DOI] [PubMed] [Google Scholar]
- 71.Rustandi RR, Loughney JW, Hamm M, Hamm C, Lancaster C, Mach A, Ha S. Qualitative and quantitative evaluation of Simon (TM), a new CE-based automated Western blot system as applied to vaccine development. Electrophoresis. 2012;33:2790–2797. doi: 10.1002/elps.201200095. [DOI] [PubMed] [Google Scholar]
- 72.Loughney JW, Lancaster C, Ha S, Rustandi RR. Residual bovine serum albumin (BSA) quantitation in vaccines using automated Capillary Western technology. Anal Biochem. 2014;461:49–56. doi: 10.1016/j.ab.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 73.Xu D, Mane S, Sosic Z. Characterization of a biopharmaceutical protein and evaluation of its purification process using automated capillary Western blot. Electrophoresis. 2015;36:363–370. doi: 10.1002/elps.201400380. [DOI] [PubMed] [Google Scholar]
- 74.Chen JQ, Wakefield LM, Goldstein DJ. Capillary nano-immunoassays: advancing quantitative proteomics analysis, biomarker assessment, and molecular diagnostics. J Transl Med. 2015;13 doi: 10.1186/s12967-015-0537-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Neill RA, Bhamidipati A, Bi XH, Deb-Basu D, Cahill L, Ferrante J, Gentalen E, Glazer M, Gossett J, Hacker K, Kirby C, Knittle J, Loder R, Mastroieni C, MacLaren M, Mills T, Nguyen U, Parker N, Rice A, Roach D, Suich D, Voehringer D, Voss K, Yang J, Yang T, Vander Horn PB. Isoelectric focusing technology quantifies protein signaling in 25 cells. P Natl Acad Sci USA. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurien BT, Scofield RH. Western blotting : methods and protocols. Humana Press; New York: 2015. pp. 465–467. [Google Scholar]
- 77.Fan XD, White IM, Shopova SI, Zhu HY, Suter JD, Sun YZ. Sensitive optical biosensors for unlabeled targets: A review. Anal Chim Acta. 2008;620:8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hunt HK, Armani AM. Label-free biological and chemical sensors. Nanoscale. 2010;2:1544–1559. doi: 10.1039/c0nr00201a. [DOI] [PubMed] [Google Scholar]
- 79.Luchansky MS, Bailey RC. High-Q Optical Sensors for Chemical and Biological Analysis. Anal Chem. 2012;84:793–821. doi: 10.1021/ac2029024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vollmer F, Arnold S. Whispering-gallery-mode biosensing: label-free detection down to single molecules. Nat Methods. 2008;5:591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 81.Wildgen SM, Dunn RC. Whispering gallery mode resonators for rapid label-free biosensing in small volume droplets. Biosensors (Basel) 2015;5:118–130. doi: 10.3390/bios5010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wade JH, Alsop AT, Vertin NR, Yang HW, Johnson MD, Bailey RC. Rapid, Multiplexed Phosphoprotein Profiling Using Silicon Photonic Sensor Arrays. Acs Central Sci. 2015;1:374–382. doi: 10.1021/acscentsci.5b00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whelan RJ, Zare RN. Surface plasmon resonance detection for capillary electrophoresis separations. Anal Chem. 2003;75:1542–1547. doi: 10.1021/ac0263521. [DOI] [PubMed] [Google Scholar]
- 84.Ly N, Foley K, Tao NJ. Integrated label-free protein detection and separation in real time using confined surface plasmon resonance imaging. Anal Chem. 2007;79:2546–2551. doi: 10.1021/ac061932+. [DOI] [PubMed] [Google Scholar]
- 85.Gaspar A, Gomez FA. Development of an ultra-low volume flow cell for surface plasmon resonance detection in a miniaturized capillary electrophoresis system. Electrophoresis. 2012;33:1723–1728. doi: 10.1002/elps.201100673. [DOI] [PubMed] [Google Scholar]
- 86.Sun YZ, Fan XD. Optical ring resonators for biochemical and chemical sensing. Anal Bioanal Chem. 2011;399:205–211. doi: 10.1007/s00216-010-4237-z. [DOI] [PubMed] [Google Scholar]
- 87.Dantham VR, Holler S, Barbre C, Keng D, Kolchenko V, Arnold S. Label-Free Detection of Single Protein Using a Nanoplasmonic-Photonic Hybrid Microcavity. Nano Lett. 2013;13:3347–3351. doi: 10.1021/nl401633y. [DOI] [PubMed] [Google Scholar]
- 88.Kim DC, Dunn RC. Integrating Whispering Gallery Mode Refractive Index Sensing with Capillary Electrophoresis Separations Using Phase Sensitive Detection. Anal Chem. 2016;88:1426–1433. doi: 10.1021/acs.analchem.5b04187. [DOI] [PubMed] [Google Scholar]
- 89.Wade JH, Bailey RC. Refractive Index-Based Detection of Gradient Elution Liquid Chromatography using Chip-Integrated Microring Resonator Arrays. Analytical Chemistry. 2014;86:913–919. doi: 10.1021/ac4035828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu HY, White IM, Suter JD, Zourob M, Fan XD. Integrated refractive index optical ring resonator detector for capillary electrophoresis. Anal Chem. 2007;79:930–937. doi: 10.1021/ac061279q. [DOI] [PubMed] [Google Scholar]
- 91.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Quantitative, Label-Free Detection of Five Protein Biomarkers Using Multiplexed Arrays of Silicon Photonic Microring Resonators. Anal Chem. 2010;82:69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu HY, Dale PS, Caldwell CW, Fan XD. Rapid and Label-Free Detection of Breast Cancer Biomarker CA15-3 in Clinical Human Serum Samples with Optofluidic Ring Resonator Sensors. Anal Chem. 2009;81:9858–9865. doi: 10.1021/ac902437g. [DOI] [PubMed] [Google Scholar]
- 93.Sheehan PE, Whitman LJ. Detection limits for nanoscale biosensors. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 94.Squires TM, Messinger RJ, Manalis SR. Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 95.Dahlin AB. Size Matters: Problems and Advantages Associated with Highly Miniaturized Sensors. Sensors-Basel. 2012;12:3018–3036. doi: 10.3390/s120303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edwards DA. Transport effects on surface reaction arrays: Biosensor applications. Math Biosci. 2011;230:12–22. doi: 10.1016/j.mbs.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 97.Huckabay HA, Wildgen SM, Dunn RC. Label-free detection of ovarian cancer biomarkers using whispering gallery mode imaging. Biosens Bioelectron. 2013;45:223–229. doi: 10.1016/j.bios.2013.01.072. [DOI] [PubMed] [Google Scholar]