Abstract
Heat treatment is a widely used process to reduce bacterial loads in the food industry or to decontaminate surfaces, e.g., in hospital settings. However, there are situations where lower temperatures must be employed, for instance in case of food production such as raw milk cheese or for decontamination of medical devices such as thermo-labile flexible endoscopes. A recently identified locus of heat resistance (LHR) has been shown to be present in and confer heat resistance to a variety of Enterobacteriaceae, including Escherichia coli isolates from food production settings and clinical ESBL-producing E. coli isolates. Here, we describe the presence of two distinct LHR variants within a particularly heat resistant E. coli raw milk cheese isolate. We demonstrate for the first time in this species the presence of one of these LHRs on a plasmid, designated pFAM21805, also encoding type 3 fimbriae and three bacteriocins and corresponding self-immunity proteins. The plasmid was highly transferable to other E. coli strains, including Shiga-toxin-producing strains, and conferred LHR-dependent heat resistance as well as type 3 fimbriae-dependent biofilm formation capabilities. Selection for and acquisition of this “survival” plasmid by pathogenic organisms, e.g., in food production environments, may pose great concern and emphasizes the need to screen for the presence of LHR genes in isolates.
Keywords: heat resistance, E. coli, transfer of heat resistance, food production, biofilms, clpK
Introduction
Heat-treatment is an efficient and widely used measure to reduce bacterial contamination. If the material to be decontaminated is heat-stable, autoclaving can be used for complete sterilization. However, there are often situations where this is not feasible, both in food production and in the medical field, and lower temperatures must be employed. This is the case for heat treatment of flexible endoscopes (where thermochemical treatment below 60°C is being used; Jørgensen et al., 2016) or in case of thermization of raw milk for the production of specific cheeses (57–68°C for 15 s or more; Peng et al., 2013a), where protection of certain enzymes is aimed for. These treatments are generally sufficient to reduce the vegetative bacterial load to safe levels, unless the contaminating bacteria are especially heat-resistant.
Exposure of bacteria to severe heat stress results in massive misfolding and aggregation of proteins, and the potential toxic effects of these aggregates, coupled with net loss of active proteins, may cause cell death. Bacteria have evolved elaborate strategies to counteract these effects. Cytosolic chaperone systems with protein folding capacities such as DnaKJE and GroESL, as well as ATP-dependent proteolytic systems such as the Clp ATPases are ubiquitous in bacterial species (Hecker et al., 1996; Frees et al., 2014; Li and Gänzle, 2016). However, we recently identified a novel Clp ATpase, ClpK, uniquely found in Gram-negative bacteria, which also confers heat resistance (Bojer et al., 2010).
The clpK gene is located within a cluster termed the locus of heat resistance (LHR) comprising up to 16 open reading frames (ORFs) (Mercer et al., 2015). Located immediately upstream of clpK is a gene encoding a small heat shock protein, sHsp20c, which—like ClpK—has been shown to contribute to heat resistance (Lee et al., 2015). The remaining ORFs remain largely uncharacterized, but some are predicted to possess functional properties such as proteases or sodium/hydrogen transporters, implying that overall, the LHR may play a more generalized role in response to external stress (Mercer et al., 2015).
The LHR was originally described in Klebsiella pneumoniae and was found in roughly 1/3 of nosocomial K. pneumoniae isolates. The high prevalence of LHR in K. pneumoniae is likely due to its plasmid-located nature in this organism (Bojer et al., 2010). Notably, LHR-encoding plasmids often also carry multidrug-resistance genes and are transferable by conjugation to other K. pneumoniae isolates (Bojer et al., 2010, 2012). The LHR was since discovered in a Cronobacter sakazakii isolate, a pathogen associated with serious infections in neonates, which are thought to be linked to contaminated dried infant milk formula (Gajdosova et al., 2011).
Quite recently, a comparative genomic analysis of 29 E. coli strains identified a putatively chromosomally located ~14 kb region with >99% identity to the LHR clusters in K. pneumoniae and C. sakazakii. In contrast to the K. pneumoniae population examined, the LHR only occurred at a frequency of ~2% among the E. coli whole genomes and genome shotgun sequences published at that time (Mercer et al., 2015). We observed a similar frequency of LHR among extended-spectrum β-lactamase (ESBL)-producing E. coli isolates collected from Danish patients in 2008–2009 (Boll et al., 2016). This could suggest that the LHR does not provide significant general benefits to E. coli, or that transfer of LHR only rarely occurs in this organism. On the other hand, previous studies have investigated E. coli strains isolated from raw milk cheeses in Switzerland and shown that many of them exhibit increased heat resistance in milk at subpasteurization temperatures and during cheese ripening (Peng et al., 2012, 2013a,b). We recently screened a total of 256 of these E. coli raw milk cheese isolates for heat resistance markers clpK and an additional LHR marker gene, demonstrating that 93 (36.3%) of them were positive for both (while 24 and 9 isolates, respectively, were positive for one marker). We hypothesize that these increased numbers reflect a thermal selection pressure in this environment (Marti et al., 2016), due to the mild heat treatment employed during processing.
In this study, we focus on a raw milk cheese isolate, FAM21805, exhibiting a significantly increased level of heat resistance. We show that FAM21805 harbors two LHR variants, both of which confer heat resistance and both of which are transferrable by means of horizontal gene transfer. Moreover, one of the LHRs is located on a conjugative plasmid belonging to the IncFII group (pFAM21805) also harboring mrkABCDF, a locus encoding type 3 fimbriae (Burmølle et al., 2008). When transferred to E. coli K-12 MG1655, the plasmid increases both the heat resistance and biofilm formation properties of that strain. Finally, the plasmid is also transferable to strains of pathogenic E. coli. To the best of the authors' knowledge, this is the first description of a plasmid-borne LHR in E. coli.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table 1. Bacteria were routinely cultured at 37°C on Luria-Bertani (LB) agar and in LB broth.
Table 1.
E. coli strains used in this study.
| Strain | Misc.a | Origin | References |
|---|---|---|---|
| FAM21805 | Heat resistant; harbors LHR1 and LHR2 | Swiss raw milk cheese | Peng et al., 2012, 2013a; Marti et al., 2016 |
| FAM21805 ΔLHR1 | Knock-out mutant, LHR1 (orf2-orf16) replaced with kanr | This study | |
| FAM21805 ΔLHR2 | Knock-out mutant, LHR2 (orf2-orfF) replaced with kanr | This study | |
| FAM21805 ΔLHR1&2 | Knock-out mutant, LHR1&2 replaced with kanr | This study | |
| FAM21805 Δmrk | Knock-out mutant, mrkABCDF replaced with tetr | This study | |
| FAM21805 ΔorfE | Knock-out mutant, orfE replaced with tetr | This study | |
| FAM21805 LHR1 kanr | LHR1 tagged with kanr without knock-out of known ORFs; donor of LHR1 in HGT experiments | This study | |
| FAM21805 LHR2 tetr | LHR2 tagged with tetr without knock-out of known ORFs; donor of LHR2 in HGT experiments | This study | |
| C604-10 | Heat resistant, ESBL-producing | Human clinical isolate | Olesen et al., 2013; Boll et al., 2016 |
| C604-10 LHR1 kanr | LHR1 tagged with kanr without knock-out of known ORFs; donor of LHR1 in HGT experiments | Boll et al., 2016 | |
| C604-10 LHR2 tetr | LHR2 tagged with tetr without knock-out of known ORFs; donor of LHR2 in HGT experiments | Boll et al., 2016 | |
| MG1655 nalr rifr | Recipient in HGT experiments, laboratory strain | Commensal K-12 strain | Møller et al., 2003 |
| MG1655 LHR2 (21805) | MG1655 transconjugant harboring LHR2 plasmid of FAM21805 tagged with tetr without knock-out of known ORFs | This study | |
| MG1655 LHR1&2 (21805) | MG1655 transconjugant harboring LHR1&2 of FAM21805; LHR1 tagged with kanr without knock-out of known ORFs | This study | |
| MG1655 LHR2 ΔorfE (21805) | MG1655 transconjugant harboring LHR2 plasmid of FAM21805 tagged by replacing a putative di-guanyl cyclase (orfE) with tetr | This study | |
| MG1655 LHR2 Δmrk (21805) | MG1655 transconjugant harboring LHR2 plasmid of FAM21805 tagged by replacing mrkABCDF with tetr | This study | |
| MG1655 LHR1 (C604-10) | MG1655 transconjugant harboring LHR1 of C604-10; tagged with kanr without knock-out of known ORFs | This study | |
| MG1655 LHR2 (C604-10) | MG1655 transconjugant harboring LHR2 of C604-10 tagged with tetr without knock-out of known ORFs | This study | |
| FAM21843 | Heat resistant; harbors LHR1 | Swiss raw milk cheese | Peng et al., 2012, 2013a; Marti et al., 2016 |
| FAM22873 | eaeA (intimin) +, stx1+, serotype O26+ | Swiss raw milk | This study |
| FAM22873 LHR2 (21805) | FAM22873 transconjugant harboring LHR2 plasmid of FAM21805 tagged with tetr without knock-out of known ORFs | This study | |
| FAM22873 ΔLHR2 (21805) | FAM22873 transconjugant harboring LHR2 plasmid of FAM21805 tagged by replacement of LHR with kanr | This study | |
| FAM23288 | stx2+ | Swiss raw milk | This study |
| FAM23288 LHR2 (21805) | FAM23288 transconjugant harboring LHR2 plasmid of FAM21805 tagged with tetr without knock-out of known ORFs | This study | |
| FAM23288 ΔLHR2 (21805) | FAM23288 transconjugant harboring LHR2 plasmid of FAM21805 tagged by replacement of LHR with kanr | This study | |
| 55989 | Enteroaggregative E. coli (EAEC) strain | Human clinical isolate | Bernier et al., 2002 |
| 55989 LHR2 (21805) | 55989 transconjugant harboring LHR2 plasmid of FAM21805 tagged with tetr without knock-out of known ORFs | This study | |
| 55989 ΔLHR2 (21805) | 55989 transconjugant harboring LHR2 plasmid of FAM21805 tagged by replacement of LHR with kanr | This study | |
| C227-11 φcu | Shiga toxin-producing EAEC strain cured of the Stx2a-bearing phage | Human clinical isolate | Zangari et al., 2013 |
| C227-11 φcu LHR2 (21805) | C227-11 φcu transconjugant harboring LHR2 plasmid of FAM21805 tagged with tetr without knock-out of known ORFs | This study | |
| C227-11 φcu ΔLHR2 (21805) | C227-11 φcu transconjugant harboring LHR2 plasmid of FAM21805 tagged by replacement of LHR with kanr | This study. |
kanr, kanamycin resistance cassette; tetr, tetracycline resistance cassette; nalr, nalidixic acid resistant; rifr, rifampicin resistant.
Screening PCRs used in this study
A total of 90 E. coli dairy isolates were screened for LHR marker genes clpK1 and clpK2, and the mrkABCDF locus encoding type 3 fimbriae using the primers and annealing temperatures indicated in Table 2.
Table 2.
Primers and annealing temperatures used in this study.
| Name | Sequence (5′-3′) | Target | Use | Tann. (°C) | References |
|---|---|---|---|---|---|
| K12-R | ATCCTGCGCACCAATCAACAA | orf264 and the IS element inserted only in K-12 strains e.g., MG1655 | Confirmation of transconjugant identity | 54 | Kuhnert et al., 1995 |
| K12IS-L | CGCGATGGAAGATGCTCTGTA | ||||
| clpK1_F | TGCTGTTGTGCGACGACCATTACC | clpK1 gene specific PCR | Screening of strains for presence of clpK1 | 64 | Boll et al., 2016 |
| clpK1_R | TTGCCGACCACCTTGCTGACCTGT | ||||
| clpK2_F | ACGATCACTATCGCCAACTG | clpK2 gene specific PCR | Screening of strains for presence of clpK2 | 64 | Boll et al., 2016 |
| clpK2_R | AGTATTTATCCAGCTCGGGCGTG | ||||
| orfE_F | CGGTCGTTCTGGCAAAGGTG | putative di-guanyl-cyclase (dgc) of FAM21805 LHR2 | PCR specific for orfE, a putative dgc, part of LHR2 plasmid | 64 | This study |
| orfE_R | CGTCCTGACGAAATCGCTCC | ||||
| mrkD_F | TCGAAGGGTCGCGCTTTACG | mrkD of the mrkABCDF locus | Screening of strains for presence mrk locus encoding type 3 fimbriae | 60 | This study |
| mrkF_R | CATGGTAGCGGTAGTGCTGGTGG |
Phenotypic heat resistance screen
E. coli dairy isolates were screened for phenotypic heat resistance by incubation of overnight cultures LB Lennox broth (LB, 10 g/L tryptone, 5 g/L yeast extract, 5 g/L sodium chloride, pH 7.0) at 55°C for 30 min. Strains were diluted 1:10 into pre-heated LB broth and sampled at 0, 15, and 30 min. (duplicate plating). Strains showing a reduction in colony forming units (CFU) of less than one log10 after 30 min. were considered phenotypically heat resistant. When comparing wildtype strains with their LHR mutants or when testing LHR transconjugant strains, a fourth time point at 45 min incubation was added and the assay was performed at least in biological triplicate. Relative survival of a strain at a given time point was calculated by dividing the CFU/ml of that time point by the initial CFU/ml (time point 0).
Genome sequencing
FAM21805 and FAM21843 genomic DNA was extracted with the GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, Buchs, Switzerland). Illumina sequencing was done in a 101-bp paired-end run (University of Bern), and de novo assembly was performed using CLCbio Genomic Workbench (v9.0.1). For all other strains, genomic DNA was extracted from isolates using a DNeasy Blood and Tissue Kit (QIAGEN, Copenhagen, Denmark). MiSeq libraries were made using the NexteraTMKit (Illumina) and sequencing was performed as 250-bp paired-end runs. Reads were assembled de novo using CLCbio Genomic Workbench (v9.5.2). The annotated sequence of LHR1FAM21805 has been deposited at GenBank under the accession KY646173. The Whole Genome Shotgun projects of FAM21805 and FAM21843 have been deposited at DDBJ/ENA/GenBank under the accessions MVEA00000000 and MVIN00000000, respectively.
Plasmid sequencing
Plasmid DNA was sequenced both on a MiSeq instrument (Illumina) and on a MinION flow cell (Oxford Nanopore Technologies). The MiSeq library was made using the Nextera XT kit (Illumina) and sequencing was performed as a paired-end 250 bp run yielding 372,720 reads with an average length of 237 bp. MinION library with native barcode (NB01 from EXP-NDB002) was prepared using the R9 Genomic Sequencing kit (SQK-NSK007) and was sequenced on a FLO-MIN105 SpotON Mk1 flow cell according to the manufacturer's instructions. Fast5 read files were subjected to base calling via a two-direction (2D) workflow using ONT's Metrichor software yielding 12,327 passed read files. Mixed assembly was performed by combining MiSeq and MinION reads using the SPAdes (v3.9.0) assembler. Finally, CLCbio Genomic Workbench (v9.5.2) was used for end trimming of the assembled plasmid and for final error correction by mapping trimmed MiSeq reads against the plasmid contig obtained after the mixed SPAdes assembly. ORFs were predicted by RAST annotation (Aziz et al., 2008) and then manually curated. The sequence has been deposited at GenBank under the accession KY416992.
Phylogenetic analysis
For phylogenetic analysis of the LHR, genomes containing homologous sequences with >80% coverage of LHR2 of raw milk cheese E. coli isolate FAM21805 were retrieved from NCBI. In addition, sequences from clinical ESBL-producing E. coli isolates C598-10 and C604-10 (Boll et al., 2016), beef E. coli isolates AW1.3, AW1.7, and GM16.6-6 (Mercer et al., 2015), raw milk cheese E. coli isolate FAM21843 (Peng et al., 2012, 2013a) as well as five clinical isolates from Statens Serum Institut were included. Single nucleotide polymorphisms (SNPs) were identified after alignment of all sequences to the LHR1FAM21805 reference using the NUCmer component (Kurtz et al., 2004) as implemented in the Northern Arizona SNP Pipeline (NASP) v1.0 (http://biorxiv.org/content/early/2016/01/25/037267). A total of 1,270 SNPs were identified from 45% of the 15 kb LHR1FAM21805 excluding any repetitive regions. The relatedness of the elements was inferred using the maximum-likelihood algorithm implemented in PhyML (http://www.atgc-montpellier.fr/phyml-sms/) with Smart Model Selection using the Bayesian Information Criterion with 100 bootstrap replicates using random starting trees.
Modification of bacterial strains
LHR1 in FAM21805 was deleted by allelic exchange with a kanamycin resistance (kanr) encoding cassette, flanked by regions homologous to sequences at the beginning and end of the loci, as previously described (Bojer et al., 2010). The kanr cassette was then removed from FAM21805ΔLHR1 using pCP20. The LHR2 locus was deleted in FAM21805 and FAM21805ΔLHR1 by allelic exchange with kanr flanked by regions homologous to sequences at the beginning and end of LHR2. Using the same technique, the mrkABCDF cluster and orfE (encoding a putative digyanylate cyclase within LHR2) in FAM21805 were deleted by allelic exchange with a tetracycline resistance (tetr)-encoding cassette. Finally, kanr or tetr were introduced immediately downstream of the last ORFs of LHR1 and LHR2 without disrupting the flanking mobile elements (Table 1).
Horizontal gene transfer experiments
Horizontal gene transfer of loci of heat resistance was assessed in plate matings as previously described (Marti et al., 2016). In short, 500 μl overnight culture of donor and recipient strains were mixed and centrifuged (12,000 × g, 2 min), the supernatant completely removed and resuspended in 50 μl NA (8 g/L NaCl, 1 g/L peptone) solution and spotted onto LB Agar plates. The rifampicin (RIF) and nalidixic acid (NAL) resistant E. coli K-12 MG1655 rifr nalr (Møller et al., 2003) was used as recipient. The donors were various heat resistant strains with LHRs tagged with either tetr or kanr cassettes (Table 1). After incubation at 37°C for 24 h (if not stated otherwise), the cells were scraped off, suspended in 3 ml NA and plated onto LB plates selective for donors (LBTET or LBKAN), recipients (LBNAL/RIF), and transconjugants (LBNAL/RIF/TET or LBKAN/NAL/RIF). After overnight incubation at 37°C, CFU were counted and transconjugant frequencies per recipient calculated. The antibiotics were used in the following concentrations: Kanamycin: 50 μg/ml, nalidixic acid: 30 μg/ml, tetracycline: 15 μg/ml, and rifampicin: 100 μg/ml (Sigma-Aldrich, Buchs, Switzerland). The assay was carried out in biological triplicate except where frequencies were too low for quantitative evaluation. Three presumptive K-12 MG1655 transconjugants per replicate were confirmed by screening with MG1655-specific primers and various combinations of specific PCRs for clpK1, clpK2, orfE, or mrkD (Table 2).
Biolog phenotype MicroArrays
Stress responses of E. coli FAM21805 wild-type and its LHR mutants, as well as E. coli K-12 transconjugants, were assessed using the Phenotype MicroArrays (PM) 9 and 10 of the Biolog system screening for growth depending on osmolytes and pH (BIOLOG, Inc., Hayward, CA, USA). Strains were prepared and plates inoculated as follows: Strains from glycerol stocks were re-activated by two overnight incubations at 37°C on blood agar plates (Columbia agar + 5% sheep blood, bioMérieux, Geneva, Switzerland). The second plate was less than 24 h old when used for inoculation of the PM plates. Inoculating fluids (IF) were prepared by addition of 25 ml sterile water to 125 ml IF-0a and addition of 1.5 ml dye mix A and 23.5 ml water to 125 ml IF-10b. Cells were inoculated into IF-0 to a transmittance of 42 ± 2% using a sterile cotton swab and diluted (1:5) into fresh IF-0 resulting in a final transmittance of 85 ± 2%. This suspension was diluted 1:200 into IF-10b + dye mix A and used to inoculate PM plates (100 μl per well). Metabolic activity was assessed at 37°C for 72 h under standard atmosphere. PM plates, IF-0 and IF-10b were obtained from Endotell AG, Allschwil, Switzerland.
Hydrogen peroxide growth challenge
In a first step, overnight cultures were diluted 1:100 into fresh tryptic soy broth (TSB, Oxoid, Pratteln, Switzerland). This was followed by a second 1:10 dilution into pre-warmed TSB containing hydrogen peroxide (H2O2) resulting in 1 ml aliquots with final concentrations of 0, 0.5, 1, 2, 5, 7.5, 10, 12.5, and 15 mM H2O2. For each concentration, a quadruplicate of 200 μl per well was added to a 96-well plate and incubated at 37°C for 24 h in a microplate reader (model: ELx808, BioTek, Luzern, Switzerland). Optical density at λ = 600 nm (OD600) was measured in intervals of 30 min. The plates were only shaken for 5 s prior to each measurement on the fast setting. The experiment was carried out in biological duplicate.
Hydrogen peroxide inactivation assay
Overnight cultures were diluted 1:10 into 0.9% NaCl solution pre-warmed to 37°C with a final H2O2 concentration of 50 mM and statically incubated at 37°C. Following 0, 15, and 30 min of incubation, 50 μl samples were taken (mixed by pipetting), and immediately diluted 1:10 into 450 μl 0.9% NaCl solution. This immediately reduced the H2O2 concentration and stopped further reduction of CFU. Once all samples were taken, further dilutions were spotted in triplicate on TSA (Oxoid, Pratteln, Switzerland) and incubated overnight at 37°C.
Crystal violet (CV) biofilm formation assay
Overnight cultures of strains grown in LB broth were diluted 1:100 in minimal media with 0.5% casamino acids as carbon source (ABTCAA, Reisner et al., 2006) and 150 μl were added per well (eight wells per strain and biological replicate) in 96-well plates (untreated PS surface, CytoOne, StarLab, Hamburg, Germany). Plates were wrapped in plastic bags and partially closed to reduce evaporation of media and incubated at 12, 28, and 37°C for 48 h. After incubation, the plates were emptied by throwing out and removing residual liquid by touching the inverted plate on paper tissue. Plates were then washed three times with 200 μl NA per well and the biofilms subsequently stained with 200 μl 0.1% crystal violet (CV) solution (Sigma-Aldrich, Buchs, Switzerland) per well for 20 min. Staining was followed by three washes with ddH2O and biofilms were dissolved in 200 μl 96% ethanol per well. Biofilm formation was assessed by measurement of OD600 values, which are reported as average and standard deviation of three biological replicates of OD600 of strains minus OD600 of the media control.
Plasmid profiling
For plasmid size determination, plasmid preparation was carried out using a modified version of the protocol by Kado and Liu (1981) as previously described (Schjørring et al., 2008). As a plasmid marker, E. coli strain 39R861 was used, containing four plasmids of 147, 63, 36, and 7 kb (Threlfall et al., 1986).
Statistical analysis
Statistical analysis of data (t-tests, Mann-Whitney rank sum tests and 1-way ANOVA) was performed using SigmaPlot 13.0 (Systat Software, San Jose, CA) as indicated in the main text and legends (α = 0.05).
Results
Two distinct heat resistance clusters are present in E. coli raw milk cheese isolates
We have recently described the presence of two loci of heat resistance (LHR1 and LHR2) in a clinical ESBL-producing E. coli isolate (C604-10), both of which contributed to a highly heat resistant phenotype (Boll et al., 2016). In a separate study, we reported a remarkably high frequency of clpK-positive E. coli raw milk or raw milk cheese isolates (93 out of 256) (Marti et al., 2016). Here, we examined a subset of these isolates (both clpK-positive and—negative) for the potential presence of both LHRs with PCRs specific for clpK1 and clpK2, the marker genes of LHR1 and LHR2, respectively. Of the 90 tested isolates, 23 contained both, 26 were positive for clpK1 only and one only for clpK2. Next, we correlated PCR results and phenotypical heat resistance to determine the predictive value of the PCRs. All of the 49 clpK1 positive strains were heat resistant and only one out of 50 heat resistant strains was clpK1 negative. Thus, there was a strong correlation of heat resistance with clpK1. Every strain positive for clpK2 (Stahlhut et al., 2013) tested phenotypically heat resistant, but only 24 of 50 heat resistant strains were clpK2 positive. It is important to note that the clpK1 negative, yet heat resistant strain was clpK2 positive. Thus, the combination of these two PCRs resulted in perfect prediction of phenotypic heat resistance in this set of strains (double PCR negative strains all tested heat sensitive).
Strains testing positive for both clpK1 and clpK2 by PCR showed significantly increased survival in our phenotypic heat resistance screening assay compared to the clpK1 single positive strains, which in turn were significantly more resistant than double negative strains. The average relative survival after 30 min at 55°C was 4.64 ± 6.61 × 10−3 for double negatives (n = 40), 3.27 ± 1.75 × 10−1 for clpK1 single positives (n = 26), and 5.39 ± 1.96 × 10−1 for clpK1 and 2 double positive strains (n = 23). The differences between groups are statistically significant (p-values < 0.001 for all pairwise comparisons, Mann-Whitney Rank Sum Test). For further characterization of the two different LHR clusters in our collection of raw milk and raw milk cheese isolates, we focused our attention on isolate FAM21805, which harbors both clpK gene variants. This strain has previously been shown to exhibit an increased degree of heat resistance compared to strains harboring a single LHR in milk at sub-pasteurization temperatures (Peng et al., 2013a).
Characterization of a plasmid-encoded LHR in raw milk cheese isolate FAM21805
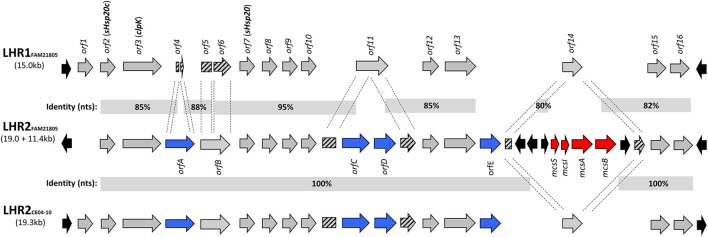
Whole genome sequencing (Illumina) analysis revealed that raw milk cheese isolate FAM21805 harbored an LHR (here designated LHR1) ~15 kb in size with a G+C content of 62% and highly similar (98–99%) to the one previously described in four heat resistant E. coli isolated from beef (Mercer et al., 2015). It contained fourteen putative ORFs and was flanked by mobile elements (Figure 1). We moreover detected additional homologs to several of these ORFs, strongly implying the presence of a second LHR in this isolate. However, de novo assembly failed to demonstrate the location of these homologs within a single genetic locus.
Figure 1.
Comparison of loci of heat resistance in FAM21805 (LHR1 and -2) and C604-10 (LHR2). ORFs present in all three LHRs are marked in gray (except for orf1 which is absent in LHR2FAM21805), ORFs unique to LHR2 are marked in blue, partially disrupted ORFs are marked with stripes, and mobile elements are marked in black. Regional nucleotide identities (%) are depicted. A 11.4 kb region containing the mcsSIAB cluster (Microcin S system, marked in red) has been inserted into LHR2FAM21805. For additional information on the ORFs, see Supplementary Table 1.
Since in K. pneumoniae, the LHR is thought to predominantly be located on plasmids, we sought to determine whether this was also the case for this putative additional LHR in E. coli isolate FAM21805. Gel electrophoresis of purified plasmids revealed that FAM21805 harbored two large plasmids of slightly different sizes around 110–120 kb (Supplementary Figure 1). We therefore introduced a kanamycin resistance encoding gene (kanr) within the clpK2 gene, purified the plasmids from the resulting FAM21805 ΔclpK2 strain and transformed the plasmids into laboratory E. coli strain NEB-10β. Plating on kanamycin-containing plates yielded several colonies, and plasmid profiling from one of these demonstrated the presence of a single plasmid in NEB-10β corresponding in size to the lower-size plasmid in isolate FAM21805 (Supplementary Figure 1), strongly suggesting that the kanr–disrupted clpK2 gene was in fact located on that plasmid.
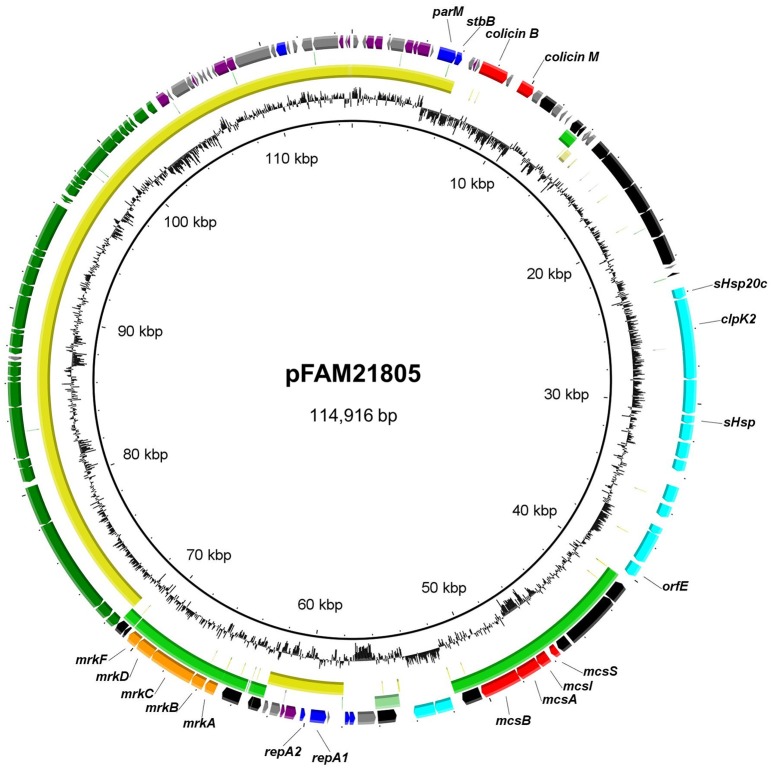
Using MinIon Nanopore R9 technology, we sequenced the kanr-tagged plasmid from the transformed NEB-10β strain. We then manually replaced the kanr–disrupted clpK2 gene with the intact clpK2 from Illumina sequencing within the complete closed sequence, thus reconstructing the original plasmid sequence. The resulting plasmid, titled pFAM21805, is 114,916 bp in length and has an average G+C content of 53.6% (Figure 2). RAST annotation predicted a total of 136 ORFs, 99 of which were functionally assigned. A single RepII replicon was identified with the F plasmid type FAB formula F96:A-:B-.
Figure 2.
Circular representation of pFAM21805 and BLASTn comparison of plasmids with shared regions. The outer ring shows predicted ORFs. Colors represent different putative functions: Gray, hypothetical proteins; blue, plasmid replication and maintenance; red, bacteriocins and accessory genes; light blue, LHR2 genes; orange, adhesins; dark blue, regulatory genes; purple, miscellaneous; and black, mobile elements. Within circles two (pCPO13026, yellow) and three (pSYM1, green), the darkest color indicates nucleotide identity exceeding 90%, whereas the lightest color represents identity exceeding 80%. Innermost circle, G+C content. The circular map was generated using BRIG.
The plasmid sequence confirmed the presence of a single LHR locus (LHR2FAM21805) ~19.0 kb in length and containing 15 putative functional ORFs (Figure 1 and Supplementary Table 1). It had a G+C content similar to LHR1 (61%) and was flanked by various mobile elements. Homologs to key elements of LHR1 were also present in LHR2FAM21805, including: (1) the Clp ATPase ClpK (orf3); (2) the two small heat shock proteins, sHsp20c (orf2) and sHsp20 (orf7); (3) a putative sodium/hydrogen exchanger with a Kef-type membrane component (orf13); (4) a putative zinc-dependent protease (orf15); and (5) a putative 2-alkenal reductase with a trypsin-like protease domain (orf16). Overall, 75% of LHR2FAM21805 was present in LHR1FAM21805 with a corresponding average identity of 88% (Figure 1).
The previously found LHR2 in ESBL-producing E. coli isolate C604-10 was highly identical to LHR2FAM21805 (Boll et al., 2016). However, whereas LHR2C604−10 (and LHR1FAM21805) was initiated by an ORF encoding a protein with a putative helix-turn-helix (HTH) motif capable of binding DNA (orf1), this ORF was absent in LHR2FAM21805. Moreover, a region spanning 11.4 kb with a G+C content of 50% had been inserted into orf14 of LHR2FAM21805 (Figures 1, 2). This region comprised the mcsSIAB gene cluster encoding an antibacterial peptide Microcin S (MccS) along with a self-immunity protein and transport apparatus recently identified on plasmid pSYM1 in probiotic E. coli strain G3/10 (Zschüttig et al., 2012).
LHR2 of FAM21805 and C604-10 both contained four ORFs not found in LHR1: (1) a putative cardiolipin synthase (orfA); (2) a putative mechanosensitive ion channel (orfC) followed by a hypothetical protein of unknown function (orfD); and (3) an ORF containing a GGDEF domain characteristic of a putative di-guanyl cyclase (orfE) (Figure 1 and Supplementary Table 1). Interestingly, remnants of the N- and C-terminal parts of orfA were present in LHR1, indicating evolutionary partial loss of this ORF in LHR1. In addition, orfC and orfD were inserted in the middle of orf11 encoding a hypothetical protein in LHR1, thereby disrupting this ORF. Finally, orfE appeared to have been inserted between orf13 and orf14 present in both LHRs (Figure 1).
In addition to LHR2 itself, pFAM21805 also contained the mrkABCDF cluster, encoding type 3 fimbriae, which mediate bacterial biofilm formation on abiotic surfaces such as plastic, and on biotic surfaces such as human epithelial cells. The mrk operon is present in nearly all K. pneumoniae isolates but is only rarely found in E. coli (Stahlhut et al., 2013). Notably, plasmid co-localization of the mrk operon and the mcs gene cluster was also observed in the probiotic G3/10 strain (Zschüttig et al., 2012). pFAM21805 also harbored a 32 kb tra region (24 tra genes, 8 trb genes, and finO), implying that the plasmid may be transferable by conjugation. Also present on the plasmid were genes associated with plasmid stability including parM and stbB as well as the genes encoding the antimicrobial compounds Colicin B and Colicin M (Figure 2). Thus, the plasmid likely confers both enhanced heat resistance, the ability to kill other E. coli and enhanced adhesive properties to FAM21805.
A large portion of pFAM21805, including the tra region, repA replicon and plasmid maintenance genes also comprised the backbone of plasmid pCPO13026 of Shiga Toxin-producing E. coli (STEC) strain 2009C-3133 isolated from a patient in New York in 2009 (Lindsey et al., 2015) (Figure 2).
Presence of loci of heat resistance in other pathogenic species
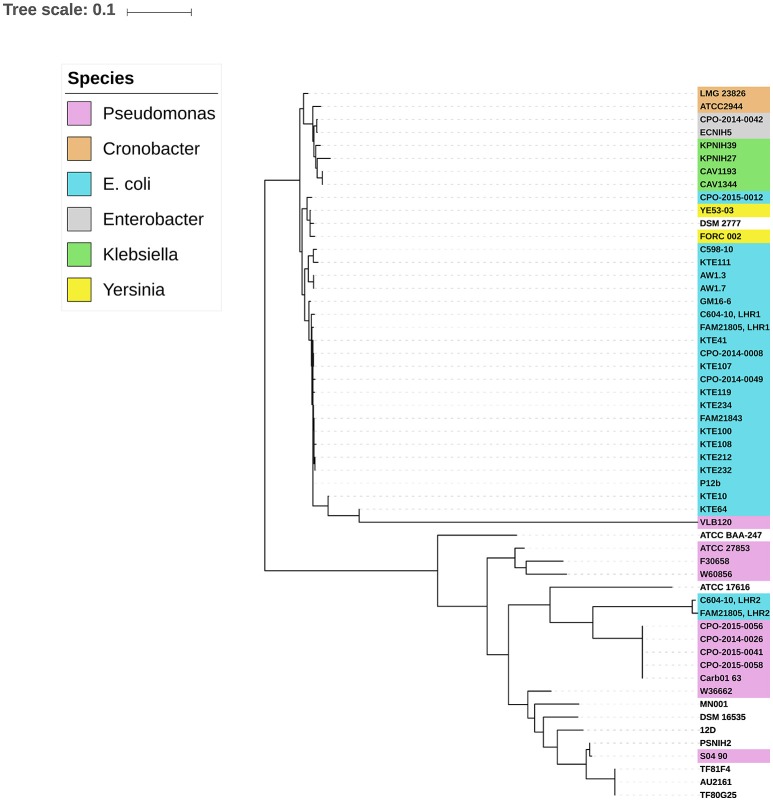
Previous studies have described the presence of LHR in several distinct pathogenic species, including E. coli, K. pneumoniae, Enterobacter species, and Pseudomonas aeruginosa (Bojer et al., 2010; Gajdosova et al., 2011; Lee et al., 2015; Mercer et al., 2015). Based on a BLAST search using the entire LHR, Mercer et al. recently observed two distinct phylogenetic groups harboring LHRs—one predominantly comprising Enterobacteriaceae and one primarily comprising P. aeruginosa (Mercer et al., 2015). We performed the same analysis using LHR2 from FAM21805 as input, which retrieved 27 sequences with more than 80% coverage. SNPs within the LHRs were identified and used to calculate a maximum-likelihood phylogenetic tree. As expected, LHR1FAM21805 and LHR1C604−10 both clustered tightly with the other E. coli LHR1s within the Enterobacteriaceae group (Figure 3). Not surprisingly, LHR2FAM21805 and LHR2C604−10 both clustered tightly together. Remarkably, however, they were located within the Pseudomonas group of the phylogenetic tree. These findings demonstrate the ability of E. coli to acquire LHRs from both phylogenetic groups.
Figure 3.
Midpoint-rooted maximum-likelihood phylogenetic tree of aligned LHR sequences (>80% coverage of LHR1FAM21805) from different bacterial species. Two distinct clusters representing predominantly Enterobacteriaceae and strains of Pseudomonas, respectively, are observed. LHR2 of FAM21805 and C604-10 cluster within the Pseudomonas group.
Both the LHR1 and LHR2 confer heat resistance and are transferable by conjugation
We have previously demonstrated that both LHRs in ESBL-producing E. coli isolate C604-10 confer heat resistance to this strain (Boll et al., 2016). To determine whether this was also the case with raw milk cheese E. coli isolate FAM21805, we generated a panel of LHR mutant strains with allelic replacements of designated regions with the kanr gene (Table 1). For FAM21805 ΔLHR1, we deleted the region spanning from orf2 (sHsp20c) to orf16. With regard to LHR2FAM21805, we deleted the region spanning from orf2 to the disrupted orf14, thereby excluding potential influence of removal of the inserted Microcin S system. As shown in Figure 4A, removal of either one of the LHRs did not affect survival of FAM21805 upon heat exposure. In contrast, survival of the FAM21805 double LHR mutant was severely reduced, thereby demonstrating that both LHRs of FAM21805 are functionally active and confer heat resistance.
Figure 4.
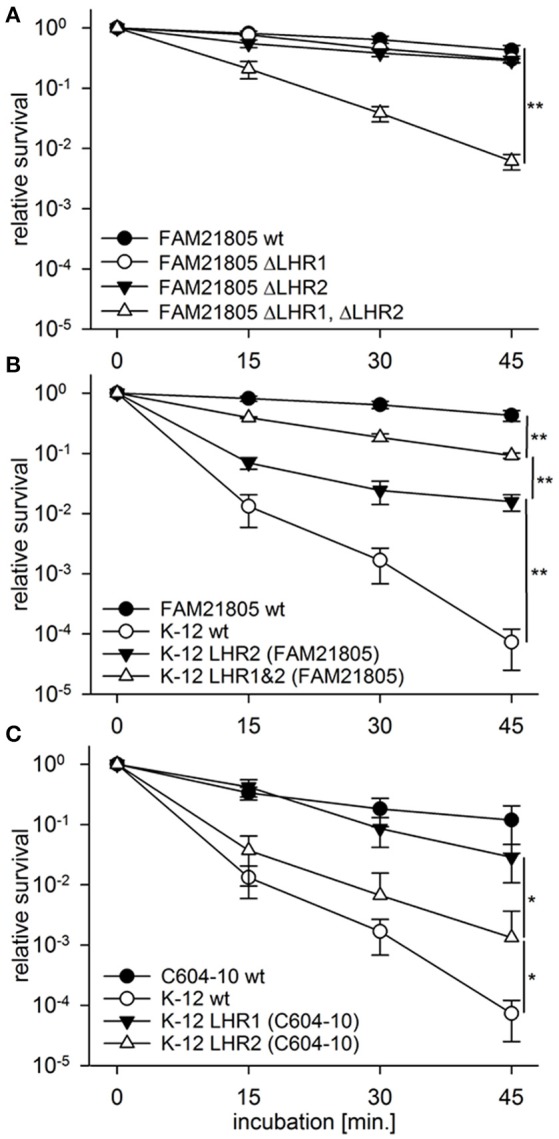
Heat resistance assays of E. coli wild-type strains, FAM21805 mutants, and K-12 MG1655 transconjugants. Relative survival compared to time point 0 of strains incubated at 55°C for 15, 30, and 45 min. (A) FAM21805 wild-type, -ΔLHR1, -ΔLHR2, and ΔLHR1ΔLHR2. (B) FAM21805 wild-type, K-12 MG1655 wild-type, and K-12 MG1655 transconjugants harboring LHR2 or both LHRs of FAM21805. (C) C604-10, K-12 MG1655 wild-type, and K-12 MG1655 transconjugants featuring LHR1 or LHR2 of C604-10. Error bars indicate standard deviations. Significances are indicated for the 45 min. time points based on one-tailed t-tests in (A,B), and Mann-Whitney rank sum tests in (C); *p < 0.05, **p < 0.01.
We next sought to determine whether the LHRs of FAM21805 and C604-10 were amenable to horizontal gene transfer. We tagged LHR1 in both FAM21805 and C604-10 as well as LHR2C604−10 (all presumably chromosomally located) with genes encoding kanr or tetr by introducing them in the region immediately downstream of orf16 (Table 1). Likewise, we inserted kanr in a non-coding region on pFAM21805. We then carried out conjugative transfer assays using either one of the four LHR-tagged strains as donors and commensal E. coli strain K-12 MG1655 as recipient. Plate mating assays resulted in very high transfer frequencies for the tagged donor LHR2FAM21805 (7.56 ± 2.00 × 10−1 transconjugants per recipient). The donor with a tagged LHR2C604−10 produced transconjugants at very low levels. Standard plate matings with tagged LHR1 of both FAM21805 and C604-10 yielded no transconjugants at all. We proceeded to increase incubation time, spotting separate plate matings for each time point and incubating up to 12 days. This way, LHR1 transconjugants of both strains were generated and confirmed by PCR, albeit once again at very low levels. Notably, all of the LHR1FAM21805 transconjugants also appeared to have acquired pFAM21805. In contrast, we did not observe co-transfer of LHRs not selected for in any other case.
Confirming retained post-transfer functionality of the LHRs, all four types of K-12 MG1655 transconjugants exhibited significantly elevated survival upon heat exposure compared to the native strain (Figures 4B,C). The highest level of survival was observed in MG1655 carrying both LHR1FAM21805 and LHR2FAM21805 (Figure 4B), illustrating that both loci contribute to heat resistance in this bacterial background. In contrast, LHR2C604−10 appeared to more modestly confer heat protection. Finally, given the relative ease by which pFAM21805 was transferable to MG1655, we examined whether the plasmid could also be transferred to isolates belonging to diarrheagenic E. coli (DEC) and Shiga-toxin (Stx)-encoding E. coli (STEC) pathotypes. Through plate mating assays, we successfully transferred pFAM21805 tagged with antibiotic resistance cassettes to two STEC strains, FAM22873, and FAM23288, as well as two enteroaggregative E. coli (EAEC) strains, 55989 and the Stx-phage-cured German outbreak strain C227-11 φcu. As shown in Table 3, pFAM21805 conferred significantly elevated heat resistance to all four pathogenic E. coli strains. Strains conjugated with pFAM21805 ΔLHR2 exhibited the same levels of heat killing as did the non-conjugated strains, confirming that this effect was attributable to LHR2 (Table 3).
Table 3.
Heat resistance assays with pathogenic E. coli (EAEC and STEC) wild-type strains and the strains conjugated with pFAM21805 or with pFAM21805 ΔLHR2.
| Strain | Relative survivala | p-valuec | |
|---|---|---|---|
| Average | SDb | ||
| FAM22873 wild-type | 5.64E-04 | 1.64E-04 | – |
| FAM22873 ΔLHR2 (21805) | 9.49E-04 | 1.08E-03 | 0.322 |
| FAM22873 LHR2 (21805) | 4.28E-02 | 2.24E-02 | 0.028 |
| FAM23288 wild-type | 9.03E-04 | 2.04E-04 | – |
| FAM23288 ΔLHR2 (21805) | 4.79E-04 | 5.38E-04 | 0.124 |
| FAM23288 LHR2 (21805) | 1.02E-02 | 8.12E-03 | 0.029 |
| 55989 wild-type | 3.60E-04 | 1.54E-04 | – |
| 55989 ΔLHR2 (21805) | 5.05E-04 | 1.98E-04 | – |
| 55989 LHR2 (21805) | 3.91E-01 | 1.23E-01 | 0.007 |
| C227-11 φcu | 3.23E-04 | 4.89E-04 | – |
| C227-11 φcuΔ LHR2 (21805) | 1.20E-04 | 1.08E-04 | – |
| C227-11 φcu LHR2 (21805) | 4.27E-01 | 1.39E-01 | 0.001 |
Relative survival at time point 45 min. compared to 0 min. incubation at 55°C.
SD, standard deviation.
p-value of direct comparison (one-tailed t-test or Mann-Whitney rank sum) against corresponding wild-type strain.
Screening for other locus of heat resistance-related stress response phenotypes
We next sought to determine whether the LHRs confer other stress-related advantages to their bacterial host in addition to enhanced thermotolerance. Since homologs of most of the ORFs are present in both of the LHR, we focused on the combined effect of removing both loci in FAM21805.
We first looked at the ability of FAM21805 wild-type and the ΔLHR1ΔLHR2 mutant to survive oxidative stress. However, neither killing nor growth challenge assays with hydrogen peroxide revealed any significant differences between the wild-type and mutant strain (data not shown). Thus, there seems to be no clear protective effect of either LHR against the oxidative action of H2O2.
To examine the potential effect of osmolarity and pH, we next screened the wild-type and ΔLHR1ΔLHR2 mutant strain using phenotypic microarrays (PM) 9 and 10 from the Biolog system, which measures activity of the bacterial metabolism via respiratory action (reduction of a redox dye). A biological triplicate with both PM 9 and 10 revealed only one consistent phenotypical difference between the two strains. PM9 contains four wells challenging bacteria with sodium benzoate at concentrations of 20, 50, 100, and 200 mM (each at pH 5.2). At a concentration of 100 mM, the wild-type strain consistently started respiring after ~36 h of incubation, while the LHR double mutant was unable to do so. Both strains were able to respire at 20 and 50 mM sodium benzoate while both were unable to do so at 200 mM over the entire 72 h incubation. FAM21805 ΔLHR1&2 complemented with either LHR2 or LHR1&2 were able to respire at 100 mM sodium benzoate (pH 5.2) after ~36 h, like the wild-type (single replicate). Notably, K-12 MG1655 transconjugants with LHR2 or LHR1&2 of FAM21805 did not show this sodium benzoate related phenotype (biological duplicate).
The pFAM21805 plasmid increases biofilm formation of E. coli MG1655
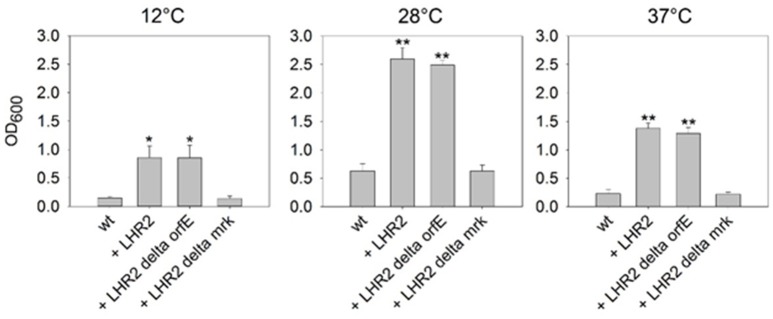
Biofilm formation is an important contributor to persistence of bacteria in both food processing and clinical settings (Abdallah et al., 2014). A combination of biofilm formation with heat resistance would increase a strain's persistence even further (Bojer et al., 2011). As described above, pFAM21805 contains the mrk gene cluster encoding type 3 fimbriae, which are strongly associated with bacterial biofilm formation (Burmølle et al., 2008; Hufnagel et al., 2015). Moreover, orfE of LHR2FAM21805 encodes a putative di-guanyl cyclase, and c-di-GMP signaling has been shown to affect biofilm formation (Burmølle et al., 2008; Schroll et al., 2010; Hufnagel et al., 2015). Thus, both factors could potentially contribute to increased biofilm formation in E. coli. To investigate if this was the case, we replaced either mrkABCDF or orfE in FAM21805 with tetr (Table 1), and transferred pFAM21805 from the two corresponding mutant strains to K-12 MG1655. We then performed crystal violet (CV) assays with K-12 MG1655 nalr rifr and its transconjugants in ABTCAA for 48 h at 12, 28, and 37°C. As shown in Figure 5, the presence of intact pFAM21805 significantly increased biofilm production of MG1655. Moreover, our results show that the mrk locus—but not the putative LHR2-encoded di-guanyl cyclase—was required to increase biofilm formation in K-12 MG1655 transconjugants. Thus, pFAM21805 has the potential to confer both enhanced survival during heat stress and adhesive properties at a wide range of temperatures to E. coli recipient strains.
Figure 5.
Impact of pFAM21805 on biofilm formation of K-12 MG1655. Biofilm formation of E. coli K-12 MG1655 wild-type and K-12 MG1655 transconjugants harboring pFAM21805, pFAM21805 ΔorfE (deletion of LHR2-encoded putative di-guanyl cyclase), or pFAM21805 Δmrk (deletion of type 3 fimbriae genes). Bacteria were grown in ABTCAA media for 48 h at 12, 28, and 37°C, after which biofilm formation was quantified by crystal violet (CV) staining. Bars represent averages and standard deviations (*p < 0.05, **p < 0.001, tested by 1-way ANOVA vs. K-12 wt control, α = 0.05, Holm-Sidak).
Discussion
Heat treatment is a commonly used process in the food industry and the main technology to reduce bacterial load. It is therefore crucially important to understand the mechanisms mediating increased heat resistance in potentially pathogenic bacteria as well as its origin, potential for distribution, and possible cross-protective effects associated with this phenotype.
We and others have recently described the LHR, which is present in and confers heat resistance to a variety of Enterobacteriaceae as well as Pseudomonas spp. (Bojer et al., 2010; Lee et al., 2015; Mercer et al., 2015; Boll et al., 2016). In contrast to K. pneumoniae, where the LHR appears to predominantly be located on plasmids, thus far, only chromosomally located LHRs have been reported in E. coli, including the fully sequenced strain P12b (Bojer et al., 2010; Liu et al., 2012; Mercer et al., 2015). This likely explains the much lower overall predicted prevalence of LHR in E. coli (~2%) compared to K. pneumoniae (1/3 of clinical isolates) (Bojer et al., 2010; Mercer et al., 2015). However, when screening for LHR marker genes in a collection of E. coli isolates from dairy production, we observed a much higher fraction of LHR-positive isolates (~36%). This most likely reflects a selection process in which LHR-positive isolates survive the thermal processing steps to a much greater extent than LHR-negative isolates (Marti et al., 2016). Further studies are needed to evaluate the occurrence of similar selection processes under sub-pasteurization heat treatments in clinical usage and food production.
In this study, we were able to more closely characterize a subset (90 isolates) of the collection of E. coli dairy isolates with regard to the presence of LHR. While distinct variants of LHRs exist in different species, the clpK gene (as well as the upstream located sHsp20c) are always present. We therefore used PCRs specifically targeting clpK1 and clpK2 as markers for E. coli LHR1 (Mercer et al., 2015) and the newly discovered variant LHR2, found in the ESBL-encoding E. coli isolate C604-10 (Boll et al., 2016), respectively. We observed a strong correlation between the presence of LHR and a heat resistant phenotype, with all of the heat resistant isolates harboring LHR1, with the exception of FAM22891, which tested clpK2 single positive. Nearly half of the LHR1-positive isolates additionally also harbored LHR2, and these isolates exhibited the highest levels of heat resistance overall. These findings stress the need for screening for both LHR1 and LHR2 to detect highly heat resistant isolates and validate the use of primers targeting the clpK genes as markers for the LHRs.
We also examined whether the LHRs would be amenable to horizontal gene transfer. Focusing on C604-10 and E. coli raw milk cheese isolate FAM21805, both of which harbor both LHR1 and LHR2, we indeed found that all four LHRs were transferrable to E. coli K-12; however, with very different rates of transfer. Interestingly, compared to the other three LHRs, LHR2FAM21805 was transferred at a much higher rate. MinIon-based sequencing revealed that, unlike the other LHRs, LHR2FAM21805 was encoded on a large IncFII-type plasmid, pFAM21805. This plasmid also contained a tra operon (Lawley et al., 2003), thus explaining why this plasmid was so readily transferred by conjugation. Importantly, pFAM21805 was also readily conjugated into STEC and EAEC, both of which are pathotypes associated with foodborne outbreaks (Rasko et al., 2011; Farrokh et al., 2013; Robertson et al., 2016). While thus far, no studies have described the natural presence of LHR in these diarrheal pathotypes, this finding highlights the potential for such cases to occur.
The fact that LHR1 was transferred at a much lower rate than the plasmid-encoded LHR2 from FAM21805 suggests that LHR1 is located either on the chromosome or on a non-conjugative plasmid in this strain. Gel electrophoresis of the LHR1 K-12 MG1655 transconjugant (also harboring pFAM21805) demonstrated the presence of a single plasmid identical in size to that of the pFAM21805 K-12 MG1655 transconjugant (data not shown). This rules out the location of LHR1 on the second large plasmid present in FAM21805. We instead aligned Illumina sequences from the LHR1 K-12 MG1655 transconjugant with the genome sequence of MG1655. Based on this, we identified 1,708 SNPs within a region spanning ~700 kb of the MG1655 genome. The same high SNP frequency occurred across the entire MG1655 genome following alignment with Illumina sequences from FAM21805. Taking together, these findings suggest that LHR1FAM21805 was transferred horizontally to MG1655 as part of a 700 kb region of FAM21805. While the underlying mechanism for this phenomenon remains to be determined, the LHR-containing DNA region could have been transferred to FAM21805 by conjugal mating with a high frequency recombinant (Hfr) donor strain with a conjugative plasmid integrated into its genome (O'Gorman et al., 1996). A similar scenario describing a chimeric K. pneumoniae strain having taken up a large portion of genomic DNA from another strain has previously been described (Struve et al., 2015). In any case, such a transfer mechanism would expectedly occur at a very low frequency, co-inciding with very low transfer frequency of LHR1FAM21805 in our study.
In addition to LHR2, pFAM21805 also contained the mrkABCDF locus encoding type 3 fimbriae, which are considered a major virulence factor of K. pneumoniae allowing the organism to produce extensive biofilm (Schroll et al., 2010; Andrade et al., 2014). In K-12 MG1655, conjugation with pFAM21805 lead to significantly increased biofilm production at 12, 28, and 37°C, which may in turn increase frequency of horizontal gene transfer (Burmølle et al., 2014; Rossi et al., 2014). The fact that the entire range of temperatures tested saw an increase in biofilm formation suggests potential beneficial effects for a host of this plasmid both in the environment and in vivo.
Moreover, the plasmid also contained three bacteriocins (along with their respective self-immunity genes): Colicin B, Colicin M, and Microcin S. Although the functionality of the bacteriocins remains to be verified, having the ability to produce multiple bacteriocins most likely provides the bacterial host of the plasmid with an expanded killing range, and thus a competitive advantage in multispecies communities (Gordon and O'Brien, 2006).
The pFAM21805 plasmid did not encode any antimicrobial resistance genes. This is in contrast to K. pneumoniae, where the LHR is often present on plasmids harboring resistances to tetracycline, neomycin, trimethoprim, sulfamethoxazole as well as encoding ESBL genes such as CTX-M-15, which confers resistance to third-generation cephalosporins (Bojer et al., 2012). Importantly, FAM21805 has been shown to be able to harbor ESBL-encoding conjugative plasmids and is able to act as donor of these (Marti et al., 2016), and the tra operon on pFAM21805 could likely enable conjugation of mobilizable (resistance) plasmids in other hosts of this LHR2 plasmid.
Interestingly, we observed a strong correlation between the presence of the mrk gene cluster and LHR in our collection of dairy E. coli isolates (all 13 mrk positive isolates also harbored clpK1 and some also clpK2). In light of this, we performed whole-genome sequencing on two mrk- and clpK2-positive isolates but found that neither isolate harbored pFAM21805 or a highly similar plasmid. Thus, in spite of the fact that the plasmid was readily transferred and provided clear benefits to its host, it did not appear to be widespread among these isolates. However, this does not rule out the possibility that other LHR-encoding plasmids exist, some of which may potentially harbor LHR1-like heat resistance clusters and mrk, similar to pFAM21805. Notably, thus far in E. coli, the mrk gene cluster has only been reported as located on conjugative plasmids (Burmølle et al., 2008), which is in agreement with our finding of mrk on pFAM21805. Further studies are needed to clarify the role of the mrk gene cluster and biofilm production capacity in E. coli isolates being persistent in hospitals or food-production.
The exact function of many of the ORFs of the LHRs remains to be unraveled. However, the fact that the majority of them are highly conserved strongly suggests that they all play a beneficial role for their host. Notably, some of them are predicted to act as ion-exchangers or proteases/peptidases suggesting that they may be involved in handling osmotic- or heat stress (Mercer et al., 2015). The ClpK chaperone itself shares many structural properties with ClpB, which is known to play a critical role in survival following various types of stress (Squires et al., 1991; Ekaza et al., 2001; Lourdault et al., 2011). We therefore endeavored to identify other stress response phenotypes associated with LHRs in FAM21805. However, both H2O2 growth challenge and killing assays revealed no significant differences between the FAM21805 wildtype and LHR double mutant strain in response to this stressor. Assays with the phenotypic microarrays 9 and 10 of the Biolog system did reveal one phenotype: The FAM21805 wildtype was able to respire in 100 mM sodium benzoate (pH 5.2) after 36 h, while its LHR1&2 double mutant could not. This phenotype could be complemented in FAM21805 ΔLHR1&2 with either LHR2 or LHR1&2, but not transferred to K-12 MG1655 by conjugation. The phenotype appears to be dependent on the genomic background around LHR2. Benzoate is being used as a preservative in a range of foods. Although a difference was only observed in one scenario here, it indicates that there could be non-thermal stresses where LHR would confer an advantage to the host isolate.
In conclusion, we have characterized in detail the presence of a recently discovered variant of LHR (LHR2) and demonstrated its presence on a plasmid in the highly heat resistant dairy E. coli isolate FAM21805. This plasmid was transferable at much higher rates than the presumably chromosomal LHRs tested, and conferred LHR2-dependent heat resistance as well as mrk-dependent biofilm formation capabilities to recipient E. coli, including pathogenic strains. In addition, the plasmid also harbored three bacteriocins and corresponding self-immunity proteins. Selection for and acquisition of this “survival” plasmid by pathogenic organisms, e.g., in food production environments, may pose great concern and emphasizes the need to screen for the presence of LHR genes in isolates.
Author contributions
EB and RM designed the work, collected, analyzed and interpreted the data and drafted the article. HH, SO, and MS analyzed and interpreted the data and critically revised the article. KN analyzed and interpreted the data. SK critically revised the article. KK and JH interpreted the data and critically revised the article. CS designed the work, interpreted the data and critically revised the article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Michala T. Sørensen (Statens Serum Institut) for carrying out plasmid profiling, Javorka Naskova (Agroscope) for technical assistance, and Daniel Wüthrich (University of Bern) for performing some of the Illumina sequencing. Ulrich Zürcher is acknowledged for the management of the Agroscope research program REDYMO. This work was financially supported by the Danish Council for Research grant DFF-1331-00161 to CS, BacFoodNet, the Agroscope research program REDYMO and in part by COST action FA1202.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00579/full#supplementary-material
References
- Abdallah M., Benoliel C., Drider D., Dhulster P., Chihib N. E. (2014). Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 196, 453–472. 10.1007/s00203-014-0983-1 [DOI] [PubMed] [Google Scholar]
- Andrade L. N., Vitali L., Gaspar G. G., Bellissimo-Rodrigues F., Martinez R., Darini A. L. (2014). Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J. Clin. Microbiol. 52, 2530–2535. 10.1128/JCM.00088-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier C., Gounon P., Le Bouguénec C. (2002). Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70, 4302–4311. 10.1128/IAI.70.8.4302-4311.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojer M. S., Hammerum A. M., Jørgensen S. L., Hansen F., Olsen S. S., Krogfelt K. A., et al. (2012). Concurrent emergence of multidrug resistance and heat resistance by CTX-M-15-encoding conjugative plasmids in Klebsiella pneumoniae. APMIS 120, 699–705. 10.1111/j.1600-0463.2012.02885.x [DOI] [PubMed] [Google Scholar]
- Bojer M. S., Krogfelt K. A., Struve C. (2011). The newly discovered ClpK protein strongly promotes survival of Klebsiella pneumoniae biofilm subjected to heat shock. J. Med. Microbiol. 60, 1559–1561. 10.1099/jmm.0.032698-0 [DOI] [PubMed] [Google Scholar]
- Bojer M. S., Struve C., Ingmer H., Hansen D. S., Krogfelt K. A. (2010). Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS ONE 5:e15467. 10.1371/journal.pone.0015467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll E. J., Frimodt-Møller J., Olesen B., Krogfelt K. A., Struve C. (2016). Heat resistance in extended-spectrum beta-lactamase-producing Escherichia coli may favor environmental survival in a hospital setting. Res. Microbiol. 167, 345–349. 10.1016/j.resmic.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Burmølle M., Bahl M. I., Jensen L. B., Sørensen S. J., Hansen L. H. (2008). Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154, 187–195. 10.1099/mic.0.2007/010454-0 [DOI] [PubMed] [Google Scholar]
- Burmølle M., Ren D., Bjarnsholt T., Sørensen S. J. (2014). Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 22, 84–91. 10.1016/j.tim.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Ekaza E., Teyssier J., Ouahrani-Bettache S., Liautard J. P., Köhler S. (2001). Characterization of Brucella suis clpB and clpAB mutants and participation of the genes in stress responses. J. Bacteriol. 183, 2677–2681. 10.1128/JB.183.8.2677-2681.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokh C., Jordan K., Auvray F., Glass K., Oppegaard H., Raynaud S., et al. (2013). Review of Shiga-toxin-producing Escherichia coli (STEC) and their significance in dairy production. Int. J. Food Microbiol. 162, 190–212. 10.1016/j.ijfoodmicro.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Frees D., Gerth U., Ingmer H. (2014). Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int. J. Med. Microbiol. 304, 142–149. 10.1016/j.ijmm.2013.11.009 [DOI] [PubMed] [Google Scholar]
- Gajdosova J., Benedikovicova K., Kamodyova N., Tothova L., Kaclikova E., Stuchlik S., et al. (2011). Analysis of the DNA region mediating increased thermotolerance at 58°C in Cronobacter sp. and other enterobacterial strains. Antonie Van Leeuwenhoek 100, 279–289. 10.1007/s10482-011-9585-y [DOI] [PubMed] [Google Scholar]
- Gordon D. M., O'Brien C. L. (2006). Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology 152, 3239–3244. 10.1099/mic.0.28690-0 [DOI] [PubMed] [Google Scholar]
- Hecker M., Schumann W., Volker U. (1996). Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19, 417–428. 10.1046/j.1365-2958.1996.396932.x [DOI] [PubMed] [Google Scholar]
- Hufnagel D. A., Depas W. H., Chapman M. R. (2015). The biology of the Escherichia coli extracellular matrix. Microbiol. Spectr. 3:MB-0014-2014. 10.1128/microbiolspec.mb-0014-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S. B., Bojer M. S., Boll E. J., Martin Y., Helmersen K., Skogstad M., et al. (2016). Heat-resistant, extended-spectrum β-lactamase-producing Klebsiella pneumoniae in endoscope-mediated outbreak. J. Hosp. Infect. 93, 57–62. 10.1016/j.jhin.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. (1981). Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert P., Nicolet J., Frey J. (1995). Rapid and accurate identification of Escherichia coli K-12 strains. Appl. Environ. Microbiol. 61, 4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., Antonescu C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Klimke W. A., Gubbins M. J., Frost L. S. (2003). F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15. 10.1016/S0378-1097(03)00430-0 [DOI] [PubMed] [Google Scholar]
- Lee C., Wigren E., Trcek J., Peters V., Kim J., Hasni M. S., et al. (2015). A novel protein quality control mechanism contributes to heat shock resistance of worldwide-distributed Pseudomonas aeruginosa clone C strains. Environ. Microbiol. 17, 4511–4526. 10.1111/1462-2920.12915 [DOI] [PubMed] [Google Scholar]
- Li H., Gänzle M. (2016). Some like it hot: heat resistance of Escherichia coli in food. Front. Microbiol. 7:1763. 10.3389/fmicb.2016.01763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey R. L., Knipe K., Rowe L., Garcia-Toledo L., Loparev V., Juieng P., et al. (2015). Complete genome sequences of two shiga toxin-producing Escherichia coli strains from serotypes O119:H4 and O165:H25. Genome Announc. 3:e01496-15. 10.1128/genomeA.01496-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Hu B., Zhou Z., Guo D., Guo X., Ding P., et al. (2012). A novel non-homologous recombination-mediated mechanism for Escherichia coli unilateral flagellar phase variation. Nucleic Acids Res. 40, 4530–4538. 10.1093/nar/gks040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourdault K., Cerqueira G. M., Wunder E. A., Jr., Picardeau M. (2011). Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 79, 3711–3717. 10.1128/IAI.05168-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti R., Muniesa M., Schmid M., Ahrens C. H., Naskova J., Hummerjohann J. (2016). Short communication: heat-resistant Escherichia coli as potential persistent reservoir of extended-spectrum beta-lactamases and Shiga toxin-encoding phages in dairy. J. Dairy Sci. 99, 8622–8632. 10.3168/jds.2016-11076 [DOI] [PubMed] [Google Scholar]
- Mercer R. G., Zheng J., Garcia-Hernandez R., Ruan L., Ganzle M. G., McMullen L. M. (2015). Genetic determinants of heat resistance in Escherichia coli. Front. Microbiol. 6:932. 10.3389/fmicb.2015.00932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller A. K., Leatham M. P., Conway T., Nuijten P. J., de Haan L. A., Krogfelt K. A., et al. (2003). An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71, 2142–2152. 10.1128/IAI.71.4.2142-2152.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman L. E., Krejany E. O., Bennett-Wood V. R., Robins-Browne R. M. (1996). Transfer of attaching and effacing from a strain of enteropathogenic Escherichia coli to E. coli K-12. Microbiol. Res. 151, 379–385. 10.1016/S0944-5013(96)80007-3 [DOI] [PubMed] [Google Scholar]
- Olesen B., Hansen D. S., Nilsson F., Frimodt-Moller J., Leihof R. F., Struve C., et al. (2013). Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J. Clin. Microbiol. 51, 1779–1785. 10.1128/JCM.00346-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Hoffmann W., Bockelmann W., Hummerjohann J., Stephan R., Hammer P. (2013b). Fate of Shiga toxin-producing and generic Escherichia coli during production and ripening of semihard raw milk cheese. J. Dairy Sci. 96, 815–823. 10.3168/jds.2012-5865 [DOI] [PubMed] [Google Scholar]
- Peng S., Hummerjohann J., Stephan R., Hammer P. (2013a). Short communication: heat resistance of Escherichia coli strains in raw milk at different subpasteurization conditions. J. Dairy Sci. 96, 3543–3546. 10.3168/jds.2012-6174 [DOI] [PubMed] [Google Scholar]
- Peng S., Stephan R., Hummerjohann J., Blanco J., Zweifel C. (2012). In vitro characterization of Shiga toxin-producing and generic Escherichia coli in respect of cheese production-relevant stresses. J. Food Safety Food Qual. 63, 136–141. 10.2376/0003-925X-63-136 [DOI] [Google Scholar]
- Rasko D. A., Webster D. R., Sahl J. W., Bashir A., Boisen N., Scheutz F., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717. 10.1056/NEJMoa1106920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner A., Krogfelt K. A., Klein B. M., Zechner E. L., Molin S. (2006). In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J. Bacteriol. 188, 3572–3581. 10.1128/JB.188.10.3572-3581.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K., Green A., Allen L., Ihry T., White P., Chen W. S., et al. (2016). Foodborne outbreaks reported to the U.S. food safety, and inspection service, fiscal years 2007 through 2012. J. Food Prot. 79, 442–447. 10.4315/0362-028X.JFP-15-376 [DOI] [PubMed] [Google Scholar]
- Rossi F., Rizzotti L., Felis G. E., Torriani S. (2014). Horizontal gene transfer among microorganisms in food: current knowledge and future perspectives. Food Microbiol. 42C, 232–243. 10.1016/j.fm.2014.04.004 [DOI] [PubMed] [Google Scholar]
- Schjørring S., Struve C., Krogfelt K. A. (2008). Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J. Antimicrob. Chemother. 62, 1086–1093. 10.1093/jac/dkn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C., Barken K. B., Krogfelt K. A., Struve C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. 10.1186/1471-2180-10-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C. L., Pedersen S., Ross B. M., Squires C. (1991). ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 173, 4254–4262. 10.1128/jb.173.14.4254-4262.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlhut S. G., Chattopadhyay S., Kisiela D. I., Hvidtfeldt K., Clegg S., Struve C., et al. (2013). Structural and population characterization of MrkD, the adhesive subunit of type 3 fimbriae. J. Bacteriol. 195, 5602–5613. 10.1128/JB.00753-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C., Roe C. C., Stegger M., Stahlhut S. G., Hansen D. S., Engelthaler D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 6:e00630. 10.1128/mBio.00630-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall E. J., Rowe B., Ferguson J. L., Ward L. R. (1986). Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J. Hyg. 97, 419–426. 10.1017/S0022172400063609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangari T., Melton-Celsa A. R., Panda A., Boisen N., Smith M. A., Tatarov I., et al. (2013). Virulence of the Shiga toxin type 2-expressing Escherichia coli O104:H4 German outbreak isolate in two animal models. Infect. Immun. 81, 1562–1574. 10.1128/IAI.01310-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschüttig A., Zimmermann K., Blom J., Goesmann A., Pohlmann C., Gunzer F. (2012). Identification and characterization of microcin S, a new antibacterial peptide produced by probiotic Escherichia coli G3/10. PLoS ONE 7:e33351. 10.1371/journal.pone.0033351 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.