Abstract
The continued growth in world population necessitates increases in both the quantity and quality of agricultural production. Triticeae members, particularly wheat and barley, make an important contribution to world food reserves by providing rich sources of carbohydrate and protein. These crops are grown over diverse production environments that are characterized by a range of environmental or abiotic stresses. Abiotic stresses such as drought, heat, salinity, or nutrient deficiencies and toxicities cause large yield losses resulting in economic and environmental damage. The negative effects of abiotic stresses have increased at an alarming rate in recent years and are predicted to further deteriorate due to climate change, land degradation, and declining water supply. New technologies have provided an important tool with great potential for improving crop tolerance to the abiotic stresses: microRNAs (miRNAs). miRNAs are small regulators of gene expression that act on many different molecular and biochemical processes such as development, environmental adaptation, and stress tolerance. miRNAs can act at both the transcriptional and post-transcriptional levels, although post-transcriptional regulation is the most common in plants where miRNAs can inhibit the translation of their mRNA targets via complementary binding and cleavage. To date, expression of several miRNA families such as miR156, miR159, and miR398 has been detected as responsive to environmental conditions to regulate stress-associated molecular mechanisms individually and/or together with their various miRNA partners. Manipulation of these miRNAs and their targets may pave the way to improve crop performance under several abiotic stresses. Here, we summarize the current status of our knowledge on abiotic stress-associated miRNAs in members of the Triticeae tribe, specifically in wheat and barley, and the miRNA-based regulatory mechanisms triggered by stress conditions. Exploration of further miRNA families together with their functions under stress will improve our knowledge and provide opportunities to enhance plant performance to help us meet global food demand.
Keywords: miRNA, Abiotic stress, Wheat, Barley, Drought
Introduction
Sustained production of food in both sufficient quality and quantity is vital for our world to meet the demands of a continuously increasing human population. Triticeae is an important tribe of the Pooideae grasses which comprises many nutritionally and economically valuable crops, such as wheat, barley, and rye (Mochida and Shinozaki 2013). The domestication of these species in western Asia resulted in the birth of the agricultural era for mankind about 10,000 years ago; from this point on, they have preserved their prominence at the top of the crop world (Feldman and Levy 2012). Wheat is the most extensively grown crop globally with more than 700 million tons of production per year, which is distributed on more than 200 million ha of land (http://faostat.fao.org/). By supplying many crucial minerals, vitamins, and essential amino acids, and owing to its relatively low fatty acid content, wheat is a widely preferred staple food for human and animal consumption (Shewry 2009). Barley also stands as an agro-ecologically essential member of the Triticeae by serving as an animal feed and fodder and a source of various fermentable beverages such as beer (Dawson et al. 2015). Additionally, it also serves as an important model for Triticeae genetics and genomics studies by courtesy of its diploid genome (Mayer et al. 2011; Mayer et al. 2012). Given the overall significance of the Triticeae tribe, many biological and physiological studies have been conducted on its members (Kantar et al. 2010; Lucas et al. 2011a; Lucas et al. 2011b; Budak et al. 2013a; Baldoni et al. 2015a; Budak et al. 2015b). However, numerous biological phenomena associated with these crops remain elusive.
Maintaining an adequate yield from planted crops is critical in satisfying the world’s food demand. Despite the advances in technology and associated increases of obtained yields via crop and agronomic improvements, a significant amount of production is lost due to adverse growth conditions (Budak et al. 2013b). In particular, climate change, which has accelerated in recent decades, has a direct negative effect on yield maintenance and stands as a major cause of abiotic stress. Abiotic stress leads to abnormalities in cellular homeostasis of plants resulting in poor growth and development (Mickelbart et al. 2015). Abiotic stress conditions such as drought, cold, heat, and salt and nutrition deficiencies are the major limiting factors in agricultural production (Akpinar et al. 2013; Budak et al. 2013b; Budak et al. 2015b). Drought, arguably the most important environmental stress, causes more than 50 % of yield loss when it occurs during the middle of the growth season (Nezhadahmadi et al. 2013). Abiotic stress-associated productivity loss is dependent on several intrinsic and extrinsic factors such as the severity and duration of stresses as well as the developmental stage of the crop (Zhang 2015). Fortunately, each plant has the plasticity to adapt to its environment which brings the ability to survive under diverse environmental conditions. The exploration and elucidation of underlying mechanisms which lead to adaptation to stressful environments are critically important for improved crop performance.
In order to preserve vitality, crop plants deploy many different cellular and molecular mechanisms in response to abiotic stress conditions. Various conserved abiotic stress-linked survival mechanisms have been identified across different plant species. In addition, genotype-specific defense is a well-known phenomenon (Mickelbart et al. 2015). Acclimation and adaptation are the major strategies in the maintenance of growth and productivity (Mickelbart et al. 2015). The underlying molecular mechanisms of these responses comprise many specific interactions and signaling networks (Cramer et al. 2011; Akpinar et al. 2012). The majority of the biochemical and physiological alterations related to abiotic stress conditions originate from the differential abundance of several transcripts and their associated proteins. Thus, the regulation of gene expression via transcriptional and post-transcriptional mechanisms is indispensable for both stress recognition and stress responses (Ergen et al. 2009; Budak et al. 2015b).
MicroRNAs (miRNAs) are small non-coding RNA molecules that act as gene expression regulators at the post-transcriptional level (Budak et al. 2015a; Budak et al. 2015c; Alptekin and Budak 2016). Plant miRNAs bind to their target transcripts in a complementary manner and inhibit their translation by cleavage and/or degradation of the target mRNA molecule (Budak and Akpinar 2015). The regulation of the expression of stress-responsive genes via the activity of miRNAs is considered to be a particular advantage in abiotic stress conditions. Several studies using the model plant Arabidopsis have revealed the importance of miRNA-based stress response where the miRNA metabolism defective mutants of hyponastic leaves-1 (hyl1), hua enhancer-1 (hen1), and dcl1 are detected as hypersensitive to abiotic stress-associated hormones such as abscisic acid (ABA) (Lu and Fedoroff 2000; Gim et al. 2014). These authors pointed out the central role of these small regulators in the survival of plants under environmental stress. Additionally, many miRNA target transcripts have been connected with stress-responsive transcription factor families, including WRKY and NAC, in the Triticeae (Kantar et al. 2010; Kantar et al. 2011a; Deng et al. 2015). The prominence of miRNA-based gene expression regulation was also shown with several phenotypic and physiological analyses after the expression of stress-responsive miRNAs and/or their target transcripts was altered (Feng et al. 2013; Kantar et al. 2010).
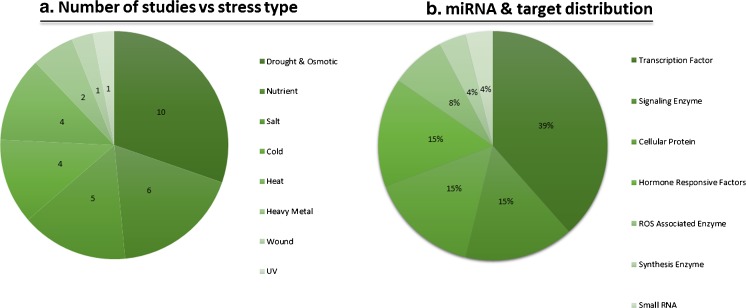
To date, several studies have been conducted to identify and characterize abiotic stress-associated miRNAs both in experimental and computational experiments (Tang et al. 2012; Gupta et al. 2014; Akpinar et al. 2015; Liu et al. 2015a; Liu et al. 2016). Most of these studies have focused on drought stress although some studies have also been performed on more specific stresses such as UV and heavy metal stresses (Wang et al. 2013; Qiu et al. 2016) (Fig. 1a). Small-RNA sequencing technology as well as quantitative real-time PCR applications have been the most frequently used experimental strategies owing to their potential to both identify and quantify miRNAs. Interestingly, some miRNA families, for example miR159, miR167, and miR169, were detected as responsive to multiple stresses (Budak et al. 2014; Sinha et al. 2015; Budak et al. 2015c). Alternatively, some miRNAs such as miR1432 and miR1137 displayed stress-specific expression patterns in several members of the Triticeae (Kantar et al. 2010; Kantar et al. 2011a; Gupta et al. 2014; Ma et al. 2015). Here, we aim to present a detailed overview of miRNA-based abiotic stress regulation in the Triticeae by critically reviewing the research on this topic. Additionally, the current understanding of molecular mechanisms and signal cascades which take place in the miRNA-associated stress responses will be explained based on association with work on several crop species. We believe that miRNA-based abiotic stress responsive mechanisms will serve as a keystone to develop plant varieties which are able to survive under changing environmental conditions.
Fig. 1.
a Number of articles found in PubMed describing work on the response to different abiotic stress conditions in the Triticeae members. b Percentage of different cellular proteins and molecules targeted by abiotic stress-responsive miRNAs. Percentages were calculated with respect to the predicted target of stress-associated miRNA families
miRNAs as regulatory keystones in crops
MicroRNAs are important regulators of many specific molecular processes such as development, environmental adaptation, and stress tolerance. In order to precisely understand the mechanisms underlying miRNA-based regulation, a deep knowledge of miRNA origin and biogenesis is necessary. Here, we summarize the common biogenesis and evolution of miRNAs in plants from an abiotic stress-associated point of view in the following section.
miRNA biogenesis
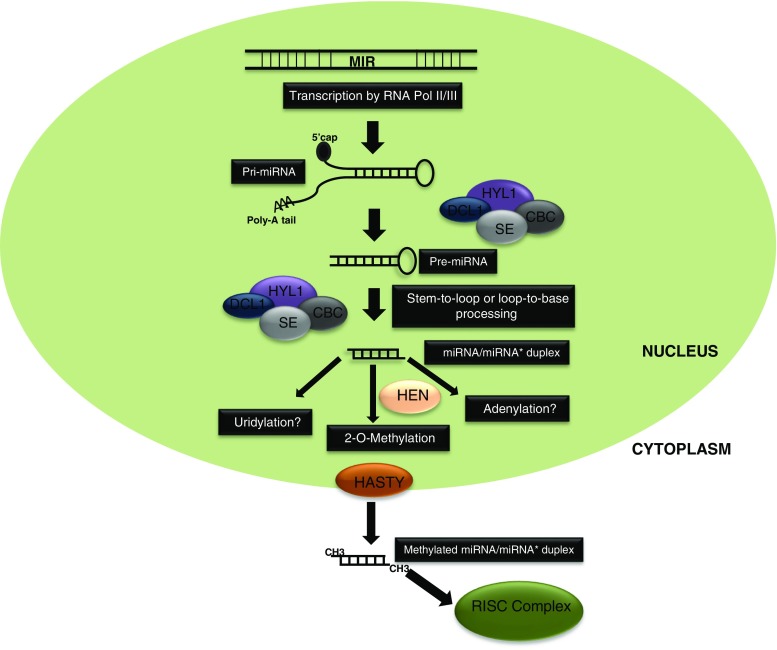
The initial step of miRNA biogenesis is transcription of the primary miRNA which mostly resides in the intergenic regions although a few miRNAs originate from the intronic or exonic sequences of protein-coding genes (Colaiacovo et al., 2012; Q. Liu, 2012). In addition, MIR loci may be located within transposable elements (TEs) where the processed miRNAs are called “transposable element-related miRNAs (TE-miRs)” (Li et al. 2011; Lucas and Budak 2012; Kurtoglu et al. 2014). Primary transcript (pri-miRNA), which is synthesized by RNA polymerase II (in some cases RNA polymerase III might take place), folds back into an imperfect hairpin, stem-loop, structure (Fig. 2). The stem-loop of the primary transcript is further recognized by the members of the DCL family of ribonucleases to form the precursor miRNAs (pre-miRNAs) and mature miRNA. The DCL1-mediated cleavage of the pri-miRNA is assisted by DCL1 interacting proteins, hyponastic leaves 1 (HYL1), serrate (SE), and nuclear cap-binding complex (CBC). While DCL1 and HYL1 are specific to the miRNA biogenesis machinery, SE and CBC have broader functions in mRNA metabolism (Voinnet, 2009). Non-lethal mutations of dcl1, hyl1, and se revealed that nuclear complex formation through these proteins is crucial for precise processing of pri-miRNAs into pre-miRNAs (Jones-Rhoades et al. 2006; Liu et al. 2011). Additionally, several studies have shown the importance of different CBP elements such as CBP20 and CBP80 together with SE in pri-miRNA processing (Kim et al. 2008; Laubinger et al. 2008). Although the contribution of these proteins in miRNA biogenesis has been found, their main functions remain elusive.
Fig. 2.
Major steps in miRNA biogenesis. The MIR loci in the genome are transcribed through the action of RNA polymerase II, or in some cases RNA polymerase III, and forms the pri-miRNA structure. Pri-miRNA is then processed into pre-miRNA through the action of DCL-1 and its interacting partners. Mature miRNA/miRNA* duplex from the pre-miRNA may be generated via two different mechanisms: stem-to-loop or loop-to-base. Mature miRNA duplex may undergo some biochemical changes before it is transported to the cytoplasm through the activity of HASTY. Mature plant miRNAs are methylated by HEN1 before they are exported to the cytoplasm. The exported mature miRNA duplex is separated, and functional mature miRNA loads onto the RISC complex in order to regulate the expression of its target transcript
Plant pre-miRNAs maturated from the pri-miRNA transcripts do not show conservation in size or sequence content that would allow proposition of a non-generalized mechanism for pre-miRNA maturation into functional miRNAs (Naqvi et al. 2012). To date, there have been two suggested models for miRNA/miRNA* duplex cleavage from the pre-miRNA sequence through the action of DLC1 and other assistant proteins which are referred to as stem-to-loop (Werner et al. 2010a) and loop-to-base processing (Bologna et al. 2009). In the stem-to-loop processing model, pre-miRNA cleavage into functional miRNA/miRNA*duplex is performed around the 15 nucleotides of the stem-loop structure via DCL1 cleavage activity (Werner et al. 2010b; Naqvi et al. 2012). This model, resembling the animal pre-miRNA maturation, represents a shared mechanism for plant pre-miRNA processing. However, observations from genesis of miR159 and miR319 have suggested another cleavage pattern. In this proposed model, initial pre-miRNA cutting is achieved near the terminal loop of pre-miRNA, the reason for referring to this model as loop-to-base processing. Multiple cutting actions are essential for functional miRNA/miRNA* duplex production via this model, and the presence of a bulge inside the pre-miRNA structure stabilizes the bona fide miRNA formation (Bologna et al. 2009).
Following the processing from pre-miRNAs, miRNA/miRNA* duplexes are exported to the cytoplasm through the activity of a homolog of the mammalian Exportin-5, HASTY (Fig. 2). In the cytoplasm, one strand of the duplex sequence, called guide strand, is recruited to members of the Argonaute (AGO) proteins which contain an sRNA binding PAZ domain and a PIWI domain that carries out the endonucleolytic cleavage of the target transcript. The guide strand, bound by AGO proteins, is then assembled into a functional RNA-induced silencing complex (RISC) that drives the mRNA cleavage and/or translational repression. The thermodynamic stability of the 5′ of each strand partially determines the selection of the guide strand, which is also assisted by accessory proteins such as HYL1 (Rogers & Chen, 2013). Accordingly, the miRNA/miRNA* duplex undergoes further biochemical changes prior to nuclear export which is directly associated with its stability, function, and quantity. The 3′ overhangs of the miRNA/miRNA* duplex are prone to uridylation by uridyl-transferases, marking the duplex for degradation by the activity of small RNA-degrading nuclease (SDN) proteins (Song et al. 2015). Adenlylation of plant miRNAs was also detected in Arabidopsis; however, its function remains elusive (Li et al. 2005). 2′-O-Methylation of the miRNA/miRNA* duplex by Hua Enhancer 1 (HEN1) prior to nuclear export is essential for protection against the possible action of exonucleases; thus, almost all plant miRNAs undergo methylation (Ren et al. 2014). Such biochemical modifications in the miRNA/miRNA* duplexes may also take place following their transfer to the cytoplasm. In addition, different AGO proteins may differ in their preferences for particular modifications. For instance, AGO1 exhibits a preference for modification of 5′ uridine residues, while AGO2 and AGO4 are mostly associated with 5′ adenosine residues (Voinnet, 2009).
miRNA origin and evolution
Understanding the evolutionary origin of molecules provides valuable insights into their precise sequence, structure, and function. Elucidating the evolutionary origin of miRNAs has relied on indirect information from comparative studies of both conserved and non-conserved miRNA species. Employment of deep sequencing for small RNA identification has revealed a myriad of non-conserved plant miRNAs together with spatio-temporal and interchangeable expression patterns of miRNAs conserved among different plant species exposed to stress conditions (Rogers and Chen 2013; Jeong and Green 2013; Budak et al. 2014; Zhang 2015). These studies have emphasized the importance of stresses that plants face on the evolution of miRNAs. Here, we have outlined the hypotheses proposed for miRNA evolution with a special focus on the relationship to abiotic stress responses.
The first hypothesis for the origin of miRNA genes was proposed by Allen et al. based on the observation that newly emerged “young” miRNAs show extended sequence homology to their targets both within and outside the mature miRNA region (Allen et al. 2004; Budak and Akpinar 2015). Accordingly, an inverted duplication event (head-to-head or tail-to-tail orientation/with or without the promoter sequence) results in the formation of a fold-back transcript that might be recognized by the DCL enzymes to generate small interfering RNAs (siRNAs) (Fig. 3a). The siRNAs may negatively regulate the expression of their founder gene and eventually adapt to the miRNA machinery through changes in their secondary structure by mutational drift. After formation of the unique target specificity followed by a duplication event that produces the mature miRNA duplexes, the sequences flanking the mature miRNA will change over evolutionary time. If the inverted duplication event includes a domain associated with a set of genes, “a family domain,” the resulting miRNA may orchestrate an extensive regulatory network which includes different members of the founder gene family (Allen et al. 2004). The targeting of several Auxin response factors by miR167 and miR160, HD-ZIP transcription factors by miR166, NAC family transcription factors by miR164, and MYB family transcription factors by miR159 in soybean (Song et al. 2011) and the miR824 targeting of MADS box genes were recently shown to be involved in drought stress responses in Brachypodium and rice, and all provide examples of how this mechanism can mediate responses to numerous abiotic stresses (Arora et al. 2007; Kutter et al. 2007; Wei et al. 2014a). While most miRNAs may have evolved through this route, accumulated mutations may lead to the silencing of transcripts from unrelated loci, which appears to have been the case for miR856 that targets both the founder gene ZAT1 and a novel gene CHX18 (Fahlgren et al. 2007; Felippes et al. 2008).
Fig. 3.
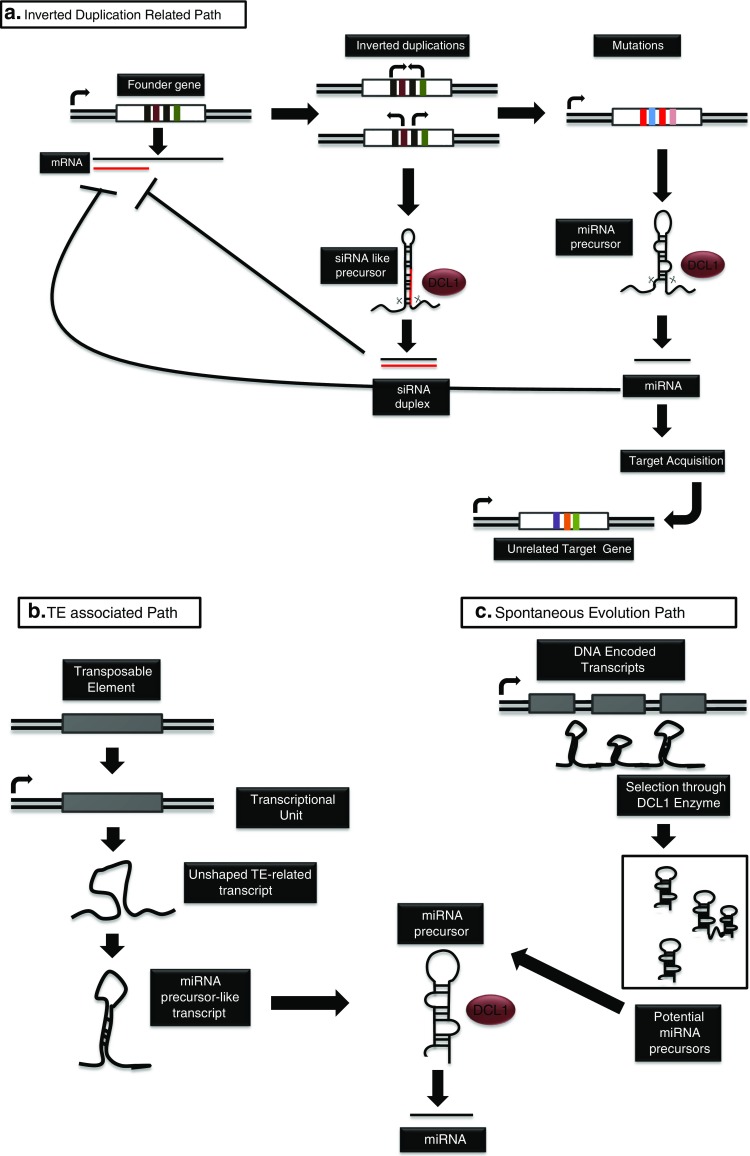
Proposed miRNA evolution paths. a Inverted duplication-related path. An inverted duplication results in the formation of a fold-back transcript that can be recognized by the DCL enzymes and generate siRNAs which can negatively regulate the expression of the founder gene. siRNAs eventually adapt to the miRNA machinery through changes in their secondary structure by mutational drift. After the formation of the unique target specificity, followed by the duplication event, a mature miRNA duplex is produced. Sequences flanking the mature miRNA region may change through evolutionary processes and target a gene that is unrelated to the founder gene. b TE-associated path. Coded transcripts from TEs may fold into unshaped RNA transcripts. Such transcripts may evolve into miRNA precursors and generate TE-derived miRNA sequences. c Spontaneous evolution path. Randomly encoded DNA transcripts may fold in on themselves. Selection of such transcripts via DCL enzymes results with the generation of new miRNA genes
Maher et al. (2006) surveyed segmental and tandem duplications in the Arabidopsis genome to reveal the evolution and expansion of miRNA gene families. This study assumed miRNAs followed a similar evolutionary path to protein-coding gene families. By analyzing non-coding flanking sequences and comparing their surrounding protein coding regions (from the same families, different families, and random genomic locations), they characterized tandemly duplicated miRNA family members that are physically linked (approximately by 1.9 kb) and resulted from large duplication events. They succeeded in showing that intra/inter-chromosomal duplications, along with subsequent inversion and rearrangement events, resulted in spawning of miR159a and miR159b in chromosome 1. They estimated that this event extends back to 30 mya based on an analysis of synonymous substitution and duplication events. miR159c, which is located in chromosome 2, was also believed to have originated from similar events. However, the evolutionary timescale could not be measured since sequence similarity in the flanking regions was lacking (Maher et al. 2006). The crucial role of these miRNA family members in plant stress response through their targeting of MYB transcription factors in several plants together with sequence variants indicates the antiquity of this family and possible role of miR159c as the founder miRNA gene (Ambawat et al. 2013; Baldoni et al. 2015b).
Aside from being unwanted parasites that are epigenetically silenced through the action of repeat associated siRNAs (Slotkin and Martienssen 2007), transposable elements (TEs) shape the eukaryotic genomes by mediating translocations and duplications. Since most TE elements have specific signals for translational regulation and splicing, their integration into protein coding genes may alter the gene expression and create new splice variants of integrated proteins (Kazazian 2004) (Fig. 3b). Through the analysis of miRNAs that neighbor the TE regions together with bona fide miRNAs that possess identical sequences to the TEs, it is suggested that some miRNAs are formed as a result of TE activity (Piriyapongsa and Jordan 2008; Li et al. 2011). Li et al. surveyed the previously annotated rice miRNAs in the miRBase and defined those that are homologous or identical to transposable elements which were named TE-miRs (Li et al. 2011). Not surprisingly, the majority of TE-miRs were derived from miniature inverted-repeat transposable elements (MITEs) reflected in their palindromic nature, while other TE-miRs are proposed to be formed through juxtaposition of inverted copies associated with the same TE. In addition, these elements are largely located in genic regions where the MITE insertion rates are high, further supporting their MITE-related origin. Expression analysis of TE-miRs showed that some are bona fide TE-miRs while others resembled the transition forms of TEs into real miRNA genes through the siRNA path since they are processed into 24-nt mature sequences. This finding supported the idea that young miRNA genes may generate heterogeneous small RNA populations that encode both miRNA and siRNA (Piriyapongsa and Jordan 2008; Li et al. 2011). In a recent study, small RNAs accompanied by high levels of miRNA* reads with anomalous putative hairpin structures seen in deep sequencing libraries have been categorized as “miRNA like siRNA loci” suggesting relationship to the TE-to-miRNA transition forms (Bertolini et al. 2013). These transition forms might incorporate into the siRNA or miRNA machinery depending on the transcription units they harbor (RNA polymerase type selectivity), invading genomic elements and selective pressure resulting from different stresses. Although TE insertion is generally deleterious for the CDS, they may generate alternative ORFs expressed from the transcriptional regulatory machinery of the CDS. This situation may explain the multi-gene targeting profile of some miRNAs where coding sequence domestication, as a result of TE insertion, can create multiple target sites for the same miRNA. However, further work will be needed to understand the mechanism completely (Li et al. 2011).
In contrast to the inverted duplication hypothesis, some of the species-specific miRNAs in Arabidopsis fail to show similarity to their genomic founder locus suggesting a different evolutionary mechanism (Fahlgren et al. 2007; Felippes et al. 2008). Therefore, an alternative miRNA evolution route has been proposed where some miRNA genes are spontaneously created from the self-complementary fold-back sequences scattered through plant genomes following capture by transcriptional regulatory units (Fig. 3c). Analysis of the flanking protein coding sequences of miRNA genes in A. lyrata revealed the absence of similar fold-back structures between the orthologous gene blocks (Felippes et al. 2008). This hypothesis suggests spontaneous arrangement for the miRNA-target pairs that are fixed through co-evolution under positive selection pressure. However, this mechanism is more suited to an explanation of animal miRNA-target pairing as the targeting does not require a near-perfect match as for plant miRNAs.
miRNAs in the regulation of stress responses
microRNAs regulate gene expression by inhibiting translation of their target transcripts into functional proteins via complementary binding which leads to translational repression or direct cleavage of associated mRNA (Budak and Akpinar 2015). Unlike animal miRNAs which specifically the target 3′-UTR of the mRNA molecule, plant miRNAs can target any location of their target transcript suggesting a more flexible target preference, but they do not show the seed region. Basically, plant miRNAs bind to their target molecules via almost full complementarity and the force coming from this binding leads to the degradation of the mRNA molecule allowing specific regulation of many molecular and biochemical processes (Zhang et al. 2006). Consequently, diverse members of the same miRNA family which show differences with only a few bases may target different mRNA transcripts and be involved in the regulation of different pathways. The expression of such miRNAs together with their associated targets may differ (Kumar et al. 2014b; Liu et al. 2015a; Liu et al. 2016) under stress conditions. Identification of such differentially expressed miRNA under stress together with their targets is critically important for the utilization of these tiny ribo-regulators for crop improvement.
To date, miRNA-based regulation of several plant genes in response to biotic and abiotic stresses has been shown with several studies (Kantar et al. 2011a; Hackenberg et al. 2014; Kumar et al. 2014a; Wang et al. 2014). During the miRNA-based stress regulation, alterations in gene expression can be achieved by direct targeting of the gene transcripts. Alternatively, indirect regulation may take place via targeting the transcription factors or hormones which then regulate the expression of specific stress-associated genes (Jones-Rhoades et al. 2011). The target preference of conserved and non-conserved miRNAs may also differ. Many conserved miRNAs across monocots and dicots, such as miR156, miR159, or miR164, have been shown to target stress-associated transcription factors such as MYB and NAC family members (Gupta et al. 2014; Qiu et al. 2016). Species and genotype specific miRNA families are more likely to target specific enzymes associated with special pathways than conserved miRNAs. Deng and colleagues showed that miR-n05 was responsive to salt stress in barley and targeted the transcript for the enzyme enolase which functions in carbohydrate metabolism in plants by catalyzing the conversion of 2-phosphoglycerate to phosphoenolpyruvate (Van der Straeten et al. 1991; Deng et al. 2015). An understanding of both conserved and genotype or species specific miRNA and their targets is crucial for interpreting abiotic stress interrelated molecular mechanisms in the Triticeae.
miRNAs associated with abiotic stress responses in the Triticeae
Several miRNAs have been described as abiotic stress regulators in the members of the Triticeae, particularly wheat and barley, by targeting several stress-associated cellular proteins and pathways (Fig. 1b). Interestingly, there is little in the literature regarding miRNAs of rye which is one of the most stress-tolerant members of the Triticeae. In the following section, we provide a detailed overview of miRNAs along with their specific target genes in the Triticeae which have been identified as “abiotic stress associated.” These miRNAs represent a substantial opportunity for crop improvement in wheat and barley.
miRNAs and drought stress responses
Drought is recognized as the most significant abiotic stress which affects crops both qualitatively and quantitatively (Lucas et al. 2011b; Budak et al. 2015b). Altered climate conditions are predicted to lead to increased rates of evaporation and decreased precipitation in many major cropping regions; these conditions are expected to lead to increased severity of drought stress. Limited availability of water and high evaporation rate affects both the vegetative and productive development of plants negatively via associated physiological, molecular, and biochemical changes. Drought causes the high economic losses directly associated with the diminution in yield (Kantar et al. 2011b; Akpinar et al. 2013). However, crop species have evolved a range of processes to provide protection against the negative effects of drought (Ferdous et al. 2015). Drought protective strategies differ according to the tissue type, timing of the stress exposure, and developmental stage of the plants when they experience the stress (Kuzuoglu-Ozturk et al. 2012). In the early stages of stress exposure, plants may escape from the stress via altering developmental stages, for example the transition from vegetative to reproductive stages. Another strategy for protection from the effect of drought supports the plant by keeping the water potential high with the aim of decreasing the hazardous effects of water loss on plant metabolism. This condition can be achieved by deepening and elongating roots in order to reach deep water sources if these are available. All drought-associated metabolic and physiological changes are based on the regulation of gene expression at the transcriptional or translational levels (Ferdous et al. 2015; Obidiegwu 2015). Consequently, miRNA-based regulation plays an important role in the drought response.
Differential expression of several miRNA families has been shown in different crops, including members of the Triticeae (Kantar et al. 2010; Budak and Akpinar 2011; Kantar et al. 2011a; Hackenberg et al. 2014; Liu et al. 2015a; Akpinar and Budak 2016) (Table 1). Observations of the expression pattern of drought-responsive miRNAs have revealed numerous significant factors. Tissue type is a determinant of miRNA expression pattern in response to drought stress. Roots and leaves are the most drought-responsive plant tissues as they directly contribute to the maintenance of high water potential and regulated osmotic pressure. Different tissue types may exhibit tissue-specific miRNA variation in the expression as is the case of miR171 from barley. The expression of miR171 targeting the Scarecrow-like transcription factor like (SCL-6) (Table 1) was found to be upregulated during drought stress in the leaves of barley while no change was detected in the root expression pattern for the same miRNA (Kantar et al. 2010). Additionally, same miRNA families may exhibit different expression patterns in diverse tissues. miR169 from barley was detected as upregulated in leaves while it is downregulated in roots. Another miRNA, miR159, showed induced expression in the leaves of wheat while it was downregulated in the roots (Gupta et al. 2014; Ma et al. 2015). Altered expression patterns of some miRNA families may directly be associated with the target specification of miRNAs since miRNAs can show multiple target preferences with respect to tissue type (Hackenberg et al. 2014). In order to understand the mechanism of tissue-specific drought-responsive miRNAs completely, further studies on tissue-specific miRNA expression along with their target transcripts is essential.
Table 1.
Drought stress-responsive miRNAs identified from several members of the Triticeae
| miRNA name | Organism | Potential target | Reference |
|---|---|---|---|
| miR1432 | T. turgidum ssp. dicocoides, T. turgidum ssp. durum, T. aestivum | Phenyl-alanine tRNA synthetase like | Liu et al. 2015a, Kantar et al. 2011a, Ma et al. 2015 |
| miR5048 | T. turgidum ssp. durum, H. vulgare | – | Liu et al. 2015a, Hackenberg et al. 2014 |
| miR5054 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR5071 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR5200 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR007 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR038 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR1029 | T. aestivum | – | Gupta et al. 2014 |
| miR109 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR1136 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR1137 | T. aestivum | – | Ma et al. 2015 |
| miR1318 | T. aestivum | – | Ma et al. 2015 |
| miR1435 | T. turgidum ssp. durum and dicocoides | – | Akpinar et al. 2015 |
| miR1450 | T. turgidum ssp. dicocoides | Mn superoxide dismutase | Kantar et al. 2011a |
| miR156 | T. turgidum ssp. dicocoides, T. turgidum ssp. durum, T. aestivum, H. vulgare | Squamosa-promoter binding protein (SBP)-like transcription factors | Kantar et al. 2011a, Liu et al. 2015a, Hackenberg et al. 2014, Kantar et al. 2010, Ma et al. 2015, Lv et al. 2012 |
| miR159 | T. turgidum ssp. durum, T. aestivum | MYB transcription factor | Gupta et al. 2014, Liu et al. 2015a, Ma et al. 2015 |
| miR166 | T. turgidum ssp. dicocoides, T. aestivum, H. vulgare | Homeodomain leucine zipper (HD-Zip) transcription factor | Kantar et al. 2011a, Hackenberg et al. 2014, Kantar et al. 2010, Ma et al. 2015 |
| miR167 | Ae. tauschii, T. turgidum ssp. durum, T. aestivum | – | Liu et al. 2015a, Ma et al. 2015, Akpinar and Budak 2016 |
| miR168 | T. aestivum | – | Gupta et al. 2014, Ma et al. 2015 |
| miR169 | H. vulgare | – | Hackenberg et al. 2014 |
| miR171 | T. turgidum ssp. dicocoides, T. aestivum, H. vulgare | Scarecrow-like transcription factor (SCL-6) | Kantar et al. 2011a, Kantar et al. 2010, Ma et al., 2015 |
| miR172 | T. aestivum, H. vulgare | – | Gupta et al. 2014, Hackenberg et al. 2014 |
| miR1867 | T. turgidum ssp. dicocoides | DUF1242 superfamily | Kantar et al. 2011a |
| miR1881 | T. turgidum ssp. dicocoides | – | Kantar et al. 2011a |
| miR319 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR393 | T. aestivum, H. vulgare | – | Gupta et al. 2014, Liu et al. 2015a, Hackenberg et al. 2014 |
| miR396 | T. turgidum ssp. dicocoides, T. turgidum ssp. durum, H. vulgare | Growth regulating factor-like (GRL) transcription factors | Kantar et al. 2011a, Liu et al. 2015a, Lv et al. 2012 |
| miR398 | T. turgidum ssp. dicocoides, T. turgidum ssp. durum, T. aestivum, H. vulgare | Copper super oxide dismutase | Kantar et al. 2011a, Liu et al. 2015a, Hackenberg et al. 2014 |
| miR398 | T. turgidum ssp. dicocoides | Cu–Zn super oxide dismutase | Kantar et al. 2011a |
| miR399 | H. vulgare | No target | Lv et al. 2012 |
| miR408 | T. turgidum ssp. durum, H. vulgare | Cu-binding domain containing chemocyanin and blue copper protein | Liu et al. 2015a, Kantar et al. 2010 |
| miR444 | H. vulgare | – | Hackenberg et al. 2014, Ma et al. 2015 |
| miR474 | T. turgidum ssp. dicocoides | Kinesin,a pentatricopeptide repeat (PPR) family protein | Kantar et al. 2011a |
| miR5024 | T. turgidum ssp. durum and dicocoides | – | Akpinar et al. 2015 |
| miR5049 | H. vulgare | – | Hackenberg et al. 2014 |
| miR528 | T. turgidum ssp. dicocoides | Similar to plantacyanin | Kantar et al. 2011a |
| miR5368 | T. aestivum | – | Ma et al. 2015 |
| miR5387 | T. turgidum ssp. durum and dicocoides | – | Akpinar et al. 2015 |
| miR5831 | T. turgidum ssp. durum and dicocoides | – | Akpinar et al. 2015 |
| miR6220 | T. turgidum ssp. durum and dicocoides | – | Akpinar et al. 2015 |
| miR6300 | T. turgidum ssp. durum | – | Liu et al. 2015a |
| miR7714 | T. turgidum ssp. durum and dicocoides | – | Akpinar et al. 2015 |
| miR827 | T. aestivum | – | Ma et al. 2015 |
| miR829 | T. aestivum | – | Ma et al., 2015 |
| miR894 | T. turgidum ssp. dicocoides | Similar to protein phosphatase PP2A-4 | Kantar et al. 2011a |
| miR896 | T. turgidum ssp. dicocoides | – | Kantar et al. 2011a |
| miR916 | T. aestivum | – | Ma et al. 2015 |
| miRn029 | H. vulgare | – | Lv et al. 2012 |
| miRn029 | H. vulgare | – | Lv et al. 2012 |
| miRn035 | H. vulgare | – | Lv et al. 2012 |
| miRX33 | H. vulgare | – | Hackenberg et al. 2014 |
| miRX34 | H. vulgare | – | Hackenberg et al. 2014 |
Drought-responsive miRNA expression patterns may also vary across different members of Triticeae. miR172 family members were shown to be upregulated in wheat leaves but downregulated in barley leaves suggesting different target regulation patterns in the two closely related species. Conversely, some miRNA families show conserved expression patterns in the same tissue of the different Triticeae members. miR398 targeting the mRNA for “Copper super oxide dismutase” was upregulated in the leaves of both durum wheat and barley (Kantar et al. 2011a; Hackenberg et al. 2014; Liu et al. 2015a). Interestingly, downregulation of the miR398 in Arabidopsis resulted in induced expression of the same target gene (Sunkar et al. 2006). Accordingly, it appears that conserved miRNA families may regulate the expression of the same gene via different signaling cascades in monocots and dicots. Hence, it is important to study the specific expression patterns of conserved miRNA families in different members of Triticeae under drought stress.
The diversity of expressed miRNA families and their expression patterns can also be regulated based on the developmental timing and duration of the stress. A miRNA with an unknown target, miR896, from tetraploid wild emmer wheat, was downregulated after 4 h of drought treatment while it is detected as upregulated after 8 h of stress (Kantar et al. 2011a) which suggests a dynamic expression pattern of drought-responsive miRNAs. The severity of stress can be effective on the expression of several different genes which might be induced or suppressed resulting from apparent disparities in the expression of regulatory miRNAs.
Crops have been selected to maintain their vitality under drought stress through a direct and dynamic interaction with their environment. However, the natural drought responsiveness of plants varies across different genotypes and cultivars of the same species. Additionally, wild species in the Triticeae display a diverse set of drought-responsive genes which might have been lost during domestication of wheat and barley (Budak et al. 2013b). To date, several different drought-responsive and susceptible wild members of the Triticeae have been identified and characterized along with their specific transcripts (Ergen et al. 2009; Ergen and Budak 2009). Such accessions are also expected to contain a diverse set of miRNAs which may have a direct association with the drought tolerance or susceptibly of the associated crop. This suggested situation was shown from a study of Akpinar and colleagues who identified different miRNA family members in drought-tolerant or susceptible durum wheats, such as miR5387 (only in TTD-22, drought susceptible), miR1435, and miR5024 (only in TR39477, drought tolerant) (Akpinar et al. 2015). Understanding the mechanisms underlying such differential expression may provide change for miRNA-based manipulation of crop species which may have direct application in the generation of drought-resistant crop varieties. Such examples of crop manipulation have already arisen from several dicot species including tomato and Arabidopsis (Zhang et al. 2011). In light of these studies, miRNA-based strategies in the Triticeae members such as overexpression or transformation of miRNA families identified from drought-tolerant genotypes may confer the drought tolerance in modern wheat and barley.
miRNAs and temperature stress responses
Temperature is an important environmental parameter affecting plant growth and productivity. Plant exposure to non-optimal temperatures leads to a decrease in the quality and quantity of grain and results in economical loss for farmers (Schlenker and Roberts 2009). Effects of temperature stress vary depending on several factors such as the period of stress exposure and the developmental stage of the plant; for instance, plants are more sensitive to high and low temperatures during their reproductive stages (Hatfield and Prueger 2015). Extreme temperatures can affect the viability of pollen and egg cells and the fertilization process. In addition to reproductive damage, non-optimal temperature conditions can cause alterations in soil water availability and mineral content which indirectly effects the plant physiology (Bita and Gerats 2013). In recent years, temperature extremes have become more frequent as a result of global warming. The global temperature is predicted to increase by over 0.2 °C per decade which is going to be hazardous for all living organisms in the world either directly or indirectly (IPCC 2014). It is predicted that an increase in the temperature may cause yield losses of more than 10 % during the twenty-first century (Hatfield et al. 2011; Hasanuzzaman et al. 2013). Economically important crops such as wheat and barley are also affected from increased non-optimal temperature conditions which have arisen recently (Högy et al. 2013; Asseng et al. 2014). In order to cope with rising temperature stress on the Triticeae members, it is essential to understand associated genetic mechanisms such as temperature-responsive gene expression along with their regulatory mechanisms.
Plants have evolved several mechanisms to preserve their vitality under temperature stress through stress avoidance, adaptation, and acclimation (Walbot 2011). Fundamentally, plant survival under heat stress is directly linked with the ability to sense temperature alteration and generate corresponding signals to maintain cell survival via molecular and physiological changes. Temperature-induced gene expression, protein translation, and metabolite synthesis have a direct effect on the degree of temperature tolerance. Increased expression of heat shock factors and proteins, chaperons, phyto-hormones as well as secondary metabolites is detected under temperature stress conditions (Liu et al. 2015b). Consequentially, regulation of temperature-associated transcript and protein expression plays an important role in the response to temperature stress. MicroRNAs are directly involved in the heat and cold stress adaptation by acting as post-transcriptional regulators in several plants (Yu et al. 2012; Jeong and Green 2013; Kruszka et al. 2014; Kumar et al. 2014a, b). Here, we summarize the information on miRNA-based regulation of temperature stress responses in the Triticeae.
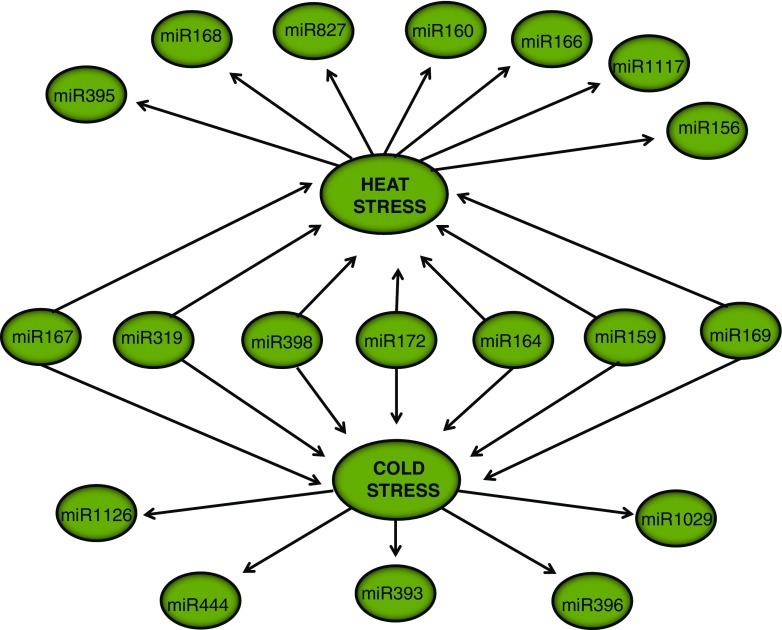
To date, several heat and cold stress-responsive miRNAs have been identified and experimentally characterized under different stress conditions in several tissues of wheat and barley (Tang et al. 2012; Wang et al. 2012; Gupta et al. 2014) (Fig. 4). Several miRNAs were defined as conserved across different species and stress conditions. These conserved miRNAs associated with temperature stress are thought to act in the regulation of general stress-responsive genes such as the target of miR398, superoxide-dismutases, which is involved in the reduction of reactive oxygen species (ROS) accumulated under stress conditions. Variation in tissue-specific expression and pattern for cold- and heat-responsive miRNAs was also observed in several studies suggesting differences in the effect of temperature stress on tissues. For example, anthesis is particularly susceptible to cold stress while heat stress mainly affects floral fertility and seed ripening (Jeong and Green 2013). In a recent study, Kumar et al. described the tissue-specific expression pattern of numerous miRNAs under heat stress in wheat where miR3466, miR5652, and miR5064 exhibited differential expression between root, stem, and leaf tissues (Kumar et al. 2014a, b). The expression of miR5652 was found to be highly cultivar dependent since its expression showed significant differences between a thermo-susceptible and tolerant variety of wheat. Further observations of the specific targets and relationship to miRNAs should improve our understanding of tissue-based temperature stress-responsive regulation of transcription and translation.
Fig. 4.
Temperature stress-responsive miRNAs in barley and wheat. Arrows indicate the stress type. miR167, miR319, miR398, miR172, miR164, miR159, and miR169 are responsive to both heat and cold stresses
Heat and cold stress result in distinct and independent modifications to cellular processes. However, several studies revealed altered expression of numerous miRNAs such as miR159, miR164, miR167, miR172, miR319, and miR398 in response to both heat and cold stresses (Tang et al. 2012; Gupta et al. 2014; Wang et al. 2014) (Fig. 4). Interestingly, several miRNAs showed reverse expression patterns under heat and cold stresses. For example, miR164 targeting heat shock protein 17 (HSP17) is upregulated during a cold stress response but downregulated in response to heat stress in wheat (Gupta et al. 2014; Kumar et al. 2014a, b). Another miRNA from wheat, miR319 targeting a MYB transcription factor, also showed induced expression under cold stress although its expression is decreased under heat stress. These observations support the view that temperature regulation mechanisms can be reversed under heat versus cold stresses although the underlying mechanisms may be similar.
Heat stress induces alterations in important cellular events such as respiration and photosynthesis along with structural damage including membrane integrity deterioration. Heat shock proteins help to maintain cellular integrity, reducing the oxidative stress activity by capturing ROS, synthesizing antioxidants, and improving protein folding under heat stress (Jiang et al. 2014). The suppressed expression of miR160 and miR164 probably results with the induction of heat shock protein expression and supports the maintenance of vitality under high temperatures. Conversely, the upregulated expression of the same miRNAs under cold stress suggests that the regulative role of heat shock proteins changes under cold stress. In addition to a role of cellular maintenance under temperature stresses, some heat shock proteins such as HSP17 were also thought to be important for normal anther development. The expression of HSP17 was upregulated under short- and long-term heat stresses in Arabidopsis, and manipulation of this gene resulted in improved heat tolerance in carrot (Malik and Hwang 1999; Löw et al. 2000). Since these examples are from dicots, further experimental studies are necessary to identify the effects of heat shock proteins and associated miRNAs in heat stress response. Regulation of such miRNA species may lead to improvements in heat/cold tolerance of wheat and barley cultivars.
Several miRNAs which are detected both in heat- and cold-stressed tissues show similar expression patterns. miR169 which is characterized as targeting nuclear transcription factor Y (NF-Y) in several plants, such as maize and Arabidopsis, is upregulated both in heat- and cold-stressed wheat (Ni et al. 2013; Gupta et al. 2014; Kumar et al. 2014b; Sorin et al. 2014; Luan et al. 2015). NF-Y, one of the most conserved transcription factors across monocots and dicots, consists of three different subunits named NF-YA, NF-YB, and NF-YC which bind to the CCAAT box in the promoter regions of target genes (Petroni et al. 2012). Although the exact mechanism of action is not known, the overexpression of the miR169/NF-YA module improved drought resistance in Arabidopsis (Li et al. 2008). In addition, miR167, which targets one of the regulators of the stress-responsive pathway, auxin responsive factor (ARF), is upregulated in both heat and cold stresses. Processing of such miRNAs and their targets might improve the overall temperature stress tolerance of crop species.
miRNAs and salinity stress responses
Salinity/salt stress due to the negative effects of excessive Na+ and Cl− ions on plant metabolism and physiology is a serious factor limiting crop growth and production in many regions of the world (Gupta and Huang 2014). Salinity stress may arise due to the accumulation of salt in the soil over time as a consequence of climatological events, primary salinity, or through human activities which cause an imbalanced soil in salt content and generate the salinity stress, secondary salinity (Parihar et al. 2015). Each year, more than 900 million ha of land is affected by salinity stress, which carries an associated yield loss (Deng et al. 2015). The effect of salinity stress on food production is predicted to increase with the effect of climate change (Wang et al. 2003). Therefore, elucidation of the molecular mechanisms associated with salinity tolerance is of growing importance.
High soil salinity can affect the plant in several ways. The osmotic effects of salt reduce the ability of plants to access water. If the accumulated salt in the soil enters the plants, it can cause cellular damage to the plasma membrane or organelles (Parihar et al. 2015). Consequently, salinity stress can directly damage important processes such as germination or photosynthesis (Deinlein et al. 2014). Despite such effects, plants are able to survive under salt stress to varying degrees through the action of several salt tolerance mechanisms. Over the past decades, we have improved our understanding of salt tolerance mechanisms in several crops leading to the description of several salt stress-responsive genes (Hamamoto et al. 2015). Salt-responsive genes mainly affect salt uptake and transport as well as acting to maintain the osmotic balance in cells (Kong 2010; Li et al. 2013a; Parihar et al. 2015; Sun et al. 2015). Here, we summarize several studies on the miRNA-based regulation of salinity stress tolerance in several members of Triticeae.
Salinity stress-associated miRNA families and their targets have been identified in several studies on barley and wheat (Table 2). Expression of some conserved miRNA families such as miR156 and miR6213 has been detected as upregulated while miR168, miR444, and miR5048 have shown suppressed expression patterns in response to salinity stress in barley (Lv et al. 2012; Deng et al. 2015). Also, many miRNA families such as miR171 and miR393 showed induced expression in wheat in response to salinity stress (Gupta et al. 2014; Wang et al. 2014). Interestingly, miR171 which targets the MYB family of transcription factors was found to be upregulated by salinity stress in both wheat and barley (Wang et al. 2014; Deng et al. 2015) (Table 2). Importantly, the same miRNA was also upregulated under salt stress in Arabidopsis (Liu et al. 2008) which suggests common regulatory mechanisms for salinity tolerance in both monocots and dicots. Since both drought and salt stresses effect the osmotic balance of plant cells, miR171 might play a role in the regulation of the osmotic balance under such stress conditions. The manipulation of miR171 under salinity and drought stresses may provide improved osmotic protection to members of the Triticeae. Further experimental characterization of this miRNA as well as other salt-responsive miRNA families may facilitate the development of salt-tolerant wheat and barley cultivars.
Table 2.
Salinity stress-responsive miRNAs from barley and wheat
| miRNA name | Organism | Tissue | Situation | Reference |
|---|---|---|---|---|
| miR1029 | T. aestivum | Seedling | Downregulated | Gupta et al. 2014 |
| miR156 | H. vulgare | Leaf | Upregulated | Lv et al. 2012 |
| miR159 | T. aestivum | Leaf | Upregulated and downregulated | Wang et al. 2014, Gupta et al. 2014 |
| miR164 | T. aestivum | Seedling | Downregulated | Gupta et al. 2014 |
| miR165 | T. aestivum | Leaf | Downregulated | Wang et al. 2014 |
| miR168 | H. vulgare | Whole plant | Downregulated | Deng et al. 2015 |
| miR171 | T. aestivum, H. vulgare | Leaf, whole plant | Upregulated | Wang et al. 2014, Deng et al. 2015 |
| miR319 | T. aestivum | Leaf | Downregulated | Wang et al. 2014 |
| miR393 | T. aestivum | Seedling | Upregulated | Gupta et al. 2014 |
| miR444 | H. vulgare | Whole plant | Downregulated | Deng et al. 2015 |
| miR5048 | H. vulgare | Whole plant | Downregulated | Deng et al. 2015 |
| miR6213 | H. vulgare | Whole plant | Upregulated | Deng et al. 2015 |
| miR855 | T. aestivum | Seedling | Downregulated | Gupta et al. 2014 |
| miRn0 | H. vulgare | Whole plant | Upregulated and downregulated | Deng et al. 2015 |
| miRn029 | H. vulgare | Leaf | Upregulated | Lv et al. 2012 |
| miRn029 | H. vulgare | Leaf | Upregulated | Lv et al. 2012 |
| miRn035 | H. vulgare | Leaf | Upregulated | Lv et al. 2012 |
| miRn2 | H. vulgare | Whole plant | Upregulated and downregulated | Deng et al. 2015 |
| miRn3 | H. vulgare | Whole plant | Upregulated and downregulated | Deng et al. 2015 |
| miRn5 | H. vulgare | Whole plant | Upregulated and downregulated | Deng et al. 2015 |
| miRn6 | H. vulgare | Whole plant | Upregulated and downregulated | Deng et al. 2015 |
The expression pattern of miRNAs under salt stress may vary depending on the duration of stress. In a recent study, a cultivar of barley, Morex, was exposed to salt stress at three different developmental time points and alterations in the expression pattern of several miRNAs were observed. For example, miR444 was upregulated in the third hour of stress but downregulated at 8 and 27 h (Deng et al. 2015). In rice, the same miRNA was detected as targeting a member of a MADS-box transcription factor, ANR1, which functions in root growth and elongation. Overexpression of miR444 induced primary root growth and caused the inhibition of lateral root propagation in rice (Yan et al. 2014). Based on these studies, it will be interesting to further characterize miR444 expression and function under salt stress conditions.
miRNAs associated with nutrient homeostasis and response to toxic metal stresses
The uptake of nutrients by diffusion or active transport from soil is essential for the maintenance of plant growth and metabolism. Nutrients are classified as macro- and micro-nutrients based on the amount required (Paul et al. 2015). Macro-nutrients include nitrogen (N), potassium (K), magnesium (Mg), phosphate (P), and sulfur (S). Micro-nutrients such as chlorine (Cl), iron (Fe), boron (B), manganese (Mn), zinc (Zn), molybdenum (Mo), and nickel (Ni) are used as small quantities as cofactors for enzymes or as components of membranes and cell walls (Powers and Salute 2011; Jeong and Green 2013). Nutrient deficiencies can lead to severe disruption of plant growth and reproduction. Consequently, plants and plant cells use numerous regulatory approaches to ensure cellular homeostasis including miRNAs. miRNA-based nutrient stress responses have been shown to operate in the Triticeae (Liang et al. 2012; Wang et al. 2015). Here, we focus on nutrient stress-responsive miRNAs together with their targets in the Triticeae.
Nitrogen is an important plant nutrient used in the synthesis of several bio-products such as nucleic acids and proteins (Kraiser et al. 2011; Sinha et al. 2015). The nitrogen requirement of a plant is generally supplied through N-rich fertilizers where the nitrogen uptake efficiency is important (Abrol et al. 2011). However, up to 60 % of applied fertilizer is lost to the environment resulting in economical loss and environmental pollution (Sinha et al. 2015). The efficiency of nitrogen uptake and transport may vary depending on the plant species and genotype. Understanding the mechanisms and regulation of nitrogen uptake, mobilization, and use is therefore vitally important for further crop improvement. miRNAs have been shown to play a major role in the regulation of N maintenance and homeostasis in numerous plant species (Liang et al. 2012; Nischal et al. 2012). However, the numbers of studies involving the Triticeae are limited. Sinha and colleagues selected 10 different miRNAs from the literature, and their expression was characterized under N-deficient conditions with Q-PCR. They observed that miR159, miR160, miR164, miR399, miR1117, and miR1120 exhibited differential expression suggesting a role in N homeostasis (Sinha et al. 2015) (Table 3). In another study, the upregulation of miR444a in roots and leaves of wheat in response to nitrogen deficiency was demonstrated (Gao et al. 2016). Overexpression of the same miRNA in tobacco improved plant growth and biomass along with increased N uptake under N deficiency and suggested that miR444a acts as a regulator of nitrate transporters (Gao et al. 2016). Further examination of genes associated with nitrate transport, the NRT genes, revealed the upregulated expression of three different members. Based on these results, it is plausible to suggest that miR444a regulates N metabolism via N transporters and manipulation of this miRNA has potential to improve crop performance under N-deficient conditions.
Table 3.
miRNAs from wheat and barley responsive to different nutrient deficiency and heavy metal stresses
| miRNA name | Stress type | Organism | Tissue | Potential target | Situation | Reference |
|---|---|---|---|---|---|---|
| miR1117 | N deficiency | T. aestivum | Seedling | – | Downregulted | Sinha et al. 2015 |
| miR1120 | N deficiency | T. aestivum | Seedling | – | Downregulted | Sinha et al. 2015 |
| miR1122 | P deficiency | T. aestivum | Root | – | Upregulated | Zhao et al. 2013 |
| miR1125 | P deficiency | T. aestivum | Root | Annexin-like protein | Upregulated | Zhao et al. 2013 |
| miR1126 | P deficiency | H. vulgare | Shoot | – | Upregulated | Hackenberg et al. 2013 |
| miR1135 | P deficiency | T. aestivum | Root | Auxin-induced protein | Upregulated | Zhao et al. 2013 |
| miR1136 | P deficiency | T. aestivum | Root | TF PWWP domain | Upregulated | Zhao et al. 2013 |
| miR1139 | P deficiency | T. aestivum | Root | NADH dehydrogenase subunit 6 | Upregulated | Zhao et al. 2013 |
| miR156 | Cd Stress | T. aestivum | Leaf and root | Squamosa promotor binding protein | Downregulted | Qui et al. 2016 |
| miR159 | N deficiency | T. aestivum | Seedling | MYB3 | Downregulted | Sinha et al. 2015 |
| miR159 | Cd Stress | T. aestivum | Leaf and root | MYB3 | Downregulted | Qui et al. 2016 |
| miR159 | P deficiency | T. aestivum | Root | MYB3 | Upregulated | Zhao et al. 2013 |
| miR160 | N deficiency | T. aestivum | Seedling | – | Downregulted | Sinha et al. 2015 |
| miR164 | N deficiency | T. aestivum | Seedling | NAC members | Downregulted | Sinha et al. 2015 |
| miR164 | Cd Stress | T. aestivum | Leaf and root | NAC members | Downregulted | Qui et al. 2016 |
| miR167 | P deficiency | T. aestivum | Root | Auxin-responsive factor | Upregulated | Zhao et al. 2013 |
| miR398 | Cd Stress | T. aestivum | Leaf and root | Cu-Zn superoxide dismutase | Downregulted | Qui et al. 2016 |
| miR399 | N deficiency | T. aestivum | Seedling | – | Downregulted | Sinha et al. 2015 |
| miR399 | P deficiency | T. aestivum | Root | – | Upregulated | Zhao et al. 2013 |
| miR399 | P deficiency | H. vulgare | Shoot | – | Upregulated | Hackenberg et al. 2013a |
| miR408 | Cd Stress | T. aestivum | Leaf and root | Plantacyanin | Downregulted | Qui et al. 2016 |
| miR408 | P deficiency | T. aestivum | Root | Plantacyanin | Downregulted | Zhao et al. 2013 |
| miR408 | Cu stress | T. aestivum | Leaf | Plantacyanin | Upregulated | Feng et al. 2013 |
| miR444 | N deficiency | T. aestivum | Leaf and root | – | Upregulated | Gao et al. 2016 |
| miR5051 | P deficiency | H. vulgare | Shoot | – | Upregulated | Hackenberg et al. 2013b |
| miR528 | P deficiency | H. vulgare | Shoot | – | Downregulted | Hackenberg et al. 2013b |
| miR827 | P deficiency | H. vulgare | Shoot | – | Upregulated | Hackenberg et al. 2013a |
Phosphate is a basic component of cells by the utilization as a part of many cell components, energy metabolism, and signal cascades. Plant phosphate requirement is high because of its extensive use, and wheat and barley account for approximately 46 % of the P fertilizers applied to cereals according to the Food and Agriculture Organization (FAO) of the United Nations (FAO Fertilizer and Plant Nutrition Bulletin 17; http://www.fao.org). Phosphate is found in a higher concentration in the soil compared to other nutrients; however, P uptake can be a problem since the orthophosphate form (Pi) is only readily used by plants (Zhang et al. 2014b). Further, the phosphate is precipitated with cations or incorporated into the organic matter which reduces its availability for plants. Under these conditions, plants adapt their metabolism to P-limited conditions via regulation of several phosphate-responsive genes at both the transcriptional and translational levels (Kuo and Chiou 2011). miRNAs that show altered expression in response to P deficiency in the Triticeae have been identified in several studies (Hackenberg et al. 2013a; Zhao et al. 2013; Hackenberg et al. 2013b). P starvation-responsive miRNA, miR399, was found to be upregulated in both barley and wheat (Zhao et al. 2013; Hackenberg et al. 2013b) (Table 3). This miRNA was also identified in Arabidopsis under conditions of P deficiency suggesting the conservation of nutrient deficiency-responsive miRNAs across monocots and dicots (Baek et al. 2013a; Baek et al. 2013b). The target of this miR399 is the mRNA encoding the enzyme “Phosphatase 2” (PHO2), and downregulation of PHO2 was shown in barley by Hackenberg and his colleagues when the miR399 was upregulated (Hackenberg et al. 2013a). Furthermore, overexpression of this miRNA in tomato resulted in the increased accumulation of Pi and secretion of phosphoric acid which facilitates the mobilization of soil organic P into Pi which is available for plant uptake (Gao et al. 2010). These observations suggest that miR399 is a candidate for manipulation of the Pi uptake pathway in wheat and barley.
miR399 is not the only miRNA detected as Pi deficiency responsive. Differential expression of miR159, miR167, miR1122, miR1125, miR1135, mir1136, miR1139, and miR408 was also observed under Pi deprivation in wheat (Zhao et al. 2013). Among these, miR159, miR167, and miR408 have orthologues from several dicot plants. However, orthologues of miR1122, miR1125, miR1135, mir1136, and miR1139 are not detected in dicots, thus suggesting that these miRNAs may be monocot-specific. These listed miRNAs target transcription factors have already been shown to function during abiotic stress.
Another well-characterized miRNA in Pi deprivation is miR827. Hackenberg and colleagues found upregulation of miR827 in response to Pi deficiency in wheat. This miRNA was shown to target a cytochrome P450-like protein. Interestingly, the upregulation of miR827 also resulted in the upregulation of a P450-like enzyme which suggests this may not be a bona fide target of this miRNA. However, as with HvPHO2, the cytochrome P450-like TBP may be protected from degradation by an unknown mimic sequence which interacts with miR827, inhibiting effective activity of the miRNA (Hackenberg et al. 2013a). This unusual situation has been proposed to explain the complex nature of Pi homeostasis in the Triticeae and reveals that further studies are required to understand the Pi regulatory pathway in wheat and barley.
Micro-nutrients are vitally important for plant metabolism even though they are required in only small quantities. Excessive amounts of some micro-nutrients such as iron and zinc can be toxic for plants (Curie and Briat 2002). Iron is generally taken up in chelating agents and is used as a cofactor for many proteins. Iron can be available for plants as Fe+2 or Fe+3. Iron deficiency results in physiological problems such as chlorosis (Briat et al. 2015). Consequently, the uptake of iron is highly regulated at both the transcriptional and translational levels to maintain iron homeostasis (Brumbarova et al. 2015).
Zinc which is taken up by zinc iron permease (ZIP) family members is required in many important molecular activities such as enzyme activation, gene expression, protein synthesis, and carbohydrate metabolism (Broadley et al. 2007). To date, several miRNAs have been associated with transporters of Zn and Fe in Arabidopsis, but no studies have been reported for the Triticeae (Kong and Yang 2010; Waters et al. 2012; Paul et al. 2015). miR159, miR164, miR172, miR173, and miR394 were detected as Fe-responsive in Arabidopsis by Kong and Yang, but the role of these miRNAs on Fe uptake and transport remains elusive (Kong and Yang 2010). Another study revealed that miR398, which is a Cu deficiency-responsive miRNA in Arabidopsis, is also regulated in Fe deficiency, but in an opposite direction to the Cu deficiency response (Buhtz et al. 2010; Waters et al. 2012; Paul et al. 2015). Additionally, miR398 was also detected as Zn deficiency responsive in sorghum suggesting its conservation across species (Li et al. 2013b). These miRNA families may be conserved in the Triticeae, but further cloning and characterization are necessary.
Heavy metals including cadmium (Cd), mercury (Hg), lead (Pb), and arsenic (As) together with some transition metals like copper (Cu) are potentially toxic for living organisms. Plants have evolved several mechanisms to survive under conditions where such metals are present. The first response of plants to toxic metal stress is avoiding the uptake. If the metal has already been absorbed, plants will try to halt the uptake process and prohibit the spread (Manara 2012).In order to achieve this regulation, plants restrict toxic metals to the apoplast or bind the metal to the cell wall. These mechanisms prevent the long-distance movement of the metal through the whole organism. When the toxic metal is already present in the cells, plants use storage and detoxification mechanisms including chelation, translocation, or oxidative defense mechanisms (Hall 2002). The role of miRNAs in response to toxic metal stress has been investigated with a few studies reported in the Triticeae (Xu et al. 2010; Feng et al. 2013; Qiu et al. 2016). Cd is one of the most toxic heavy metals for plants. Its accumulation in cells reduces growth, cell proliferation, and photosynthesis. Cd inhibits the reproduction of cells both in roots and stems along with a decrease in chlorophyll content. Several miRNAs have been detected in response to Cd stress and underlined as potential regulators of the Cd stress response (Xu et al. 2010; Qiu et al. 2016). Downregulation of miR156, miR159, miR164, miR398, and miR408 was observed under Cd stress while their targets were mostly upregulated. Interestingly, the target of miR164, a member of the NAC transcription factor family, was positively correlated with miRNA expression suggesting an indirect role in the regulatory pathway (Qiu et al. 2016). In another study, Feng and colleagues showed the differential expression of miR408 which targets the TaCLP1, which encodes a chemoxyanin belonging to blue copper proteins under high Cu condition (Table 3). Corresponding to the Feng study, miR408 downregulation also led to upregulation of the target chemocyanin gene in response to Cd stress (Table 3). Based on these results, it suggests that a shared protective mechanism for dealing with different toxic metals might be present in crops. miR408 was also found to target cadmium and copper transport elements together with their associated transcription factors in Arabidopsis. However, the expression pattern was reversed in response to copper deficiency. This may reflect the conserved miRNA species functioning in heavy metal stress conditions across different species even though their expression may exhibit different patterns.
miRNAs associated with wound and light stresses
Plants are susceptible to injuries which may be caused by herbivores, pathogens, and mechanical agents such as wind. Consequently, they have evolved different barriers and protection strategies to avoid such stress conditions. Physical barriers against mechanical and wound stresses are exemplified by the cuticle, cell wall, and hardened/woody surfaces (Minibayeva et al. 2015). The Triticeae are grasses and face frequent mechanical stress (Mochida and Shinozaki 2013). When tissue damage occurs, crops initiate a signaling cascade to maintain their normal physiology. Firstly, they attempt to heal the damaged tissues and, at the same time, they produce chemicals toxic to predators to inhibit further damage (Vaughan et al. 2015). Additionally, plants may show a systematic response to the stress condition by activating the expression of genes via specific transcription factors which leads to alteration inside of the plant cell.
Several miRNAs have been reported as responding to wound stress in wheat. Wang and collogues identified induced expression of miR159 and miR399 upon wounding while miR164, miR167, miR393, and miR398 were detected as downregulated by the same process. Among these, miR398 targets members of the CDS gene family which are involved in protection of cells from oxidative stress (Wang et al. 2014). Wounding leads to an oxidative stress response to protecting the cell from ROS generated in the process. This explains the miR398-based regulation of CDS gene expression.
Another wound-responsive miRNA, miR159, targets a member of the MYB transcription factor family, MYB3. The upregulation of miR159 results in the reduction of MYB3 transcripts. MYB3 is an inhibitor of secondary metabolite biosynthesis. Consequently, downregulation of MYB3 may lead to upregulation of secondary metabolites such as jasmonic acid (Plett et al. 2014). Several studies from dicots, such as tomato and Nicotiana, revealed jasmonic acid-associated differential expression of several miRNAs such as miR319 (Bozorov et al. 2012; Zhao et al. 2015). The same miRNA was also detected as slightly upregulated in Wang’s study suggesting a jasmonic acid-responsive wheat orthologue (Wang et al. 2014).
Light is both an important energy source and a developmental regulator in higher plants. However, under high light intensity, particularly high UV, light induces stress by photo-bleaching and damaging DNA and proteins via the generation of ROS species (Müller-Xing et al. 2014). In response to UV stress, plants induce the expression of several genes and regulate the expression of others. The role of miRNAs in UV stress response was characterized by Wang and colleagues in wheat (Wang et al. 2013). In their study, they found that the expression of miR164, miR395, and miR156 was downregulated while miR159, miR167, and miR171 expression was upregulated in leaf tissues of wheat. The differentially expressed miRNAs were predicted to target transcripts from several stress-associated genes. For instance, miR395 targeted the APS genes which are responsible for regulation of the sulfur assimilation pathway. In the same study, miR6000 targeted a hypothetical protein, Ta74774, which was significantly upregulated (Wang et al. 2013). Experimental characterization of this protein might be beneficial for UV stress-associated improvement of wheat and other Triticeae.
miRNA and stress regulation mechanisms
So far, we have described the involvement of several miRNAs in the response to different abiotic stress conditions. Abiotic stress-associated miRNA families are associated with numerous transcription factors which are associated with stress-responsive gene expression. The connection between miRNA and transcription factors during abiotic stress response will be summarized with examples from several species. The relationship between small interfering RNA (siRNA) and miRNA in response to abiotic stress will be explained since siRNAs are highly similar to miRNAs. Additionally, an emerging topic from the small RNA world, miRNA inheritance, will be described with respect to its potential use for crop improvement.
The relationship between miRNAs and transcription factors during an abiotic stress response
microRNAs have the ability to control stress responses through their relationship with transcription factors (TFs) (Rhoades et al. 2002). miRNA associations with several stress-associated TFs such as NAC, WRKY, and DREB have been described and characterized in response to abiotic stress (Fang et al. 2014a; Zhang et al. 2014a). The plant-specific NAC family, comprising NAM, ATAF, and CUC TFs, has been implicated in the regulation of various abiotic stress responses including drought and salinity (Nakashima et al. 2012). However, there are only a few reports on the relationship between miRNAs and NAC TFs under abiotic stress conditions, specifically in the Triticeae. Among these studies, the miR164 family was reported to target six NAC family members, of which four had negative effects on drought tolerance in rice seedlings (Fang et al. 2014b).In Brassica, miR164 was shown to target a NAC TF, whose expression was negatively correlated with miR164 under drought, salinity, and high-temperature stresses (Bhardwaj et al. 2014). Additionally, maize miR164 contributed to lateral root development through cleavage of a target NAC TF (J. Li et al. 2012), which also suggests a role in abiotic stress responses, given the role of roots in drought and salinity responses. Interestingly, AsNAC60, the NAC TF homolog of rice ONAC60, was downregulated by the overexpression of miR319 which enhanced drought and salinity tolerance in creeping bent-grass (Zhou et al. 2013). Furthermore, ONAC60 is targeted by miR164 in rice (Wu et al. 2009b). Additionally, a recent study from Feng and colleagues showed miR164 regulation of TaNAC21/22 gene under infection by wheat stripe rust (Feng et al. 2014). This study supports a NAC/miRNA association under stress in a member of the Triticeae. The involvement of NAC members such as NAC2a, NAC4a, and NAC6a in the regulation of transcription under abiotic stress such as drought has also been demonstrated (Budak et al. 2013b).
Another example of the miRNA-TF relationship comes from the SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) family of TFs. SPL is a conserved family of transcription factors and has been detected in all green plants including single-celled algae (Preston and Hileman 2013). SPL was detected in the modulation of transition from juvenile phase to adult phase in the shoot development process in Arabidopsis, where miR156 regulates the expression of miR172 through several members of this TF (Wu et al. 2009a, b). Also, the miR156-SPL association was shown to be effective in grain development of rice and barley (Miura et al. 2010; Curaba et al. 2012). These are several examples of the same miRNA/target module in abiotic stress responses from a few plants. The miR156-SPL module is associated with heat stress response and memory to delay flowering by the repressing of the expression of SPL TFs in Arabidopsis (Stief et al., 2014) and also the regulation of lateral root development which determines the efficiency of water and nutrient uptake in Brassica (Yu et al. 2015). Similar miRNA/target associations may be present in the Triticeae and may be useful for crop improvement. Opportunities such as these have also been outlined in the recent review by Liu and colleagues (Liu et al. 2016).
Another important transcription factor family in plants, WRKY, is also targeted by several microRNAs. In potato, a WRKY TF, TC199112, was predicted as a target of miR4398 and its expression was negatively correlated with miR4398 expression under drought stress (Zhang et al. 2014a). Similarly, the expression of the validated target of miR396, HaWRKY6, exhibited an opposite profile to miR396 under high-temperature stress in sunflower. Transgenic Arabidopsis lines carrying miR396-resistant HaWRKY6 had increased sensitivity to heat treatment, further pointing to a miRNA-mediated response to abiotic stress for this member of the WRKY family (Giacomelli et al. 2012). The role of WRKY transcription factor family members in the biotic and abiotic stress responses is well characterized in the members of the Triticeae with several studies (Chen et al. 2012; Satapathy et al. 2014).
MYB is a well-described TF family in eukaryotes including plants, and they function in the regulation of a wide range of molecular events such as cell cycle, hormone, or stress-related responses (Ambawat et al. 2013). The differential expression of numerous MYB TFs in the Triticeae was shown with several studies in response to abiotic stress conditions such as drought and salt stresses (Rahaie et al. 2010; Zhang et al. 2012). Association of the MYB family with miRNAs has also been shown in different plants (Zhang et al. 2010). Differential expression of the MYB-miR159 module was observed in response to aluminum toxicity in soybean (Qiao-Ying et al. 2012). miR811 relationship with MYB TF, CV431094, was characterized in potato under drought stress conditions with downregulation of miR811 coupled with the upregulation of its target (Zhang et al. 2014a, b). Even though there is no study suggesting the miRNA/MYB association in wheat and barley, it seems reasonable to propose that miRNAs in wheat and barley may regulate the abiotic stress response in a similar manner to that observed in dicots.
Another widespread TF in the plant kingdom with regulatory roles in many pathways including growth, environmental adaptation, and stress response is DEHYDRATION RESPONSIVE ELEMENT BINDING (DREB) family members (Licausi et al. 2013). DREB1 and DREB2 are integral components of the ABA-independent signal transduction pathway that initiates downstream stress responses in plants (Agarwal et al. 2006). Although the co-regulation of certain DREB family members’ expression together with miRNAs has been observed under drought stress (Hackenberg et al. 2014), DREB TFs have not been predicted or validated as miRNA targets to date, in contrast to ARF, NAC, SPL, and MYB TFs (Qiao-Ying et al. 2012). Despite some evidence for miRNA-MYB and miRNA-DREB association under abiotic stress conditions, details of fundamental mechanisms are required.
miRNA and siRNA interaction during an abiotic stress response
Small interfering RNAs (siRNAs) are members of the RNAi mechanism and resemble miRNAs in many aspects despite differences in their biogenesis and target preferences (Axtell 2013). Plant miRNAs may interact with siRNAs under abiotic stress and provide a comprehensive response network. miRNA and siRNA relationships may involve both co-regulation or direct triggering of particular miRNA/siRNA in response to several stresses. Co-regulation of miRNAs and siRNAs was detected in Arabidopsis under drought stress. siRNAs generated from the cis-natural antisense transcript of drought-responsive NFYA5, NERF, were as effective as miR169-mediated repression of NFYA5 (Gao et al. 2015). Elucidating such co-regulatory miRNA and siRNA associations might improve the likelihood of manipulating plant metabolism to generate more stress-tolerant plant varieties.
In addition to the co-regulatory networks, miRNAs can also trigger the biogenesis of phased siRNAs from TAS loci. These siRNAs, called trans-acting siRNAs or ta-siRNAs, are produced by DCL4 from dsDNA that is generated by the action of Suppressor of Gene Silencing 3 (SGS3) on the miRNA-cleaved TAS transcript following its transcription by RNA-dependent RNA polymerase 6 (RDR6) (Liu et al. 2014; Rock 2013). The miRNA-mediated cleavage of the TAS gene transcript sets the registration point for phasing. Accurate phasing is crucial for the targeting ability of the resulting siRNAs (Howell et al. 2007). Phased ta-siRNAs triggered by miRNAs may act on related genes affecting their biogenesis (Allen et al. 2005). Therefore, the most prevalent mode of ta-siRNA biogenesis, the two-hit trigger model, may involve a single miRNA cleaving the TAS transcript at two distinct sites or a miRNA acting together with the ta-siRNAs originating from the same locus (Rajeswaran et al. 2012).
In Arabidopsis, four miRNA-targeted TAS loci have been described; TAS1 and TAS2 are targeted by miR173 (Allen et al. 2005; Rajeswaran et al. 2012), TAS3 targeted by miR390 (Allen et al. 2005), and TAS4 targeted by miR828 (Rajagopalan et al. 2006). While TAS3 and TAS4 loci spawn ta-siRNAs targeting ARF and MYB TFs, respectively (Allen et al. 2005), phased ta-siRNAs generated from TAS1 and TAS2 loci have been associated with the regulation of Pentatricopeptide Repeat (PPR) genes (Howell et al. 2007; Xia et al. 2013). In plants, the PPR family is a major gene family comprising several hundred members, although only a few have been characterized so far. Most PPR proteins localize to mitochondria and chloroplasts where they regulate the expression of organellar transcripts (Saha et al. 2007). In addition to their roles in developmental processes, PPR proteins are also involved in biotic and abiotic stress responses (Laluk et al. 2011; Tan et al. 2014; Yuan & Liu 2012). Overexpression of PPR40/At3g16890 enhanced salinity tolerance in Arabidopsis (Zsigmond et al. 2012). Similarly, TAS1-ta-siRNA targets, At4g29760, At4g29770, and At5g18040 (designated as Heat Induced TAS1 Target genes, HTT4, HTT1 and HTT2, respectively), were responsive to heat stress and downregulation of these targets reduced heat tolerance in Arabidopsis (Li et al. 2014). Of these targets, At4g29760 and At5g18040, along with another potential TAS1-ta-siRNA target, At1g51670, also showed contrasting expression levels compared to TAS1 originating ta-siRNAs in response to cold stress (Kume et al. 2010). This result suggests that miRNA-triggered ta-siRNA generation from the TAS1 loci can contribute to stress responses.
Targeted by TAS4 originating ta-siRNAs, PAP1/MYB75 and PAP2/MYB90 TFs are involved in anthocyanin/flavonoid biosynthesis (Rajagopalan et al. 2006). Flavonoid accumulation has been linked to enhanced abiotic stress tolerance through ROS scavenging (Nakabayashi et al. 2014; Ilk et al. 2015). The ARF family TFs, of which ARF1-4 is targeted by phased ta-siRNAs processed from TAS3 loci, mediate auxin signaling during a range of developmental processes although differential expression patterns of certain members have been observed also in response to abiotic stress conditions (Aglawe et al. 2012; Allen et al. 2005; Matsui et al. 2014). Importantly, the involvement of multiple small RNAs in the biogenesis of ta-siRNAs has been suggested as a tool to restrain ta-siRNA-mediated silencing of endogenous transcripts (Howell et al. 2007). Stress conditions often impede normal developmental processes and arrest growth. Therefore, it is tempting to speculate that miRNA-triggered ta-siRNAs provide a network of stress response regulation, so that endogenous transcripts with crucial roles in development and growth are not silenced for too long.
miRNA inheritance
Acquired tolerance to abiotic and biotic stresses across generations constitutes a pre-defense mechanism for the progeny of the stressed plant (Molinier et al. 2006; Kou et al. 2011; Ou et al. 2012). As a candidate mediator of this resistance strategy, inheritance of microRNA expression is a new and exciting research avenue. The transmission of acquired stress response mediators to the next generation would create a strong positive selection advantage by providing a reversible regulatory mechanism to the progeny. Several lines of evidence suggested that miRNAs, along with siRNAs, might be epigenetic vectors for establishing this trans-generational stress memory. Small RNAs may inherit the parental transcriptional states either by triggering DNA methylation (transcriptionally) or by being transferred to the zygote through uptake by the germ line cells (post-transcriptionally) (Castel and Martienssen 2013; Rechavi 2014).
To date, siRNAs have been the major RNAi actors in terms of inheritance as they play a critical role in maintaining genome integrity by immobilizing transposable elements through RNA-directed DNA methylation (RdDM) (Matzke et al. 2009), a process which was detected in pollen and embryo, in addition to somatic cells (Calarco et al. 2012). Evidence for the developmental role of parentally inherited mRNAs for the development in Drosophila melanogaster (Chang et al. 2011; Johnson and Spence 2011) suggested a putative and functionally important transfer mechanism for sRNAs to progeny through the germ line. Additionally, the mobility of miRNAs and siRNAs between different cells and even in different tissues of plants further supports this idea (Molnar et al. 2010; Carlsbecker et al. 2011; Melnyk et al. 2011). Nevertheless, epiallele inheritance in plants indicates that this reprogramming is speculative and many details require clarification (Riddle 2014).
TE silencing activity of siRNAs has been shown in Arabidopsis pollen. TE-derived siRNAs, produced in the vegetative nucleus (germ-line supporting companion cells) which lack the heterochromatin remodeler DDM1 protein (DECREASE IN DNA METHYLATION 1), have been detected in sRNA libraries of sperm cells. These siRNAs were proposed to silence TE activity in the sperm (Slotkin et al. 2009). In another study, the miRNA repertoire of Arabidopsis pollen was examined by pyrosequencing and qRT-PCR experiments and miR173 cleavage products confirmed the functionality of the miRNA machinery inside the pollen. Among the sperm miRNAs, miR159a was enriched more than fivefold compared to the total pollen expression levels (Grant-Downton et al. 2009; Borges et al. 2011). Though research in plant systems is lagging, maternally inherited miRNAs in D. melanogaster, zebrafish, and mouse have been demonstrated (Soni et al. 2013; Lee et al. 2014). Support for functionality of the miRNA machinery during reproduction was found by Olmedo-Monfil and her colleagues who found AGO9 (At5g21150) mRNA expression and immunolocalization in somatic companion cells of Arabidopsis and vegetative cells in the pollen grain but not in the associated gamete cells. Moreover, sRNA sequencing of Ago9 co-immunoprecipitated sRNAs showed their TE origin and targeting to the egg cell. These findings suggested Ago9 TE silencing activity in plants as in invertebrates and mammals. Mutants lacking Ago9, SUPPRESSOR OF GENE SILENCING 3 and RNA-DEPENDENT RNA POLYMERASE 6, all involved in sRNA silencing, revealed the importance of sRNA transfer for normal gametogenesis (Olmedo-Monfil et al. 2010). Likewise, transcriptome analysis of Arabidopsis egg cells showed the upregulation in AGO9 expression along with other proteins that have roles in the RNAi pathway: AGO1, AGO2, AGO5, DCL1, HYL1, AT4G00420 (RNASE THREE-LIKE PROTEIN 1 paralog), and AT5G21030 (AGO8), indicating sRNA-mediated epigenetic regulation is an ongoing process in the female gametes (Wuest et al. 2010). Shotgun bisulfide sequencing in companion cells (central cell and vegetative cell) of Arabidopsis egg and sperm also indicated a role for small RNAs from neighboring cells, as TE silencers in gametes. In the same study, DEMETER (DME) DNA glycosylase expression was upregulated while Met1 methyltransferase was downregulated in diploid central cells. DME DNA glycosylase-mediated demethylation in the companion cells results in elevated sRNA levels which subsequently percolate into gametes to control TE activity by CG methylation (Ibarra et al. 2012). Moreover, sRNA transfer to the gametes was shown to be facilitated by the iso-miRNAs (isomiRs) of known miRNAs. IsomiRs of some canonical miRNAs: miR156h, miR167d, miR173, miR393, miR447, miR771, miR773, miR825*, and miR828 are preferentially accumulated inside the pollen and sperm cells. Among them, 22-nt-long isomiRs with C extensions at the 3′- end are proposed as triggers for siRNA production such as miR156h which is found to associate with AGO5, rather than AGO1 as its canonical counterpart (Ebhardt et al. 2010; Borges et al. 2011). In the male germline of Arabidopsis, the numbers of such isomiRs are shown to be increased together with their associated ARGONAUTE family members which is suitable for related isomiR generation. While AGO1 and AGO10, the major miRNA-directed mRNA cleavage mediators, are downregulated, a phylogenetically close relative, AGO5, and TE silencing mediator, AGO9, are significantly upregulated in the sperm cells of Arabidopsis. Abundance of AGO5-associated isomiRs of miR156, miR158, and miR845 in the pollen and sperm, combined with the findings described above, suggests a novel and intricate miRNA-mediated regulation mechanism for reproductive cells relative to somatic cells (Borges et al. 2011).
sRNA translocation to the gametes has been shown to occur to some extent, and there are several examples for transgenerationally inherited resistance memory of abiotic stress in plants (Molinier et al. 2006; Kathiria et al. 2010; Boyko and Kovalchuk 2010). However, the molecular mechanisms of inheritance and the persistency of this memory over generations have not been explained. As previously stated, transgenerational silencing effects of small RNAs can be observed both post-transcriptionally or transcriptionally via RdDM and chromatin modification (Castel and Martienssen 2013). In the second route, inheritance of silencing was shown in Caenorhabditis elegans that were fed bacteria expressing dsRNA which target host mRNA transcripts. siRNA-mediated DNA methylation that produces the H3K9me3 chromatin mark was inherited to the progeny of the organisms exposed to dsRNA. As H3K9 trimethylation appeared after the intermediatory siRNAs were detected in a time-course experiment, it was suggested that the chromatin mark is re-established by small RNAs in the next generation rather than direct inheritance (Burton et al. 2011). Several RNAi pathway elements were found to be essential for this inheritance (Grishok et al. 2000). Likewise, RNAi-mediated transcriptional gene silencing through methylation of specific cytosines after infection with plant RNA virus was shown to be heritable. Though not necessary for initiation, inheritance of this RdDM mechanism was dependent on Met1 methyltransferase expression (Jones et al. 2001). Accordingly, it was proposed that an effective trans-generational silencing mechanism must amplify the inherited small RNA to overcome the dilution effect during gametogenesis. In line with this hypothesis, long-term RNAi inheritance was hindered by RNA-dependent RNA polymerase (RdRP) RRF-1 defective C. elegans mutants. Though RdRP-mediated siRNA amplification does not continue this silencing indefinitely, Alcazar et al. suggested that either the siRNA levels fail to meet a threshold after each generation or another signal which controls the amplification is diluted over generations (Alcazar et al. 2008).
A recent study on Brassica rapa plants delineated the role of the miRNA-mediated transgenerational inheritance in the abiotic stress response. Transcriptome and small RNAome sequencing of somatic and reproductive tissues of heat-shock-stressed plants and their non-treated progeny were analyzed. Strikingly, the highest expression fluctuations were seen in the sRNAome of the progeny and reproductive tissues which were not directly exposed to the stress. Common miRNA families that are differentially expressed in pollen, endosperm, and leaf tissues of treated parents and non-treated progenies included bra-miR167, bra-miR390, and bra-miR168. As the braAGO1 expression levels were also significantly different between embryos of stressed and control plants, AGO1-regulating activity of miR168 highlighted the potential role of this miRNA as a putative epigenetic stress memory messenger (Bilichak et al. 2015). Based on this information, it is tempting to speculate the miRNA inheritance via gametes might be possible in plants and could be used to improve stress tolerance.
Conclusion and future remarks
Abiotic stresses are major limitations to crop production and represent significant obstacles to maintenance of adequate food supplies. To meet the demands of an increasing world population, we need varieties that show high, stable yield across diverse environments. Over the two last decades, there has been considerable advance in the identification of stress-responsive genes and their associated proteins. Modification of the expression of these genes has shown promising results for improving the stress tolerance of wheat and barley. However, bringing these lines to farmers’ fields has been a problem since no transgenic wheat or barley lines have been commercially released and consumer resistance to the application of this technology has not diminished. Emphasis has shifted to the use of variation within the cultivated, land race and wild germplasm pools for these crops and in exploring the modification of regulatory pathways (Tester and Langridge 2010; Langridge and Reynolds 2015). miRNAs have emerged as exciting targets for enhancing the abiotic stress tolerance of crops. A clear role for miRNAs in regulating stress tolerance has been demonstrated, and there is now good evidence that variation exists in the level and timing of miRNA expression providing an opportunity for selection for specific alleles or variants in breeding programs. However, despite the great progress that has been made, many issues regarding the function and utilization of miRNAs in crop improvement require clarification. Some of the major issues facing the further development and application of miRNA to crop improvement are:
Existing studies aimed at using miRNAs to improve crop tolerance to abiotic stresses have focused on the identification of differentially expressed miRNA families. Through these efforts, several comprehensive miRNA databases have been developed. However, the targets for many of miRNAs have not been established. Additionally, the pattern of regulation for many of these differentially expressed miRNAs has been only poorly characterized. Since miRNA-based regulation of gene expression relies on the translational inhibition of mRNA targets, fully characterized targets of miRNAs provide a route to the manipulation of plant metabolism. Therefore, studies need to focus on the relationship between miRNA and their target mRNA.
Although only a few miRNA/target mRNA relationships have been well characterized, several important examples from wheat and barley have been described as outlined above. However, the signaling cascades that transmit the stress-associated signal to the synthesis of miRNAs and regulate the differential expression patterns are largely unknown. Deep and comprehensive understanding of these signaling cascades may provide new routes to the manipulation of miRNA-based abiotic stress regulation. The relationship between stress-associated pathways and stress-responsive miRNAs needs to be elucidated together with their physiological and biochemical effects on cell metabolism and plant development.
Domestication of the Triticeae crops resulted with the loss of some stress-associated genes and alleles as well as genomic loci associated with the maintenance of miRNA expression. The wild relatives and land races of these species provide a huge source of novel diversity (Able et al. 2007). For instance, the wheat genome is derived from three different species (Triticum urartu, Aegilops speltoides, and Aegilops tauschii) and its ancestors are still present in nature (Pont et al. 2013). To date, little effort has been made to identify abiotic stress-associated miRNA from the wild relatives and land races although we know these species are valuable sources of variation for structural genes (Kantar et al. 2011a; Akpinar et al. 2015; Liu et al. 2015a; Akpinar and Budak 2016). New sequencing technologies and breeding strategies offer great improvements in both the discovery and use of variation in miRNA sequence and expression pattern from unadapted lines. However, introgression of novel variation from the relatives of wheat and barley remains difficult and time consuming. This difficulty may be overcome through novel gene-editing technologies since these methods should allow us to induce small sequence changes of alterations in the expression pattern of a given miRNA. A detailed knowledge of the miRNA sequence and expression from Triticeae relatives that have adapted to adverse environments should therefore provide targets for editing the miRNA coding sequences in wheat and barley.
Although miRNAs have great potential for crop improvement, other small RNA species may also be used to enhance abiotic stress tolerance. The association of small RNAs such as siRNAs, snoRNAs, or snRNAs with stress tolerance opens additional opportunities for modifying regulatory pathways. The interaction between miRNAs with other small RNA species may confer new approaches to improving abiotic stress tolerance. However, our knowledge of these mechanisms and processes is still in its infancy and more work is needed.
Rye is an important member of the Triticeae and is particularly tolerant to environmental extremes. The high diversity and stress tolerance of this crop are likely due to its outbreeding nature compared to the strong inbreeding system in wheat and barley (Martis et al. 2013). Importantly, rye can be readily crossed with wheat and the wheat/rye crop, Triticale, has been successful in many regions where wheat and barley are marginal. However, we know little about the mechanisms of abiotic stress tolerance in rye and virtually nothing about the role of miRNA in this species. There is clearly scope to explore the role of small RNAs from rye and in regulating stress tolerance and root development and then applying these results to rye’s close relatives, wheat and barley.
Footnotes
This article forms part of a special issue of Functional & Integrative Genomics entitled “miRNA in model and complex organisms” (Issue Editors: Hikmet Budak and Baohong Zhang).
References
- Able JA, Langridge P, Milligan AS. Capturing diversity in the cereals: many options but little promiscuity. Trends Plant Sci. 2007;12:71–79. doi: 10.1016/j.tplants.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Abrol YP, Chatterjee SR, Kumar PA, Jain V. Improvement in nitrogen use efficiency: physiological and molecular approaches. Curr Sci. 2011;76:1357–1364. [Google Scholar]
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006;25:1263–1274. doi: 10.1007/s00299-006-0204-8. [DOI] [PubMed] [Google Scholar]
- Aglawe SB, Fakrudin B, Patole CB, et al. Quantitative RT-PCR analysis of 20 transcription factor genes of MADS, ARF, HAP2, MBF and HB families in moisture stressed shoot and root tissues of sorghum. Physiol Mol Biol Plants. 2012;18:287–300. doi: 10.1007/s12298-012-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar BA, Budak H. Dissecting miRNAs in wheat D genome progenitor, Aegilops tauschii. Front Plant Sci. 2016;7:1–17. doi: 10.3389/fpls.2016.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar BA, Avsar B, Lucas SJ, Budak H. Plant abiotic stress signaling. Plant Signal Behav. 2012;7:1450–1455. doi: 10.4161/psb.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar BA, Lucas SJ, Budak H. Genomics approaches for crop improvement against abiotic stress. Sci World J. 2013 doi: 10.1155/2013/361921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar BA, Kantar M, Budak H. Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct Integr Genomics. 2015 doi: 10.1007/s10142-015-0453-0. [DOI] [PubMed] [Google Scholar]
- Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Alptekin B, Budak H. Wheat miRNA ancestors: evident by transcriptome analysis of A, B, and D genome donors. Funct Integr Genomics. 2016 doi: 10.1007/s10142-016-0487-y. [DOI] [PubMed] [Google Scholar]
- Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asseng S, Ewert F, Martre P, et al. Rising temperatures reduce global wheat production. Nat Clim Chang. 2014;5:143–147. doi: 10.1038/nclimate2470. [DOI] [Google Scholar]
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- Baek D, Kim MC, Chun HJ, et al. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 2013;161:362–373. doi: 10.1104/pp.112.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Park HC, Kim MC, Yun D-J. The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal Behav. 2013;8:e23488. doi: 10.4161/psb.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni E, Genga A, Cominelli E, et al. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci. 2015;5:1–16. doi: 10.3389/fpls.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini E, Verelst W, Horner DS, et al. Addressing the role of microRNAs in reprogramming leaf growth during drought stress in brachypodium distachyon. Mol Plant. 2013;6:423–443. doi: 10.1093/mp/sss160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj AR, Joshi G, Pandey R, et al. A genome-wide perspective of miRNAome in response to high temperature, salinity and drought stresses in Brassica juncea (Czern) L. PLoS One. 2014;9:e92456. doi: 10.1371/journal.pone.0092456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak A, Ilnytskyy Y, Wóycicki R, et al. The elucidation of stress memory inheritance in Brassica rapa plants. Front Plant Sci. 2015;6:5. doi: 10.3389/fpls.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita CE, Gerats T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Mateos JL, Bresso EG, Palatnik JF. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009;28:3646–3656. doi: 10.1038/emboj.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Pereira PA, Slotkin RK, et al. MicroRNA activity in the Arabidopsis male germline. J Exp Bot. 2011;62:1611–1620. doi: 10.1093/jxb/erq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A, Kovalchuk I. Transgenerational response to stress in Arabidopsis thaliana. Plant Signal Behav. 2010;5:995–998. doi: 10.4161/psb.5.8.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozorov TA, Baldwin IT, Kim S-G. Identification and profiling of miRNAs during herbivory reveals jasmonate-dependent and -independent patterns of accumulation in Nicotiana attenuata. BMC Plant Biol. 2012;12:209. doi: 10.1186/1471-2229-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J-F, Dubos C, Gaymard F. Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 2015;20:33–40. doi: 10.1016/j.tplants.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Broadley MR, White PJ, Hammond JP, et al. Zinc in plants: Tansley review. New Phytol. 2007;173:677–702. doi: 10.1111/j.1469-8137.2007.01996.x. [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015;20:124–133. doi: 10.1016/j.tplants.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Budak H, Akpinar A. Dehydration stress-responsive miRNA in Brachypodium distachyon: evident by genome-wide screening of microRNAs expression. OMICS. 2011;15:791–799. doi: 10.1089/omi.2011.0073. [DOI] [PubMed] [Google Scholar]
- Budak H, Akpinar BA. Plant miRNAs: biogenesis, organization and origins. Funct Integr Genomics. 2015 doi: 10.1007/s10142-015-0451-2. [DOI] [PubMed] [Google Scholar]
- Budak H, Akpinar BA, Unver T, Turktas M. Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI-MS/MS. Plant Mol Biol. 2013;83:89–103. doi: 10.1007/s11103-013-0024-5. [DOI] [PubMed] [Google Scholar]
- Budak H, Kantar M, Yucebilgili Kurtoglu K. Drought tolerance in modern and wild wheat. Sci World J. 2013 doi: 10.1155/2013/548246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H, Khan Z, Kantar M (2014) History and current status of wheat miRNAs using next-generation sequencing and their roles in development and stress. Brief Funct Genomics elu021–. doi: 10.1093/bfgp/elu021 [DOI] [PubMed]
- Budak H, Hussain B, Khan Z, et al. From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front Plant Sci. 2015;6:1012. doi: 10.3389/fpls.2015.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H, Kantar M, Bulut R, Akpinar BA. Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 2015;235:1–13. doi: 10.1016/j.plantsci.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Budak H, Bulut R, Kantar M, Alptekin B (2015a) MicroRNA nomenclature and the need for a revised naming prescription. Brief Funct Genomics elv026. doi: 10.1093/bfgp/elv026 [DOI] [PubMed]
- Buhtz A, Pieritz J, Springer F, Kehr J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 2010;10:64. doi: 10.1186/1471-2229-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci. 2011;108:19683–19688. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MTA, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee J-Y, Roberts CJ, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Cell. 2011;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Nashchekin D, Wheatley L, et al. Anterior-posterior axis specification in drosophila oocytes: identification of novel bicoid and Oskar mRNA localization factors. Genetics. 2011;188:883–896. doi: 10.1534/genetics.111.129312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, et al. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta Gene Regul Mech. 2012;1819:120–128. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M, Lamontanara A, Bernardo L, et al. On the complexity of miRNA-mediated regulation in plants: novel insights into the genomic organization of plant miRNAs. Biol Direct. 2012;7:15. doi: 10.1186/1745-6150-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Urano K, Delrot S, et al. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Spriggs A, Taylor J, et al. miRNA regulation in the early development of barley seed. BMC Plant Biol. 2012;12:120. doi: 10.1186/1471-2229-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat J-F. Iron transport and signaling in plants. Annu Rev Cell Dev Biol. 2002;18:53–80. doi: 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- Dawson IK, Russell J, Powell W, et al. Barley: a translational model for adaptation to climate change. New Phytol. 2015;206:913–931. doi: 10.1111/nph.13266. [DOI] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, et al. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19:371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P, Wang L, Cui L, et al. Global identification of microRNAs and their targets in barley under salinity stress. PLoS One. 2015;10:e0137990. doi: 10.1371/journal.pone.0137990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Fedynak A, Fahlman RP. Naturally occurring variations in sequence length creates microRNA isoforms that differ in argonaute effector complex specificity. Silence. 2010;1:12. doi: 10.1186/1758-907X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergen NZ, Budak H. Sequencing over 13 000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ. 2009;32:220–236. doi: 10.1111/j.1365-3040.2008.01915.x. [DOI] [PubMed] [Google Scholar]
- Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H. Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics. 2009;9:377–396. doi: 10.1007/s10142-009-0123-1. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy AA. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192:763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FFDE, Schneeberger K, Dezulian T, et al. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA. 2008;14:2455–2459. doi: 10.1261/rna.1149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhang Q, Wang Q, et al. Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol Biol. 2013;83:433–443. doi: 10.1007/s11103-013-0101-9. [DOI] [PubMed] [Google Scholar]
- Feng H, Duan X, Zhang Q, et al. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol Plant Pathol. 2014;15:284–296. doi: 10.1111/mpp.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous J, Hussain SS, Shi BJ. Role of microRNAs in plant drought tolerance. Plant Biotechnol J. 2015;13:293–305. doi: 10.1111/pbi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Su Y, Min J, et al. Transgenic tomato overexpressing ath-miR399d has enhanced phosphorus accumulation through increased acid phosphatase and proton secretion as well as phosphate transporters. Plant Soil. 2010;334:123–136. doi: 10.1007/s11104-009-0219-3. [DOI] [Google Scholar]
- Gao W, Liu W, Zhao M, Li W-X. NERF encodes a RING E3 ligase important for drought resistance and enhances the expression of its antisense gene NFYA5 in Arabidopsis. Nucleic Acids Res. 2015;43:607–617. doi: 10.1093/nar/gku1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Guo C, Zhang Y, et al. Wheat microRNA member tamir444a is nitrogen deprivation-responsive and involves plant adaptation to the nitrogen-starvation stress. Plant Mol Biol Report. 2016 [Google Scholar]
- Giacomelli JI, Weigel D, Chan RL, Manavella PA. Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 2012;195:766–773. doi: 10.1111/j.1469-8137.2012.04259.x. [DOI] [PubMed] [Google Scholar]
- Gim J-A, Ha H-S, Ahn K, et al. Genome-wide identification and classification of microRNAs derived from repetitive elements. Genomics Inform. 2014;12:261–267. doi: 10.5808/GI.2014.12.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R, Le Trionnaire G, Schmid R, et al. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics. 2009;10:643. doi: 10.1186/1471-2164-10-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Gupta B, Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics. 2014;2014:701596. doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta OP, Meena NL, Sharma I, Sharma P. Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat. Mol Biol Rep. 2014;41:4623–4629. doi: 10.1007/s11033-014-3333-0. [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Shi B-J, Gustafson P, Langridge P. Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol. 2013;13:214. doi: 10.1186/1471-2229-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg MI, Huang POUNG, Huang CHUNUAN, et al. (2013b) A comprehensive expression profile of microRNAs and other classes of non-coding small RNAs in barley under phosphorous-deficient and -sufficient conditions 109–125 [DOI] [PMC free article] [PubMed]
- Hackenberg M, Gustafson P, Langridge P, Shi BJ (2014) Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol J. 2–13. doi: 10.1111/pbi.12220 [DOI] [PMC free article] [PubMed]
- Hall JL. Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot. 2002;53:1–11. doi: 10.1093/jxb/53.366.1. [DOI] [PubMed] [Google Scholar]
- Hamamoto S, Horie T, Hauser F, et al. HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol. 2015;32:113–120. doi: 10.1016/j.copbio.2014.11.025. [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M, Nahar K, Alam MM, et al. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int J Mol Sci. 2013;14:9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield JL, Prueger JH. Temperature extremes: effect on plant growth and development. Weather Clim Extrem. 2015;10:4–10. doi: 10.1016/j.wace.2015.08.001. [DOI] [Google Scholar]
- Hatfield JL, Boote KJ, Kimball BA, et al. Climate impacts on agriculture: implications for crop production. Agron J. 2011;103:351–370. doi: 10.2134/agronj2010.0303. [DOI] [Google Scholar]
- Högy P, Poll C, Marhan S, et al. Impacts of temperature increase and change in precipitation pattern on crop yield and yield quality of barley. Food Chem. 2013;136:1470–1477. doi: 10.1016/j.foodchem.2012.09.056. [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilk N, Ding J, Ihnatowicz A, et al. Natural variation for anthocyanin accumulation under high-light and low-temperature stress is attributable to the ENHANCER OF AG-4 2 (HUA2) locus in combination with PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2. New Phytol. 2015;206:422–435. doi: 10.1111/nph.13177. [DOI] [PubMed] [Google Scholar]
- IPCC (2014) Climate change 2014 synthesis report. Contrib Work Groups I, II III to Fifth Assess Rep Intergov Panel Clim Chang 1–151
- Jeong DH, Green PJ. The role of rice microRNAs in abiotic stress responses. J Plant Biol. 2013;56:187–197. doi: 10.1007/s12374-013-0213-4. [DOI] [Google Scholar]
- Jiang S, Lu Y, Li K, et al. Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol Plant Pathol. 2014;15:907–917. doi: 10.1111/mpp.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Spence AM. Epigenetic licensing of germline gene expression by maternal RNA in C. elegans. Science. 2011;333:1311–1314. doi: 10.1126/science.1208178. [DOI] [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe DC. RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol. 2001;11:747–757. doi: 10.1016/S0960-9822(01)00226-3. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Jones-rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Plant Biol. 2011;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kantar M, Unver T, Budak H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics. 2010;10:493–507. doi: 10.1007/s10142-010-0181-4. [DOI] [PubMed] [Google Scholar]
- Kantar M, Lucas SJ, Budak H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta. 2011;233:471–484. doi: 10.1007/s00425-010-1309-4. [DOI] [PubMed] [Google Scholar]
- Kantar M, Lucas SJ, Budak H. Drought stress. Molecular genetics and genomics approaches. Adv Bot Res. 2011;57:445–493. doi: 10.1016/B978-0-12-387692-8.00013-8. [DOI] [Google Scholar]
- Kathiria P, Sidler C, Golubov A, et al. Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 2010;153:1859–1870. doi: 10.1104/pp.110.157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, et al. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y. Differential expression of microRNAs in maize inbred and hybrid lines during salt and drought stress. Am J Plant Sci. 2010;01:69–76. doi: 10.4236/ajps.2010.12009. [DOI] [Google Scholar]
- Kong WW, Yang ZM. Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol Biochem. 2010;48:153–159. doi: 10.1016/j.plaphy.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Kou HP, Li Y, Song XX, et al. Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.) J Plant Physiol. 2011;168:1685–1693. doi: 10.1016/j.jplph.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Kraiser T, Gras DE, Gutiérrez AG, et al. A holistic view of nitrogen acquisition in plants. J Exp Bot. 2011;62:1455–1466. doi: 10.1093/jxb/erq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka K, Pacak A, Swida-Barteczka A, et al. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J Exp Bot. 2014;65:6123–6135. doi: 10.1093/jxb/eru353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Singh D, Kanodia P, et al. Discovery of novel leaf rust responsive microRNAs in wheat and prediction of their target genes. J Nucleic Acids. 2014 doi: 10.1155/2014/570176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RR, Pathak H, Sharma SK, et al. Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.) Funct Integr Genomics. 2014;15:323–348. doi: 10.1007/s10142-014-0421-0. [DOI] [PubMed] [Google Scholar]
- Kume K, Tsutsumi K-I, Saitoh Y. TAS1 trans-acting siRNA targets are differentially regulated at low temperature, and TAS1 trans-acting siRNA mediates temperature-controlled At1g51670 expression. Biosci Biotechnol Biochem. 2010;74:1435–1440. doi: 10.1271/bbb.100111. [DOI] [PubMed] [Google Scholar]
- Kuo H-F, Chiou T-J. The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 2011;156:1016–1024. doi: 10.1104/pp.111.175265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtoglu KY, Kantar M, Budak H. New wheat microRNA using whole-genome sequence. Funct Integr Genomics. 2014;14:363–379. doi: 10.1007/s10142-013-0357-9. [DOI] [PubMed] [Google Scholar]
- Kutter C, Schöb H, Stadler M, et al. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuoglu-Ozturk D, Yalcinkaya OC, Akpinar BA, et al. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta. 2012;236:1081–1092. doi: 10.1007/s00425-012-1657-3. [DOI] [PubMed] [Google Scholar]
- Laluk K, Abuqamar S, Mengiste T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 2011;156:2053–2068. doi: 10.1104/pp.111.177501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge P, Reynolds MP. Genomic tools to assist breeding for drought tolerance. Curr Opin Biotechnol. 2015;32:130–135. doi: 10.1016/j.copbio.2014.11.027. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Choi Y, Kim K, et al. Adenylation of maternally inherited microRNAs by Wispy. Mol Cell. 2014;56:696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, et al. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-X, Oono Y, Zhu J, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li C, Xia J, Jin Y. Domestication of transposable elements into microrna genes in plants. PLoS One. 2011 doi: 10.1371/journal.pone.0019212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo G, Guo W, et al. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.) BMC Plant Biol. 2012;12:220. doi: 10.1186/1471-2229-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Duan H, Li J, et al. Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol Biol. 2013;81:525–539. doi: 10.1007/s11103-013-0010-y. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Shi D, et al. Spatial-temporal analysis of zinc homeostasis reveals the response mechanisms to acute zinc deficiency in Sorghum bicolor. New Phytol. 2013;200:1102–1115. doi: 10.1111/nph.12434. [DOI] [PubMed] [Google Scholar]
- Li S, Liu J, Liu Z, et al. HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell. 2014;26:1764–1780. doi: 10.1105/tpc.114.124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Yu D. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One. 2012 doi: 10.1371/journal.pone.0048951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199:639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- Liu Q. Novel miRNAs in the control of arsenite levels in rice. Funct Integr Genomics. 2012;12:649–658. doi: 10.1007/s10142-012-0282-3. [DOI] [PubMed] [Google Scholar]
- Liu H-H, Tian X, Li Y-J, et al. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia L, Wang H, He Y. HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J Exp Bot. 2011;62:4367–4381. doi: 10.1093/jxb/err167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YX, Wang M, Wang XJ. Endogenous small RNA clusters in plants. Genomics, Proteomics Bioinformatics. 2014;12:64–71. doi: 10.1016/j.gpb.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Searle IR, Watson-Haigh NS, et al. Genome-wide identification of microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. PLoS One. 2015;10:e0142799. doi: 10.1371/journal.pone.0142799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Feng L, Li J, He Z. Genetic and epigenetic control of plant heat responses. Front Plant Sci. 2015;06:267. doi: 10.3389/fpls.2015.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Able AJ, Able JA (2016) Water-deficit stress-responsive microRNAs and their targets in four durum wheat genotypes. Funct Integr Genomics 1–15. doi: 10.1007/s10142-016-0515-y [DOI] [PubMed]
- Löw D, Brändle K, Nover L, Forreiter C. Cytosolic heat-stress proteins Hsp17.7 class I and Hsp17.3 class II of tomato act as molecular chaperones in vivo. Planta. 2000;211:575–582. doi: 10.1007/s004250000315. [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12:2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan M, Xu M, Lu Y, et al. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene. 2015;555:178–185. doi: 10.1016/j.gene.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Lucas SJ, Budak H. Sorting the wheat from the Chaff: identifying miRNAs in genomic survey sequences of Triticum aestivum chromosome 1AL. PLoS One. 2012 doi: 10.1371/journal.pone.0040859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S, Dogan E, Budak H. TMPIT1 from wild emmer wheat: first characterisation of a stress-inducible integral membrane protein. Gene. 2011;483:22–28. doi: 10.1016/j.gene.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Lucas S, Durmaz E, Akpnar BA, Budak H. The drought response displayed by a DRE-binding protein from Triticum dicoccoides. Plant Physiol Biochem. 2011;49:346–351. doi: 10.1016/j.plaphy.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Lv S, Nie X, Wang L, et al. Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by high-throughput sequencing. Int J Mol Sci. 2012;13:2973–2984. doi: 10.3390/ijms13032973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Xin Z, Wang Z, et al. Identification and comparative analysis of differentially expressed miRNAs in leaves of two wheat (Triticum aestivum L.) genotypes during dehydration stress. BMC Plant Biol. 2015;15:21. doi: 10.1186/s12870-015-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16:510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Hwang Z. Modified expression of a carrot small heat shock protein gene, hsp17. 7, results in increased or decreased thermotolerance double dagger. Plant J. 1999;20:89–99. doi: 10.1046/j.1365-313X.1999.00581.x. [DOI] [PubMed] [Google Scholar]
- Manara A (2012) Plants and heavy metals. Signal Transduct 27–54. doi: 10.1007/978-94-007-4441-7
- Martis MM, Zhou R, Haseneyer G, et al. Reticulate evolution of the rye genome. Plant Cell. 2013;25:3685–3698. doi: 10.1105/tpc.113.114553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Mizunashi K, Tanaka M, et al. tasiRNA-ARF pathway moderates floral architecture in Arabidopsis plants subjected to drought stress. Biomed Res Int. 2014;2014:303451. doi: 10.1155/2014/303451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, et al. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Martis M, Hedley PE, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Waugh R, Langridge P, et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Molnar A, Bassett A, Baulcombe DC. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol. 2011;21:1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16:237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Minibayeva F, Beckett RP, Kranner I. Roles of apoplastic peroxidases in plant response to wounding. Phytochemistry. 2015;112:122–129. doi: 10.1016/j.phytochem.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Unlocking Triticeae genomics to sustainably feed the future. Plant Cell Physiol. 2013;54:1931–1950. doi: 10.1093/pcp/pct163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- Müller-Xing R, Xing Q, Goodrich J. Footprints of the sun: memory of UV and light stress in plants. Front Plant Sci. 2014;5:474. doi: 10.3389/fpls.2014.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, et al. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta Gene Regul Mech. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Naqvi AR, Sarwat M, Hasan S, Roychodhury N. Biogenesis, functions and fate of plant microRNAs. J Cell Physiol. 2012;227:3163–3168. doi: 10.1002/jcp.24052. [DOI] [PubMed] [Google Scholar]
- Nezhadahmadi A, Prodhan ZH, Faruq G. Drought tolerance in wheat. ScientificWorldJournal. 2013;2013:610721. doi: 10.1155/2013/610721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol. 2013;82:113–129. doi: 10.1007/s11103-013-0040-5. [DOI] [PubMed] [Google Scholar]
- Nischal L, Mohsin M, Khan I, et al. Identification and comparative analysis of microRNAs associated with low-n tolerance in rice genotypes. PLoS One. 2012 doi: 10.1371/journal.pone.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obidiegwu JE. Coping with drought: stress and adaptive responses in potato and perspectives for improvement. Front Plant Sci. 2015;6:1–23. doi: 10.3389/fpls.2015.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Duran-Fiueroa N, Arteaga-Vazquez M, et al. Control of female gamete fromation by a small RNA pathway in Arabidopsis. Plant Signal Behav. 2010;5:1476–1479. doi: 10.4161/psb.5.11.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, Zhang Y, Xu C, et al. Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice (Oryza sativa L.) PLoS One. 2012;7:e41143. doi: 10.1371/journal.pone.0041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parihar P, Singh S, Singh R, et al. Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res. 2015;22:4056–4075. doi: 10.1007/s11356-014-3739-1. [DOI] [PubMed] [Google Scholar]
- Paul S, Datta SK, Datta K. miRNA regulation of nutrient homeostasis in plants. Front Plant Sci. 2015;6:232. doi: 10.3389/fpls.2015.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, et al. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012;24:4777–4792. doi: 10.1105/tpc.112.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett JM, Khachane A, Ouassou M, et al. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol. 2014;202:270–286. doi: 10.1111/nph.12655. [DOI] [PubMed] [Google Scholar]
- Pont C, Murat F, Guizard S, et al. Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J. 2013;76:1030–1044. doi: 10.1111/tpj.12366. [DOI] [PubMed] [Google Scholar]
- Powers JS, Salute S. Macro- and micronutrient effects on decomposition of leaf litter from two tropical tree species: inferences from a short-term laboratory incubation. Plant Soil. 2011;346:245–257. doi: 10.1007/s11104-011-0815-x. [DOI] [Google Scholar]
- Preston JC, Hileman LC. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front Plant Sci. 2013;4:1–13. doi: 10.3389/fpls.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao-Ying Z, Cun-Yi Y, Qi-Bin M, et al. Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 2012;12:182. doi: 10.1186/1471-2229-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Hai B, Yuan M, et al. Characterization of wheat miRNAs and their target genes responsive to cadmium stress. Plant Physiol Biochem. 2016 doi: 10.1016/j.plaphy.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Rahaie M, Xue GP, Naghavi MR, et al. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010;29:835–844. doi: 10.1007/s00299-010-0868-y. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R, Aregger M, Zvereva AS, et al. Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 2012;40:6241–6254. doi: 10.1093/nar/gks242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O. Guest list or black list: heritable small RNAs as immunogenic memories. Trends Cell Biol. 2014;24:212–220. doi: 10.1016/j.tcb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Chen X, Yu B. Small RNAs meet their targets: when methylation defends miRNAs from uridylation. RNA Biol. 2014;11:1099–1104. doi: 10.4161/rna.36243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, et al. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Riddle NC. Heritable generational epigenetic effects through RNA. In: Tollefsbol T, editor. Transgenerational epigenetics. Birmingham, Alabama: Elsevier; 2014. pp. 105–119. [Google Scholar]
- Rock CD. Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development? Trends Plant Sci. 2013;18:601–610. doi: 10.1016/j.tplants.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Prasad AM, Srinivasan R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol Biochem. 2007;45:521–534. doi: 10.1016/j.plaphy.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Satapathy L, Singh D, Ranjan P, et al. Transcriptome-wide analysis of WRKY transcription factors in wheat and their leaf rust responsive expression profiling. Mol Genet Genomics. 2014;289:1289–1306. doi: 10.1007/s00438-014-0890-9. [DOI] [PubMed] [Google Scholar]
- Schlenker W, Roberts MJ. Nonlinear temperature effects indicate severe damages to U.S. crop yields under climate change. Proc Natl Acad Sci. 2009;106:15594–15598. doi: 10.1073/pnas.0906865106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR. Wheat. J Exp Bot. 2009;60:1537–1553. doi: 10.1093/jxb/erp058. [DOI] [PubMed] [Google Scholar]
- Sinha SK, Rani M, Bansal N, et al. Nitrate starvation induced changes in root system architecture, carbon: nitrogen metabolism, and miRNA expression in nitrogen-responsive wheat genotypes. Appl Biochem Biotechnol. 2015;177:1299–1312. doi: 10.1007/s12010-015-1815-8. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q-X, Liu Y-F, Hu X-Y, et al. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11:5. doi: 10.1186/1471-2229-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Song J, Mo B, Chen X. Uridylation and adenylation of RNAs. Sci China Life Sci. 2015;58:1057–1066. doi: 10.1007/s11427-015-4954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni K, Choudhary A, Patowary A, et al. MiR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res. 2013;41:4470–4480. doi: 10.1093/nar/gkt139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Declerck M, Christ A, et al. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014;202:1197–1211. doi: 10.1111/nph.12735. [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, et al. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26:1792–1807. doi: 10.1105/tpc.114.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xu L, Wang Y, et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.) BMC Genomics. 2015;16:1–16. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu J-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Tan Z, Wu F, et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol Plant. 2014;7:1329–1349. doi: 10.1093/mp/ssu054. [DOI] [PubMed] [Google Scholar]
- Tang Z, Zhang L, Xu C, et al. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012;159:721–738. doi: 10.1104/pp.112.196048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327:818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- Van der Straeten D, Rodrigues-Pousada RA, Goodman HM, Van Montagu M. Plant enolase: gene structure, expression, and evolution. Plant Cell. 1991;3:719–735. doi: 10.2307/3869253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MM, Christensen S, Schmelz EA, et al. Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 2015 doi: 10.1111/pce.12482. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Walbot V. How plants cope with temperature stress. BMC Biol. 2011;9:79. doi: 10.1186/1741-7007-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun F, Cao H, et al. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One. 2012 doi: 10.1371/journal.pone.0048445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Sun YF, Song N, et al. Identification of UV-B-induced microRNAs in wheat. Genet Mol Res. 2013;12:4213–4221. doi: 10.4238/2013.October.7.7. [DOI] [PubMed] [Google Scholar]
- Wang B, Sun YF, Song N, et al. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol Biochem. 2014;80:90–96. doi: 10.1016/j.plaphy.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Wang L, ZengJ HQ, Song J, et al. miRNA778 and SUVH6 are involved in phosphate homeostasis in Arabidopsis. Plant Sci. 2015;238:273–285. doi: 10.1016/j.plantsci.2015.06.020. [DOI] [PubMed] [Google Scholar]
- Waters BM, McInturf SA, Stein RJ. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J Exp Bot. 2012;63:5903–5918. doi: 10.1093/jxb/ers239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B, Zhang R, Guo J, et al. Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS One. 2014;9:e84781. doi: 10.1371/journal.pone.0084781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, et al. Rice microRNA effector complexes and targets. Plant Cell. 2009;21:3421–3435. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, et al. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- Xia R, Meyers BC, Liu Z, et al. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell. 2013;25:1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yin H, Liu X, Li X. Salt affects plant Cd-stress responses by modulating growth and Cd accumulation. Planta. 2010;231:449–459. doi: 10.1007/s00425-009-1070-8. [DOI] [PubMed] [Google Scholar]
- Yan Y, Wang H, Hamera S, et al. MiR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014;78:44–55. doi: 10.1111/tpj.12446. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang H, Lu Y, et al. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot. 2012;63:1025–1038. doi: 10.1093/jxb/err337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Niu Q, Ng K, Chua N. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015;83:673–685. doi: 10.1111/tpj.12919. [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu D. Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J. 2012;70:432–444. doi: 10.1111/j.1365-313X.2011.04883.x. [DOI] [PubMed] [Google Scholar]
- Zhang B. MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot. 2015;66:1749–1761. doi: 10.1093/jxb/erv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhou J, Han S, et al. Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem Biophys Res Commun. 2010;398:355–360. doi: 10.1016/j.bbrc.2010.06.056. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Gong P, et al. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol Lett. 2011;33:403–409. doi: 10.1007/s10529-010-0436-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Jia J, et al. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot. 2012;63:203–214. doi: 10.1093/jxb/err264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yang J, Wang Z, et al. Identification of novel and conserved microRNAs related to drought stress in potato by deep sequencing. PLoS One. 2014;9:e95489. doi: 10.1371/journal.pone.0095489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol. 2014;56:192–220. doi: 10.1111/jipb.12163. [DOI] [PubMed] [Google Scholar]
- Zhao X, Liu X, Guo C, et al. Identification and characterization of microRNAs from wheat (Triticum aestivum L.) under phosphorus deprivation. J Plant Biochem Biotechnol. 2013;22:113–123. doi: 10.1007/s13562-012-0117-2. [DOI] [Google Scholar]
- Zhao W, Li Z, Fan J, et al. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot. 2015;66:4653–4667. doi: 10.1093/jxb/erv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Li D, Li Z, et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013;161:1375–1391. doi: 10.1104/pp.112.208702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsigmond L, Szepesi A, Tari I, et al. Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant Sci. 2012;182:87–93. doi: 10.1016/j.plantsci.2011.07.008. [DOI] [PubMed] [Google Scholar]