Abstract
Here, we present a mouse brain protein atlas that covers 17 surgically distinct neuroanatomical regions of the adult mouse brain, each less than 1 mm3 in size. The protein expression levels are determined for 6,500 to 7,500 gene protein products from each region and over 12,000 gene protein products for the entire brain, documenting the physiological repertoire of mouse brain proteins in an anatomically resolved and comprehensive manner. We explored the utility of our spatially defined protein profiling methods in a mouse model of Parkinson's disease. We compared the proteome from a vulnerable region (substantia nigra pars compacta) of wild type and parkinsonian mice with that of an adjacent, less vulnerable, region (ventral tegmental area) and identified several proteins that exhibited both spatiotemporal- and genotype-restricted changes. We validated the most robustly altered proteins using an alternative profiling method and found that these modifications may highlight potential new pathways for future studies. This proteomic atlas is a valuable resource that offers a practical framework for investigating the molecular intricacies of normal brain function as well as regional vulnerability in neurological diseases. All of the mouse regional proteome profiling data are published on line at http://mbpa.bprc.ac.cn/.
Recently, Allen Brain Atlas reported genome-wide gene expression patterns in the brains of adult and developing mice using high throughput in situ hybridization (ISH) 1 (1, 2), which provides mRNA expression information on murine brain anatomy at the single cell level, and this increased our understanding of the brain's architecture and function. In other studies, brain-region mRNAs were quantitatively measured by DNA microarray (3, 4). Because mRNA levels are not necessarily proportional to protein levels (5), transcript profiling must be cross-validated by protein profiling. Until recently, several attempts have been made to identify region-specific distribution of proteins by proteomic profiling (6–8); however, these studies suffered from inadequate protein coverage with each covering only 1,000 to 2,000 proteins. In 2015, Sharma et al. (9) reported a complete mouse cell type and brain regional proteome that can cover 12,000 gene products in as short as 4 h of MS instrument running time. This study thus provided an entry point into brain-wide profiling methodology and opened the doors for applications related to this profiling. One particular value of a region-specific proteome stems from the idea that most known brain disorders are often specific to a particular brain region. For instance, Parkinson's disease (PD) and Huntington's disease (HD) are both disorders driven by abnormal protein accumulation and toxicity, yet in PD, dopaminergic SNc neurons degenerate, whereas in HD, medium spiny neurons in the striatum degenerate (10). The proteins responsible for such regional and selective neuronal vulnerability in most cases remain elusive to investigators. Therefore, proteome profiling of the brain with a spatial resolution that distinguishes surgically distinctive neuroanatomy relevant to regions most affected by neurological disorders will provide valuable information about disease pathogenesis and potentially uncover pathways for therapeutic intervention. In this study, we set out to test whether miniaturized proteomic profiling is possible in as low as 5 μg of tissue lysate with 1 mm diameter of spatial resolution and whether we could use this methodology to glean insight from a mouse model of Parkinsonism.
EXPERIMENTAL PROCEDURES
Animals
Mice used in this study were housed in a room with a 12-h light/12-h dark cycle with ad libitum access to standard rodent chow and water. Mouse housing was done in an air-controlled ith mice housed up to five per cage. All procedures involving mice were approved by the Institutional Animal Care and Use Committee for Baylor College of Medicine and Affiliates. The 6–8-week-old C57Bl/6 wild type male mice were used for brain regional proteome profiling study. For the Parkinson's disease model, α-synuclein (α-syn) transgenic mice (mThy-Syn, line 61) were a generous gift from Dr. Marie-Françoise Chesselet and Dr. Eliezer Masliah (11, 12). The α-syn transgenic female mice were used for mating, and males were used for experimentation due to the insertion of the transgene on the x chromosome (13). The Parkinsonian mouse model study was carried out on age- and sex-matched littermates at 3 weeks, 3 months, and 7 months of age as detailed below.
Mouse Brain Sub-structural Region Sample Collection
Mice were deeply anesthetized using an intraperitoneal injection of ketamine/xylazine/acepromazine given at 50 mg/kg, 10 mg/kg, and 2 mg/kg body weight, respectively. Depth of anesthesia was monitored by absence of withdrawal reflex to strong toe pinch and relaxation of extremities when placed on the back. To avoid contamination of mouse brain tissues with mouse blood proteins, mice were treated by intra-cardiac perfusion with Krebs-Ringer solution containing 20 mm glucose and 0.2 mm EGTA, pH 7.4, prior to collection of mouse brains. When the animal was adequately anesthetized, it was pinned to a corkboard, and the incision site was clipped and cleaned using 70% ethanol to prevent hair and topical bacteria from contaminating wet tissue samples. Briefly, incision of the abdominal wall immediately below the thoracic cavity was followed by incision of the diaphragm and rib cage. After exposure of the heart, a 2-mm partial-thickness scalpel incision was made in the left ventricle to allow insertion of a blunted wide-bore cannula (2-mm internal diameter) into the lumen of the ascending aorta. After cannulation of the heart, the valve to the Krebs-Ringer solution reservoir was opened, and a 1- to 2-mm cut was immediately made in the wall of the right atrium to allow blood and perfusate to escape. The volume of perfusate required per adult mouse is ∼30 ml at 5 ml/min perfusion rate controlled by height of reservoir. Intact mouse brains were removed and placed on a stainless steel adult mouse brain slicer matrix to make 1.0-mm coronal section slice intervals (Zivic Instruments, BSMAA001-1). A total of 17 selected mouse brain regions were then harvested using a blunt 18-gauge needle (1-mm internal diameter) punch. Chosen samples were dispersed by pipetting in 10 sample volumes of lysis buffer (50 mm ammonium bicarbonate, 1 mm CaCl2) and then snap-frozen using liquid nitrogen and thawed at 37 °C three times. Proteins were then boiled at 95 °C for 3 min. All freeze-thaw denaturation procedures were repeated twice. Protein concentration was measured using a Bradford reagent, and 20 μg of proteins were digested with 200 ng of trypsin (T9600, GenDepot, Houston, TX) overnight at 37 °C. After the first digestion, an additional 100 ng of trypsin was added to the samples, which were then incubated for 4 h at 37 °C. Double-digested peptides were extracted by 50% acetonitrile, 2% formic acid, and peptide supernatant was taken after spin-down at 10,000 × g for 1 min. The remaining pellet was extracted with 80% acetonitrile, 2% formic acid once again and pooled into previous extract after spin down. Pooled peptide supernatant was dried using vacuum drier and stored at −20 °C until further procedure.
High pH C18 Reverse Phase Sample Preparation
Vacuum-dried peptides were dissolved in pH 10 (10 mm ammonium bicarbonate, pH 10, adjusted by NH4OH) buffer and subjected to pH 10 C18 reverse phase column chromatography. A micro-pipette tip C18 column was made from a 200-μl pipette tip by layering 6 mg of C18 matrix (Reprosil-Pur Basic C18, 3 μm, Dr. Maisch GmbH, Germany) on top of the C18 disk (3M, EmporeTM C18) plug. Vacuum-dried peptides were dissolved with 150 μl of pH 10 buffer and loaded on the C18 tip equilibrated with pH 10 solution. Bounded peptide was eluted with step gradient of 150 μl of 6, 9, 12, 15, 18, 21, 25, 30, 35% ACN (pH 10) and pooled into 6 pools (6% eluent combined with 25% eluent, 9% plus 30%, and 12% plus 35%) and vacuum-dried for nano-HPLC-MS/MS.
Nano-HPLC-MS/MS Analysis
Vacuum-dried peptide was dissolved in 20 μl of loading solution (5% methanol containing 0.1% formic acid), and one-fourth of the reconstituted samples were subjected to nano-LC-MS/MS analysis with a nano-LC 1000 (Thermo Fisher Scientific) coupled to Orbitrap Velos Pro, Orbitrap Elite or Orbitrap FusionTM TribridTM mass spectrometer (Thermo Fisher ScientificTM) mass spectrometer with NSI source. From a starting amount of 20 μg, each subdivision of high pH RP is regarded as containing 3 μg of peptides. We loaded one-fourth of each pool, so that 0.7–0.8 μg of peptide was used for one single mass run. The peptides were loaded onto an in-house Reprosil-Pur Basic C18 (3 μm) trap column, which was 2 cm × 100 μm. Then the trap column was washed with loading solution and switched in-line with an in-house 6-cm × 150-μm column packed with Reprosil-Pur Basic C18 (2 μm) equilibrated in 0.1% formic acid/water. The peptides were separated with a 75-min discontinuous gradient of 2–24, 4–24, or 8–26% acetonitrile, 0.1% formic acid at a flow rate of 800 nl/min. Separated peptides were directly electro-sprayed into a mass spectrometer. A brain region profiling study was done on Thermo Velos Pro and Thermo Elite. The instrument was operated in the data-dependent mode acquiring fragmentation under direct control of Xcalibur software (Thermo Fisher Scientific). Precursor MS spectrum was scanned at 375–1300 m/z with 240,000 (Elite) or 100,000 (Velos) resolution at 400 m/z, 2 × 106 AGC target (10 ms maximum injection time) by Orbitrap. Then, the top 25 strongest ions were fragmented by collision-induced dissociation with 35 normalized collision energy and 1 m/z isolation width and detected by ion trap with 30 s of dynamic exclusion time, 1 × 104 AGC target, and 100 ms of maximum injection time. Parkinson's disease mouse model profiling was done by Thermo Fusion Orbitrap. Precursor MS spectrum was scanned at 300–1400 m/z 120,000 resolution at 400 m/z, 5 × 105 AGC target (50-ms maximum injection time) by Orbitrap. Then, the top 50 strongest ions were scanned by Quadrupole with 2 m/z isolation window, 18-s exclusion time (±7 ppm), fragmented by High-energy collisional dissociation (HCD) with 32 normalized collision energy, and detected by ion trap with rapid scan range, 5 × 103 AGC target, and 35 ms of maximum injection time.
Protein Identification and Label-free Quantification
Obtained MS/MS spectra were searched against target-decoy Mouse refseq database (release 2015_06, containing 58,549 entries) in Proteome Discoverer 1.4 interface (PD1.4, Thermo Fisher Scientific) with Mascot algorithm (Mascot 2.4, Matrix Science). Dynamic modifications of acetylation of the N terminus and oxidation of methionine were allowed. The precursor mass tolerance was confined within 20 ppm with fragment mass tolerance of 0.5 dalton, and a maximum of two missed cleavages was allowed. Assigned peptides were filtered with 1% false discovery rate (FDR) using percolator validation based on q-value. iBAQ algorithm was used to calculate protein abundance. The spectral assignments from PD1.4 were then converted to the MS-platform independent mzXML format and channeled through an in-house pipeline for peptide quantification (iPAC) and protein identification and quantification (grouper, which utilizes iPAC results). iPAC (integrated Peak Alignment Corrector) is a program to obtain optimal areas-under-the-curve (AUC) estimates for the detected peptide peaks, which extract candidate peptide information from the searching result list, including peptide, protein ID, modification, charge, m/z, retention time (RT), and scan number. These intensity values can be constructed into extracted ion chromatogram (XIC) peak for the peptide along the RT axis. The XIC were smoothed by Savitzky-Golay filter (14), and peak areas were calculated by trapezoidal numerical integrate (15). It is positive correlation between the peak area values and the protein abundance. We built a dynamic regression function based on those common identified peptides, according to correlation value R2; our program chose linear or quadratic function. We then calculated RT of corresponding hidden peptides, and checked the existence of the XIC based on the m/z and calculated RT. Finally, the program evaluated the peak area values of those existing XICs. These peak area values were considered part of corresponding proteins. Our previous studies proved this approach could enhance the accuracy of protein quantification. Grouper is a program built in-house that assigns detected peptides into gene products and tags corresponding experimental measurements (sum spectral counts, sum protein areas, qualitative bins by peptide FDR and Mascot ion scores, and homology groups by distribution of unique and shared peptides). The spectral matches were assigned into eight confidence bins, termed IDGroups, based on a combination of their Mascot ion scores and Percolator q values. The even IDGroups have q values of 0.01–0.05, and odd IDGroups have <0.01 q values. Peptide-spectrum match (PSM) IDGroups 1–2 have ion scores of >30; groups 3–4 have ion scores in 20–30 range, groups 5–6 have 10–20 ion scores, and groups 7–8 have 7–10 ion scores. The identifications with ion scores of <7 were filtered out. A special case of IDGroup 9 was given to peptide peaks that were identified by m/z and RT alignment to different runs but without a spectral match. The protein identifications (at gene locus level per NCBI GeneIDs) were assigned into three confidence bins as follows: “strict,” “relaxed,” and “all.” These bins were defined based on the shared peptide distributions and quality of the best PSM as measured by IDgroups. The protein products that have unique-to-gene peptides (unambiguous assignments) or have the largest distinct set of shared peptides were assigned into the strict bin if they had at least one spectral match with IDGroup <4 and the relaxed bin if the best spectral evidence has IDGroup 5–6. The rest of possible protein products (proteins identified by smaller subsets of peptides belonging to strict and relaxed bins and/or proteins with poor IDGroups of 7–9) were marked all. The resulting assignments can then be viewed at three levels, with each level defining the minimal confidence allowed (e.g. relaxed level included strict and relaxed confidence identifications). The two main advantages of this program are as follows: 1) it retains maximum possible gene product assignments while specifying quality of protein identification in a straightforward way, and 2) it corrects quantification issues associated with shared peptides by distributing them between proteins according to their corresponding unique peptide area ratios. Overall, this approach allows researchers to visualize not only quantitative but also qualitative differences in protein measurements with ease. It is suitable for relaying information-rich results to both experts and clients with minimal training. The align! displays both qualitative and quantitative parameters for MS identifications and calculates quantitative ratios for control-paired datasets. Furthermore, align! allows bioinformatics integration by including functional classification for proteins, expression from BioGPS and protein profiling studies, annotation of significant cancer mutations, and a function to display protein interaction networks within this program. This information is displayed in parallel with experiment-specific mass spectrometry data, and all parameters are fully searchable.
Parallel Reaction Monitoring (PRM)
To validate regional specific outlier gene products (region-specific proteins) from 17 regions profiling and outlier or specific target proteins from Parkinson's disease model SNc/VTA profiling, we utilized PRM outliers using Orbitrap FusionTM TribridTM mass spectrometer. Depends on unique peptide availability, 2 or 3 unique peptides for each target proteins were selected for PRM analysis. 500 ng of digested peptide were analyzed at each machine run with 4–24% of acetonitrile gradient for 35 min for 17 regions profiling and 75-min gradients for SNc/VTA profiling of Parkinson's disease model. Pre-selected precursor ions were scanned with a 5–10-min predicted elution window with 120,000 of resolution and 2.0e5 of AGC targets by Orbitrap and isolated by quadrupole followed by collision-induced dissociation/MS2 analysis. Product ions (MS2) were scanned at 350–1400 m/z with 1.0e4 of AGC target in rapid mode by Ion Trap. For relative quantification, the raw spectrum file was crunched to .mgf format by PD1.4 and then imported to Skyline with raw data file. We validated each result by deleting non-identified spectrum and adjusting the AUC range. Finally, the sum of the area of the three strongest product ions for each precursor ion (each peptide) was used for the result. In the case of 17 regions profiling/PRM comparison to compare the data between PRM and Profile, row values (protein groups) were normalized as a percentile of each protein group's maximum value and visualized by color scale. The protein list was arranged by descending order of percentile value from OLF to MY. Briefly, first the protein group list was arranged by OLF percentile values with descending order, and RSOPs of OLF were excluded for further procedures. The remaining list was arranged again by Striatum (STR) values as before and by MY sequentially. The arranged protein group list was used for PRM data arrangement with the same order without any modification.
Clustering
To determine relationship between 17 different regions, all correlation values were clustered by Qlucore Omics Explorer version 3.1. Euclidean average linkage hierarchical clustering values were applied without any normalization or filtration.
Comparison of iFOT and ISH
To obtain adult mouse ISH-based expression data from the Allen Institute for Brain Science projects, we queried Brain Atlas databases through their Restful Model Access (RMA) interface. First, we downloaded records for 39,116 section DataSets in Product ID 1 (“genome-wide high resolution ISH data detailing gene expression throughout the adult mouse brain”; the URL was http://www.brain-map.org/api/v2/data/query.xml?criteria=model::SectionDataSet,rma::criteria,products[id$eq1],rma::include,genes,rma::options[only$eq'genes.entrez_id']). Of these, 25,527 datasets had GeneID assignments. To get the structure-unionize expression energy values for the 12 brain structures in “Mouse-Coarse” structure dataset (structure-set = 2) for these sectionDataSets, the following query was used: http://www.brain-map.org/api/v2/data/query.xml?criteria=model::Structure,rma::criteria,structure_sets[id$eq2],pipe::list[xstructures$eq'id'],model::StructureUnionize,rma::criteria,[structure_id$in$xstructures],rma::include,structure,rma::options[only$eq'structure_unionizes.expression_energy,structure_unionizes.section_data_set_id,structures.acronym']. This query resulted in 366,763 structure-unionize expression energy values. 306,108 of these records matched to the final set of 25,509 section DataSets that had semi-quantitative ISH-based expression-energy value for all 12 brain structures in Mouse-Coarse structure dataset. The number of unique gene products in these 25,509 section DataSets (by entrez-id) in this set was 19,420. Ten brain structures in the Allen Mouse Brain ISH data match our sectioning schema for proteomic profiling. Therefore, we were able to perform pairwise comparison of ISH-based expression and mass spectrometry-based iFOT protein levels for these 10 brain regions.
To compare proteomic and transcriptomic datasets, the pool of genes and gene products was reduced to those occurring in both ISH and FOT, respectively. Extreme outliers (quantile borders of 0.005 and 0.995) were removed from both datasets, and genes without any abundance value were omitted, leading to two matrices with 5,264 gene products and 12 brain structures. For better comparability, the ISH dataset was log scaled with a base of 4 and the FOT dataset with a base of 10. We categorized the genes based on their standard deviations across brain regions and on the cosine distance between FOT and ISH.
The cosine distance between the datasets for each gene was calculated as
where A and B are the 12 values for one gene in FOT and ISH, respectively. We selected this distance method because it is more robust to high absolute values than other correlation types as it represents the angle between the two vectors A and B, not their length difference. We added the standard deviations to the classification criteria because the cosine distance alone is not always relevant, especially for ubiquitous genes and genes where either expression level or protein abundance is constant across brain regions. The standard deviation borders for “low” and “high” were set as terciles for each dataset individually (i.e. low is within the 33% lowest values). The borders for the cosine distance were set to 20° for low and 60° for high (Fig. 3D). We categorized ubiquitous (good RNA/protein correlation, expressed highly everywhere), RNA-regulated (good RNA/protein correlation with differential expression in brain regions), post-RNA-regulated (similar RNA signals with differential protein levels), stable protein (similar protein levels with different RNA staining), and irregular (strong anti-correlation between expression and protein abundance) as shown in Fig. 3B. Finally, we systematically categorized RNA and protein abundance levels for each gene and brain region to note for trends in regulation. From these datasets, we found that each protein could be grouped into subgroups based on their relative RNA/protein abundance measurements. We highlighted some of these groups based on their regression lines. For clarity, we did not include proteins without categories in the plot. For ubiquitous and negatively regulated genes, only one dot represents the gene in all brain regions (supplemental Fig. 5).
Fig. 3.
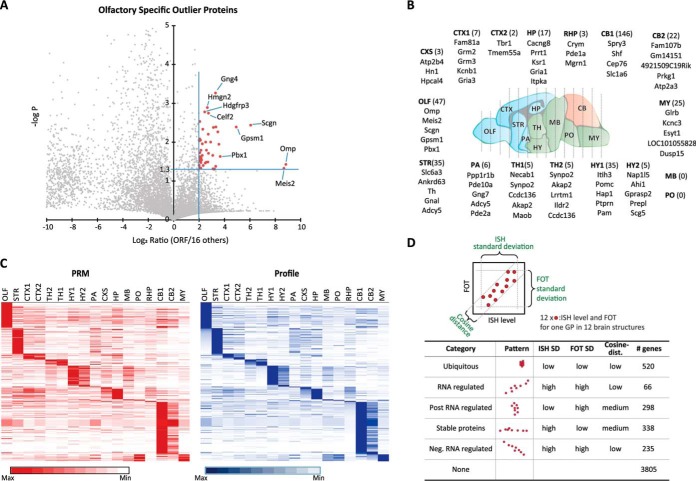
Statistical analysis, confirmation of RSOPs, and comparison of GP level with RNA expression level. A, example of RSOP calculation in olfactory bulb region. GPs show over a 4-fold increase and Student's t test p value lower than 0.05 compared with all other regions considered as RSOP (red rectangles). B, top five of RSOP listed. Total number of ROSP from each region is in parentheses. C, validation of profiling quantification data for RSOP by PRM. D, five distinctive categories of ISH and GP correlation.
Statistics for RSOP Calculation
Any GPs missed in over 40% of samples (>40% missingness) was excluded from the calculation. Of 19,246 total proteins, 3,952 were selected for downstream analysis following this criterion. Next, the remaining missing values in the filtered dataset were replaced with half of the minimally detected value in the entire dataset. Following log2 transformation of this dataset, the differential analysis (t test) was performed comparing one specific group against all the samples combined. This step was repeated for all the different experimental groups concerned. The resulting p values were adjusted using Benjamini-Hochberg method to correct for multiple hypothesis testing and to obtain a q-value. Any protein was deemed RSOPs if it has a q-value of <0.05 and greater than 4 linear fold change.
Experimental Design, Statistical Rationale, and Data Deposition
For brain regional proteome profiling, three biological samples from the 17 regions were measured with Thermo Orbitrap EliteTM and VelosTM MS to achieve MS-independent results. Six highly correlated (R >0.8) measurements from biological and instrumental replicates were taken (supplemental Fig. 2). The MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the MASSIVE repository (MSV000079789) with the dataset identifier PXD004263. For the Parkinsonian mouse model study, three biological samples from the SNc and VTA region of wild type and transgenic mouse were measured with Orbitrap Fusion MS. Parkinsonian mouse data have been separately deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the MASSIVE repository (MSV000080437) with the dataset identifier PXD005626.
RESULTS
Fast Proteome Profiling
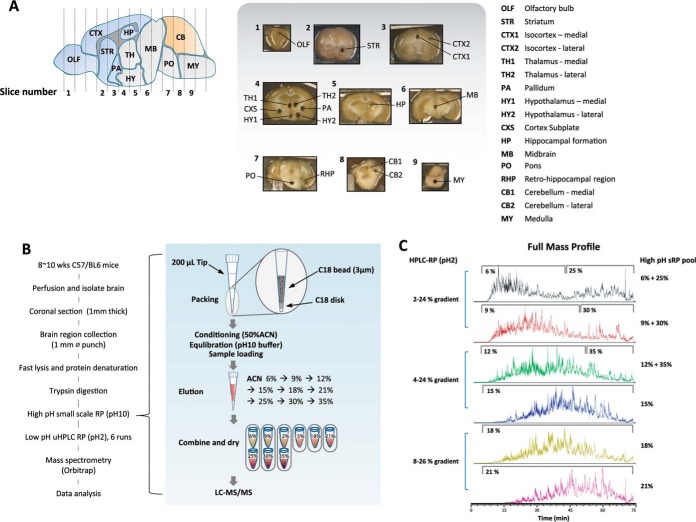
We generated a regionally resolved proteome library using 17 brain regions (Fig. 1A). We used the center position of each of the 13 distinctive brain regions, and for comparatively larger regions, we included both medial and lateral portions of the region to account for diversity of proteome composition. We picked a cylindrical piece of tissue (∼1 mm in diameter and 1 mm thick) to be the representative of the particular region. We developed a procedure that allows us to efficiently extract proteins from this small amount of tissue by digesting the tissue directly with trypsin without any prior protein extraction process, and then we extracted the tryptic peptides for further analysis. We miniaturized a two-dimensional orthogonal pH reverse phase chromatography (2D sRP-RP) (Fig. 1B) to avoid sample loss and to increase the detection efficiency. sRP-RP is a newly established method compared with gel-based protein separation system and ensures a high level of reproducibility between biological replicates (16). The acetonitrile gradient of the second low pH nano-HPLC was adjusted according to the hydrophobicity of the high pH eluent to make even distribution of peptide elution across the entire HPLC elution time to ensure maximum coverage (Fig. 1C). Mass spectrometry and peptide identification conditions are highlighted under “Experimental Procedures.” For label-free quantification fraction of total (iFOT), an AUC curating program to be more precise than conventional iBAQ (17) was used. The principle and details of the iFOT calculation program are also described under “Experimental Procedures.” The repeatability slope from a set of technical repeats of label-free quantification was greatly improved from 0.47 to 0.90 once the iBAQ calculation was re-defined with an in-house iFOT correction program (supplemental Fig. 1). These results indicate the effectiveness of in-house AUC curating program.
Fig. 1.
Sample preparation for regional analysis of proteins. A, 17 surgically distinctive mouse brain regions were isolated using 1-mm inner diameter punches from coronal section. B, simplified sample preparation method. Mouse brain regional samples were isolated from 1-mm-thick coronal sections followed by in-solution digestion. sRP-RP was used for separate and concentrate-digested peptide. Orbitrap instrument was used for MS analysis. First dimensional separation is done with high pH reverse phase micro-column using C18 beads. Stepwise ACN gradient was applied, and eluent was pooled into six fractions depending on ACN concentration before MS/MS. C, different continuous ACN gradient applied for different high pH eluent pools based on hydrophobicity nature of each pools.
We measured three biological samples from the 17 regions individually to account for all operational variations. To account for potential differences between mass spectrometer (MS) instruments, we measured the same sample with Thermo Orbitrap EliteTM and VelosTM MS to achieve MS-independent results. We continued measuring biological repeats until we acquired six highly correlated (R > 0.8) measurements from biological and instrumental replicates. Most of the data were acquired using three mice, some of them used as many as five mice, and 10 measurements, including the two MS measurements. The full reproducibility correlation table is shown in supplemental Fig. 2.
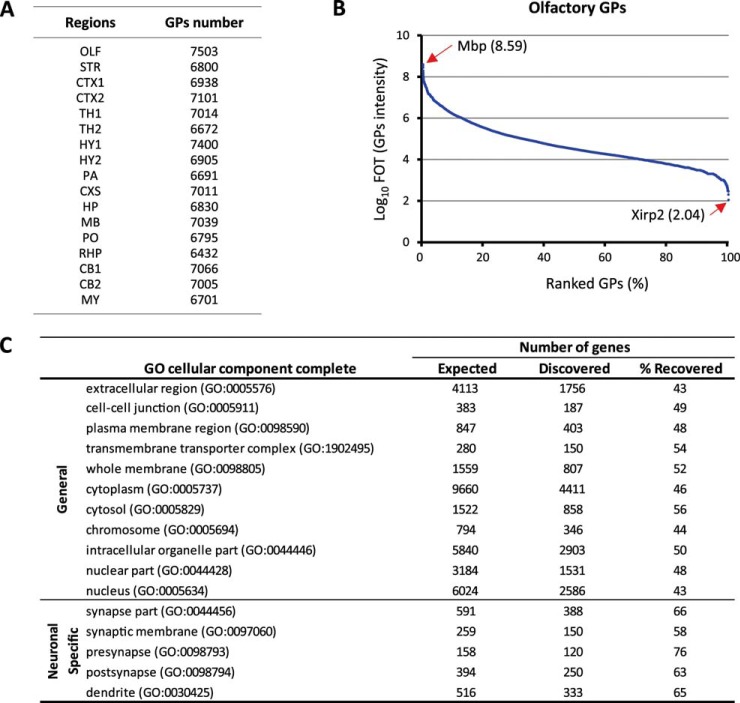
With these improvements, we measured 102 individual proteomes and were able to create a quantitative mouse brain atlas covering 17 regions of distinct anatomy, revealing protein distribution ranges using a total of 986 h of MS running time. The number of gene protein products (GPs) ranged from 6,432 to 7,503 for each region (Fig. 2A). Collectively, 12,000 GPs were identified for the 17 dissected brain regions. The dynamic range of the relative protein abundance in the proteome from each mouse brain region spanned almost 7 orders of magnitude (Fig. 2B). Gene ontology analysis showed recovered GPs located in various intra- and extracellular domains ranging from extracellular matrix to nuclear compartments with around 50% of coverage from expected genes. Moreover, synapse-specific genes showed a little bit higher recovery rate (60–70%) as exemplified in the olfactory region (Fig. 2C).
Fig. 2.
Proteome coverage. A, total GP numbers recovered from each brain region after six repeatable analyses. B, normalized protein abundances (FOT) from the highest to the lowest in olfactory sample. C, proportions of olfactory GPs annotated to various cellular compartments according to Gene Ontology.
Characterization of Mouse Brain Region-specific Outlier Gene Protein Products
We first searched for region-specific outlier gene products. Outliers were identified based on Student's t test significance (p value<0.05) and fold change (>4-fold) of the average value of each region against the average of all other regions. The example of olfactory outliers is shown in Fig. 3A. A total of 352 proteins were identified as RSOPs. The top five RSOPs in each region are shown in Fig. 3B, and the full list is presented as supplemental Table 1. To test for concordance in a previously published dataset, we overlaid identified GPs from our data set with those identified by Sharma et al. (9) in a similarly dissected region, the olfactory, striatum, and cerebellum. We found a high level of concordance of these datasets (R = 0.84, 0.85, and 0.82, respectively, see supplemental Table 2) thus further validating this in-house pipeline.
To further solidify the definition of RSOPs, we retraced RSOP abundance with a more accurate targeted MS analysis method, PRM, using a third type of Orbitrap Fusion TribridTM mass spectrometer (18). To compare PRM data with proteomic profiling data, we normalized by maximum value at each protein group over all regions. Profiling data were sorted in descending order from OLF to MY sequentially, and PRM data were matched in the same order. As a result of relative comparison, 69% of RSOP identified by profiling showed highly correlated concordance (Pearson's R >0.8) with measurement of PRM (Fig. 3C, supplemental Fig. 3, and supplemental Table 3). Together, these data add another level of confidence to this streamlined procedure for label-free quantitative proteome profiling.
Brain regions widely differ in their cellular identities and firing properties. One way to cluster these has been via transcriptomic signatures (1, 19, 20). However, whether these transcriptomic signatures directly correlate to protein abundance remains unclear. To address general trends in correlation between mRNA and corresponding protein products, we compared our proteomics profiling toolkit to the Allen Mouse Brain Atlas (1). Although we acknowledge differences in quantitation methods and sample collection scheme between these two studies, we were able to perform pairwise comparisons for 10 regions (supplemental Fig. 4 and supplemental Table 4). This comparison was not expected to be quantitative, because ISH-based expression energy values (referred to as “ISH values” herein) for RNA expression across a brain section are inherently semi-quantitative. This analysis was rather a qualitative comparison method to test to generalizability of concordance between RNA and protein levels. We performed pairwise comparison of 5,264 gene products found both in our profiling results and Allen Brain Atlas ISH. We based the classification boundaries on the standard deviation of ISH and iFOT value distributions and on their correlation, accepting that not all genes can be categorized. We found that 27% of RSOPs showed tight correlation trends between ISH and proteins (supplemental Table 4). However, the majority of RSOPs had lower correlation (R <0.8), indicating a discordance between RNA expression and protein abundance for region-specific proteins. In addition to RSOPs, around 40% of all, unfiltered, proteins from our datasets correlate with Allen Brain Atlas's ISH data upon pairwise comparisons (supplemental Fig. 5). We found that expression and abundance of proteins strongly occur throughout the whole range of mRNA expression levels, suggesting that the levels of their mRNA directly regulate many proteins. However, certain gene products observed differential regulation as categorized in Fig. 3D. We found that these differentially regulated gene products followed trends that were directly proportional to their transcript expression or their protein abundance. By separating each transcript-gene product pairs according to their abundance, we found that each could be classified into five broad categories as follows: ubiquitous; RNA-regulated; post-RNA-regulated; stable proteins, and negative RNA-regulated. These are summarized in Fig. 3D, and an example of each is highlighted in supplemental Fig. 5. Notably, although the classification schema did not depend directly on absolute expression level or protein abundance, gene products of different categories clearly accumulate in specific regions as shown in supplemental Fig. 5. As expected, ubiquitous proteins of constant abundance and expression occur mostly at high expression values. The opposite group with irregular abundances is generally of low RNA and protein levels. This may be due to technical noise associated with the low value measurements.
Functional Analysis of Brain Region-specific Outlier Gene Protein Products
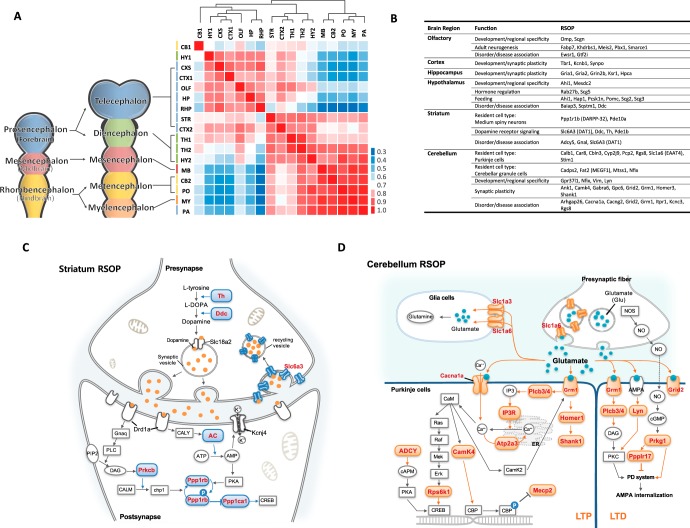
In calculating the correlation of protein abundance across the 17 regions, we found that relative protein abundance clustered in an anatomical order, reflecting the rostro-caudal development of these regions (Fig. 4A). Specifically, we found the following three distinct clusters, which roughly correspond to the brain regions observed in vertebrate CNS development (21): the proencephalon (telencephalon and diencephalon), the mesencephalon, and the rhombencephalon (metencephalon and myelencephalon). The remarkable conservation of proteomic content in the clusters clearly reflects the developmental origins of the sampled brain regions. Moreover, we found that the nature of region-specific proteins was tightly coupled to cellular identity and function (Fig. 4B). For example, genes that regulate dopaminergic synapses (22) and genes implicated in movement disorders (23) were highly represented by the RSOPs identified in the striatum punches (Fig. 4C). Moreover, cell type-specific markers such as Ppp1r1b (DARPP-32) correctly identified the main resident cell type found in the striatum, medium spiny neurons (23), and also identified proteins that are enriched at dopaminergic termini, a major synaptic input to the striatum, such as Slc6A3(DAT1), Ddc (AADC), and Th (22). Similar observations could be made in the context of cerebellar outliers, where Purkinje cells (24) and cerebellar granule cells (25) are well represented by several markers (Fig. 4D). Finally, the hypothalamic RSOPs were predominantly characterized as proteins involved in hormone regulation and control of feeding behavior and thus confirm the functional concordance of these RSOPs to different anatomical regions (26, 27). Taken together, RSOPs can be used as a proteomic signature that allows for faster and more insightful sample-to-sample comparisons and may ultimately serve as a foundation through which RSOP-based sub-typing of anatomical regions may be possible.
Fig. 4.
Functional annotation of RSOP. A, Pearson correlation of total gene protein product distribution was compared with brain anatomical boundaries. B, list of RSOP related with region-specific cellular identity and function. C, dopaminergic specific gene products found in striatum RSOP shown in red in blue shapes. D, cerebellum RSOP identified major glutamate receptor signaling molecules, here shown in red and pink shapes.
Validation of Proteome Profiling Platform in a Model of Parkinson's Disease
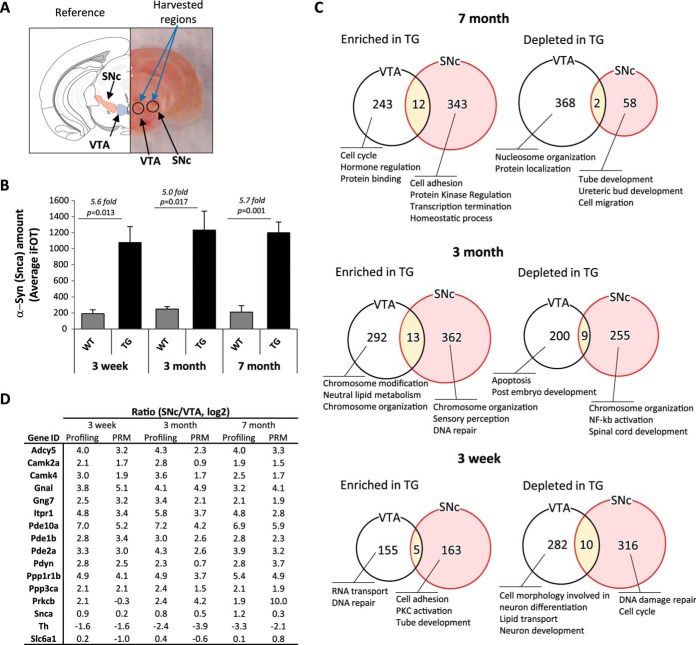
To test the utility of our dataset, we set out to determine specific anatomically resolved proteomes for an established mouse model of Parkinson's disease. We chose α-syn transgenic mice (α-syn TG, mThy1-Syn “line 61”) as a model as they display progressive accumulation of α-syn and age-dependent motor abnormalities (12). Furthermore, we focused on the substantia nigra pars compacta (SNc) as the target “susceptible” region to examine, due to its vulnerability in human PD. Moreover, given its close proximity and neurochemical similarity to the SNc, we chose the VTA as a control region, which is largely spared in PD (Fig. 5A) (28). To have a proper handle on the temporal nature of proteomic changes between brain regions in these transgenic mice, we performed proteomic profiling on mice aged 3 weeks, 3 months, and 7 months of age. We picked these time points as early-, mid-, and end-stage disease models to test whether we could identify the incipient protein changes that may be mediated by aberrant α-syn overexpression. Profiling of the SNc and VTA was done by Thermo Orbitrap Fusion and revealed ∼7,500–9,500 GPs per tissue punch when using a high stringency peptide identification filter (supplemental Fig. 6). We found that transgene overexpression at different ages corresponded to a 5–6-fold α-syn enrichment in the SNc, consistent with previous reports using alternative methods (Fig. 5B) (13). Overall, gene protein product quantification correlation was very high between α-syn TG and WT mice at all time points tested, which suggesting that overexpression of α-syn induces only subtle changes in the overall protein distribution pattern (supplemental Fig. 7). However, we did find a larger dysregulation of up-regulated gene products in the Parkinsonian mice at later time points (375 and 355 GPs enriched in the TGs at 3 months and 7 months, respectively) compared with the early time point (168 GPs enriched in the TGs at 3 weeks) consistent with larger global alterations following transgene expression (Fig. 5C and supplemental Table 5).
Fig. 5.
Comparative deep proteome analysis between the SNc and the VTA of α-syn transgenic mice. A, isolation of the SNc from 2-mm-thick coronal section containing midbrain (right). B, Snca iFOT distribution of SNc regions from Snca wild type and transgenic mouse. C, numbers and summarized GO term biological function of SNc-specific proteins regulated by α-syn overexpression compared with VTA. D, normal wild type SNc and VTA site-specific proteins age-dependent abundancy changes.
We next explored the α-syn overexpression driven contributions in a spatiotemporal context. First, we looked at the regional differences between two neurochemically similar regions, the SNc and the VTA. Although several proteomic changes were observed at all time points, we were intrigued to find a strong enrichment for membrane-signaling proteins such as GnaI, Pde10a, and Itpr1 in the SNc compared with the VTA in all tested ages (Fig. 5D).To confirm the validity of these findings, we further validated a subset of these GPs using PRM and noticed a high validation rate and degree of concordance between our results (validation rate >92%; r = 0.74, comparing profiling with PRM values) (Fig. 5D).
We next looked at the genotype-specific differences observed between mice overexpressing α-syn over time and their wild type littermates. We found that gene ontology categories varied substantially between different time points suggesting that the effects of α-syn-mediated toxicity are highly dynamic (Fig. 5C). However, we especially found that α-syn TG mice largely showed increased markers of extracellular matrix remodeling and inflammation (i.e. Hspg2, Lama5, and Lamb2) specifically at a late stage of disease (7 months) (Fig. 6A). These findings, also validated using PRM, would be consistent with the accumulation of inflammatory glial cell processes that occur in these mice over the course of their life span (Fig. 6B) (29, 30).
Fig. 6.
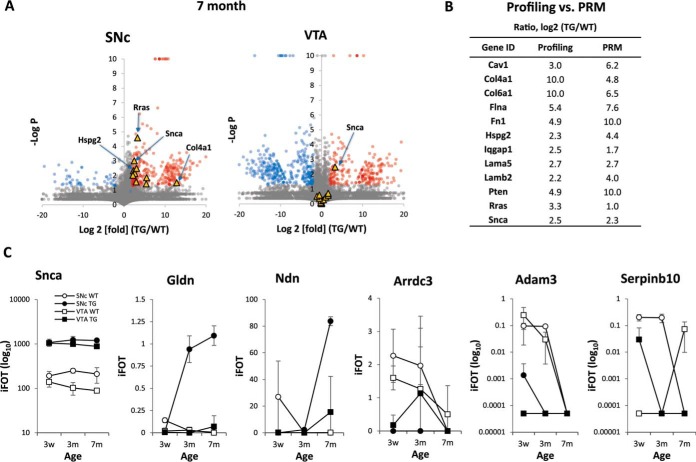
Age-dependent changes of Snca transgenic dependent RSOP. A, plot of relative quantification change of proteins depending on α-syn overexpression in 7-month-old mouse, a few outliers of relevance to PD pathogenesis are highlighted (yellow triangles). GPs show over 4-fold increase (red circle) or decrease (blue circle), and Student's t test p value lower than 0.05 is also indicated. B, PRM confirmation of 7-month SNc regional Snca transgenic mouse RSOPs. C, age-dependent relative quantification change for several representative RSOPs.
Finally, we looked to see whether we could combine our newly acquired tridimensional data (genotype, time, and anatomical location) to identify targets that may be important for disease pathogenesis. We first looked for GPs that may be contributing to the disease process in the SNc region of the α-syn transgenic mice over time. We found that gliomedin (Gldn) and necdin (Ndn) followed trends where their relative levels specifically increased in the SNc over time in the context of α-syn overexpression (Fig. 6C and supplemental Fig. 8). We next looked to see whether resident neuroprotective genes may exist in the SNc and VTA but are disturbed upon α-syn overexpression. In this case, we found that GPs such as Arrdc3, Adam3, and Serpinb10 observed trends where they are highly expressed in the WT mice but are lost in cases of α-syn overexpression and aging (Fig. 6C and supplemental Fig. 8). Altogether, these findings suggest that there are tightly controlled spatiotemporal changes that occur following α-syn overexpression in mice and that the earliest events may be worth pursuing as novel pathways for the earliest events contributing to α-syn-mediated toxicity.
DISCUSSION
In this study, we generated a spatially defined proteomic map of the mouse brain and used this tool to glean insight into biology and disease. We streamlined the 2D sRP-RP-MS procedure, allowing for proteome profiling in fewer than 8 h of mass spectrometer running time to detect 6,500 to 7,500 gene protein products from one brain sample (Fig. 2A, supplemental Table 7). During the writing of this paper we learned of a study that looked at cell type- and brain region-resolved mouse brain proteome (9). In that paper (9), they dissected the mouse brain into 10 surgically distinct parts and used these parts as a whole for protein profiling. They report relative quantification of around 12,000 proteins per each brain region. Building on their findings, we generated maps of 17 distinct brain regions and confirmed the concordance between our two studies. The throughput of one proteome per machine per day for triple bio-repeats makes large scale measurement feasible. In addition to high throughput capacity, our label-free proteome-profiling platform has a high sensitivity that requires as little as 5 μg of tissue lysate. The high sensitivity of this system allows for proteome profiling of small samples from highly organized and structurally complex regions, where sample collection amount is limited. Indeed, we found that the dynamic range of the proteins we identified spanned nearly 7 orders of magnitude (Fig. 2B). This range corresponds well with most of the previously published proteome profiling experiments reporting 7 orders of dynamic range of proteome with various organisms (31, 32). This indicates our method has no limitation to detect proteins of a wide range of abundance. Gene ontology enrichment search for the cellular component also shows our method has no limitation to detect proteins from various cellular compartments, outer membrane to nucleus and chromatin (Fig. 2C). Because our protein extraction procedure was mild compared with typical approaches using detergents (9), it is important to check recovery of integral membrane proteins. We found >75% (983 of 1,296) of Surfaceome Protein list from the Cell Surface Protein Atlas (33). Most of the previously reported membrane-bound proteins could be identified using this approach (supplemental Table 6). This finding indicates that our system has no problem to recover membrane proteins. With this in mind, the generation of a spatially restricted matrix-based map of the entire mouse brain (in 0.5 or 1 mm size increments) could be feasible in the foreseeable future (29, 30). The remarkable conservation of proteomic content in the clusters clearly reflects the developmental origins of the bulk of the brain regions sampled in this study. Our approach therefore provides an accurate and efficient way to identify proteins that are region-specific and tightly coupled to cellular identity and function (Fig. 4B). The utility of the mouse brain proteome can also be seen in light of its similarities and differences to brain-wide RNA datasets such as the ones generated by the Allen Brain Atlas. Coupled to large RNA expression-based datasets, our RSOPs reveal both correlated and non-correlated RNA-protein findings. Indeed, the top RSOPs identified in our platform include the following: 1) some that were not previously found in the same brain region by RNA profiling; 2) some that are expressed by major resident cell types; 3) some that perform known functions of the resident cells of that particular region; and 4) some that are associated with disease in the queried brain region. Thus, these complementary approaches offer new insight as well as further validation of the role of a particular gene product in the context of neurobiology and disease.
Finally, we adapted our approach to answer the long-asked biological question. Why are two adjacent brain regions differentially susceptible to neurotoxicity in disease? Given our success in miniaturization and spatial resolution, we decided to compare the proteome between two spatially confined and neurochemically similar (both are dopaminergic and have similar cellular morphology) regions that are differentially susceptible in Parkinson's disease, the SNc and the VTA. Moreover, given the rapid output of this approach, we were able to provide a temporal dimension to this anatomical dissection by testing animals at three stages of disease. With this multidimensional workflow, we were able to pinpoint several compelling groups of proteins whose abundance was aberrantly altered specifically in the SNc over the VTA. At the same time, we could internally control for dissection efficiency by looking at markers of dopaminergic neurons such as TH and DAT (Slc6a1). The identified “hits” thus serve as a promising resource to the PD field and a good entry point for hypothesis-driven research in the context of selective neuronal vulnerability in PD. Looking first at spatiotemporal changes in gene products between the SNc and the VTA, we found that membrane-bound signaling proteins such as G-protein-coupled receptor subunits/effectors and calcium-dependent signaling mediators were altered at an early stage. These are consistent with the idea that α-syn is primarily associated with membranes, and thus the first proteins to sense a change in its overexpression may indeed be associated there (34). Interestingly, these “mis-expressed” proteins remained unaltered in the VTA and could therefore be a sensitizing factor in disease progression. Follow-up studies that directly target a subset of these altered membrane proteins will surely shed insight into the first steps of disease pathogenesis. When we next looked at genotype-specific differences, we found that α-syn-driven proteomic changes were highly dynamic. Indeed, gene ontology revealed differential category enrichment over time (Fig. 5C). With regard to a later stage of disease pathogenesis, we found that α-syn overexpression promoted the accumulation of immune and extracellular matrix proteins at the level of the SNc. This is consistent with the large role played by the immune system and the extracellular matrix in the context of age-dependent neurodegeneration. Indeed, a recent report suggested that one of the top misregulated proteins in our dataset, Hspg2, abnormally accumulates in another neurodegenerative disease, Alzheimer's disease, and accelerates the aggregation of its primary driver of pathology, β-amyloid (35, 36). Finally, we combined all measurements to identify GPs that may be contributing to PD pathogenesis in a spatiotemporal and genetic manner. We found that Gldn and Ndn could serve as contributing factors as they exhibited an α-syn-dependent increase in the SNc over time, whereas Arrdc3, Adam3, and Serpinb10 could serve as neuroprotective factors as they exhibited an α-syn-dependent decrease in the SNc over time. Although the function of these proteins in the context of PD are not clear at this point, careful examination of their role in neuronal function in the context of aging will surely shed light onto mechanisms of α-syn toxicity in the context of PD.
Taken together, our mouse brain proteome opens the gates for further investigation into individual proteins or groups of aberrantly regulated gene products to elucidate new disease pathways for therapeutic intervention. Profiling of individual candidate genes that are differentially expressed in specific brain regions could in turn provide insight into the selective vulnerability of neurons in models of neurological disease.
Supplementary Material
Acknowledgments
We thank Catherine Gillespie for helpful comments and critical reading of the manuscript.
Footnotes
Author Contributions: S. Y. J., J. M. C., and M. W. C. R. performed the experiments; S. Y. J., A. M., S. M., and J. K. performed the statistical analysis; W. Z., and Y. H. developed the data webpage; S. Y. J., M. W. C. R., H. Y. Z., Y. W., J. J. K., and J. Q. wrote the manuscript; S. Y. J. and J. Q. supervised and conceived the project.
* This work was supported in part by Cancer Prevention and Research Institute of Texas Grants RP110784 and RP120092, National Institutes of Health Center Grant P30CA125123 from NCI (to J. Q.), Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology Grant 2015R1A6A3A03019779 (to J. M. C.), the Michael J. Fox Foundation (Target Validation Program 2014), the Huffington Foundation, the Howard Hughes Medical Institute (to H. Y. Z.), Canadian Institutes of Health Research Fellowship 201210MFE-290072-173743 (to M. W. C. R.), and Baylor College of Medicine Intellectual and Developmental Disabilities Research Center National Institutes of Health Grant U54 HD083092 from the Eunice Kennedy Shriver NICHD. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ISH
- in situ hybridization
- AUC
- area under the curve
- FDR
- false discovery rate
- HD
- Huntington's disease
- iBAQ
- intensity-based absolute quantification
- iFOT
- fraction of total
- iPAC
- integrated peak alignment corrector
- PD
- Parkinson's disease
- PD1.4
- Proteome Discoverer 1.4 interface
- PRM
- parallel reaction monitoring
- RMA
- restful model access
- RP
- reverse phase
- RSOP
- region-specific outlier proteins
- RT
- retention time
- SNc
- substantia nigra pars compacta
- sRP
- small scale reverse phase
- TG
- transgenic
- VTA
- ventral tegmental area
- XIC
- extracted ion chromatogram
- GP
- gene protein product
- XIC
- extracted ion chromatogram
- α-syn
- α-synuclein
- ACN
- acetonitrile
- Gldn
- gliomedin
- Ndn
- necdin
- HCD
- High-energy collisional dissociation
- OLF
- Olfactory bulb
- MY
- Medulla
- STR
- Striatum
- PSM
- Peptide-spectrum match.
REFERENCES
- 1. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T. M., Chin M. C., Chong J., et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 [DOI] [PubMed] [Google Scholar]
- 2. Thompson C. L., Ng L., Menon V., Martinez S., Lee C. K., Glattfelder K., Sunkin S. M., Henry A., Lau C., Dang C., Garcia-Lopez R., Martinez-Ferre A., Pombero A., Rubenstein J. L., Wakeman W. B., et al. (2014) A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ning K., and Fermin D. (2010) SAW: a method to identify splicing events from RNA-Seq data based on splicing fingerprints. PLoS One 5, e12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ning K., Fermin D., and Nesvizhskii A. I. (2012) Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-Seq gene expression data. J. Proteome. Res. 11, 2261–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maier T., Güell M., and Serrano L. (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 [DOI] [PubMed] [Google Scholar]
- 6. Dagley L. F., White C. A., Liao Y., Shi W., Smyth G. K., Orian J. M., Emili A., and Purcell A. W. (2014) Quantitative proteomic profiling reveals novel region-specific markers in the adult mouse brain. Proteomics 14, 241–261 [DOI] [PubMed] [Google Scholar]
- 7. Eberlin L. S., Liu X., Ferreira C. R., Santagata S., Agar N. Y., and Cooks R. G. (2011) Desorption electrospray ionization then MALDI mass spectrometry imaging of lipid and protein distributions in single tissue sections. Anal. Chem. 83, 8366–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heijs B., Carreira R. J., Tolner E. A., de Ru A. H., van den Maagdenberg A. M., van Veelen P. A., and McDonnell L. A. (2015) Comprehensive analysis of the mouse brain proteome sampled in mass spectrometry imaging. Anal. Chem. 87, 1867–1875 [DOI] [PubMed] [Google Scholar]
- 9. Sharma K., Schmitt S., Bergner C. G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U. K., Philips M. A., Rossner M. J., Mann M., and Simons M. (2015) Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saxena S., and Caroni P. (2011) Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron 71, 35–48 [DOI] [PubMed] [Google Scholar]
- 11. Fleming S. M., Salcedo J., Fernagut P. O., Rockenstein E., Masliah E., Levine M. S., and Chesselet M. F. (2004) Early and progressive sensorimotor anomalies in mice overexpressing wild-type human α-synuclein. J. Neurosci. 24, 9434–9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rockenstein E., Mallory M., Hashimoto M., Song D., Shults C. W., Lang I., and Masliah E. (2002) Differential neuropathological alterations in transgenic mice expressing α-synuclein from the platelet-derived growth factor and Thy-1 promoters. J. Neurosci. Res. 68, 568–578 [DOI] [PubMed] [Google Scholar]
- 13. Chesselet M. F., Richter F., Zhu C., Magen I., Watson M. B., and Subramaniam S. R. (2012) A progressive mouse model of Parkinson's disease: the Thy1-aSyn (“Line 61”) mice. Neurotherapeutics 9, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savitzky A., and Golay M. J. (1964) Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 [Google Scholar]
- 15. Weideman J. A. (2002) Numerical integration of periodic functions: a few examples. Am. Mathematical Monthly 109, 21–36 [Google Scholar]
- 16. Ding C., Jiang J., Wei J., Liu W., Zhang W., Liu M., Fu T., Lu T., Song L., Ying W., Chang C., Zhang Y., Ma J., Wei L., Malovannaya A., et al. (2013) A fast workflow for identification and quantification of proteomes. Mol. Cell. Proteomics 12, 2370–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 18. Peterson A. C., Russell J. D., Bailey D. J., Westphall M. S., and Coon J. J. (2012) Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 11, 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hawrylycz M. J., Lein E. S., Guillozet-Bongaarts A. L., Shen E. H., Ng L., Miller J. A., van de Lagemaat L. N., Smith K. A., Ebbert A., Riley Z. L., Abajian C., Beckmann C. F., Bernard A., Bertagnolli D., Boe A. F., et al. (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., and Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kandel E. R., Schwartz J. H., Jessell T. M., Siegelbaum S. A., and Hudspeth A. J. (2013) Principles of Neural Science, 5th Ed., pp. 1165–1186, McGraw-Hill, NY [Google Scholar]
- 22. Aumann T., and Horne M. (2012) Activity-dependent regulation of the dopamine phenotype in substantia nigra neurons. J. Neurochem. 121, 497–515 [DOI] [PubMed] [Google Scholar]
- 23. Yger M., and Girault J. A. (2011) DARPP-32, jack of all trades. Master of which? Front. Behav. Neurosci. 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ito-Ishida A., Okabe S., and Yuzaki M. (2014) The role of Cbln1 on Purkinje cell synapse formation. Neurosci. Res. 83, 64–68 [DOI] [PubMed] [Google Scholar]
- 25. Nakayama M., Nakajima D., Yoshimura R., Endo Y., and Ohara O. (2002) MEGF1/fat2 proteins containing extraordinarily large extracellular domains are localized to thin parallel fibers of cerebellar granule cells. Mol. Cell. Neurosci. 20, 563–578 [DOI] [PubMed] [Google Scholar]
- 26. Krashes M. J., Shah B. P., Madara J. C., Olson D. P., Strochlic D. E., Garfield A. S., Vong L., Pei H., Watabe-Uchida M., Uchida N., Liberles S. D., and Lowell B. B. (2014) An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valassi E., Scacchi M., and Cavagnini F. (2008) Neuroendocrine control of food intake. Nutr. Metab Cardiovasc. Dis. 18, 158–168 [DOI] [PubMed] [Google Scholar]
- 28. Brichta L., and Greengard P. (2014) Molecular determinants of selective dopaminergic vulnerability in Parkinson's disease: an update. Front. Neuroanat. 8, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watson M. B., Richter F., Lee S. K., Gabby L., Wu J., Masliah E., Effros R. B., and Chesselet M. F. (2012) Regionally-specific microglial activation in young mice over-expressing human wild type α-synuclein. Exp. Neurol. 237, 318–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rousseaux M. W., de Haro M., Lasagna-Reeves C. A., De Maio A., Park J., Jafar-Nejad P., Al-Ramahi I., Sharma A., See L., Lu N., Vilanova-Velez L., Klisch T. J., Westbrook T. F., Troncoso J. C., Botas J., and Zoghbi H. Y. (2016) TRIM28 regulates the nuclear accumulation and toxicity of both α-synuclein and τ. Elife 5, e19809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Azimifar S. B., Nagaraj N., Cox J., and Mann M. (2014) Cell-type-resolved quantitative proteomics of murine liver. Cell Metab. 20, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 32. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., and Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bausch-Fluck D., Hofmann A., Bock T., Frei A. P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R. L., Yoon C., Schiess R., Schmidt A., Mirkowska P., Härtlová A., Van Eyk J. E., et al. (2015) A mass spectrometric-derived cell surface protein atlas. PLoS One 10, e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Snead D., and Eliezer D. (2014) Alpha-synuclein function and dysfunction on cellular membranes. Exp. Neurobiol. 23, 292–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castillo G. M., Ngo C., Cummings J., Wight T. N., and Snow A. D. (1997) Perlecan binds to the β-amyloid proteins (Aβ) of Alzheimer's disease, accelerates Aβ fibril formation, and maintains Aβ fibril stability. J. Neurochem. 69, 2452–2465 [DOI] [PubMed] [Google Scholar]
- 36. Snow A. D., Sekiguchi R., Nochlin D., Fraser P., Kimata K., Mizutani A., Arai M., Schreier W. A., and Morgan D. G. (1994) An important role of heparan sulfate proteoglycan (Perlecan) in a model system for the deposition and persistence of fibrillar Aβ-amyloid in rat brain. Neuron 12, 219–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.