Abstract
Epidermal homeostasis relies on a well-defined transcriptional control of keratinocyte proliferation and differentiation, which is critical to prevent skin diseases such as atopic dermatitis, psoriasis or cancer. We have recently shown that the homeobox transcription factor DLX3 and the tumor suppressor p53 co-regulate cell cycle-related signaling and that this mechanism is functionally involved in cutaneous squamous cell carcinoma development. Here we show that DLX3 expression and its downstream signaling depend on protein kinase C α (PKCα) activity in skin. We found that following 12-O-tetradecanoyl-phorbol-13-acetate (TPA) topical treatment, DLX3 expression is significantly upregulated in the epidermis and keratinocytes from mice overexpressing PKCα by transgenic targeting (K5-PKCα), resulting in cell cycle block and terminal differentiation. Epidermis lacking DLX3 (DLX3cKO), which is linked to the development of a DLX3-dependent epidermal hyperplasia with hyperkeratosis and dermal leukocyte recruitment, displays enhanced PKCα activation, suggesting a feedback regulation of DLX3 and PKCα. Of particular significance, transcriptional activation of epidermal barrier, antimicrobial peptide and cytokine genes is significantly increased in DLX3cKO skin and further increased by TPA-dependent PKC activation. Furthermore, when inhibiting PKC activity, we show that epidermal thickness, keratinocyte proliferation and inflammatory cell infiltration are reduced and the PKC-DLX3-dependent gene expression signature is normalized. Independently of PKC, DLX3 expression specifically modulates regulatory networks such as Wnt signaling, phosphatase activity and cell adhesion. Chromatin immunoprecipitation sequencing analysis of primary suprabasal keratinocytes showed binding of DLX3 to the proximal promoter regions of genes associated with cell cycle regulation, and of structural proteins and transcription factors involved in epidermal differentiation. These results indicate that Dlx3 potentially regulates a set of crucial genes necessary during the epidermal differentiation process. Altogether, we demonstrate the existence of a robust DLX3–PKCα signaling pathway in keratinocytes that is crucial to epidermal differentiation control and cutaneous homeostasis.
Balanced proliferation and differentiation of keratinocytes is essential to cutaneous homeostasis and prevention of skin diseases associated with epidermal barrier defects and cancer. In this context, the coordinated activity of specific enzymes and transcription factors define a complex signaling network that regulates keratinocyte maturation toward terminal differentiation.1, 2
The homeobox transcription factor DLX3 has emerged as a key factor in the control of epidermal homeostasis.3, 4 DLX3 is expressed during the calcium (Ca2+)-dependent epidermal differentiation process.5, 6 Mice lacking DLX3 in keratinocytes (DLX3cKO) display epidermal hyperplasia accompanied by barrier disruption with associated development of an inflammatory response,3 while DLX3 ectopic expression in the basal layer promotes premature keratinocyte differentiation and severely affects the skin phenotype.5 DLX3 is a target of the p53 family member p63 during ectodermal development and is involved in a regulatory feedback loop with p63 which is crucial for the maintenance of the stratified epithelia.7, 8 More recently, we demonstrated that DLX3 exerts cell cycle regulatory activity in the epidermis by cooperating with the tumor suppressor p53 and is potentially associated with cutaneous squamous cell carcinoma (cSCC) development by controlling p53 and p63 target genes in keratinocytes.4 However, the specific signaling pathways regulating DLX3 expression during keratinocyte differentiation remains to be fully determined.
In skin, protein kinase C (PKC) signaling is implicated in several pathways leading to epidermal differentiation,9 with the identification of specific transcriptional PKC effectors in keratinocytes, such as the AP1 complex10 and other recently determined.11, 12, 13, 14 Among the PKC isozymes, PKCα is activated in response to differentiation and pro-inflammatory stimuli, such as calcium or 12-O-tetradecanoyl-phorbol-13-acetate (TPA),9 and acts by inhibiting early differentiation marker expression in favor of late differentiation markers.15 We previously showed that PKC isoforms are able to phosphorylate DLX3 protein in vitro, primarily by PKCα.16 DLX3 protein was phosphorylated in nuclear extracts of mouse keratinocytes and this phosphorylation was inhibited by the PKC inhibitor GF109203X, indicating that DLX3 protein could be potentially phosphorylated by PKC in vivo. The phosphorylation event of the DLX3 full-length protein by PKC altered its DNA binding activity.16
Herein we provide evidence of a robust DLX3–PKCα axis in keratinocytes, where PKCα also directly regulates DLX3 expression. Gene expression profiles associated with DLX3 epidermal loss, as well as the DLX3cKO skin phenotype, are partially reversed by PKC activity inhibition, emphasizing the functional relevance of the DLX3–PKC-dependent signaling in skin homeostasis. Moreover, we identified a DLX3-dependent, PKC-independent gene signature that is transcriptionally modulated in response to epidermal differentiation.
Results
DLX3 is a downstream effector of PKCα activity
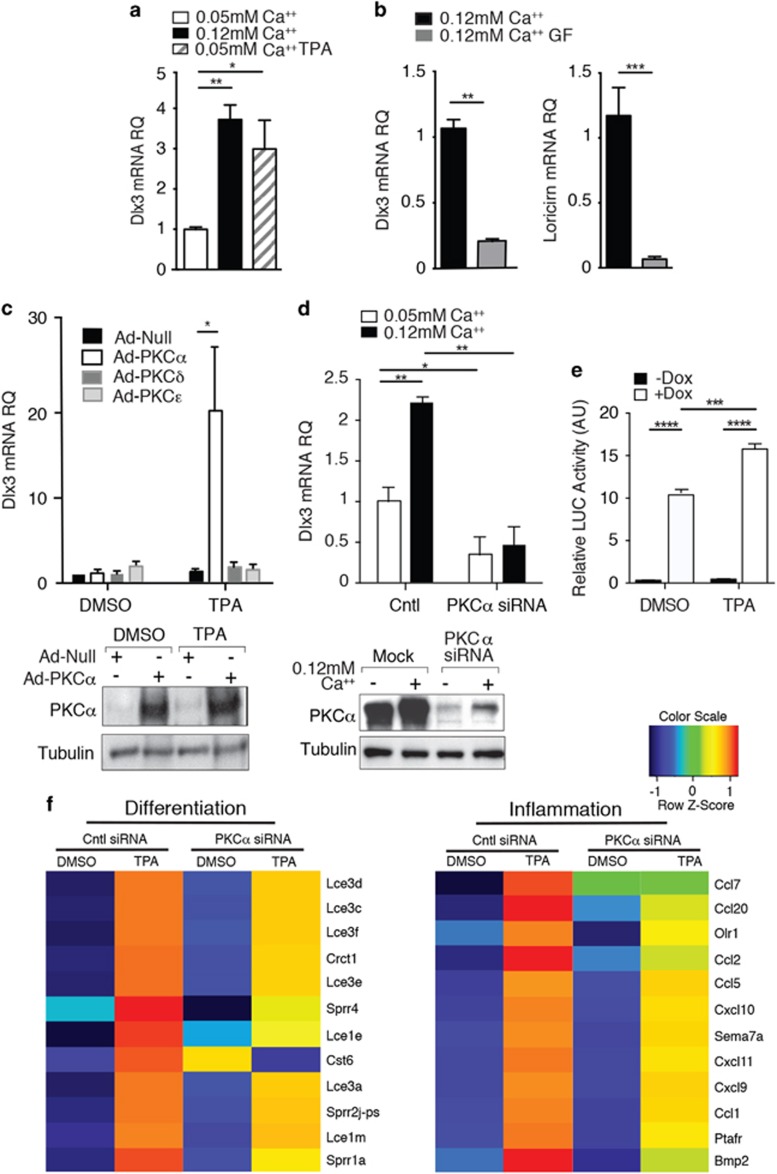
DLX3 expression is Ca2+ dependent in human and mouse epidermis and crucial for cell cycle exit and differentiation of keratinocytes.4, 6 To determine the signaling mechanisms involved in the induction of DLX3 expression by Ca2+ in keratinocytes, we assessed whether this expression was dependent on PKC activity. We demonstrated that Ca2+ treatment and TPA, a direct PKC activator, increased DLX3 mRNA (Figure 1a) in cultured primary keratinocytes. Conversely, PKC inhibition by GF109203X (hereafter referred to as GF) reduced the Ca2+-induced DLX3 mRNA levels, as well as the expression of the late differentiation markers loricrin and filaggrin, and of the cell cycle inhibitor and DLX3 target, p21 (Figure 1b and Supplementary Figure S1).
Figure 1.
PKCα triggers DLX3 expression in keratinocytes. (a) DLX3 expression in primary keratinocytes maintained in proliferative (0.05 mM Ca2+) and differentiating media (0.12 mM Ca2+) or treated with TPA for 24 h. (b) DLX3 and Loricrin expression in primary keratinocytes maintained in 0.12 mM Ca2+ media +/− GF109203X at 24 h. (c) Relative expression level of DLX3 in primary keratinocytes transduced with Adeno-Null or Adeno-PKCα, δ or ɛ viruses and treated with TPA for 1 h in 0.05 mM Ca2+ media. Bottom panel, western blot performed with protein extracts from cells transduced with Ad-Null and Adeno-PKCα, with or without TPA treatment. (d) DLX3 expression in primary keratinocytes transfected with PKCα siRNA at 48 h in 0.05 mM Ca2+ or 0.12 mM Ca2+ media. Bottom panel, western blot performed with protein extracts from cells transduced with PKCα siRNA. (e) Luciferase reporter assays were performed in PAM212-DLX3 tet-on cells transfected with a Firefly luciferase reporter construct containing the concatemerized canonical DLX3 binding. Cells were grown with or without Doxycyclin for 24 h to induce DLX3 expression. Significantly higher reporter activity was observed in cells treated with Dox after 6 h of TPA treatment. For all panels, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 and the results are mean ± SD of three experiments. (f) Heatmaps of differentially expressed mRNAs in PKCα versus Scramble siRNA treated keratinocytes after TPA or DMSO (control) stimulus for differentiation (left) or inflammation (right) clusters. Expression values are colored based on their z-score after normalization across treatments
Among the epidermal-specific PKC isoforms, the activation of PKCα is Ca2+-regulated during epidermal differentiation.15 We demonstrated that the overexpression and activation of PKCα, but not PKCδ or ɛ, induced DLX3 expression (Figure 1c), while DLX3 transcription was specifically reduced by PKCα silencing either in proliferative or in differentiating conditions (Figure 1d). In contrast, DLX3 mRNA levels significantly increased by PKCδ and PKCɛ silencing, with PKCδ silencing detected only under high-Ca2+ differentiating conditions (Supplementary Figure S1c). This effect could be potentially associated with the roles of PKCɛ and PKCδ in keratinocyte proliferation and differentiation17, 18 through a mechanism that remains to be characterized.
To evaluate the effect of PKCα activation by TPA on DLX3 transcriptional activity, luciferase reporter assays were performed utilizing PAM-TetOn-V5DLX3/GFP cells that are an immortalized murine keratinocyte cell line that express DLX3 upon addition of doxycycline (Dox).19 PAM-TetOn-V5DLX3/GFP cells were transfected with a Firefly luciferase reporter construct pGL3-3xDLX3, which contains three concatenated canonical DLX3 binding sites.19 Cells were grown with or without Dox for 24 h to induce DLX3 expression, and with DMSO or TPA. Significantly higher reporter activity was observed in cells treated with Dox in addition to TPA (Figure 1e). No significant difference was found when there was no addition of Dox. These results confirm that the induction of DLX3 transcription is directly dependent on PKCα activation in keratinocytes in vitro and support that DLX3 transcriptional activity is also PKC dependent.
To determine the biological processes associated with PKCα-DLX3 signaling, we performed a global transcriptome analysis by DAVID Functional Annotation tool (https://david.ncifcrf.gov) of cultured keratinocytes after PKCα siRNA and subsequent TPA treatment for 24 h. Gene Ontology (GO) term analysis shows that PKCα silencing, and the consequent DLX3 downregulation, leads to a significant enrichment in Regulation of transcription, Keratinocyte differentiation, Phosphorylation and Cell-adhesion Biological Process (BP) terms (Supplementary Figure S1d). When cells are TPA treated to induce PKC activation, PKCα silencing results in a significant enrichment in Immune response, T-cell chemotaxis, angiogenesis and negative regulation of cell proliferation GO BP terms (Supplementary Figure S1d). Interestingly, the marked induction of differentiation (Lce and Sprr late differentiation-associated genes) and inflammation related genes was substantially diminished with PKCα siRNA in the presence of TPA (Figure 1f). Moreover, by comparing genes downregulated in DMSO PKCα siRNA versus DMSO control siRNA treated cells, we found an enrichment in genes significantly related to protein kinase signaling and cell adhesion (Supplementary Figure S1e). Globally, these results confirm the relevance of PKCα signaling in keratinocyte differentiation and its contribution to the TPA-induced inflammatory response.20 These results also support the existence of a PKCα–DLX3 axis, which is involved in the regulation of epidermal differentiation by the modulation of PKC signaling and in the regulation of cutaneous homeostasis.
Our previous work demonstrated that PKC isoforms, primarily PKCα, phosphorylate recombinant DLX3 protein in vitro.16 Herein, we demonstrated that DLX3 protein levels are upregulated by PKC activation, either by calcium or by TPA treatment in primary cultured keratinocytes (Supplementary Figure S2a). The analysis of DLX3 phosphorylation by the use of Phos-tag acrylamide technology (Wako Pure Chemical Industries, Ltd, Richmond, VA, USA) on keratinocytes transduced with the Adeno-DLX3 expression vector and then treated with TPA confirmed that DLX3 protein is phosphorylated at 1-h post TPA treatment in the absence of protein phosphatase inhibitor (Supplementary Figure S2).
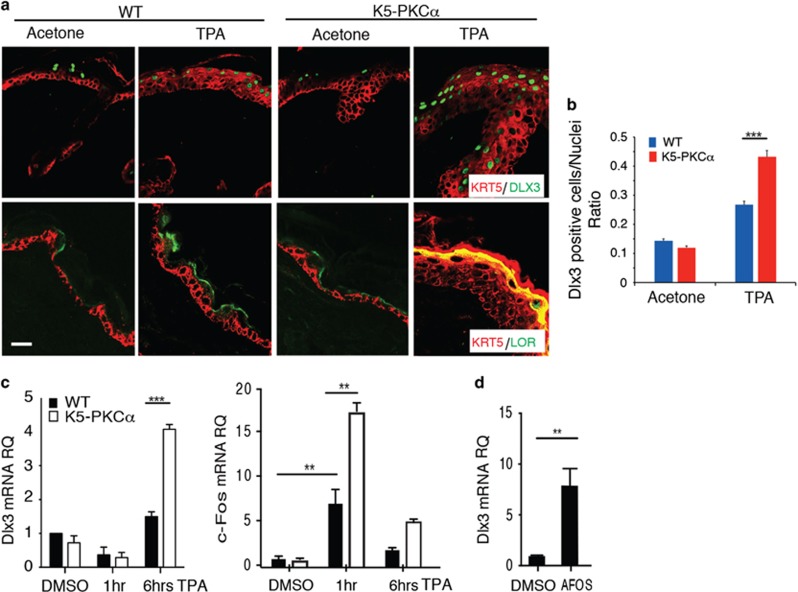
The biological relevance of the PKCα–DLX3 connection was tested in vivo using an epidermal-targeted, TPA-inducible K5-PKCα transgenic mouse model.20 We found a significant increase in DLX3-expressing cells, as well as higher loricrin expression, in TPA-treated transgenic epidermis (Figures 2a and b). Cultured TPA-treated K5-PKCα keratinocytes displayed DLX3 mRNA induction 6 h after treatment (Figure 2c), which is preceded by the PKC effector c-Fos at 1 h post TPA treatment. The temporal upregulation of c-Fos expression preceding DLX3 was also confirmed by cycloheximide (CHX) treatment of keratinocytes in a time course experiment (Supplementary Figure S3).
Figure 2.
DLX3 expression is upregulated by PKCα activation in vivo. (a) Immunohistochemical staining of TPA-treated WT and K5-PKCα transgenic skin with antibodies against KRT5 and DLX3 or KRT5 and Loricrin (LOR). Scale bar, 20 μm. (b) Bar graph showing the number of DLX3-positive cells normalized versus the nuclei in WT and K5-PKCα skin treated with TPA or acetone (control). DLX3-positive cells and the number of nuclei in each section were determined by ImageJ software analysis of the labeled particles. Data are presented as mean±S.D. of three different areas from three independent mice for each condition. ***P<0.001. (c) Relative expression level of DLX3 and c-Fos in WT or K5-PKCα primary keratinocytes treated with TPA at 1 and 6 h. (d) DLX3 expression level in cultured keratinocytes transduced with Ad-AFOS or Ad-Cntl vectors at 48 h. For all panels: **P<0.01, ***P<0.001 and the results are shown as mean±S.D. of three independent experiments
Increased AP1 activity is associated with PKCα activation and its inhibition prevents TPA-dependent cell death of K5-PKCα keratinocytes.20 Keratinocytes transduced with a dominant negative to AP1 activation (AFOS) in proliferative conditions (0.05 mM Ca2+) showed significantly increased DLX3 expression (Figure 2d), leading to the hypothesis that basal (non TPA driven) AP1 activity is negatively regulating Dlx3 mRNA expression. Altogether, these results support that DLX3 expression requires PKCα activation and is a downstream target and effector of PKCα signaling in skin.
The DLX3–PKC interplay control of epidermal homeostasis
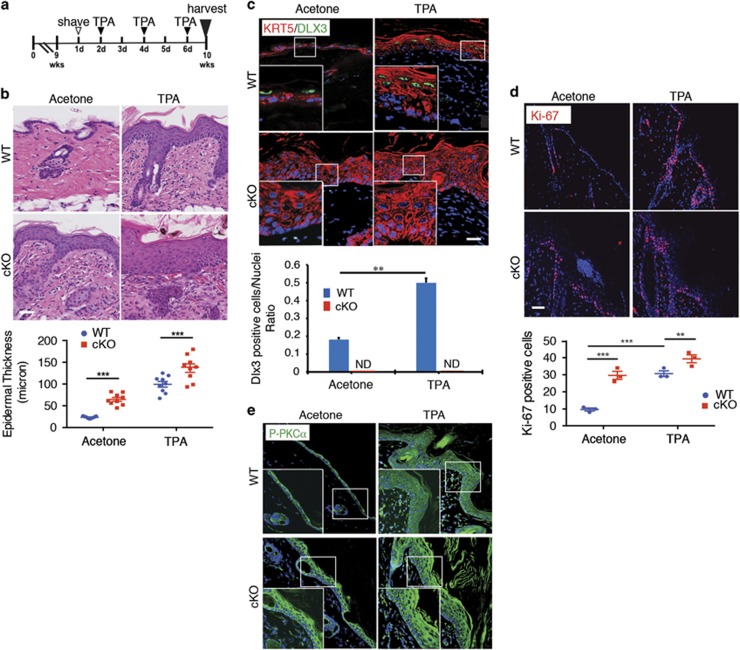
To functionally evaluate the facets of DLX3 and PKCα axis, we used TPA topical application to activate PKC in a mouse model with DLX3 epidermal deletion (K14Cre;DLX3Kin/flox; hereafter referred as DLX3cKO 3) and in WT skin (Figure 3a). TPA treatment led to heightened hyperplasia and hyperkeratosis with significantly increased epidermal thickness and dermal cell infiltration in the DLX3cKO skin when compared with untreated DLX3cKO or TPA-treated WT skin (Figure 3b).
Figure 3.
PKC activation exacerbates DLX3cKO epidermal hyperproliferation. (a) Schematic representation of TPA treatment of WT or DLX3cKO mice. (b) Hematoxylin and eosin staining of TPA-treated WT or DLX3cKO transgenic skin. Scale bar, 20 μm. Epidermal thickness was measured and data are presented as mean±S.D. of three different areas from three different mice for each condition. ***P<0.001. (c) Immunohistochemical staining of TPA-treated WT and DLX3cKO transgenic skin with antibodies against KRT5 and DLX3. Nuclei were stained with DAPI. Scale bar, 20 μm. Bottom panel, bar graph showing the number of DLX3-positive cells normalized versus the nuclei in WT and DLX3cKO skin treated with TPA or acetone (control). DLX3-positive cells and the number of nuclei in each section were determined by ImageJ software analysis of the labeled particles. Data are presented as mean±S.D. of three different areas from three independent mice for each condition. **P<0.01. (d) Immunohistochemical staining of TPA-treated WT and DLX3cKO transgenic skin with antibodies against Ki-67. Nuclei were stained with DAPI. Scale bar, 20 μm. Bottom panel, Ki-67-positive cell count in WT and DLX3cKO epidermis +/− TPA. Data are presented as mean±S.D. of three different areas from three different mice for each condition. (e) Immunohistochemical staining of TPA-treated WT and DLX3cKO mice with antibodies against P-PKCα. Nuclei were stained with DAPI. Scale bar, 20 μm
Analogous to the results obtained with the K5-PKCα transgenic mouse model (Figure 2a), treatment with TPA strongly increased the number of DLX3-expressing cells in WT skin (Figure 3c). Increased cell proliferation, determined by higher number of Ki-67-positive cells, was assessed for acetone (vehicle) and TPA-treated DLX3cKO epidermis when compared with WT in the same conditions (Figure 3d). Moreover, immunohistochemical analysis of PKCα phosphorylation in acetone and TPA-treated WT and DLX3cKO skin showed heightened detection of p-PKCα in untreated DLX3cKO epidermis as well as in the differentiated compartment of the TPA-treated DLX3cKO epidermis when compared with WT (Figure 3e).
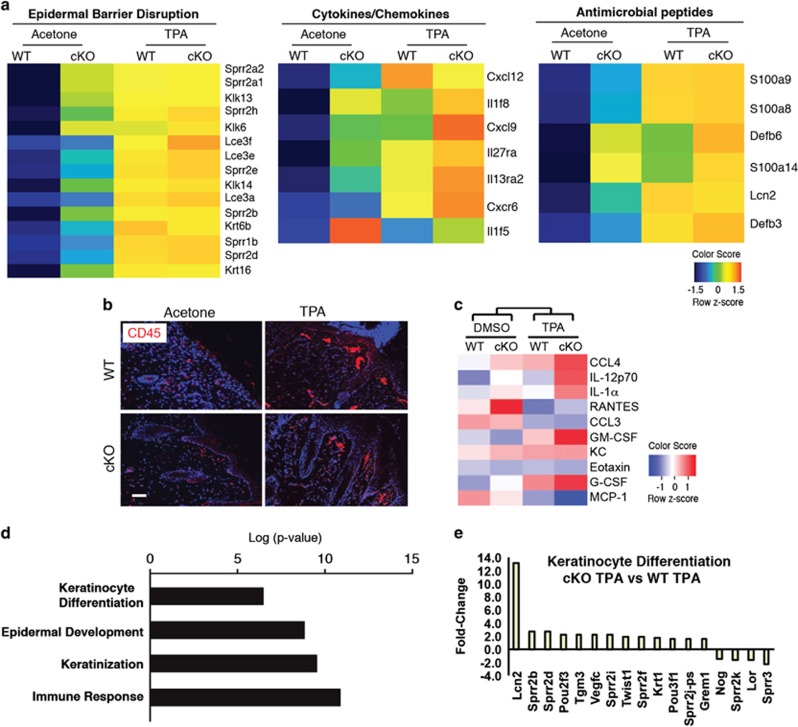
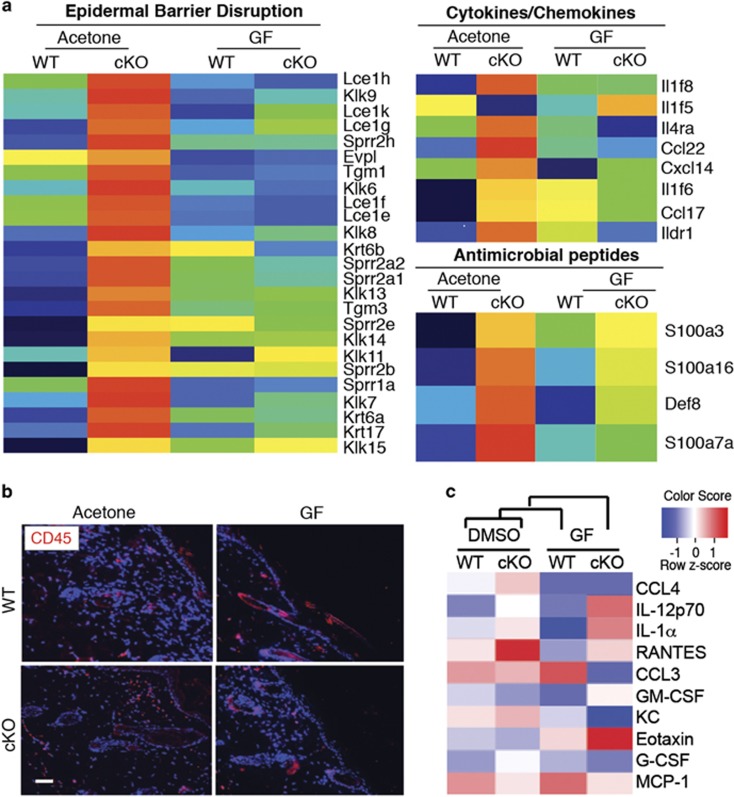
A global transcriptome analysis by DAVID Functional Annotation tool, of acetone and TPA-treated DLX3cKO or WT skin revealed that TPA treatment in a DLX3-ablated epidermis leads to the upregulation of gene expression profiles related to epidermal barrier disruption, cytokines/chemokines and antimicrobial peptides (Figure 4a). Particularly relevant in the TPA-treated DLX3cKO skin is the upregulation of expression of a subset of Lce, Sprr, S100 and kallikrein genes, as well as beta-defensin, lipocalin-2, Krt6b and Krt16 genes, which are strongly linked to epidermal hyperproliferative inflammatory diseases.21, 22, 23, 24, 25
Figure 4.
PKC activation raises the DLX3cKO epidermal barrier alteration and inflammatory phenotype. (a) Heatmaps of differentially expressed mRNAs in WT or DLX3cKO skin treated with TPA or acetone (control) represent the top affected clusters by DAVID functional annotation analysis: epidermal barrier disruption (left), cytokine/chemokine (middle) and antimicrobial peptides (right). Expression values are colored based on their z-score after normalization across treatments. (b) Immunohistochemical staining of TPA-treated WT and DLX3cKO transgenic skin with antibodies against CD45. Nuclei were stained with DAPI. Scale bar, 20 μm. (c) Heatmap of the cytokines released by WT or DLX3cKO keratinocytes after treatment with TPA in culture for 24 h. DMSO was used as a control. Cytokine concentration values are colored based on their z-score after normalization across treatments. (d) Bar graph showing GO term Biological Process (BP) enrichment by DAVID Functional Annotation tool in DLX3cKO versus WT keratinocytes treated with TPA for 36 h. (e) Bar graph showing the fold change for keratinocyte differentiation-related genes in TPA-treated DLX3cKO versus WT keratinocytes
Given that the epidermal deletion of DLX3 is linked to an IL-17-associated skin inflammation,3 we analyzed leukocyte accumulation in the dermis by staining for CD45 in TPA-treated DLX3cKO skin. CD45-positive cell count was determined by measuring the total labeled particles and area, as described in Materials and Methods. We found that TPA treatment further increased the detection of CD45-positive cells in DLX3cKO skin compared with the TPA-treated WT or untreated DLX3cKO (Figure 4b and Supplementary Figure S4). We also assessed the inflammatory response of DLX3-null keratinocytes in culture by treatment with TPA and analysis of cytokine release utilizing a profiler array. Z-score evaluation of the cytokine concentrations confirmed a correlation with the release of the inflammatory mediators associated with leukocyte chemotaxis26, 27, 28, 29, 30 in TPA-treated DLX3cKO keratinocytes when compared with DMSO (control vehicle)-treated DLX3cKO cells, or TPA and DMSO-treated WT cells (Figure 4c).
Transcriptome analysis of TPA-treated DLX3cKO versus WT cultured keratinocytes revealed significant enrichment in epidermal differentiation, keratinization, epidermal development and immune response GO Biological Process terms (Figure 4d), with differential expression of the epidermal differentiation-related genes such as Sprr, Tgm3 and Lcn2 (Figure 4e). These results correlate with those obtained in TPA-treated keratinocytes in culture (Figure 1f) and in TPA-treated DLX3cKO skin (epidermal barrier disruption and antimicrobial peptides; Figure 4a). Interestingly, the expression of Loricrin, which is a downstream target of PKCα-dependent differentiation,31 was specifically reduced in DLX3-null keratinocytes compared with WT in the presence of PKC activation. These data support a functional and integrated DLX3–PKC pathway required for proper transcriptional control of keratinocyte proliferation and differentiation, as well as a homeostatic maintenance of the epidermal barrier.
PKC inhibition restores a more normal phenotype to DLX3cKO epidermis
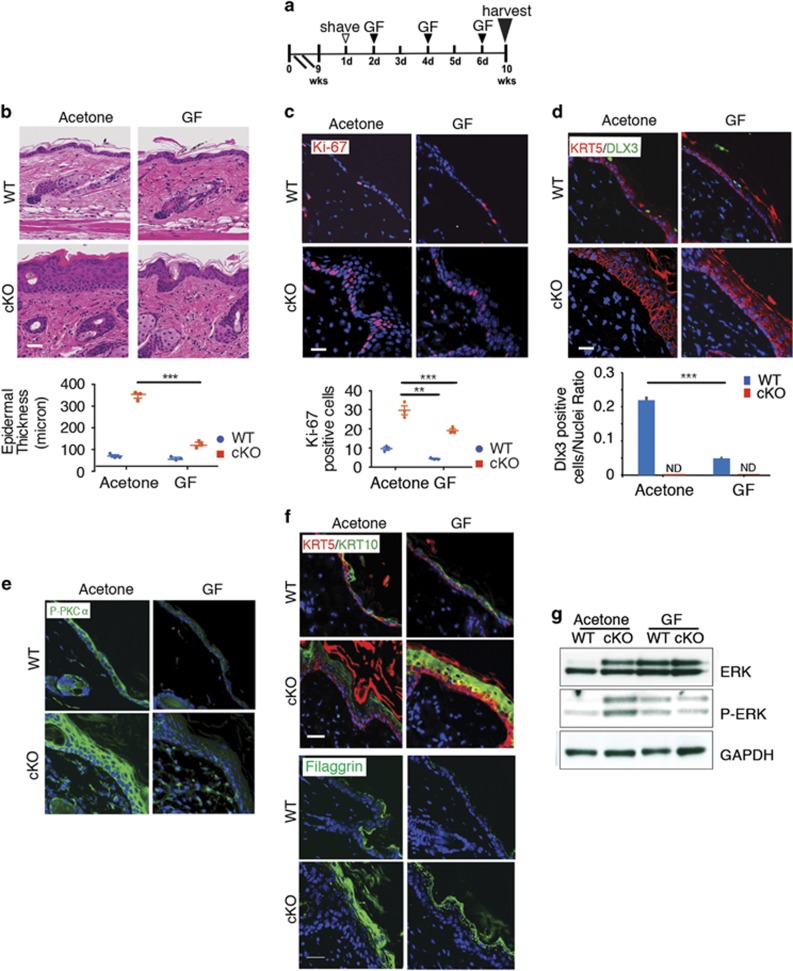
Next, we investigated the effect of PKC inhibition in DLX3cKO epidermis by treatment with GF following the same protocol used for TPA treatments (Figure 5a). GF application significantly decreased epidermal thickness compared with the untreated mice (Figure 5b), with decreased hyperplasia and hyperkeratosis and a reduction of Ki-67-positive cells (Figure 5c). A reduced number of DLX3-positive cells in GF-treated WT skin was detected (Figure 5d). The epidermal hyperplasia in DLX3cKO skin was also normalized by the specific PKCα inhibitor Gö6976 (Supplementary Figure S5).
Figure 5.
PKC inhibition partially restores epidermal thickness and keratinocyte proliferation in DLX3cKO skin. (a) Schematic representation of GF109203X (GF) treatment of WT or DLX3cKO mice. (b) H&E staining of GF-treated WT and DLX3cKO skin. Epidermal thickness was measured and data are presented as mean±S.D. of three different areas from three different mice per condition. (c) Immunohistochemical staining of GF-treated WT and DLX3cKO transgenic skin with antibodies against Ki-67. Nuclei were stained with DAPI. Scale bar, 20 μm. Bottom panel, Ki-67-positive cell count in WT and DLX3cKO epidermis +/− GF. Data are presented as mean±S.D. of three different areas from three different mice for each condition. (d) Immunohistochemical staining of GF-treated WT and DLX3cKO transgenic skin with antibodies against KRT5 and DLX3. Nuclei were stained with DAPI. Scale bar, 20 μm. Bottom panel, bar graph showing the number of DLX3-positive cells normalized versus the nuclei in WT and DLX3cKO skin treated with GF or acetone (control). DLX3-positive cells and the number of nuclei in each section were determined by ImageJ software analysis of the labeled particles. Data are presented as mean±S.D. of three separate areas from three different mice for each condition. ***P<0.001. (e) Immunohistochemical staining of GF-treated WT and DLX3cKO mice with antibodies against P-PKCα and (f) against KRT5 and KRT10 (upper panel) and Filaggrin (lower panel). Nuclei were stained with DAPI. Scale bar, 20 μm. (g) Total ERK and phosphor-ERK (P-ERK) expression levels determined by western blot of protein extracts from GF-treated WT and DLX3cKO transgenic skin. GAPDH was used as a loading control
The inhibition of PKCα phosphorylation after GF treatment was confirmed by immunohistochemistry (Figure 5e). In addition, the inhibition of PKC normalizes the expression of filaggrin and recovers the expression of keratin 10 in GF-treated DLX3cKO epidermis (Figure 5f) underlining the role of activated PKCα in DLX3cKO epidermis. Moreover, we found increased mRNA levels for p53, p21 and CcnE1 in GF-treated DLX3cKO skin compared with vehicle-treated skin (Supplementary Figure S6), suggesting a potential cell cycle rescue in DLX3cKO skin.
ERK is a well-known downstream effector of PKCα activity in keratinocytes32 and we previously reported that loss of DLX3 in cultured keratinocytes is associated with activation of ERK.4 Here, we corroborated this effect in vivo, and showed that DLX3cKO epidermis presented higher level of the phosphorylated ERK and that it decreased in GF-treated DLX3cKO skin, with total ERK remaining unchanged (Figure 5g). Altogether, these results support that PKCα signaling is connected with DLX3-dependent regulation of epidermal growth and absence of DLX3 function in keratinocytes causes a PKCα-dependent alteration of the epidermal phenotype.
We conducted a global transcriptomic analysis of the GF-treated and untreated DLX3cKO and WT skin and found that relative to untreated DLX3cKO skin, GF treatment downregulated a subset of barrier-associated markers, cytokine/chemokines and antimicrobial peptides (Figure 6a), previously found to be affected by activated PKCα in DLX3cKO skin (Figure 4a). We show that GF treatment decreased the detection of CD45-positive cells in DLX3cKO skin compared with the GF-treated WT or untreated DLX3cKO (Figure 6b and Supplementary Figure S7).
Figure 6.
PKC inhibition restores the DLX3cKO epidermal barrier alteration and inflammatory phenotype. (a) Heatmaps of differentially expressed mRNAs in WT or DLX3cKO skin treated with GF109203X or acetone (control) related to the top affected clusters by DAVID functional annotation analysis: epidermal barrier disruption (left), cytokine/chemokine (middle) and antimicrobial peptides (right). Expression values are colored based on their z-score after normalization across treatments. (b) Immunohistochemical staining of GF109203X-treated WT and DLX3cKO transgenic skin with antibodies against CD45. Nuclei were stained with DAPI. Scale bar, 20 μm. Data are presented as mean±S.D. of three different areas from three different mice for each condition. (c) Heatmap of the cytokines released by WT or DLX3cKO keratinocytes after treatment with GF109203X in culture for 24 h. DMSO was used as a control. Cytokine concentration values are colored based on their z-score after normalization across treatments
GF treatment also specifically decreased the level of cytokines released by DLX3cKO cells in vitro (Figure 6c). Interestingly, the greater IL-1α release by TPA-treated DLX3-null keratinocytes compared with WT in the same condition was not inhibited by GF, indicating that this mechanism is independent of the PKC–DLX3 axis, but is associated with the DLX3-null keratinocyte phenotype.4 In contrast, the release of G-CSF, KC, CCL3 and CCL4 was specifically inhibited in the DLX3cKO cells and the secretion of GM-CSF, RANTES and MCP-1 was more comparable to the WT level after GF treatment (Figure 6c). Other cytokines such as Eotaxin and IL-12p70 showed upregulation after GF treatment in a process that might involve signals distinct from the PKC–DLX3 pathway.
DLX3-dependent control of epidermal homeostasis by PKC-independent signaling
The analysis of differentially expressed genes between DLX3cKO and WT skin, both treated with TPA or GF, or untreated, allowed the identification of specific signaling pathways within cutaneous homeostasis whose regulation is DLX3 dependent and not linked to PKC activity (Figure 7a).
Figure 7.
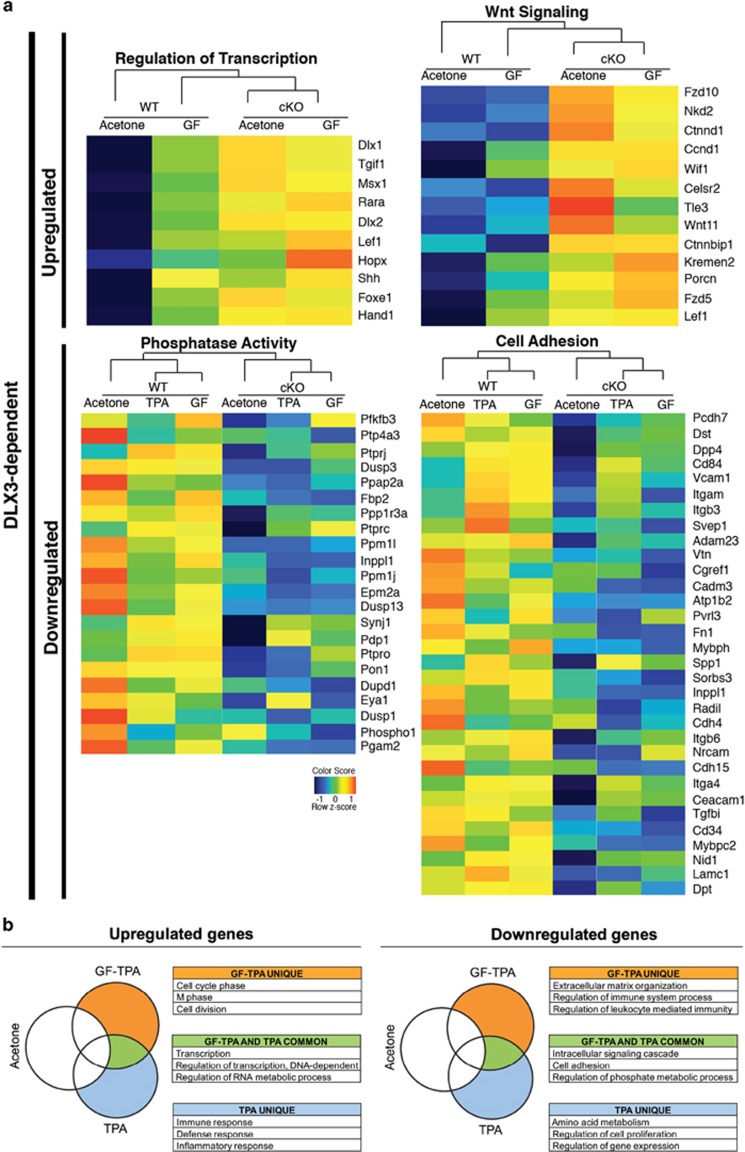
DLX3-dependent regulation of epidermal homeostasis-related signaling pathways. (a) Heatmaps showing the DLX3-dependent upregulation of the 'Regulation of Transcription' and 'Wnt Signaling' pathways between DLX3cKO and WT skin treated with GF109203X or acetone (control). Bottom: Heatmaps showing the DLX3-dependent downregulation of the 'Phosphatase Activity' and 'Cell Adhesion' signaling pathways between DLX3cKO and WT skin treated with TPA or GF109203X or acetone (control). Expression values are colored based on their z-score after normalization across treatments. (b) Schematic representation by Venny diagrams of the significantly upregulated (left) or downregulated (right) genes in DLX3cKO skin after treatment with GF109203X prior to TPA (GF-TPA), TPA or acetone (control). Top significant GO Biological Process (BP) terms are indicated in tables: GF-TPA unique (orange), TPA unique (light blue) and GF-TPA and TPA common (green)
In DLX3-null skin, we found that the expression level of Msx1, Rara, Shh, Lef1 and Foxe1 (Regulation of transcription; Figure 7a) was upregulated and PKC inhibition did not significantly modulate their expression level (Figure 7a, upper left heatmap). These genes are expressed during epidermal differentiation, target-specific aspects of keratinocyte maturation and hair follicle development.33, 34, 35 DLX3 is expressed during hair morphogenesis in the hair matrix at the beginning of hair shaft differentiation and its deletion results in impaired expression of hair keratins leading to alopecia.36 The modulation of a subset of genes, for example, Lef1, Shh, Msx1, Foxe1 that are regulated by DLX3 independently of PKC, could also be attributed to DLX3 function in keratinocytes of the hair follicle.36 DLX3 function was also linked to the Wnt signaling pathway, with all of the genes included in this pathway being significantly upregulated in DLX3cKO skin. Wnt signaling pathway showed little alteration after GF treatment (Figure 7a, upper right heatmaps).
Transcripts for genes encoding for protein phosphatases37 and adhesion molecules, such as integrins and CAM38, 39 (Figure 7a, lower heatmaps) were decreased in the absence of DLX3 and expression was not affected by PKC inhibition. Remarkably, the dual specificity phosphatases, which present downregulation during cSCC progression,37 are downregulated in DLX3-null cells.
TPA treatment induces skin inflammation and hyperproliferation through different mechanisms.9 In order to define what aspects of these processes are PKC dependent in DLX3cKO skin, we performed treatments with GF prior to TPA application (hereafter referred to as GF-TPA treatment), following the same protocol used in Figure 3a. RNA-seq analysis determined the upregulated and downregulated genes in acetone (control vehicle), TPA or GF-TPA-treated DLX3cKO skin, which were normalized against WT. Unique and common genes were determined by Venny comparative tool (http://bioinfogp.cnb.csic.es/tools/venny) and each list was analyzed with DAVID Functional Annotation tool to obtain GO BP term enrichment. When PKC signaling is inhibited in DLX3cKO skin, there is a subset of specific genes involved in cell cycle (cell cycle phase, M phase, cell division) (Figure 7b, left panel) that is upregulated by TPA, while genes within regulation of the immune system process and regulation of leukocyte-mediated immunity clusters were downregulated in the same conditions (Figure 7b, right panel). Therefore, PKC activation in DLX3cKO skin is linked to the TPA-induced phenotype. Interestingly, the analysis of the genes common between GF-TPA and TPA treatments corroborated that the regulation of transcription, the intracellular signaling cascade, cell adhesion and the regulation of phosphate metabolic processes are DLX3 -dependent signaling pathways.
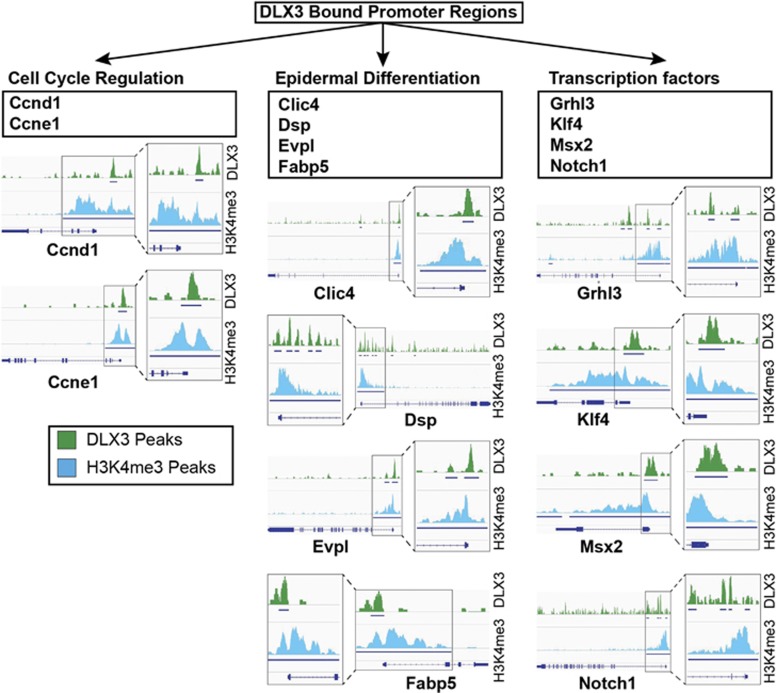
In order to determine potential direct targets of DLX3 regulation, we performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis on epidermal suprabasal cells. ChIP-seq analysis of DLX3 and H3K4me3 DNA binding domains identified DLX3-bound promoter regions of genes involved in cell cycle regulation during differentiation, such as Ccnd1, Ccne1;41, 42 and epidermal structure, such as Clic4, Dsp, Evpl, Fabp5,43, 44, 45, 46 and transcription factors such as Grhl3, Klf4, Msx2 and Notch147, 48, 49, 50 (Figure 8). Together with the transcriptomic analysis, the ChIP-seq analysis of DLX3 and H4K4m3 binding supports a potential DLX3-dependent transcriptional regulation of these genes during epidermal differentiation.
Figure 8.
DLX3 transcriptional control of cell cycle and epidermal differentiation. ChIP-seq analysis of DLX3 and H3K4me3 DNA binding domains in suprabasal epidermal keratinocytes, showing DLX3 binding within the promoter regions of genes involved in cell cycle regulation (Ccnd1, Ccne1), epidermal differentiation (Clic4, Dsp, Evpl, Fabp5) and transcription factors (Grhl3, Klf4, Msx2, Notch1). Integrative Genomics Viewer and PAPST59 were used to visualize the peak regions from Transcription Start Sites (TSS)
Discussion
The homeobox transcription factor DLX3 is well known for its implication in developmental regulation of several tissues. 51, 52 However, its role as a 'switch molecule' regulating the balance between differentiation and proliferation of keratinocytes in epidermis has recently emerged.4 DLX3 is expressed in the nuclei of the suprabasal epidermal cells of healthy skin.3 In contrast, in skin disorders associated with altered differentiation and barrier defects, such as atopic dermatitis or psoriasis, and squamous cell carcinoma, its expression pattern is altered with a higher number of DLX3-positive cells in atopic dermatitis compared with no detectable expression in psoriasis and cSCC.3, 4 These findings and observations indicate that nuclear suprabasal DLX3 expression is strongly associated with the maintenance of epidermal homeostasis, by potentially regulating multiple pathways in skin either directly or indirectly through other transcription factors. In fact, we previously showed that a conditional epidermal deletion of DLX3 in mouse skin induces skin phenotype alterations with epidermal hyperplasia, barrier defects and the development of an IL-17 dependent inflammatory response,3 overall supporting the relevance of the DLX3 signaling pathways in epidermal homeostasis.
We report that DLX3 is a downstream signaling effector of PKC and highlight the importance of a functional PKC–DLX3 axis during epidermal differentiation to preserve the cutaneous homeostatic balance. Previous work demonstrates that PKC isoforms are able to interact and phosphorylate DLX3 in vitro. Our results here confirm this data and show a direct link between PKCα and DLX3, establishing DLX3 as a target of PKCα signaling. Furthermore, while PKCα regulates DLX3, there is heightened detection of p-PKCα in DLX3cKO epidermis evoking a potential negative feedback loop that regulates PKCα activity. We found no putative DLX3 binding site in the PKCα promoter and no significant variation of PKCα mRNA expression in any of the data set profiles analyzed (Supplementary Table S1). Nevertheless, p-PKCα is elevated in epidermis genetically deleted of DLX3 and the hyperproliferative response to TPA is increased, suggesting that the homeobox protein indirectly regulates the activity in the pathway, possibly through an effect on reduced phosphatase expression (Figure 7a). The reduction in phosphatase activity could also explain the increase in p-ERK previously reported in the DLX3cKO epidermis.4
Increasing PKCα activity in skin is linked to altered terminal differentiation and intraepidermal inflammation.20 Here we demonstrate that, in absence of DLX3, the expression of late differentiation markers is favored and that recruitment of inflammatory cells, as well as release of the PKCα-dependent inflammatory mediators, increased. Taken together, these results indicate that altered PKC signaling is at least partially determinant in the development of the DLX3cKO phenotype.
Therefore, we propose a model (Figure 9) where in response to differentiation or pro-inflammatory stimuli, such as calcium or TPA, DLX3 partially regulates the expression of a PKC-dependent gene signature characterized by epidermal differentiation and barrier formation defects, inflammatory mediator release, antimicrobial peptides and cell cycle. However, other pathways related to the control of epidermal homeostasis (i.e. regulation of transcription, Wnt signaling, phosphatase activity and cell adhesion) result from DLX3-dependent, PKC-independent mechanisms and are directly or indirectly mediated by still unknown signaling intermediates. Other signaling pathways have been determined to be indispensable for keratinocyte differentiation and barrier function.53, 54 It remains to be determined which other DLX3-dependent signaling pathway will prove to be essential in epidermal homeostasis. Taken together, the work presented here reveals a DLX3–PKC axis in the regulation of keratinocyte proliferation and differentiation balance.
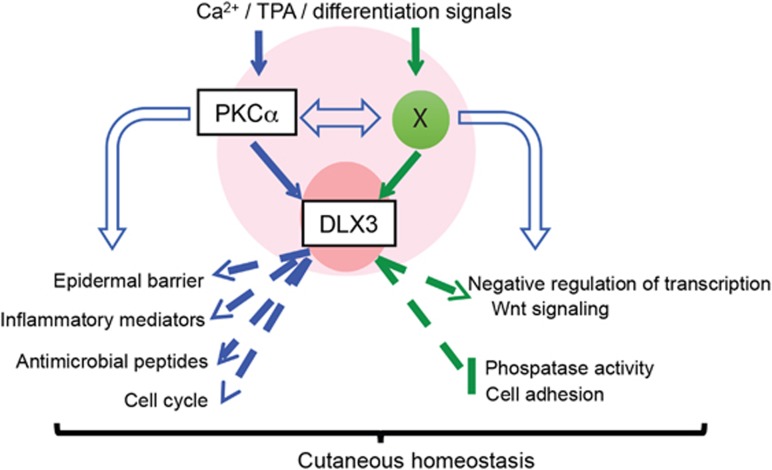
Figure 9.
PKC–DLX3-dependent control of cutaneous homeostasis. Schematic working model of PKC–DLX3 signaling in keratinocytes displaying that, in response to differentiation or pro-inflammatory stimuli, such as calcium or TPA, DLX3 regulates PKC-dependent gene expression signature associated with barrier defects, inflammatory mediator release, antimicrobial peptides and cell cycle. Independently of PKC, DLX3 is implicated in the transcriptional control of other pathways, such as negative regulation of transcription, Wnt signaling, phosphatase activity and cell adhesion. Globally this signaling is determinant for the maintenance of the cutaneous homeostasis
Materials and methods
Generation and analysis of transgenic mice
Mouse experiments were performed under a protocol approved by the National Cancer Institute (NCI) and the NIAMS Animal Care and Use Committee. DLX3cKO mice were generated and genotyped as previously reported.36 TPA-inducible K5-PKCα mice were generated as previously reported.20 Tissue was dissected 3 days after TPA treatment (2 μg/200 μl acetone) for IHC. Acetone (200μl)-treated mice were used as a control.
For GF109203X, Gö6976 and TPA treatments, 9 weeks WT and DLX3cKO mice were shaved one day prior to topical application. Mice were topically treated with TPA (5 μg/100 μl acetone), GF (50 μg/100 μl), or acetone (100 μl) every 2 days for 6 days for a total of three treatments. Tissues were dissected the day after the last treatment for IHC, RNA or protein extraction.
Primary keratinocyte culture, cell culture and transduction
Normal human epidermal keratinocytes (NHEK) were obtained from surgically resected human foreskins from the NIH Clinical Center, Dermatology Branch, after provision of written consent. NHEK were isolated and maintained in KGM (Lonza, Walkersville, MD, USA), with Ca2+ concentration of 0.05 mM (proliferating media). To induce keratinocyte differentiation, a final concentration of 1.8 mM Ca2+ was used. Primary murine keratinocytes (PMK) were isolated from TPA-inducible K5-PKCα, DLX3cKO and WT mice as previously described.55 PMK were cultured in S-MEM (Gibco, Waltham, MA, USA) with 8% fetal bovine serum (FBS; Atlanta Bio, Flowery Branch, GA, USA), 1% antibiotic (PSA; Sigma, St. Louis, MO, USA), with Ca2+ concentration of 0.05 mM (proliferating media). To induce keratinocyte differentiation, a final concentration of 0.12 mM Ca2+ was used. For gene and protein expression studies, cells were transfected with scramble siRNA or PKCα siRNA (50 nM; Qiagen, Hilden, Germany); transduced with Null or PKCα, PKCδ, PKCɛ adenoviruses (5 MOI) or transduced with Control or AFOS adenoviruses (5 MOI). Adenoviral particles were generated as previously described.56
Reporter assays
To determine the transcriptional activity, a synthetic oligonucleotide containing three tandem copies of the DLX3 responsive element (DRE; GCGATAATTGCGGCGATAATTGCGGCGATAATTGCG)57 followed by the hamster sarcoma virus thymidine kinase proximal promoter region was inserted into the pGL3-basic vector (pGL3-3XDRE) driving a Firefly luciferase reporter cassette. PAM-Tet-ON DLX3 cells were transiently co-transfected with the reporter construct and the pRL-TK vector (Renilla luciferase used for normalization). Twenty-four hours after transfection, relative luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Chemical treatments in vitro
Cells were treated with DMSO (control), TPA (5 ng/ml) (Sigma) or GF109203X (10 μM) (Calbiochem, San Diego, CA, USA) and harvested at different time points according to the specific experiment.
Western blot analysis
Lysates of cells and tissues were prepared in SDS sample buffer (60 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS and 5% 2-mercaptoethanol). Equal amounts of lysate proteins (25 μg) were separated on 4–12% SDS-PAGE gels, transferred to PVDF membranes (Invitrogen, Carlsbad, CA, USA), and incubated with primary antibodies specific to DLX3 (Epitomics, Burlingame, CA, USA), PKCα (Sigma), P-ERK (Cell Signaling, Danvers, MA, USA), T-ERK (Cell Signaling), Vinculin (Sigma) and β-actin (Sigma). Blots were then rinsed in TBST and further incubated in peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies, respectively. Proteins were visualized using an ECL detection system (Pierce Biotech, Waltham, MA, USA) and exposed to film. All experiments were repeated in triplicate.
Histology and immunohistochemistry
For histological and immunohistochemical analysis, tissues were fixed with 4% PFA and embedded in paraffin. Five micrometer sections were prepared for specific staining. Each section was incubated overnight at 4 °C with the primary antibodies which included: DLX3 (Epitomics), P-PKCα (Abcam, Cambridge, UK), KRT5 and KRT10 (Covance, Princeton, NJ, USA), Filaggrin (Covance), Ki-67 (BD Bioscience, San Jose, CA, USA) and CD45 (BD Biosciences). The Alexa Fluor 488 and 555-conjugated were used as secondary antibodies. Epidermal thickness was determined by ImageJ software analysis throughout each image. DLX3 and CD45-positive cells were determined by ImageJ software through the use of the cell counter function or by integrating particle analysis with the measure of the labeled area, respectively.
RNA extraction and qRT-PCR
Total RNA was extracted from cells and tissues using RNeasy kit (Qiagen) according to the maufacturer's instructions. cDNA was prepared using the ImProm-II Reverse Transcription System (Promega). For each gene qRT-PCR analysis was done in triplicate using the iQSYBR Green Supermix (Bio-Rad, Hercules, CA, USA). The following genes were analyzed: DLX3, p21, Flg (Filaggrin) and Lor (Loricrin), c-Fos, p53, CCNE1. Primer sequences used in qRT-PCR are indicated in Supplementary Table S2. Relative expression was normalized against the housekeeping gene RPLPO. Fold changes were calculated by the Ct method as previously described.4
Cytokine array
A Luminex Cytokine Array (Bio-Rad) was used to determine cytokine release by cells. Briefly, WT and DLX3cKO cells were treated with TPA or GF for 24 h before supernatant was collected for analysis. DMSO was used as a control. Cytokine concentration in the supernatant was determined according to the manufacturer's protocol.
Expression profiling
RNA sequencing was performed in the NIAMS Genome Core Facility at the NIH. Mondrian SP was used for the cDNA library preparation and the HiSeq 2000 was used for sequencing. Significantly affected genes (P<0.05 and fold change>1.5) were determined on ANOVA.
Computational analysis
RNA-Seq expression fold changes were calculated using Partek Genomics Suite (http://www.partek.com). Pathway enrichment of significantly modulated genes (q<0.05 and Max RPKM>0) was generated through DAVID Functional Annotation Tools (https://david.ncifcrf.gov/). All heatmaps were generated with the R programming environment using Wards method with Euclidean distance (http://www.r-project.org/).
Suprabasal epidermal cells ChIP-seq analysis
Suprabasal epidermal cells were obtained after epidermis–dermis separation of neonatal skins by Percoll-gradient.58 Briefly, the epidermal cells were resuspended in a mix of 50% Percoll, 40% high calcium media and 10% 10 × PBS and then centrifugated to collect the suprabasal fraction. Chromatin crosslinking of the isolated suprabasal keratinocytes was performed with formaldehyde 1% for 10 min at 37C, followed by neutralization with 125 mM glycine. Fixed cells were suspended in shearing buffer (50 mM Tris at pH 7.6 and 0.2% Triton X-100) and proteinase inhibitors, and then sonicated using Bioruptor (Diagenode Inc., Denville, NJ, USA). Detergents (1% Triton X-100, 0.1% SDS and 0.1% sodium deoxycholate (DOC) and 0.1 mM EDTA solution were added and the suspension was centrifuged at 13 000 r.p.m. for 10 min to collect the supernatant. After overnight incubation of supernatant with protein A Dynabeads (Thermo Fisher Scientific, Waltham, MA, USA) and either (Anti-DLX3 (Abcam ab178428) or Anti-H3K4me3 (Diagenode C15410003), the beads were washed with a series of washing buffer solutions (two times for each buffer): RIPA buffer, RIPA buffer containing 0.3M NaCl, LiCl buffer containing 0.5% NP40 and 0.5% NaDOC, Tris-EDTA buffer containing 0.2% Triton-X-100 and Tris-EDTA. DNA fragments were obtained by incubation for 4 h at 65C with Tris-EDTA solution containing 0.3% SDS and 1 mg/ml proteinase K. The DNA Library was prepared after fragment purifications and sequenced using Illumina HiSeq 2000 (Illumina, San Diego, CA, USA). MACS 1.4 was used for peak calling and Integrative Genomics Viewer and PAPST was used to visualize the peak regions from Transcription Start Sites (TSS).
Statistical analysis
The data were analyzed by Prism Software (GraphPad Software, Inc, La Jolla, CA, USA) and significance was assessed by the Student's t-test. For all experiments with error bars, standard deviations were calculated to indicate variations within each experiment and data, and values represent mean±standard deviation of mean. P<0.05, P<0.01, P<0.001, P<0.0001 were considered significant.
Acknowledgments
We thank members of the Laboratory of Skin Biology (LSB) and of the Laboratory of Cancer Biology and Genetics (LCBG), specially Olivier Duverger, Juliane Lessard and Veronica Nagle (LSB) and Brandi Carofino (LCBG) for assistance, helpful suggestions and discussions. We also thank Gustavo Gutierrez-Cruz of the NIAMS Genome Analysis Core Facility and the NIAMS Light Imaging Core Facility. This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (M.I.M AR041124-15). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Completed RNA-Seq and ChIP-seq data have been deposited in the Gene Expression Omnibus (GEO) site GSE63049 and GSE89884, respectively.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
The authors declare no conflict of interest.
Supplementary Material
References
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 2005; 6: 328–340. [DOI] [PubMed] [Google Scholar]
- Radoja N, Gazel A, Banno T, Yano S, Blumenberg M. Transcriptional profiling of epidermal differentiation. Physiol Genomics 2006; 27: 65–78. [DOI] [PubMed] [Google Scholar]
- Hwang J, Kwon KR, Choi HS, Lee EH, Udey SH, Morasso MC et al. Epidermal ablation of Dlx3 is linked to IL-17-associated skin inflammation. Proc Natl Acad Sci USA 2011; 108: 11566–11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo E, Kellett M, Cataisson C, Gormley A, Bible PW, Pietroni V et al. The homeoprotein DLX3 and tumor suppressor p53 co-regulate cell cycle progression and squamous tumor growth. Oncogene 2016; 35: 3114–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morasso MI, Markova NG, Sargent TD. Regulation of epidermal differentiation by a distal-less homeodomain gene. J Cell Biol 1996; 135: 1879–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GT, Morasso MI. Regulation of the Dlx3 homeobox gene upon differentiation of mouse keratinocytes. J Biol Chem 1999; 274: 26599–26608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoja N, Guerrini L, Lo Iacono N, Merlo GR, Costanzo A, Weinberg WC et al. Homeobox gene Dlx3 is regulated by p63 during ectoderm development: relevance in the pathogenesis of ectodermal dysplasias. Development 2007; 134: 13–18. [DOI] [PubMed] [Google Scholar]
- Di Costanzo A, Festa L, Duverger O, Vivo M, Guerrini L, La Mantia G et al. Homeodomain protein Dlx3 induces phosphorylation-dependent p63 degradation. Cell Cycle 2009; 8: 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer 2007; 7: 281–294. [DOI] [PubMed] [Google Scholar]
- Rutberg SE, Saez E, Glick A, Dlugosz AA, Spiegelman BM, Yuspa SH. Differentiation of mouse keratinocytes is accompanied by PKC-dependent changes in AP-1 proteins. Oncogene 1996; 13: 167–176. [PubMed] [Google Scholar]
- Jerome-Morais A, Rahn HR, Tibudan SS, Denning MF. Role for protein kinase C-alpha in keratinocyte growth arrest. J Invest Dermatol 2009; 129: 2365–2375. [DOI] [PubMed] [Google Scholar]
- Helfrich I, Schmitz A, Zigrino P, Michels C, Haase I, le Bivic A et al. Role of aPKC isoforms and their binding partners Par3 and Par6 in epidermal barrier formation. J Invest Dermatol 2007; 127: 782–791. [DOI] [PubMed] [Google Scholar]
- Chew YC, Adhikary G, Xu W, Wilson GM, Eckert RL. Protein kinase C delta increases Kruppel-like factor 4 protein, which drives involucrin gene transcription in differentiating keratinocytes. J Biol Chem 2013; 288: 17759–17768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You HL, Eng HL, Hsu SF, Chen CM, Ye TC, Liao WT et al. A PKC-Sp1 signaling pathway induces early differentiation of human keratinocytes through upregulation of TSG101. Cell Signal 2007; 19: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Denning MF, Dlugosz AA, Williams EK, Szallasi Z, Blumberg PM, Yuspa SH. Specific protein kinase C isozymes mediate the induction of keratinocyte differentiation markers by calcium. Cell Growth Differ 1995; 6: 149–157. [PubMed] [Google Scholar]
- Park GT, Denning MF, Morasso MI. Phosphorylation of murine homeodomain protein Dlx3 by protein kinase C. FEBS Lett 2001; 496: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma AK, Wheeler DL, Aziz MH, Manoharan H. Protein kinase C epsilon and development of squamous cell carcinoma, the nonmelanoma human skin cancer. Mol Carcinog 2006; 45: 381–388. [DOI] [PubMed] [Google Scholar]
- Breitkreutz D, Braiman-Wiksman L, Daum N, Denning MF, Tennenbaum T. Protein kinase C family: on the crossroads of cell signaling in skin and tumor epithelium. J Cancer Res Clin Oncol 2007; 133: 793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB et al. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J Biol Chem 2008; 283: 20198–20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataisson C, Murillas JE, Wang R, Atwell A, Torgerson C, Gerdes S et al. Activation of cutaneous protein kinase C alpha induces keratinocyte apoptosis and intraepidermal inflammation by independent signaling pathways. J Immunol 2003; 171: 2703–2713. [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet 2009; 41: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard JC, Piña-Paz S, Rotty JD, Hickerson RP, Kaspar RL, Balmain A et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci USA 2013; 110: 19537–19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R, Wolf D. Abnormal epidermal barrier in the pathogenesis of atopic dermatitis. Clin Dermatol 2012; 30: 329–334. [DOI] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest 2006; 116: 1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darido C, Georgy SR, Jane SM. The role of barrier genes in epidermal malignancy. Oncogene 2016; 35: 5705–5712. [DOI] [PubMed] [Google Scholar]
- Cataisson C, Pearson AJ, Tsien MZ, Mascia F, Gao JL, Pastore S et al. CXCR2 ligands and G-CSF mediate PKCalpha-induced intraepidermal inflammation. J Clin Invest 2006; 116: 2757–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwehand K, Scheper RJ, de Gruijl TD, Gibbs S. Epidermis-to-dermis migration of immature Langerhans cells upon topical irritant exposure is dependent on CCL2 and CCL5. Eur J Immunol 2010; 40: 2026–2034. [DOI] [PubMed] [Google Scholar]
- Liu J, Li F, Ping Y, Wang L, Chen X, Wang D et al. Local production of the chemokines CCL5 and CXCL10 attracts CD8+ T lymphocytes into esophageal squamous cell carcinoma. Oncotarget 2015; 6: 24978–24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik-Dasthagirisaheb YB, Conti P. Letter to Editor: Chemokine network involved in inflammatory skin diseases. Ann Clin Lab Sci 2015; 45: 452–457. [PubMed] [Google Scholar]
- Tan SY, Roediger B, Weninger W. The role of chemokines in cutaneous immunosurveillance. Immunol Cell Biol 2015; 93: 337–346. [DOI] [PubMed] [Google Scholar]
- Dlugosz AA, Yuspa SH. Coordinate changes in gene expression which mark the spinous to granular cell transition in epidermis are regulated by protein kinase C. J Cell Biol 1993; 120: 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz AA, Cheng C, Williams EK, Dharia AG, Denning MF, Yuspa SH. Alterations in murine keratinocyte differentiation-induced by activated Ras(Ha) genes are mediated by protein-kinase C-alpha. Cancer Res 1994; 54: 6413–6420. [PubMed] [Google Scholar]
- Bryan JT, Morasso MI. The Dlx3 protein harbors basic residues required for nuclear localization, transcriptional activity and binding to Msx1. Journal of cell science 2000; 113(Pt 22): 4013–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum A, El-Sabban ME, Khoury J, Yuspa SH, Darwiche N. Overexpression of retinoic acid receptors alpha and gamma into neoplastic epidermal cells causes retinoic acid-induced growth arrest and apoptosis. Carcinogenesis 2001; 22: 1955–1963. [DOI] [PubMed] [Google Scholar]
- Eichberger T, Regl G, Ikram MS, Neill GW, Philpott MP, Aberger F et al. FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J Invest Dermatol 2004; 122: 1180–1187. [DOI] [PubMed] [Google Scholar]
- Hwang J, Mehrani T, Millar SE, Morasso MI. Dlx3 is a crucial regulator of hair follicle differentiation and cycling. Development 2008; 135: 3149–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piipponen M, Nissinen L, Farshchian M, Riihilä P, Kivisaari A, Kallajoki M et al. Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J Invest Dermatol 2016; 136: 1701–1710. [DOI] [PubMed] [Google Scholar]
- Hegde S, Raghavan S. A skin-depth analysis of integrins: role of the integrin network in health and disease. Cell Commun Adhes 2013; 20: 155–169. [DOI] [PubMed] [Google Scholar]
- Sumigray KD, Lechler T. Cell adhesion in epidermal development and barrier formation. Curr Top Dev Biol 2015; 112: 383–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler S, Mueller MM. Inflammation and skin cancer old pals telling new stories. Cancer J 2013; 19: 517–524. [DOI] [PubMed] [Google Scholar]
- Freije A, Ceballos L, Coisy M, Barnes L, Rosa M, De Diego E et al. Cyclin E drives human keratinocyte growth into differentiation. Oncogene 2012; 31: 5180–5192. [DOI] [PubMed] [Google Scholar]
- Fernandez-Hernandez R, Rafel M, Fusté NP, Aguayo RS, Casanova JM, Egea J et al. Cyclin D1 localizes in the cytoplasm of keratinocytes during skin differentiation and regulates cell-matrix adhesion. Cell Cycle 2013; 12: 2510–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh KS, Mutoh M, Gerdes M, Yuspa SH. CLIC4, an intracellular chloride channel protein, is a novel molecular target for cancer therapy. J Invest Dermatol Symp Proc 2005; 10: 105–109. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol 2007; 176: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinin AE, Idler WW, Marekov LN, McPhie P, Bowers B, Steinert PM et al. Co-assembly of envoplakin and periplakin into oligomers and Ca-2 +-dependent vesicle binding—implications for cornified cell envelope formation in stratified squamous epithelia. J Biol Chem 2004; 279: 22773–22780. [DOI] [PubMed] [Google Scholar]
- Dallaglio K, Marconi A, Truzzi F, Lotti R, Palazzo E, Petrachi T et al. E-FABP induces differentiation in normal human keratinocytes and modulates the differentiation process in psoriatic keratinocytes in vitro. Exp Dermatol 2013; 22: 255–261. [DOI] [PubMed] [Google Scholar]
- Gordon WM, Zeller MD, Klein RH, Swindell WR, Ho H, Espetia F et al. A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. J Clin Invest 2014; 124: 5205–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault MP, Weinblatt D, Shaverdashvili K, Yang YZ, Katz JP. KLF4 transcriptionally activates non-canonical WNT5A to control epithelial stratification. Sci Rep 2016; 6: 26130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama R, Tagami H, Aiba S. Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases. J Dermatol Sci 2008; 49: 187–194. [DOI] [PubMed] [Google Scholar]
- Palazzo E, Morandi P, Lotti R, Saltari A, Truzzi F, Schnebert S et al. Notch cooperates with Survivin to maintain stemness and to stimulate proliferation in human keratinocytes during ageing. Int J Mol Sci 2015; 16: 26291–26302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morasso MI, Radoja N. Dlx genes, p63, and ectodermal dysplasias. Birth Defects Res C Embryo Today 2005; 75: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger O, Morasso MI. Role of homeobox genes in the patterning, specification, and differentiation of ectodermal appendages in mammals. J Cell Physiol 2008; 216: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayedyahossein S, Rudkouskaya A, Leclerc V, Dagnino L. Integrin-linked kinase is indispensable for keratinocyte differentiation and epidermal barrier function. J Invest Dermatol 2016; 136: 425–435. [DOI] [PubMed] [Google Scholar]
- Perez White BE, Getsios S. Eph receptor and ephrin function in breast, gut, and skin epithelia. Cell Adhes Migr 2014; 8: 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U, Anders J, Yuspa S. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Proc 2008; 3: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataisson C PA, Torgerson S, Nedospasov SA, Yuspa SH. Protein kinase C alpha-mediated chemotaxis of neutrophils requires NF-kappa B activity but is independent of TNF alpha signaling in mouse skin in vivo. J Immunol 2005; 174: 1686–1692. [DOI] [PubMed] [Google Scholar]
- Feledy JA, Morasso MI, Jang SI, Sargent TD. Transcriptional activation by the homeodomain protein distal-less 3. Nucleic Acids Res 1999; 27: 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc 2008; 3: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bible PW, Kanno Y, Wei L, Brooks SR, O'Shea JJ, Morasso MI et al. PAPST, a user friendly and powerful Java platform for ChIP-Seq peak co-localization analysis and beyond. PLos ONE 2015; 10: e0127285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.