Abstract
Sensory system plays important roles in a wide array of insect’s behavior and physiological events, including the host landing and locating, feeding, flying, sex responding, mating and oviposition which happen independently and in sequence. The armyworm Mythimna separata (Lepidoptera: Noctuidae) of migratory insect is destructive for alimentarn crop and economic crop throughout the world. Here we present the high throughput sequencing of the head transcriptome and identify members of the major sensory genes which are crucial for armyworm’s success worldwide, including 8 opsins, 22 chemosensory proteins, 50 odorant binding proteins, 60 odorant receptors, 8 gustatory receptors, 24 ionotropic receptors, and 2 sensory neuron membrane proteins. It is worth noting that a duplication of the LW opsin gene exists in this insect. Several genes were clustered with functionally validated genes, such as Co-receptors of OR and IR, PBPs, PRs, CO2 GRs, bitter GRs and sweet GRs, were also identified. The transcriptome gene library provided the basis for further studies that elucidate the fundamental molecular mechanism of biology and control in M. separata. Our research exhibits the first comprehensive catalogue of the sensory genes fundamental for success and distribution in M. separata, which are potential novel targets for pest control strategies.
Insects are the most successful living creature consideration of the numbers, the biomass, and their distribution, in which the sensory systems play a significant role in their success. The armyworm Mythimna separata (Lepidoptera: Noctuidae) is destructive for crop production in Asia and Oceania with wide plant hosts, such as wheat, maize, rice and many other cereal crops. It is a typical pest that migrate seasonally in long distance of 500–1,000 km and the migration happens at night with downwind1,2. In china, there are at least four times of migration for M. separata which always lead to disastrous pest outbreaks cross generations in massive scales3,4. For example, the control area of armyworm was up to 8,455,000 hm2 and the yearly actual loss of main crops by armyworm reached 692,000 tons during 2012–20135. Therefore, many studies were conducted on the biological and ecological habits, population monitoring, prediction and integrated control of M. separata.
It has been known that migration of M. separata is an interaction between environment and genetic. Olfactory, visuosensory, and gustatory system are important contributors for M. separata to feel, handle and respond to environment cues. These may help us understand the intrinsic factor of its migration. However, the fundamental mechanisms are not well understood. Insect head is often equipped with compound eyes, antennae, mouthpart, and related neurons. Antenna structure of M. separata was introduced to explain the behavior mechanism in chemecological level6. Recent studies of our group, compound eye fine structures of M. separata, would help us to understand the visual-based behavior of M. separata (unpublished data).
Transcriptome analysis has been applied in a large number of insects for gene mining and different expressed gene identification during various life processes7. Taking the importance of heads in M. separata and the completeness of sequence information into consideration, heads from 1–6th larvae, female and male pupae, and female and male adults of M. separata were sampled in respective stages. The mRNA of mixed heads was applied for transcriptome analysis. The transcriptome gene library obtained in this study provided the basis for further studies that elucidate the fundamental molecular mechanism of biology and control in M. separata.
Results
Head transcriptome
Illumina sequencing of the cDNA library prepared from the mRNA of the larvae, pupae and Adults Mythimna separata head totally present 123,444,756 raw reads. Trimming adaptor sequences and eliminating low quality reads produced 102,202,512 reads (Supplementary Data S1). After assembly, with scaffolding of the head transcriptomes, there were 228,197 transcripts, from which 156,254 unigenes were predicted, and average length of 416 bp and a maximum length of 29,803 bp. (Supplementary Data S1). The assembled transcripts were used as queries in BLASTx against the database Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), GO (Gene Ontology), PFAM (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), and KO (KEGG Orthology), with an e-value cut-off of 10E−5. In general, the sequences had e-values between 1.0E–4 and 1.0E–10 (Fig. S1). Searches against the databases returned 111,124 transcripts showing sequence similarity to known proteins (Supplementary Data S1). The head transcripts of M. separata was most similar to the sequences of Danaus plexippus, followed by the sequences of Bombyx mori (Fig. S2).
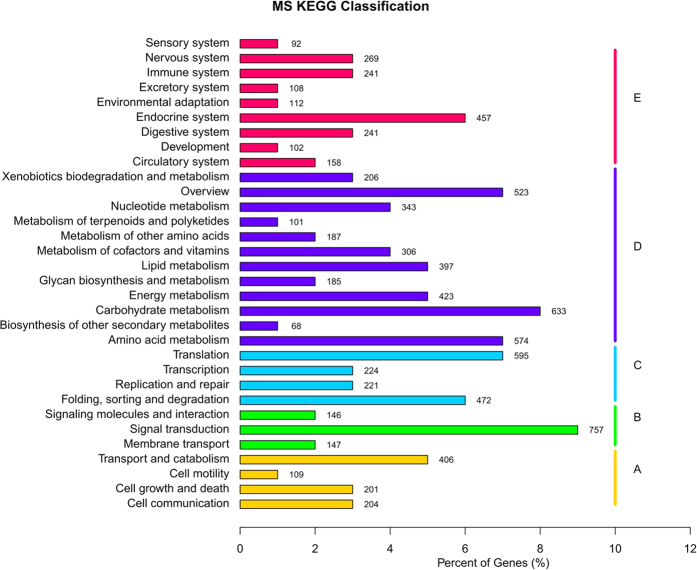
All 7976 unigenes encoded in The KEGG returned cut-off BLAST hits > 1.0E–5. A KEGG metabolic pathway analysis present 5461 transcripts that could generate 136 predicted pathways (Fig. 1). The top 14 KEGG pathways contained over 120 unigenes. For example, 280 sequences belonged to the class “Purine metabolism” (PATH: ko00230), the class “Carbon metabolism” (PATH: ko01200) have 280 sequences too, followed by 260 in the “Biosynthesis of amino acids” (PATH: ko01230) and 212 in “Oxidative phosphorylation” (PATH: ko00190) (Supplementary Data S1).
Figure 1. Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation summary.
KEGG distribution of the Mythimna separata unigenes were annotated by 136 pathways in 5 major groups.
Of these transcripts, 22,949 (~20.65% of all predicted proteins) were classifies as at least one GO term (Fig. S3). The largest number of GO term associations were related to the fundamental functions of cell; GO terms related to sensory such as binding, stress response, receptor activity and signal transduction as well as enzyme activity were also included in the data sets. A lot of transcripts uncorrelated with GO terms (88,175 transcripts, ~ 79.35%) probably existed as orphan genes.
A summary of the highly-expressed transcripts in the M. separata head is present in Supplementary Data S2. The highly-expressed transcripts consisted of cuticular proteins, troponin, OPSINs, CSPs and OBP. Chitin and certain proteins constitute the insect cuticle, of which different properties (flexible or stiff) are manifested according to the developmental stage of insect. Troponin which is composed of three regulatory proteins participates in skeletal muscle and myocardial contraction. However, the contraction mechanism of smooth muscle which is necessary for movement do not require the involvement of troponin. The Opsin (MS|comp149511_c0), OBP (MS|comp141270_c0), and CSPs (MS|comp154133_c0, MS|comp149520_c0, and MS|comp156988_c0) were highly expressed in the head with 5,344.8, 4,073.9, 8,937.6, 7,433.8 and 5,835.5 FPKMs, respectively, which means that they play significant roles in odorant reception (Supplementary Data S2). Major ribosomal protein genes such as ribosomal protein S27a (Rps27a), Rps2, Rps17, Rpl10, Rpl23, and Rpl39 were highly expressed in the head of M. separata (Supplementary Data S2). Interestingly, heat shock protein 19.8 was highly expressed in the M. separata head, and it showed 77% similarity to Cydia pomonella (ADX96000.1).
Identification of candidate OPSINs
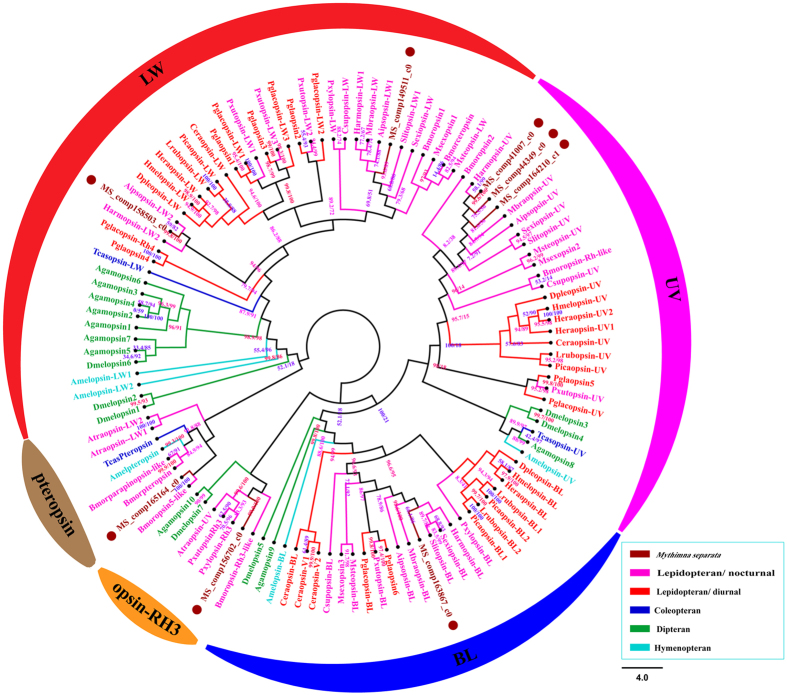
Eight genes were identified as vision-related opsin and four opsins likely represent full-length sequences (Supplementary Data S3). A phylogenetic tree based on the maximum likelihood method was constructed used opsin protein sequences from Lepidopteran, Coleopteran, Dipteran and Hymenopteran insects of 25 species (Fig. 2). Two long wavelength light-sensitive opsins genes were found in several Lepidopterans, which resulted from the early duplication of gene LW18,9. Interestingly, we found two LW opsins genes in M. separata, LW1 opsin (MS|comp149511_c0) was expressed in higher level (FPKM: 5344.8) than LW2 opsin (MS|comp158503_c0) (FPKM: 30.3). FPKM of blue light-sensitive (BL) opsin (MS|comp163867_c0), ultraviolet light-sensitive (UV) opsin (MS|comp164210_c1), and peropsin (MS|comp165164_c0) were 135.9, 537.4, and 42.1, respectively. Two UV opsin (MS|comp41007_c0, MS|comp44349_c0) and one opsin-RH3-like (MS|comp156702_c0) were also opsin genes with weak expression levels (FPKM < 5) (Supplementary Data S5). Sequence alignment revealed that M. separata opsins shared strong homology with opsins identified from other Lepidopteran insects, such as Helicoverpa armigera, Plutella xylostella, B. mori and D. plexippus. The observed homology was 34.7% ~ 86.0% between opsins in M. separata (Supplementary Data S4).
Figure 2. Phylogenetic tree of vision-related opsins (opsins).
Included are opsins from Lepidopteran, Coleopteran, Dipteran and Hymenopteran insects of 25 species. M. separata opsins are marked with a “•”. The specific clades are marked. Node support was assessed with 1000 bootstrap replicates.
Identification of candidate chemosensory proteins
Twenty-two transcripts encoding candidate chemosensory proteins (CSPs) were identified (Fig. S4), sixteen of which likely represent full-length proteins. MS|comp154133_c0 and MS|comp149520_c0 are the most highly expressed transcripts (FPKM > 7000) (Supplementary Data S3). A total of the identified full-length proteins included a signal peptide and the highly conserved four-cysteine profile (C1-X6-C2-X18-C3-X2-C4, where X represents any amino acid) (Fig. S4 and Supplementary Data S5). The agreement rate of the deduced CSPs ranged from 2% to 72%, which indicated that they belong to the different CSP protein families (Supplementary Data S4). The protein sequence alignment shows that aromatic residues are highly conserved within the M. separata CSP protein family (Fig. S4). hydrophobic residues and alpha helical domains of lepidopteran CSPs were totally retained (Fig. S4).
The CSP sequences of seven lepidopteran species, Tribolium castaneum and Drosophila melanogaster, were aligned to build an ML tree, which represent three orders (Fig. S4). The lepidopteran specific CSP lineage that diverged from dipterans and coleopterans were formed (Fig. S4). MS|comp149055_c0 and MS|comp158640_c0 are clustered with HarmCSP4. MS|comp154133_c0, MS|comp149520_c0, MS|comp155046_c0 and MS|comp149356_c0 are clustered with BmorCSP1, BmorCSP2, BmorCSP6, HarmCSP6, respectively (Fig. S4).
Identification of putative odorant binding proteins
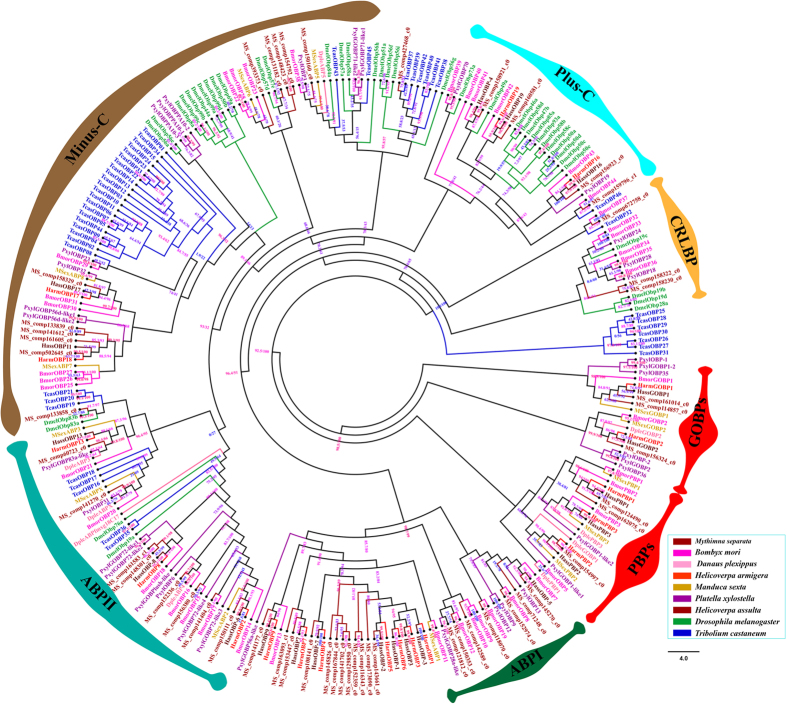
We analyzed 50 odorant binding proteins (OBPs) transcripts. Of the 30 full-length OBPs of M. separata, 15 exhibited the classic arrangement of conserved six-cysteines, 12 was the Plus-C gene motif, and 3 were Minus-C (Fig. 3). Table 1 summarized transcript name, length, Signal Peptide, Cysteine Number and FPKM in three biological replicates of putative odorant binding proteins. We also detected low amino acid sequence identity among the full-length OBPs of M. separata, which ranged from 5.1% to 68.5%, with an average of 17.9%. Among the 50 OBPs of M. separata, MS|comp141270_c0 and MS|comp116343_c0 showed the highest expression level (FPKM > 3000) (Supplementary Data S3).
Figure 3. Phylogenetic tree of odorant binding proteins (OBPs).
Included are OBPs from M. separata (MS), B. mori (Bmor), D. plexippus (Dple), H. armigera (Harm), M. sexta (Msex), P. xylostella (Pxyl), H. assulta (Hass), D. melanogaster (Dmel) and T. castaneum (Tcas). The specific clades are marked. Node support was assessed with 1000 bootstrap replicates.
Table 1. Detailed information on the OBP ungenes of Mythimna separata.
| Unigene ID | Unigene Length (bp) | ORF Length (aa) | Complete ORF | 5′ or 3′ Terminus Lost | Signal Peptide | Signal Peptide (aa) | Cysteine Number | FPKM (mean) | FPKM1 | FPKM2 | FPKM3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MS|comp116343_c0 | 605 | 146 | YES | — | YES | 21 | 8 | 3092.00 | 3070.29 | 1416.29 | 4789.41 |
| MS|comp118070_c0 | 500 | 148 | YES | — | YES | 19 | 6 | 1.86 | 1.35 | 0.00 | 4.22 |
| MS|comp129850_c0 | 759 | 145 | YES | — | YES | 21 | 7 | 12.64 | 15.43 | 11.03 | 11.45 |
| MS|comp141177_c0 | 651 | 144 | YES | — | YES | 23 | 8 | 425.24 | 482.35 | 341.87 | 451.49 |
| MS|comp141270_c0 | 638 | 137 | YES | — | YES | 20 | 6 | 4073.95 | 3613.61 | 2183.50 | 6424.75 |
| MS|comp141612_c0 | 743 | 153 | YES | — | YES | 17 | 5 | 191.25 | 219.91 | 105.38 | 248.47 |
| MS|comp141702_c0 | 628 | 139 | YES | — | YES | 21 | 9 | 42.02 | 58.06 | 29.15 | 38.85 |
| MS|comp142589_c0 | 645 | 154 | YES | — | YES | 23 | 10 | 10.64 | 13.69 | 4.42 | 13.82 |
| MS|comp148301_c0 | 565 | 139 | YES | — | YES | 18 | 7 | 127.82 | 118.57 | 113.56 | 151.34 |
| MS|comp150111_c0 | 556 | 142 | YES | — | YES | 21 | 7 | 278.95 | 313.43 | 270.81 | 252.62 |
| MS|comp150160_c0 | 711 | 168 | YES | — | YES | 20 | 7 | 348.76 | 332.15 | 247.67 | 466.46 |
| MS|comp150353_c0 | 673 | 153 | YES | — | YES | 21 | 6 | 4.09 | 9.30 | 1.86 | 1.10 |
| MS|comp152336_c0 | 630 | 164 | YES | — | NO | — | 9 | 25.05 | 27.53 | 21.22 | 26.40 |
| MS|comp152359_c0 | 1482 | 149 | YES | — | YES | 21 | 7 | 119.11 | 113.53 | 84.42 | 159.38 |
| MS|comp152974_c0 | 646 | 147 | YES | — | YES | 15 | 8 | 13.53 | 16.61 | 11.15 | 12.83 |
| MS|comp153447_c0 | 662 | 148 | YES | — | YES | 21 | 7 | 760.80 | 798.02 | 762.58 | 721.81 |
| MS|comp156324_c0 | 669 | 162 | YES | — | YES | 21 | 7 | 1929.52 | 1859.69 | 1742.96 | 2185.91 |
| MS|comp156380_c0 | 827 | 136 | YES | — | NO | — | 7 | 10.89 | 10.72 | 12.70 | 9.25 |
| MS|comp156923_c0 | 675 | 146 | YES | — | YES | 16 | 10 | 47.20 | 49.32 | 17.95 | 74.33 |
| MS|comp158230_c0 | 1253 | 252 | YES | — | YES | 19 | 3 | 78.99 | 115.66 | 66.80 | 54.51 |
| MS|comp158329_c0 | 815 | 133 | YES | — | YES | 16 | 5 | 587.29 | 704.26 | 393.22 | 664.38 |
| MS|comp158921_c0 | 677 | 183 | YES | — | YES | 17 | 16 | 248.53 | 294.46 | 143.34 | 307.79 |
| MS|comp159796_c1 | 613 | 165 | YES | — | YES | 16 | 12 | 437.41 | 400.40 | 328.19 | 583.65 |
| MS|comp160581_c0 | 794 | 197 | YES | — | YES | 17 | 13 | 46.84 | 59.44 | 38.85 | 42.24 |
| MS|comp161605_c0 | 953 | 154 | YES | — | NO | — | 6 | 2638.01 | 2474.12 | 1167.94 | 4271.96 |
| MS|comp162075_c0 | 1055 | 170 | YES | — | YES | 27 | 7 | 1919.57 | 1612.91 | 1276.67 | 2869.14 |
| MS|comp163093_c1 | 3862 | 515 | YES | — | NO | — | 10 | 268.11 | 172.50 | 439.16 | 192.67 |
| MS|comp167844_c0 | 609 | 146 | YES | — | YES | 23 | 8 | 299.55 | 314.18 | 148.05 | 436.42 |
| MS|comp173090_c0 | 648 | 149 | YES | — | YES | 22 | 8 | 68.25 | 74.26 | 29.43 | 101.07 |
| MS|comp60723_c0 | 587 | 141 | YES | — | YES | 18 | 8 | 1436.46 | 1557.41 | 1258.45 | 1493.51 |
| MS|comp108141_c0 | 440 | 138 | NO | 3′ | YES | 22 | 7 | 5.16 | 4.32 | 0.00 | 11.16 |
| MS|comp114857_c0 | 245 | 68 | NO | 3′ | YES | 17 | 2 | 694.08 | 649.36 | 839.41 | 593.47 |
| MS|comp121812_c0 | 275 | 90 | NO | 3′ | NO | — | 3 | 2.48 | 5.45 | 0.00 | 1.99 |
| MS|comp124490_c0 | 219 | 72 | NO | 5′and 3′ | YES | 17 | 3 | 3.69 | 5.95 | 2.84 | 2.29 |
| MS|comp131182_c0 | 345 | 96 | NO | 5′and 3′ | NO | — | 3 | 4.58 | 0.00 | 10.38 | 3.36 |
| MS|comp133839_c0 | 470 | 128 | NO | 5′ | NO | — | 5 | 2.65 | 3.87 | 2.51 | 1.56 |
| MS|comp133858_c0 | 254 | 82 | NO | 5′ | NO | — | 5 | 1.71 | 0.00 | 0.00 | 5.13 |
| MS|comp141404_c0 | 436 | 137 | NO | 3′ | YES | 25 | 8 | 119.67 | 122.50 | 121.69 | 114.82 |
| MS|comp143661_c0 | 563 | 145 | NO | 5′ | YES | 19 | 8 | 1.49 | 1.57 | 2.21 | 0.70 |
| MS|comp145270_c0 | 370 | 117 | NO | 3′ | YES | 17 | 7 | 3.31 | 5.92 | 0.64 | 3.38 |
| MS|comp145826_c0 | 410 | 129 | NO | 3′ | YES | 22 | 6 | 1.91 | 1.88 | 2.65 | 1.19 |
| MS|comp154997_c0 | 532 | 165 | NO | 3′ | YES | 23 | 8 | 1115.64 | 1056.35 | 1259.04 | 1031.54 |
| MS|comp158322_c0 | 1517 | 20 | NO | 5′ | NO | — | 0 | 454.46 | 514.82 | 529.24 | 319.32 |
| MS|comp161014_c0 | 1133 | 91 | NO | 5′′ | NO | — | 4 | 348.36 | 381.04 | 382.50 | 281.53 |
| MS|comp161583_c1 | 487 | 60 | NO | 5′ | NO | — | 2 | 3.01 | 2.53 | 4.75 | 1.76 |
| MS|comp393373_c0 | 444 | 77 | NO | 5′ | NO | — | 3 | 1.26 | 0.33 | 1.38 | 2.06 |
| MS|comp427468_c0 | 210 | 64 | NO | 3′ | YES | 21 | 2 | 10.98 | 9.54 | 20.62 | 2.79 |
| MS|comp502645_c0 | 284 | 92 | NO | 3′ | YES | 16 | 3 | 1.22 | 2.49 | 1.18 | 0.00 |
| MS|comp672758_c0 | 297 | 75 | NO | 5′ | NO | — | 3 | 2.04 | 3.70 | 0.00 | 2.41 |
| MS|comp71248_c0 | 233 | 43 | NO | 5′ | NO | — | 3 | 4.84 | 4.65 | 9.86 | 0.00 |
The 50 OBPs of M. separata along with 230 OBPs from 8 other species (including B. mori, P. xylostella, D. plexippus, H. armigera, Helicoverpa assulta, Manduca sexta, T. castaneum and D. melanogaster) were chosen to construct a phylogenetic tree based on the amino acid sequences. The tree was classified into several distinct branches: the GOBP/PBP family, the CRLBP family, the ABPI family, the ABPII family, the Plus-C OBP family and the Minus-C OBP family (Fig. 3). As expected, the lepidopteran specific GOBP/PBP family were clustered into separate clades away from other OBPs. We also found some OBPs of M. separata sharing high homology and closely clustered with DmelOBP49a, DmelOBP57e, DmelOBP49a, BmorPBP1 and HarmPBP1 which have been functionally characterized.
Identification of candidate odorant receptors
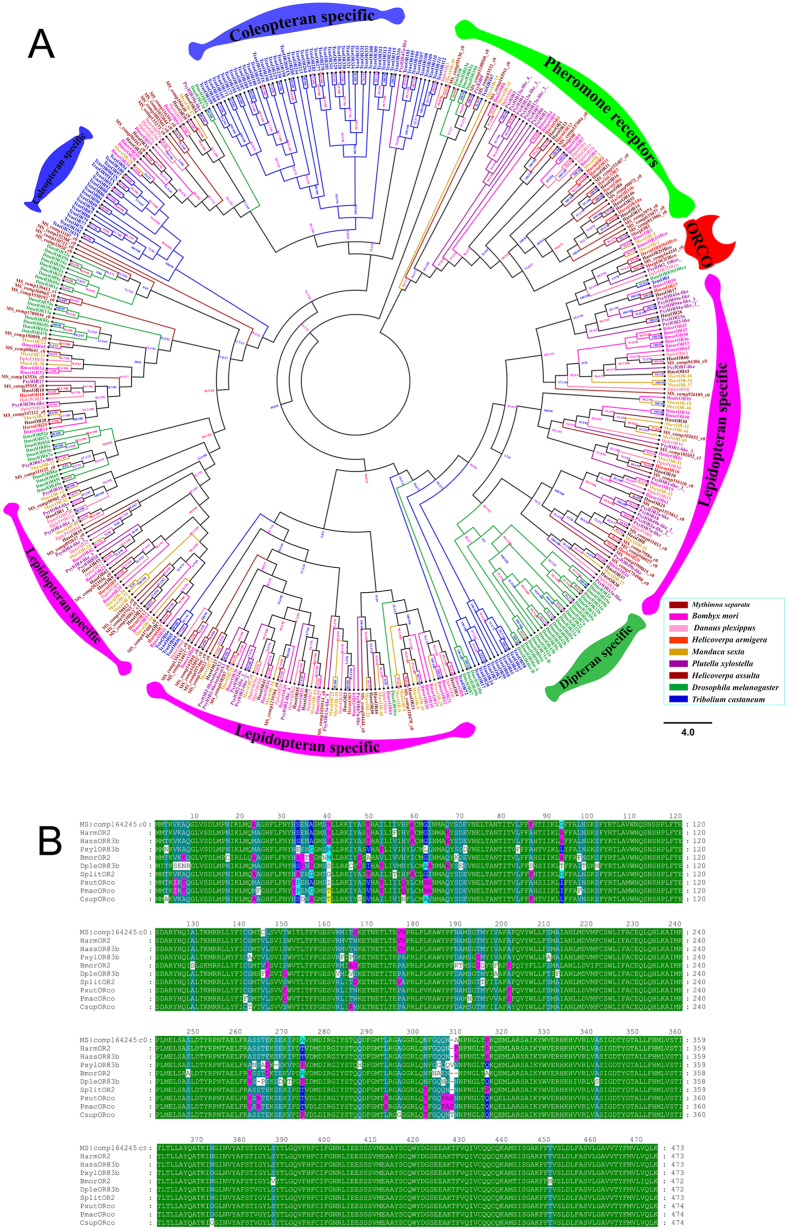
Sixty candidate OR proteins were identified from the data sets. Supplementary Data S5 summarized transcript name, length, Signal Peptide, TMD Number and FPKM in three biological replicates of the OR genes. The average sequence length was 327 bp. Four ORs likely represent full-length sequences. The highest FPKM value is 267.0 (MS|comp165911_c0) and the average FPKM value of the ORs was just 7.9. Most of partial length transcripts likely represent separate individual protein, as overlapping regions among them showed low identity of amino acid sequence. The amino acid identity of full length putative M. separata ORs ranged from 11.2% to 21.6% (average 17.5%), which was consistent with the diversity of the OR gene family (Supplementary Data S4). According to the analysis of Predictive software, there might be zero to seven transmembrane domains represent in full-length candidate M. separata OR transcripts. Consistent with the length of the partial transcripts, the remaining M. separata ORs may posse between zero and three transmembrane domains. It was worth noting that the highly-conserved co-receptor Orco which showed 87% to 97% amino acid sequence identity with Orco from B. mori, P. xylostella, D. plexippus and other Lepidopterans (Fig. 4 and Supplementary Data S5). A phylogenetic tree of ORs based on the maximum likelihood method was constructed using protein sequences of ORs from M. separata, six other lepidopteran species, T. castaneum and D. melanogaster (Fig. 4). ORs are divided into several subgroups of various size and content in the phylogenetic tree, the odorant co-receptor (Orco) and pheromone receptor (PR) families were highly conserved. Interestingly, MS|comp50905_c0 sharing high homology and closely clustered with MsexOR42, which detects cis-3-hexenyl acetate thereby affect oviposition site location10.
Figure 4. Phylogenetic tree of odorant receptors (ORs) and Alignment of the Odorant receptor co-receptor (Orco).
Phylogenetic tree of the ORs (A). ORs from Lepidopteran, Coleopteran, Dipteran. The specific clades are marked. Node support was assessed with 1000 bootstrap replicates. Alignment of the Orco (B). Orco proteins from M. separata (MS), H. armigera (Harm), H. assulta (Hass), B. mori (Bmor), D. plexippus (Dple), P. xylostella (pxyl), S. litura (Split), P. xuthus (Pxut), P. machaon (Pmac) and C. suppressalis (Csup).
Identification of candidate gustatory receptors
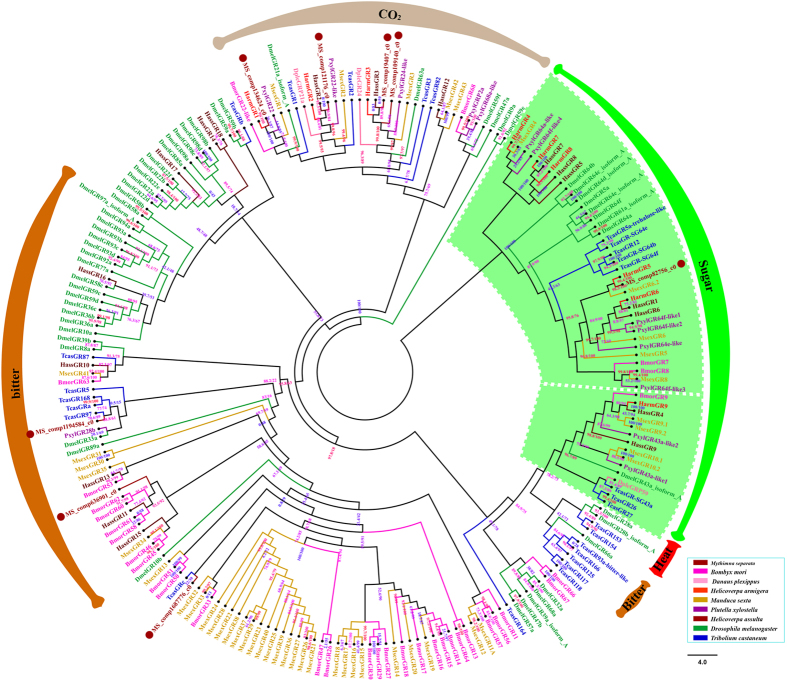
Eight candidate gustatory receptors (GRs) transcripts with very low expression levels (Transcript abundance levels ranged from 0.46 to 1.94 FPKM) were identified (Supplementary Data S3). Only one candidate M. separata GRs represent full-length protein and most of them represent partial length fragments. They encoded overlapping but distinct sequences, which established the proteins as components of independent genes. As expected with other insect GRs, transmembrane domain and topology predictions indicated that full-length transcripts most likely possess between zero and two domains with an intracellular N-terminus and extracellular C-terminus (Supplementary Data S5). A phylogenetic tree was constructed using M. separata GRs (8), B. mori GRs (33), P. xylostella GRs (14), D. plexippus GRs (3), H. armigera GRs (9), H. assulta GRs (18), M. sexta GRs (45), T. castaneum GRs (57) and D. melanogaster GRs (27) (Fig. 5). All candidate M. separata GRs cluster with CO2, sugar, and putative bitter receptor families.
Figure 5. Phylogenetic tree of gustatory receptors (GRs).
GRs from Lepidopteran, Coleopteran, Dipteran. The specific clades are marked. Node support was assessed with 1000 bootstrap replicates.
Identification of candidate ionotropic receptors
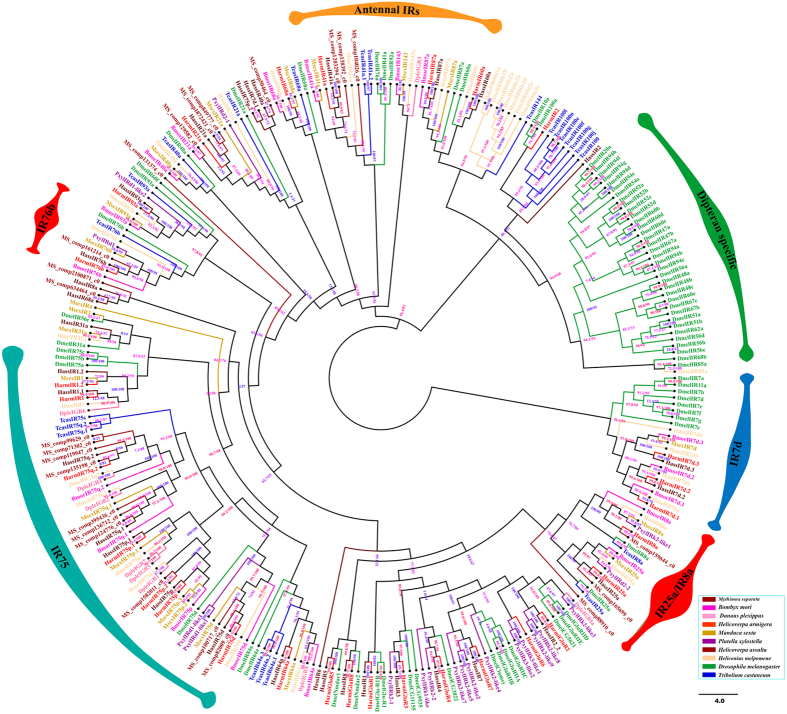
Twenty-four candidate ionotropic receptors were identified with very low expression levels in the head (Supplementary Data S5). However, Only MS|comp165699_c0, MS|comp135198_c0, MS|comp82009_c0 have relatively highly expressed (FPKM: 21, 27, 15, respectively.) (Supplementary Data S5). Three IRs likely represent full-length sequences and ware longer than 1600 bp in general. The most conserved sequence exhibited in three transmembrane domains and the ion channel pore (Fig. S5)11,12 and characteristic variability of the glutamate-binding residues in the ligand-binding S1 and S2 domains (Fig. S5). These Twenty-four IRs together with seven lepidopteran species, T. castaneum and D. melanogaster were used for a phylogenetic analysis (Fig. 6). The conserved “antennal IRs”, three highly conserved co-receptors (IR76b, IR8a and IR25a) and the species-specific “divergent IRs” were present here as well as the large sub-families of IR75 clades.
Figure 6. Phylogenetic tree of ionotropic receptors (IRs).
IRs from Lepidopteran, Coleopteran, Dipteran. The specific clades are marked. Node support was assessed with 1000 bootstrap replicates.
Identification of candidate sensory neuron membrane proteins
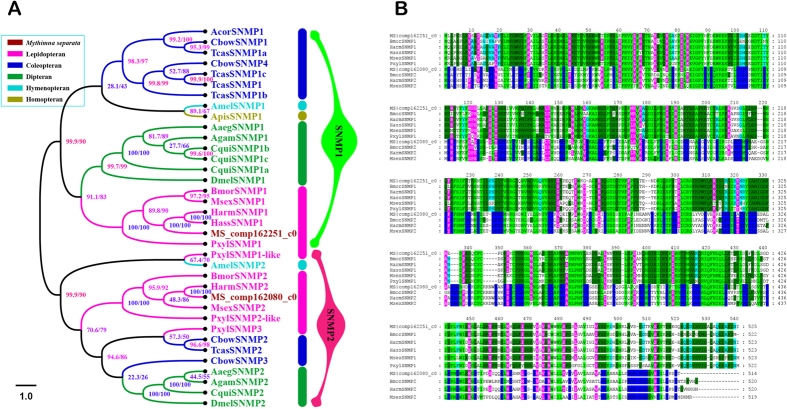
Two candidate SNMPs (MS|comp162251_c0 and MS|comp162080_c0) were identified in the transcriptomes. Both likely represent full-length genes (exceeded 1,000 bp in size; Supplementary Data S5). The FPKM results showed that MS|comp162080_c0 displayed a higher expression level (FPKM: 74.5) to MS|comp162251_c0(FPKM: 18.8) (Supplementary Data S3). The two candidate SNMPs were very similar to the HarmSNMP1 and HarmSNMP1 published in GenBank, with 89.3% and 85.6% identity, respectively. In addition, which shared more than 60% (73.0% and 625.6%) identity with the corresponding SNMPs in B. mori (Supplementary Data S4). We also detected a mean amino acid sequence identity among M. separata SNMPs of 28.9%. Phylogenetic analysis showed that MS|comp162251_c0 clustered with the insect SNMP1 group, and MS|comp162080_c0 clustered with the insect SNMP2 group. They belonged to Lepidopteran clade in the phylogenetic tree, respectively (Fig. 7).
Figure 7. Sensory neuron membrane proteins.
Phylogenetic tree of the SNMPs (A). SNMPs from Lepidopteran, Coleopteran, Dipteran, Hymenoptern and Homopteran. The specific clades are marked. Node support was assessed with 1000 bootstrap replicates. Alignment of the SNMPs (B). SNMPs from M. separata (MS), H. armigera (Harm), H. assulta (Hass), B. mori (Bmor), P. xylostella (Pxyl), M. sexta (Msex).
Discussion
Through the analysis of head transcriptome from larvae, pupae and adults of Mythimna separata, the major opsins and chemosensory genes involved in sensory mechanism were first reported in the research, which further enriched the molecular biological fundament of armyworm sensory. As sensory systems played significant roles in M. separata behavior, the identified genes could be potential novel targets for future pest control methods.
The GO assignment results of classification of predicted functions in M. Separata were consistent with that of other invertebrates13,14,15,16,17,18,19. As regard to the number of the identified sensory genes, there was no statistical difference among M. Separata and other Lepidopteran, Dipteran, and Coleopteran species with identified transcriptome11,17,18. Without considering the potential significance of individual genes to each species studied, the similarity of the different data sets does indicate a certain level of sensory gene conservation with respect to their expression.
Vision deeply involved in the regulatory mechanism of insect behaviors, such as food and mates searching, predators avoiding12,20,21. Thousands of ommatidia composed Compound eyes and the later converted lights into visual images22. The compound eyes were one of the most important parts of M. separata’s head23. Opsins played significant roles in molecular detection of photons and the transduction of visual images deeply depended on amino acid sequences of opsins24. The evolution of opsin genes provided a solid molecular basis for color vision adaption25. The three major subtypes of opsins were UV, blue and LW opsin group and It has been reported that LW opsin group had the most variety: the nymphalid butterfly Heliconius erato uses only one LW opsin to discriminate colors in wide LW ranges with the help of filtering pigments26; Helicoverpa armigera owns two LW opsins27 and Papilio glaucus four LW opsins8. We also found two LW opsins in M. Separata. In accordance with the phylogenetic analysis, the two candidate LWs were very similar to the Harmopsin_LW1 and Harmopsin_LW2 published in GenBank, with 97.9% and 93.1% identity, respectively. And the homology was 85.9% between LW opsins in M. separata (Supplementary Data S4). The second LW opsin was classified as a member of the LW2 gene family of lepidopteran species28, which indicated that LW duplication also occurred in M. separata. In addition, LW1 (MS|comp149511_c0) of M. Separata was found to be the most abundant opsin genes (FPKM: 5344.8), and more than 170 times higher expression to LW2 opsin (MS|comp158503_c0) (FPKM: 30.3), this is close to the reported in H. armigera29. It is possible that LW opsin is important to nocturnal insects because the long-wavelength light is the strongest at night in three different wavelength lights, Thus, the elevated expression of LW opsin might be associated with the nocturnal activities of insects27. Amelpteropsin expression in the brain was previously observed in honeybees30. These results indicated that opsin genes might mediate not only visual function but also nonvisual function. Interestingly, (MS|comp165164_c0) clustered with the pteropsin group in the phylogenetic tree (Fig. 2). Nonvisual function of opsin has been reported, such as thermosensation, photoperiodic responses, long-distance migration, mechanotransduction of auditory organ and egg-laying31,32,33,34,35,36.
CSPs contain a signal peptide and four conserved cysteines that are capable of forming two disulfide bridges to stabilize the tertiary structure37,38. which have soluble nature, flexible polypeptide folding, compact structure and a smaller molecular weight (10–16 kDa), permit them to bind a variety of ligands and therefore could undertake several tasks in the biological process39. CSPs were highly and almost ubiquitously distributed in chemosensory tissues as well as in non-chemosensory tissues, suggesting that CSPs in insects may also participate in other functions in addition to chemosensation, such as limb regeneration, female survival and reproduction, embryo development, recognition of sex pheromone, sucking and migratory behavioral40,41,42,43,44,45,46. Fortunately, MS|comp154133_c0 sharing 66.9% homology and closely clustered with BmorCSP1, and MS|comp149520_c0 sharing 65.8% homology and closely clustered with BmorCSP2. The two most abundant CSPs have different sex-specific expression patterns in antennae of Bombyx mori47. HarmCSP6, sharing 84.4% homology and closely clustered with MS|comp149356_c0, was reported to be highly transcribed in pheromone glands and display high binding affinity for pheromone components48. MS|comp158640_c0 and MS|comp149055_c0 shared 58.9% and 89.7% identity with HarmCSP4 which was detected to be exclusively present in proboscis and could help solubilizing terpenoids present in flower nectar45. We identified 22 CSPs in the M. separata transcriptome, comparing with the number of CSPs identified from other Lepidoptera: B. mori (24)37, Danaus plexippus (30), H. armigera (19)49, Manduca sexta (21)50, Sesamia inferens (24)51, we may have missed some CSPs in this transcriptome.
The OBP is soluble protein with a molecular weight of 12–20 kDa, ferrying the hydrophobic semiochemicals across the sensilla lymph to olfactory receptors, and has a signal peptide sequence of about 20 amino acids at the N-terminus52,53,54. Insect OBPs can be classified into classical OBPs (six-cysteine conserved signature) and Minus-C (missing C2 and C5) and Plus-C (carry additional conserved cysteine located between C1 and C2 and after C6). In addition, the classical insect OBPs include GOBP/PBP, CRLBP, ABPI and ABPII53,55. A phylogenetic tree based on the maximum likelihood method and used the amino acid sequences of OBPs from 9 species, show that the 50 OBPs of M. separata belong to six insect OBP subfamilies, respectively (Fig. 4). OBPs appear in olfactory and gustatory system and have reported olfactory and gustatory functionally characterized in Recent studies56,57,58,59. Interestingly, MS|comp158921_c0 closely clustered with the Drosophila melanogaster OBP49a is indispensable for the suppression of sweet taste by bitter chemicals60. MS|comp154792_c0, MS|comp148423_c0, MS|comp131182_c0 closely clustered with the DmelOBP57e and DmelOBP57d which are not only involved in taste perception but could also change the behavioral response to the host odors61. In Lepidoptera, two subfamilies of GOBPs and PBPs, are responsible for recognizing and transporting host odorants and pheromones, respectively62,63. GOBPs are a subfamily of OBPs, consisting of two members, GOBP1 and GOBP2 in most Lepidopterans. MS|comp161014_c0 and MS|comp114857_c0 closely clustered with the GOBP1, and MS|comp156324_c0 closely clustered with the GOBP2. PBPs are a subfamily of OBPs and constituted of three members in Lepidopteran, and we found MS|comp124490_c0, MS|comp162075_c0 and MS|comp154997_c0 closely clustered with the lepidopteran specific PBP family in the phylogenetic tree (Fig. 4). While the PBP1 of B.mori is capable of enhancing sensitivity and selectively mediating the response to bombykol64. The PBP1 of H. armigera binds strongly with two principal pheromone components (Z)-11-tetradecenal and (Z)-9-hexadecenal, seems that HarmPBP1 plays a key role in sex pheromone recognition65.
Two ORs are required in order to transduce odor-evoked signals, a highly-conserved co-receptor (Orco) and a specific OR, which varies according to ORN type66,67,68,69. The number of OR genes varies from 10 to 35070,71. Insect ORs was determined that ORs display a high degree of divergence, both within and across species due to gene duplications and deletion events72. These variations represent the olfaction sensing ability of insects with high level odor detection in insects harboring more odor-specific subunits. The 60 sequences numbers are comparable to the reported numbers in H. armigera49, M. sexta50, Cydia pomonella73 and Athetis dissimilis74. ORs in moths contain pheromone receptors (PRs) detecting sex pheromone and non-PR ORs, which respond to a variety of volatile chemicals, including plant- and microbe-derived compounds75,76,77.
In the phylogenetic tree of ORs, the specific Orco lineage contained MS|comp164245_c0, which shows that it has high similarity with six lepidopteran Orco, TcasOrco and DmelOrco, and that MS|comp164245_c0 could be the Orco of M. separata. The female M. separata moths emit the main component of the sex pheromone, Z11–16:OAc78. Lepidoptera sex pheromones produced by females may attract males for mating opportunities79,80,81. Based on phylogenetic tree analyzes, six M. separata ORs (MS|comp813906_c0, MS|comp533077_c0, MS|comp3974_c0, MS|comp90072_c0, MS|comp152487_c0 and MS|comp157094_c0) clustered in a conserved clade of PRs found in Lepidopteran insects (Fig. 4). We, therefore, hypothesize that some or all of them appear to be dedicated to sex pheromone detection. In addition, MS|comp1239304_c0 clustered with MsexOR18 and BmorOR29 which responds to linalool, citral and linalyl acetate82. BmorOR-24, which is broadly tuned and detects82, has three orthologues in M. separata, which could detect similar ligands. MS|comp892037_c0 clustered with MsexOR24 and BmorOR42 which responds to linalool and linalyl acetate82. The heterologous expression system has established to further investigate the functional characteristics of M. separata ORs.
Only eight GR-encoding transcripts were identified from the head transcriptome, Due to gustatory sensory neurons are primarily found in chemosensory sensilla on antennal, head tissues, leg tarsi and ovipositors and the low expression level of GR83,84. GRs play an important role in the detection of taste chemicals and ultimately influence an insect’s decisions about food, mates and egg deposition sites52,85. The GR family of insect includes receptors for sugars and bitter compounds, as well as cuticular hydrocarbons and odorants (CO2). GRs perceive essential nutrients whose chemical structures remain constant such as sugars and CO2 receptors. Thus, sugar and CO2 receptor genes are relatively highly conserved in most of the insect genomes that have been sequenced to date11,86,87,88. Based upon phylogeny, MS_comp121176_c0, MS_comp134624_c0, MS_comp109140_c0, MS_comp19407_c0 in M. separata grouped together with TcasGR1, TcasGR2, TcasGR3, DmelGR63a and DmelGR21a, which function as CO2 receptors, MS_comp82756_c0 grouped together with DmelGR5a, DmelGR61a, TcasGR64 and DmelGR64, which function as sugar receptors, MS_comp1194584_c0 and MS_comp636901_c0 grouped together with DmelGR10a, DmelGR33a and BmorGR60, which function as bitter receptors, none of “GR43-like” receptors were identified. So far, insect GRs have been identified as sugar receptors in B. mori89, H. armigera90 and D. melanogaster85. As CO2, fructose and bitter receptors in D. melanogaster91,92. However, putative bitter GRs have not been functionally characterized in moths.
In insects, IRs are a conserved family of synaptic ligand-gated ion channels that evolved from ionotropic glutamate receptors (iGluRs) and includes the conserved “antennal IRs” having an olfactory function, and the species-specific “divergent IRs” having gustatory function93,94. Among “antennal IRs”, one or two of the broadly expressed coreceptors (IR8a, IR25, and IR76b) in one IR-expressing neuron95. IRs belong to an ancient chemoreceptor family, and most of the IRs in Drosophila have clear orthologs with genus and Lepidoptera49,50,51,96. In this research, 24 candidate IRs with very low expression were identified in the M. separata head transcriptome, which is similar to observations in Plutella xylostella (19), H. armigera (29) and M. sexta (21)97. Some IRs have been functionally characterized. i.e. IR co-receptors respond to odorant stimulation94, IR40a, which detects DEET and is a target of insect repellents98, IR64a, which is involved in acid detection99, IR94b involved in auditory sense36 and IR76b involved in low-salt tasting100. But moth IRs have not been functionally investigated. The three candidate co-receptors: MS|comp159644_c0, MS|comp165699_c0 and MS|comp161214_c0. They shared 49%, 67% and 42% amino acid identities with D. melanogaster IR8a, IR25a and IR76b, respectively, and showed a higher amino acid identity of over 75% with the co-receptors of other lepidopteran species (B. mori, P. xylostella, D. plexippus, Heliconius melpomene, H. armigera, Helicoverpa assulta and M. sexta). The phylogenetic analysis proved that the large sub-families of IR75 clades contain ten candidate IRs of M. separata while the large sub-families of IR7d contain none (Fig. 6). The relationship between the evolution of these novel receptors and the ecology of the species require further research which will ultimately reveal the manipulation mechanism of this novel receptor family.
SNMPs are members of the CD36 family of proteins and associated with pheromone-sensitive neurons in Lepidoptera and Diptera101,102. The insect SNMP family consists of two subfamilies, SNMP1 and SNMP2, which were first identified from Antheraea polyphemus103 and M. sexta63, respectively. Both are expressed in the sensillum trichodeum, but they differ in location and level of expression. Currently, the general mechanism of insect SNMP function is still poorly understood. In D. melanogaster, SNMP1 is necessary for proper OSN responses to the pheromone compound, cis-vaccenyl acetate104. Lepidopteran SNMPs contain two conserved groups of SNMP1 and SNMP2101,105. While more than two SNMPs has been reported in coleopteran, lepidopteran and dipteran species11,96,106,107. In the moth, SNMP1 was primarily expressed in antennae and SNMP2 was abundant expressed in antennae as well as in legs18. In the phylogenetic tree, all SNMPs from Lepidoptera, Coleoptera, Diptera, Hymenoptera and Homoptera clustered into two clades, SNMP1 and SNMP2. Two candidate SNMPs of M. separata (MS|comp162251_c0 and MS|comp162080_c0) were belong to Lepidopteran sub-clades in each clade, respectively (Fig. 7). The large diversity of SNMP1 and 2 proteins within insect orders suggests that they contribute to the specificity of odour recognition52.
Conclusion
The armyworm Mythimna separata is a specialist insect that feeds mainly on maize, sorghum and rice, causing large economic losses. We first obtained abundant biology information on the transcriptome of M. separata head using high-throughput sequencing technology with the aim of identifying of the genes potentially involved in the sensory process. A total of 174 transcripts encoding putative sensory proteins from the seven major opsins and olfactory gene families were annotated: 8 opsins, 22 CSPs, 50 OBPs, 60 ORs, 8 GRs, 24 IRs, and 2 SNMPs. Comparative analysis with other Lepidopteran species suggests that near complete information regarding the molecular basis of M. separata perception was obtained. As the first step towards understanding gene functions, we conducted a comprehensive and comparative phylogenetic analysis. Several genes were clustered with functionally validated genes from other insects, such as Co-receptors of OR and IR, PBPs, PRs, CO2 GRs, bitter GRs and sweet GRs. Our findings made it possible for future research on the molecular level of olfactory system of M. separata, and in particular, the discovery of receptor genes will also contribute to the identification of novel volatile host compounds, which would gain novel targets for the pest management with semiochemicals.
Methods
Insects and sampling
Lab species of Mythimna separata was reared under controlled conditions: 25 ± 1 °C, 70~80% RH, and photoperiod of 14 L: 10D. Heads of M. separata were harvested from 1–6th instar larvae, 1–10 day-old pupae (half male and female), and 0–5 day-old adults (half male and female), respectively. Three repetitions were conducted. Detailed composition of mixed heads was provided in the Table S1.
RNA preparation and quality control
Total RNA was extracted from mixed heads. NanoPhotometer® spectro-photometer (IMPLEN, CA, USA) and Nano6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) were applied for checking the purity and integrity of total RNA, respectively. Qualified mRNA was purified from total RNA using poly-Toligo-attached magnetic beads.
Library preparation and sequencing
Sequencing libraries were generated using NEBNext® Ultra ™ RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq2000 platform and 100 paired-end reads were generated.
Transcriptome assembly
Transcriptome assembly was accomplished based on the left.fq and right.fq using Trinity with min_kmer_cov set to 2 by default and all other parameters set default108,109.
Gene functional annotation
Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (Gene Ontology).
Sequence alignment and phylogenetic analysis
The signal peptides were predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/)110. The Trans-Membrane Domains(TMDs) were predicted using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM)111. Amino acid sequence alignment was performed using the ClustalW method implemented in the Mega v7.0 and visualized with genedoc112,113.
The M. separata OPSIN, OBP, CSP, OR, SNMP, GR and IR nucleotide sequences were used as queries (BLASTx) to the GenBank database, and sequences from different insect species and their amino acids were retrieved and used to construct a phylogenetic tree. Amino acid sequences were aligned using the Muscle method implemented in the Mega v7.0113. The resulting alignment was manually curated to remove gap-rich regions. Maximum-likelihood trees (for OPSIN, OBP, CSP, OR, SNMP, GR and IR) were constructed using IQ-TREE with the best-fitting substitution-model114 and subsequently viewed and graphically edited in FigTree v1.4.313 and Adobe Illustrator. Branch support was assessed using the bootstrap method based on 1000 replicates.
Additional Information
How to cite this article: Liu, Z. et al. Sensory genes identification with head transcriptome of the migratory armyworm, Mythimna separata. Sci. Rep. 7, 46033; doi: 10.1038/srep46033 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The study was financially supported by Natural Science Foundation of China (NSFC) (Grant No. 31572017) and special fund for agro-scientific research in the public interest, the Chinese ministry of agriculture (201403031). We appreciated the help of the Novogene at Beijing for its assistance in sequencing and data analysis.
Footnotes
The authors declare no competing financial interests.
Author Contributions This experiment was conceived by Z.X.L., F.Z., X.Y.W. and C.C.L. The experiment was performed by Z.X.L. Data analysis was performed by X.Y.W. and F.Z. Z.X.L. and F.Z. wrote the manuscript. All authors read and approved the final version of the manuscript.
References
- Chen R. L. et al. Radar observations of the spring migration into northeastern China of the oriental armyworm moth, Mythimna separata, and other insects. Ecol. Entomol. 17, 1714–1715 (1998). [Google Scholar]
- Sharma H. & Davies J. The oriental armyworm, Mythimna separata (Wlk.) distribution, biology and control: a literature review. Miscellaneous Reports-Centre for Overseas Pest Research 59, 1–24 (1983). [Google Scholar]
- Jiang X., Zhang L., Cheng Y. & Luo L. Current status and trends in research on the oriental armyworm, Mythimna separata (Walker) in China. Chin. J. Appl. Entomol. 51, 881–889 (2014). [Google Scholar]
- Wang L., Hui C., Sandhu H. S., Li Z. & Zhao Z. Population dynamics and associated factors of cereal aphids and armyworms under global change. Sci. Rep. 5, 18801; 10.1038/srep18801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Y., Li C. G., Zeng J. & Liu J. Population dynamics of the armyworm in China: A review of the past 60 years’ research. Chin. J. Appl. Entomol. 51, 890–898, 10.7679/j.issn.2095-1353.2014.109 (2014). [DOI] [Google Scholar]
- Chang X. Q., Zhang S., Lv L. & Wang M. Q. Insight into the ultrastructure of antennal sensilla of Mythimna separata (Lepidoptera: Noctuidae). J. Insect Sci. 15, 124, 10.1093/jisesa/iev103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang L. H. & Zhang A. B. High-throughput transcriptome sequencing technology and its applications in Lepidoptera. Acta Entomol. Sinica 57, 991–1000 (2014). [Google Scholar]
- Briscoe A. D. Six opsins from the butterfly Papilio glaucus: molecular phylogenetic evidence for paralogous origins of red-sensitive visual pigments in insects. J. Mol. Evol. 51, 110–121 (2000). [DOI] [PubMed] [Google Scholar]
- Spaethe J. & Briscoe A. D. Early duplication and functional diversification of the opsin gene family in insects. Mol. Biol. Evol. 21, 1583–1594 (2004). [DOI] [PubMed] [Google Scholar]
- Allmann S. et al. Feeding-induced rearrangement of green leaf volatiles reduces moth oviposition. Elife 2, e00421, 10.7554/eLife.00421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch O., Papanicolaou A., Lennard C., Kirkbride K. P. & Anderson A. Chemosensory genes identified in the antennal transcriptome of the blowfly Calliphora stygia. Bmc Genomics 16, 255, 10.1186/S12864-015-1466-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bybee S. M., Johnson K. K., Gering E. J., Whiting M. F. & Crandall K. A. All the better to see you with: a review of odonate color vision with transcriptomic insight into the odonate eye. Org. Divers. Evol. 12, 241–250 (2012). [Google Scholar]
- Wu Z. Z. et al. Differential expression analysis of chemoreception genes in the striped flea beetle Phyllotreta striolata using a transcriptomic approach. PloS One 11, e0153067, 10.1371/journal.pone.0153067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. et al. Identification and comparative study of chemosensory genes related to host selection by legs transcriptome analysis in the tea geometrid Ectropis obliqua. PloS One 11, 10.1371/journal.pone.0149591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W. B., Gonzalez F., Garczynski S. F. & Witzgall P. The chemosensory receptors of codling moth Cydia pomonella-expression in larvae and adults. Sci. Rep. 6, 23518, 10.1038/Srep23518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Wang J. Z., Cui M. M., Tao J. & Luo Y. Q. Antennal transcriptome analysis of the Asian longhorned beetle Anoplophora glabripennis. Sci. Rep. 6, 26652, 10.1038/Srep26652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony B. et al. Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. Bmc Genomics 17, 69, 10.1186/s12864-016-2362-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H-assulta. PloS One 10, e0117054, 10.1371/journal.pone.0117054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. Y., Xu W., Papanicolaou A., Dong S. L. & Anderson A. Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. Bmc Genomics 15, 597, 10.1186/1471-2164-15-597 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelber A., Vorobyev M. & Osorio D. Animal colour vision-behavioural tests and physiological concepts. Biol. Rev. 78, 81–118 (2003). [DOI] [PubMed] [Google Scholar]
- Zhao H. B. et al. The evolution of color vision in nocturnal mammals. P. Natl. Acad. Sci. USA 106, 8980–8985 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. H., Xu H. H., Munch T. A., Bennett R. R. & Grable E. A. The retina of Manduca sexta: rhodopsin expression, the mosaic of green-, blue- and UV-sensitive photoreceptors, and regional specialization. J. Exp. Biol. 206, 3337–3348 (2003). [DOI] [PubMed] [Google Scholar]
- Everett A., Tong X. L., Briscoe A. D. & Monteiro A. Phenotypic plasticity in opsin expression in a butterfly compound eye complements sex role reversal. Bmc Evol. Biol. 12, 232; 10.1186/1471-2148-12-232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F. D. et al. Adaptive evolution of color vision as seen through the eyes of butterflies. P. Natl. Acad. Sci. USA 104, 8634–8640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe A. D. & Chittka L. The evolution of color vision in insects. Annu. Rev. Entomol. 46, 471–510 (2003). [DOI] [PubMed] [Google Scholar]
- Zaccardi G., Kelber A., Sison-Mangus M. P. & Briscoe A. D. Color discrimination in the red range with only one long-wavelength sensitive opsin. J. Exp. Biol. 209, 1944–1955 (2006). [DOI] [PubMed] [Google Scholar]
- Yan S. et al. The Expression of three opsin genes from the compound eye of Helicoverpa armigera (Lepidoptera: Noctuidae) is regulated by a circadian clock, light conditions and nutritional status. PloS One 9, e111683, 10.1371/journal.pone.0111683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F. D. et al. Opsin clines in butterflies suggest novel roles for insect photopigments. Mol. Biol. Evol. 32, 368–379, 10.1093/molbev/msu304 (2015). [DOI] [PubMed] [Google Scholar]
- Xu P. et al. Functional opsin retrogene in nocturnal moth. Mobile DNA 7, 18, 10.1186/s13100-016-0074-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde R. A. & Al E. Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Molec. Biol. 35, 1367–1377 (2005). [DOI] [PubMed] [Google Scholar]
- Zhu Edward Y., Guntur Ananya R., He R., Stern U. & Yang C.-H. Egg-Laying Demand Induces Aversion of UV Light in Drosophila Females. Curr. Biol. 24, 2797–2804, http://dx.doi.org/10.1016/j.cub.2014.09.076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Takemoto S., Uryu O., Kamae Y. & Tomioka K. Opsins are involved in nymphal photoperiodic responses in the cricket Modicogryllus siamensis. Physiol. Entomol. 38, 163–172, 10.1111/Phen.12015 (2013). [DOI] [Google Scholar]
- Pennisi E. Opsins: Not just for eyes. Science 339, 754–755, 10.1126/science.339.6121.754 (2013). [DOI] [PubMed] [Google Scholar]
- Senthilan P. R. et al. Drosophila Auditory Organ Genes and Genetic Hearing Defects. Cell 150, 1042–1054, doi: 10.1016/j.cell.2012.06.043 (2012). [DOI] [PubMed] [Google Scholar]
- Zhan S., Merlin C., Boore J. L. & Reppert S. M. The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185, 10.1016/j.cell.2011.09.052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. L. et al. Function of Rhodopsin in temperature discrimination in Drosophila. Science 331, 1333–1336, 10.1126/science.1198904 (2011). [DOI] [PubMed] [Google Scholar]
- Gong D. P. et al. Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem. Molec. Biol. 37, 266 (2007). [DOI] [PubMed] [Google Scholar]
- Pelosi P., Zhou J. J., Ban L. P. & Calvello M. Soluble proteins in insect chemical communication. Cell. Molec. life Sci 63, 1658 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue A. et al. X-ray structure and ligand binding study of a moth chemosensory protein. J. Biol. Chem. 277, 32094–32098 (2002). [DOI] [PubMed] [Google Scholar]
- Gong L., Luo Q., Rizwan-ul-Haq M. & Hu M. Y. Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. B. Entomol. Res. 102, 600–609 (2012). [DOI] [PubMed] [Google Scholar]
- Gu S. H. et al. Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition. PloS One 7, e42871, 10.1371/journal.pone.0042871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PloS Genet. 7, e1001291, 10.1371/journal.pgen.1001291 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszka J., Forêt S., Saint R. & Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 217, 189–196 (2007). [DOI] [PubMed] [Google Scholar]
- Nomura A., Kawasaki K., Kubo T. & Natori S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int. J. Dev. Biol. 36, 391–398 (1992). [PubMed] [Google Scholar]
- Liu Y. L., Guo H., Huang L. Q., Pelosi P. & Wang C. Z. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217, 1821 (2014). [DOI] [PubMed] [Google Scholar]
- Jacquinjoly E., Vogt R. G., François M. C. & Nagnanle M. P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. senses 26, 833–844 (2001). [DOI] [PubMed] [Google Scholar]
- Dani F. R. et al. Odorant-binding proteins and chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori. Chem.senses 36, 335–344 (2011). [DOI] [PubMed] [Google Scholar]
- Li Z. Q. et al. Expression analysis and binding assays in the chemosensory protein gene family indicate multiple roles in Helicoverpa armigera. J. Chem. Ecol. 41, 473–485 (2015). [DOI] [PubMed] [Google Scholar]
- Liu Y., Gu S., Zhang Y., Guo Y. & Wang G. Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PloS One 7, e48260, 10.1371/journal.pone.0048260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E. et al. Antennal transcriptome of Manduca sexta. P. Natl. Acad. Sci. USA 108, 7449–7454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. N. et al. Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PloS One 8, e69715, 10.1371/journal.pone.0069715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373 (2013). [DOI] [PubMed] [Google Scholar]
- Pelosi P. & Maida R. Odorant-binding Proteins in insects. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 111, 503–514 (1995). [DOI] [PubMed] [Google Scholar]
- Vogt R. G. & Riddiford L. M. Pheromone binding and inactivation by moth antennae. Nature 293, 161–163 (1981). [DOI] [PubMed] [Google Scholar]
- Leal W. S., Nikonova L. & Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. Febs Lett. 464, 85–90 (1999). [DOI] [PubMed] [Google Scholar]
- Yasukawa J., Tomioka S., Aigaki T. & Matsuo T. Evolution of expression patterns of two odorant-binding protein genes, Obp57d and Obp57e in Drosophila. Gene 467, 25–34 (2010). [DOI] [PubMed] [Google Scholar]
- Ozaki M., Morisaki K., Idei W., Ozaki K. & Tokunaga F. A putative lipophilic stimulant carrier protein commonly found in the taste and olfactory systems. A unique member of the pheromone-binding protein superfamily. Eur. J. Biochem. 230, 298–308 (1995). [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Hasan G., Rouyer F. & Rosbash M. Members of a family of drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 12, 35–49, 10.1016/0896-6273(94)90150-3 (1994). [DOI] [PubMed] [Google Scholar]
- Mckenna M. P., Hekmatscafe D. S., Gaines P. & Carlson J. R. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 269, 16340–16347 (1994). [PubMed] [Google Scholar]
- Jeong Y. T. et al. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron 79, 725–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Sugaya S., Yasukawa J., Aigaki T. & Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PloS Biol. 5, 985–996 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist G. & Vogt R. Insect pheromone biochemistry and molecular biology: the biosynthesis and detection of pheromones and plant volatiles. (Elsevier, 2003). [Google Scholar]
- Robertson H. M. et al. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol. 8, 501–518 (1999). [DOI] [PubMed] [Google Scholar]
- He X. et al. Binding of the general odorant binding protein of Bombyx mori BmorGOBP2 to the moth sex pheromone components. J. Chem. Ecol. 36, 1293–1305 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang T. T. et al. Characterization of three pheromone-binding proteins (PBPs) of Helicoverpa armigera (Hübner) and their binding properties. J. Insect Physiol. 58, 941–948 (2012). [DOI] [PubMed] [Google Scholar]
- Bengtsson J. M. et al. Putative Chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PloS One 7, e31620, 10.1371/journal.pone.0031620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D. et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011 (2008). [DOI] [PubMed] [Google Scholar]
- Hallem E. A., Ho M. G. & Carlson J. R. The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979 (2004). [DOI] [PubMed] [Google Scholar]
- Jones P. L., Pask G. M., Rinker D. C. & Zwiebel L. J. Functional agonism of insect odorant receptor ion channels. P. Natl. Acad. Sci. USA 108, 8821–8825 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-secific signatures of odor coding. PloS Genet. 8, e1002930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness E. F. et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. P. Natl. Acad. Sci. USA 107, 12168–12173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B. S. & Stensmyr M. C. Evolution of insect olfaction. Neuron 72, 698–711 (2011). [DOI] [PubMed] [Google Scholar]
- Cao D. et al. Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int. J. Biol. Sci. 10, 846–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J. F., Song Y. Q., Li W. L., Shi J. & Wang Z. Y. Identification of putative chemosensory receptor genes from the Athetis dissimilis antennal transcriptome. PloS One 11, e0147768; 10.1371/journal.pone.0147768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr M. C. et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357 (2012). [DOI] [PubMed] [Google Scholar]
- Carey A. F., Wang G., Su C. Y., Zwiebel L. J. & Carlson J. R. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T. et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. P. Natl. Acad. Sci. USA 101, 16653–16658 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuno H. et al. Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang D. D. & Löfstedt C. Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PloS One 8, e77345, 10.1371/journal.pone.0077345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner K. W. et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PloS One 5, e8685; 10.1371/journal.pone.0008685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J. et al. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). P. Natl. Acad. Sci. USA 101, 11845–11850 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Acta Agronomica Hungarica 19, 881–890 (2009). [DOI] [PubMed] [Google Scholar]
- Scott K. et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661 (2001). [DOI] [PubMed] [Google Scholar]
- Clyne P. J., Warr C. G. & Carlson J. R. Candidate taste receptors in Drosophila. Science 287, 1830–1834 (2000). [DOI] [PubMed] [Google Scholar]
- Freeman E. G., Wisotsky Z. & Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. P. Natl. Acad. Sci. USA 111, 1598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M. & Kent L. B. Evolution of the gene lineage encoding the carbon dioxide receptor in insects. J. Insect Sci. 9, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner K. W. & Robertson H. M. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629 (2009). [DOI] [PubMed] [Google Scholar]
- Lu T. et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. J. et al. Topological and functional characterization of an insect gustatory receptor. PloS One 6, e24111; 10.1371/journal.pone.0024111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Zhang H. J. & Alisha A. A Sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 38, 1513–1520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L. et al. A gustatory receptor paralog controls rapid warmth avoidance in Drosophila. Nature 500, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe A. D. et al. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PloS Genet. 9, e1003620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz R., Croset V. & Benton R. Ionotropic Receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Molec. Biol. 43, 888–897 (2013). [DOI] [PubMed] [Google Scholar]
- Croset V. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PloS Genet 6, e1001064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., Gomezdiaz C. & Vosshall L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. N. et al. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). Bmc Genomics 14, 198; 10.1186/1471-2164-14-198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig C. et al. A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem. Molec. Biol. 66, 51–63 (2015). [DOI] [PubMed] [Google Scholar]
- Kain P. et al. Odour receptors and neurons for detection of DEET and new insect repellents. Nature 502, 507–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ai M. et al. Acid sensing by the Drosophila olfactory system. Nature 468, 691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. V., Ni J. & Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt R. G. et al. The insect SNMP gene family. Insect Biochem. Molec. Biol. 39, 448–456 (2009). [DOI] [PubMed] [Google Scholar]
- Nichols Z. & Vogt R. G. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Molec. Biol. 38, 398–415 (2008). [DOI] [PubMed] [Google Scholar]
- Rogers M. E., Sun M., Lerner M. R. & Vogt R. G. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J. Biol. Chem. 272, 14792–14799 (1997). [DOI] [PubMed] [Google Scholar]
- Benton R., Vannice K. S. & Vosshall L. B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293 (2007). [DOI] [PubMed] [Google Scholar]
- Rogers M. E., Krieger J. & Vogt R. G. Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J. neurobiol. 49, 47–61 (2001). [DOI] [PubMed] [Google Scholar]
- Liu N. Y., Zhang T., Ye Z. F., Li F. & Dong S. L. Identification and characterization of candidate chemosensory gene families from Spodoptera exigua developmental transcriptomes. Int J Biol Sci 11, 1036–1048, 10.7150/ijbs.12020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidala P. et al. Identification of odor-processing genes in the Emerald Ash Borer, Agrilus planipennis. PloS One 8, e56555, 10.1371/journal.pone.0056555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512, 10.1038/nprot.2013.084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G. & Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. molec. Biol. 305, 567–580, 10.1006/jmbi.2000.4315 (2001). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J. & Higgins D. G. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics/editoral board, Andreas D. Baxevanis … [et al.] Chapter 2, Unit 2 3, 10.1002/0471250953.bi0203s00 (2002). [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L. T., Von H. A. & Minh B. Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likel ihood analysis. Nucleic Acids Res. 44, 232–235, 10.1093/nar/gkw256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.