Abstract
Pelizaeus-Merzbacher disease (PMD) is a pediatric disease of myelin in the central nervous system and manifests with a wide spectrum of clinical severities. Although PMD is a rare monogenic disease, hundreds of mutations in the X-linked myelin gene proteolipid protein 1 (PLP1) have been identified in humans. Attempts to identify a common pathogenic process underlying PMD have been complicated by an incomplete understanding of PLP1 dysfunction and limited access to primary human oligodendrocytes. To address this, we generated panels of human induced pluripotent stem cells (hiPSCs) and hiPSC-derived oligodendrocytes from 12 individuals with mutations spanning the genetic and clinical diversity of PMD—including point mutations and duplication, triplication, and deletion of PLP1—and developed an in vitro platform for molecular and cellular characterization of all 12 mutations simultaneously. We identified individual and shared defects in PLP1 mRNA expression and splicing, oligodendrocyte progenitor development, and oligodendrocyte morphology and capacity for myelination. These observations enabled classification of PMD subgroups by cell-intrinsic phenotypes and identified a subset of mutations for targeted testing of small-molecule modulators of the endoplasmic reticulum stress response, which improved both morphologic and myelination defects. Collectively, these data provide insights into the pathogeneses of a variety of PLP1 mutations and suggest that disparate etiologies of PMD could require specific treatment approaches for subsets of individuals. More broadly, this study demonstrates the versatility of a hiPSC-based panel spanning the mutational heterogeneity within a single disease and establishes a widely applicable platform for genotype-phenotype correlation and drug screening in any human myelin disorder.
Keywords: human induced pluripotent stem cell, hiPSC, Pelizaeus-Merzbacher disease, PMD, leukodystrophy, proteolipid protein 1, PLP1, oligodendrocyte, myelin, genotype-phenotype correlation, endoplasmic reticulum stress
Introduction
Leukodystrophies are a set of rare genetic disorders characterized by developmental delay and motor impairment due to deficits in myelin, also called “white matter,” in the central nervous system (CNS).1, 2 Myelin is a highly structured membrane that ensheathes neuron axons to provide ancillary support and promote proper coordination of electric impulses. Most leukodystrophies have an onset in early childhood, and many are fatal. Although individuals are routinely diagnosed on the basis of symptoms and genetic testing, most leukodystrophies are still poorly understood, and treatment options are largely limited to palliative symptom management.3, 4, 5
Pelizaeus-Merzbacher disease (PMD [MIM: 312080]) is an X-linked leukodystrophy that affects approximately 1,000 children in the United States6, 7, 8, 9 (also see GeneReviews in Web Resources). First described in 1885,10, 11 PMD was mapped to proteolipid protein 1 (PLP1 [MIM: 300401]) in the late 1980s.12, 13, 14, 15 PLP1 and its splice isoform, DM20, are predominantly expressed by oligodendrocytes—the myelin-producing cell of the CNS—and their progenitors (oligodendrocyte progenitor cells [OPCs]), respectively.16 PLP1 is the major protein component of myelin and has been found to compose as much as 50% of myelin’s total protein content.17 PLP1’s amino acid composition is 100% conserved among humans, rats, and mice,17, 18 there is only a single variant in dogs (p.Val161Ile), and mutations that cause PMD-like symptoms have been identified in each of these species.19, 20, 21, 22, 23 PLP1’s strong cell-type specificity, abundance in myelin, and inter-species conservation all suggest that it fills an indispensable role in oligodendrocyte and myelin biology. However, PLP1’s function in oligodendrocytes and myelin has only recently begun to be elucidated24, 25, 26, 27 and is still very much in question. As a result, there is no current consensus on the pathogenic processes by which PLP1 mutations cause PMD.
Although PMD is a monogenic disease, affected individuals present with a surprising spectrum of onset, disability, and mortality, which have been grouped into three categories. The common “classic” form manifests as a constellation of hypotonia, nystagmus, and/or motor delay in early childhood and the development of progressive spasticity, ataxia, and/or choreiform movements through adolescence and early adulthood.28, 29, 30, 31 Some individuals live into their seventh decade, but many develop fatal complications of hypotonia and spasticity by their late 20s. In the more severe “connatal” form, symptoms arise early in infancy and are typically fatal within the first few years of life. Lastly, a few males and most of the exceedingly rare females who present with PMD can develop mild, late-onset spasticity in the legs or assorted mild peripheral neuropathies with minimal CNS deficits32 (also see GeneReviews in Web Resources).
This significant clinical heterogeneity has been attributed to hundreds of different mutations of PLP1. A majority of PMD cases (70%) are caused by duplications of the PLP1 locus and manifest with classic PMD of mild to moderate severity.7 Rare triplications (<1%) cause severe connatal disease, whereas full gene deletions (1%–2%) are associated with mild, late-onset symptoms, often termed “null syndrome”18, 33, 34, 35, 36 (also see GeneReviews in Web Resources). Additionally, over 200 unique point mutations have been identified in individuals (25%) presenting with PMD across the entire range of severity.18 Point mutations and indels have been found throughout PLP1’s coding sequence, splice sites, and introns. PLP1 has one splice isoform, DM20, created by a cryptic splice signal in exon 3 and exclusion of the latter 105 nucleotides in that exon (exon 3b).37 In the oligodendrocyte lineage, DM20 is the first isoform expressed in developing OPCs, whereas expression and upregulation of the full-length PLP1 occur coincidently with the maturation of OPCs to oligodendrocytes. Of note, mutations in the PLP1-specific region of exon 3 often manifest as mild PMD. However, apart from this observation, there are no clear correlations between mutation locus and disease severity (see GeneReviews in Web Resources).
This surfeit and variety of human mutations suggest that multiple pathogenic processes could be responsible for the diverse manifestations of PMD. In prior literature, five possible molecular defects have been ascribed to certain PLP1 mutations: reduced expression, overexpression,38 direct disruption of protein functional domains,26 protein mistrafficking,25, 39, 40 and protein misfolding leading to endoplasmic reticulum (ER) stress.38, 41, 42, 43 The occurrence of these defects individually or in combination most likely accounts for much of the clinical heterogeneity observed in PMD. However, because prior studies have largely focused on mutations one at a time, it is difficult to ascribe any findings to a mutation apart from that in which it was originally observed.
Replicating the efforts of the past 30 years of PMD research for each new PLP1 mutation is a daunting proposition if left to traditional cellular approaches. PLP1 trafficking and membrane dynamics can be modeled to an extent with the use of immortalized cells, but the myelin sheath is a highly specialized membrane that cannot truly be recapitulated apart from oligodendrocytes. Access to primary human oligodendrocytes is severely lacking, however, because brain biopsies are implicitly dangerous and, in the case of developmental myelin disorders, the relevant stages of PMD pathogenesis have already occurred by the time a clinical diagnosis is made, let alone autopsy. As a result, animal models have proven indispensable for in vivo studies of myelin development but would be prohibitively expensive and time consuming on the scale necessary to span the genetic diversity of PMD.
Instead of attempting to adapt surrogate systems to model PMD heterogeneity, the advent of human induced pluripotent stem cell (hiPSC) and cell-fate reengineering technologies now provide us with robust methods for generating oligodendrocytes for large-scale studies directly in disease-relevant human-derived cells.44, 45 In the current study, we developed a hiPSC-based platform to efficiently model and functionally assess point mutations and duplication, triplication, and deletion of PLP1 across 12 individuals with PMD. We utilized these hiPSCs to generate OPCs and oligodendrocytes from all 12 individuals in parallel for comparative molecular and cellular assessments. These studies establish a framework for classifying PMD subgroups on the basis of defects observed in disease-relevant cells, inform personalized therapeutics testing, and demonstrate the power of using hiPSC panels to model heterogeneity in a monogenic disease.
Material and Methods
Generation of hiPSCs
Skin fibroblast samples, de-identified except for mutation and clinical severity, were obtained from Coriell (PMD6), James Garbern, and G.M.H. Before receipt, fibroblasts had been isolated from skin biopsies, cultured for one to seven passages, and frozen. Upon receipt, samples were assigned arbitrary identifications (PMD1–PMD12) according to statutes for exempt human subjects research outlined by the institutional review board of Case Western Reserve University. In preparation for hiPSC generation, fibroblasts were thawed, expanded for two passages, and tested mycoplasma free.
hiPSCs were generated by standard approaches, including either a floxed polycistronic lentivirus (hSTEMCCA) encoding the pluripotency factors OCT3/4, SOX2, KLF4, and c-MYC (PMD1–PMD4, PMD6, and PMD8–PMD10)46 or non-integrating episomal vectors encoding OCT3/4, SOX2, KLF4, L-MYC, LIN28, and a p53 shRNA (PMD5, PMD7, PMD11, and PMD12).47 At least three independent hiPSC colonies were selected for clonal expansion according to colony morphology and OCT3/4 immunocytochemistry. Clones were subsequently split 1:6 every 4 or 5 days until they expanded sufficiently for the collection of DNA and RNA and freeze-down stocks.
Two independent clonal lines derived from each PMD sample were ultimately selected for further characterization and inclusion in these studies. Seven control human pluripotent lines (designated “NC”) were also included: three approved human embryonic stem cell (hESC) lines from the NIH hESC Registry (NC1, “H1” NIHhESC-10-0043; NC2, “H7” NIHhESC-10-0061; NC3, “H9” NIHhESC-10-0062)48 and four in-house-derived hiPSC lines from healthy donors (NC4–NC7).
DNA Isolation and Analyses
Genomic DNA was extracted from each pluripotent cell line either one or two passages before initiation of the oligodendrocyte differentiation protocol (see below) with the DNeasy Blood & Tissue Kit (69504, QIAGEN). Isolation was performed according to the manufacturer’s protocol for cultured cells (July 2006 Handbook).
For Sanger sequencing of the PLP1 coding sequence, individual PCR primer pairs were designed to encompass each exon of PLP1 with NCBI Primer-BLAST (Table S1). Each exon was amplified with KAPA HiFi HotStart ReadyMix (07958935001, Roche) at the manufacturer’s suggested reaction concentrations and cycling conditions: annealing temperature of 61°C, extension time of 15 s, and 30 cycles. PCR products were purified with the QIAquick PCR Purification Kit (28104, QIAGEN) and sequenced at the Case Western Reserve University Genomics Core facility. Reported PLP1 mutations were validated in PMD samples, and all sequences were compared against the consensus human sequence (UCSC Genome Browser hg19).
High-density whole-genome SNP genotyping was performed with the Illumina Infinium Omni5 DNA Analysis BeadChip. Log R values were adjusted with the genomic.wave.pl program within PennCNV.49 Ordered log R values of every coordinate were plotted for visualization of any large-scale copy-number variations present in each line.
For clonal confirmation and future disambiguation, cell-line identity was established and confirmed by short-tandem-repeat-based DNA fingerprinting in fibroblasts and derived hiPSC lines, as well as in control pluripotent lines (Cell Line Genetics).
RNA Isolation, RNA-Seq, and Expression and Splicing Analyses
Pluripotent cells were collected for RNA isolation simultaneously with initial passaging for oligodendrocyte differentiation (see below). 500,000 cells were pelleted at 1,200 rpm for 4 min, resuspended in 1 mL TRIzol (15596026, ThermoFisher), and immediately frozen at −80°C. Total RNA was isolated with the QIAGEN RNeasy Mini Kit (74104, QIAGEN), including minor modifications to the beginning of the manufacturer’s protocol (“Purification of Total RNA from Animal Cells using Spin Technology,” Fourth Edition, June 2012 RNeasy Mini Handbook). In brief, cells in TRIzol were thawed on ice, vortexed until homogeneous, and incubated at room temperature for 5 min. 200 μL chloroform was added and mixed vigorously. The sample was transferred to PhaseLock gel tubes (2302830, 5 Prime), incubated at room temperature for 3 min, and then centrifuged at 12,000 × g for 15 min at 4°C. The aqueous phase was collected, and 1.5 volumes of 100% ethanol was added and mixed thoroughly. The sample was then transferred to an RNeasy Mini column, and the remainder of the protocol, including the recommended DNase digest, was followed as written.
For generation of the cDNA library for RNA sequencing (RNA-seq), 1 μg of each sample was rRNA depleted (Ribo-Zero Gold rRNA Removal Kit, MRZG12324, Illumina), fragmented, and indexed with the TruSeq Stranded mRNA Library Preparation Kit (RS-122-2103, Illumina) per the manufacturer’s protocol. 100 bp paired-end reads were generated for each sample with an Illumina HiSeq 2500 at the Case Western Reserve University Genomics Core facility. Output FASTQ files were aligned to the hg19 genome with TopHat (version 2.0.8) with default settings.50 Data were normalized, and fragments per kilobase per million reads (FPKM) were calculated for known RefSeq genes with Cufflinks (version 2.0.2).51 With the “heatmap.2” function of the gplots R package, Pearson’s correlation distance was calculated for comparing transcriptome similarities between individual cell lines. For analysis of PLP1 mRNA splicing, aligned BAM files were loaded into the Integrated Genomics Viewer (version 2.3.68 [97]) and visualized with the “sashimi plot” function.
OPC and Oligodendrocyte Differentiation
Differentiation of OPCs and oligodendrocytes was performed on two independently derived hiPSC clones for each of the 12 PMD samples (n = 2 biological replicates per mutation) and three hESC and four hiPSC normal controls (n = 7 biological replicates identical in PLP1 sequence). The entire panel of 31 lines was differentiated simultaneously with two technical replicates per line.
OPCs and oligodendrocytes were generated from pluripotent cells according to a pre-publication version of the Douvaras et al. protocol.44, 52 Minor variations from the published protocol are noted here.
Immediately before differentiation, cells were incubated in 10 μM Y-27632, dissociated with collagenase and dispase, and plated at 200,000 cells per 9.5 cm2 Matrigel-coated well in mTeSR1 medium and 10 μM Y-27632. Cells were cultured for 2 days, during which mTeSR1 was changed each day. Differentiation (protocol day 0) began on the third day after passaging and was conducted as follows:
Days 0–7: cells underwent daily complete media changes of DMEM/F12 (11320-033, GIBCO), 1× high-insulin N2 supplement (N2; AR009, R&D Systems), 10 μM SB431542 (04-0010, Stemgent), 250 nM LDN189193 (04-0074, Stemgent), 100 nM all-trans retinoic acid (RA; R2625, Sigma), and 5 U/mL penicillin and streptomycin (PenStrep).
Days 8–11: cells underwent daily complete medium changes of DMEM/F12, 1× low-insulin N2 (17502048, Life Technologies), 100 nM RA, 1 μM smoothened agonist (SAG; 566660, EMD Millipore), and 5 U/mL PenStrep.
Days 12–19: on day 12, cells were manually lifted with a cell scraper, broken into approximately 10- to 50-cell clusters, and plated into ultra-low attachment plates (3471, Corning) for promoting the formation of free-floating neurospheres. Every other day, spheres underwent two-thirds medium changes of DMEM/F12, 1× N2, 1× B27 supplement without vitamin A (B27; 12587010, Life Technologies), 100 nM RA, 1 μM SAG, and 5 U/mL PenStrep.
Days 20–30: every other day, cells underwent two-thirds medium changes of DMEM/F12, 1× N2, 1× B27, 10 ng/mL platelet-derived growth factor (PDGF; 221-AA, R&D Systems), 10 ng/mL insulin-like growth factor 1 (IGF; 291-G1, R&D Systems), 5 ng/mL hepatocyte growth factor (HGF; 294-HG, R&D Systems), 10 ng/mL neurotrophin-3 (NT3; GF031, EMD Millipore), 60 ng/mL 3,3′,5-triiodothronine (T3; ST2877, Sigma), 100 ng/mL biotin (4639, Sigma), 1 μM cyclic-AMP (cAMP; D0260, Sigma), 25 μg/mL insulin (I9278, Sigma), and 5 U/mL PenStrep.
Days 30–60: on day 30, neurospheres were plated onto 0.1 mg/mL poly-l-ornithine- (P3655, Sigma) and 10 μg/mL laminin-coated (L2020, Sigma) 9.5 cm2 plates and allowed to attach. Every other day, cells underwent two-thirds medium changes of DMEM/F12, 1× N2, 1× B27, 10 ng/mL PDGF, 10 ng/mL IGF, 5 ng/mL HGF, 10 ng/mL NT3, 60 ng/mL T3, 100 ng/mL biotin, 1 μM cAMP, 25 μg/mL insulin, and 5 U/mL PenStrep.
After Day 60, cultures could either be maintained in PDGF-containing medium for the promotion of OPC proliferation or be transitioned to PDGF-, IGF-, or HGF-free medium for further differentiation of oligodendrocytes.
For oligodendrocyte phenotyping of the entire panel of PMD and control cell lines in Figure 4, on day 96, cultures were dissociated with Accutase (AT-104, Innovative Cell Technologies) for 40 min and split 1:6 into 96-well poly-d-lysine VisiPlates (1450-605 PerkinElmer) pre-incubated for 1 hr with 10 μg/mL laminin. Every other day, cells underwent two-thirds medium changes of DMEM/F12, 1× N2, 1× B27, 10 ng/mL NT3, 60 ng/mL T3, 100 ng/mL biotin, 1 μM cAMP, 25 μg/mL insulin, and 5 U/mL PenStrep.
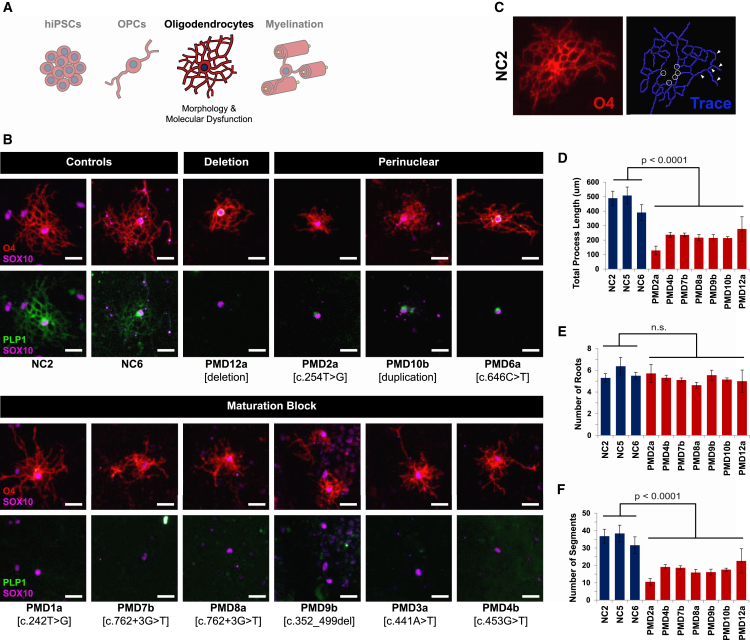
Figure 4.
Classifications of Oligodendrocyte Phenotypes
(A) Schematic of the experimental stage, oligodendrocyte morphology, and insights presented in this figure.
(B) OPCs from one line per PMD sample (n = 1 biologic replicate) were differentiated to oligodendrocytes (n = 2 technical replicates) and immunostained for oligodendrocyte markers O4, SOX10, and PLP1. Shown here are representative images from each culture. PMD5 and PMD11 failed to produce any O4+ cells. PMD2, PMD6, and PMD10 demonstrated perinuclear retention of PLP1. The remaining lines produced O4+ cells but failed to produce a PLP1 signal. Scale bars represent 25 μm.
(C) A representative immunofluorescence image of an O4+ oligodendrocyte from the NC2 control line (left) and a trace of its processes (right) generated by an oligodendrocyte identification, tracing, and quantification algorithm derived with PerkinElmer Harmony software. White circles highlight examples of “roots” where processes contact the cell body. White arrows indicate examples of individual “segments” between process branch points.
(D) The processes of O4+ oligodendrocytes were traced and measured by a machine-learning algorithm derived in house. Total process length was calculated as an average across individually measured oligodendrocytes. Error bars indicate standard error of the mean.
(E) Junctions of the oligodendrocyte cell body and extending processes were identified and counted by the tracing algorithm. The total number of roots was calculated as an average across individually traced oligodendrocytes. No significant difference was found between control and PMD lines. Error bars indicate standard error of the mean.
(F) Segments, defined as a linear portion of process between any two intersections (branches) in the trace, were identified and counted by the tracing algorithm. The total number of segments was calculated as an average across individually traced oligodendrocytes. Error bars indicate standard error of the mean.
For oligodendrocyte differentiation of NC2, PMD2, and PMD10 for time-lapse imaging (Movies S1, S2, and S3), small-molecule testing (Figure 5), and neuron co-culture (Figure 6), differentiation was repeated as above through day 60. After day 60, cultures were transitioned to two-thirds medium changes of DMEM/F12, 1× N2, 1× B27, 60 ng/mL T3, 100 ng/mL biotin, 1 μM cAMP, 25 μg/mL insulin, 10 mM HEPES sodium salt (H3784, Sigma), 20 μg/mL l-ascorbic acid (A4544, Sigma), and 5 U/mL PenStrep every third day.
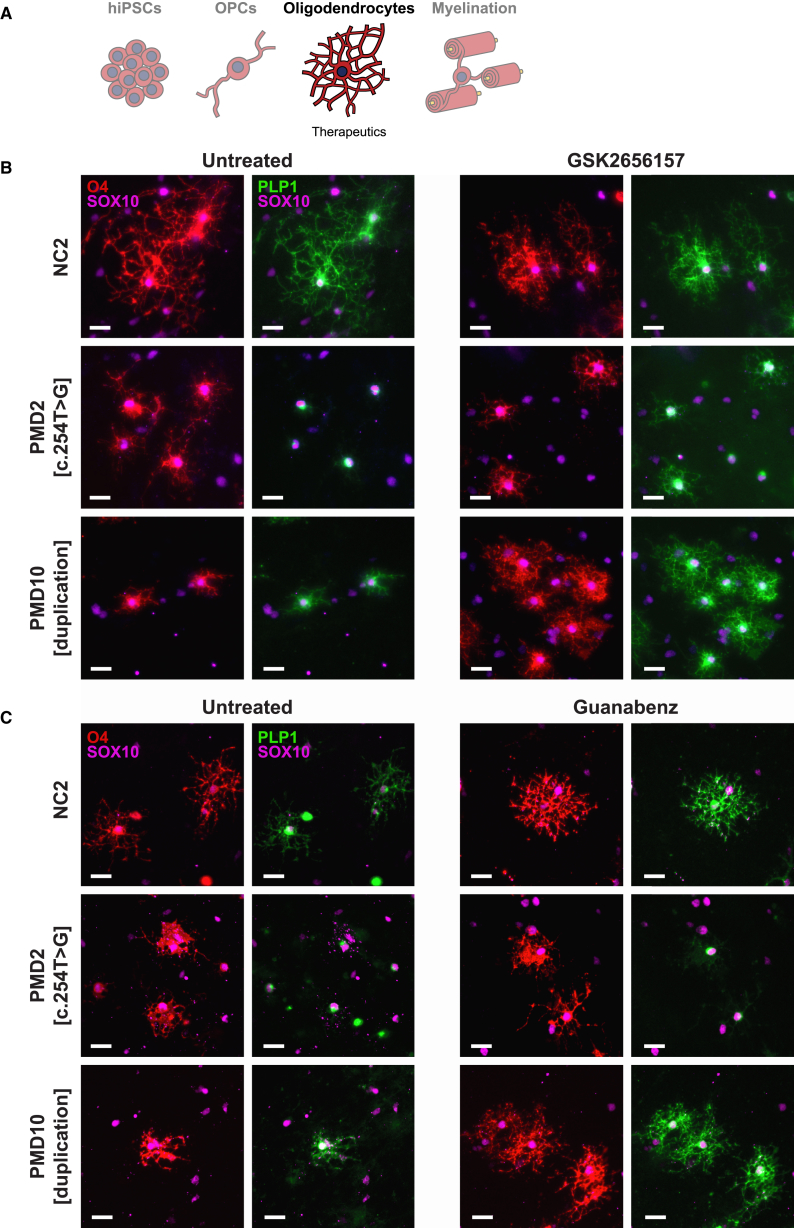
Figure 5.
Modulation of the ER Stress Response Improves PLP1 Perinuclear Retention
(A) Schematic of the experimental stage, oligodendrocyte morphology, and insights presented in this figure.
(B) Representative images of oligodendrocytes after 28 days of treatment with 1 μM GSK2656157 (n = 2 technical replicates) and immunostaining for O4, SOX10, and PLP1. Note the rescue of PLP1 distribution in treated PMD2-derived oligodendrocytes and the improvement of oligodendrocyte morphology in PMD10. Scale bars represent 25 μm.
(C) Representative images of oligodendrocytes after 35 days of treatment with 2.5 μM guanabenz (n = 2 technical replicates) and immunostaining for O4, SOX10, and PLP1. Note the improvement of cell morphology in PMD10-derived, but not PMD2-derived, oligodendrocytes. Also, compared with those in Figure 4B, untreated PMD10 cells here demonstrate PLP1 diffusing throughout the cell body in addition to intense perinuclear signal. This appears to be due to a longer period of culture between passaging and immunostaining, which possibly allowed the cells a degree of recovery from the added extrinsic stress of passaging. Scale bars represent 25 μm.
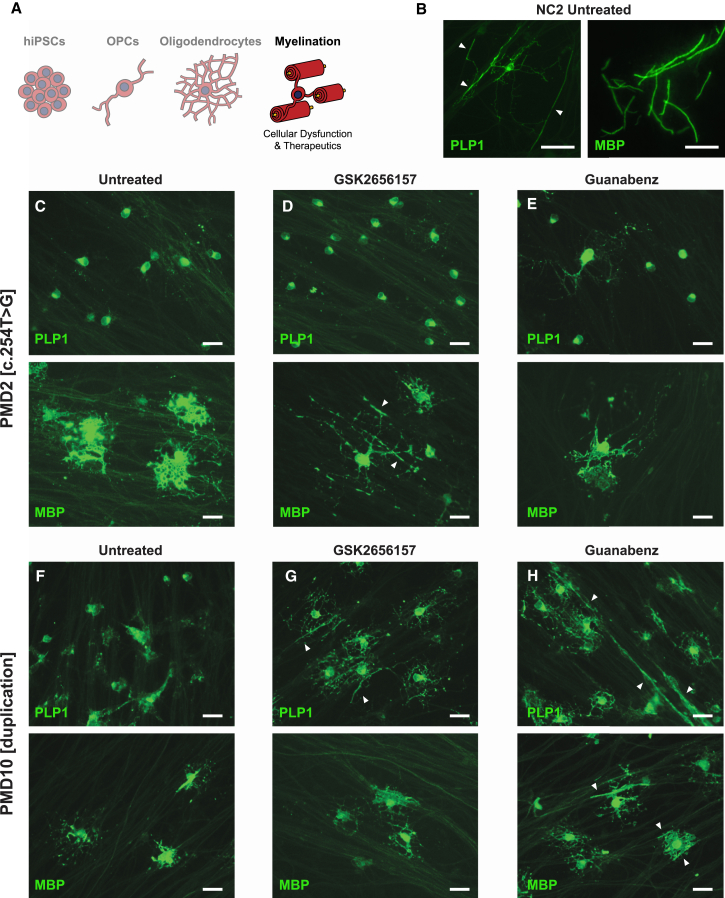
Figure 6.
Modulation of the ER Stress Response in Oligodendrocytes Co-cultured with DRG Neurons
(A) Schematic of the experimental stage, oligodendrocyte morphology, and insights presented in this figure.
(B) Representative images of untreated NC2 oligodendrocytes demonstrating the two distinct morphologies appreciable in co-culture by immunostaining with PLP1 versus MBP. PLP1 signal marks the oligodendrocyte cell body, processes, and early tracts extending along neurons (white arrowheads). MBP signal is restricted to the tracts and is indicative of early axonal ensheathment. Scale bars represent 50 μm.
(C–E) Representative PLP1 (top) and MBP (bottom) immunofluorescence images of PMD2 oligodendrocytes co-cultured with neurons and untreated (C) or treated with 100 nM GSK2656157 (D) or 2.5 μM guanabenz (E) (n = 2 technical replicates). Note the rescue of MBP-positive cell processes’ morphology when treated with GSK26157. Scale bars represent 25 μm.
(F–H) Representative PLP1 (top) and MBP (bottom) immunofluorescence images of PMD10 oligodendrocytes co-cultured with neurons and untreated (F) or treated with 100 nM GSK2656157 (G) or 2.5 μM guanabenz (H) (n = 2 technical replicates). Note the rescue of PLP1 distribution in treated oligodendrocytes (G and H). White arrowheads depict tracts of myelin along neuronal axons (G and H). Scale bars represent 25 μm.
Flow Cytometry
OPC differentiation cultures were incubated in pre-warmed Accutase for 40 min at 37°C until cells lifted off the plate. Lifted cultures were diluted with DMEM/F12 supplemented with 1% bovine serum albumin (BSA; 15260-037, GIBCO) and gently pipetted with a 1,000 μL capacity tip for dissociating the cells that had grown out from neurospheres without shearing the OPCs. The spheres themselves remained intact and were removed and discarded. Remaining single cells were counted and centrifuged at 200 × g for 5 min at room temperature. Cells were resuspended at up to 10 × 106 cells per 100 μL in DMEM/F12 supplemented with 5% donkey serum and PE-conjugated anti-PDGFRA (CD140a, 1:50; 556002, BD Biosciences) and incubated on ice for 45 min. Cells were washed three times with DMEM/F12 supplemented with 1% BSA, centrifuged at 200 × g for 5 min at room temperature, resuspended in 100 μL DMEM/F12 supplemented with 5% donkey serum and APC-conjugated anti-A2B5 (1:11; 130093582, Miltenyi), and incubated on ice for 45 min. Cells were washed three times with DMEM/F12 and 1% BSA; centrifuged at 200 × g for 5 min at room temperature; resuspended in 500 μL DMEM/F12, 1× N2, 1× B27, 10 ng/mL PDGF, 10 ng/mL IGF, 5 ng/mL HGF, 10 ng/mL NT3, 60 ng/mL T3, 100 ng/mL biotin, 1 μM cAMP, 25 μg/mL insulin, and 5 U/mL PenStrep; and filtered through a cell strainer. Cells were flowed with a 100 μm nozzle on a FACS-Aria (BD Biosciences) at 25 psi and 1,500–1,800 events per second. 10,000 events were recorded. Data were analyzed with WinList 3D (version 7.0). Initial debris and doublet gates were set with unstained NC2-derived cultures and validated against unstained NC6-derived cultures. Gates were set on the basis of side scatter (SSC-A) or forward scatter (FSC-A) for distinguishing live cells from dead cells and debris and then side-scatter width (SSC-W) or side-scatter height (SSC-H) for excluding cell doublets. A bona fide, CD140a+ OPC population was initially gated on the basis of immunostained NC2-derived cultures, validated against the remaining six immunostained control cultures, and only then used for evaluating the number of derived OPCs in each PMD culture.
Small Molecules
GSK2656157 (5046510001, EMD Millipore) 10 mM stock solution in DMSO was prepared, aliquoted, and stored at −20°C. Guanabenz (0885, Tocris Bioscience) 20 mM stock solution in DMSO was prepared, aliquoted, and stored at −20°C. Small molecules were warmed to 37°C for 20 min before being added to pre-warmed medium. Frozen aliquots were thawed no more than twice before being discarded. During treatments, every 3 days, cells underwent a two-thirds medium change that included 100 nM GSK2656157, 1 μM GSK2656157, or 2.5 μM guanabenz.
Oligodendrocyte and Dorsal Root Ganglion Neuron Co-culture
24-well VisiPlates (1450-605, PerkinElmer) were pre-incubated with 125 μL rat tail collagen and allowed to air dry for 72 hr. 50,000 dorsal root ganglion neurons (DRGs)53 were plated in 100 μL of M1 medium (MEM [11095-080, GIBCO], 10% FBS, and 2% glucose [G7528, Sigma]) with 100 ng/mL NGF (556-NG, R&D Systems) and 5 U/mL PenStrep in the center of the well; bubbles were added with a p100 pipette; and DRGs were allowed to attach for 24 hr at 37°C. The next day, wells were flooded with 500 μL E2F medium (MEM, 2% glucose, 1× N2 supplement, 245 ng/mL FDU [F0503, Sigma], and 245 ng/mL uridine [U3003, Sigma]) with 100 ng/mL NGF and 5 U/mL PenStrep. Medium was changed every second or third day thereafter (M1, NGF, and PenStrep on days 3, 7, 11, 14, and 17; E2F, NGF, and PenStrep on days 5 and 9). On day 20, one-half of a 9.5 cm2 well of differentiating OPC cultures was seeded onto DRGs in DMEM/F12, 1× N2, 1× B27, and 5 U/mL PenStrep. Medium was changed every other day for 20 days and then fixed for immunostaining.
Live-Cell Imaging
Cultures of NC2, PMD2, and PMD10 OPCs were differentiated to oligodendrocytes over the course of 20 days as specified above. On day 20, cultures were dissociated and plated onto 0.1 mg/mL poly-l-ornithine- and 10 μg/mL laminin-coated 4 cm2 plates at low density (split 1:6 by surface area) to permit observation of individual cells. The next day, cultures were transferred to a humidity-, temperature-, and CO2-controlled chamber on an inverted microscope (DMI6000, Leica) for phase imaging. Regions of interest were identified manually and marked with Leica Application Suite X software, after which they were automatically imaged every 10 min for the next 60 hr. Static image series were stitched into a movie with Microsoft Windows Movie Maker (version 2012).
Immunocytochemistry
Cultures for immunocytochemistry were initially fixed with 4% ice-cold paraformaldehyde for 15 min.
O4 immunostaining was performed on live cells prior to fixation. O4 antibody supernatant was added to cultures and incubated for 30 min at 37°C. Wells were washed three times with room-temperature DMEM/F12 and then immediately fixed. Additional immunostaining was performed as below.
Monolayer cell cultures (e.g., hiPSCs and OPCs) were permeabilized with 0.2% Triton X for 10 min at room temperature, blocked in 10% donkey serum in PBS for 1 hr at room temperature, incubated in primary antibody in blocking buffer for 1 hr at room temperature (typically) or overnight at 4°C (for PLP1 [AA3] antibody only), washed three times with PBS, incubated in secondary antibody in blocking buffer for 45 min at room temperature, washed three times (with DAPI, when used, in the first wash to stain nuclei), and imaged with an Operetta High-Content Imaging System with Harmony Analysis Software (HH12000000, PerkinElmer) and standard fluorescence settings.
DRG co-cultures to be immunostained for PLP1 were permeabilized with 10% Triton X for 30 min at room temperature, washed three times with PBS, blocked in 5% donkey serum and 0.1% Triton X in PBS for 1 hr at room temperature, incubated in PLP1 antibody in blocking buffer overnight at 4°C, and then incubated in neurofilament (NF) and SOX10 antibodies in blocking buffer for 1 hr at room temperature, washed three times with PBS, incubated in secondary antibodies in blocking buffer for 45 min at room temperature, washed three times, and imaged with an inverted fluorescence microscope (Leica DM IL LED), 12-bit monochrome camera (QIC-F-M-12-C, QImaging), and QCapture Pro imaging software (version 6.0.0.605, QImaging).
DRG co-cultures to be immunostained for myelin basic protein (MBP) were washed three times in PBS, permeabilized with 100% ice-cold methanol for 30 min at −20°C, washed three times with PBS, blocked in 5% donkey serum and 0.1% Triton X in PBS for 1 hr at room temperature, incubated in MBP, NF, and SOX10 antibodies in 2% donkey serum and 0.1% saponin overnight at 4°C, washed three times with PBS, incubated in secondary antibodies in 10% donkey serum and 0.1% Triton X for 1 hr at room temperature, washed three times, and imaged as above.
Primary antibodies included mouse-anti-O4 (1:10 unconcentrated supernatant, generously provided by Bruce Trapp, Robert Miller, and Wendy Macklin), OCT3/4 (400 ng/mL; SC-5279, Santa Cruz), Pax6 (6.67 μg/mL; PRB-278P, Covance), SOX1 (1 μg/mL; AF3369, R&D Systems), OLIG2 (1:1,000; AB9610, Millipore), NKX2.2 (1:100; 74.5A5, Developmental Studies Hybridoma Bank), rat-anti-PLP1 (1:100; AA3, generously provided by Bruce Trapp), rat-anti-MBP (1:100; AB7349, Abcam), goat-anti-SOX10 (2 μg/mL; AF2864, R&D Systems), mouse-anti-pan-axonal NF (1:1,000; SMI311, Covance), mouse-anti-pan-neuronal NF (500 ng/mL; SMI312, Covance), and DAPI (1 μg/mL; D8417, Sigma).
All secondary antibodies were Alexa-Fluor-conjugated secondary antibodies (Life Technologies) used at a dilution of 1:500.
Results
Assembly, Generation, and Characterization of a Panel of PMD-Derived hiPSCs
The goal of this study was to establish a platform for assessing the developmental, cellular, and molecular defects caused by PMD-relevant PLP1 mutations in human-derived oligodendrocytes. In order to capture the genetic and phenotypic heterogeneity found in PMD, we selected samples for inclusion according to three criteria: type of mutation, distribution of point mutations throughout PLP1, and reported clinical severity. Before being included in this study, all individuals had been diagnosed with PMD clinically and had PLP1 mutations confirmed by genetic testing. We obtained primary fibroblasts de-identified except for their mutation and clinical severity impression (mild, moderate, or severe) or functional disability score (ranging from 1 [most severe] to 32 [least severe]).54 Our panel ultimately consisted of 12 lines with various PLP1 mutations (Figures 1A and 1B and Table S2) and seven normal control lines (Table S3).
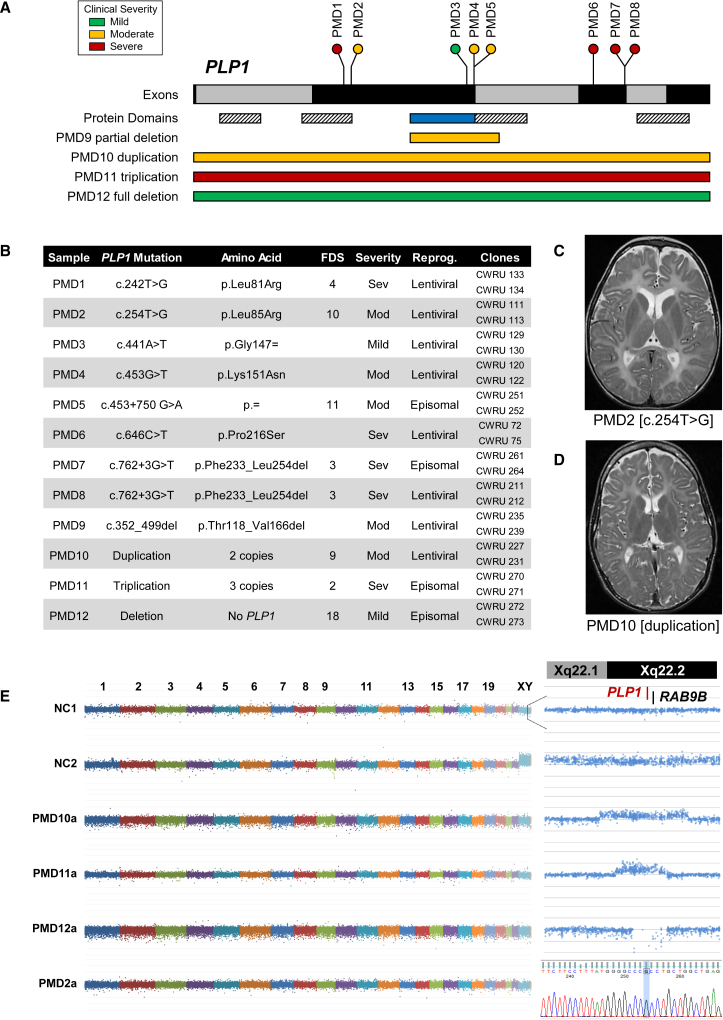
Figure 1.
Genetic Characterization of a PMD hiPSC Panel
(A) A schematic of PLP1 and the 12 mutations included in this study. Full-length PLP1 consists of seven exons (black and gray bars), whereas its splice isoform, DM20, results from exclusion of the PLP1-specific domain (blue bar). Both isoforms contain four putative transmembrane domains (striped bars). The locations of individual mutations are indicated as lollipop plots (point mutations) or bars (partial deletion and copy-number variants). Relative clinical severities are indicated by color (green, mild; yellow, moderate; red, severe).
(B) Individuals were selected for this study with the intent of maximizing genetic and phenotypic diversity. Clinical severities had been previously assessed and reported by functional disability score (FDS) and/or clinical impression. Skin fibroblasts were reprogrammed to hiPSCs with either a lentiviral or episomal construct. Two independently derived clonal hiPSC lines were selected for characterization and inclusion in subsequent studies. PLP1 mutations were confirmed by Sanger sequencing of each hiPSC line.
(C) Taken at age 4 years, T2-weighted MRI of PMD2 demonstrates increased signal intensity throughout white-matter structures and enlargement of the lateral ventricles.
(D) Taken at age 10 years, T2-weighted MRI of PMD10 demonstrates increased signal intensity and distinct atrophy of white-matter structures.
(E) Plots demonstrating the gross genomic integrity of derived pluripotent lines. Relative copy number was calculated for each SNP in a high-density SNP microarray and plotted as a normalized log R ratio. (Left) Plots of every 100th SNP, arranged by ranked genomic coordinate and colored by chromosome. NC1 (male) and NC2 (female) demonstrate the relative enrichment of the X chromosome in NC2. (Top right) Plots of each SNP within a 2 Mb region surrounding PLP1 on the X chromosome, arranged by relative genomic coordinate. For PMD10, PMD11, and PMD12, the SNP array delineates the region of chromosome X duplication, triplication, and deletion, respectively. (Bottom right) A Sanger sequencing trace showing the T-to-G substitution found in PMD2.
For two individuals, PMD2 and PMD10, axial T2-weighted MRI taken when they were ages 4 and 12 years, respectively, was also available (Figures 1C and 1D). Both children exhibited diffusely increased signal intensity in white-matter structures and atrophy of the subcortical white matter. The gross reduction in white matter, particularly in PMD2, resulted in moderate enlargement of the lateral ventricles. Such MRI is highly representative of children with moderate to severe PMD and demonstrates both the ambiguity and convergence of clinical presentations across people with disparate PLP1 mutations.54, 55, 56
To generate a renewable source of PMD-derived cells, we reprogrammed fibroblasts from all 12 individuals to hiPSCs (see Material and Methods). Two independently derived hiPSC lines per individual were ultimately selected for rigorous characterization. These 24 PMD lines (PMD1–PMD12, A and B), along with four hiPSC lines from healthy individuals (NC4–NC7) and three normal human embryonic stem cell (hESC) lines (NC1–NC3), constitute the “panel” of 31 pluripotent stem cell lines used throughout the following experiments.
Initially, we rigorously characterized each line in the panel to ensure its identity, pluripotency, and genomic integrity. To first validate the PLP1 point mutations reported for each PMD line and to confirm that no additional mutations were present in any PMD or control line, we Sanger sequenced all seven exons of PLP1 for each line in the panel. PMD7 and PMD8 were found to contain a nonpathogenic synonymous SNP (c.609T>C [p.Asp203=]) that is common in the general population (rs1126707, C = 22.6%).57 All other lines conformed to the consensus human sequence (GenBank: NM_000533.4).58
Because the process of hiPSC reprogramming can occasionally induce chromosomal defects, copy-number variation in each cell line was evaluated at fine resolution with a high-density SNP microarray. Comparison of the relative copy number of each SNP confirmed the absence of any gross chromosomal duplications or deletions in all lines (Figures 1E and S1). Furthermore, this resolution allowed delineation of the relative sizes and boundaries of the PLP1 locus duplication, triplication, and deletion in PMD10, PMD11, and PMD12, respectively (Figure 1E, right).
To confirm that our cell lines had been completely reprogrammed and retained their pluripotent properties throughout subsequent expansion and characterization, one passage before oligodendrocyte differentiation, we isolated, sequenced, and compared RNA from each cell line against the RNA profiles of primary fibroblasts from which control hiPSCs had been derived. Hierarchical clustering demonstrated close association of all pluripotent lines, and there was no significant distinction between PMD and control lines, hiPSCs and hESCs, or out-grouping of both PMD-derived lines from any single individual (Figure 2B). All pluripotent lines also showed robust and consistent expression of canonical pluripotency markers whose expression correlates with complete reprogramming and acquisition of pluripotent identity (Figure 2C).59 Using this rigorous pipeline, we generated and characterized a diverse panel of hiPSCs to provide a cellular resource for the study of PMD.
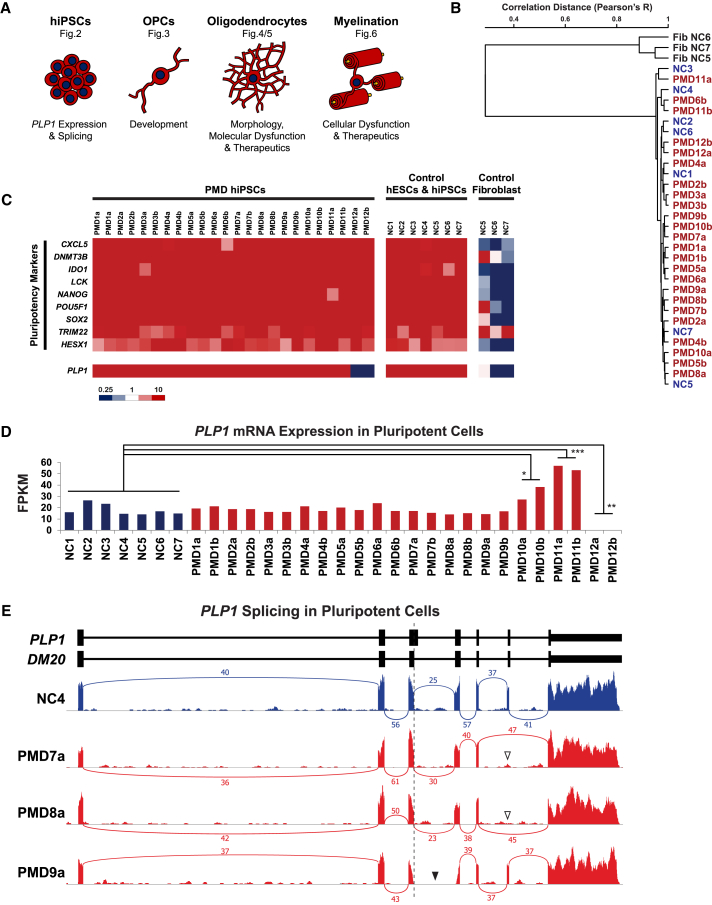
Figure 2.
RNA Characterization of PMD hiPSCs and PLP1 Transcript Defects
(A) A schematic of the major cell types derived in this study, the stereotypic morphologic appearance of each cell type when cultured in vitro, some of the insights these cells can provide, and the figure(s) in which they feature.
(B) Dendrogram depicting hierarchical clustering analysis of stranded RNA-seq. RNA was isolated from each pluripotent line one passage before initiation of the OPC differentiation protocol and compared against RNA isolated from primary fibroblasts corresponding to NC5–NC7.
(C) A heatmap depicting the FPKM of canonical pluripotency genes and PLP1 across all PMD hiPSCs, normal controls, and primary fibroblasts corresponding to NC5–NC7.
(D) A bar graph comparing levels of PLP1 mRNA expression in hiPSCs between various PMD cultures and controls (∗p = 0.0134, ∗∗p = 0.0017, ∗∗∗p < 0.0001).
(E) Sashimi plots of RNA-seq transcripts aligning to PLP1 (hg19) quantify PLP1 and DM20 mRNA splicing events. Numeric labels indicate the number of quality-filtered transcripts (sequencing depth) that span the indicated exon-exon junction. Exclusion of the distal portion of PLP1 exon 3 (dotted vertical line) in control transcripts indicates that DM20 is the solely expressed isoform in pluripotent cells. PMD7 and PMD8 demonstrate skipping of exon 6 (white arrowheads). The exon 3–4 junction cannot be annotated in PMD9 (black arrowhead) because of its partial deletion, which spans the PLP1-specific region of exon 3 and proximal portion of exon 4.
Assessment of PLP1 Transcript Dynamics and Defects in hiPSCs
Endogenous expression of PLP1 and DM20 is restricted to oligodendrocytes and OPCs, respectively. As a result, studies on the effects of specific human mutations on protein structure and expression have previously been limited to post mortem tissue or transgenic overexpression. However, although the protein is not translated, DM20 mRNA is robustly transcribed in pluripotent stem cells.60 Serendipitously, this provides an opportunity for rapid assessment of specific transcript defects without the protracted differentiation of oligodendrocytes. To begin to characterize the effects of mutations in our panel, we used our RNA-seq dataset to interrogate DM20 mRNA expression and splicing directly in hESCs and hiPSCs (Figure 2A).
Comparison of mRNA transcript levels between control and PMD-derived pluripotent lines provides a glimpse into the effects of copy-number variations at the PLP1 locus. Control cultures, both hiPSC and hESC, displayed an average DM20 expression level of 18.1 ± 4.5 FPKM (Figure 2D). Similarly, all point mutations (PMD1–PMD8) and the partial deletion (PMD9) showed no significant differences in transcript levels (average FPKMs of 18.1 ± 2.5 and 15.5 ± 1.2, respectively; Figure 2D). However, PMD10 and PMD11 expressed 2- and 3-fold more DM20 than controls (32.7 ± 5.5 and 55.1 ± 1.8, respectively), consistent with their respective duplication and triplication of the PLP1 locus (Figure 2D). As expected, the deletion (PMD12) showed no expression of DM20 (Figure 2D).
In addition to permitting quantification of expression levels, the presence of DM20 mRNA in pluripotent cells also allows for identification of mutation-specific splicing defects. In all controls, exon 3 terminated at the internal DM20 splice site (exon 3a), indicating that only the shorter, OPC-specific isoform is present in pluripotent stem cells (Figure 2E). This preempts appreciation of any splicing defects in PMD3–PMD5, whose mutations fall in exon 3b and intron 3 (Figure S2). Additionally, PMD1, PMD2, PMD6, and PMD10–PMD12 showed no defects or alternative splicing (Figure S2) but would not particularly be expected to, considering the nature of their exon and copy-number mutations. However, splicing analysis in PMD7 and PMD8, brothers with an intronic mutation outside the canonical splice site, presented with complete skipping of the exon preceding their PLP1 mutation (Figure 2E), confirming prior analyses from autopsy tissues.61 Lastly, PMD9, a partial deletion spanning exons 3b, intron 3, and part of exon 4, demonstrated the expected in-frame deletion of the proximal portion of exon 4, and no additional splicing defects were found (Figure 2E).
Assessment of OPC Production in PMD Cultures
We next wanted to determine whether we could use our genetically diverse panel of PMD-derived hiPSCs to garner insights into the clinical variability of PMD in disease-relevant cells (Figure 3A). Our pluripotent stem cell panel was differentiated to OPCs over a 90 day time course according to a protocol that recapitulates in vivo neurodevelopmental transitions including patterning of neuroectoderm, ventralization, OPC specification, and OPC proliferation (Figure 3B).52 We performed differentiation and all subsequent experiments in parallel for all 12 PMD samples (in duplicate with two independently derived hiPSC lines per individual) and seven control samples to enable direct comparison of data and minimize the influence of variables, such as reagent lots, ambient conditions, or handling.
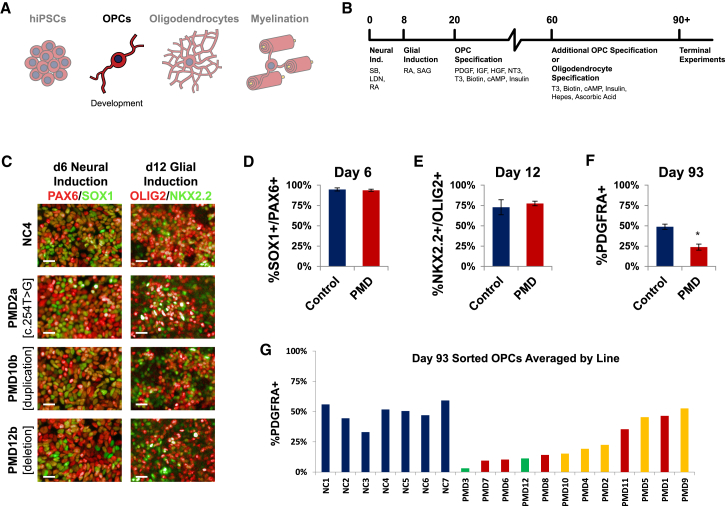
Figure 3.
Differentiation to OPCs Demonstrates PMD Variability
(A) Schematic of the experimental stage, OPC morphology, and insights presented in this figure.
(B) Overview of the timeline, small molecules, and growth factors used for generating OPCs and oligodendrocytes.
(C) Representative immunofluorescence images comparing stage-specific transcription factors in PMD and control cultures on day 6 (immunostained for neural lineage markers PAX6 and SOX1) and day 12 (immunostained for glial lineage markers OLIG2 and NKX2.2) of the differentiation protocol. Scale bars represent 25 μm.
(D) Quantification of the percentage of PAX6+SOX1+ cells as of 6 days in culture. Shown here are the averages of all controls (n = 7) versus all PMD lines (n = 24, including n = 2 biologic replicates per PMD line). Two independently differentiated wells per line were immunostained (n = 2 technical replicates). Error bars indicate standard error of the mean. No significant difference was found between control and PMD lines.
(E) Quantification of the percentage of OLIG2+NKX2.2+ cells as of 12 days in culture. Shown here are the averages of all controls (n = 7) versus all PMD lines (n = 24, including n = 2 biologic replicates per PMD line). Two independently differentiated wells per line were immunostained (n = 2 technical replicates). Error bars indicate standard error of the mean. No significant difference was found between control and PMD lines.
(F) Day 93 cultures were immunostained for the OPC-specific marker PDGFRA and counted by flow cytometry. Shown here are the averages of all controls (n = 7) versus all PMD lines (n = 24, including n = 2 biologic replicates per PMD line). One well per line was counted (n = 1 technical replicate). Error bars indicate standard error of the mean (∗p = 0.0016).
(G) The same results from (F), plotted here as individual controls versus the average of both hiPSC lines derived from a given PMD sample. PMD results are rank ordered by average number of OPCs and colored according to clinical severity (green, mild; yellow, moderate; red, severe).
Throughout the process of differentiation, cultures were immunostained for markers of key stages in the development of oligodendrocytes. Day 6 immunostaining for the neural lineage transcription factors PAX6 and SOX1 demonstrated efficient induction across both control (95% ± 1.9%) and PMD (94% ± 1.4%) cultures (Figures 3C and 3D). Day 12 immunostaining for the early glial lineage transcription factors OLIG2 and NKX2.2 also showed strong induction consistent across control (73% ± 9.2%) and PMD (77% ± 2.8%) cultures (Figures 3C and 3E). These data demonstrate that despite the presence of PLP1 mRNA transcripts in hiPSCs, early neurodevelopmental differentiation appears to be unaffected by mutations in PLP1.
We maintained cultures an additional 11 weeks to allow OPC specification, at which point we quantified the cultures by flow cytometry for the percentage of platelet-derived growth factor receptor alpha (PDGFRA)+ OPCs in each culture.62 The average proportion of PDGFRA+ OPCs was significantly lower in PMD cultures (24% ± 3.7%) than in controls (49% ± 3.3%) (Figure 3F). However, although the proportion of OPCs was generally consistent between both hiPSC clones derived from any given individual, there was substantial variability between cultures derived from separate people (Figure 3G). OPC numbers were strikingly reduced in a majority of PMD cultures (PMD2–PMD4, PMD6–PMD8, and PMD10–PMD12), whereas only four cultures (PMD1, PMD5, PMD9, and PMD11) contained OPCs at proportions comparable to those in controls. Intriguingly, PMD3, from a mildly affected individual with a synonymous substitution, demonstrated the greatest depletion of OPCs. Similarly, PMD12, from a mildly affected individual with a PLP1 deletion, also displayed poor OPC numbers, whereas more severely affected individuals possessing duplication and triplication of PLP1 (PMD10 and PMD11) trended toward normal numbers of OPCs.
Classes of Cellular and Molecular Defects in PMD Oligodendrocytes
We next wanted to determine whether specific defects would manifest as the OPCs matured into oligodendrocytes (Figure 4A). We induced PMD and control OPCs to mature into pre-myelinating oligodendrocytes and assessed cell morphology by immunostaining for O4 antigen (an early oligodendrocyte-specific surface sulfatide) and PLP1 (with a C-terminal antibody that defines a more mature oligodendrocyte stage). A typical wild-type oligodendrocyte generated in cell culture has a readily identifiable morphology consisting of a round, central cell body with multiple branching processes that extend symmetrically outward and lend the oligodendrocyte a spider-in-a-web-like appearance (Figure 4B, “controls”).
We made two major observations in these cultures. First, despite OPC deficits, most PMD cultures (except PMD5 and PMD11) were still capable of producing oligodendrocytes. However, all PMD-derived oligodendrocytes were noticeably defective (Figure 4B). In order to elucidate the defects in these cells, we developed a machine-learning-based algorithm by using PerkinElmer Harmony High-Content Imaging and Analysis software to trace and measure oligodendrocyte processes and identify branch points (Figure 4C). In PMD oligodendrocytes, total process length was significantly lower than in controls (Figure 4D). Although PMD and control oligodendrocytes extended a similar number of primary processes from the cell body (Figure 4E), the number of distal process branches was severely reduced (Figure 4F). Interestingly, oligodendrocytes of PMD12, the full PLP1 deletion with a mild phenotype, demonstrated both the highest average and widest range of total process length and branches (Figures 4B, 4D, and 4F). Collectively, these data suggest that PMD oligodendrocytes suffer either a PLP1-induced defect of process extension and branching or a non-specific arrest of maturation as a result of general disruption of cellular homeostasis. Time-lapse imaging of maturing PMD2 and PMD10 cultures captured oligodendrocytes producing short processes that failed to extend or branch distally and ultimately resulted in cell death (Movies S1, S2, and S3).
In addition to these shared cellular defects, two molecular defects were observed in subsets of the PMD hiPSC-derived oligodendrocytes. First, in the majority of the cultures (PMD1, PMD3, PMD4, PMD7–PMD9, and PMD12), O4+ oligodendrocytes were present, but PLP1 expression was not detectable (Figure 4B). This was expected for PMD12, a complete PLP1 deletion, but not the additional cultures that failed to mature to a PLP1+ stage. Interestingly, although the individual with the PLP1 deletion has a mild phenotype, other cultures that failed to express PLP1 were derived from individuals presenting with some of the most clinically severe presentations of the panel (Figure 1B). Second, in PMD2, PMD6, and PMD10, cells matured to a PLP1+ stage; however, PLP1 signal was completely restricted to the perinuclear region of the cell body, and no signal was evident in the processes (Figure 4B).
Mobilization of PLP1 into Oligodendrocyte Processes by Small-Molecule Modulators of ER Stress Pathways
Perinuclear retention of a misfolded protein is a hallmark of ER stress. Our comparative hiPSC panel identified that only a subgroup of PMD cultures (PMD2, PMD6, and PMD10) exhibited perinuclear retention of PLP1, so we selected PMD2 and PMD10, from individuals with genetically distinct point and duplication mutations, to explore strategies for modulating the ER stress response (Figure 5A).
We tested two small molecules that specifically inhibit or enhance the ER stress response. GSK2656157 is a recently described inhibitor of protein kinase R-like ER kinase (PERK), which senses misfolded proteins and initiates a response to ER stress.63 Guanabenz is an inhibitor of the protein phosphatase 1 regulatory subunit GADD34, which allows normal cellular functions to recommence once the stressor has been resolved.64
We assessed the effects of GSK2656157 and guanabenz on oligodendrocyte cultures from control NC2 and PLP1 mutants PMD2 and PMD10. PMD10 oligodendrocytes demonstrated remarkable restoration of cell morphology under both conditions (Figures 5B and 5C). However, when treated with GSK2656157, PMD2 showed modest mobilization of PLP1 into cell processes but had no response to guanabenz (Figures 5B and 5C). Neither small molecule caused appreciable changes in the morphology of control NC2 cells.
Modulation of ER Stress Phenotypes in Oligodendrocyte-Neuron Co-Cultures
Oligodendrocytes in vivo do not exist in a state of homogeneous, monolayer culture, and although exceptional for identifying cell-intrinsic deficits, phenotyping in this system does not capture defects of myelination. In order to create a more physiologically relevant model of the defects caused by PLP1 mutations, we adapted a protocol for co-culturing human oligodendrocytes on rat dorsal root ganglion neurons (Figure 6A)53 to assess oligodendrocyte maturation, axonal tracking, and ensheathment (in vitro “myelination”). In these conditions, control oligodendrocytes extend processes that search out and travel along individual neuron axons (Figure 6B), forming long linear tracts as opposed to the branching, web-like morphology seen in monoculture. PLP1 signal is present throughout the cell, including the cell body, processes, and tracts. Meanwhile, MBP, a structural protein in the myelin sheath, is restricted to the cell body and tracts, identifying regions in the early stages of myelination.
NC2, PMD2, and PMD10 oligodendrocytes were seeded onto neurons in basal medium supplemented with GSK2656157 or guanabenz. Similar to monocultures, untreated PMD2 oligodendrocytes showed prominent PLP1 perinuclear retention and no PLP1 immunofluorescence in the processes (Figure 6C, top). Meanwhile, MBP immunofluorescence delineated the entirety of the oligodendrocytes, including extensive, matted processes, but showed no tracking with neurons (Figure 6C, bottom). Interestingly, GSK2656157-treated PMD2 oligodendrocytes did not show mobilization of PLP1 into the processes, as they had in monoculture (Figure 6D, top). Despite this, the process matting seen in MBP largely resolved, and short lengths of tracking were clearly visible (Figure 6D, bottom). On the other hand, guanabenz promoted a degree of PLP1 mobilization into oligodendrocyte processes (Figure 6E, top) but did not resolve the MBP matting to the same extent as GSK2656157 (Figure 6E, bottom).
Untreated PMD10 oligodendrocytes recapitulated the PLP1 perinuclear retention observed in monoculture (Figure 6F, top), but MBP immunofluorescence was present throughout the processes instead of being restricted to defined tracts (Figure 6F, bottom). Where present, tracts were also shorter than for controls. Treatment with GSK2656157 completely restored PLP1 mobilization into processes (Figure 6G, top) but did not improve MBP signal or tracking over untreated conditions (Figure 6G, bottom). Guanabenz, however, drastically increased PLP1 mobilization into processes, rescued MBP fluorescence intensity, and increased the prevalence of tracts, although they were still less prevalent and shorter than for controls (Figure 6H).
Discussion
Historically, PMD has been a challenging disease to parse. Affected individuals present with symptoms across a spectrum of disease severity, and there are no direct links between an individual’s unique PLP1 mutation and the etiology or course of their disease. The inaccessibility of primary oligodendrocytes severely limits direct studies in humans, and it would be prohibitively expensive and time consuming to model the hundreds of different PMD-linked PLP1 mutations in animals. In this study, by taking advantage of natural mutational diversity and recent advancements in hiPSC-based disease modeling, we demonstrated the feasibility of generating, differentiating, and assessing a panel of hiPSCs that capture the full spectrum of PMD’s clinical and genetic heterogeneity. We characterized hiPSCs, OPCs, and oligodendrocytes from 12 individuals with PMD and identified shared and individual defects spanning PLP1 expression, PLP1 splicing, OPC production, oligodendrocyte morphology, and response to small-molecule therapeutics.
Although PLP1 is restricted to the oligodendrocyte lineage, DM20 mRNA is robustly transcribed in pluripotent cells,60 providing an opportunity for rapid assessment of mutation-specific transcript defects in a scalable, homogeneous population of cells. DM20, the OPC-specific isoform of PLP1 and the sole isoform expressed in hiPSCs, limited our ability to interpret the effects of mutations in the PLP1-specific region of exon 3b. However, we were interested to discover that, although neither the point mutations nor the partial deletion showed any effect on DM20 expression levels, the duplication and triplication showed 2- and 3-fold higher expression, respectively, than did controls. Given the fact that PLP1 in a normal hemizygous individual constitutes as much as 50% of myelin’s total protein, we speculate that this linear relationship between copy number and mRNA expression could lead to excess protein abundance in oligodendrocytes of individuals with supernumerary copies of PLP1. This linear trend has not been reported previously in animals.65 However, those studies were performed on whole brain tissue, wherein OPCs are but a small fraction of the total cell population. Alternately, when protein is actually translated in OPCs and oligodendrocytes, it is possible that PLP1’s overabundance could trigger a degree of feedback regulation that we cannot appreciate in hiPSCs.
The presence of DM20 mRNA also enables analysis of splicing and structural defects in hiPSCs. Importantly, whereas protein levels vary by cell type and can be inferred only from hiPSC studies, splicing defects have a direct and immutable impact on protein structure across cell types. We confirmed a prior report of PLP1 exon 6 skipping caused by a mutation of the +3 nucleotide of intron 5.61 Skipping of exon 6 causes an in-frame deletion of 22 amino acids, including part of PLP1’s putative fourth transmembrane domain. Next, we demonstrated that our partial deletion causes an in-frame fusion of exon 3a to the distal portion of exon 4 with no additional mis-splicing or decrease in expression. Similarly, we found that a mutation at the +750 nucleotide of intron 3 does not appear to affect splicing or expression in hiPSCs. Collectively, mRNA analyses in hiPSCs provide a rapid and minimally invasive means of identifying convergent structural and expression defects caused by disparate PLP1 mutations. As more mutations are characterized, this approach could eventually allow subgrouping of mutations by mRNA defect and aid in predicting individuals’ prognoses.
Differentiating hiPSCs to OPCs provides important insight into cell-intrinsic pathologic consequences of disparate PLP1 mutations. Although we anticipated that certain mutations could lead to an OPC defect, we were surprised that two-thirds of the PMD cultures were severely depleted of OPCs. On the basis of prior imaging and pathology studies, PMD has traditionally been considered a disease of myelin production and structure and thus predominantly a defect of oligodendrocytes.17 Our findings contradict this generalization and suggest that for at least a subset of individuals with PMD, a precedent defect at the OPC stage would limit subsequent oligodendrocyte production and thus preempt myelination. Of particular note, mutations within the PLP1-specific region of exon 3b (e.g., PMD3 and PMD4) would not be expected to manifest in OPCs, given the predicted expression of the DM20 splice isoform. Yet here, PMD3 cultures were some of the most severely depleted in the entire panel. This apparent paradox would have been difficult to appreciate without the ability to directly compare against the spectrum of other PMD-derived cultures. It is important to acknowledge that this single time point is not sufficient for discerning whether relative reductions in OPCs are due to a block of proliferation, failure of migration out of neurospheres, premature cell maturation and loss of the PDGFRA marker, or outright cell death. Nonetheless, our collective results suggest that prevalent, OPC-intrinsic defects strongly contribute to phenotypic variability. Most importantly, these findings suggest that future therapeutic development could necessitate earlier intervention than previously appreciated.
On the basis of simultaneous comparative phenotyping of the 12 individuals in our panel, we identified three distinct classes of cellular defects in PMD-derived oligodendrocytes: failure to produce oligodendrocytes, failure to produce PLP1+ oligodendrocytes, and perinuclear retention of PLP1. A point mutation in intron 3 and a PLP1 triplication were the only cultures that did not generate any O4+ or PLP1+ oligodendrocytes, even though these cultures had previously generated robust numbers of OPCs. This could be due to either a block in the maturation of OPCs to oligodendrocytes or rapid death of newly formed oligodendrocytes as PLP1 production is upregulated. Of the remaining cultures, oligodendrocytes were either O4+PLP1− or O4+PLP1+, but the PLP1 signal was completely retained perinuclearly. Perinuclear localization is a hallmark of protein misfolding and ER retention and has previously been demonstrated in vitro in two human oligodendrocyte lines with PLP1 point mutations, one of which corresponds with our PMD6 mutation.41 Additionally, across both of these categories, all oligodendrocytes presented with severe defects of process extension and branching. The function of PLP1 has not been fully established. However, PLP1 has been implicated in both OPC migration26 and as a bridge between the membrane and cytoskeleton.66 It is possible that, in the absence of PLP1, cell processes become insensitive to stimuli that would normally trigger them to extend. Similarly, observed deficits in distal branching could be secondary to failed process extension or result from loss of PLP1’s structural cytoskeletal support. Identification of a mutation where cells extend processes that are of normal length but completely unbranched could elucidate this further. Ultimately, the fact that an individual’s mutation and clinical presentation alone were not predictive of their cellular phenotype highlights the power of this platform to categorize disparate PLP1 mutations, enabling the development of PMD-subgroup-specific prognoses and therapeutic plans.
The genetic breakdown of our perinuclear retention cohort was intriguing because it contained a substitution of a proline in an extracellular loop, a substitution of a leucine in a transmembrane domain, and a full PLP1 duplication. Prior studies in animal and human models have implicated ER stress as a pathogenic result of particular PLP1 mutations,41, 67, 68, 69, 70, 71 but the larger PMD community has struggled to leverage these findings to treat the general PMD population. We suspect that this is due to intrinsic differences in the types of mutations being targeted. To address this, we treated our duplication and transmembrane point mutations with two small molecules that modulate ER stress in two completely opposite manners. GSK2656157, a newly described PERK inhibitor, targets the initiation of the pathway, averting the negative downstream effects of a continuous ER stress response, particularly apoptosis. At the other end, guanabenz, a GADD34 inhibitor, deliberately prolongs the stress response, maintaining expression of molecular chaperones to promote clearance of misfolded protein aggregates. The point mutation and duplication responded variably to these two approaches. In the case of the duplication, both inhibiting and enhancing the ER stress pathway had a drastic positive effect, relieving the stress sufficiently for oligodendrocytes to reestablish normal morphologies. The point mutation, however, contains a gain-of-function mutation that shows only partial response to inhibition of ER stress and no response to enhanced refolding. It is possible that this mutation is simply refractory to refolding. However, because PLP1 was observed to mobilize into the processes with GSK2656157 treatment yet the cells did not fully recover morphologically, it is more likely that the position of this mutation disrupts either membrane integration or a functional domain of PLP1. Further characterization and titration of GSK2656157 will provide a foundation for assessment of this and other small-molecule therapeutics for personalized applications in the future.
The neuron-oligodendrocyte co-culture system provides a model of the endogenous structural environment that influences oligodendrocyte and myelin biology in the brain, providing additional insight into the nuances of PMD pathogenesis. Using this in vitro system, we demonstrated that individual PMD phenotypes can be modulated to restore oligodendrocyte morphology and axonal tracking. As opposed to our initial monoculture system, the co-culture system revealed more appreciable differences between PMD oligodendrocytes’ responses to GSK2656157 and guanabenz. It was interesting to observe that untreated cells’ morphologies were improved in this system, presumably as a result of supportive factors released by the neurons that were absent from our media conditions and less frequent passaging and the extrinsic stress that entails. The transmembrane point mutation again demonstrated improvement of cell morphology when the ER stress response was completely shut down by GSK2656157 but nonetheless could not associate with neurons. This further confirms that ER stress exacerbates the pathogenesis of this mutation, but persistence of the mutation itself prevents true functional restoration by GSK2656157 alone. On the other hand, the duplication showed a dramatic response when the ER stress response was prolonged by guanabenz. Presumably, whereas inhibition of ER stress leaves misfolded protein stuck in the ER, the prolonged action of chaperones and other protective molecules triggered by ER stress promotes folding and mobilization of PLP1 to its ultimate destination in the emerging myelin sheath.
Since their first report a decade ago, hiPSCs have been transformed into a multitude of different cell types, providing invaluable insights into human health and disease. hiPSC technologies are a particular boon to the study of PMD and other leukodystrophies, obviating many of the challenges that have previously limited our ability to model and investigate oligodendrocyte dysfunction. Using hiPSC technologies, we can now generate the entire oligodendrocyte lineage in the laboratory, model the full progression of disease pathology, and observe pathogenesis in real time. Importantly, many of the defects we report here could not have been predicted by individuals’ clinical histories or mutations alone. However, characterization of these defects across all samples in parallel enabled identification of distinct subclasses of cellular and molecular pathogeneses that now link disparate PLP1 mutations. We hope this work will serve as a foundation for the assessment of oligodendrocyte dysfunction throughout the greater community of genetic myelin diseases.
Conflicts of Interest
P.J.T. is on the scientific advisory board of Cell Line Genetics.
Acknowledgments
This research was supported by grants from the Pelizaeus-Merzbacher Disease Foundation (P.J.T. and G.M.H.), NIH grants R01NS093357 (P.J.T. and M.W.) and R01NS058978 (G.M.H.), the New York Stem Cell Foundation (P.J.T. and V.F.), NIH predoctoral training grants T32GM007250 and F30HD084167 (Z.S.N.), and the Adelson Medical Research and Novo Nordisk Foundations (S.A.G. and M.S.W.). P.D. is a NYSCF-Druckenmiller fellow. Additional support was provided by the Cytometry & Imaging Microscopy and Genomics core facilities of the Case Comprehensive Cancer Center (P30CA043703). We are grateful to the late James Garbern for providing PMD samples; Leslie Cooperman, Elizabeth Shick, and William Qu for technical assistance; and Peter Scacheri, Anthony Wynshaw-Boris, Nancy Bass, Marius Wernig, and the Tesar Lab for discussion and comments on the manuscript.
Published: March 30, 2017
Footnotes
Supplemental Data include two figures, three tables, and three movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.03.005.
Accession Numbers
The accession number for the raw RNA-seq datasets reported in this paper is GEO: GSE96049.
Web Resources
Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/
GeneReviews, Hobson, G.M., and Kamholz, J. (1993). PLP1-Related Disorders, https://www.ncbi.nlm.nih.gov/books/NBK1182/
OMIM, http://www.omim.org
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
Oligodendrocytes were differentiated from NC2 OPCs over the course of 20 days and then passaged at low density to allow imaging of individual cells every 10 min for 60 hr.
Oligodendrocytes were differentiated from PMD2 OPCs over the course of 20 days and then passaged at low density to allow imaging of individual cells every 10 min for 60 hr.
Oligodendrocytes were differentiated from PMD10 OPCs over the course of 20 days and then passaged at low density to allow imaging of individual cells every 10 min for 60 hr.
References
- 1.Parikh S., Bernard G., Leventer R.J., van der Knaap M.S., van Hove J., Pizzino A., McNeill N.H., Helman G., Simons C., Schmidt J.L., GLIA Consortium A clinical approach to the diagnosis of patients with leukodystrophies and genetic leukoencephelopathies. Mol. Genet. Metab. 2015;114:501–515. doi: 10.1016/j.ymgme.2014.12.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kevelam S.H., Steenweg M.E., Srivastava S., Helman G., Naidu S., Schiffmann R., Blaser S., Vanderver A., Wolf N.I., van der Knaap M.S. Update on Leukodystrophies: A Historical Perspective and Adapted Definition. Neuropediatrics. 2016;47:349–354. doi: 10.1055/s-0036-1588020. [DOI] [PubMed] [Google Scholar]
- 3.Helman G., Van Haren K., Escolar M.L., Vanderver A. Emerging treatments for pediatric leukodystrophies. Pediatr. Clin. North Am. 2015;62:649–666. doi: 10.1016/j.pcl.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helman G., Van Haren K., Bonkowsky J.L., Bernard G., Pizzino A., Braverman N., Suhr D., Patterson M.C., Ali Fatemi S., Leonard J., GLIA Consortium Disease specific therapies in leukodystrophies and leukoencephalopathies. Mol. Genet. Metab. 2015;114:527–536. doi: 10.1016/j.ymgme.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Haren K., Bonkowsky J.L., Bernard G., Murphy J.L., Pizzino A., Helman G., Suhr D., Waggoner J., Hobson D., Vanderver A., Patterson M.C., GLIA Consortium Consensus statement on preventive and symptomatic care of leukodystrophy patients. Mol. Genet. Metab. 2015;114:516–526. doi: 10.1016/j.ymgme.2014.12.433. [DOI] [PubMed] [Google Scholar]
- 6.Hobson G.M., Garbern J.Y. Pelizaeus-Merzbacher disease, Pelizaeus-Merzbacher-like disease 1, and related hypomyelinating disorders. Semin. Neurol. 2012;32:62–67. doi: 10.1055/s-0032-1306388. [DOI] [PubMed] [Google Scholar]
- 7.Inoue K. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 2005;6:1–16. doi: 10.1007/s10048-004-0207-y. [DOI] [PubMed] [Google Scholar]
- 8.Bonkowsky J.L., Nelson C., Kingston J.L., Filloux F.M., Mundorff M.B., Srivastava R. The burden of inherited leukodystrophies in children. Neurology. 2010;75:718–725. doi: 10.1212/WNL.0b013e3181eee46b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Numata Y., Gotoh L., Iwaki A., Kurosawa K., Takanashi J., Deguchi K., Yamamoto T., Osaka H., Inoue K. Epidemiological, clinical, and genetic landscapes of hypomyelinating leukodystrophies. J. Neurol. 2014;261:752–758. doi: 10.1007/s00415-014-7263-5. [DOI] [PubMed] [Google Scholar]
- 10.Pelizaeus F. Uber eine eigenthümliche Form Spastischer Lähmung mit Cerebralerschinungen auf hereditärer Grundlage (Multiple Sklerose) Arch. Psychiatr. Nervenkr. 1885;16:698–710. [Google Scholar]
- 11.Merzbacher L. Eine eigenarige familiär-hereditare Erkrankungsform (Aplasia axialis extra-corticalis congenita) Z. Neurol. Psychiatr. 1910;3:1–138. [Google Scholar]
- 12.Willard H.F., Riordan J.R. Assignment of the gene for myelin proteolipid protein to the X chromosome: implications for X-linked myelin disorders. Science. 1985;230:940–942. doi: 10.1126/science.3840606. [DOI] [PubMed] [Google Scholar]
- 13.Dautigny A., Mattei M.G., Morello D., Alliel P.M., Pham-Dinh D., Amar L., Arnaud D., Simon D., Mattei J.F., Guenet J.L. The structural gene coding for myelin-associated proteolipid protein is mutated in jimpy mice. Nature. 1986;321:867–869. doi: 10.1038/321867a0. [DOI] [PubMed] [Google Scholar]
- 14.Koeppen A.H., Ronca N.A., Greenfield E.A., Hans M.B. Defective biosynthesis of proteolipid protein in Pelizaeus-Merzbacher disease. Ann. Neurol. 1987;21:159–170. doi: 10.1002/ana.410210208. [DOI] [PubMed] [Google Scholar]
- 15.Gencic S., Abuelo D., Ambler M., Hudson L.D. Pelizaeus-Merzbacher disease: an X-linked neurologic disorder of myelin metabolism with a novel mutation in the gene encoding proteolipid protein. Am. J. Hum. Genet. 1989;45:435–442. [PMC free article] [PubMed] [Google Scholar]
- 16.LeVine S.M., Wong D., Macklin W.B. Developmental expression of proteolipid protein and DM20 mRNAs and proteins in the rat brain. Dev. Neurosci. 1990;12:235–250. doi: 10.1159/000111853. [DOI] [PubMed] [Google Scholar]
- 17.Baumann N., Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 18.Garbern J.Y. Pelizaeus-Merzbacher disease: Genetic and cellular pathogenesis. Cell. Mol. Life Sci. 2007;64:50–65. doi: 10.1007/s00018-006-6182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips R.J. Jimpy, a new totally sexlinked gene in the house mouse. Z. Indukt. Abstamm. Vererbungsl. 1954;86:322–326. doi: 10.1007/BF00312228. [DOI] [PubMed] [Google Scholar]
- 20.Eicher E.M., Hoppe P.C. Use of chimeras to transmit lethal genes in the mouse and to demonstrate allelism of the two X-linked male lethal genes jp and msd. J. Exp. Zool. 1973;183:181–184. doi: 10.1002/jez.1401830205. [DOI] [PubMed] [Google Scholar]
- 21.Duncan I.D., Hammang J.P., Trapp B.D. Abnormal compact myelin in the myelin-deficient rat: absence of proteolipid protein correlates with a defect in the intraperiod line. Proc. Natl. Acad. Sci. USA. 1987;84:6287–6291. doi: 10.1073/pnas.84.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths I.R., Duncan I.D., McCulloch M., Harvey M.J. Shaking pups: a disorder of central myelination in the Spaniel dog. Part 1. Clinical, genetic and light-microscopical observations. J. Neurol. Sci. 1981;50:423–433. doi: 10.1016/0022-510x(81)90154-4. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths I.R., Scott I., McCulloch M.C., Barrie J.A., McPhilemy K., Cattanach B.M. Rumpshaker mouse: a new X-linked mutation affecting myelination: evidence for a defect in PLP expression. J. Neurocytol. 1990;19:273–283. doi: 10.1007/BF01217305. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths I., Klugmann M., Anderson T., Thomson C., Vouyiouklis D., Nave K.A. Current concepts of PLP and its role in the nervous system. Microsc. Res. Tech. 1998;41:344–358. doi: 10.1002/(SICI)1097-0029(19980601)41:5<344::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 25.Hüttemann M., Zhang Z., Mullins C., Bessert D., Lee I., Nave K.A., Appikatla S., Skoff R.P. Different proteolipid protein mutants exhibit unique metabolic defects. ASN Neuro. 2009;1:1. doi: 10.1042/AN20090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow D.E., Saul K.E., Komuro H., Macklin W.B. Myelin Proteolipid Protein Complexes with αv Integrin and AMPA Receptors In Vivo and Regulates AMPA-Dependent Oligodendrocyte Progenitor Cell Migration through the Modulation of Cell-Surface GluR2 Expression. J. Neurosci. 2015;35:12018–12032. doi: 10.1523/JNEUROSCI.5151-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laukka J.J., Kamholz J., Bessert D., Skoff R.P. Novel pathologic findings in patients with Pelizaeus-Merzbacher disease. Neurosci. Lett. 2016;627:222–232. doi: 10.1016/j.neulet.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulloche J., Aicardi J. Pelizaeus-Merzbacher disease: clinical and nosological study. J. Child Neurol. 1986;1:233–239. doi: 10.1177/088307388600100310. [DOI] [PubMed] [Google Scholar]
- 29.Hodes M.E., Pratt V.M., Dlouhy S.R. Genetics of Pelizaeus-Merzbacher disease. Dev. Neurosci. 1993;15:383–394. doi: 10.1159/000111361. [DOI] [PubMed] [Google Scholar]
- 30.Cailloux F., Gauthier-Barichard F., Mimault C., Isabelle V., Courtois V., Giraud G., Dastugue B., Boespflug-Tanguy O., Clinical European Network on Brain Dysmyelinating Disease Genotype-phenotype correlation in inherited brain myelination defects due to proteolipid protein gene mutations. Eur. J. Hum. Genet. 2000;8:837–845. doi: 10.1038/sj.ejhg.5200537. [DOI] [PubMed] [Google Scholar]
- 31.Hudson L.D. Pelizaeus-Merzbacher disease and spastic paraplegia type 2: two faces of myelin loss from mutations in the same gene. J. Child Neurol. 2003;18:616–624. doi: 10.1177/08830738030180090801. [DOI] [PubMed] [Google Scholar]
- 32.Hurst S., Garbern J., Trepanier A., Gow A. Quantifying the carrier female phenotype in Pelizaeus-Merzbacher disease. Genet. Med. 2006;8:371–378. doi: 10.1097/01.gim.0000223551.95862.c3. [DOI] [PubMed] [Google Scholar]
- 33.Klugmann M., Schwab M.H., Pühlhofer A., Schneider A., Zimmermann F., Griffiths I.R., Nave K.A. Assembly of CNS myelin in the absence of proteolipid protein. Neuron. 1997;18:59–70. doi: 10.1016/s0896-6273(01)80046-5. [DOI] [PubMed] [Google Scholar]
- 34.Inoue K., Osaka H., Thurston V.C., Clarke J.T., Yoneyama A., Rosenbarker L., Bird T.D., Hodes M.E., Shaffer L.G., Lupski J.R. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am. J. Hum. Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Q., Thurston V.C., Vance G.H., Dlouhy S.R., Hodes M.E. Genetic diagnosis of PLP gene duplications/deletions in patients with Pelizaeus-Merzbacher disease. Clin. Genet. 2005;68:466–467. doi: 10.1111/j.1399-0004.2005.00522.x. [DOI] [PubMed] [Google Scholar]
- 36.Harlow D.E., Saul K.E., Culp C.M., Vesely E.M., Macklin W.B. Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination. J. Neurosci. 2014;34:1333–1343. doi: 10.1523/JNEUROSCI.2477-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nave K.A., Lai C., Bloom F.E., Milner R.J. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc. Natl. Acad. Sci. USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karim S.A., Barrie J.A., McCulloch M.C., Montague P., Edgar J.M., Kirkham D., Anderson T.J., Nave K.A., Griffiths I.R., McLaughlin M. PLP overexpression perturbs myelin protein composition and myelination in a mouse model of Pelizaeus-Merzbacher disease. Glia. 2007;55:341–351. doi: 10.1002/glia.20465. [DOI] [PubMed] [Google Scholar]
- 39.Simons M., Kramer E.M., Macchi P., Rathke-Hartlieb S., Trotter J., Nave K.A., Schulz J.B. Overexpression of the myelin proteolipid protein leads to accumulation of cholesterol and proteolipid protein in endosomes/lysosomes: implications for Pelizaeus-Merzbacher disease. J. Cell Biol. 2002;157:327–336. doi: 10.1083/jcb.200110138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gow A., Friedrich V.L., Jr., Lazzarini R.A. Many naturally occurring mutations of myelin proteolipid protein impair its intracellular transport. J. Neurosci. Res. 1994;37:574–583. doi: 10.1002/jnr.490370504. [DOI] [PubMed] [Google Scholar]
- 41.Numasawa-Kuroiwa Y., Okada Y., Shibata S., Kishi N., Akamatsu W., Shoji M., Nakanishi A., Oyama M., Osaka H., Inoue K. Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Reports. 2014;2:648–661. doi: 10.1016/j.stemcr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southwood C.M., Garbern J., Jiang W., Gow A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron. 2002;36:585–596. doi: 10.1016/s0896-6273(02)01045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gow A., Sharma R. The unfolded protein response in protein aggregating diseases. Neuromolecular Med. 2003;4:73–94. doi: 10.1385/NMM:4:1-2:73. [DOI] [PubMed] [Google Scholar]
- 44.Douvaras P., Wang J., Zimmer M., Hanchuk S., O’Bara M.A., Sadiq S., Sim F.J., Goldman J., Fossati V. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports. 2014;3:250–259. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S., Bates J., Li X., Schanz S., Chandler-Militello D., Levine C., Maherali N., Studer L., Hochedlinger K., Windrem M., Goldman S.A. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somers A., Jean J.C., Sommer C.A., Omari A., Ford C.C., Mills J.A., Ying L., Sommer A.G., Jean J.M., Smith B.W. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 48.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 49.Diskin S.J., Li M., Hou C., Yang S., Glessner J., Hakonarson H., Bucan M., Maris J.M., Wang K. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douvaras P., Fossati V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 2015;10:1143–1154. doi: 10.1038/nprot.2015.075. [DOI] [PubMed] [Google Scholar]
- 53.Chan J.R., Watkins T.A., Cosgaya J.M., Zhang C., Chen L., Reichardt L.F., Shooter E.M., Barres B.A. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron. 2004;43:183–191. doi: 10.1016/j.neuron.2004.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laukka J.J., Stanley J.A., Garbern J.Y., Trepanier A., Hobson G., Lafleur T., Gow A., Kamholz J. Neuroradiologic correlates of clinical disability and progression in the X-linked leukodystrophy Pelizaeus-Merzbacher disease. J. Neurol. Sci. 2013;335:75–81. doi: 10.1016/j.jns.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laukka J.J., Makki M.I., Lafleur T., Stanley J., Kamholz J., Garbern J.Y. Diffusion tensor imaging of patients with proteolipid protein 1 gene mutations. J. Neurosci. Res. 2014;92:1723–1732. doi: 10.1002/jnr.23458. [DOI] [PubMed] [Google Scholar]
- 56.Sumida K., Inoue K., Takanashi J., Sasaki M., Watanabe K., Suzuki M., Kurahashi H., Omata T., Tanaka M., Yokochi K. The magnetic resonance imaging spectrum of Pelizaeus-Merzbacher disease: A multicenter study of 19 patients. Brain Dev. 2016;38:571–580. doi: 10.1016/j.braindev.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shimojima K., Inoue T., Imai Y., Arai Y., Komoike Y., Sugawara M., Fujita T., Ideguchi H., Yasumoto S., Kanno H. Reduced PLP1 expression in induced pluripotent stem cells derived from a Pelizaeus-Merzbacher disease patient with a partial PLP1 duplication. J. Hum. Genet. 2012;57:580–586. doi: 10.1038/jhg.2012.71. [DOI] [PubMed] [Google Scholar]
- 61.Hobson G.M., Davis A.P., Stowell N.C., Kolodny E.H., Sistermans E.A., de Coo I.F., Funanage V.L., Marks H.G. Mutations in noncoding regions of the proteolipid protein gene in Pelizaeus-Merzbacher disease. Neurology. 2000;55:1089–1096. doi: 10.1212/wnl.55.8.1089. [DOI] [PubMed] [Google Scholar]
- 62.Sim F.J., McClain C.R., Schanz S.J., Protack T.L., Windrem M.S., Goldman S.A. CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nat. Biotechnol. 2011;29:934–941. doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Axten J.M., Romeril S.P., Shu A., Ralph J., Medina J.R., Feng Y., Li W.H., Grant S.W., Heerding D.A., Minthorn E. Discovery of GSK2656157: An Optimized PERK Inhibitor Selected for Preclinical Development. ACS Med. Chem. Lett. 2013;4:964–968. doi: 10.1021/ml400228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsaytler P., Harding H.P., Ron D., Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- 65.Clark K., Sakowski L., Sperle K., Banser L., Landel C.P., Bessert D.A., Skoff R.P., Hobson G.M. Gait abnormalities and progressive myelin degeneration in a new murine model of Pelizaeus-Merzbacher disease with tandem genomic duplication. J. Neurosci. 2013;33:11788–11799. doi: 10.1523/JNEUROSCI.1336-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamazaki R., Ishibashi T., Baba H., Yamaguchi Y. Knockdown of Unconventional Myosin ID Expression Induced Morphological Change in Oligodendrocytes. ASN Neuro. 2016;8:8. doi: 10.1177/1759091416669609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Numata Y., Morimura T., Nakamura S., Hirano E., Kure S., Goto Y.I., Inoue K. Depletion of molecular chaperones from the endoplasmic reticulum and fragmentation of the Golgi apparatus associated with pathogenesis in Pelizaeus-Merzbacher disease. J. Biol. Chem. 2013;288:7451–7466. doi: 10.1074/jbc.M112.435388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roboti P., Swanton E., High S. Differences in endoplasmic-reticulum quality control determine the cellular response to disease-associated mutants of proteolipid protein. J. Cell Sci. 2009;122:3942–3953. doi: 10.1242/jcs.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhaunchak A.S., Nave K.A. A common mechanism of PLP/DM20 misfolding causes cysteine-mediated endoplasmic reticulum retention in oligodendrocytes and Pelizaeus-Merzbacher disease. Proc. Natl. Acad. Sci. USA. 2007;104:17813–17818. doi: 10.1073/pnas.0704975104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gow A., Southwood C.M., Lazzarini R.A. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J. Cell Biol. 1998;140:925–934. doi: 10.1083/jcb.140.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gow A., Lazzarini R.A. A cellular mechanism governing the severity of Pelizaeus-Merzbacher disease. Nat. Genet. 1996;13:422–428. doi: 10.1038/ng0896-422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligodendrocytes were differentiated from NC2 OPCs over the course of 20 days and then passaged at low density to allow imaging of individual cells every 10 min for 60 hr.
Oligodendrocytes were differentiated from PMD2 OPCs over the course of 20 days and then passaged at low density to allow imaging of individual cells every 10 min for 60 hr.
Oligodendrocytes were differentiated from PMD10 OPCs over the course of 20 days and then passaged at low density to allow imaging of individual cells every 10 min for 60 hr.