Abstract
Several genes and risk factors are associated with Parkinson’s disease (PD). Although many of the genetic markers belong to a common pathway, a unifying pathogenetic mechanism is yet to be found. Also, missing heritability analyses have estimated that only part of the genetic influence contributing to PD has been found. Here, we carried out whole-exome sequencing (WES) on 438 Finnish patients with early-onset PD. We also re-analyzed previous data from genome-wide association studies (GWAS) on the same cohort. Variants in the CEL gene/locus were associated with PD in both GWAS and WES analysis. Exome-wide gene-based association tests also identified the MPHOSPH10, TAS2R19 and SERPINA1 genes in the discovery dataset (p<2.5E-6). MPHOSPH10 had estimated odds ratio (OR) of 1.53 and the rs141620200 variant in SERPINA1 had OR of 1.27. We identified several candidate genes, but further investigation is required in order to determine the role of these genes in PD.
Keywords: GWAS, WXS, WES, Finnish, PD
Introduction
Several genes and risk loci have been associated with Parkinson’s disease (PD) (see e.g. (Singleton and Hardy, 2016)). A recent meta-analysis on genome-wide association studies (GWAS) from 13,708 cases and 95,282 controls has identified 28 independent risk alleles at 24 gene loci associated with risk for PD (Nalls et al., 2014). Risk loci found in GWAS studies have been estimated to explain about 5% of the genetic variance of PD, while a missing heritability analysis has estimated that the common heritable component of PD is 27% (Keller et al., 2012). These proportions suggest a substantial amount of genetic influence in PD remains to be discovered.
We have previously ascertained a nationwide cohort of patients with early-onset PD (EOPD) (Ylikotila et al., 2015). In this cohort we have found that the birthplaces of patients with PD among first-degree relatives are clustered within the country and that the distance between the birthplaces of their parents was shorter than that among patients with negative family history. The presence of autosomal recessive susceptibility genes for PD in the population could provide an explanation for our findings. In order to characterize the genetic basis of Finnish EOPD, we carried out exome sequencing (WES) on 438 Finnish patients with early-onset PD (EOPD). In addition to WES analysis, we re-analyzed the previous GWAS data on the same cohort (Hernandez et al., 2012) using an extended set of controls.
Subjects and methods
DNA of 438 Finnish EOPD patients was subject to exome sequencing and DNA of 403 patients was subject to genome-wide genotyping. WES data from STAMPEED and FUSION studies on Finnish samples were included into the study as population controls (for details, see supplementary material).
Sample preparation and pre-processing
WES was performed in two sets. The first set included 170 EOPD samples prepared by using Nextera Rapid Capture Expanded Exome Kit and the second set included 268 EOPD samples prepared by using TruSeq Exome Library Prep Kit. The discovery dataset used 170 samples from the first set, 68 samples from the second set and 563 samples from the STAMPEED study. The replication dataset contained sequence data from 200 EOPD samples from the second set.
Analysis
After genotype and variant quality control, rare-variant association analysis and gene-level analysis were carried out using either EPACTS or Raremetal (Feng et al., 2014) with default settings. Gene-based analyses employed emmaxCMC (called here as burden test), sequence kernel association test (SKAT) with SKAT-O, Madsen-Browning weighted burden test (MB) and variable threshold test (VT). Variants resulting in significant p values in single variant association (SVA) test (P < 5E-8) and gene-based analysis (P < 0.05) in discovery dataset were studied in the replication dataset. Variants found in both datasets were characterized further as candidate variants with possible association to PD. Power calculations were performed using the R package ‘CaTS’ v1.02 (Skol et al., 2006) with following parameters: Risk Ratios were calculated using allele frequencies; prevalence=0.02; additive model; pimarkers=1; ncases=185; ncontrols=563; alpha=4.15e-7. Plots for estimating the power of the study and methods used in the re-analysis of the GWAS data, see supplementary material.
Results and discussion
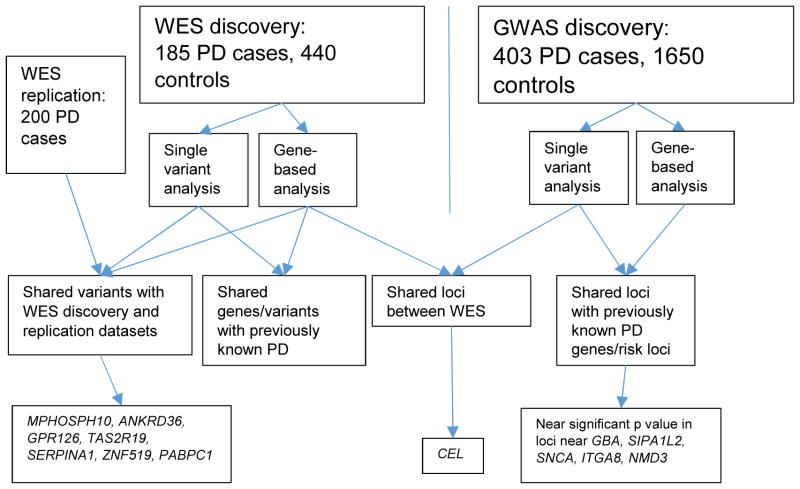
WES study compared 185 cases and 480 controls in the discovery dataset (supplementary table S1 and S2). An overview on the analysis and the results is shown in figure 1. SVA test of rare variants revealed several significant associations with PD (supplementary figure S2 and supplementary table S5), none of which had previously been identified. Variants in the CEL locus were associated with PD in GWAS (supplementary material) and those in the CEL gene in WES analysis. Eight EOPD cases had CEL variants in common in the GWAS and WES datasets and six of them had the CEL variant chr9:135955826 (human genome assembly 19) in the two datasets. The CEL gene harbors a region rich in indel polymorphisms (Taylor et al., 1991), which may point to the possibility that this is a false positive finding but, nevertheless, the role of this gene in PD warrants further investigation.
Figure 1.
Analysis overview.
Key: WES=Whole-exome sequencing; GWAS=Genome wide association study.
A recent study has suggested that variants in the TNR and TNK2 genes may have role in familial PD (Farlow et al., 2016). We found that four EOPD cases (0.91 %) but none of the 563 controls had the rs61731112 variant in TNR, and one case and three controls had the rs147204644 variant. Furthermore, four EOPD cases and three controls had the rs112384084 variant in the TNK2 gene.
Gene-based association tests had exome-wide significant p values (p<2.5E-6) for the genes MPHOSPH10, TAS2R19 and SERPINA1 (table 1, supplementary figure S3 and supplementary table S7). Effect of the gene variation on PD was highest in the case of MPHOSPH10 with an estimated odds ratio (OR) of 1.53 (95% CI; 1.27–1.84). The OR of the rs141620200 variant in SERPINA1 in the SVA test was 1.27 (95% CI; 1.18–1.36). The quality scores of SERPINA1 variants were relatively low suggesting that this association may be a false positive finding. Notably, seven (25 %) out of the 28 cases with the rs141620200 variant had a positive family history with respect to PD, while a family history was reported by 45 cases (11 %) among the remaining EOPD patients (Ylikotila et al., 2015). Interestingly, a recent study including Finnish PD patients has suggested that certain serpinA1 isoforms differ between PD patients with or without dementia (Halbgebauer et al., 2016). Detailed information on characteristics of the genes and variants found both in the discovery and replication datasets is shown in supplementary tables S6–S8 and supplementary text.
Table 1.
Characterization of the statistically significant variants in WES gene-based tests.
| #DISCOVERY DATASET | REPLICATION DATASET | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GENE | CHR | POS | ID | AF | ExAC_AF | SISU_AF | Gene_P | Gene_P_Bonf | Gene_OR | Gene_OR_ci | Single_Var_OR | Single_Var_OR_ci | AC | AF | AC | POWER |
| MPHOSPH10 | 2 | 71361156 | rs143555311 | 1.27E-03 | 0.002 | 0.0013 | 3.46E-06 | 0.044 | 1.526 | 1.266–1.839 | NA | (NA-NA) | 2 | 0.005 | 2 | 3.9e-06 |
| ANKRD36 | 2 | 97858622 | rs201600563 | 1.27E-03 | 0.0014 | 0.0003 | 1.58E-04 | 1.000 | 1.022 | 0.998–1.047 | NA | (NA-NA) | 2 | 0.0076 | 3 | NA |
| GPR126 | 6 | 142758601 | . | 1.27E-03 | 4.14E-05 | 0.0008 | 1.42E-06 | 1.000 | 1.056 | 1.016–1.098 | NA | (NA-NA) | 2 | 0.005 | 2 | 1.4e-04 |
| TAS2R19 | 12 | 11174795 | rs12424373 | 4.44E-03 | 0.05 | 0.0039 | 8.53E-07 | 0.011 | 1.043 | 1.012–1.075 | 0.995 | 0.869–1.139 | 7 | 0.0125 | 5 | 5.3e-7 |
| SERPINA1 | 14 | 94845944 | rs141620200 | 0.019670051 | 0.0023 | 0.0019 | 9.23E-11 | 1.18E-06 | 1.075 | 1.035–1.117 | 1.268 | 1.179–1.364 | 31,1 | 0.01 | 4 | 0.29 |
| SERPINA1 | 14 | 94847262 | rs17580 | 8.25E-03 | 0.02 | 0.0083 | 9.23E-11 | 1.18E-06 | 1.075 | 1.035–1.117 | 0.972 | 0.888–1.063 | 13 | 0.0125 | 5 | 1.4e-06 |
| ZNF519 | 18 | 14105770 | rs61730995 | 0.011435832 | 0.0051 | 0.0053 | 5.05E-06 | 1.000 | 1.123 | 1.063–1.185 | 0.963 | 0.896–1.036 | 18 | 0.01 | 4 | 4.7e-06 |
Table shows variants found in both the discovery and replication datasets. Discovery dataset; cases, N=185; controls, N=480. Replication dataset; cases, N=200.
Key: Gene, gene symbol where variant is located; Chr, chromosome; Pos, Genome position (Grch37); ID, variant RS-number; AF, minor allele frequency; ExAC_AF, Exome aggregate consortium allele frequency for all populations; SISU AF, SISU project (Lim et al., 2014) database allele frequencies; Gene_P, P value in Burden test; Gene_P_Bonf, Bonferroni correction for P value; Gene_OR, estimated odds ratio in Burden test; Gene_OR_ci, Confidence interval for Gene OR; Single_Var_OR, estimated odds ratio for single variant; Single_Var_OR_ci, Confidence interval for single variant OR; AC, minor allele count; Power, statistical power of single variant given known allele frequency.
The results of the WES study correlated poorly with those of the GWAS study probably because of low statistical power. Indeed, our post-hoc power analysis of the discovery dataset estimated that the power to observe associations between known PD variants and risk loci was at best 11% at the significance level of p=1E-6 (supplementary text and supplementary table S12), although no known PD variants were excluded from the PD sequencing cohort. However, it is good to keep in mind that the Finnish population is genetically less diverse (Andersen et al., 2016) than an average European population and, therefore, the complexity of PD might have reduced increasing the overall power to detect novel associated variants.
The poor correlation between results from WES and GWAS might also be considered an indicator of false positives in either of the studies. In addition, the low Ti/Tv value in the discovery dataset may indicate a relatively high number of false positive findings. On the other hand, differences between the WES and GWAS results could also indicate that the variants responsible for the significant p values in the GWAS study might be located outside exonic regions or that, due to the sample size, GWAS is unable to identify the variants.
We identified novel candidate variants and genes that were associated with early-onset PD. Some caution is advised in the interpretation of these findings, because the study setup had limitations such as low power, low Ti/Tv quality and insufficient replication of the findings. Furthermore, because the study has several issues regarding sample size, stage separation and different technologies used, we would like to emphasize that this is a hypothesis generating study. The results deviate from previous studies on patients with European ancestry, but warrant further investigation in order to determine the role of these genes in Parkinson’s disease.
Supplementary Material
Highlights.
Report on exome sequencing of 438 Finnish patients with early-onset Parkinson’s disease
Identified novel candidate genes associated with early-onset Parkinson’s disease
Acknowledgments
For funding details and acknowledgments, please see the supplementary material.
Sources of financial support
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services [project Z01 AG000958-09]. The study was supported by grants from the Council for Health Research of the Academy of Finland [project 127764]; Sigrid Juselius Foundation; Asla-Fulbright Foundation; Orion-Farmos Foundation; The Finnish Parkinson Foundation; Medical Research Center Oulu Doctoral Programme; The Finnish Medical Foundation; Oulun Duodecim-seura; University of Oulu Graduate School; and Oulun Lääketieteellinen Tutkimussäätiö.
Michael A. Nalls’ participation is supported by a consulting contract between Kelly Services and the National Institute on Aging, NIH, Bethesda, MD, USA.
Footnotes
Additional information about the study is provided in supplementary material.
Declaration of interest
Conflicts of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen MK, Pedersen CE, Moltke I, Hansen T, Albrechtsen A, Grarup N. Genetics of Type 2 Diabetes: the Power of Isolated Populations. Curr Diab Rep. 2016;16 doi: 10.1007/s11892-016-0757-z. 65-016-0757-z. [DOI] [PubMed] [Google Scholar]
- Farlow JL, Robak LA, Hetrick K, Bowling K, Boerwinkle E, Coban-Akdemir ZH, Gambin T, Gibbs RA, Gu S, Jain P, Jankovic J, Jhangiani S, Kaw K, Lai D, Lin H, Ling H, Liu Y, Lupski JR, Muzny D, Porter P, Pugh E, White J, Doheny K, Myers RM, Shulman JM, Foroud T. Whole-Exome Sequencing in Familial Parkinson Disease. JAMA Neurol. 2016;73:68–75. doi: 10.1001/jamaneurol.2015.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Liu D, Zhan X, Wing MK, Abecasis GR. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics. 2014;30:2828–2829. doi: 10.1093/bioinformatics/btu367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbgebauer S, Nagl M, Klafki H, Haussmann U, Steinacker P, Oeckl P, Kassubek J, Pinkhardt E, Ludolph AC, Soininen H, Herukka SK, Wiltfang J, Otto M. Modified serpinA1 as risk marker for Parkinson’s disease dementia: Analysis of baseline data. Sci Rep. 2016;6:26145. doi: 10.1038/srep26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Ylikotila P, Keller M, Hardy JA, Majamaa K, Singleton AB. Genome wide assessment of young onset Parkinson’s disease from Finland. PLoS One. 2012;7:e41859. doi: 10.1371/journal.pone.0041859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MF, Saad M, Bras J, Bettella F, Nicolaou N, Simon-Sanchez J, Mittag F, Buchel F, Sharma M, Gibbs JR, Schulte C, Moskvina V, Durr A, Holmans P, Kilarski LL, Guerreiro R, Hernandez DG, Brice A, Ylikotila P, Stefansson H, Majamaa K, Morris HR, Williams N, Gasser T, Heutink P, Wood NW, Hardy J, Martinez M, Singleton AB, Nalls MA International Parkinson’s Disease Genomics Consortium (IPDGC), Wellcome Trust Case Control Consortium 2 (WTCCC2) Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum Mol Genet. 2012;21:4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Wurtz P, Havulinna AS, Palta P, Tukiainen T, Rehnstrom K, Esko T, Magi R, Inouye M, Lappalainen T, Chan Y, Salem RM, Lek M, Flannick J, Sim X, Manning A, Ladenvall C, Bumpstead S, Hamalainen E, Aalto K, Maksimow M, Salmi M, Blankenberg S, Ardissino D, Shah S, Horne B, McPherson R, Hovingh GK, Reilly MP, Watkins H, Goel A, Farrall M, Girelli D, Reiner AP, Stitziel NO, Kathiresan S, Gabriel S, Barrett JC, Lehtimaki T, Laakso M, Groop L, Kaprio J, Perola M, McCarthy MI, Boehnke M, Altshuler DM, Lindgren CM, Hirschhorn JN, Metspalu A, Freimer NB, Zeller T, Jalkanen S, Koskinen S, Raitakari O, Durbin R, MacArthur DG, Salomaa V, Ripatti S, Daly MJ, Palotie A Sequencing Initiative Suomi (SISu) Project. Distribution and medical impact of loss-of- function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R, Ikram MA, Ioannidis JP, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefansson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T, Singleton AB International Parkinson’s Disease Genomics Consortium (IPDGC), Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI), 23andMe GenePD, NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator Cohorts for Health, Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC), United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium, Alzheimer Genetic Analysis Group. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton A, Hardy J. The Evolution of Genetics: Alzheimer’s and Parkinson’s Diseases. Neuron. 2016;90:1154–1163. doi: 10.1016/j.neuron.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Taylor AK, Zambaux JL, Klisak I, Mohandas T, Sparkes RS, Schotz MC, Lusis AJ. Carboxyl ester lipase: a highly polymorphic locus on human chromosome 9qter. Genomics. 1991;10:425–431. doi: 10.1016/0888-7543(91)90328-c. [DOI] [PubMed] [Google Scholar]
- Ylikotila P, Tiirikka T, Moilanen JS, Kaariainen H, Marttila R, Majamaa K. Epidemiology of early-onset Parkinson’s disease in Finland. Parkinsonism Relat Disord. 2015;21:938–942. doi: 10.1016/j.parkreldis.2015.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.