Abstract
The herpes simplex virus type 1 (HSV-1) immediate-early (IE) regulatory protein infected-cell protein 0 (ICP0) is a strong and global transactivator of both viral and cellular genes. In a previous study, we reported that ICP0 is highly phosphorylated and contains at least seven distinct phosphorylation signals as determined by phosphotryptic peptide mapping (D. J. Davido et al., J. Virol. 76:1077-1088, 2002). Since phosphorylation affects the activities of many viral regulatory proteins, we sought to determine whether the phosphorylation of ICP0 affects its functions. To address this question, it was first necessary to identify the regions of ICP0 that are phosphorylated. For this purpose, ICP0 was partially purified, and phosphorylation sites were mapped by microcapillary high-pressure liquid chromatography tandem mass spectrometry. Three phosphorylated regions containing 11 putative phosphorylation sites, all within or adjacent to domains important for the transactivating activity of ICP0, were identified. The 11 sites were mutated to alanine as clusters in each of the three regions by site-directed mutagenesis, generating plasmids expressing mutant forms of ICP0: Phos 1 (four mutated sites), Phos 2 (three mutated sites), and Phos 3 (four mutated sites). One-dimensional phosphotryptic peptide analysis confirmed that the phosphorylation state of each Phos mutant form of ICP0 is altered relative to that of wild-type ICP0. In functional assays, the ICP0 phosphorylation site mutations affected the subcellular and subnuclear localization of ICP0, its ability to alter the staining pattern of the nuclear domain 10 (ND10)-associated protein PML, and/or its transactivating activity in Vero cells. Only mutations in Phos 1, however, impaired the ability of ICP0 to complement the replication of an ICP0 null mutant in Vero cells. This study thus suggests that phosphorylation is an important regulator of ICP0 function.
Phosphorylation is a universal posttranslational modification that alters the activities of many viral regulatory proteins. Among the best examples of this effect is large T antigen of simian virus 40, a multifunctional 708-amino-acid nuclear phospho- and oncoprotein required for the replication of simian virus 40 (reviewed in references 61 and 76). Many phosphorylation sites have been identified on T antigen, and several lie adjacent to its nuclear localization signal and origin-binding domain, modulating simian virus 40 DNA replication (reviewed in reference 61). Specifically, mutation of these sites alters T antigen's origin-binding activity, hexamer-hexamer interaction, nuclear localization, and/or DNA replication activity, affecting both its biochemical and biological functions (11, 14, 41, 50-52, 54, 78).
In herpes simplex virus type 1 (HSV-1) infection, the first genes to be expressed are the immediate-early (IE) genes (39). These genes encode infected-cell protein (ICPs) 0, 4, 22, 27, and 47, which collectively exhibit diverse regulatory and immunomodulatory functions. At least four of the five IE regulatory proteins (ICPs 0, 4, 22, and 27) are known to be phosphorylated, suggesting that phosphorylation is an important modulator of the functions of these proteins (1, 64, 82). In support of this hypothesis, studies by Xia et al. demonstrated that phosphorylation of the N-terminal region of ICP4, the major transcriptional activator of HSV genes, is required for efficient viral replication in cells of neuronal lineage and in a mouse ocular model of HSV-1 latency (83). A second IE protein, ICP22, essential for replication in selected cell types (73), is phosphorylated either directly or indirectly by the delayed-early viral kinase and structural protein UL13 (63, 64). Notably, phosphorylation of ICP22 is required to activate the normal program of viral gene expression in permissive cells (48, 66). A third IE protein, ICP27, an essential gene involved in transport and posttranslational processing of viral transcripts, is also phosphorylated (1, 85). In the study that mapped and mutated phosphorylation sites on ICP27, however, the mutants isolated and characterized were able to complement the growth of an ICP27 null mutant in cell culture (85).
ICP0, the focus of this study, is a 110-kDa nuclear phosphoprotein that transactivates all classes of HSV-1 genes, IE, early (E), and late (L), as well as numerous cellular genes and genes of other viruses (9, 21, 27, 28, 34-36, 55, 65, 80). Mutant viruses affected in ICP0's transactivating activity replicate, establish latency, and reactivate from latency inefficiently and are sensitive to the cellular antiviral factors interferons (6, 23, 37, 38, 47, 56, 68, 75). Although the precise mechanism by which ICP0 mediates its strong and broad transactivating activity is unclear, this activity requires the cellular ubiquitin-proteasome pathway and correlates with the dispersal and/or degradation of cellular proteins linked to cellular transcription, proliferation, differentiation, and apoptosis (reviewed in reference 26). Many but not all of these proteins associate with nuclear structures termed ND10s, and a subset of these proteins are upregulated by interferons (reviewed in reference 57). Thus, ICP0's ability to impair the interferon response during HSV infection appears to be associated with its transactivating activity.
Although early studies demonstrated that the phosphorylation of ICP0 is a dynamic process (1, 80), the role of phosphorylation in ICP0 function, the location of specific phosphorylation sites on ICP0, and the kinases that phosphorylate ICP0 are largely unknown. To date, two types of kinases have been shown to affect the phosphorylation state of ICP0: cellular cyclin-dependent kinases (cdks) and the viral kinase, UL13. One report demonstrated that cdk-1 can phosphorylate the second exon (residues 20 to 241) of ICP0 in vitro (3). In the same study, the authors showed that a dominant-negative form of cdk-1 inhibited the expression of a late gene in HSV-infected cells; however, the phosphorylation state of ICP0 was not examined in these cells. We and others have shown that the posttranslational modification of ICP0, including its phosphorylation state, is altered in the presence of the cdk inhibitor roscovitine, which inhibits the activities of cdks-1, -2, likely -3, -5, -7, and -9 (2, 16, 53, 72, 79). This alteration correlated with a significant decrease in ICP0's transactivating activity, suggesting a link between ICP0's posttranslational modification state (including but not exclusively related to phosphorylation) and its transactivating activity (16). UL13, on the other hand, is required to achieve maximal levels of ICP0 phosphorylation during viral infection and can phosphorylate ICP0 in vitro (58). Notably, it has not yet been established that UL13-mediated phosphorylation of ICP0 is involved in ICP0 function. This study also established that ICP0 is phosphorylated to significant levels in the absence of UL13, indicating that other kinases, most likely cellular kinases, also phosphorylate ICP0. The latter possibility was confirmed by Isler and Schaffer, who demonstrated that ICP0 expressed in cell culture in the absence of other HSV proteins is extensively phosphorylated (40). Thus, although much is known about the phosphorylation of ICP0, the functional relevance of ICP0 phosphorylation to HSV replication has not been analyzed systematically.
The purpose of the present study was to determine which regions and sites on ICP0 are phosphorylated and assess what role(s) these regions and sites play in the many functions of the protein. To identify which of ICP0's 133 potential phosphoacceptor sites are actually phosphorylated, microcapillary high-pressure liquid chromatography tandem mass spectrometry (μLC-MS/MS) analysis was performed on ICP0 isolated from infected cell extracts. μLC-MS/MS analysis identified three major regions of phosphorylation containing 11 potential phosphorylation sites. Mutations which alter the phosphorylation state of each of the three regions affect one or more of the activities of ICP0. This study therefore suggests a significant role for phosphorylation in ICP0 function.
MATERIALS AND METHODS
Cells and viruses.
Vero cells, an African green monkey kidney cell line, were obtained from the American Type Culture Collection (Manassas, Va.) and propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum as described previously (71). L7 cells, Vero cells stably transformed with the ICP0 gene of HSV-1, were passaged as described previously (69). HSV-1, strain KOS (passage 11), was used as the wild-type virus and propagated as described (71). An ICP0 null mutant derived from KOS, 7134, was propagated in Vero cells as previously described (7), and titers of 7134 were determined on L7 cells.
Partial purification of ICP0 for μLC-MS/MS sequencing.
Four 100-mm dishes were seeded with 2 × 106 Vero cells, and 23 h after plating, cells were treated with cycloheximide (50 μg/ml) for 1 h and infected with 5 PFU of KOS per cell for 1 h at 37°C in the presence of cycloheximide. After 1 h of adsorption, the inoculum was removed, and the cells were washed three times with phosphate-buffered saline (PBS) containing cycloheximide; 5 ml of Vero cell medium containing cycloheximide was then added to each dish, and the cells subsequently were incubated for 5 h (t = 6 h postinfection) at 37°C. At t = 6 h postinfection, the medium was removed, and infected cells were washed three times with PBS and incubated for an additional 6 h. At t = 12 h postinfection, cells were washed twice with ice-cold PBS, scraped into 2 ml of ice-cold PBS, and pelleted at 800 × g at 4°C. The supernatant fluid was removed, and the resulting cell pellets were stored at −80°C. Subsequently, samples were thawed on ice, and resuspended in 2 ml of radioimmunoprecipitation assay (RIPA) lysis buffer [150 mM NaCl, 50 mM Tris (pH 7.5), 0.1% sodium dodecyl sulfate (SDS), 1% Nonidet P-40, and 0.5% deoxycholic acid] containing the protease inhibitors phenylmethylsulfonyl fluoride (1 mM), 1 μg of leupeptin per ml, and 1 μg of aprotinin per ml; 8 μl of J17, an ICP0 polyclonal rabbit antibody (86), was added per sample, and the suspensions were gently agitated overnight at 4°C.
The next day, 160 μl of protein A-agarose (Invitrogen Life Technologies, Carlsbad, Calif.) was added to each sample and agitated for 2 h at 4°C. Immune complexes were pelleted by centrifugation for 2 min at 3,300 × g at 4°C. The supernatant fluid was removed, and each pellet was washed and repelleted three times in RIPA buffer plus protease inhibitors. The resulting pellet was resuspended in 60 μl of 1x Laemmli buffer plus 1 mM phenylmethylsulfonyl fluoride (46). Samples were heated at 100°C for 5 min, placed on ice for 2 min, and centrifuged to pellet the protein A-agarose. The supernatants were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 0.75-mm 6% acrylamide gel. Proteins were visualized by Coomassie blue staining, and an estimate of the total amount of ICP0 protein isolated (1.5 μg) was determined relative to a standard curve of bovine serum albumin. Gel pieces containing ICP0 bands were marked, destained for Coomassie blue dye, excised, and stored at −20°C until further analysis.
Identification of phosphorylation sites by μLC-MS/MS.
Phosphorylated residues in ICP0 were detected with μLC-MS/MS. With an in-house program, Enzyme Optimizer, the ICP0 sequence was evaluated for a dual enzyme strategy which would optimize for coverage of S, T, and Y residues. The program considers peptide properties and experimental conditions that influence the recovery and detection of a predicted peptide rather than simple protein coverage. The band corresponding to ICP0 was then split in half for separate in-gel trypsin and chymotrypsin digestions after reduction and carboxyamidomethylation. The resultant digests were pooled just prior to μLC-MS/MS injection. Phosphorylated peptide sequences were determined with a 75-μm reverse-phase microcolumn terminating in a custom nanoelectrospray source directly coupled to a Finnigan LCQ DECA XP+ quadrupole ion trap mass spectrometer (Thermo Electron). The flow rate was nominally 250 nl/min. The ion trap repetitively surveyed the range m/z 395 to 1,600, executing data-dependent MS/MS on the four most abundant ions in each survey scan. MS/MS spectra were acquired with a relative collision energy of 30%, a 2.5-Da isolation width, and recurring ions dynamically excluded. Preliminary sequencing of peptides was facilitated by database correlation with the algorithm SEQUEST (19). The discovery of peptides carrying phosphorylation and subsequent manual validation of their MS/MS spectra were aided by the in-house programs Muquest and FuzzyIons, respectively (15).
Plasmids.
Plasmid pIE3-CAT, which expresses the chloramphenicol acetyltransferase (CAT) gene under the control of the HSV-1 IE ICP4 promoter, was constructed as previously described (18). Plasmid pAlter-1+ICP0 was constructed by isolating a 4.6-kb EcoRI-HindIII fragment containing the ICP0 gene from the plasmid pSH (9) and cloned into the vector pAlter-1 (Promega Corp. Madison, Wis.), with the same restriction enzyme sites. pAlter-1+ICP0 was subsequently used as the parental vector to mutate the putative phosphorylation sites of ICP0 to alanine with mutagenic primers (IDT, Coralville, Iowa) according to the manufacturer's protocol (Promega Corp., Madison, Wis.). The primers used for the mutagenesis are Phos 1 (S224A, T226A, T231A, T232A), 5′-CTGGGGGGGCACACGGTGAGGGCCCTagCGCCggCCCACCCTGAGCCggCCgCGGACGAGGATGACGACGACCTGGAC-3′; Phos 2 (S365A, S367A, S371A), 5′-GCAAACAACAGAGACCCCATAGTGATCgcCGAtgCCCCCCCGGCCgCTCCCACAGGCCCCCCGCGGCGCCC-3′; and Phos 3 (S508A, S514A, S517A, T518A), 5′-GCGGTGCGTCCGAGGAAGAGGCGCGGGgCcGGCCAGGAAAACCCCgCCCCgCAGgCCgCGCGTCCCCCCCTCGCGCCGGCAGGGG-3′. Lowercase letters indicate the nucleotides mutated relative to the wild-type (strain KOS) ICP0 sequences. Putative mutants were identified by restriction enzyme analysis (Fig. 1B) and confirmed by DNA sequencing.
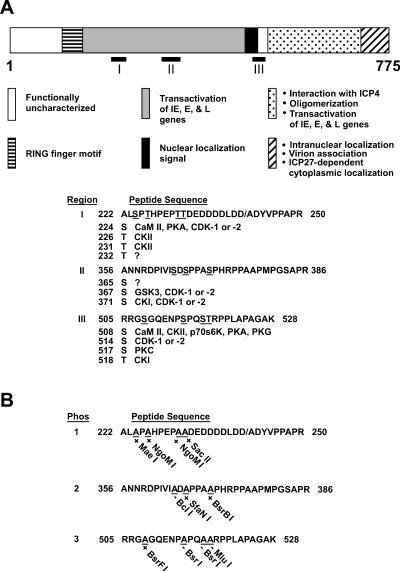
FIG. 1.
Putative phosphorylated sequences on ICP0. (A) Location of phosphorylated sequences on ICP0 as determined by μLS-MS/MS analysis. The 775 amino acids of ICP0, the locations of the major functional domains of ICP0, and the locations of the 11 putative phosphorylation sites in regions I, II, and III are shown. Beneath the diagram of ICP0, the amino acid sequences of regions I, II, and III are shown. The slash mark between D and A in region I represents the amino acid boundary between the second and third exons of ICP0. The codon numbers of the putatively phosphorylated serine (S) and threonine (T) residues are listed beneath the peptide sequence in each region. To the right of each serine or threonine are listed the cellular kinases which most likely target these residues, as determined by computer-based modeling with NetPhos 2.0 (Center for Biological Sequence Analysis, Technical University of Denmark), ScanProsite (Swiss Institute for Bioinformatics), and MacVector 7.1.1 (GCG, Madison, Wis.). Kinases include calmodulin kinase II (CaM II), protein kinase A (PKA), cyclin-dependent kinase 1 (cdk-1) and 2, casein kinase I (CKI), glycogen synthase kinase 3 (GSK3), casein kinase II (CKII), p70S6K kinase, protein kinase G (PKG), and protein kinase C (PKC). (B) Mutations in putative ICP0 phosphorylation sites. Serines and threonines in each phosphorylated region were mutated to alanine (A), resulting in the gain (+) or loss (−) of a specific restriction enzyme cleavage site, as shown beneath each substituted alanine residue. Forms of ICP0 in which phosphorylated serine (S) or threonine (T) residues in regions I, II and III have been changed to alanine (A) are designated Phos 1, 2, and 3, respectively, in the text.
Transfection/infection and 32P and 35S labeling of wild-type ICP0 and its mutant forms.
Vero cells were plated at 5 × 105 cells per 60-mm dish, and at 2 h prior to transfection (22 h postplating), the medium was changed. At 24 h after plating, transfections were performed with Lipofectamine 2000 (Invitrogen Corp., Carlsbad, Calif.) according to the manufacturer's protocol with 8 μg of plasmid or salmon sperm DNA and 16 μl of Lipofectamine 2000 diluted in Opti-MEM (Invitrogen Corp.). The DNA/Lipofectamine 2000 mixture was subsequently added to plates containing 5 ml of Opti-MEM in a dropwise manner and left on the cells for 4 h at 37°C. Four hours after transfection, cells were treated for 1 h with 50 μg of cycloheximide per ml and mock infected or infected with 5 PFU of KOS or 7134 per cell for 1 h at 37°C in the presence of cycloheximide. After 1 h of adsorption, the inoculum was removed, the cells were washed three times with PBS containing cycloheximide, 4 ml of Vero cell medium plus cycloheximide was added per dish, and the dishes were incubated for 4.5 h postinfection at 37°C. At t = 5.5 h postinfection, the cells were incubated for 0.5 h in phosphate- or methionine- and cysteine-free DMEM containing cycloheximide. At t = 6 h postinfection, the medium was removed, and the infected cells were washed three times with phosphate- or methionine- and cysteine-free DMEM containing 1% fetal bovine serum. The cells were then labeled with 500 μCi of 32Pi or 100 μCi of [35S]methionine/cysteine (PerkinElmer Life Sciences, Inc., Boston, Mass.) in phosphate- or methionine- and cysteine-free DMEM containing 1% fetal bovine serum, respectively, for an additional 6 h. At t = 12 h postinfection, cells were washed twice with ice-cold PBS, scraped into 1 ml of ice-cold RIPA lysis buffer containing protease inhibitors as described above. Extract preparation, immunoprecipitation of ICP0, and SDS-PAGE analysis were performed as described previously (16). Proteins were visualized, and their signal intensities were quantified by PhosphorImager analysis (Amersham Biosciences, Piscataway, N.J.).
ICP0 phosphotryptic peptide digestion and one-dimensional alkaline gel electrophoresis.
32P-labeled bands of wild-type ICP0 or its mutant forms were excised from the SDS-PAGE gel described above and washed two times for 5 min in fresh ammonium bicarbonate (50 mM). Each gel piece was brought to a final volume of 500 μl with sodium bicarbonate, homogenized with a pestle grinder, and treated with 40 μg of TPCK [l-(tosyl/amido-z-phenyl)ethyl ketone)-treated trypsin (Worthington Biochemicals, Lakewood, N.J.) while gently agitating at 34°C overnight. An additional 25 μg of TPCK-treated trypsin was added per sample, incubating for a second time overnight at 34°C. Samples were spun at 20,800 × g for 10 min at room temperature, and the resulting supernatant was removed. Remaining protein in the gel pieces was extracted twice by incubating the pieces in 400 μl of acetonitrile-formic acid (1:1) for 20 min with gentle rocking at room temperature. The acetonitrile-formic acid extracts and the aqueous supernatant of the phosphotryptic digests were pooled (≈1,200 μl) and dried in a SpeedVac (Savant Instruments, Inc., Farmingdale, N.Y.). Phospholabeled peptides of ICP0 were suspended in 25 μl of sample buffer [0.125 M Tris-HCl buffer (pH 6.8) and 6 M urea], loaded with equal Cherenkov counts (≈1,150 cpm), and separated on a 27-cm 30% (wt/vol) alkaline acrylamide gel as described previously (81) at 10 mA for 44 h. Phosphotryptic peptides from the alkaline gel electrophoresis were visualized by PhosphorImager analysis.
Immunofluorescence.
Vero cells were plated on coverslips in 12-well plates, and 22 h later, fresh Vero cell medium was added to each well 2 h prior to transfection. Transfections were performed with Lipofectamine 2000 according to the manufacturer's protocol with 3 μg of plasmid DNA and 6 μl of Lipofectamine 2000 diluted in Opti-MEM (Invitrogen Corp.). The DNA-Lipofectamine 2000 mixture was then added to each well containing 500 μl of Opti-MEM in a dropwise manner and left on the cells for 5 h at 37°C. The medium was removed, and fresh medium was added, incubating the monolayers for an additional 19 h. Twenty-four hours posttransfection, the medium was removed, and the coverslips were washed twice with PBS. Transfected cells were fixed and permeabilized by formaldehyde and acetone treatments according to Zhu et al. (86).
Immunofluorescence staining for ICP0, its mutant forms, and PML was performed as previously described (17). The primary antibodies and the dilutions used for the dual staining of ICP0 and PML were ICP0 at 1:500 (H1112; mouse monoclonal antibody; Rumbaugh-Goodwin Institute for Cancer Research, Plantation, Fla.) and PML-14 at 1:500 (rabbit polyclonal antibody; Gerd Maul, Wistar Institute, Philadelphia, Pa.). The following secondary antibodies and dilutions were used for each primary antibody: ICP0 (H1112) at 1:100 (goat anti-mouse immunoglobulin G conjugated with fluorescein isothiocyanate) and PML-14 at 1:100 (goat anti-rabbit immunoglobulin G conjugated with rhodamine red X). All secondary antibodies were purchased from Jackson Immunoresearch (West Grove, Pa.). Following incubations with the primary and secondary antibodies, the coverslips were washed, and 7 μl of Prolong antifade solution (Molecular Probes, Eugene, Oreg.) was added per coverslip. Cells were viewed by fluorescence microscopy with a Nikon Eclipse TE300 fluorescence microscope at ×400 magnification and photographed with an RT Slider digital camera (Diagnostic Instruments, Sterling Heights, Mich.), and images were processed in Adobe Photoshop (Adobe Systems Inc., Mountain View, Calif.). Images were assembled and labeled in Canvas 8 (Deneba Systems, Miami, Fla.).
At least 150 ICP0-stained cells from random fields were examined in each preparation and categorized as having nuclear only, nuclear and cytoplasmic, or cytoplasmic only staining. The percentage of cells in each category was determined by dividing the number of cells in a given category by the total number of cells counted in all three categories.
Transient transfections and CAT assays.
Vero cells (5 × 105 cells per 60-mm dish) were plated, and 2 h before transfection (22 h postplating), the medium was changed. Twenty-four hours after plating, the transfections were performed with Lipofectamine 2000 according to the manufacturer's protocol with a total of 8 μg of DNA at 1 μg of CAT expression vector, salmon sperm testis DNA, and/or increasing amounts of plasmid DNA (as indicated in Fig. 5) and 16 μl of Lipofectamine 2000 diluted in Opti-MEM per dish. DNA-Lipofectamine 2000 was then added to dishes containing 5 ml of Opti-MEM in a dropwise manner and left on cells for 5 h at 37°C. The medium was removed, and fresh Vero cell medium was added, incubating the monolayers for an additional 43 h. At 48 h posttransfection, cells were washed three times with Tris-buffered saline (TBS), harvested in 2 ml of TBS, and pelleted at 800 × g; the supernatant was removed. The resulting cell pellets were stored at −80°C. Samples were thawed on ice, resuspended in 150 μl of TBS, sonicated for 20 s at 80% power in a Misonix Sonicator 3000 (Misonix, Inc., Farmingdale, N.Y.), and cell debris was pelleted for 10 min at 4°C at 20,800 × g. The resulting supernatant was assayed for CAT activity as performed by Seed and Sheen (74).
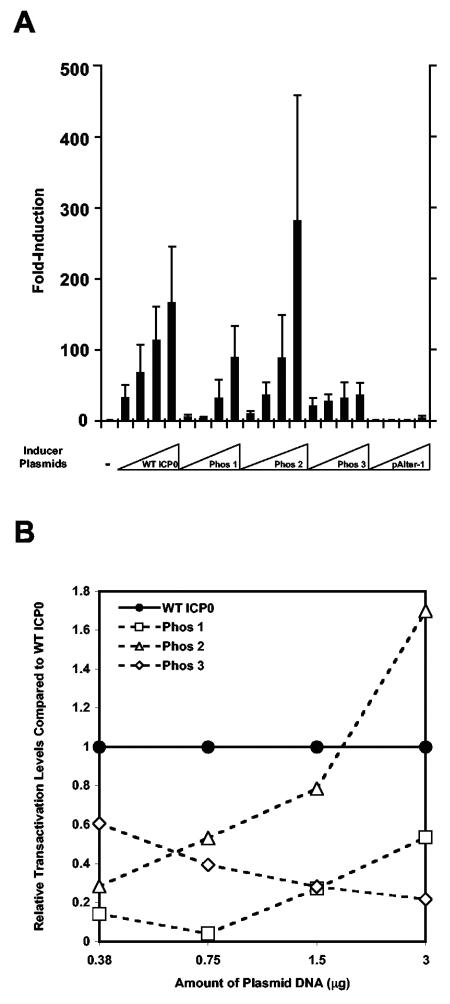
FIG. 5.
Transactivating activity induced by wild-type (WT) and Phos mutant forms of ICP0. (A) Vero cells were plated and 24 h later cotransfected with 1 μg of pIE3-CAT reporter plasmid (ICP4 promoter-CAT construct) (18) or with increasing amounts of inducer plasmids (0.375, 0.75, 1.5, and 3 μg) expressing wild-type ICP0 (ICP0), Phos 1, 2, and 3, or the cloning vector alone (pAlter-1). Forty-eight hours after transfection, cells were harvested, extracts were prepared, and CAT assays were performed. The induction (fold) of CAT activity relative to the basal CAT activity of the pIE3-CAT reporter plasmid transfected alone, given the arbitrary value of 1 (bar on the far left), is shown. (B) Transactivating activity of Phos mutants relative to wild-type ICP0 as a function of the amount of inducer DNA transfected. The relative level of transactivation of each Phos mutant was determined by dividing the CAT activity for a given amount of expression plasmid transfected over the CAT activity of an equivalent of amount of wild-type ICP0-expressing plasmid transfected, based on the data presented in panel A. In all cases, wild-type ICP0 was given the arbitrary value of 1 for each amount of wild-type ICP0-expressing plasmid transfected.
Complementation and plating efficiency of 7134.
Vero cells were plated at 2 × 105 cells per 35-mm dish. Twenty-two hours later, fresh medium was added to each well. Twenty four hours postplating, the transfections were performed with Fugene 6 (Roche Diagnostic Corporation, Indianapolis, Ind.) with a total of 3 μg of DNA (0.5 μg of infectious KOS or 7134 viral DNA) (9), salmon sperm DNA, and/or 0.075 μg of plasmid DNA from pAlter-1 or wild-type ICP0- or Phos mutant-expressing plasmids and 12 μl of Fugene 6 diluted in Opti-MEM. The DNA/Fugene 6 mixture was divided in half, added dropwise to 35-mm dishes (for duplicate samples) containing 2 ml of Opti-MEM, and left on the cells for 5 h at 37°C. The medium was removed, fresh Vero cell medium was added, and the monolayers were incubated for an additional 43 h. Transfected cells were harvested 48 h posttransfection and assayed for infectious virus by standard plaque assays on Vero cells for KOS or on L7 cells for 7134.
For the 7134 plating efficiency experiments, cells were transfected as described for the 7134 complementation experiments with the following modifications (8). Five hours posttransfection, the medium was removed from each culture and replaced with 2.5 ml of medium containing 0.5% methylcellulose and 10% fetal bovine serum in DMEM. Three days posttransfection, the methylcellulose-containing medium was removed from each plate, and the cells were washed twice with PBS, fixed with 2% formaldehyde, and stained for β-galactosidase activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as described by Sanes et al. (70). X-Gal staining was performed to note the spread of 7134 infection (7134 contains the lacZ gene in place of both copies of ICP0) by counting cytopathic blue foci (CBF; clusters of eight or more cells exhibiting cytopathic effect) per quadrant (one-quarter of the area of each dish).
RESULTS
Identification of putative phosphorylation sites on ICP0 by μLC-MS/MS analysis.
To identify the sites on ICP0 that are phosphorylated, μLC-MS/MS analysis was performed on ICP0 partially purified from HSV-1-infected cells. ICP0 synthesized after removal of a cycloheximide block was immunoprecipitated from extracts of 8 × 106 cells and separated by standard SDS-PAGE. ICP0 was digested with trypsin and chymotrypsin in the gel and subjected to μLC-MS/MS analysis. The results of μLC-MS/MS analysis are summarized in Fig. 1A. Three phosphorylated regions on ICP0 were identified and designated regions I, II, and III. Three peptides were identified in region I, four in region II, and seven in region III. Each peptide was phosphorylated at one or two phosphoacceptor sites.
The largest peptide sequence is shown for each region (Fig. 1A). Each region lies within or adjacent to domains of ICP0 known to be important for its transactivating activity. Computer-based modeling indicated that 11 putative phosphorylation sites (four within a 9-amino-acid stretch, three within a 7-amino-acid stretch, and four within an 11-amino-acid stretch for regions I, II, and III, respectively) are present on ICP0. Based on recognized kinase substrate/consensus motifs, these peptides and their putative phosphorylation sites were further analyzed by NetPhos 2.0, ScanProsite, and MacVector 7.1.1 to identify cellular kinases that might phosphorylate these sites. These analyses indicated that 9 of the 11 phosphoacceptor sites likely serve as targets of one or more cellular kinases (Fig. 1A). No cellular kinases were identified for Thr-232 and Ser-365 (Fig. 1A).
The 11 putative phosphorylation sites were mutated to alanine in clusters by site-directed mutagenesis, resulting in the loss or acquisition of a restriction enzyme cleavage site (Fig. 1B). Substitution of alanine for serine or threonine at similar sites in other proteins has been shown to inhibit the phosphorylation of these phosphoacceptor sites (31, 41). By clustering the mutations, we were able to determine which regions of phosphorylation are not required for ICP0 function, eliminating the need to mutate and analyze individual sites. Multiply mutated ICP0 genes were screened for the presence of the mutations by restriction enzyme analysis and DNA sequencing. This mutagenesis strategy generated three mutant plasmids: Phos 1, containing four mutated sites; Phos 2, containing three mutated sites; and Phos 3, containing four mutated sites. These mutations were present in regions I, II, and III (Fig. 1B). The Phos 1, 2, and 3 plasmids were then tested in assays for ICP0 function.
Levels of ICP0 synthesis and phosphorylation state of ICP0 Phos mutants.
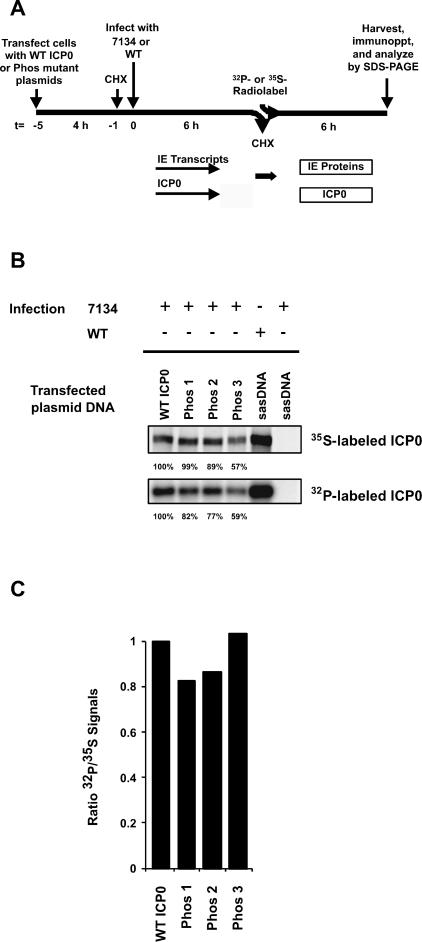
We first examined the levels and phosphorylation states of the wild-type and Phos mutant forms of ICP0 by SDS-PAGE with the protocol illustrated in Fig. 2A. The results of these tests indicate that the level of Phos 3 protein was modestly reduced as determined by 35S labeling, whereas the levels of the Phos 1 and 2 proteins were similar to that of wild-type ICP0 (Fig. 2B). The levels of phosphorylation of the Phos mutants were modestly reduced relative to that of wild-type ICP0, as determined by 32P labeling (Fig. 2B). Compared with wild-type ICP0, the relative level of phosphorylation of each Phos mutant as determined by 32P labeling corresponded closely with the level of each mutant protein as determined by 35S labeling (Fig. 2C). As expected, wild-type virus-infected cells expressed high levels of phosphorylated ICP0, whereas mock-transfected and 7134-infected cells expressed no ICP0 (Fig. 2B).
FIG. 2.
Protein levels and phosphorylation state of the Phos mutant forms of ICP0. (A) Experimental design. Vero cell monolayers plated 24 h previously were transfected with plasmids expressing wild-type ICP0, Phos 1, 2, or 3, or salmon sperm DNA (sasDNA) for 4 h. At 4 h posttransfection, transfected cells were treated for 1 h (t = −1) with the protein synthesis inhibitor cycloheximide (CHX), mock infected or infected with 5 PFU/cell of wild-type (WT) or ICP0 null mutant (7134) (t = 0), and incubated in the presence of cycloheximide for 6 h (t = 6). IE transcripts were synthesized during the 6-h period. Half of the cultures were then released from the cycloheximide block into phosphate-free medium containing [32P]orthophosphate. The remaining cultures were released into methionine- and cysteine-free medium containing [35S]methionine/cysteine. Radiolabeling was carried out for 6 h (t = 12). (B) Immunoprecipitation and SDS-PAGE analysis of radiolabeled ICP0. At t = 6 h postlabeling, extracts were prepared, ICP0 was immunoprecipitated with an ICP0 rabbit poly-clonal antibody (J17), and resolved by SDS-6% PAGE. 35S- and 32P-specific ICP0 bands were visualized by PhosphorImager analysis. Band intensities of wild-type ICP0 and Phos mutants were measured by PhosphorImager analysis, and the intensities of Phos mutant forms of ICP0 relative to wild-type ICP0 (expressed as 100%) for each radiolabeled band were determined. (C) Ratio of the relative 32P to 35S bands intensities of wild-type ICP0 and Phos 1, 2, and 3. The ratios of band intensities of wild-type and each Phos mutant form of ICP0 were determined by dividing the relative 32P band intensity by the 35S band intensity (shown in B) for wild-type ICP0 or Phos 1, 2, and 3.
One-dimensional phosphotryptic peptide analysis.
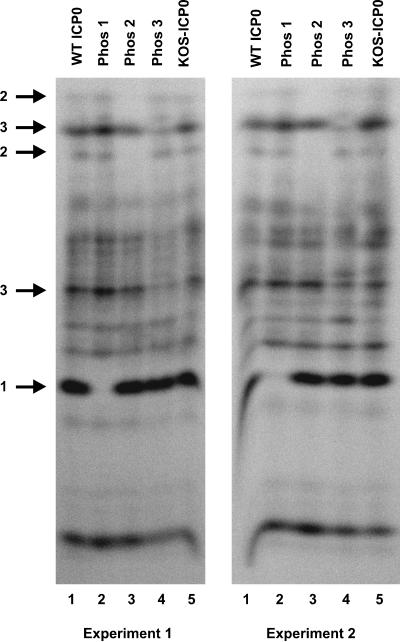
Among the 133 potential phosphorylation sites on ICP0, μLC-MS/MS analysis identified subsets of sites in three regions that are actually phosphorylated. To confirm that mutagenesis resulted in the loss of phosphorylation of the Phos 1, 2, and 3 mutant forms of ICP0, we performed one-dimensional phosphotryptic peptide analysis with the phosphorylated samples shown in Fig. 2B. In two independent experiments, multiple phosphotryptic peptides were observed in gels of extracts of cells transfected with plasmids expressing wild-type ICP0 or the three Phos mutants and infected with 7134 (Fig. 3, lanes 1, 2, 3, and 4) or mock transfected and infected with wild-type virus (lane 5). As indicated by the arrows on the left of the figure, a loss or reduction in the signal of one or two specific phosphotryptic peptides was evident for each of the three mutant proteins. In contrast, wild-type ICP0 isolated from KOS-infected cells was essentially identical to the phosphotryptic peptide profile of wild-type ICP0 synthesized in transfected and infected cells (Fig. 3, lanes 1 and 5). Thus, the mutagenesis strategy was successful in altering the phosphorylation state of ICP0 expressed from each of the mutant plasmids. As evident from the multiple phosphorylated bands not affected by any of the 11 alanine substitution mutations, μLC-MS/MS analysis identified only selected phosphorylation sites on ICP0.
FIG. 3.
One-dimensional phosphotryptic peptide analysis of wild-type and Phos mutant forms of ICP0. 32P-labeled wild-type (WT) and Phos 1, 2, and 3 mutant forms of ICP0 expressed from Vero cells transfected with the corresponding plasmids and infected with 7134 or ICP0 expressed from KOS-infected cells was isolated following SDS-PAGE from the gels shown in Fig. 2B and digested with trypsin. The resulting phosphopeptides were separated on a 30% alkaline polyacrylamide gel (81) and visualized by PhosphorImager analysis. The results of two independent experiments are shown. Numbered arrows on the left identify specific phosphotryptic peptides present in wild-type ICP0 that are absent or reduced in the Phos 1, 2, or 3 mutant forms of ICP0.
Immunofluorescence assays. (i) Subcellular localization of wild-type and Phos mutant forms of ICP0 in transfected cells.
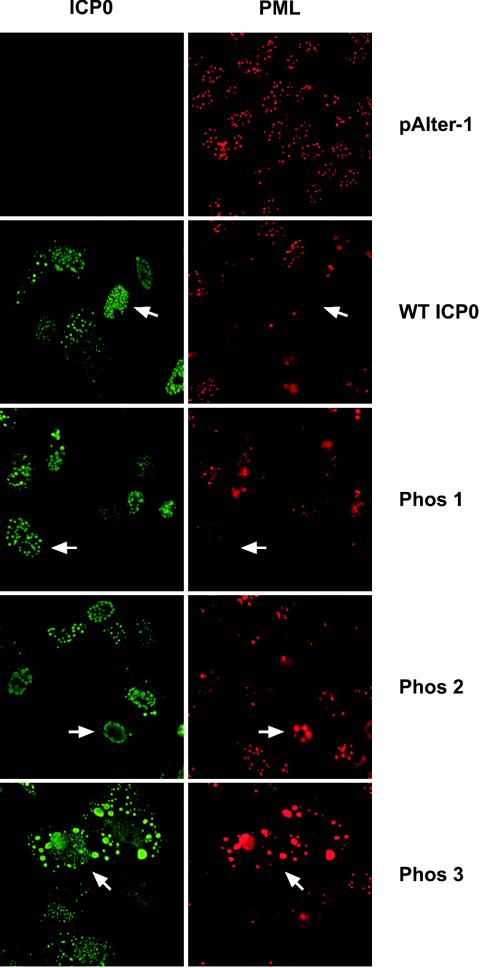
Having established that the phosphorylation states of the Phos 1, 2, and 3 mutant forms of ICP0 were altered with respect to wild-type ICP0, the effects of these clustered mutations on the subcellular localization of ICP0 and ICP0's ability to alter the staining pattern of the ND10-associated protein promyelocytic leukemia (PML) were examined by immunofluorescence microscopy. Focusing first on the subcellular localization of ICP0, transfected cells were classified in one of three categories based on their subcellular staining patterns: nuclear only, nuclear and cytoplasmic, and cytoplasmic only. The staining patterns of wild-type ICP0, Phos 1, and Phos 2 were predominantly nuclear and punctate (Fig. 4 and Table 1) (an example of each is indicated by the white arrow), as previously reported for wild-type ICP0 by Zhu and Schaffer (86). Specifically, 84 to 90% of transfected cells expressing these forms of ICP0 stained only in the nucleus, 8 to 15% stained both in the nucleus and in the cytoplasm, and less than 4% stained in the cytoplasm only. In contrast, the predominant subcellular location of the Phos 3 form of ICP0 was both nuclear and cytoplasmic (72.6% of cells; Fig. 4, white arrow), whereas ≈25% of cells exhibited staining exclusively in the nucleus (Table 1). Furthermore, Phos 3-expressing cells exhibited finer punctate, nuclear staining and large cytoplasmic globular staining relative to cells expressing wild-type ICP0. Thus, one or more of the four substitution mutations in Phos 3 are responsible for the altered subcellular and subnuclear staining pattern of ICP0.
FIG. 4.
ICP0- and PML-specific immunofluorescence in Vero cells transfected with plasmids expressing wild-type (WT) ICP0 or Phos 1, 2, or 3 mutant forms of ICP0. Vero cells were grown on coverslips in 12-well plates. Twenty-four hours after plating, cells were transfected with 3 μg of plasmid DNA expressing wild-type ICP0, Phos 1, 2, or 3 mutant forms of ICP0, or the cloning vector alone (pAlter-1). Twenty four hours posttransfection, cells were washed, fixed, permeabilized, and probed with primary and secondary antibodies to detect ICP0 and PML by immunofluorescence. The white arrows show ICP0- and PML-specific staining in individual cells positive for ICP0 staining. Cells were viewed by fluorescence microscopy with a Nikon Eclipse TE300 Fluorescence microscope at ×400 magnification and photographed with a digital camera.
TABLE 1.
Subcellular localization of wild-type and mutant forms of ICP0a
| Plasmid | % of cells with staining pattern:
|
||
|---|---|---|---|
| N | N + C | C | |
| Wild-type ICP0 | 83.7 | 15.1 | 1.2 |
| Phos 1 | 90.3 | 8.4 | 1.3 |
| Phos 2 | 86.1 | 10.6 | 3.3 |
| Phos 3 | 24.8 | 72.6 | 2.6 |
The subcellular localization of wild-type and Phos mutant forms of ICP0 in individual Vero cells 24 h after transfection was determined. Cells that stained positive for ICP0 were classified into one of three groups based on the subcellular localization of ICP0: cells that displayed only nuclear staining (N), cells that displayed both nuclear and cytoplasmic staining (N + C), and cells that displayed only cytoplasmic staining (C). One hundred fifty or more ICP0-stained cells in each preparation were analyzed, and the percentage of cells exhibiting a specific pattern of subcellular localization was calculated by dividing the number of ICP0-expressing cells in a given category by the total number of cells counted in all three categories for each protein.
(ii) Do the Phos mutations affect the staining pattern of PML?
PML-specific immunofluorescence assays were performed with a primary rabbit polyclonal antibody which recognizes PML. PML-specific staining in the absence of ICP0 (pAlter-1) was uniformly nuclear and punctate (Fig. 4). In cells that expressed wild-type ICP0 or an ICP0 Phos mutant, a reduction in PML-specific staining or an alteration in the PML staining pattern was observed. PML-specific staining was less intense in almost all wild-type ICP0- and Phos 1-expressing cells (Fig. 4, white arrows). PML staining was also less intense in many Phos 2-expressing cells; however, some large nuclear PML-specific bodies were present in nuclei expressing Phos 2 (white arrows), but they did not colocalize with Phos 2-specific staining, which was in the nuclear margin. In Phos 3-expressing cells, PML-specific nuclear staining was less intense overall, although PML-specific staining colocalized with large globular Phos 3 cytoplasmic bodies (white arrows) in a subset of cells.
Transactivating activity of Phos mutants.
To examine the transactivating activity of the Phos mutants, Vero cells were cotransfected with wild-type ICP0, Phos 1-, 2-, or 3-expressing plasmids, or pAlter-1 and the HSV-1 ICP4 promoter-CAT reporter construct (pIE3-CAT) or pIE3-CAT alone. As shown in Fig. 5A, CAT activity increased as a function of increasing amounts of wild-type ICP0- and Phos mutant-expressing plasmids transfected. Relative to wild-type ICP0, Phos 1 transactivating activity was impaired 7- to 20-fold at the two lowest amounts of plasmid transfected and only 2- to 3-fold at the two highest amounts transfected (Fig. 5A and B). The CAT activity induced by Phos 2 was reduced threefold at the lowest amount of plasmid transfected, but was similar (≤2-fold differences) to wild-type ICP0 at the three highest amounts of the inducer plasmid tested (Fig. 5A and B). Phos 3 induced similar levels of CAT activity (≤2-fold differences) to wild-type ICP0 at the two lowest amounts of plasmid transfected but was impaired 3- to 5-fold at the two highest amounts (Fig. 5A and B). Phos 3 was nonetheless able to induce limited CAT activity at least 20-fold above the basal level of the pIE3-CAT vector alone. Relative to wild-type ICP0, the altered levels of activity observed with the ranges of DNAs transfected may be a consequence of the residual transactivating activities of mutant forms of ICP0 and their interactions with other regulatory proteins.
7134-complementing activity of the Phos mutants.
To determine whether the Phos mutants are able to complement the replication of the ICP0 null mutant 7134 in Vero cells, infectious wild-type or 7134 DNA was transfected alone or infectious 7134 DNA was cotransfected with pAlter-1 or a plasmid expressing a wild-type or Phos mutant form of ICP0 as described by Cai and Schaffer (9). Forty-eight hours after transfection, cells were harvested and lysates were assayed for the presence of infectious wild-type and 7134 virus by standard plaque assays in Vero and L7 cells, respectively. In these assays, high titers of wild-type virus were detected (Table 2). In contrast, 7134 virus titers were low (≈6 orders of magnitude lower than titers of wild-type virus) when 7134 DNA was transfected alone or in combination with pAlter-1. Complementation of 7134 replication was achieved by transfection of infectious 7134 viral DNA with the wild-type ICP0-expressing plasmid (≈1,230-fold enhancement compared with 7134 DNA alone) (Table 2). 7134 titers were 20-fold lower when cotransfected with the Phos 1-expressing plasmid than with the plasmid expressing wild-type ICP0, whereas viral titers following cotransfection of 7134 DNA with Phos 2 and Phos 3 were only 1.6- and 1.2-fold lower, respectively.
TABLE 2.
Complementation of 7134 replication by ICP0 Phos mutant plasmidsa
| Virus + plasmid DNAs | Titerb (PFU/ml) | Difference relative to WT ICP0c (fold) |
|---|---|---|
| KOS | 1.33 × 107 | |
| 7134 | 5 | |
| 7134 + pAlter-1 | 5 | |
| 7134 + WT ICP0 | 6,150 | 1 |
| 7134 + Phos 1 | 315 | ↓20 |
| 7134 + Phos 2 | 3,925 | ↓1.6 |
| 7134 + Phos 3 | 5,000 | ↓1.2 |
Vero cells were transfected with 0.25 μg of KOS or 7134 infectious viral DNA and 0.0375 μg of pAlter-1 or a plasmid expressing wild-type (WT) ICP0 or a Phos mutant. Transfected cells were harvested 48 h posttransfection and assayed for infectious virus. The experiment was repeated twice, and results from one experiment are shown.
Average viral titers from duplicate samples were determined on Vero cells (for KOS) or L7 cells (for 7134).
Difference in viral titers for each Phos mutant-expressing plasmid relative to the wild-type ICP0-expressing plasmid.
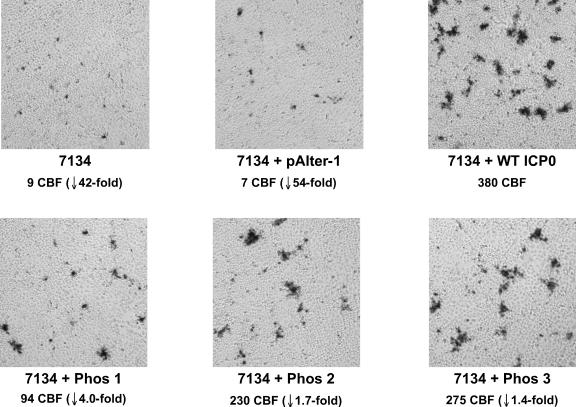
A second series of experiments tested the ability of Phos mutant and wild-type forms of ICP0 to enhance the replication of infectious 7134 DNA as measured by the production of cytopathic blue foci (CBF) in Vero cells. The ability of the plasmids to induce foci (clusters of eight or more cells exhibiting cytopathic effect) when cotransfected with 7134 DNA was assessed 3 days posttransfection and visualized by X-Gal staining of the cultures. X-Gal staining was performed to visualize the spread of infection with 7134, which contains the lacZ gene in place of both copies of ICP0. The production of CBF results primarily from complementation but may also result from recombination (in this case rare [<5%] large plaques were observed). The ability of infectious 7134 viral DNA or 7134 DNA cotransfected with pAlter-1 to form CBF was limited, whereas cotransfection of 7134 DNA with the plasmid expressing wild-type ICP0 increased the number of CBF by at least 42-fold (Fig. 6). The ability of Phos 1 to induce CBF was reduced 4-fold relative to wild-type ICP0, and Phos 2 and 3 reduced the number of CBF by ≈1.7- and 1.4-fold, respectively. Furthermore, the size of the Phos 1-induced CBF was smaller than that of those induced by wild-type ICP0, Phos 2, or Phos 3. Thus, the mutations in the Phos 1 form of ICP0 render this protein less capable of complementing the replication and plating efficiency of 7134 infectious DNA than the mutations in the Phos 2 or 3 forms of ICP0 in Vero cells.
FIG. 6.
Complementation of 7134 as determined by cytopathic blue focus (CBF) formation by wild-type ICP0 and Phos mutant forms of ICP0. Vero cells were transfected with 0.25 μg of 7134 infectious viral DNA alone or cotransfected with 0.0375 μg of pAlter-1 or plasmids expressing wild-type ICP0 or mutant forms of ICP0 (Phos 1, 2, and 3) per dish. 7134 contains the lacZ gene in place of both copies of ICP0. Five hours after transfection, monolayers were overlaid with methylcellulose-containing medium. Three days posttransfection, monolayers were washed, fixed, and stained for β-galactosidase activity with X-Gal and incubated for an additional 14 to 16 h at room temperature. The number and the differences (fold) in CBF (clusters of eight or more rounded, cytopathic cells) produced on monolayers transfected with 7134 infectious viral DNA alone, 7134 infectious viral DNA and pAlter-1, or 7134 and individual Phos mutant-expressing plasmids relative to 7134 cotransfected with a plasmid expressing wild-type ICP0 are shown beneath the label for each photo. Monolayers were viewed with a Nikon light microscope at ×80 magnification and photographed with a digital camera.
DISCUSSION
This study demonstrates that ICP0 is phosphorylated in at least three regions which collectively contain 11 putative phosphorylation sites as determined by μLC-MS/MS analysis. With site-directed mutagenesis, potentially phosphorylated serine and threonine residues in these regions were changed to alanine. The resulting mutations in the three regions affect ICP0's subcellular and subnuclear localization (Phos 3), ND10-disrupting activity in a subset of cells (Phos 2 and 3), and transactivating activity (Phos 1 and 3) in Vero cells. Despite the altered phenotypes of all three Phos mutant forms of ICP0, only the Phos 1 form of ICP0 exhibited significantly reduced ability to complement the replication and plating efficiency of infectious 7134 DNA. This study represents the first attempt to fine map and mutate known posttranslational modification sites on ICP0 and determine their roles in ICP0 function, as the great majority of ICP0 structure-function analyses have employed nonsense, insertion, or deletion mutants. It is also acknowledged that introducing alanines in place of putative phosphoacceptor sites in clusters may alter the folding or confirmation of ICP0 molecules and thus affect ICP0 function. Future studies designed to identify the specific phosphorylation sites utilized in regions I, II, and III will allow us to prove or refute this possibility. While μLC-MS/MS analysis of the peptides generated by trypsin and chymotrypsin digestion identified three phosphorylated regions on ICP0 that are important for its diverse biological activities, this analysis clearly identified only a subset of phosphorylated regions on ICP0, as illustrated in Fig. 3. The location of additional regions and their role(s) in the functions of ICP0 will be determined by μLC-MS/MS with alternative enzymes to generate phosphopeptides.
Do the locations of the mutations in Phos 1, 2, and 3 correspond with domains or interaction sites known to be important for the activities of ICP0? (i)Phos 1.
Based on μLC-MS/MS analysis, each of the three phosphorylated regions of ICP0 identified lies within or overlaps domains reported to be important for ICP0's transactivating activity. Thus, the four mutations in Phos 1 at positions 224, 226, 231, and 232 are adjacent to the RING finger motif of ICP0, which is required for its E3 ubiquitin ligase and transactivating activities (Fig. 1A) (5, 22, 24). This study has shown that the Phos 1 form of ICP0 is impaired in its transactivating activity and in its ability to complement the replication of an ICP0 null mutant but not its ability to degrade and/or disperse ND10. Insertion and deletion mutations in region I or between region I and ICP0's RING finger have been shown to diminish both the transactivating activity of ICP0 and its ability to colocalize with conjugated ubiquitin (20, 22, 24, 25); the latter activity is consistent with ICP0's E3 ubiquitin ligase activity.
Mutations in the RING finger motif (residues 116 to 156) and an adjacent region (residues 162 to 188) show a range of ND10-disrupting phenotypes that are distinct from the phenotype of wild-type ICP0 (20, 29, 49). To date, however, the ND10-disrupting activities of the majority of ICP0 mutants with mutations that lie in region I and between region I and the RING finger motif have not been reported. Additionally, mutation of Asp-199 (which lies between region I and the RING finger motif) to alanine negates the binding of ICP0 to cyclin D3, accelerating the destabilization of cyclin D3 (44). This mutation attenuates the pathogenesis of HSV-1 (77).
Residues 20 to 241 of ICP0 have been shown to interact with the cellular transcription factor BMAL1 (43). The interaction of ICP0 with BMAL1 is thought to facilitate synergistic transactivation of BMAL-1 responsive genes. Thus, it is possible that the effects that we observed with the Phos 1 mutations in Vero cells result from alterations in these adjacent regions. Of interest is the fact that the phenotypes of Phos 1 are strikingly similar to the phenotypes of ICP0 synthesized in the presence of the cdk inhibitor roscovitine (17). Specifically, ICP0 synthesized in the presence of roscovitine is impaired in its transactivating activity but not its ND10-disrupting activity. Thus, roscovitine-sensitive cdk-mediated phosphorylation of phosphoacceptor sites in region I of ICP0, especially Ser-224 which is a potential cdk-1 or -2 phosphorylation site, may contribute to ICP0's transactivating activity.
(ii) Phos 2.
The mutations in Phos 2 at positions 365, 367, and 371 lie within a large proline-rich domain important for ICP0's transactivating activity. (Fig. 1A) (80). Phos 2 was affected only in its capacity to disperse or degrade ND10-associated PML in a subset of Phos 2-expressing cells; the transactivating- and 7134-complementing activities of Phos 2 were only minimally affected relative to wild-type ICP0. The study of Chen et al. noted a reduction in ICP0's transactivating activity in a mutant form of ICP0 lacking residues 263 to 448, which includes all of region II (13). Consistent with the transactivating potential of Phos 2, a report by Everett noted that an insertion in region II at residue 370 and deletion of residues 341 to 374 had marginal effects on ICP0-mediated transactivation in Vero cells (22). The ability of these deletion and insertion mutations to affect the dispersal or degradation of ND10-associated proteins (including PML) has not been reported. Notably, the magnitude of the impairment in the transactivating activity of these mutants correlates with the severity of their mutations.
(iii) Phos 3.
Mutations in Phos 3 at positions 508, 514, 517, and 518 overlap the putative nuclear localization signal of ICP0 and are adjacent to its multifunctional C-terminal domain (9, 12, 13, 24, 27, 29, 49). Phos 3 was affected in its subcellular and nuclear localization and its ability to disperse or degrade PML in a subset of cells expressing Phos 3; however, these mutations only minimally affected its ability to complement 7134 in Vero cells. The diminished transactivation potential of Phos 3 may also be due to its altered subcellular and/or subnuclear localization. Similar observations have been reported for deletion mutations of ICP0 which eliminate its putative nuclear localization signal (24, 27). Notably, a proportion of cells expressing these deletion mutants colocalized with ND10-associated proteins in the cytoplasm, similar to our observations with PML and Phos 3 (Fig. 4) (29, 49).
Because the mutations in Phos 3 are directly adjacent to the putative nuclear localization signal of ICP0 and because phosphorylation of the nuclear localization signal may affect nuclear import, Phos 3 may be inefficiently transported to or from the nucleus, as our immunofluorescence data suggest (Fig. 4). In support of this possibility, mutagenesis of potential phosphorylation sites has been shown to regulate the subcellular localization of viral regulatory proteins from DNA-containing viruses, including the T antigen of simian virus 40 (41, 84), pp65 of human cytomegalovirus (32), latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus (60), IE63 protein of varicella zoster virus (4), and US11, an RNA-binding protein (67), and ICP27 of HSV-1 (10, 85).
Of the viral regulatory proteins just mentioned with the exception of US11, casein kinase II, cdk-1, and protein kinase A have been implicated in their subcellular localization, and putative phosphorylation sites for these cellular kinases have been identified in region III of ICP0. The subcellular localization of Phos 3 may also indirectly affect the phosphorylation state of a portion of ICP0 molecules. For example, if ICP0 is efficiently phosphorylated by nuclear kinases which are required for its biological activities, then impairment of ICP0's nuclear translocation would prevent its phosphorylation by such kinases. Although the mutations in Phos 3 reduced its transactivating activity, they affected its 7134-complementating activity in Vero cells only minimally. These observations demonstrate that Phos 3 possesses sufficient transactivating activity to stimulate the replication of an ICP0 null mutant to levels similar to that of wild-type ICP0. This possibility is reinforced by two studies demonstrating that mutant forms of ICP0 exhibiting impaired transactivating activity are capable of supporting significant complementation of an ICP0 null mutant virus (9, 12). Finally, although we examined the phenotypic effects of the Phos 1, 2, and 3 mutations in Vero cells, it is conceivable that these mutations have cell type-specific effects in the functional assays used in this study, alter other known activities of ICP0 (e.g., E3 ubiquitin ligase activity, accumulation of conjugated ubiquitin, or cell cycle-blocking activity), and/or have distinct phenotypes in the context of viral infection in vivo. These possibilities warrant further investigation.
Role of cellular and viral kinases in ICP0 function.
While this study has identified 11 putative phosphorylation sites on ICP0, whether these sites are actually phosphorylated and, if they are, the identity of the cellular and viral kinases that phosphorylate these sites and the roles of specific phosphorylation events in the regulation of ICP0 function remain to be determined. Of the three regions identified, our results indicate that the Phos 1 mutations had the greatest effect on ICP0-induced transactivation and viral replication, suggesting that the kinases that phosphorylate one or more of these sites are necessary for these activities of ICP0. From computer-based modeling, cellular kinases that potentially phosphorylate Ser-224, Thr-226, and Thr-231 are calmodulin kinase II, protein kinase A, cdk-1 and -2, and casein kinase II (Fig. 1A). No cellular kinases were identified for T232 in our computer analysis.
Do these cellular kinases play a role in the functions of HSV IE proteins, including ICP0? Indeed, several studies indicate that cellular kinases most likely modulate the functions of IE proteins through phosphorylation. As noted in the introduction, phosphorylation of a serine-rich region of ICP4 from residues 140 to 210 by protein kinase A appears to be required for efficient HSV replication in PC12 cells (a rat cell line of neural lineage), efficient replication in the tear film and trigeminal ganglia of mice during acute infection, and efficient reactivation from latency in the mouse ocular model (83). ICP27 has been shown to be phosphorylated on at least four major peptides in infected cell cultures (85). Two of the four peptides are phosphorylated by protein kinase A and casein kinase II in vitro. Mutation of these phosphorylation sites did not affect the ability of ICP27 to complement the growth of a virus lacking ICP27, indicating that these sites have no apparent role in viral replication in cell culture. The effects of these mutations in vivo will require further testing. A recent report noted that casein kinase II binds to ICP27, stimulating casein kinase II activity during viral infection (45). Casein kinase II subsequently phosphorylates ICP27, affecting its interaction with another ICP27-interacting cellular protein, heterogeneous ribonucleoprotein, which has been implicated in transcript transport. Cdk-1 has been shown to phosphorylate the second exon (residues 20 to 241) of ICP0 in vitro (3), and the portion of region I that lies within this exon contains an aforementioned putative cdk-1 or -2 phosphorylation site at Ser-224 (Fig. 1A; left of the hatchmark in region I). Consequently, cdk-1 or -2 may phosphorylate Ser-224, contributing to the biological activities of ICP0. The roles of the remaining cellular kinases identified in our computer analysis in ICP0 function (Fig. 1A; glycogen synthase kinase 3, casein kinase I, p70S6K kinase, protein kinase G, and protein kinase C) remain to determined.
Based on sequence analysis and functional studies, HSV encodes three viral kinases: the products of the UL13 gene, a delayed-early tegument protein (64); the UL39 gene (ICP6), an E protein and the large subunit of ribonucleotide reductase (36, 59); and the US3 gene, an E protein implicated in cell survival (30, 33, 62). Of the three viral kinases, UL13 is the most extensively studied with regard to IE protein function. As previously discussed, ICP22 is phosphorylated, either directly or indirectly, by UL13, and this phosphorylation is required for ICP22 function in selected cell types (48, 63, 64, 66, 73). The phosphorylation of ICP0 is also mediated in part by UL13 in infected cells and in vitro with infected cell extracts, but the significance of this phosphorylation for the activities of ICP0 is unknown (58). Interestingly, one study reported that UL13 and cdk-1 can phosphorylate the same serine residue on the cellular translation protein elongation factor 1δ (42). Consequently, UL13 may also phosphorylate the putative cdk-1 or -2 consensus motifs on ICP0 identified in our study.
Future studies of ICP0 phosphorylation will entail identifying the individual phosphorylation site or sites that affect the activities of each Phos mutant form of ICP0 and the roles of these sites in ICP0 functions as they relate to HSV-1 replication. This fine-mapping approach will permit us to determine whether the effects of the clustered mutations in Phos 1, 2, and 3 are due to specific alterations in the phosphorylation state of ICP0 or to alterations in the structure of ICP0. Ultimately, identifying the cellular and viral kinases that phosphorylate ICP0 and elucidating the roles of these kinases in the regulation of ICP0 function should provide insight into how virus-cell interactions influence the HSV life cycle.
Acknowledgments
This work was supported by Public Health Service grant RO1CA20260 from the National Cancer Institute (to P.A.S.).
We thank Gerd Maul for providing the PML antibody, Joe Orlando for providing technical assistance with site-directed mutagenesis, and other members of the Schaffer lab and the Boston area herpesvirus group for helpful comments and suggestions.
REFERENCES
- 1.Ackermann, M., D. K. Braun, L. Pereira, and B. Roizman. 1984. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J. Virol. 52:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Advani, S. J., R. Hagglund, R. R. Weichselbaum, and B. Roizman. 2001. Posttranslational processing of infected cell proteins 0 and 4 of herpes simplex virus 1 is sequential and reflects the subcellular compartment in which the proteins localize. J. Virol. 75:7904-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2000. The role of cdc2 in the expression of herpes simplex virus genes. Proc. Natl. Acad. Sci. USA 97:10996-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bontems, S., E. Di Valentin, L. Baudoux, B. Rentier, C. Sadzot-Delvaux, and J. Piette. 2002. Phosphorylation of varicella-zoster virus IE63 protein by casein kinases influences its cellular localization and gene regulation activity. J. Biol. Chem. 277:21050-21060. [DOI] [PubMed] [Google Scholar]
- 5.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., and P. A. Schaffer. 1991. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J. Virol. 65:4078-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catez, F., M. Erard, N. Schaerer-Uthurralt, K. Kindbeiter, J. J. Madjar, and J. J. Diaz. 2002. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol. Cell. Biol. 22:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cegielska, A., I. Moarefi, E. Fanning, and D. M. Virshup. 1994. T-antigen kinase inhibits simian virus 40 DNA replication by phosphorylation of intact T antigen on serines 120 and 123. J. Virol. 68:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., C. Panagiotidis, and S. Silverstein. 1992. Multimerization of ICP0, a herpes simplex virus immediate-early protein. J. Virol. 66:5598-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, J. X., X. X. Zhu, and S. Silverstein. 1991. Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type 1. Virology 180:207-220. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y. R., S. P. Lees-Miller, P. Tegtmeyer, and C. W. Anderson. 1991. The human DNA-activated protein kinase phosphorylates simian virus 40 T antigen at amino- and carboxy-terminal sites. J. Virol. 65:5131-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chittum, H. S., W. S. Lane, B. A. Carlson, P. P. Roller, F. D. Lung, B. J. Lee, and D. L. Hatfield. 1998. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37:10866-10870. [DOI] [PubMed] [Google Scholar]
- 16.Davido, D. J., D. A. Leib, and P. A. Schaffer. 2002. The cyclin-dependent kinase inhibitor roscovitine inhibits the transactivating activity and alters the posttranslational modification of herpes simplex virus type 1 ICP0. J. Virol. 76:1077-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davido, D. J., W. F. Von Zagorski, G. G. Maul, and P. A. Schaffer. 2003. The differential requirement for cyclin-dependent kinase activities distinguishes two functions of herpes simplex virus type 1 ICP0. J. Virol. 77:12603-12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca, N. A., and P. A. Schaffer. 1985. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol. Cell. Biol.. 5:1197-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng, J. K., A. L. McCormack, and J. R. Yates, III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 20.Everett, R., P. O'Hare, D. O'Rourke, P. Barlow, and A. Orr. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol. 69:7339-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett, R. D. 1985. Activation of cellular promoters during herpes virus infection of biochemically transformed cells. EMBO J. 4:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J. Mol. Biol. 202:87-96. [DOI] [PubMed] [Google Scholar]
- 23.Everett, R. D. 1989. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J. Gen. Virol. 70:1185-1202. [DOI] [PubMed] [Google Scholar]
- 24.Everett, R. D. 1987. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 6:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 27.Everett, R. D. 1988. Promoter sequence and cell type can dramatically affect the efficiency of transcriptional activation induced by herpes simplex virus type 1 and its immediate-early gene products Vmw175 and Vmw110. J. Mol. Biol. 203:739-751. [DOI] [PubMed] [Google Scholar]
- 28.Everett, R. D. 1984. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 3:3135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frame, M. C., F. C. Purves, D. J. McGeoch, H. S. Marsden, and D. P. Leader. 1987. Identification of the herpes simplex virus protein kinase as the product of viral gene US3. J. Gen. Virol. 68:2699-2704. [DOI] [PubMed] [Google Scholar]
- 31.Fu, X. D., P. T. Tuazon, J. A. Traugh, and J. Leis. 1988. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein, pp 12, at Serine 40, the primary site of phosphorylation in vivo. J. Biol. Chem. 263:2134-2139. [PubMed] [Google Scholar]
- 32.Gallina, A., E. Percivalle, L. Simoncini, M. G. Revello, G. Gerna, and G. Milanesi. 1996. Human cytomegalovirus pp65 lower matrix phosphoprotein harbours two transplantable nuclear localization signals. J. Gen. Virol. 77:1151-1157. [DOI] [PubMed] [Google Scholar]
- 33.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelman, I. H., and S. Silverstein. 1985. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 82:5265-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gius, D., and L. A. Laimins. 1989. Activation of human papillomavirus type 18 gene expression by herpes simplex virus type 1 viral transactivators and a phorbol ester. J. Virol. 63:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein, D. J., and S. K. Weller. 1988. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62:196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halford, W. P., and P. A. Schaffer. 2000. Optimized viral dose and transient immunosuppression enable herpes simplex virus ICP0-null mutants to establish wild-type levels of latency in vivo. J. Virol. 74:5957-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isler, J. A., and P. A. Schaffer. 2001. Phosphorylation of the herpes simplex virus type 1 origin binding protein. J. Virol. 75:628-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jans, D. A., M. J. Ackermann, J. R. Bischoff, D. H. Beach, and R. Peters. 1991. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J. Cell Biol. 115:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 77:2359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koffa, M. D., J. Kean, G. Zachos, S. A. Rice, and J. B. Clements. 2003. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J. Virol. 77:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 47.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 50.McVey, D., L. Brizuela, I. Mohr, D. R. Marshak, Y. Gluzman, and D. Beach. 1989. Phosphorylation of large tumour antigen by cdc2 stimulates SV40 DNA replication. Nature 341:503-507. [DOI] [PubMed] [Google Scholar]
- 51.McVey, D., S. Ray, Y. Gluzman, L. Berger, A. G. Wildeman, D. R. Marshak, and P. Tegtmeyer. 1993. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol. 67:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McVey, D., B. Woelker, and P. Tegtmeyer. 1996. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J. Virol. 70:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meijer, L., A. Borgne, O. Mulner, J. P. Chong, J. J. Blow, N. Inagaki, M. Inagaki, J. G. Delcros, and J. P. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527-536. [DOI] [PubMed] [Google Scholar]
- 54.Montenarh, M., and D. Muller. 1987. The phosphorylation at Thr 124 of simian virus 40 large T antigen is crucial for its oligomerization. FEBS Lett. 221:199-204. [DOI] [PubMed] [Google Scholar]
- 55.Mosca, J. D., D. P. Bednarik, N. B. Raj, C. A. Rosen, J. G. Sodroski, W. A. Haseltine, G. S. Hayward, and P. M. Pitha. 1987. Activation of human immunodeficiency virus by herpesvirus infection: identification of a region within the long terminal repeat that responds to a trans-acting factor encoded by herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 84:7408-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 58.Ogle, W. O., T. I. Ng, K. L. Carter, and B. Roizman. 1997. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology 235:406-413. [DOI] [PubMed] [Google Scholar]
- 59.Paradis, H., P. Gaudreau, B. Massie, N. Lamarche, C. Guilbault, S. Gravel, and Y. Langelier. 1991. Affinity purification of active subunit 1 of herpes simplex virus type 1 ribonucleotide reductase exhibiting a protein kinase activity. J. Biol. Chem. 266:9647-9651. [PubMed] [Google Scholar]
- 60.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prives, C. 1990. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell 61:735-738. [DOI] [PubMed] [Google Scholar]
- 62.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinlan, M. P., and D. M. Knipe. 1985. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol. Cell. Biol.. 5:957-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rice, S. A., M. C. Long, V. Lam, P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samaniego, L. A., N. Wu, and N. A. DeLuca. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanes, J. R., J. L. Rubenstein, and J. F. Nicolas. 1986. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 5:3133-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schaffer, P. A., G. M. Aron, N. Biswal, and M. Benyesh-Melnick. 1973. Temperature-sensitive mutants of herpes simplex virus type 1: isolation, complementation and partial characterization. Virology 52:57-71. [DOI] [PubMed] [Google Scholar]
- 72.Schang, L. M., A. Bantly, M. Knockaert, F. Shaheen, L. Meijer, M. H. Malim, N. S. Gray, and P. A. Schaffer. 2002. Pharmacological cyclin-dependent kinase inhibitors inhibit replication of wild-type and drug-resistant strains of herpes simplex virus and human immunodeficiency virus type 1 by targeting cellular, not viral, proteins. J. Virol. 76:7874-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the alpha 22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seed, B., and J. Y. Sheen. 1988. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene 67:271-277. [DOI] [PubMed] [Google Scholar]
- 75.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66:179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Sant, C., Y. Kawaguchi, and B. Roizman. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc. Natl. Acad. Sci. USA 96:8184-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Virshup, D. M., A. A. Russo, and T. J. Kelly. 1992. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol. Cell. Biol. 12:4883-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, D., C. de la Fuente, L. Deng, L. Wang, I. Zilberman, C. Eadie, M. Healey, D. Stein, T. Denny, L. E. Harrison, L. Meijer, and F. Kashanchi. 2001. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J. Virol. 75:7266-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weber, P. C., and B. Wigdahl. 1992. Identification of dominant-negative mutants of the herpes simplex virus type 1 immediate-early protein ICP0. J. Virol. 66:2261-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.West, M. H. P., W. S. Wu, and W. M. Bonner. 1984. Polyacrylamide gel electrophoresis of small peptides. Electrophoresis 5:133-138. [Google Scholar]
- 82.Wilcox, K. W., A. Kohn, E. Sklyanskaya, and B. Roizman. 1980. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J. Virol. 33:167-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia, K., D. M. Knipe, and N. A. DeLuca. 1996. Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J. Virol. 70:1050-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao, C. Y., S. Hubner, R. M. Elliot, A. Caon, and D. A. Jans. 1996. A consensus cAMP-dependent protein kinase (PK-A) site in place of the CcN motif casein kinase II site simian virus 40 large T-antigen confers PK-A-mediated regulation of nuclear import. J. Biol. Chem. 271:6451-6457. [DOI] [PubMed] [Google Scholar]
- 85.Zhi, Y., and R. M. Sandri-Goldin. 1999. Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J. Virol. 73:3246-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu, Z., W. Cai, and P. A. Schaffer. 1994. Cooperativity among herpes simplex virus type 1 immediate-early regulatory proteins: ICP4 and ICP27 affect the intracellular localization of ICP0. J. Virol. 68:3027-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]