Abstract
Sleep deprivation impairs cognitive performance and reliably alters brain activation in wakefulness and sleep. Nevertheless, the molecular regulators of prolonged wakefulness remain poorly understood. Evidence from genetic, behavioral, pharmacologic and imaging studies suggest that dopaminergic signaling contributes to the behavioral and electroencephalographic (EEG) consequences of sleep loss, although direct human evidence thereof is missing. We tested whether dopamine neurotransmission regulate sustained attention and evolution of EEG power during prolonged wakefulness. Here, we studied the effects of functional genetic variation in the dopamine transporter (DAT1) and the dopamine D2 receptor (DRD2) genes, on psychomotor performance and standardized waking EEG oscillations during 40 hours of wakefulness in 64 to 82 healthy volunteers. Sleep deprivation consistently enhanced sleepiness, lapses of attention and the theta-to-alpha power ratio (TAR) in the waking EEG. Importantly, DAT1 and DRD2 genotypes distinctly modulated sleep loss-induced changes in subjective sleepiness, PVT lapses and TAR, according to inverted U-shaped relationships. Together, the data suggest that genetically determined differences in DAT1 and DRD2 expression modulate functional consequences of sleep deprivation, supporting the hypothesis that striato-thalamo-cortical dopaminergic pathways modulate the neurobehavioral and neurophysiological consequences of sleep loss in humans.
Sleep deprivation enhances sleepiness, impairs performance and alters electrical brain activity in a highly predictable fashion1,2,3,4. Modern lifestyle means that these sleep-loss related cognitive deficits have developed to a public health concern. In the laboratory, neurobehavioral deficits of sleep deprivation are accurately indexed by lapses of attention on the psychomotor vigilance task (PVT), which is considered a gold-standard measurement of sustained vigilant attention3,5. Neurophysiological consequences of sleep loss on the other hand, are reflected by distinct oscillations in the waking electroencephalogram (EEG)6,7. Importantly, these neurobehavioral and neurophysiological consequences vary widely among individuals. Ample evidence now demonstrates that genetic influences strongly modulate waking EEG oscillations and alters PVT lapses across prolonged waking, even within consecutive test sessions8,9,10,11,12. Consistent with a genetic contribution, the impact of sleep loss on distinct subjective, PVT, and neurophysiological markers of alertness, is trait-like and highly robust within individuals. Intriguingly, however, these variables don’t typically show a clear association with each other, but rather develop seemingly independently13,14. Here, we hypothesized that functional variation in genes impacting local dopaminergic neurotransmission affects sleep loss-induced changes in PVT lapses and the waking EEG along a similar trajectory.
The neurotransmitter dopamine contributes to the regulation of different brain functions, including sustained attention15 and EEG oscillations during wakefulness16,17. Wakefulness-promoting medications typically enhance dopaminergic neurotransmission and improve performance on the PVT during simulated night shifts and after sleep deprivation18,19,20. Nevertheless, a distinct role for dopamine in the proper regulation of sleep wake-states has long been controversial21. Intriguing recent data obtained with chemogenetic and optogenetic methods in flies and mice have started to change this view22,23,24. Moreover, genetically modified animals lacking a functional dopamine re-uptake transporter (DAT) exhibit prolonged wakefulness and shortened sleep25,26,27,28,29. In humans, a variable-number-tandem-repeat (VNTR) polymorphism (SNP-ID: rs28363170) of the gene encoding DAT (SLC6A3 or DAT1) modulates individual effects of sleep loss on sleep rebound and sensitivity to caffeine, as well as rewards and punishments30,31. Homozygous ten-repeat allele (10R/10R) carriers of this polymorphism presumably have 15–20% reduced DAT protein expression compared to nine-repeat (9R) allele carriers32,33. Compared to the 9R carriers, 10R/10R homozygotes exhibits a clearly and more pronounced increase in sleep intensity after extended waking (as measured by EEG slow-wave activity [SWA] in non-rapid-eye-movement [NREM] sleep)30. This measure is the best established biomarker of increased sleep need after sleep loss34.
Dopamine D2 receptors were recently proposed to be an important part of the network that regulates sleep and wakefulness35,36. Nevertheless, the exact role of D2 receptor agonist and antagonists in modulating behavioral states remain controversial37. Alike DAT, these receptors are primarily expressed in the striatum. Common variants of the gene (DRD2) encoding the D2 receptor in humans influence reported habitual sleep duration38. Molecular imaging studies revealed that sleep deprivation reduces D2 receptor availability in the striatum39,40,41, and suggest that these receptors are involved in modulating visual attention in rested wakefulness and after sleep deprivation42. The DRD2 gene includes a functional 957 C > T single nucleotide polymorphism (SNP-ID: rs6277) that affects levels and stability of mRNA and has been associated with 15–20% enhanced striatal D2 receptor availability in C-allele carriers compared to T-allele carriers43. Importantly, functional interactions between DAT1 and DRD2 genes44,45 appear to modulate dopamine-dependent neuronal activity according to an inverse U-shaped relationship46.
Based on the evidence presented above, we aimed at investigating the impacts of functional DAT1 and DRD2 polymorphisms on sleep deprivation-induced changes in subjective sleepiness, attentional lapses and the waking EEG in humans. We hypothesized that these genetic variants impact individual consequences of sleep loss and expected that they interact to modulate vigilant attention and EEG power during prolonged wakefulness according to U-shaped relationships.
Results
The impact of sleep deprivation on PVT performance and EEG oscillations are likely governed by separate mechanisms
To assess the consequences of sleep deprivation, performance on the PVT, subjective sleepiness and the waking EEG were quantified in 14 equally-spaced sessions across the 40 hours of prolonged wakefulness. From the baseline to the sleep deprivation day, PVT performance deteriorated (factor ‘day’: lapse frequency:  = 0.312; response variability:
= 0.312; response variability: 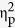 = 0.109), whereas the sleepiness score increased (
= 0.109), whereas the sleepiness score increased (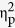 = 0.805; 2.43 ± 0.08 vs. 4.67 ± 0.09) (Fig. 1A–D; for detailed statistics, see Supplementary Table 1). The changes from sleep deprivation in lapse frequency and sleepiness revealed a tight association (rs2 = 0.110, p < 0.008, n = 64). Failed responses also increased with time on task within PVT sessions (
= 0.805; 2.43 ± 0.08 vs. 4.67 ± 0.09) (Fig. 1A–D; for detailed statistics, see Supplementary Table 1). The changes from sleep deprivation in lapse frequency and sleepiness revealed a tight association (rs2 = 0.110, p < 0.008, n = 64). Failed responses also increased with time on task within PVT sessions (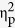 = 0.212), and this increase was amplified by sleep deprivation (‘day’ × ‘time on task’:
= 0.212), and this increase was amplified by sleep deprivation (‘day’ × ‘time on task’: 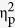 = 0.157). By contrast, response variability was virtually unaffected by time on task (
= 0.157). By contrast, response variability was virtually unaffected by time on task (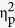 = 0.011). Significant interactions between ‘day’ and ‘clock time’ were observed for both neurobehavioral performance (lapse frequency:
= 0.011). Significant interactions between ‘day’ and ‘clock time’ were observed for both neurobehavioral performance (lapse frequency: 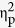 = 0.083; response variability:
= 0.083; response variability: 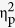 = 0.017) and sleepiness (
= 0.017) and sleepiness (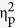 = 0.135), confirming that these state variables not only depend on sleep-wake history but also on diurnal fluctuations.
= 0.135), confirming that these state variables not only depend on sleep-wake history but also on diurnal fluctuations.
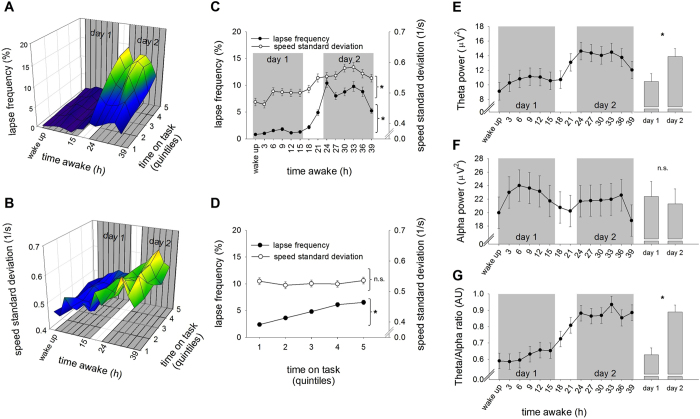
Figure 1. Psychomotor vigilance task (PVT) performance and waking EEG oscillations across prolonged wakefulness.
Evolution of psychomotor vigilance task (PVT) performance and waking EEG oscillations across prolonged wakefulness. (A,B) Vertical axes (y-axes) on the 3D plots represent attention lapses (lapse frequency) and response variability (standard deviation of response speed), as a function of time awake (x-axis) and time-on-task (z-axis). Warmer colors represent higher lapse frequency and increased response variability. (C,D) 2D plots illustrating increased lapse frequency and response variability by sleep deprivation and increased lapse frequency by time-on-task. (E–G) Effects of sleep deprivation on waking EEG theta activity, alpha activity, and theta/alpha ratio (TAR). The gray background shows the data included in the statistical analyses, categorized as day 1 (baseline) and day 2 (sleep deprivation). Stars indicate least significant difference (LSD) between day 1 and day 2 (p < 0.0001).
Because it was previously concluded that the behavioral consequences of sleep deprivation can dissociate from neurophysiological indices of elevated sleep pressure47, the evolution of electrical brain activity in five predefined frequency bands and the theta-to-alpha ratio (TAR) was quantified at 3-hour intervals across prolonged wakefulness. By determining the TAR, the signal to noise ratio (SNR) between-subjects (TAR: 1.76 ± 0.10) was enhanced compared to the theta (1.17 ± 0.20; t13 = −10.43, p < 0.0001) and alpha frequency bands (1.06 ± 0.08; t13 = −17.2, p < 0.0001), which improved the capability to detect genotype-dependent differences (see below). By contrast, within-subject SNR was only slightly reduced (TAR: 4.25 ± 2.13; theta: 4.55 ± 2.01; alpha: 4.83 ± 2.09; TAR vs. theta: t80 = 1.22, p > 0.22; TAR vs. alpha t80 = 2.11, p < 0.04).
Sleep deprivation increased theta activity (‘day’: 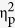 = 0.548) and TAR (
= 0.548) and TAR (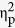 = 0.519), while alpha activity was unaffected (
= 0.519), while alpha activity was unaffected (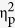 = 0.011, Fig. 1E–G, Supplementary Table 1). On the other hand, time of day affected theta (‘clock time’:
= 0.011, Fig. 1E–G, Supplementary Table 1). On the other hand, time of day affected theta (‘clock time’: 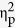 = 0.062; ‘clock time’ × ‘day’:
= 0.062; ‘clock time’ × ‘day’: 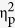 = 0.075) and alpha activity (‘clock time’:
= 0.075) and alpha activity (‘clock time’: 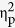 = 0.067; ‘clock time’ × ‘day’:
= 0.067; ‘clock time’ × ‘day’: 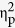 = 0.049), and also tended to modulate TAR (‘clock time’:
= 0.049), and also tended to modulate TAR (‘clock time’: 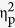 = 0.017; ‘clock time’ × ‘day’:
= 0.017; ‘clock time’ × ‘day’: 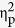 = 0.015). Consistent with the literature, the sleep deprivation induced changes in theta power and TAR were not associated with increased PVT lapse frequency (rall < 0.076, pall > 0.55), suggesting that these markers are governed by separate mechanisms.
= 0.015). Consistent with the literature, the sleep deprivation induced changes in theta power and TAR were not associated with increased PVT lapse frequency (rall < 0.076, pall > 0.55), suggesting that these markers are governed by separate mechanisms.
DAT1 and DRD2 genotypes affect PVT performance and subjective sleepiness
In the next step, the effects of the functional polymorphisms rs28363170 (‘DAT1’), rs6277 (‘DRD2’) and their combination (‘DAT1-DRD2 combined’) on the evolution of PVT performance during prolonged wakefulness were investigated. The p-values of all ANOVA’s investigating genetic effects were corrected for by multiple comparison using false discovery rate (FDR) correction (see Supplementary Table 2).
When PVT response speed was considered, complex interactions between the genotypes, sleep deprivation and diurnal rhythmicity were observed (‘DAT1’ × ‘clock time’ × ‘day’: 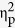 = 0.020; ‘DAT1-DRD2 combined’ × ‘clock time’ × ‘day’:
= 0.020; ‘DAT1-DRD2 combined’ × ‘clock time’ × ‘day’: 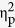 = 0.066). When investigating the sleep-deprivation induced change in PVT lapses, a significant modulation by the ‘DRD2’ genotype was observed, yet with a minuscule effect size (
= 0.066). When investigating the sleep-deprivation induced change in PVT lapses, a significant modulation by the ‘DRD2’ genotype was observed, yet with a minuscule effect size (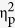 = 0.008). When ‘DAT1’ and ‘DRD2’ genotypes were combined, however, much larger effect sizes could be identified. The statistical analyses demonstrated a main effect of genotype (‘DAT1-DRD2 combined’:
= 0.008). When ‘DAT1’ and ‘DRD2’ genotypes were combined, however, much larger effect sizes could be identified. The statistical analyses demonstrated a main effect of genotype (‘DAT1-DRD2 combined’: 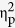 = 0.156), as well as interactions with time awake and clock time (‘DAT1-DRD2 combined’ × ‘clock time’:
= 0.156), as well as interactions with time awake and clock time (‘DAT1-DRD2 combined’ × ‘clock time’: 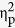 = 0.080; ‘DAT1-DRD2 combined’ × ‘day’:
= 0.080; ‘DAT1-DRD2 combined’ × ‘day’: 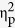 = 0.055; ‘DAT1-DRD2’ × ‘clock time’ × ‘day’:
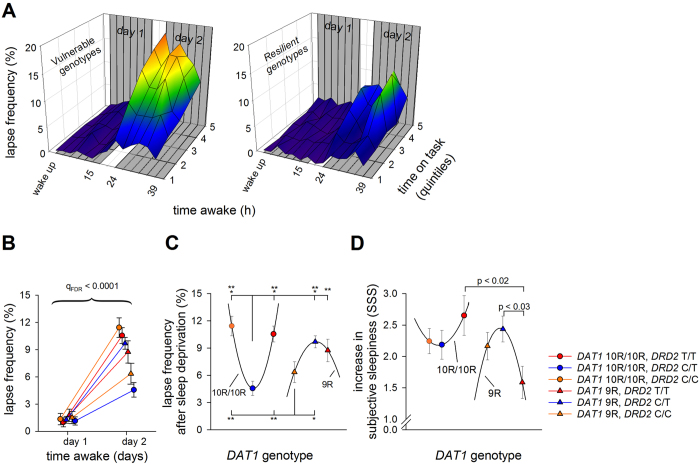
= 0.055; ‘DAT1-DRD2’ × ‘clock time’ × ‘day’: 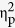 = 0.080) (Fig. 2 and Supplementary Figure 1). Post hoc testing revealed no significant differences at baseline (day 1), yet multiple significant effects on day 2 after sleep deprivation (Fig. 2B,C). These findings show that the 10R/10R-C/T and 9R-C/C genotypes had significantly fewer lapses after sleep deprivation than the remaining four DAT1-DRD2 genotypes. Taken together, these results indicate that the DAT1-DRD2 10R/10R-C/T and 9R-C/C genotypes maybe more resilient to deteriorating psychomotor vigilance by sleep deprivation (Fig. 2A and Supplementary Figure 1). It also should be noted, that the U-shaped relationship observed on day 2 for PVT lapse frequency, closely mirrored the relative increase in PVT lapses after sleep deprivation (Supplementary Figure 1B).
= 0.080) (Fig. 2 and Supplementary Figure 1). Post hoc testing revealed no significant differences at baseline (day 1), yet multiple significant effects on day 2 after sleep deprivation (Fig. 2B,C). These findings show that the 10R/10R-C/T and 9R-C/C genotypes had significantly fewer lapses after sleep deprivation than the remaining four DAT1-DRD2 genotypes. Taken together, these results indicate that the DAT1-DRD2 10R/10R-C/T and 9R-C/C genotypes maybe more resilient to deteriorating psychomotor vigilance by sleep deprivation (Fig. 2A and Supplementary Figure 1). It also should be noted, that the U-shaped relationship observed on day 2 for PVT lapse frequency, closely mirrored the relative increase in PVT lapses after sleep deprivation (Supplementary Figure 1B).
Figure 2. The neurobehavioral consequences of sleep deprivation split by the DAT1-DRD2 combined genotypes.
Effect of 40 hours sustained wakefulness on PVT lapses and subjective sleepiness (Stanford Sleepiness Scale [SSS]), split by the DAT1-DRD2 combined genotypes. (A) Evolution of PVT lapses (y-axis) across time awake (x-axis) and time-on-task (z-axis) split into the most vulnerable DAT1-DRD2 genotypes (10R/10R-C/C, 10R/10R-T/T, 9R-T/T and 9R-C/T; left) compared to the two more resilient genotypes (10R/10R-C/T and 9R-C/C; right). Warmer colors refers to higher lapse frequency. (B,C) Illustration of the highly significant interaction between the DAT1-DRD2 genotypes and sleep deprivation for lapses of attention. (B) The evolution of lapses from baseline (day 1) to sleep deprivation (day 2). Significant differences between genotypes were only observed on day 2. (C): PVT lapse frequency on day 2 can be described by a U-shaped curve with an arbitrary horizontal axis split by the DAT1-DRD2 combined genotypes. (D) Illustration of the interaction between the DAT1-DRD2 genotypes and sleep deprivation (qFDR < 0.02) for subjective sleepiness (SSS). Similar to (C), the genotypes are plotted on a U-shaped curve, split by DAT1 genotype. Colors represent the six genetic groups (orange: DRD2 C/C, blue: DRD2 C/T, red: DRD2 T/T, circles: DAT1 10R/10R, triangles: DAT1 9R) and are identical in panels B through D. Group sizes: DAT1 10R/10R, DRD2 C/C (lapses: n = 7, SSS: n = 9), DAT1 10R/10R, DRD2 C/T (lapses: n = 14, SSS: n = 18), DAT1 10R/10R, DRD2 T/T (lapses: n = 11, SSS: n = 13), DAT1 9R, DRD2 C/C (lapses: n = 6, SSS: n = 8), DAT1 9R, DRD2 C/T (lapses: n = 20, SSS: n = 24), DAT1 9R, DRD2 T/T (lapses: n = 6, SSS: n = 9). Significant post-hoc comparisons are illustrated by horizontal lines, whereas long vertical lines represent the genetic group investigated. Stars and p-values represent the least significant difference (LSD) following corresponding ANOVAs. Triple stars: p < 0.0001, double stars: p < 0.005, single stars: p < 0.02.
Similar to PVT performance, the increase in subjective sleepiness by sleep deprivation was not modulated by DAT1 and DRD2 genotypes alone, yet by the combined DAT1-DRD2 genotypes (‘DAT1-DRD2 combined’ × ‘day’: 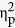 = 0.080). Intriguingly, the increase in subjective sleepiness resembled a similar U-shaped relationship, split by DAT1 genotype (Fig. 2D).
= 0.080). Intriguingly, the increase in subjective sleepiness resembled a similar U-shaped relationship, split by DAT1 genotype (Fig. 2D).
DAT1 and DRD2 genotypes affect the waking EEG during prolonged wakefulness
Next, it was investigated whether the dopaminergic genotypes also influence waking EEG oscillations during prolonged wakefulness. Three-way ANOVAs revealed no effects of ‘genotype’, or their interaction, on the evolution of delta, theta, low-beta and high-beta frequency activity during prolonged waking (see Supplementary Table 2 for statistics). By contrast, DAT1 (‘DAT1’ × ‘day’: 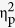 = 0.059) and DAT1-DRD2 combined genotypes (‘DAT1-DRD2 combined’ × ‘day’:
= 0.059) and DAT1-DRD2 combined genotypes (‘DAT1-DRD2 combined’ × ‘day’: 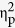 = 0.109) affected the change in alpha activity by sleep deprivation. Similarly, DAT1 (‘DAT1’ × ‘day’:
= 0.109) affected the change in alpha activity by sleep deprivation. Similarly, DAT1 (‘DAT1’ × ‘day’: 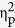 = 0.049), DRD2 (‘DRD2’ × ‘day’:
= 0.049), DRD2 (‘DRD2’ × ‘day’: 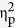 = 0.071) and DAT1-DRD2 combined genotypes (‘DAT1-DRD2 combined’ × ‘day’:
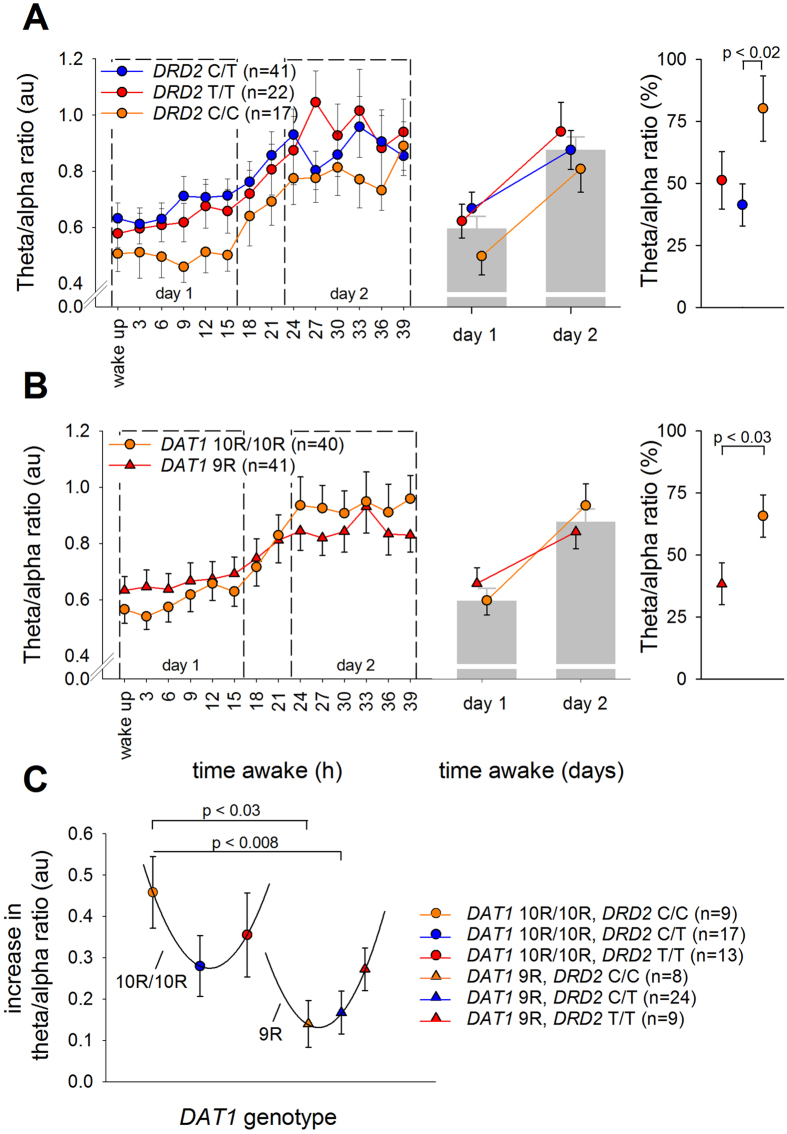
= 0.071) and DAT1-DRD2 combined genotypes (‘DAT1-DRD2 combined’ × ‘day’: 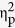 = 0.137) modulated TAR (Fig. 3). Specifically, the percentage increase in TAR after sleep deprivation was blunted in DAT1 9R-allele carriers when compared to DAT1 10R/10R homozygotes (Fig. 3A) and in DRD2 C/T heterozygotes compared to C/C homozygotes (Fig. 3B).
= 0.137) modulated TAR (Fig. 3). Specifically, the percentage increase in TAR after sleep deprivation was blunted in DAT1 9R-allele carriers when compared to DAT1 10R/10R homozygotes (Fig. 3A) and in DRD2 C/T heterozygotes compared to C/C homozygotes (Fig. 3B).
Figure 3. The neurophysiological consequences of sleep deprivation split by the DAT1-DRD2 combined genotypes.
Effect of 40 hours prolonged wakefulness on the increase in EEG theta-to-alpha ratio (TAR), split by DAT1, DRD2, and combined DAT1-DRD2 genotypes, all showing significant genotype x sleep deprivation interactions (qFDR < 0.02). The portion of the waking EEG recordings with eyes open was analyzed. (A,B) Left: TAR quantified at 3-hour intervals for DAT1 (panel A; 10R/10R: orange, 9R: red) and DRD2 (panel B; C/C: orange, C/T: blue, T/T: red) genotypes. Right: Change in TAR from day 1 to day 2 in DAT1 and DRD2 genotypes. Statistics revealed a significant effect of sleep deprivation on TAR, which was stronger in 10R/10R homozygotes than 9R-allele carriers of DAT1 and in DRD2 C/C homozygotes compared to C/T heterozygotes. (C) Increase in TAR from day 1 to day 2 in the combined DAT1-DRD2 genotype groups. The sleep deprivation-induced increase in TAR is described by a U-shaped curve with an arbitrary horizontal axis, split by DAT1 genotype. P-values represent the least significant difference (LSD) following the corresponding ANOVA.
Similar to PVT lapses, the increase in TAR after sleep deprivation may be described by a U-shaped relationship, split by DAT1 genotype (Fig. 3C). Intriguingly, however, whereas the curve associated with DAT1 10R/10R was reminiscent of the sleep deprivation-induced consequences on sleepiness and neurobehavioral performance, the curve associated with the DAT1 9R genotype appeared inverted.
Discussion
Modulation of dopaminergic neurotransmission in Drosophila and mouse mutants are associated with profound modulations of wakefulness and sleep26,27,28,29. Recent chemogenetic and optogenetic studies in flies and mice confirmed this crucial role of dopamine in regulating sleep-wake behaviors22,23,24. Nevertheless, the contribution of the dopaminergic system to sleep-wake regulation in humans is still only poorly described. The present study revealed that functional polymorphisms of DAT1 and DRD2 together modulate the neurobehavioral, subjective and neurophysiological consequences of sleep deprivation. Nevertheless, the distinct contributions of DAT1 and DRD2 genotypes to wakefulness-induced changes in physiological fatigue and sustained attention on one hand48,49, and EEG-derived markers of vigilance on the other hand6,7,50 appear to differ.
The relationships between dopaminergic neurotransmission and brain functions are complex. They are task specific and depend on the different dopamine receptor families and dopaminergic pathways involved51,52. Furthermore, epistasis between genetic variants of DAT1 and DRD2 genes has been described44. Functional magnetic resonance imaging data suggested that activation of the striatum in a monetary reward paradigm depends on genetic variants of both DAT1 and DRD2 genes. Indeed, Li et al. showed in more than 1200 healthy adults of European decent that individuals with elevated levels of dopaminergic transmission, associated with DAT1 9R and DRD2 C/C genotypes, showed better performance on a serial memory task than individuals with lower dopaminergic transmission45. Similarly, activation of the prefrontal cortex during a working memory and recognition task could be described by a U-shaped relationship depending on functional variants of DAT1 and DR2D genes modifying dopaminergic neurotransmission46. Consistent with these data, we found that increased lapse frequency on the PVT, subjective sleepiness, and EEG TAR after sleep deprivation can be described by U-shaped curves split by DAT1 genotype. The curves reflecting PVT performance and subjective sleepiness were similar, suggesting that the observed effects may reflect inherent consequences of sleep deprivation. This is intriguing, especially when considering that the sample sizes differed by 20% (between 64 and 80 individuals) for PVT recordings and subjective sleepiness ratings. Considering PVT lapse frequency, our data suggests that subjects carrying the DAT1-DRD2 10R/10R-C/T or the DAT1-DRD2 9R-C/T genotypes, are more resilient to sleep deprivation than the remaining four genotypes. When inspecting the increase in subjective sleepiness, however, the DAT1-DRD2 9R-C/C genotype is significantly less affected by prolonged wakefulness than the DAT1-DRD2 10R/10R-T/T and 9R-C/T genotypes. These effects could not be attributed to either the DAT1 or the DRD2 genotypes alone, and show that genotype dependent discrepancies between subjective and objective markers of sleep loss are present.
The statistical analyses revealed that the combined effects of DAT1 and DRD2 genotypes explained about 8% of the observed variability in subjective sleepiness, 15% in PVT lapse frequency, and 13% in EEG TAR. The magnitude of these effect sizes can be considered moderate to large53,54. They appear considerably larger than what was previously reported for other genetic variants11, which emphasizes the robustness of the findings. The direction by which the DAT1 and DRD2 genotypes modulated sustained attention and subjective sleepiness after sleep deprivation should also be noted (Fig. 2). Interestingly, the effect of the DRD2 genotypes was almost inverted in 10R/10R homozygotes and 9R allele carriers of DAT1. The different relationships highlight the importance of considering related signaling pathway components, rather than single genetic variants, when examining possible genetic influences on the vulnerability to sleep deprivation. Future studies may investigate the relative quantitative contributions of DAT and D2 protein expression on neurobehavioral and neurophysiological consequences of sleep deprivation.
Not only the neurobehavioral consequences of sleep deprivation, but also those of ‘time-on-task’ in rested and sleep deprived subjects were associated with common genetic variants55. Lim and colleagues investigated the impact of six polymorphisms related to dopaminergic neurotransmission on time-on-task-dependent decrements in PVT performance in 350 adults in the absence of sleep deprivation12. The authors concluded that the VNTR in DAT1, but not the ANKK1 polymorphism of DRD2 (SNP-id: rs1800497), modulated the decline in reaction speed associated with increasing task duration. Although both lapse frequency and response variability increased with time-on-task, our repeated measure analyses did not corroborate a DAT1 genotype-dependent modulation. Further studies are needed to explore whether the discrepancy reflects differences in study design or reduced power because of the smaller sample size in the present study.
Previous work stressed the lack of clear associations between PVT performance, sleepiness and neurophysiological markers of vigilance13,14,47,56. Our analyses corroborate these discrepancies, but only when the 9R-allele carriers of the DAT1 gene are considered. In 10R/10R homozygotes, the suggested U-shaped relationships between DRD2 genotypes and increased lapse frequency, subjective sleepiness, and EEG TAR after sleep deprivation were remarkably similar. Given the lack of association between increased lapse frequency and theta activity or TAR, however, the genotypes may independently affect behavioral and EEG measures. This implies that they are likely governed by separate mechanisms, which are influenced by dopaminergic neurotransmission.
The present study indicates that TAR may be a promising novel physiological marker of reduced vigilance associated with sleep deprivation. Indeed, work investigating the action of adrenergic and dopaminergic compounds on the multiple sleep latency test suggested that changes in TAR may predict vigilance6. In addition, based on comparisons of 85 electrophysiological EEG measures in 20 healthy adults it was concluded that the TAR is the best EEG marker to predict changes in vigilance from wakefulness to sleep, and from wakefulness to superficial stage 1 sleep50. As reported here, the TAR enhanced between-subjects signal-to-noise-ratio when compared to either alpha or theta activity alone. Furthermore, the TAR showed a robust increase with sleep deprivation. Especially the impact of DAT1 genotype on increased TAR after sleep deprivation should be highlighted. Previous findings of our group revealed that homozygous 10R/10R carriers exhibited a more pronounced increase than 9R-allele carriers in nocturnal slow wave sleep and EEG slow-wave activity after prolonged wakefulness, suggesting that 10R/10R homozygotes show a more pronounced homeostatic response to sleep loss30. The data presented here support this notion and show that 10R/10R homozygotes of DAT1 display a stronger increase in TAR by sleep deprivation. Moreover, the data also reveal that DRD2 C/C homozygotes show a stronger increase in TAR after sleep-loss than the C/T heterozygotes, which suggests that also the DRD2 polymorphism is involved in homeostatic sleep-wake regulation. Nocturnal sleep EEG recordings before and after sleep deprivation are needed to further corroborate these findings.
The DAT and dopamine D2 receptors are primarily expressed in the striatum. This brain region may be an important part of the intrinsic system that controls sleep and wakefulness35,36. The present genetic findings support data collected in animals24,25,26,27,28,57 and observations from brain imaging30,39,40,42 and epidemiological38 studies in humans, suggesting prominent roles for DAT and D2 receptors in sleep-wake regulation. They highlight the importance of the striatum in regulating changes in vigilant attention, subjective sleepiness, and neurophysiological EEG oscillations during prolonged wakefulness. It is likely that striatal dopaminergic signaling, together with the prefrontal cortex and related brain neurotransmitter- and neuromodulator- systems, contributes to the control of the sleep-wake continuum58. The data presented here suggests that common functional genetic variants linked to altered striatal dopaminergic neurotransmission contribute to individual neurobehavioral consequences of sleep loss. These genetic variants could help define trait-like risks for sleep deprivation-related motor vehicle- and work-related accidents. Moreover, the genetic variants may help explain individual differences in the efficacy of dopaminergic psychostimulants, used to mitigate the detrimental effects of sleep-wake disorders and sleep deprivation.
Methods
Study participants and sleep deprivation protocol
Eighty-two healthy right-handed volunteers (12 females) between 19 and 35 years completed the sleep deprivation experiment (see refs 30,59, for further details on the study protocols). All volunteers reported to be good sleepers, adhere to regular bedtimes, be in good physical health, and have no history of neurological or psychiatric disorders. Two months prior to enrollment, subjects did not consume any medication or illicit drugs, did not pass through time zones and consumed only moderate amounts of caffeine and alcohol. Three participants were moderate smokers (<10 cigarettes per day). Good sleep quality (no undiagnosed sleep disorders) and efficiency (>85%) were confirmed in a screening night in the sleep laboratory prior to study inclusion. At least three days before study initiation, participants were instructed to wear a wrist activity monitor on their non-dominant arm, fill in a sleep-wake diary, refrain from caffeine and alcohol, and strictly maintain 8 hours’ time in bed, corresponding to the scheduled bedtimes of the study.
All participants completed 40 hours of constantly supervised wakefulness, preceded by two consecutive 8-hour sleep episodes in the sleep laboratory (adaptation and baseline nights). During prolonged waking, the consequences of sleep loss were monitored at 3 hour intervals with standardized test sessions, including subjective sleepiness ratings, PVT and waking EEG recordings.
The study protocols were approved by the ethics committee of the Canton of Zürich for research on human subjects. Written informed consent was obtained from all participants prior to the experiments, as required according to the principles in the Declaration of Helsinki. All subjects received financial compensation for their participation.
Genotyping
Genomic DNA was extracted from 3 ml fresh EDTA-blood (Wizard® Genomic DNA Purification Kit, Promega, Madison, WI). The rs28363170 polymorphism of DAT1 was determined by allele-specific PCR on an MJ Research PTC-225 thermal cycler (MJ Research/Bio-Rad, Reno, NV) at an annealing temperature of 67 °C. Forward primer, 5′-tgtggtgtagggaacggcctga-3′, and reverse primer, 5′-cttcctggaggtcacggctcaa-3′, with HOT FIREPol® DNA Polymerase was used. The 430–480 bp PCR products were analyzed by agarose gel electrophoresis. The functional polymorphisms rs6277 of DRD2 was determined using a Taqman® SNP Genotyping Assay (Life Technologies Europe B.V.; probe number: C__11339240_10). Allelic discrimination analysis was performed with SDS v2.2.2 software (Applied Biosystems, Foster City, CA, USA). All genotypes were replicated at least once for independent confirmation of results. The DRD2 genotype of one individual is lacking due to missing genetic material. Comparisons of demographic variables between DAT1 and DRD2 genotypes included weight, height, body mass index (BMI), age, gender, habitual alcohol and caffeine consumption, self-reported daily sleep duration, daytime sleepiness and trait anxiety (Table 1). No variable revealed a difference between the genotypes. Fishers exact test revealed that the DRD2 and DAT1 genetic groups were similarly distributed, both in the full (n = 81, p > 0.39) and in the subsample (n = 64; p > 0.30).
Table 1. Demographic characteristics of study participants.
| DRD2 genotypes | F2,77 (p) | DAT1 genotypes | F2,79 (p) | |||
|---|---|---|---|---|---|---|
| Weight (kg) | T/T | 70.4 ± 1.81 | 0.01 (0.99) | 9R | 69.4 ± 1.48 | 0.63 (0.42) |
| C/T | 70.6 ± 1.32 | 10R/10R | 73.7 ± 1.74 | |||
| C/C | 70.4 ± 2.05 | |||||
| Height (cm) | T/T | 178.6 ± 1.54 | 0.42 (0.66) | 9R | 177.9 ± 1.12 | 0.08 (0.78) |
| C/T | 177.0 ± 1.13 | 10R/10R | 177.4 ± 1.14 | |||
| C/C | 178.2 ± 1.75 | |||||
| BMI (kg/m2) | T/T | 22.0 ± 0.39 | 0.50 (0.69) | 9R | 22.1 ± 0.29 | 1.98 (0.16) |
| C/T | 22.5 ± 0.27 | 10R/10R | 22.6 ± 0.29 | |||
| C/C | 22.1 ± 0.42 | |||||
| Age (years) | T/T | 23.5 ± 0.65 | 0.97 (0.39) | 9R | 24.3 ± 0.48 | 0.07 (0.79) |
| C/T | 24.3 ± 0.48 | 10R/10R | 24.1 ± 0.48 | |||
| C/C | 24.8 ± 0.74 | |||||
| Alcohol consumption (drinks/week) | T/T | 3.39 ± 0.61 | 0.48 (0.63) | 9R | 3.00 ± 0.44 | 0.23 (0.63) |
| C/T | 3.27 ± 0.45 | 10R/10R | 3.30 ± 0.45 | |||
| C/C | 2.56 ± 0.69 | |||||
| (mg/day) | T/T | 119.1 ± 28.07 | 0.45 (0.65) | 9R | 114.7 ± 20.46 | 0.81 (0.37) |
| C/T | 138.5 ± 20.56 | 10R/10R | 140.9 ± 20.72 | |||
| C/C | 104.2 ± 31.93 | |||||
| Habitual sleep duration (hours) | T/T | 7.15 ± 0.15 | 1.49 (0.24) | 9R | 7.31 ± 0.11 | 1.11 (0.29) |
| C/T | 7.36 ± 0.11 | 10R/10R | 7.14 ± 0.11 | |||
| C/C | 7.03 ± 0.17 | |||||
| Daytime Sleepiness (ESS) | T/T | 7.32 ± 0.59 | 1.91 (0.16) | 9R | 7.31 ± 0.50 | 0.70 (0.40) |
| C/T | 7.46 ± 0.43 | 10R/10R | 7.04 ± 0.59 | |||
| C/C | 5.94 ± 0.67 | |||||
| Trait Anxiety (TAI) | T/T | 34.0 ± 1.65 | 0.58 (0.57) | 9R | 35.5 ± 1.39 | 0.30 (0.58) |
| C/T | 35.9 ± 1.21 | 10R/10R | 35.0 ± 1.64 | |||
| C/C | 36.48 ± 1.88 | |||||
| DRD2 genotypes | (p) | DAT1 genotypes | (p) | |||
| Gender ratio (% females) | T/T | 13.6 | (1.00) | 9R | 12.2 | (0.75) |
| C/T | 14.6 | 10R/10R | 15.4 | |||
| C/C | 11.8 | |||||
Demographic variables compared between DRD2 and DAT1 genotypes based on validated self-report questionnaires. Data represent means ± SEM; Statistics originate from one-way ANOVAs and Fisher’s exact test (gender ratio only). Caffeine consumption per serving was calculated based on self-reported values64,66. Similarly, as reported in the table, no significant difference between genotypes were observed in the subsample of 64 individuals (data not shown). TAI: Trait Anxiety Inventory67; ESS: Epworth Sleepiness Scale68.
Psychomotor vigilance task (PVT) and subjective sleepiness
Vigilant attention during sleep deprivation was assessed by a PC-implemented e-Prime version (Psychology Software Tools Inc., Pittsburgh, PA) of the psychomotor vigilance task60. Eighteen individuals were excluded from the analyses because they performed a different, non-computerized version of the task, resulting in a sample size of 64 subjects for the PVT task analyses.
The PVT is a simple reaction time task. When a digital millisecond counter starts to scroll in the center of the computer screen, subjects have to press a button with their right index finger on a response box (standardized keyboard) connected to the PC. Subjects received oral instructions and performed one training session on the evening prior to the baseline night. For each PVT trial, 100 stimuli were presented (random inter-stimulus intervals: 2–10 s). To assess performance as a function of ‘time on task’ (TOT), the task was divided into five quintiles of 20 trials. Two extensively validated PVT variables were quantified48,56,61,62: ‘lapses of attention’ (defined as the number of trials with reaction times longer than 500 ms) and ‘standard deviation of response speed’ (calculated based on inverse reaction times and referred to as response variability). Both these variables are prominent markers of sleep loss and sensitive to TOT.
Immediately prior to all PVT assessments, a validated German version of the Stanford Sleepiness Scale was administered63. The sleepiness ratings of all 82 subjects were included in the analyses.
Waking EEG recordings
At 3-hour intervals throughout prolonged wakefulness, standardized waking EEG recordings with polygraphic PSA24 (Braintronics Inc., Almere, The Netherlands) (n = 18; see: ref. 2) or Artisan (Micromed, Mogliano Veneto, Italy) (n = 64; see e.g.: ref. 64) amplifiers were performed. The EEG data recorded with PSA24 amplifiers, were analogue band-pass filtered (−3 dB at 0.16 Hz; −3 dB at 102 Hz) and sampled at 512 Hz, then digitally low-pass filtered (−3 dB at 49 Hz) and stored with a resolution of 128 Hz. The EEG data recorded with Artisan amplifiers, were analogue band-pass filtered (−3 dB at 0.15 Hz; −3 dB at 67.2 Hz), sampled, and stored with a resolution of 256 Hz. Note that DAT1 and DRD2 genotypes were similarly distributed between PSA24 and Artisan amplifiers (pall > 0.87, Fishers exact test). The standard EEG derivation for sleep state scoring, C3M2, was analyzed.
At least 1 hour before each recording, subjects were confined to the sleep-laboratory (constant temperature: 19–21 °C; light intensity: <150 lux). Fifteen minutes before each recording, subjects stayed in their own private bedroom. The recordings consisted of an initial 3-min recording period with eyes closed, followed by a 5-min period with eyes open, while fixating a black dot on the wall. During all recordings, study participants comfortably relaxed in a chair, and placed their chin on an individually adjusted chinrest. Artifacts were visually identified and excluded. Due to a technical problem, the data of one subject needed to be excluded. The power spectra of artifact-free, 2-s EEG epochs (50% overlap, Hanning window, frequency resolution 0.5 Hz) recorded with eyes open were computed and analyzed between 0–30 Hz.
To investigate changes in the waking EEG across prolonged time awake, spectral power in the standard EEG bands, delta (1–4.5 Hz), theta (5–7.5 Hz), alpha (8–11.5 Hz), low-beta (12–19.5 Hz), and high-beta (20–30 Hz) was computed. Previous research has shown that the wakefulness-induced changes are particularly pronounced in theta (~4–8 Hz) and alpha (~8–12 Hz) ranges, as well as in the theta-to-alpha ratio (TAR)6,7,50. The EEG power and ratio values were transformed by a base 10 logarithm, to approximate a Gaussian distribution.
Statistical analyses
Spearman’s rank-correlation coefficients (rs), as well as two-, three- and four-way mixed-model ANOVAs (analyses of variance) for repeated measures were computed. The covariance matrix applied was first order autoregressive and subjects were considered as random effect. To estimate the influences of circadian and homeostatic sleep regulatory processes34, the effect of time awake was analyzed by considering the within-subjects’ factors ‘clock time’ (8:00, 11:00, 14:00, 17:00, 20:00, 23:00) and ‘day’ (baseline vs. sleep deprivation day). In 22 participants, the assessments at 17:00 and 20:00 were missing due to neuroimaging59. For the analyses of PVT performance, an additional factor ‘time on task (TOT)’ (quintiles 1–5) was added. Within each quintile, lapse frequency was defined as the percentage of lapse trials and response variability as the standard deviation of the mean response time. The ANOVAs testing overall effects of sleep loss independently of genotype are shown in Supplementary Table 1. To assess genetic effects, separate ANOVAs with the following genotypes were performed: ‘DAT1’ (9R; 10R/10R); ‘DRD2’ (C/C; C/T; T/T); ‘DAT1-DRD2 combined’ (9R-C/C; 9R-C/T; 9R-T/T; 10R/10R-C/C; 10R/10R-C/T; 10R/10R-T/T). Effect sizes (partial eta squared: 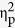 ) were calculated from corresponding ANOVA F-values and degrees of freedom. Effect sizes of 0.0099, 0.0588 and 0.1379 are considered small, moderate and large, respectively53,54. To correct for multiple comparisons, p-values originating from ANOVAs investigating genetic effects were corrected by false discovery rate (FDR) correction65. Corrected p-values (qFDR-values) below 0.05 were considered significant; post-hoc analyses were only performed when the respective main effect and/or interactions of the ANOVA withstood FDR correction. Results of all FDR corrected ANOVAs containing the factor ‘genotype’ are listed in Supplementary Table 2. Unless otherwise specified, only significant effects and interactions are mentioned. Following FDR corrected significant ANOVAs, least significant difference (LSD) post-hoc comparisons were performed.
) were calculated from corresponding ANOVA F-values and degrees of freedom. Effect sizes of 0.0099, 0.0588 and 0.1379 are considered small, moderate and large, respectively53,54. To correct for multiple comparisons, p-values originating from ANOVAs investigating genetic effects were corrected by false discovery rate (FDR) correction65. Corrected p-values (qFDR-values) below 0.05 were considered significant; post-hoc analyses were only performed when the respective main effect and/or interactions of the ANOVA withstood FDR correction. Results of all FDR corrected ANOVAs containing the factor ‘genotype’ are listed in Supplementary Table 2. Unless otherwise specified, only significant effects and interactions are mentioned. Following FDR corrected significant ANOVAs, least significant difference (LSD) post-hoc comparisons were performed.
Signal-to-noise ratios (SNR) were defined as group means (μ) divided by the standard deviation (σ) of EEG power in a given frequency band:
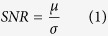 |
Following this classification, between-subject SNR was defined as the EEG band-power mean and standard deviation per session across the 81 subjects:
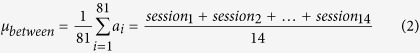 |
 |
On the other hand, within-subject SNR was defined as the EEG band-power mean and standard deviation per subject across the 14 sessions:
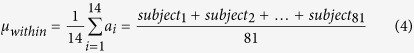 |
 |
Signal to noise ratios across EEG bands, were compared using paired student t-tests.
Statistical analyses were performed with SAS 9.4 software (SAS Institute, Cary, NC), whereas PVT performance was assessed using IBM SPSS Statistics 22 (IBM Corp., Armonk, USA). Throughout, estimated means and standard errors of the respective ANOVAs are presented. For illustrative purposes, genetically-determined higher dopaminergic signaling (DAT1 10R/10R and DRD2 C/C genotypes) was illustrated in orange color, intermediate dopaminergic signaling (DRD2 C/T genotype) in blue color, and lower dopaminergic signaling (DAT1 9R and DRD2 T/T genotypes) in red color when appropriate.
Additional Information
How to cite this article: Holst, S. C. et al. Functional Polymorphisms in Dopaminergic Genes Modulate Neurobehavioral and Neurophysiological Consequences of Sleep Deprivation. Sci. Rep. 7, 45982; doi: 10.1038/srep45982 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (grant # 320030-135414), the University Research Priority Program “Integrative Human Physiology” (University of Zürich), the Clinical Research Priority Program “Sleep & Health” (University of Zürich), and the Forschungskredit of the University of Zurich (grant # FK-15-039). We thank Dr. S. Bodenmann, Dr. V. Bachmann, Dr. J. Rétey, Dr. K. Stingelin, Dr. C. Klein, Mrs. S. Roethlisberger, Mrs. M. Rötlisberger and Mr. W. Schakel for their help with data collection and Mr. S. Vogt, and Dr. R. Dürr for their help with data analyses.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.C.H. and H.-P.L. planed the study. S.C.H., T.M., A.V. and H.-P.L. conducted the study. S.C.H., T.M., A.V., and B.S. conducted the analyses. S.C.H. drafted the main manuscript text. H.-P.L. provided extensive feedback. All authors reviewed and approved the final version of the manuscript.
References
- Pilcher J. J. & Huffcutt A. I. Effects of sleep deprivation on performance: a meta-analysis. Sleep 19, 318–26 (1996). [DOI] [PubMed] [Google Scholar]
- Retey J. V. et al. Adenosinergic Mechanisms Contribute to Individual Differences in Sleep Deprivation-Induced Changes in Neurobehavioral Function and Brain Rhythmic Activity. J. Neurosci. 26, 10472–10479 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. & Dinges D. F. A meta-analysis of the impact of short-term sleep deprivation on cognitive variables. Psychol. Bull. 136, 375–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt H.-P., Sousek A & Holst S. C. In ESR. S. Sleep Medicine Textbook (eds Bassetti C. L., Dogas Z. & Peigneux P.) 49–52 (2014). [Google Scholar]
- Corsi-Cabrera M., Arce C., Ramos J., Lorenzo I. & Guevara M. a. Time course of reaction time and EEG while performing a vigilance task during total sleep deprivation. Sleep 19, 563–569 (1996). [DOI] [PubMed] [Google Scholar]
- Goldenberg F., Weil J. S. & Von Frenckell R. Utilization of theta/alpha spectral range as an indicator of vigilance: pharmacological applications during repeated sleep latency tests. Neurophysiol. Clin. Clinical Neurophysiol. 18, 433–445 (1988). [DOI] [PubMed] [Google Scholar]
- Cajochen C., Wyatt J. K., Czeisler C. A. & Dijk D. J. Separation of circadian and wake duration-dependent modulation of EEG activation during wakefulness. Neuroscience 114, 1047–1060 (2002). [DOI] [PubMed] [Google Scholar]
- Van Dongen H. P. a, Baynard M. D., Maislin G. & Dinges D. F. Systematic interindividual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep 27, 423–433 (2004). [PubMed] [Google Scholar]
- Bodenmann S. et al. Polymorphisms of ADORA2A modulate psychomotor vigilance and the effects of caffeine on neurobehavioural performance and sleep EEG after sleep deprivation. Br. J. Pharmacol. 165, 1904–1913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo J. C. et al. Effects of Partial and Acute Total Sleep Deprivation on Performance across Cognitive Domains, Individuals and Circadian Phase. PLoS One 7, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield B. C., Wisor J. P., Field S. A., Schmidt M. A. & Van Dongen H. P. A. TNFα G308A polymorphism is associated with resilience to sleep deprivation-induced psychomotor vigilance performance impairment in healthy young adults. Brain, Behav. Immun. 47, 66–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. et al. Dopaminergic polymorphisms associated with time-on-task declines and fatigue in the psychomotor vigilance test. PLoS One 7, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen H. P. A., Maislin G., Mullington J. M. & Dinges D. F. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep 26, 117–126 (2003). [DOI] [PubMed] [Google Scholar]
- Leproult R. et al. Individual differences in subjective and objective alertness during sleep deprivation are stable and unrelated. Am. J. Physiol. Regul. Integr. Comp. Physiol. 284, R280–R290 (2003). [DOI] [PubMed] [Google Scholar]
- Nieoullon A. Dopamine and the regulation of cognition and attention. Progress in Neurobiology 67, 53–83 (2002). [DOI] [PubMed] [Google Scholar]
- Sebban C., Zhang X. Q., Tesolin-Decros B., Millan M. J. & Spedding M. Changes in EEG spectral power in the prefrontal cortex of conscious rats elicited by drugs interacting with dopaminergic and noradrenergic transmission. Br. J. Pharmacol. 128, 1045–54 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwan S. J., Ji Y. K., Sang H. K. & Lee M. G. Role of dopamine receptors on electroencephalographic changes produced by repetitive apomorphine treatments in rats. Korean J. Physiol. Pharmacol. 13, 147–151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J. K., Randazzo A. C., Stone K. L. & Schweitzer P. K. Modafinil improves alertness, vigilance, and executive function during simulated night shifts. Sleep 27, 434–9 (2004). [DOI] [PubMed] [Google Scholar]
- Bodenmann S. et al. The Functional Val158Met Polymorphism of COMT Predicts Interindividual Differences in Brain Oscillations in Young Men. J. Neurosci. 29, 10855–10862 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M. et al. Effects of noradrenaline and dopamine on supraspinal fatigue in well-trained men. Med. Sci. Sports Exerc. 44, 2299–2308 (2012). [DOI] [PubMed] [Google Scholar]
- Saper C. B., Fuller P. M., Pedersen N. P., Lu J. & Scammell T. E. Sleep state switching. Neuron 68, 1023–1042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eban-Rothschild A., Rothschild G., Giardino W. J., Jones J. R. & de Lecea L. VTA dopaminergic neurons regulate ethologically relevant sleep-wake behaviors. Nat. Neurosci. 1–14, doi: 10.1038/nn.4377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D. et al. Operation of a homeostatic sleep switch. Nature 536, 333–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y. et al. Activation of ventral tegmental area dopamine neurons produces wakefulness through dopamine D2-like receptors in mice. Brain Struct. Funct. 1–9, doi: 10.1007/s00429-017-1365-7 (2017). [DOI] [PubMed] [Google Scholar]
- Wu M. N., Koh K., Yue Z., Joiner W. J. & Sehgal A. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep 31, 465–472 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisor J. P. et al. Dopaminergic role in stimulant-induced wakefulness. J. Neurosci. 21, 1787–1794 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giros B., Jaber M., Jones S. R., Wightman R. M. & Caron M. G. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature 379, 606–612 (1996). [DOI] [PubMed] [Google Scholar]
- Kume K., Kume S., Park S. K., Hirsh J. & Jackson F. R. Cellular/Molecular Dopamine Is a Regulator of Arousal in the Fruit Fly. J. Neurosci. 25, 7377–7384 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R., Van Swinderen B. & Greenspan R. J. Dopaminergic modulation of arousal in Drosophila. Curr. Biol. 15, 1165–1175 (2005). [DOI] [PubMed] [Google Scholar]
- Holst S. C. et al. Dopaminergic Role in Regulating Neurophysiological Markers of Sleep Homeostasis in Humans. J. Neurosci. 34, 566–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S. M., Goldstein A. N., Knutson B. & Walker M. P. A Genetic Polymorphism of the Human Dopamine Transporter Determines the Impact of Sleep Deprivation on Brain Responses to Rewards and Punishments. J. Cogn. Neurosci. 28, 803–810 (2016). [DOI] [PubMed] [Google Scholar]
- Costa A., Riedel M., Müller U., Möller H. J. & Ettinger U. Relationship between SLC6A3 genotype and striatal dopamine transporter availability: A meta-analysis of human single photon emission computed tomography studies. Synapse 65, 998–1005 (2011). [DOI] [PubMed] [Google Scholar]
- Spencer T. J. et al. Functional genomics of attention-deficit/hyperactivity disorder (adhd) risk alleles on dopamine transporter binding in ADHD and healthy control subjects. Biol. Psychiatry 74, 84–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achermann P. & Borbély A. A. In Principles and Practice of Sleep Medicine (eds Kryger M. H., Roth T. & Dement W. C.) 431–444 (2011). [Google Scholar]
- Lazarus M., Huang Z.-L., Lu J., Urade Y. & Chen J.-F. How do the basal ganglia regulate sleep wake behavior? Trends Neurosci. 35, 723–732 (2012). [DOI] [PubMed] [Google Scholar]
- Holst S. C. & Landolt H. P. Sleep Homeostasis, Metabolism, and Adenosine. Curr. Sleep Med. Reports 1, 27–37 (2015). [Google Scholar]
- Herrera-Solís A., Arias-Carrión O., Sarro-Ramírez A., Salas-Crisóstomo M. & Murillo-Rodríguez E. In Dopamine and Sleep (eds Monti J. M., Pandi-Perumal S. R. & Chokroverty S.) 39–46 (Springer International Publishing, 2016). [Google Scholar]
- Cade B. E. et al. Common variants in DRD2 are associated with sleep duration: The CARe consortium. Hum. Mol. Genet. 25, 167–179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N. D. et al. Sleep Deprivation Decreases [11 C]Raclopride’s Binding to Dopamine D2/D3 Receptors in the Human Brain. J. Neurosci. 28, 8454–8461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N. D. et al. Hyperstimulation of striatal D2 receptors with sleep deprivation: Implications for cognitive impairment. Neuroimage 45, 1232–1240 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N. D. et al. Evidence that sleep deprivation downregulates dopamine D2R in ventral striatum in the human brain. J. Neurosci. 32, 6711–6717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D., Wang G.-J. & Volkow N. D. Association between striatal dopamine D2/D3 receptors and brain activation during visual attention: effects of sleep deprivation. Transl. Psychiatry 6, e828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen M. M. et al. C957T polymorphism of dopamine D2 receptor gene affects striatal DRD2 in vivo availability by changing the receptor affinity. Synapse 63, 907–912 (2009). [DOI] [PubMed] [Google Scholar]
- Forbes E. E. et al. Genetic variation in components of dopamine neurotransmission impacts ventral striatal reactivity associated with impulsivity. Mol. Psychiatry 14, 60–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C. et al. Aging magnifies the effects of dopamine transporter and D2 receptor genes on backward serial memory. Neurobiol. Aging 34, (2013). [DOI] [PubMed] [Google Scholar]
- Bertolino A. et al. Genetically determined interaction between the dopamine transporter and the D2 receptor on prefronto-striatal activity and volume in humans. J. Neurosci. 29, 1224–1234 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliaud E. et al. Sharp and sleepy: Evidence for dissociation between sleep pressure and nocturnal performance. J. Sleep Res. 17, 11–15 (2008). [DOI] [PubMed] [Google Scholar]
- Doran S. M., Van Dongen H. P. A. & Dinges D. F. Sustained attention performance during sleep deprivation: Evidence of state instability. Arch. Ital. Biol. 139, 253–267 (2001). [PubMed] [Google Scholar]
- Lee I. S., Bardwell W. A., Ancoli-Israel S. & Dimsdale J. E. Number of lapses during the psychomotor vigilance task as an objective measure of fatigue. J. Clin. Sleep Med. 6, 163–168 (2010). [PMC free article] [PubMed] [Google Scholar]
- Šušmáková K. & Krakovská a. Classification of waking, sleep onset and deep sleep by single measures. Meas. Sci. Rev. 7, 34–38 (2007). [Google Scholar]
- Cools R. & D’Esposito M. Inverted-U-shaped dopamine actions on human working memory and cognitive control. Biological Psychiatry 69, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floresco S. B. Prefrontal dopamine and behavioral flexibility: Shifting from an ‘inverted-U’ toward a family of functions. Frontiers in Neuroscience, doi: 10.3389/fnins.2013.00062 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences Acad. Press (2013). [Google Scholar]
- Richardson J. T. E. Eta squared and partial eta squared as measures of effect size in educational research. Educational Research Review 6, 135–147 (2011). [Google Scholar]
- Maire M. et al. Time-on-task decrement in vigilance is modulated by inter-individual vulnerability to homeostatic sleep pressure manipulation. Front. Behav. Neurosci. 8, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. & Dinges D. F. Sleep deprivation and vigilant attention. in Annals of the New York Academy of Sciences 1129, 305–322 (2008). [DOI] [PubMed] [Google Scholar]
- Qiu M. H., Yao Q. L., Vetrivelan R., Chen M. C. & Lu J. Nigrostriatal Dopamine Acting on Globus Pallidus Regulates Sleep. Cereb. Cortex 26, 1430–1439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst S. C., Valomon A. & Landolt H.-P. H.-P. Sleep Pharmacogenetics: Personalized Sleep-Wake Therapy. Annu. Rev. Pharmacol. Toxicol. 56, 577–603 (2016). [DOI] [PubMed] [Google Scholar]
- Hefti K. et al. Increased metabotropic glutamate receptor subtype 5 availability in human brain after one night without sleep. Biol. Psychiatry 73, 161–168 (2013). [DOI] [PubMed] [Google Scholar]
- Durmer J. S. & Dinges D. F. Neurocognitive consequences of sleep deprivation. Semin. Neurol. 25, 117–129 (2005). [DOI] [PubMed] [Google Scholar]
- Dinges D. F. & Powell J. W. Microcomputer analyses of performance on a portable, simple visual RT task during sustained operations. Behav. Res. Methods, Instruments, Comput. 17, 652–655 (1985). [Google Scholar]
- Graw P., Kräuchi K., Knoblauch V., Wirz-Justice A. & Cajochen C. Circadian and wake-dependent modulation of fastest and slowest reaction times during the psychomotor vigilance task. Physiol. Behav. 80, 695–701 (2004). [DOI] [PubMed] [Google Scholar]
- Sturm A., Clarenbach P., Largiadèr F. & Wicki O. Checklisten der aktuellen Medizin, Checkliste Schlafstörungen (Thieme, 1997). [Google Scholar]
- Bachmann V. et al. Functional ADA polymorphism increases sleep depth and reduces vigilant attention in humans. Cereb. Cortex 22, 962–970 (2012). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- Valomon A. et al. Genetic polymorphisms of DAT1 and COMT differentially associate with actigraphy-derived sleep–wake cycles in young adults. Chronobiol. Int. 31, 705–714 (2014). [DOI] [PubMed] [Google Scholar]
- Spielberger C. D., Gorsuch R. L. & Lushene R. E. Manual for the State-Trait Anxiety Inventory. Palo Alto, CA Consult. Psychol. Press 1–23 (1970). [Google Scholar]
- Johns M. W. A new method for measuring daytime sleepiness: the Epworth sleepiness scale. Sleep 14, 540–545 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.