Abstract
The p6 domain of the human immunodeficiency virus type 1 (HIV-1) Gag polyprotein mediates virion budding from infected cells via protein-protein contacts with the class E vacuolar protein sorting factors, Tsg101 and AIP1/ALIX. Interaction with Tsg101 is strengthened by covalent attachment of monovalent ubiquitin to HIV-1 p6. To identify additional host factors that bind to HIV-1 p6, a human cDNA library was screened in the yeast two-hybrid system. HIV-1 p6 was found to interact with small ubiquitin-like modifier 1 (SUMO-1) as well as the E2 SUMO-1 transfer enzyme, Ubc9. Interaction with p6 was also detected with Daxx, a cellular protein to which SUMO-1 is sometimes covalently attached. SUMO-1 was incorporated into HIV-1 virions where it was protected within the virion membrane from digestion by exogenous protease. Of the two lysine residues in p6, lysine 27 uniquely served as a site of covalent SUMO-1 attachment. As previously reported, though, HIV-1 bearing the p6-K27R mutation replicated just like the wild type. Overproduction of SUMO-1 in HIV-1 producer cells had no apparent effect on virion release or on virion protein or RNA content. Infectivity of the resulting virions, though, was decreased, with the defect occurring after membrane fusion, at the time of viral cDNA synthesis. HIV-1 bearing the p6-K27R mutation was insensitive to SUMO-1 overexpression, suggesting that covalent attachment of SUMO-1 to p6 is detrimental to HIV-1 replication.
Human immunodeficiency virus type 1 (HIV-1) assembly is driven by the Gag polyprotein, which, concurrent with the release of particles from infected cells, is processed by the viral protease into structural proteins that perform essential functions in the mature virion: matrix (MA) forms a protein layer beneath the virion envelope, nucleocapsid (NC) coats the viral RNA, and capsid (CA) forms the shell that encases the viral ribonucleoprotein complex (14).
In addition to the MA, CA, and NC domains that are found in all retroviruses, the carboxyl terminus of the HIV-1 Gag polyprotein contains a small proline-rich domain that matures with cleavage by the viral protease into free p6. The carboxyl-terminal p6 domain is required for late steps in virion budding, in which the membrane of the nascent virion pinches off from the host plasma membrane (18), and for incorporation into virions of the viral accessory protein Vpr (24, 40). Of the HIV-1-encoded proteins in the virion, p6 is the major phosphoprotein (33), though the significance of p6 phosphorylation has not been determined, and it is not known what role p6 plays during the early steps of HIV-1 infection. The exact location of p6 within intact virions is unknown, though p6 readily dissociates from other virion components when HIV-1 cores are purified (1, 51).
A highly conserved PT/SAP amino acid sequence located near the N terminus of p6 was shown to be important for virion budding (22). Similar motifs, such as PPXY or YPXL/LXXLF, are found in many retroviruses as well as other enveloped viruses, and mutations in these motifs often arrest a late stage in virion release (for a review, see reference 15). The observation that these tetrapeptide motifs remain functional when moved to heterologous locations within Gag, and in some cases when swapped among viruses (26, 29, 30, 39, 54), suggests that HIV-1 p6 serves as a docking site for host factors required for virion release from producer cells.
Monoubiquitination of plasma membrane receptors regulates their internalization into the endocytic pathway and serves as a sorting signal to direct the movement of internalized receptors between different endocytic compartments (21). HIV-1 p6 is also monoubiquitinated (35), and virion budding is inhibited by proteasome inhibitors, drugs which are thought to deplete the intracellular pool of free ubiquitin (45). It was therefore hypothesized that monoubiquitinated Gag directs virion release by recruiting the same cellular factors that direct the trafficking of endocytic vesicles. Indeed, p6 interacts with Tsg101 (50), which has a ubiquitin binding domain that associates with the PTAP motif, and this interaction is enhanced if p6 is ubiquitinated (42, 43). In addition, Tsg101 is a component of the endosomal sorting complex known as ESCRT-I that recognizes ubiquitinated endosomal cargo and is required for vesicle budding events that result in formation of multivesicular bodies in a manner topologically similar to retrovirus budding from the cell membrane.
The ESCRT-I complex activates the formation of yet another complex, ESCRT-II, which in turn recruits ESCRT-III, the core of the machinery that drives multivesicular-body formation. Via a YPXL motif at its carboxyl terminus, the p6 domain associates with AIP1/ALIX (47), a host protein which also interacts with Tsg101 and components of the human ESCRT-III complex. These results support a model in which the formation of the complex of HIV-1 p6, Tsg101, and AIP1/ALIX is required for ESCRT-III recruitment to the viral budding site.
In this study we report the isolation of SUMO-1 (small ubiquitin-like modifier 1) and its E2 conjugation enzyme Ubc9 as interaction partners of p6. We demonstrate that p6 is covalently modified by SUMO-1 at amino acid residue K27. Furthermore, we show that overproduction of SUMO-1 in virion producer cells reduces the infectivity of the released HIV-1 virions and that this inhibitory effect of SUMO-1 is dependent upon the presence of the sumoylated residue K27 in p6.
MATERIALS AND METHODS
Plasmid DNAs.
PCR was performed using Pfu DNA polymerase (Stratagene). HIV-1HXB2 p6 was amplified and subcloned into pBSII KS(−) (Stratagene) using primers 1 and 2 (Table 1). All mutations in the p6 coding sequence were engineered in this subclone. The four conservative base substitutions for disruption of p6 RNA secondary structure were introduced using primers 3 and 4. The PTAP mutation, LIRL, was engineered with primers 5 and 2. The K27R and K33R single mutations and the K27/33R double mutation were made using primers 6 and 7, 8 and 9, and 10 and 11, respectively.
TABLE 1.
Primers used for cloning and mutagenesis
| Primer | Sequence |
|---|---|
| 1 | 5′-GCGGAATTCCTTCAGAGCAGACCAGAG-3′ |
| 2 | 5′-GCGCGCTCGAGTTATTGTGACGAGGGGTCG-3′ |
| 3 | 5′-CTCCCCCTCAAAAACAAGAACCTATAGACAAGGAACTG-3′ |
| 4 | 5′-GTTCCTTGTCTATAGGTTCTTGTTTTTGAGGGGGAGTTG-3′ |
| 5 | 5′-GCGGAATTCCTTCAGAGCAGACCAGAGCTAATTCGCCTACCAGAAGAGAGCTTCAGG-3′ |
| 6 | 5′-CCCCCTCAAAGACAAGAACCTATAGAC-3′ |
| 7 | 5′-AGGTTCTTGTCTTTGAGGGGGAGTTG-3′ |
| 8 | 5′-CCTATAGACAGAGAACTGTATCCTTTAAC-3′ |
| 9 | 5′-AGGATACAGTTCTCTGTCCACAGGTTCTTG-3′ |
| 10 | 5′-CCCCCTCAAAGACAAGAACCTATAGACAGAGAACTGTATCCTTTAAC-3′ |
| 11 | 5′-AGGATACAGTTCTCTGTCTATAGGTTCTTGTCTTTGAGGGGGAGTTG-3′ |
| 12 | 5′-CCCGAATTCACCATGGGTTACCCATACGATGTTCCAGATTACGCTCTTCAGAGCAGACCAGAG-3′ |
| 13 | 5′-CCCGTGGATCCGTTGTGACGAGGGGTCGTT-3′ |
| 14 | 5′-TTTGGATCCTCTGACCAGGAGGCAAAACC-3′ |
| 15 | 5′-GCTCTCGAGCTAAACTGTTGAATGAGCCGCCGTTTGTTCCTG-3′ |
| 16 | 5′-GCTCTCGAGCTAAACTGTTGAATGACC-3′ |
Green fluorescent protein (GFP)-p6 fusions were constructed by subcloning the wild-type or mutant p6 coding sequences into pBSIIKS-bearing enhanced GFP (eGFP) (Clontech). For expression in mammalian cells, the GFP-p6 coding sequences were subcloned into pcDNA3.1(+) (Invitrogen). To create the various HAp6-LexA fusions for yeast two-hybrid assays, primers 12 and 13 were used to amplify and add an amino-terminal hemagglutinin (HA) tag; the products were cloned into pNLexA (OriGene Technologies, Rockville, Md.).
HA-tagged SUMO-1 (HASUMO-1) and HASUMO-1AA were amplified using primers 14 and 16 and 15 and 16, respectively. The PCR products were subcloned into pBS-HA (12). For expression in 293T cells, these HA-tagged proteins were cloned into pEF/myc/cyto (Invitrogen). The same primer sets were used to amplify and clone the wild-type and mutant SUMO-1 proteins into pGADNOT (27), which fused them to the carboxyl termini of the GAL4 activation domain coding sequences. The pGAL4 AD-Ubc9 fusion was obtained from Stephen Goff. pNL4-3 contains a complete infectious HIV-1 provirus.
Yeast two-hybrid screen and β-galactosidase assay.
The yeast two-hybrid system and the HeLa cDNA library used in this study have been described elsewhere (20). This system identifies bait-interacting proteins by selection for leucine prototrophy and screening for β-galactosidase activity in Saccharomyces cerevisiae strain EGY48. The bait consisted of the entire HIV-1 p6 fused at its amino terminus to an HA tag and fused at its carboxyl terminus to LexA using the plasmid pNLexA. A total of 5 × 105 colonies were screened according to published protocols (10). Interactions between bait and prey proteins were quantitated using the standard o-nitrophenyl-β-d-galactopyranoside liquid assay.
Cell lines and transfections.
Human 293T embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum and transfected by calcium phosphate precipitation. GHOST cells (catalog number 3942; National Institutes of Health AIDS Research and Reference Reagent Program) were maintained in DMEM supplemented with 10% fetal bovine serum, 500 μg of G418/ml, 100 μg of hygromycin/ml, and 1 μg of puromycin/ml; these HOS cell derivatives are transduced with the MV7neo-T4 retroviral vector and stably transfected with an HIV-2 LTR-GFP construct. Coreceptor cDNAs CXCR4 and CCR5 were introduced via retroviral transduction.
Antibodies and Western blot analysis.
Murine monoclonal anti-SUMO-1 antibody was purchased from Zymed Laboratories. Rabbit polyclonal antibody against GFP was purchased from Clontech, and rabbit polyclonal anti-HA and goat polyclonal anti-Tsg101 antibodies were purchased from Santa Cruz Biotech. Rabbit polyclonal antibodies against HIV-1 p6 and HIV-1 reverse transcriptase (RT) were acquired from the National Institutes of Health AIDS Research and Reference Reagent Program. Transfected 293T cells were lysed 48 h posttransfection with radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate) containing 1 mM phenylmethylsulfonyl fluoride and 10 mM N-ethylmaleimide as protease inhibitors. Lysate was cleared by centrifugation at 16,000 × g for 15 min at 4°C, and Western blot analysis was done as described previously (27). To probe virions for the presence of SUMO-1, 293T cells were cotransfected with pNL4-3 or pNL4-3K27R and a plasmid encoding HASUMO-1. Culture supernatant was harvested 48 h posttransfection, syringe filtered (0.45-μm-pore-size Acrodisc), and ultracentrifuged through a 25% sucrose cushion. Virions were subjected to a subtilisin protease protection assay (37), and protein content was analyzed by immunoblotting.
Dot blot analysis.
Virions produced by calcium phosphate transfection of 293T cells were purified by ultracentrifugation through 25% sucrose, resuspended, and normalized by exogenous RT activity. Virions were then transferred to a nylon membrane using the Minifold I dot blot apparatus (Schleicher & Schuell) and probed with a 32P-end-labeled DNA oligonucleotide as described previously (11).
Assays for virion production and infectivity.
Virus stocks for infection were produced in 293T cells by cotransfection of pNL4-3 or pNL4-3/K27R and a plasmid encoding HASUMO-1. Forty-eight hours posttransfection, the culture supernatant was harvested, syringe filtered (0.45-μm-pore-size Acrodisc), and stored at −80°C. Stocks were normalized according to RT activity (6). To assess virion infectivity, 4 × 104 GHOST cells per well were seeded in 24-well plates the day before infection. The cells were exposed to HIV-1 stock solutions for 16 h, after which the medium was replaced with fresh DMEM with dextran sulfate to block syncytia formation and further rounds of infection (25). Forty-eight hours postinfection, the cells were fixed with 3% formaldehyde, and the number of GFP-positive cells was determined by flow cytometry (FACScalibur; Becton Dickinson).
Monitoring HIV-1 cDNA synthesis by real-time PCR.
GHOST cells (106) were infected with HIV-1 stock solutions at a multiplicity of infection of 1. The cells were washed with phosphate-buffered saline 24 h postinfection, and total cellular DNA was extracted using the Blood & Cell Culture DNA Midi kit (QIAGEN). PCR products (40 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 30 s) were quantitated using an ABI Prism 7700 Sequence Detector (Applied Biosystems) as previously described (2). Products were detected using SYBR green (1:100,000; Molecular Probes) and quantitated by comparison with standard curves generated by serial dilution of plasmid template containing the relevant amplicon. Full-length linear viral cDNA was detected using a primer pair which distinguishes between plasmid DNA and viral DNA synthesized de novo; this was possible due to nucleotide mismatches between the two LTRs of pNL4-3 (6). 2-LTR circle DNA was detected using a previously published primer set (5).
RESULTS
Optimization of HIV-1 p6 expression.
To identify HIV-1 p6-interacting proteins, we set out to screen a human cDNA library using the yeast two-hybrid system. Initial attempts to express p6 fusion proteins in yeast were unsuccessful. We suspected that the efficiency of p6 translation was decreased due to the extensive secondary structure of HIV-1 RNA. Four conservative mutations (G78A, G81A, G84A, and G87A) that were predicted to reduce the thermodynamic stability of the p6 mRNA secondary structure were engineered in the p6 coding sequence (www.bioinfo.rpi.edu/applications/mfold/). This new p6 sequence produced a LexA fusion protein that was detectable by immunoblotting (data not shown).
HIV-1 p6 interacts with SUMO-1 and Ubc9 in cells.
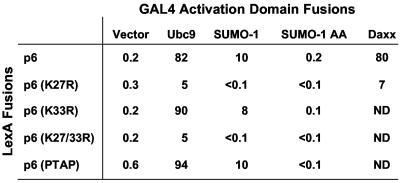
HIV-1 p6-LexA was used as bait in a yeast two-hybrid screen against fusion proteins encoded by a HeLa cDNA library. A total of 5 × 105 tranformants were screened, and six positive clones were found to bear plasmids that retained activity upon retesting. Of these six clones, two bore cDNAs encoding full-length SUMO-1 protein, while a third cDNA clone encoded SUMO-1 that lacked the first five amino acids. Many proteins that interact with SUMO-1 also interact with the SUMO-1 conjugating enzyme, Ubc9 (32). We did not isolate Ubc9 in the two-hybrid library screen but we obtained a full-length Ubc9 cDNA fused to a GAL4 activation domain and found that this protein also interacts with p6 (Fig. 1).
FIG. 1.
HIV-1 p6 interacts with SUMO-1 and Ubc9. An S. cerevisiae strain bearing a lacZ reporter under the control of multimerized LexA binding sites was transformed with plasmids encoding LexA or GAL4 activation domain fusions with the indicated proteins. β-Galactosidase activity of transformants is reported in Miller units.
The remaining three positive clones bore cDNAs encoding Daxx with various amino-terminal truncations, though all of them encoded the carboxyl-terminal Fas binding domain. Daxx was originally identified as an adapter protein involved in apoptosis via its interaction with the Fas death domain and Jun C-terminal protein kinase, ASK1 (9, 52). Daxx also associates with subnuclear structures called promyelocytic leukemia (PML) bodies or ND10s by binding to sumoylated PML protein (23).
Covalent modification of HIV-1 p6 by SUMO-1.
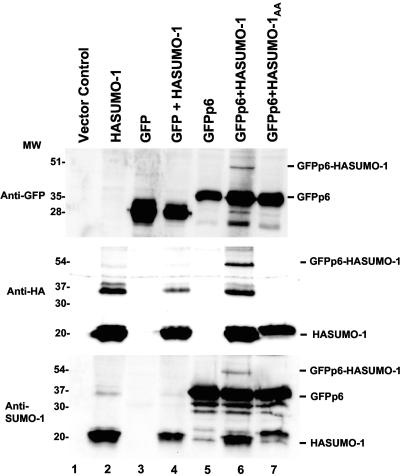
To test whether HIV-1 p6 is modified by SUMO-1 in mammalian cells, we created two p6 expression constructs, one with an HA tag at the amino terminus of p6 and one with an HA tag at the carboxyl terminus of p6. These plasmids were transfected into 293T cells, but we could not detect expression of either protein by Western blotting using antibodies against the HA epitope or against p6. A potential explanation for our inability to detect p6 expression was provided by the identification of a putative PEST region between residues 4 and 15 using PESTfind (http://emb1.bcc.univie.ac.at/embnet/tools/bio/PESTfind/). This region of p6 also contains the PTAP motif which mediates p6's interaction with Tsg101 and is necessary for virion budding (16, 18, 22, 30, 50), but we were unable to detect expression of p6 mutant protein in which PTAP was mutated to LIRL (data not shown). Finally, we created a mammalian expression construct with p6 fused to the carboxyl terminus of GFP. Upon transfection into 293T cells, this protein was detected by Western blotting using antibodies against either GFP or p6 (Fig. 2, lane 5).
FIG. 2.
SUMO-1 covalently modifies p6. Human 293T cells were transfected with plasmids encoding the proteins indicated above each lane. Cell lysates were analyzed by Western blotting with antibodies against GFP (top), the HA epitope tag (middle), or SUMO-1 (bottom). The positions of migration of size markers (in kilodaltons) are indicated on the left of the panels. The positions of migration of the expressed proteins, as well as of GFPp6 covalently attached to HASUMO-1, are indicated on the right of the blots. Note that the anti-SUMO-1 antibody seems to nonspecifically recognize GFP-p6.
No low-mobility forms of GFP-p6 indicative of covalent attachment to endogenous SUMO-1 were detected (Fig. 2, lane 5). Many reports in the literature indicate that biochemical detection of sumoylated substrates often requires SUMO-1 overproduction (31, 53). 293T cells were therefore cotransfected with plasmids expressing GFP-p6 and HASUMO-1. Western blot analysis with antibodies against GFP, SUMO-1, HA, or p6 detected a 54-kDa protein specific to the cells expressing both GFPp6 and HA-SUMO-1 (Fig. 2, lane 6, and data not shown), consistent with covalent modification of GFPp6 by HASUMO-1. To confirm that the 54-kDa protein was indeed due to covalent attachment of SUMO-1 to p6, we mutated the two carboxyl-terminal glycines of SUMO-1 to alanines to create a molecule that cannot be ligated to its targets (SUMO-1AA). When SUMO-1AA was coexpressed with GFPp6, no 54-kDa protein was detected (Fig. 2, lane 7).
Mapping the SUMO-1 modification site of HIV-1 p6.
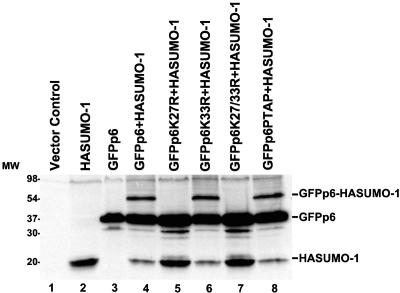
Having determined that HIV-1 p6 is sumoylated, we sought to identify the p6 residues that are modified by SUMO-1. Since SUMO-1 is conjugated to substrate lysine residues, we mutated the two lysines in p6 to arginine (K27R and K33R) and examined the effect of these mutations on GFPp6 sumoylation. 293T cells were transfected with HASUMO-1 in combination with GFPp6, GFPp6K27R, GFPp6K33R, or GFPp6K27/33R. Western blot analysis of the cell lysates revealed that the K33R mutant was modified by SUMO-1 as efficiently as wild-type p6, while the K27R mutation or the K27R/K33R double mutation abolished sumoylation (Fig. 3). We also examined the p6 mutant in which PTAP residues were changed to LIRL. This mutant was sumoylated as efficiently as the wild-type GFPp6 (Fig. 3, lane 8), indicating that the PTAP motif, which is required for virion budding from cells, does not play a role in p6 sumoylation.
FIG. 3.
HIV-1 p6 lysine 27 is required for covalent attachment of SUMO-1. Human 293T cells were transfected with plasmids encoding the proteins indicated above each lane. Cell lysates were probed by immunoblotting with anti-SUMO-1 antibody. Data are presented as described in the legend to Fig. 2. MW, molecular size in kilodaltons.
In vivo interaction of p6 with SUMO-1, Ubc9, or Daxx is dependent on covalent conjugation to SUMO-1.
We next examined whether the p6 K27R mutation, which disrupts p6 sumoylation, affects the interaction of HIV-1 p6 with SUMO-1, Ubc9, or Daxx in the yeast two-hybrid system. Whereas the K33R and PTAP mutations did not affect interaction with SUMO-1 or Ubc9, the K27R mutation abolished association with either of these two proteins (Fig. 1) as well as with Daxx. Furthermore, the SUMO-1AA mutant, which cannot be covalently attached to substrates, failed to interact with HIV-1 p6, even though its expression was comparable to that of wild-type SUMO-1 (data not shown). These results suggest that the interaction between p6 and SUMO-1 in yeast is dependent upon covalent conjugation of SUMO-1 to p6 and that recognition of p6 by Ubc9 or Daxx also requires sumoylation.
SUMO-1 incorporation into HIV-1 virions.
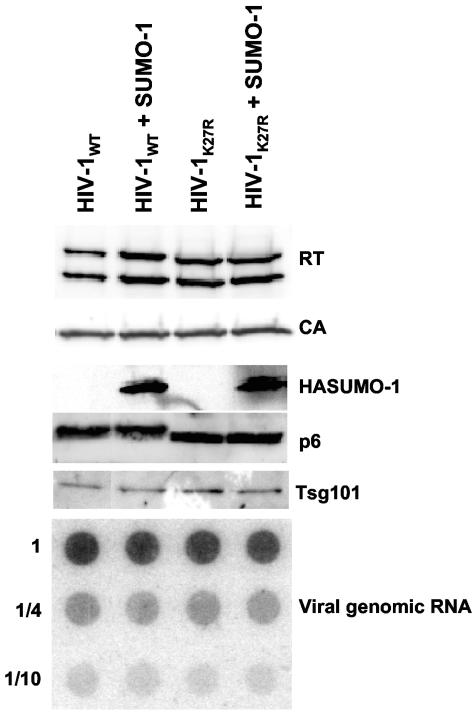
To determine whether SUMO-1 is packaged into HIV-1 virions during assembly, we cotransfected 293T cells with pNL4-3 and a HASUMO-1 expression plasmid. Virions were collected from the supernatant by acceleration through a sucrose cushion. These virions were then incubated with subtilisin to digest any proteins exposed on the virion surface (19, 37). Proteins that were protected within the virion membrane were then analyzed by Western blotting. SUMO-1 was incorporated into virions (Fig. 4, lane HIV-1WT + SUMO-1), though sumoylated p6 was not detected in these preparations. SUMO-1 incorporation into K27R (HIV-1K27R) or K27R/K33R mutant virions was as efficient as with the wild-type virus (HIV-1WT) (Fig. 4, lane HIV-1K27R + SUMO-1, and data not shown). Thus, SUMO-1 is incorporated within the HIV-1 virion membrane via a mechanism that is independent of any p6 lysine residues.
FIG. 4.
SUMO-1 is incorporated into HIV-1 virions. Virions produced by 293T cells cotransfected with pNL4-3 or pNL4-3K27R and a HASUMO-1 expression plasmid were purified through a 25% sucrose cushion. After subtilisin digestion, virions were analyzed by Western blotting using the indicated antibodies. To measure viral genomic RNA incorporation, virions were normalized by exogenous RT assay, loaded onto a nylon membrane, and probed with a 32P-end-labeled DNA oligonucleotide specific for HIV-1 genomic RNA.
Mutation of lysine residues in p6 does not change HIV-1 infectivity in vitro.
Several approaches were taken to determine whether SUMO-1 plays a role in HIV-1 replication. We attempted to disrupt expression of SUMO-1 or Ubc9 with RNA interference, but in neither case did we succeed in disrupting expression. SUMO-1 was overexpressed in susceptible target cells, with or without overexpression of Ubc9, and, though a modest decrease in infectivity was observed, the same magnitude reduction was observed with HIV-1 bearing the p6 K27R mutation (data not shown).
The p6 K27R, K33R, and K27R/K33R mutations were engineered within the context of an infectious provirus. As reported previously (35), these mutations had no observable effect on virion assembly or infectivity, as measured by spreading infection in Jurkat T cells. In addition, no abnormality in infectivity was observed in single-cycle assays with GHOST or MAGI cells, even when the cell cycle of the target cells was arrested with aphidicolin.
Overproduction of SUMO-1 has no detectable effect on virion assembly.
We then examined the effect of SUMO-1 overexpression in virion producer cells. 293T cells were cotransfected with pNL4-3 and a SUMO-1 expression vector. Overproduction of SUMO-1 had no significant effect on the yield of virion particles, as determined by measuring RT activity in the culture supernatant and in virus preparations concentrated through 25% sucrose. No defects were observed when the released virions were probed in immunoblots with antibodies against RT, CA, p6, or TM and in dot blots with a DNA probe specific for viral genomic RNA (Fig. 4).
We detected covalent attachment of SUMO-1 to HIV-1 p6 residue K27 (Fig. 2 and 3). Residues K27 and K33 are sites of attachment for monoubiquitin (35). This raises the possibility that SUMO-1 competes with ubiquitin for covalent attachment to lysines on p6. Overexpression of SUMO-1 had no detectable effect on the pattern of Gag ubiquitination or on the incorporation of free ubiquitin into HIV-1 virions (data not shown). Ubiquitin strengthens the interaction of p6 with Tsg101 (16, 42, 43), but we saw no effect of SUMO-1 overexpression on Tsg101 incorporation into virions (Fig. 4).
HIV-1 infectivity is reduced by SUMO-1 overexpression in virion producer cells.
The HIV-1 virions released from cells that were cotransfected with the SUMO-1 expression plasmid had fourfold-reduced infectivity in a single-cycle assay using GHOST reporter cells (Fig. 5). Overproduction of Ubc9 during virus production had no effect on the infectivity of the released virions. When Ubc9 and SUMO-1 were both overproduced, the infectivity was reduced to the same extent as it was with SUMO-1 overproduction alone (data not shown).
FIG. 5.
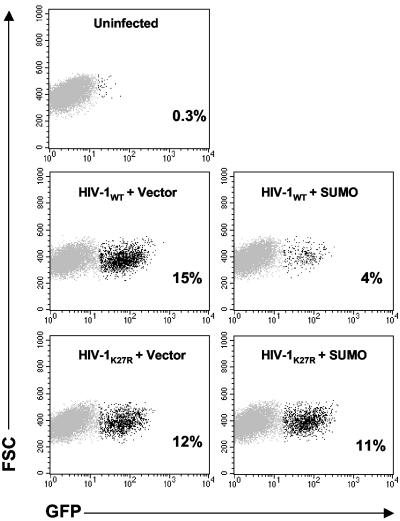
SUMO-1 overproduction during HIV-1 virion assembly reduces subsequent infectivity of the virions that are produced. 293T cells were transfected with HIV-1NL4-3 wild-type or HIV-1NL4-3 K27R mutant proviral DNAs, in combination with empty vector or a plasmid expressing HASUMO-1. Virions harvested from the supernatant were normalized by exogenous RT activity and used to infect cells bearing an LTR-GFP reporter. Cells were fixed 2 days later and analyzed by flow cytometry for GFP expression; the darker dots indicate the GFP-positive cells. In each case, the percentage of infected cells is indicated. The results shown are representative of three independent experiments. FSC, forward scatter.
The infectivity of the p6 K33R mutant virions was inhibited by SUMO-1 overproduction, similar to wild-type virions. However, the infectivity of the p6 K27R and the K27/33R double mutant virions was resistant to overproduction of HASUMO-1, consistent with a regulatory role for p6 sumoylation in virion infectivity (Fig. 5).
To determine why virions produced in the presence of excess SUMO-1 had reduced infectivity, we performed experiments to determine whether virion fusion with the target cells was affected. Using an enzymatic assay in which a Vpr-β-lactamase fusion is delivered into target cells (8), we determined that the block to infection was not at the entry step (data not shown).
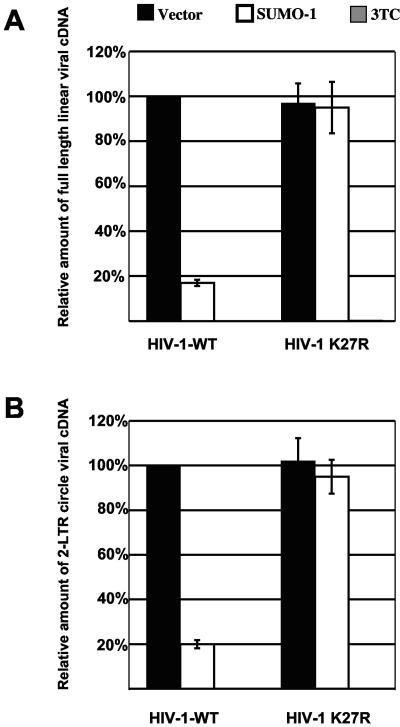
Next we examined whether reverse transcription was affected by SUMO-1 overproduction. HIV-1WT and HIV-1K27R virions produced in the presence of HASUMO-1 were normalized by exogenous RT assay and used to infect GHOST cells at a multiplicity of infection of 1. Total cellular DNA was subjected to real-time PCR using primer sets that monitor the synthesis of full-length linear viral cDNA as well as 2-LTR DNA circles. Wild-type virions produced in the presence of overexpressed HASUMO-1 had a fivefold reduction in the number of full-length linear and 2-LTR circle DNAs (Fig. 6) compared to control virions, which explains the reduction in infectivity we have seen with the GHOST single-cycle infectivity assay. Overproduction of SUMO-1 did not affect reverse transcription of the HIV-1K27R virions postentry.
FIG. 6.
Virions released from SUMO-1-overproducing cells have defects in reverse transcription. HIV-1WT and HIV-1K27R virions produced by cells in which SUMO-1 was overexpressed, as indicated, were used to infect GHOST cells. As a control, the HIV-1 RT inhibitor 3TC was added to target cells 2 h prior to infection. Eighteen hours postinfection, total cellular DNA was isolated, and reverse transcription was monitored using real-time PCR with primers specific for full-length linear cDNA (A) or 2-LTR circles (B). The results shown are representative of three independent experiments.
DISCUSSION
In this study we identified SUMO-1, Ubc9, and Daxx as proteins that interact with HIV-1 p6. We have shown that SUMO-1 is covalently attached to HIV-1 p6 lysine 27 and that this residue is required for interaction with Ubc9 or Daxx. In some cases, sumoylation of a protein regulates its interaction with other proteins. Daxx, for example, is recruited to ND10 nuclear bodies via interaction with sumoylated PML (23). HIV-1 p6 interaction with Daxx and Ubc9 is also dependent upon sumoylation of p6 (Fig. 1).
Overproduction of SUMO-1 decreased HIV-1 virion infectivity, and the K27R mutant virus was resistant to this effect, suggesting that modification of HIV-1 p6 by SUMO-1 regulates HIV-1 infectivity. Consistent with the relevance of these findings, HIV-1 p6 K27 and the minimal consensus sumoylation sequence (ψKXE, where ψ is generally a hydrophobic residue and X is any amino acid) (31, 46) are conserved in nearly all HIV-1 isolates, with exceptions in some A2 and K isolates (HIV Sequence Database, Los Alamos National Laboratory [http://www.hiv.lanl.gov]).
The first SUMO-1 substrate to be identified was RanGAP (28, 44), the GTPase-activating protein of the Ran GTPase that regulates nucleocytoplasmic transport. Since then, many sumoylated nuclear proteins have been identified, including transcription factors, components of PML nuclear bodies, and chromatin-associated proteins (31, 46). SUMO-1 E2 and E3 conjugases and isopeptidases localize to the nucleus or to the nuclear pore complex (17, 34, 38, 41, 46, 55), indicating that SUMO-1 attachment and removal take place at the nuclear pore in connection with nucleocytoplasmic transport. Indeed, the heterogenous nuclear ribonucleoproteins C and M are sumoylated as they traverse the nuclear pore, resulting in the release of their RNA cargo into the cytoplasm (49).
SUMO-1 overproduction did not appear to reduce the efficiency of viral cDNA import into the nucleus. Though there was a reduction in the quantity of viral cDNA circles produced, this reduction was identical to the reduction in reverse transcription (Fig. 6). In addition, when expressed in 293T cells, both GFP-p6 and GFP-p6(K27R) fusion proteins were located diffusely throughout the cell (data not shown). Hence, our experiments do not suggest that sumoylation of p6 alters its subcellular localization or that sumoylated p6 plays a role in nuclear import of the viral preintegration complex. It has also been reported that HIV-1 transduction disrupts nuclear structures called PML bodies (48), though others have failed to observe this effect (3, 4). p6 does not localize to PML bodies and does not disrupt them (data not shown).
Since SUMO-1 is covalently attached to lysine residues, this modification can block ubiquitination. Sumoylation of critical lysine residues in IκΒα or Mdm2 blocks polyubiquitination and protects these proteins from degradation (7, 13). Ubiquitination of p6 strengthens its interaction with Tsg101 (16), but it is not necessary for virus replication, since a p6 mutant with both lysines substituted by arginines does not have any assembly defects and replicates like wild-type HIV-1 in tissue culture and in peripheral blood mononuclear cells (reference 35 and data not shown). If sumoylation of p6 inhibited ubiquitination and led to a significant decrease in its interaction with Tsg101, then one would expect to observe budding defects in cells overproducing SUMO-1. However, overproduction of SUMO-1 had no effect on virion assembly (Fig. 4) or on the level of ubiquitinated Gag proteins associated with released virions (data not shown).
The only detectable phenotype resulting from SUMO-1 overproduction in HIV-1 producer cells was reduction in infectivity of the released virions (Fig. 5) at an early step during reverse transcription (Fig. 6). Perhaps overproduction of SUMO-1 reduces virion infectivity by modulating the level of incorporation of a cellular protein that is used by the virion at an early step during infection. Another possibility is that incorporation of excess SUMO-1 into the virions directly reduces infectivity. However, HIV-1 p6 K27R incorporates HASUMO-1 just as efficiently as wild-type virus (Fig. 4, compare lanes HIV-1WT + SUMO-1 and HIV-1K27R + SUMO-1), and the K27R mutant is resistant to HASUMO-1 overproduction. Thus, reduction in virion infectivity appears not to be explained by excess free SUMO-1 in virions but is perhaps due to sumoylation of p6.
We have not been able to detect sumoylated p6 within HIV-1 virions. This may be due to the fact that only a small portion of p6 is sumoylated. Free ubiquitin is incorporated into HIV-1 virions just like free SUMO-1, and only 2% of p6 in virions is monoubiquitinated, as determined by high-performance liquid chromatography analysis (35, 36). Hence, if only a small percentage of p6 is sumoylated within the virions, it would not be surprising that this modification would be difficult to detect.
Acknowledgments
We thank David Ott and Stephen Goff for providing reagents.
This work was supported by National Institutes of Health grant RO1AI41857 to J.L. and utilized core facilities of the Columbia-Rockefeller Center for AIDS Research. L.B. was an Elizabeth Glaser Pediatric AIDS Foundation Scholar.
REFERENCES
- 1.Accola, M. A., A. Ohagen, and H. G. Gottlinger. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6(gag). J. Virol. 74:6198-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asmal, M., J. Colgan, F. Naef, B. Yu, Y. Lee, M. Magnasco, and J. Luban. 2003. Production of ribosome components in effector CD4+ T cells is accelerated by TCR stimulation and coordinated by ERK-MAPK. Immunity 19:535-548. [DOI] [PubMed] [Google Scholar]
- 3.Bell, P., L. J. Montaner, and G. G. Maul. 2001. Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 75:7683-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthoux, L., G. J. Towers, C. Gurer, P. Salomoni, P. P. Pandolfi, and J. Luban. 2003. As2O3 enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J. Virol. 77:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buschmann, T., S. Y. Fuchs, C. G. Lee, Z. Q. Pan, and Z. Ronai. 2000. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell 101:753-762. [DOI] [PubMed] [Google Scholar]
- 8.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 9.Chang, H. Y., H. Nishitoh, X. Yang, H. Ichijo, and D. Baltimore. 1998. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281:1860-1863. [DOI] [PubMed] [Google Scholar]
- 10.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Rescue of multiple viral functions by a second-site suppressor of a human immunodeficiency virus type 1 nucleocapsid mutation. J. Virol. 74:4273-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colgan, J., H. E. Yuan, E. K. Franke, and J. Luban. 1996. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J. Virol. 70:4299-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 14.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 17.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 18.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 76:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 21.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 22.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, E., and H. G. Gottlinger. 1996. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J. Virol. 70:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lederman, S., R. Gulick, and L. Chess. 1989. Dextran sulfate and heparin interact with CD4 molecules to inhibit the binding of coat protein (gp120) of HIV. J. Immunol. 143:1149-1154. [PubMed] [Google Scholar]
- 26.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 31.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 32.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2000. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275:36316-36323. [DOI] [PubMed] [Google Scholar]
- 33.Muller, B., T. Patschinsky, and H. G. Krausslich. 2002. The late-domain-containing protein p6 is the predominant phosphoprotein of human immunodeficiency virus type 1 particles. J. Virol. 76:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 35.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 38.Panse, V. G., B. Kuster, T. Gerstberger, and E. Hurt. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5:21-27. [DOI] [PubMed] [Google Scholar]
- 39.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 42.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 43.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh, H., R. Pu, M. Cavenagh, and M. Dasso. 1997. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. USA 94:3736-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 47.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 48.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 49.Vassileva, M. T., and M. J. Matunis. 2004. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 24:3623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. G. Krausslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, X., R. Khosravi-Far, H. Y. Chang, and D. Baltimore. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]
- 54.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]