Abstract
Epidermal growth factor receptor (EGFR) mutation is a reliable and sensitive biomarker for EGFR-TKI therapy in non-small-cell lung cancer (NSCLC). However, detection of EGFR mutation in tissues has obvious limitations. Circulating free DNA (cfDNA) has been reported as an alternative approach for the detection of EGFR mutations. This systematic review and meta-analysis was designed to assess the diagnostic performance of cfDNA, compared with tissues. True-positive (TP), false-positive (FP), false-negative (FN), and true-negative (TN) values were extracted or calculated for each study. Pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) were calculated. A summary receiver operating characteristic curve (SROC) and area under curve (AUC) were used to evaluate the overall diagnostic performance. 20 eligible studies involving 2012 cases were included in this meta-analysis. The pooled sensitivity, specificity, PLR, NLR, andDORwere 0.674 (95%CI: 0.517–0.800), 0.935 (95%CI: 0.888–0.963), 10.307 (95%CI: 6.167–17.227), 0.348 (95%CI: 0.226–0.537), and 29.582 (95%CI: 4.582–60.012), respectively. The AUC was 0.93 (95% CI: 0.90–0.95). The meta-analysis suggests that detection of EGFR mutation by cfDNA is of adequate diagnostic accuracy and cfDNA analysis could be a promising screening test for NSCLC.
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for about 80% cases of lung cancer, and most NSCLC patients are at advanced stage when diagnosed1. Epithermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs), such as gefitinib and erlotinib have been used as targeted therapy in NSCLC. However, only a part of NSCLC patients with female gender, never smokers, adenocarcinoma and Asian ethnicity are sensible to EGFR-TKIs2. In 2009, the landmark clinical trial, IPASS (Iressa Pan-Asia Study) demonstrated that gefitinib showed better survival in NSCLC patients with activating EGFR mutations3. After that, many clinical trials confirmed that the selection of EGFR-TKIs should be based on EGFR mutation status not on clinical characteristics4,5,6.
EGFR mutation status is a sensible and reliable biomarker for the responsiveness to EGFR-TKIs7,8. The deletion in exon 19 and point mutation in exon 21 (L858R) predict good response to EGFR-TKIs8, while the point mutation in exon 20 (T790M) indicates resistance to EGFR-TIKs and poor prognosis9. Additionally, it was found that chemotherapy could affect EGFR mutation status, and patients whose EGFR mutations switched from positive to negative after chemotherapy had a better partial response10. Thus, detection of EGFR mutation status is critical for the application of EGFR-TKIs and monitoring chemotherapy response in clinical practice.
Since most NSCLC patients are diagnosed at advanced stage, surgery is no longer possible and it is hard to get sufficient tissues for molecular testing. On the other hand, for real-time monitoring of EGFR mutation status, repeat biopsy is impossible. Thus, it is needed for a feasible and sensitive biomarker for the detection of EGFR mutation. Circulating free DNA (cfDNA) has been proposed as an alternative approach for the detection of EGFR mutation11,12. Numerous studies have investigated the diagnostic performance of cfDNA and a wide range of the concordance rates between cfDNA and tissues have been reported13,14,15,16.
With accumulating evidence, varied results raise concern about the diagnostic value of cfDNA for the detection of EGFR mutation. To address this issue, we performed this meta-analysis and systematic review to compare the diagnostic accuracy of cfDNA to tissues for the detection of EGFR mutations.
Methods
Literature search
This meta-analysis was performed and reported according to the guideline about diagnostic studies17. Potentially relevant studies were identified by searching PubMed, EMBASE, and the Cochrane library. A systematic and comprehensive search was performed for the 3 databases using combination of key words and medical subheadings: “lung neoplasms” or “lung cancer”, “Epidermal Growth Factor Receptor” or “erbB1”, “serum” or “plasma” or “circulating”, and “mutations”. Alternative spellings and abbreviations were also considered. To identify additional studies, reference lists of included studies and relevant reviews were also manually searched. The literature search was conducted without any limitations and the last search was performed on March 3, 2014.
Inclusion and exclusion criteria
Records retrieved from databases and reference lists were first screened by titles and abstracts and then full-text articles of relevant studies were retrieved for further review. Eligible studies were selected according to the following inclusion criteria: 1) all NSCLC patients involved should be diagnosed histopathologically or cytologically; 2) EGFR mutation status should be detected by circulating free DNA; 3) EGFR mutations were verified by detection of tumor tissues; 4) enough data to construct the diagnostic 2 × 2 table.
Studies met the following criteria were excluded: 1) tumor tissues and cfDNA were not paired; 2) EGFR mutation status were not verified by detection of tumor tissues; 3) insufficient data to construct the 2 × 2 table; 4) duplicate reports from the same patients (the latest or the one with most NSCLC patients were included). All records were reviewed by two authors independently and reached consensus at each eligible study.
Data extraction
The following data were extracted by 2 authors independently: name of author, year of publication, country where the study was conducted, percentage of female, percentage of ever-smokers, histological type, TNM stage, methods for EGFR mutation status detection in cfDNA, true positive (TP), false positive (FP), false negative (FN), and true negative (TN). When multiple methods were used for EGFR mutation detection in cfDNA, the methods with best sensitivity or specificity was extracted. According to the media of sample size, eligible studies were classified as large (≥median sample size) or small (<median sample size). The third author assessed the data and resolved the disagreement.
Quality assessment
Methodological quality of eligible studies was evaluated by QUADAS-2 (quality assessment of diagnostic accuracy studies 2) by two investigators18. QUADAS-2 is an improved tool, designed to evaluate the quality of primary diagnostic accuracy studies, which consists of 4 key domains (patient selection, index test, reference standard, and flow and timing). With signaling questions, risk of bias and concerns regarding applicability (except for the “flow and timing” domain) were judged as “low”, “high”, or “unknown”. Summary of QUADAS plot was generated by Review Manager software (version 5.2.9, the Cochrane Collaboration).
Statistical analysis
The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), positive predicted value, negative predicted value, diagnostic odds ratio (DOR) and corresponding 95% confidence intervals (95% CI) were calculated by the accuracy data (TP, FP, FN, and TN) extracted from each eligible studies. The PLR is calculated as: sensitivity/(1-specificity) and the NLR is calculated as: (1-sensitivity)/specificity. A clinically useful test was defined with a PLR > 5.0 and a NLR < 0.2. DOR is a measure that combined sensitivity and specificity, which is calculated as: PLR/NLR19. The summary receiver operative curve (SROC) was generated and the area under the curve (AUC) was calculated.
The Spearman correlation between the logit of sensitivity and logit of 1-specificity was calculated to determine the effect of threshold, and a P value < 0.05 indicated significant threshold effect. The heterogeneity caused by non-threshold effect was measured by Q test and the inconsistency index (I2), and a P value ≤ 0.05 and a I2 value ≥50% indicated significant heterogeneity caused by non-threshold effect. In the presence of significant heterogeneity, the DerSimonian Laird method was used to calculate the estimates20 and meta-regression was performed to detect the source. Sub-group analyses were performed for sample size, countries, detection methods, and TNM stages. Publication bias was detected by the Deek's funnel plot21 and a P vale< 0.05 indicated the presence of publication bias.
All statistical analyses were performed using the STATA software (version 11.2, STATA Corp., Texas USA) with the MIDAS module and Meta-DiSc.
Results
Study selection
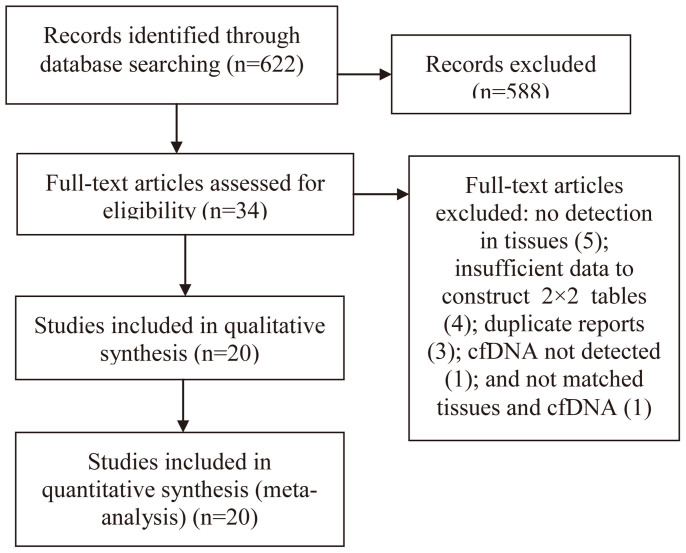
As shown in Figure 1, after primary screening, 34 full-text articles10,13,14,15,16,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50 were selected for further evaluation of eligibility. By rigorous evaluation, 20 eligible studies were identified and included in meta-analysis13,14,15,16,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37. The main reasons for exclusion were: EGFR mutation status was not detected in tissues10,38,39,40,41(5), insufficient data to construct 2 × 2 tables42,43,44,45(4), duplicate reports46,47,48(3), cfDNA not detected49 (1), and not matched tissues and cfDNA50(1). No additionally studies were identified by searching the references of eligible studies or relevant review.
Figure 1. Flow diagram of study selection.
Characteristics of eligible studies
Baseline characteristics of eligible studies were shown in Table 1. All eligible studies were published between 2006 and 2013. 2012 NSCLC patients were included in this meta-analysis, and most of them were at advanced stage (TNM III–IV) with adenocarcinoma. There were various kinds of methods applied for detection of EGFR mutation in cfDNA, while the ARMS was the most common method. Notably, only one study was carried in USA33and the other 19 studies were all performed in Asia. QUADAS-2 summary plot was presented in Figure S1. As shown, methodological quality of eligible studies were adequate and not significantly affected by bias.
Table 1. Characteristics of eligible studies.
| Author | Year | Country | Ethnicity | Number | Female | Ever Smoker | AC | TNM (I/II/III/IV) | Assay |
|---|---|---|---|---|---|---|---|---|---|
| Wang S | 2013 | China | Asian | 74 | 48.5% | 46.3% | 80.6% | 0/0/9/115 | ARMS |
| Jing CW | 2013 | China | Asian | 120 | 42.5% | NA | 58.3% | 38/82† | HRM |
| Zhang H | 2013 | China | Asian | 86 | 43.0% | 51.2% | 75.6% | 0/0/16/70 | MEL |
| Kim HR | 2013 | Korea | Asian | 40 | 35.0% | 28.3% | 88.3% | 0/0/7/53 | PNA |
| Liu X | 2013 | China | Asian | 86 | 34.9% | 54.7% | 98.8% | 0/0/4/82 | ARMS |
| Lv C | 2013 | China | Asian | 6 | 54.5% | 45.5% | NA | 0/0/11/0 | DHPLC |
| Kim ST | 2013 | Korea | Asian | 57 | 38.6% | 56.1% | 70.2% | 0/0/7/50 | PNA-LNA |
| Xu F | 2012 | China | Asian | 34 | 39.2% | NA | 84.3% | 0/0/6/45 | ARMS |
| Hu C | 2012 | China | Asian | 24 | 50.0% | 45.8% | 58.3% | 11/8/4/1 | HRM |
| Zhao X | 2012 | China | Asian | 111 | 31.5% | 51.4% | 65.8% | 22/10/33/46 | ME-PCR |
| Yam I | 2012 | China | Asian | 35 | 60.0% | 14.3% | 94.3% | 0/0/8/27 | AS-APEX |
| Goto K | 2012 | Japan | Asian | 86 | 87.6% | 9.0% | NA | NA | ARMS |
| Jiang B | 2011 | China | Asian | 58 | 31.0% | 62.1% | 72.4% | NA | ME sequencing |
| He C | 2009 | China | Asian | 18 | 36.6% | 53.0% | 75.4% | 19/115† | ME-PCR |
| Bai H | 2009 | China | Asian | 230 | 46.5% | 44.8% | 74.3% | 0/0/80/150 | DHPLC |
| Kuang Y | 2009 | USA | Caucasian | 43 | 81.5% | NA | NA | NA | ARMS |
| Yung TK | 2009 | China | Asian | 29 | NA | NA | NA | NA | digital PCR |
| Kimura H | 2007 | Japan | Asian | 42 | 33.3% | 66.7% | 73.8% | NA | ARMS |
| Kimura H | 2006 | Japan | Asian | 11 | 37.3% | NA | 85.2% | 0/0/3/24 | ARMS |
| Huang Z | 2012 | China | Asian | 822 | 46.7% | 41.4% | 78.0% | NA | DHPLC |
AC: adenocarcinoma; NA: not available; † data present as I–II/III–IV; ARMS: Scorpion Amplification Refractory Mutation System; HRM: High Resolution Melting; MEL: Mutant Enriched Liquidchip; PNA: Peptide Nucleic Acid (PN A)-mediated PCR clamping; DHPLC: Denaturing High-Performance Liquid Chromatography; PNA-LNA: Peptide Nucleic Acid-Locked Nucleic Acid PCR; ME-PCR: mutant-enriched PCR; AS-APEX: Allele-Specific Arrayed Primer Extension.
Accuracy of cfDNA for the detection of EGFR mutation
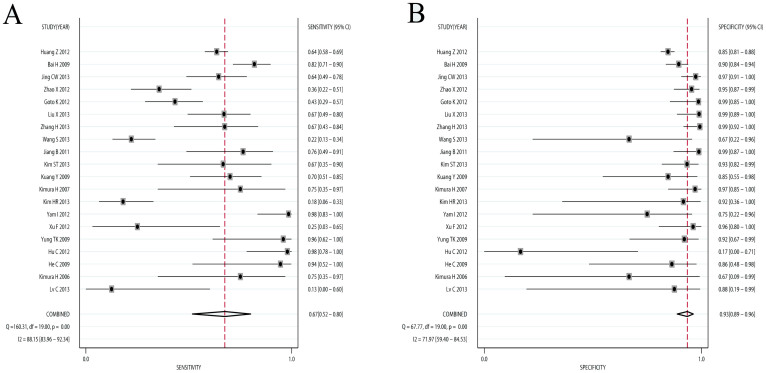
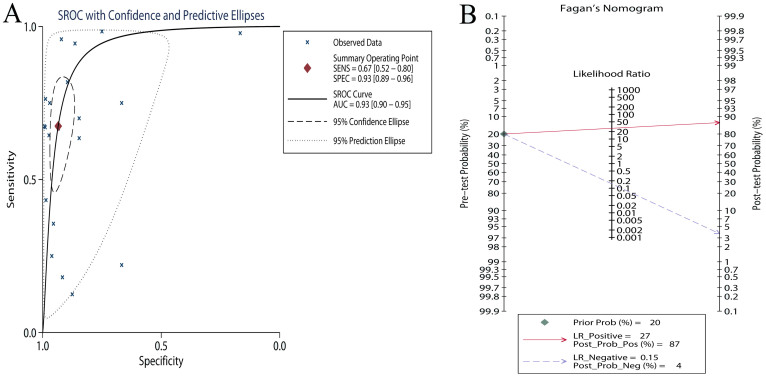
Results of this meta-analysis were shown in Table 2. Compared with NSCLC tumor tissues, the pooled sensitivity and specificity of cfDNA for the detection of EGFR mutation status were 0.674 (95%CI: 0.517–0.800) and 0.935 (95%CI: 0.888–0.963), respectively (Figure 2). The PLR and NLR of cfDNA were 10.307 (95%CI: 6.167–17.227) and 0.348 (95%CI: 0.226–0.537), respectively (Figure S2). The DOR was 29.582 (95%CI: 4.582–60.012) (Figure S3). Figure 3A showed the SROC with AUC of 0.93 (95% CI: 0.90–0.95), indicating cfDNA had high diagnostic accuracy. Fagan plot was generated for the visual presentation of diagnostic performance (Figure 3B). Sub-group analyses were performed to assess the influence of sample size, countries, detection methods, and TNM stages (Table 2). The diagnostic accuracy data were consistent across different sub-groups.
Table 2. Meta-analyses Results.
| Sensitivity | Specificity | PLR | NLR | DOR | AUC | |
|---|---|---|---|---|---|---|
| Overall | 0.674 (0.517–0.800) | 0.935 (0.888–0.963) | 10.307 (6.167–17.227) | 0.348 (0.226–0.537) | 29.582 (4.582–60.012) | 0.93 (0.90–0.95) |
| Sample Size | ||||||
| Small | 0.776 (0.459–0.934) | 0.898 (0.785–0.955) | 7.621 (3.906–14.871) | 0.250 (0.088–0.709) | 30.509 (9.471–98.279) | 0.92 (0.90–0.94) |
| Large | 0.591 (0.461–0.709) | 0.954 (0.901–0.979) | 12.820 (5.494–29.915) | 0.429 (0.312–0.590) | 29.897 (10.474–85.340) | 0.89 (0.86–0.91) |
| Country | ||||||
| China | 0.721 (0.511–0.865) | 0.930 (0.861–0.966) | 10.349 (5.336–20.069) | 0.300 (0.159–0.566) | 34.526 (13.371–89.151) | 0.93 (0.91–0.95) |
| Japan | 0.590 (0.349–0.794) | 0.962 (0.824–0.993) | 15.591 (3.319–73.233) | 0.426 (0.243–0.749) | 36.556 (6.948–192.339) | 0.87 (0.84–0.90) |
| Korea | 0.378 (0.111–0.747) | 0.928 (0.758–0.981) | 5.240 (0.966–28.417) | 0.670 (0.363–1.238) | 7.816 (0.874–69.889) | 0.92 (0.90–0.94) |
| Detection Methods | ||||||
| ARMS | 0.525 (0.357–0.688) | 0.947 (0.860–0.981) | 9.948 (3.181–31.107) | 0.501 (0.342–0.734) | 19.855 (4.839–81.468) | 0.86 (0.83–0.89) |
| DHPLC | 0.658 (0.396–0.849) | 0.854 (0.783–0.904) | 4.499 (2.119–9.550) | 0.401 (0.186–0.865) | 11.221 (2.474–50.894) | 0.86 (0.83–0.89) |
| HRM | 0.887 (0.402–0.989) | 0.736 (0.042–0.994) | 3.360 (0.197–57.314) | 0.153 (0.035–0.673) | 21.974 (1.522–317.223) | 0.91 (0.88–0.93) |
| TNM Stages | ||||||
| III–IV | 0.576 (0.333–0.787) | 0.934 (0.862–0.970) | 8.702 (3.760–20.144) | 0.454 (0.255–0.807) | 19.163 (5.560–66.042) | 0.92 (0.89–0.94) |
| I–IV | 0.831 (0.380–0.975) | 0.883 (0.563–0.978) | 7.072 (1.904–26.260) | 0.192 (0.040–0.929) | 36.856 (9.199–147.668) | 0.93 (0.90–0.95) |
Data were present as accuracy data with 95% confidence intervals. PLR: positive likelihood ratio; NLR: negative likelihood ratio; DOR: diagnostic odds ratio; AUC: area under curve; ARMS: Scorpion Amplification Refractory Mutation System; DHPLC: Denaturing High-Performance Liquid Chromatography; HRM: High Resolution Melting.
Figure 2. Forest plots of sensitivity and specificity of cfDNA.
The pooled sensitivity was 0.691 (95% CI: 0.569–0.790) and the pooled specificity was 0.922 (95% CI: 0.878–0.951).
Figure 3. The summary operative receiver characteristic curve indicated high diagnostic accuracy (A, the area under summary receiver characteristics curve was) and Fagan plot presents the clinical utility of cfDNA (B).
Threshold effect and heterogeneity
Threshold effect is a major source of between study heterogeneity. Visual assessment of ROC plane revealed no significant threshold effect (Figure S4). Spearman correlation coefficient and P value were calculated to assess the threshold effect. The Spearman correlation coefficient was 0.114 and the P value was 0.652 (>0.05), confirming that the threshold effect was not significant. As shown in the forest plots of accuracy data (sensitivity, specificity, PLR, NLR, and DOR), significant heterogeneity was detected. Thus, meta-regression was performed to detect the source of heterogeneity, and sample size, TNM stage, detection methods, and country were analyzed for each accuracy data. However, none of the above covariates contributed heterogeneity.
Sensitivity analysis and publication bias
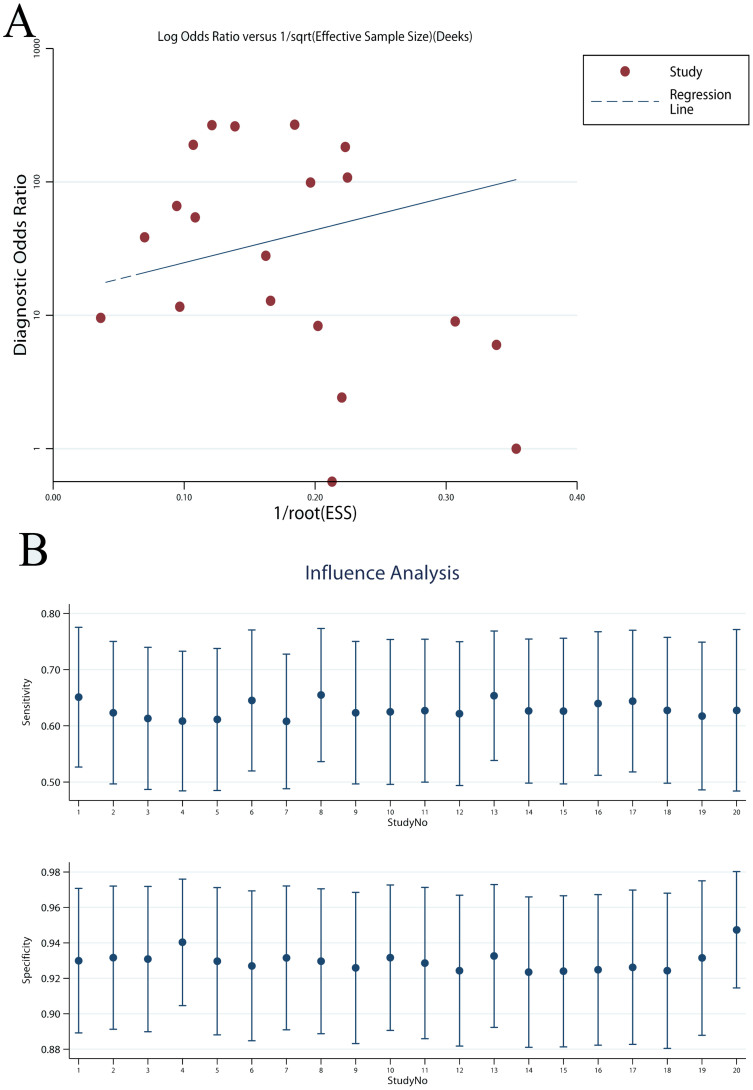
Publication bias was tested by the Deek's funnel plot. As shown, the funnel plot and P value 0.243 (>0.05) suggested no evidence of publication bias (Figure 4A). Sensitivity analysis was performed and the results showed the pooled results were not affected by individual studies (Figure 4B).
Figure 4. Deek's funnel plot showed no significant publication bias (A) and sensitivity analysis showed that the pooled results were robust and not affected by individual studies (B).
The P value of Deek's funnel plot was 0.243, indicating no significant publication bias.
Discussion
The need for a feasible, reliable, and minimally invasive approach for EGFR mutation detection has been a limiting factor in clinical research and practice. Although EGFR mutation could be detected by tumor tissues, its limitations are well known. As a more feasible and less invasive alternative, cfDNA has received more and more interest51,52. However, a wide range of diagnostic accuracy values of cfDNA have been reported.
We performed this meta-analysis and systematic review to determine the diagnostic accuracy of cfDNA for EGFR mutation detection. The pooled sensitivity for cfDNA was 0.674 and the specificity was 0.935. The sensitivity of cfDNA is not high enough as a diagnostic method. However, as a cancer screening test, sensitivity is not vital and a high specificity is more important if it triggers invasive diagnostic procedures53. In the circumstance of real-time monitoring of EGFR mutation status in regimens of NSCLC, cfDNA might be a suitable screening test for EGFR mutation status, due to the high specificity and non-invasive nature33. It is worth noting that the high DOR and AUC indicate an overall high diagnostic accuracy of cfDNA.
Under the background that a feasible biomarker with optimal performance is needed and the uncertainty of whether cfDNA provides satisfying diagnostic accuracy, this meta-analysis adds important evidence to the literature.
This is the first meta-analysis of the diagnostic performance of cfDNA for the detection of EGFR mutation status and represents an attempt to provide guidance for future studies. However, some issues worth noting when reviewed current literature. First, most studies were retrospective studies and the tissue samples were formalin-fixed paraffin-embedded, which lead to significant DNA degradation and increase detection bias52. Second, chemotherapy could affect EGFR mutation status, thus, the timing of tissue collection and peripheral blood collection matters the concordance rate. It is believed that more studies using standardized handling and detecting procedures may contribute to more robust diagnostic performance.
A lot of method have been developed to detect EGFR mutations in cfDNA, such as direct sequencing, the scorpion-amplified refractory mutation system (ARMS), denaturing high-performance liquid chromatography (DHPLC), peptide nucleic acid mediated polymerase chain reaction clamping method, high resolution melting (HRM), digital PCR, and so on. The diagnostic performance has increased with the development of detection methods. Our sub-group analyses also confirmed that DHPLC and HRM showed higher sensitivity than ARMS. It is highly possible that the diagnostic performance of cfDNA will improve in the future.
Limitation of this meta-analysis should also be highlighted. First, most studies analyzed were small-sized, which might lead to bias. To evaluate the effects of small-sized studies, sub-group and sensitivity analyses were performed and results revealed that the pooled results were stable and not affected by bias. Second, significant heterogeneity was observed. Spearman correlation and ROC plane suggested that the heterogeneity was not caused by threshold effect. Multi-variables meta-regression was also performed, and unfortunately, none of the analyzed covariates was the source of heterogeneity. Third, only English databases were searched in this meta-analysis, while most eligible studies were conducted in Asia (China, Japan, and Korea). Thus, it was possible that there were some non-English studies which were not included in this meta-analysis. On the other hand, PubMed and EMBASE are the two most comprehensive databases of medicine, and most high power full trials tend to publish in English. Additionally, Deek's funnel plot was applied to detect the publication bias and no evidence of publication bias was found.
In conclusion, in our meta-analysis of 20 studies including >2000 participants, detection EGFR mutations in cfDNA appears to be of adequate diagnostic value in NSCLC. Due to its high specificity and non-invasive nature, cfDNA might be a promising screening test for NSCLC.
Supplementary Material
supplementary information
Footnotes
The authors declare no competing financial interests.
Author Contributions J.L., L.S. and D.Z. designed this study; J.L. and L.S. searched databases and collected full-text papers; J.L., L.S. and D.Z. extracted and analyzed data; J.L., L.S. and D.Z. wrote the manuscript.
References
- Govindan R. et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 24, 4539–4544 (2006). [DOI] [PubMed] [Google Scholar]
- Shigematsu H. et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 97, 339–346 (2005). [DOI] [PubMed] [Google Scholar]
- Mok T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361, 947–957 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 12, 735–742 (2011). [DOI] [PubMed] [Google Scholar]
- Mitsudomi T. et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11, 121–128 (2010). [DOI] [PubMed] [Google Scholar]
- Rosell R. et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 361, 958–967 (2009). [DOI] [PubMed] [Google Scholar]
- Hirsch F. R. et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol 24, 5034–5042 (2006). [DOI] [PubMed] [Google Scholar]
- Sharma S. V., Bell D. W., Settleman J. & Haber D. A. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 7, 169–181 (2007). [DOI] [PubMed] [Google Scholar]
- Engelman J. A. et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 116, 2695–2706 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H. et al. Influence of chemotherapy on EGFR mutation status among patients with non-small-cell lung cancer. J Clin Oncol 30, 3077–3083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito A. et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: Clinical perspectives and research opportunities. Cancer Treat Rev 40, 648–655 (2014). [DOI] [PubMed] [Google Scholar]
- Murtaza M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta 430C, 63–70 (2013). [DOI] [PubMed] [Google Scholar]
- Jing C. W., Wang Z., Cao H. X., Ma R. & Wu J. Z. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev 14, 6619–6623 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang H. et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn 15, 819–826 (2013). [DOI] [PubMed] [Google Scholar]
- Kim H. R. et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res 32, 50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang M. M., Deeks J. J., Gatsonis C. & Bossuyt P. M. Systematic reviews of diagnostic test accuracy. Ann Intern Med 149, 889–897 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting P. F. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155, 529–536 (2011). [DOI] [PubMed] [Google Scholar]
- Glas A. S., Lijmer J. G., Prins M. H., Bonsel G. J. & Bossuyt P. M. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 56, 1129–1135 (2003). [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks J. J., Macaskill P. & Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58, 882–893 (2005). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 66, 1065–1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C. et al. A pilot study: sequential gemcitabine/cisplatin and icotinib as induction therapy for stage IIB to IIIA non-small-cell lung adenocarcinoma. World J Surg Oncol 11, 96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. T. et al. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Med Oncol 30, 328 (2013). [DOI] [PubMed] [Google Scholar]
- Xu F. et al. Comparison of different methods for detecting epidermal growth factor receptor mutations in peripheral blood and tumor tissue of non-small cell lung cancer as a predictor of response to gefitinib. Onco Targets Ther 5, 439–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. et al. Direct serum and tissue assay for EGFR mutation in non-small cell lung cancer by high-resolution melting analysis. Oncol Rep 28, 1815–1821 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao X. et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration 85, 119–125 (2013). [DOI] [PubMed] [Google Scholar]
- Yam I. et al. EGFR array: uses in the detection of plasma EGFR mutations in non-small cell lung cancer patients. J Thorac Oncol 7, 1131–1140 (2012). [DOI] [PubMed] [Google Scholar]
- Goto K. et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 7, 115–121 (2012). [DOI] [PubMed] [Google Scholar]
- Jiang B. et al. Serum detection of epidermal growth factor receptor gene mutations using mutant-enriched sequencing in Chinese patients with advanced non-small cell lung cancer. J Int Med Res 39, 1392–1401 (2011). [DOI] [PubMed] [Google Scholar]
- He C. et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer 125, 2393–2399 (2009). [DOI] [PubMed] [Google Scholar]
- Bai H. et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 27, 2653–2659 (2009). [DOI] [PubMed] [Google Scholar]
- Kuang Y. et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res 15, 2630–2636 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung T. K. et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 15, 2076–2084 (2009). [DOI] [PubMed] [Google Scholar]
- Kimura H. et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 97, 778–784 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 12, 3915–3921 (2006). [DOI] [PubMed] [Google Scholar]
- Huang Z. et al. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer 3, 334–340 (2012). [DOI] [PubMed] [Google Scholar]
- Li L. et al. [A study on the long-term non-small cell lung cancer survivors in the Expand Access Program of gefitinib in China]. Zhongguo Fei Ai Za Zhi 15, 332–339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Detection of EGFR somatic mutations in non-small cell lung cancer (NSCLC) using a novel mutant-enriched liquidchip (MEL) technology. Curr Drug Metab 13, 1007–1011 (2012). [DOI] [PubMed] [Google Scholar]
- Tiseo M. et al. Epidermal growth factor receptor intron-1 polymorphism predicts gefitinib outcome in advanced non-small cell lung cancer. J Thorac Oncol 3, 1104–1111 (2008). [DOI] [PubMed] [Google Scholar]
- Taniguchi K. et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 17, 7808–7815 (2011). [DOI] [PubMed] [Google Scholar]
- Brevet M., Johnson M. L., Azzoli C. G. & Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer 73, 96–102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Guerra H. et al. Phase II study of preoperative gefitinib in clinical stage I non-small-cell lung cancer. J Clin Oncol 27, 6229–6236 (2009). [DOI] [PubMed] [Google Scholar]
- Yeo C. D. et al. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer 81, 207–212 (2013). [DOI] [PubMed] [Google Scholar]
- Nakamura T. et al. Application of a highly sensitive detection system for epidermal growth factor receptor mutations in plasma DNA. J Thorac Oncol 7, 1369–1381 (2012). [DOI] [PubMed] [Google Scholar]
- Kim S. T. et al. Can serum be used for analyzing the EGFR mutation status in patients with advanced non-small cell lung cancer? Am J Clin Oncol 36, 57–63 (2013). [DOI] [PubMed] [Google Scholar]
- Bai H. et al. [The detection by denaturing high performance liquid chromatography of epidermal growth factor receptor mutation in tissue and peripheral blood from patients with advanced non-small cell lung cancer]. Zhonghua Jie He He Hu Xi Za Zhi 31, 891–896 (2008). [PubMed] [Google Scholar]
- Kimura H. et al. EGFR mutation of tumor and serum in gefitinib-treated patients with chemotherapy-naive non-small cell lung cancer. J Thorac Oncol 1, 260–267 (2006). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. [Detection of epidermal growth factor receptor mutations in non-small cell lung cancer tumor specimens from various ways by denaturing high-performance liquid chromatography]. Zhongguo Fei Ai Za Zhi 13, 850–855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti E. et al. Association of polymorphisms in AKT1 and EGFR with clinical outcome and toxicity in non-small cell lung cancer patients treated with gefitinib. Mol Cancer Ther 9, 581–593 (2010). [DOI] [PubMed] [Google Scholar]
- Crowley E., Di Nicolantonio F., Loupakis F. & Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 10, 472–484 (2013). [DOI] [PubMed] [Google Scholar]
- Diaz L. A. Jr & Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32, 579–586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley O. W. & Kramer B. S. Cancer screening in theory and in practice. J Clin Oncol 23, 293–300 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information