Abstract
Background
microRNA (miR)-181a has been reported to be downregulated in Parkinson’s disease (PD), but the regulatory mechanism of miR-181a on neuron apoptosis and autophagy is still poorly understood. We aimed to investigate the neuroprotective effects of miR-181a on PD in vitro.
Material/Methods
Human SK-N-SH neuroblastoma cells were incubated with different concentrations of 1-methyl-4-phenylpyridinium ion (MPP+) to induce the PD model. The expression of miR-181a was then analyzed. After transfection with miR-181a mimic or scramble following MPP+ treatment, the expression of autophagy protein markers (LC3II, LC3I, and Beclin 1) and p38 mitogen-activated protein kinase (MAPK)/c-Jun N-terminal kinases (JNK) signaling proteins (p-p38, p38, p-JNK, and JNK) and cell apoptosis were detected. Furthermore, the cells were transfected with miR-181a inhibitor and cultured in the presence or absence of p38 inhibitor SB203582 or JNK inhibitor SP600125, and the cell apoptosis was tested again.
Results
The expression of miR-181a was gradually decreased with the increase of MPP+ concentration (P<0.05, P<0.01, or P<0.001). Overexpression of miR-181a significantly decreased the LC3II/LC3I ratio, Beclin 1 expression, cell apoptosis, and the expression of p-p38 and p-JNK compared to the MPP+ + miR-181a scramble group (all P<0.05). In addition, we observed that SB203582 or SP600125 showed no effects on cell apoptosis, but the effects of miR-181a inhibitor on cell apoptosis were reversed by administration of SB203582 or SP600125 compared to the scramble group (P<0.05).
Conclusions
Our results suggest that miR-181a regulates apoptosis and autophagy in PD by inhibiting the p38 MAPK/JNK pathway.
MeSH Keywords: Apoptosis, Autophagy, MAP Kinase Signaling System, MicroRNAs, p38 Mitogen-Activated Protein Kinases, Parkinson Disease
Background
Parkinson’s disease (PD) is the second most common neurodegenerative disease following Alzheimer disease (AD), which is characterized by massive degeneration and progressive loss of dopaminergic (DA) neurons in the midbrain substantia nigra pars compacta (SNpc) of the basal ganglia [1,2]. It has been reported that the prevalence of PD is approximately 0.3% of the population in developed countries [3]. Unfortunately, there are no available therapies to inhibit the degenerative process of the disease, and only symptomatic treatment exists to improve the quality of life in patients with PD due to the unclear causes [3]. Although the pathogenesis of the disease is not completely understood, aging, genetic susceptibility, mitochondrial dysfunction, inflammation, oxidative stress, and protein aggregation are responsible for PD, leading to cell apoptosis [4–6]. Recently, a growing body of evidence suggests that autophagy plays a critical role in the progression of PD [7–10]. Thus, a better understanding of cell apoptosis and autophagy involved in PD might provide an effective and efficient treatment for PD patients.
MicroRNAs (miRNAs) are small, non-coding, and highly conserved RNAs that post-transcriptionally regulate gene expression by targeting mRNAs and prompting either translation repression or RNA degradation [11]. It has been well acknowledged that miRNAs play significant roles in many human diseases, including PD [12–14]. Recent studies have suggested that a number of miRNAs such as miR-34c, miR-107, miR-133b, miR-205, and miR-433 are aberrantly expressed in PD and are involved in several cellular processes including cell replication, mature neurons’ survival, neuron differentiation, neurite outgrowth, apoptosis, and autophagy [15–18]. The potential usefulness of a miRNA-based therapy in PD has been considered as a powerful tool [17]. However, the relevant mechanisms are far more complicated. Among the miRNAs, miR-181a is a conserved miRNA that is highly expressed in the brain [19] and has been reported to be increased during maturation of hippocampal neurons [20]. Besides, overexpression of miR-181 increases astrocyte death [21], and suppression of miR-181a can decrease evidence of astrocyte dysfunction [22]. In addition, a recent study revealed that the serum level of miR-181a is downregulated in PD and it could serve as a potential biomarker for the diagnosis of PD [23]. However, little information is available about the regulatory mechanism of miR-181a on DA neuron apoptosis and autophagy.
Therefore, in the present study, we aimed to investigate the regulatory mechanism of miR-181a on DA neuron apoptosis and autophagy, as well as potential signaling pathways. Human SK-N-SH neuroblastoma cells were incubated with 1-methyl-4-phenylpyridin’ium ion (MPP+) to induce the PD model. The expression of miR-181a was endogenously altered, and then the effects of miR-181a on DA neuron apoptosis and autophagy were analyzed. Our study might provide new insight into novel therapeutic targets for treatment of PD.
Material and Methods
Cell culture
Human SK-N-SH neuroblastoma cells were obtained from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL, Grand Island, New York, USA) containing 10% fetal bovine serum (FBS; Sigma Chemical Co., St. Louis, Missouri), penicillin 100 U/mL, and streptomycin 100 U/mL (Sigma) in an atmosphere of 5% CO2 at 37°C.
Cell treatment and transfection
For cell treatment, SK-N-SH cells were seeded in plates and grown to 80–90% confluency. Thereafter, the cells were incubated with different concentrations of MPP+ (0, 30, 50, 100, 200, or 400 μM), SB203582 (25 μM), or SP600125 (25 μM) for 24 h and collected for further analysis. SB203582 is a selective inhibitor of p38 mitogen-activated protein kinase (MAPK) that inactivates MAPK-activated protein kinase-K2 (MAPKAP K2) [24]. SP600125 is a selective inhibitor of c-Jun N-terminal kinase (JNK) that suppresses the phosphorylation of JNK by competitive binding to the JNK ATP-binding site [25]. For cell transfection, miR-181a mimic, inhibitor, and scramble were purchased from Sangon Biotech (Shanghai, China). Briefly, the cells (2×105/well) were seeded in 96-well plates and then transiently transfected with miR-181a mimic, inhibitor, or scramble according to the manufacturer’s instructions. Cell transfection was performed using the Lipofectamine 2000 according to the manufacture’s protocol (Invitrogen, Carlsbad, California, USA).
Cell apoptosis
Cell apoptosis was determined by flow cytometry using the Annexin V-FITC cell apoptosis kit (Abcam, Cambridge, Massachusetts, USA) according to the manufacturer’s protocol. Briefly, the cells were treated with 0 or 100 μM MPP+ and/or transfected with miR-181a mimic, inhibitor, or scramble for 48 h, and then the cells were cultured with fresh serum-free DMEM medium. Afterwards, the cells were washed three times with phosphate-buffered saline (PBS) buffer (pH 7.4), re-suspended in the staining buffer, and then mixed with annexin-V-FITC (5 μL) and propidium iodide (PI, 5 μL) at room temperature for 15 min. Labeled cells were analyzed by using the FACScan flow cytometry (Becton Dickinson, Franklin Lakes, New Jersey, USA). Each condition was repeated at least 3 times.
Quantitative real time PCR (qRT-PCR) analysis
After 48 h of treatment with different concentrations of MPP+ (0, 30, 50, 100, 200, or 400 μM) and/or transfection with miR-181a mimic, inhibitor, or scramble, total mRNA was isolated from the cells using TRIzol (Invitrogen) according to the manufacturer’s protocol. First-strand complementary DNA (cDNA) was produced using reverse transcriptase (iScript™ cDNA Synthesis Kit; Bio-Rad Laboratories, Hercules, CA, USA). The expression levels of mRNAs were measured by PrimeScript RT Reagent Kit (TaKaRa, Tokyo, Japan) and SYBR green-based quantitative RT-PCR (SYBR Green Master mix; Thermo Scientific, Waltham, Massachusetts, USA) based on the manufacturer’s instructions. The expression levels of miRNA were measured by TaqMan miRNA assays (ABI, Forest City, California). Gene and miRNA expression was normalized by GAPDH and U6 snRNA, respectively. All primers were synthesized by Sangon Biotech. Co., Ltd. (Shanghai, China).
Western blotting
After 48 h of treatment with 0 or 100 μM MPP+ and/or transfection with miR-181a mimic, inhibitor, or scramble, protein was extracted from the cells and the protein concentration was determined by using Bio-Rad protein assay reagent (Bio-Rad). The protein samples (30 μg per lane) were separated on a 10–12% sodium dodecyl sulfate (SDS)-polyacrylamide gel, blotted onto polyvinylidene difluoride membranes (GE Healthcare, Little Chalfont, England), and blocked in 5% non-fat dried milk in Tris-buffered saline with Tween (TBST) for 2 h. Then the membranes were probed with the following primary antibodies overnight at 4°C: anti-LC3 A/B (ab128025, Abcam), anti-Beclin-1 (ab55878, Abcam), anti-phospho-p38 (anti-p-p38; #4511, Cell Signaling Technology Inc., Beverly, Massachusetts, USA), anti-p38 (#8690, Cell Signaling Technology), anti-phospho-JNK (anti-p-JNK; #4668, Cell Signaling Technology), or anti-JNK (#9252, Cell Signaling Technology). The membranes were then incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. The immunoreactive protein bands were analyzed using enhanced chemiluminescence (Pierce, Rockford, Illinois, USA). GAPDH was used as the internal control.
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Student’s t test was performed to analyze the significant difference between two groups. Statistical differences between gene expression were assessed using a one-way variance analysis (ANOVA), followed by Tukey’s multiple comparison test. For measurements of protein expression, post hoc testing was employed when there was a significant difference. Statistical analyses were performed using Graph prism 6.0 software (GraphPad Prism, San Diego, California, USA). P<0.05 was considered as statistically significant.
Results
MiR-181a was downregulated in MPP+-treated SK-N-SH cells
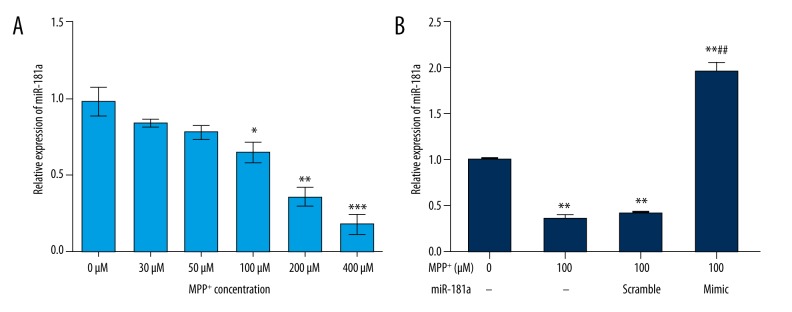
To investigate the functional role of miR-181a in PD, we first evaluated the expression levels of miR-181a in an in vitro MPP+ model of PD. SK-N-SH cells were maintained in different concentrations of MPP+ (0, 30, 50, 100, 200, or 400 μM), and then the expression levels of miR-181a in SK-N-SH cells were assessed by qRT-PCR. As shown in Figure 1A, we observed that the relative expression of miR-181a was gradually decreased with the increase of MPP+ concentration. However, significance was reached by 100 (P<0.05), 200 (P<0.01), and 400 μM (P<0.001) MPP+. The results suggested that miR-181a was downregulated in PD. Then we transfected miR-181a mimic or scramble into MPP+-treated (100 μM) SK-N-SH cells. The untreated cells were regarded as a control group. As expected, the relative levels of miR-181a were significantly decreased by MPP+ and MPP+ + miR-181a scramble compared to the control group (both P<0.05), while the relative levels were statistically increased by miR-181a mimic compared to the control group (P<0.01) or 100 μM MPP+-treated group (P<0.01) (Figure 1B), indicating that all the vectors were successfully transfected into SK-N-SH cells.
Figure 1.
MiR-181a is downregulated in MPP+-treated SK-N-SH cells. The SK-N-SH cells were incubated with different concentrations of MPP+ (0, 30, 50, 100, 200, or 400 μM) and/or transfected with miR-181a mimic or scramble. The expression of miR-181a was then analyzed. Non-treated cells were considered as a control group. (A) The relative expression of miR-181a was gradually decreased with the increase of MPP+ concentration. (B) The relative expression of miR-181a was significantly increased by miR-181a mimic. * P<0.05, ** P<0.01, or *** P<0.01 compared to the control group; ## P<0.01 compared to the MPP+ + miR-181a scramble group. MiR – microRNA; MPP+ – 1-methyl-4-phenylpyridinium ion.
Overexpression of miR-181a inhibited autophagy
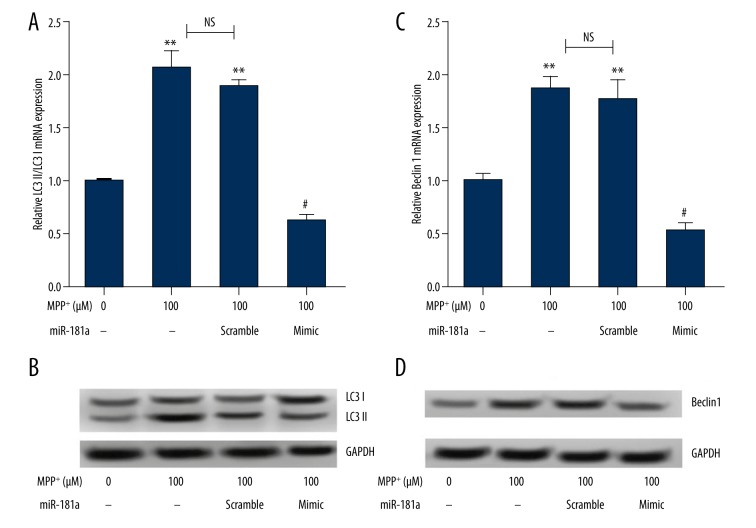
It is well known that autophagy is tightly linked to PD. To explore the effects of miR-181a overexpression on autophagy, the expressions of autophagy protein markers (LC3II, LC3I, and Beclin 1) were detected after transfection with miR-181a mimic by Western blotting. As indicated in Figure 2A, 2B, a significant increase in LC3II/LC3I ratio expressions was found in the MPP+ and MPP+ + miR-181a scramble group compared to the control group (both P<0.01). However, there was no significant difference between the MPP+ group and MPP+ + miR-181a scramble group. Interestingly, we found that the LC3II/LC3I ratio was statistically reduced by overexpression of miR-181a compared to the MPP+ + miR-181a scramble group (P<0.05). In addition, the expression of Beclin 1 revealed results similar to those of the LC3II/LC3I ratio (Figure 2C, 2D). These results demonstrated that overexpression of miR-181a could significantly inhibit autophagy in PD.
Figure 2.
Overexpression of miR-181a inhibits autophagy. The mRNA and protein expressions of LC3II, LC3I, and Beclin 1 were detected after administration with MPP+ and/or transfection with miR-181a mimic or scramble. Non-treated cells were considered as a control group. (A, B) The mRNA and protein expressions of the LC3II/LC3I ratio were statistically decreased by MPP+ + miR-181a mimic. (C, D) The mRNA and protein expressions of Beclin 1 were significantly reduced by MPP+ + miR-181a mimic. ** P<0.01 compared to the control group; # P<0.05 compared to the MPP+ + miR-181a scramble group. MiR – microRNA; MPP+ – 1-methyl-4-phenylpyridinium ion; NS, no significance.
Overexpression of miR-181a reduced neuron apoptosis
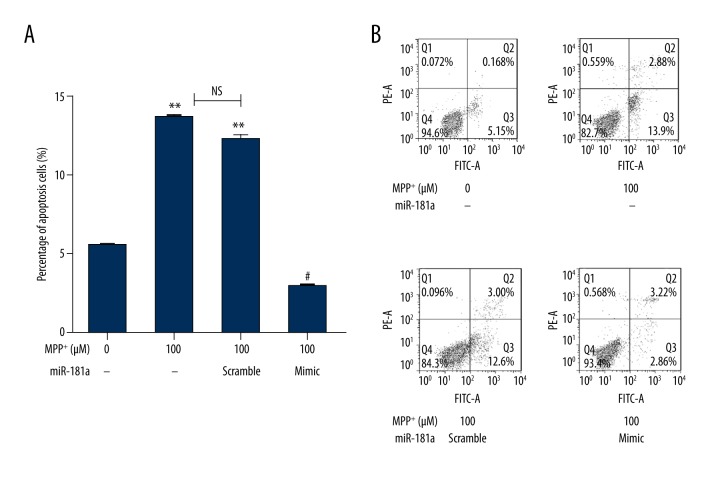
Next, we analyzed the effects of miR-181a overexpression on neuron apoptosis by flow cytometry using the Annexin V-FITC cell apoptosis kit. The results showed that the percentages of cell apoptosis were markedly increased by MPP+ and MPP+ + miR-181a scramble compared to the control group (both P<0.01). No significant differences were observed between the two groups. However, the percentages of cell apoptosis were distinctly decreased by overexpression of miR-181a compared to the MPP+ + miR-181a scramble group (P<0.05) (Figure 3A, 3B). The results indicated that overexpression of miR-181a could significantly prevent neuron apoptosis in PD.
Figure 3.
Overexpression of miR-181a reduces neuron apoptosis. The percentages of cell apoptosis were analyzed after administration with MPP+ and/or transfection with miR-181a mimic or scramble. Non-treated cells were considered as a control group. (A, B) The percentages of cell apoptosis were distinctly decreased by MPP+ + miR-181a mimic. ** P<0.01 compared to the control group; # P<0.05 compared to the MPP+ + miR-181a scramble group. MiR – microRNA; MPP+ – 1-methyl-4-phenylpyridinium ion; NS – no significance.
Overexpression of miR-181a inhibited p38 MAPK/JNK signal activation
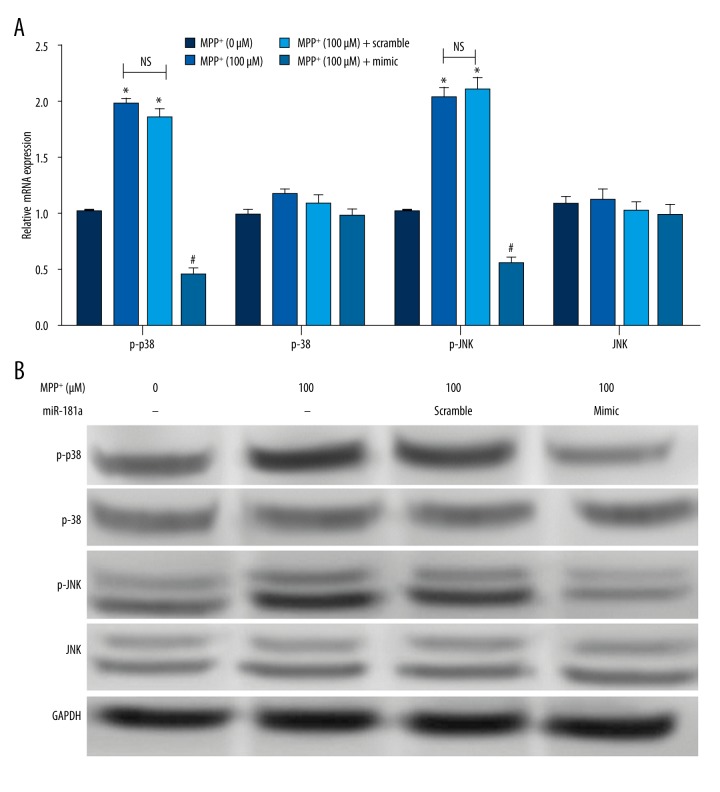
Activation of p38MAPK/JNK has been reported to be associated with PD. Therefore, we analyzed the effects of miR-181a overexpression on p38 MAPK and JNK phosphorylation by qRT-PCR and Western blotting. As demonstrated in Figure 4A, the results revealed that both the relative mRNA expression levels of p-p38 and p-JNK were significantly elevated by MPP+ and MPP+ + miR-181a scramble group compared to the control group (P<0.05). Also, we found that there was no distinct difference in levels of p-p38 and p-JNK between the MPP+ group and MPP+ + miR-181a scramble group. Besides, the levels of p-38 and JNK were not dramatically changed. However, the levels of p-p38 and p-JNK were remarkably lowered by overexpression of miR-181a compared to the MPP+ + miR-181a scramble group (P<0.05). The protein expression of p-p38 and p-JNK showed similar results (Figure 4B). These results demonstrated that overexpression of miR-181a could activate p38 MAPK/JNK pathways.
Figure 4.
Overexpression of miR-181a inhibits p38 MAPK/JNK signal activation. The mRNA and protein expressions of p-p38, p38, p-JNK, and JNK were determined after administration with MPP+ and/or transfection with miR-181a mimic or scramble. Non-treated cells were considered as a control group. (A, B) The mRNA and protein expressions of p-p38 and p-JNK were dramatically decreased by MPP+ + miR-181a mimic, and no significant difference was found in levels of p38 and JNK. * P<0.05 compared to the control group; #P<0.05 compared to the MPP+ + miR-181a scramble group. MiR – microRNA; MPP+ – 1-methyl-4-phenylpyridinium ion; NS – no significance.
Suppression of miR-181a promoted p38 MAPK/JNK signal activation
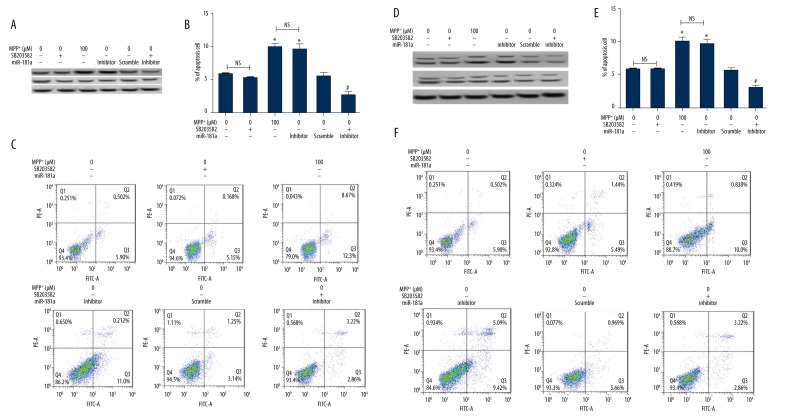
To further confirm whether miR-181a regulates neuron apoptosis by modulating p38 MAPK/JNK pathways, we administered the inhibitor of p38 MAPK (SB203582) and the inhibitor of JNK (SP600125), and then analyzed the effects of miR-181a suppression on neuron apoptosis. As expected, the expression of p-p38 was markedly decreased by its inhibitor SB203582 but was elevated by MPP+ and miR-181a inhibitor compared to the control group. It was noteworthy that the effects of SB203582 on the expression of p-p38 were eliminated by miR-181a inhibitor, indicating that miR-181a inhibitor could activate the p38 MAPK signaling pathway (Figure 5A). Furthermore, we observed that SB203582 had no significant effect on cell apoptosis. The percentages of cell apoptosis were significantly increased by MPP+ and miR-181a inhibitor compared to the control group (P<0.05), but the effects of miR-181a inhibitor on apoptosis were reversed by administration with SB203582 compared to the scramble group (P<0.05) (Figure 5B, 5C), demonstrating that miR-181a inhibitor promotes cell apoptosis by activating the p38 MAPK signaling pathway. Meanwhile, the results showed that the JNK signaling pathway could also be activated by miR-181a inhibitor (Figure 5D). The effects of miR-181a inhibitor on cell apoptosis were also reversed by administration with SP600125 (Figure 5E, 5F). These results suggested that suppression of miR-181a could promote the cell apoptosis by activation of p38 MAPK/JNK pathways.
Figure 5.
Suppression of miR-181a promotes p38 MAPK/JNK signal activation. The inhibitor of p38 MAPK (SB203582) and the inhibitor of JNK (SP600125) were added to the cells, and then the effects of miR-181a suppression on neuron apoptosis were analyzed. Non-treated cells were considered as a control group. (A) The protein expression of p-p38 was inhibited by SB203582 but promoted by MPP+ and miR-181a inhibitor, and the effects of miR-181a inhibitor on p-p38 expression were reversed by SB203582. (B, C) The percentages of cell apoptosis were significantly increased by MPP+ and miR-181a inhibitor, but the effects of miR-181a inhibitor on apoptosis were reversed by SB203582. (D) The protein expression of p-JNK was decreased by SP600125 but increased by MPP+ and miR-181a inhibitor, and the effects of miR-181a inhibitor on p-JNK expression were reversed by SP600125. * P<0.05 compared to the control group; # P<0.05 compared to the scramble group. MiR – microRNA; MPP+ – 1-methyl-4-phenylpyridinium ion; NS – no significance.
Discussion
In the present study, we found that the expression of miR-181a was significantly downregulated in the PD model. Overexpression of miR-181a inhibited the expression of autophagy protein markers (LC3II/LC3I ratio and Beclin 1) and reduced the percentages of cell apoptosis. Further, the results demonstrated that overexpression of miR-181a inhibited the activation of p38MAPK/JNK pathways, while suppression of miR-181a promoted the activation of p38MAPK/JNK pathways. Additionally, suppression of miR-181a statistically induced cell apoptosis, but the effects were prominently alleviated by application of the inhibitor of p38 MAPK (SB203582) and the inhibitor of JNK (SP600125).
Growing evidence suggests that miRNAs play a significant role in the regulation of a variety of basic biological and pathological processes in many brain diseases, including PD. Several miRNAs have been identified to be abnormally expressed in the brain tissues of patients or animals with PD and are responsible for the development of DA neurons [12,26–28]. Thus, many miRNAs are regarded as novel biomarkers for molecular diagnosis and potential therapy targets. Among the miRNAs, miR-181a is a member of miR-181 family, which shows a robust enrichment in the brain [19,29]. It has been reported that miR-181a is increased during maturation of hippocampal neurons and in the infarct core, but decreased in the penumbra after focal ischemia [20,30]. MiR-181a has been established to regulate the functions of synapsis, and miR-181a expression could be induced by DA signaling in primary neurons [20]. Besides, suppression of miR-181a reduced astrocyte dysfunction and increased cornu ammonis (CA1) neuronal survival [22]. Recently, it has been demonstrated that miR-181a is involved in PD [23,31]. However, the clear mechanism for the effects of miR-181a on PD remains to be identified. In the present study, we focused on the functional role of miR-181a in DA neuron apoptosis and autophagy.
Apoptosis and autophagy are basic physiologic processes that are responsible for the maintenance of cellular homeostasis [32]. They are independent processes but inversely related. Emerging evidence supports the view that neuron apoptosis and autophagy play critical roles in the development of PD [8–10,33,34]. MiRNAs have been reported to be involved in PD by regulating the cell apoptosis and autophagy. For example, miR-124 could regulate apoptosis and autophagy processes in the 1-methyl-4-pheny-1, 2, 3, 6-tetrahydropyridine (MPTP) model of PD by targeting Bim [35] and by regulating the adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK)/mammalian target of rapamycin (mTOR) pathway [36]. To confirm the functional role of miR-181a in apoptosis and autophagy, we used MPP+, a frequently used neurotoxin [37], to induce cellular models of PD. SK-N-SH neuroblastoma cells were incubated with MPP+, and then the expression of miR-181a was analyzed. Consistent with Ding et al. [23], the expression of miR-181a in MPP+ treated cells was significantly downregulated compared to the non-treated cells. Subsequently, the effects of overexpression of miR-181a on cell apoptosis and autophagy were determined. As expected, MPP+ caused higher percentages of apoptotic cells, but the effects were alleviated by overexpression of miR-181a, demonstrating the inhibitory effects of miR-181a overexpression on cell apoptosis.
Our results were in line with those of a previous study, in which the results showed that apoptosis was inhibited by overexpression of miR-181a in osteosarcoma cells [38]. In the scenario of autophagy, the expression of LC3II, LC3I, and Beclin 1 was measured after application of MPP+ and transfected with miR-181a mimic. LC3II is the cleaved form of LC3I, and the ratio of LC3II/LC3I demonstrates the degree of autophagosome formation and is also a marker widely used to indicate autophagy [39]. An increase of the LC3II/LC3I ratio reflects active autophagy. Beclin 1 is an essential autophagy-related protein that has been linked to apoptosis and autophagy [40]. In spite of many studies supporting the protective role of autophagy in PD, extreme autophagy activation is related to neuronal loss [41]. Therefore, reduction of autophagy may be of benefit in PD. Our results revealed that the ratio of LC3II/LC3I and the expression of Beclin 1 were statistically downregulated by overexpression of miR-181a compared to the scramble group, indicating the autophagy inhibition was generated bymiR-181a upregulation.
p38 MAPK and JNK are two important components of MAPK cascades, which play significant roles in regulation of diversiform cellular activities such as cell survival, cell proliferation, differentiation, and apoptosis [42]. Recent studies have highlighted that p38 MAPK and JNK are activated by neurotoxicants, stress, and/or inflammation and may be responsible for PD, and that blocking of the p38 MAPK and JNK pathways may help for the development of therapeutic approaches that could benefit PD patients [43,44]. Interestingly, our results showed that miR-181a upregulation caused lower levels of p-p38 and p-JNK. The results implied that overexpression of miR-181a inactivated the p38 MAPK and JNK pathways. The results were partly similar to those of Song et al. [45], in which miR-181a achieved its functionalities by regulating the p38 MAPK pathway. The present study also showed, for the first time to the best of our knowledge, that miR-181a regulated the JNK pathway. To further confirm that the effects of miR-181a on cell apoptosis might be via the p38 MAPK and JNK pathways, we administered the inhibitors of the two pathways, SB203582 and SP600125, and then analyzed the suppression of miR-181a on cell apoptosis. The results revealed that suppression of miR-181a activated the p38 MAPK and JNK pathways, which reversely verified that overexpression of miR-181a inactivated the two pathways. Moreover, we observed that suppression of miR-181a significantly increased the percentages of cell apoptosis; however, these effects were statistically relieved by application of SB203582 and SP600125.
Conclusions
In conclusion, our results suggest that miR-181a is downregulated in MPP+-intoxicated SK-N-SH neuroblastoma cells. Overexpression of miR-181a inhibits autophagy and reduces apoptosis. The effects of miR-181a on autophagy and apoptosis might be achieved by regulation of the p38 MAPK and JNK pathways.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
Source of support: Departmental sources
References
- 1.Gao HM, Jiang J, Wilson B, et al. Microglial activation-mediated delayed and progressive degeneration of rat nigral dopaminergic neurons: Relevance to Parkinson’s disease. J Neurochem. 2002;81:1285–97. doi: 10.1046/j.1471-4159.2002.00928.x. [DOI] [PubMed] [Google Scholar]
- 2.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–22. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 3.Dexter DT, Jenner P. Parkinson disease: From pathology to molecular disease mechanisms. Free Radic Biol Med. 2013;62:132–44. doi: 10.1016/j.freeradbiomed.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci. 2006;7:207–19. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Huang Y, Przedborski S. Oxidative stress in Parkinson’s disease. Ann NY Acad Sci. 2008;1147:93–104. doi: 10.1196/annals.1427.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNaught KSP, Olanow CW. Protein aggregation in the pathogenesis of familial and sporadic Parkinson’s disease. Neurobiol Aging. 2006;27:530–45. doi: 10.1016/j.neurobiolaging.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–97. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 8.Anglade P, Vyas S, Javoy-Agid F, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–32. [PubMed] [Google Scholar]
- 9.Pan T, Kondo S, Le W, Jankovic J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson’s disease. Brain. 2008;131:1969–78. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]
- 10.Nixon RA. Autophagy in neurodegenerative disease: Friend, foe or turncoat? Trends Neurosci. 2006;29:528–35. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson’s disease. J Chem Neuroanat. 2011;42:127–30. doi: 10.1016/j.jchemneu.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouradian MM. MicroRNAs in Parkinson’s disease. Neurobiol Dis. 2012;46:279–84. doi: 10.1016/j.nbd.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Rapp J, Rainone S, Hébert SS. MicroRNAs underlying memory deficits in neurodegenerative disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2016 doi: 10.1016/j.pnpbp.2016.04.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Qiu L, Zhang W, Tan EK, Zeng L. Deciphering the function and regulation of microRNAs in Alzheimer’s disease and Parkinson’s disease. ACS Chem Neurosci. 2014;5:884–94. doi: 10.1021/cn500149w. [DOI] [PubMed] [Google Scholar]
- 16.Vallelunga A, Ragusa M, Di Mauro S, et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Wei L, Wu F, et al. Advances with microRNAs in Parkinson’s disease research. Drug Des Devel Ther. 2013;7:1103–13. doi: 10.2147/DDDT.S48500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Erviti L, Seow Y, Schapira AH, et al. Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell Death Dis. 2013;4:e545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 20.Saba R, Störchel PH, Aksoy-Aksel A, et al. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32:619–32. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouyang Y-B, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12:213–19. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon J-m, Xu L, Giffard RG. Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J Cereb Blood Flow Metab. 2013;33:1976–82. doi: 10.1038/jcbfm.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding H, Huang Z, Chen M, et al. Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Parkinsonism Relat Disord. 2016;22:68–73. doi: 10.1016/j.parkreldis.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Jiang MS, Adams JL, Lee JC. Pyridinylimidazole compound SB 203580 inhibits the activity but not the activation of p38 mitogen-activated protein kinase. Biochem Biophys Res Commun. 1999;263:825–31. doi: 10.1006/bbrc.1999.1454. [DOI] [PubMed] [Google Scholar]
- 25.Bogoyevitch MA, Arthur PG. Inhibitors of c-Jun N-terminal kinases: JuNK no more? Biochim Biophys Acta. 2008;1784:76–93. doi: 10.1016/j.bbapap.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu L, Zhang W, Tan EK, Zeng L. Deciphering the function and regulation of microRNAs in Alzheimer’s disease and Parkinson’s disease. ACS Chem Neurosci. 2014;5:884–94. doi: 10.1021/cn500149w. [DOI] [PubMed] [Google Scholar]
- 27.Filatova E, Alieva AK, Shadrina M, Slominsky P. MicroRNAs: Possible role in pathogenesis of Parkinson’s disease. Biochemistry (Moscow) 2012;77:813–19. doi: 10.1134/S0006297912080020. [DOI] [PubMed] [Google Scholar]
- 28.Hao B, Chen X, Dai D, et al. Bioinformatic analysis of microRNA expression in Parkinson’s disease. Mol Med Rep. 2015;11:1079–84. doi: 10.3892/mmr.2014.2837. [DOI] [PubMed] [Google Scholar]
- 29.Miska EA, Alvarez-Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang Y-B, Lu Y, Yue S, et al. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012;45:555–63. doi: 10.1016/j.nbd.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botta-Orfila T, Morató X, Compta Y, et al. Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson’s disease. J Neurosci Res. 2014;92:1071–77. doi: 10.1002/jnr.23377. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay S, Panda PK, Sinha N, et al. Autophagy and apoptosis: Where do they meet? Apoptosis. 2014;19:555–66. doi: 10.1007/s10495-014-0967-2. [DOI] [PubMed] [Google Scholar]
- 33.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 34.Ghavami S, Shojaei S, Yeganeh B, et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Ye Y, Zhu Z, et al. MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s Disease by targeting to Bim. Brain Pathol. 2016;26(2):167–76. doi: 10.1111/bpa.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong X, Wang H, Ye Y, et al. miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am J Transl Res. 2016;8:2127–37. [PMC free article] [PubMed] [Google Scholar]
- 37.Kalivendi SV, Kotamraju S, Cunningham S, et al. 1-Methyl-4-phenylpyridinium (MPP+)-induced apoptosis and mitochondrial oxidant generation: role of transferrin-receptor-dependent iron and hydrogen peroxide. Biochem J. 2003;371:151–64. doi: 10.1042/BJ20021525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jianwei Z, Fan L, Xiancheng L, et al. MicroRNA 181a improves proliferation and invasion, suppresses apoptosis of osteosarcoma cell. Tumor Biol. 2013;34:3331–37. doi: 10.1007/s13277-013-0902-0. [DOI] [PubMed] [Google Scholar]
- 39.Karim MR, Kanazawa T, Daigaku Y, et al. Cytosolic LC3 ratio as a sensitive index of macroautophagy in isolated rat hepatocytes and H4-II-E cells. Autophagy. 2007;3:553–60. doi: 10.4161/auto.4615. [DOI] [PubMed] [Google Scholar]
- 40.He C, Levine B. The beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–49. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xilouri M, Vogiatzi T, Vekrellis K, et al. Abberant α-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One. 2009;4:e5515. doi: 10.1371/journal.pone.0005515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol cell Biol. 2002;3:663–72. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- 43.Peng J, Andersen J. The Role of c-Jun N-Terminal Kinase (JNK) in Parkinson’s Disease. IUBMB Life. 2003;55:267–71. doi: 10.1080/1521654031000121666. [DOI] [PubMed] [Google Scholar]
- 44.Karunakaran S, Ravindranath V. Activation of p38 MAPK in the substantia nigra leads to nuclear translocation of NF-κB in MPTP-treated mice: Implication in Parkinson’s disease. J Neurochem. 2009;109:1791–99. doi: 10.1111/j.1471-4159.2009.06112.x. [DOI] [PubMed] [Google Scholar]
- 45.Song M-K, Park Y-K, Ryu J-C. Polycyclic aromatic hydrocarbon (PAH)-mediated upregulation of hepatic microRNA-181 family promotes cancer cell migration by targeting MAPK phosphatase-5, regulating the activation of p38 MAPK. Toxicol Appl Pharmacol. 2013;273:130–39. doi: 10.1016/j.taap.2013.08.016. [DOI] [PubMed] [Google Scholar]