Abstract
The molecular complexes formed by specific members of the family of CARMA proteins, the CARD domain-containing adapter molecule BCL10 and MALT1 (CBM complex) represent a central hub in regulating activation of the pleiotropic transcription factor NF-κB. Recently, missense mutations in CARMA2sh have been shown to cause psoriasis in a dominant manner and with high penetrancy. Here, we demonstrate that in human keratinocytes CARMA2sh plays an essential role in the signal transduction pathway that connects pathogen-associated molecular patterns recognition to NF-κB activation. We also find that the serine/threonine kinase ULK2 binds to and phosphorylates CARMA2sh, thereby inhibiting its capacity to activate NF-κB by promoting lysosomal degradation of BCL10, which is essential for CARMA2sh-mediated NF-κB signaling. Remarkably, CARMA2sh mutants associated with psoriasis escape ULK2 inhibition. Finally, we show that a peptide blocking CARD-mediated BCL10 interactions reduces the capacity of psoriasis-linked CARMA2sh mutants to activate NF-κB. Our work elucidates a fundamental signaling mechanism operating in human keratinocytes and opens to novel potential tools for the therapeutical treatment of human skin disorders.
Psoriasis is a debilitating skin disease affecting ~2–3% of human population.1 The disease is considered to have key genetic underpinnings, and genome-wide association studies and meta-analysis have identified more than 40 susceptibility loci for psoriasis.1, 2, 3, 4, 5 Recently, missense mutations in the CARD14/CARMA2/Bimp2 (CARMA2) gene products have been shown to dominantly transmit the psoriatic trait with high penetrancy.6, 7, 8 Mutations in CARMA2 are also found in individuals affected by familial pityriasis rubra pilaris (PRP), a skin disorder phenotypically related to psoriasis.9 When introduced in CARMA2sh, the prominent CARMA2 isoform expressed in the human skin,7, 10, 11 most CARMA2 mutations associated with psoriasis and PRP cause a deregulated, enhanced activity of NF-κB, a transcription factor that catalyzes expression of inflammatory genes.6, 7, 9, 10 Indeed, similarly to classic psoriasis, transcriptomes of psoriatic patients carrying mutations in CARMA2 are significantly enriched in transcripts functioning in inflammatory pathways including the LPS/IL-1 pathways.10 CARMA2sh activates NF-κB through assembly of the CARMA/BCL10/MALT1 (CBM) complex, a molecular complex that in addition to CARMA2sh also includes the paracaspase MALT1 and the adapter molecule BCL10.11, 12, 13 BCL10 is a critical mediator of NF-κB activation in immune cells, and genetic deficiency of BCL10 results in combined immunodeficiency in both humans and mice, due to impaired NF-κB activation following antigen receptor stimulation.14, 15, 16 In addition, in lymphoid cells selective degradation of BCL10 represents an homeostatic mechanism that prevents unrestrained NF-κB signaling.17, 18, 19 More recent reports have also identified BCL10 as a positive regulator of inflammatory cascades in non-immune cells, including the LPS-toll-like receptor (TLR)4-induced pathway of NF-κB activation.20, 21, 22 Here, we find that the serine/threonine kinase ULK2 binds to and phosphorylates CARMA2sh, thereby inhibiting its capacity to activate NF-κB by promoting lysosomal degradation of BCL10. Remarkably, CARMA2sh mutants associated with psoriasis escape ULK2 inhibition. We also show that in human keratinocytes the CBM complex plays an essential role in the signal transduction pathway that links pathogen-associated molecular patterns (PAMPs) recognition on the cell membrane to NF-κB activation.
Results
ULK2 binds to and phosphorylates CARMA2sh
Most of the studies conducted to elucidate the functional role of CARMA2 and its mutated forms associated with psoriasis have been carried out using CARMA2sh,6, 7, 8, 9, 11 the prominent isoform of CARMA2 expressed in the human skin.7, 10 Thus, to search for functional regulators of CARMA2sh we performed a yeast two-hybrid screen using as a bait CARMA2sh fused to the GAL4 DNA-binding domain. A total of 14 yeast clones were isolated that activated the β-galactosidase reporter gene when ~2 × 106 library plasmids were screened. Sequence analysis of these CARMA2sh putative interactors revealed that one isolated plasmid encoded for amino acids Leu805–Val1036 of ULK2, a serine/threonine kinase involved in autophagy.23, 24 As summarized in Table 1, this library clone did not activate the yeast GAL4-reporter genes by itself or when co-expressed with the empty GAL4BD vector, or with a control plasmid. Conversely, it strongly interacted with GAL4BD-CARMA2sh fusion protein.
Table 1. Interaction of CARMA2sh with ULK2 in the yeast two-hybrid assay.
|
Protein fused to GAL4 domain |
Yeast growth on selective media | |
|---|---|---|
| DNA binding | Activating | |
| — | ULK2805–1036 | − |
| Vector | ULK2805–1036 | − |
| FADD | ULK2805–1036 | − |
| CARMA2sh | ULK2805–1036 | +++ |
| CARMA2sh | — | − |
Yeast AH109 was transformed with CARMA2sh fused to the GAL4-activating domain together with the indicated cDNAs fused to GAL4 DNA-binding domain. The cDNA encoding for FADD served as a putative negative control. Interactions were examined by yeast growth on selective media; assays were done for 10 independent transformants. Yeast colonies were scored as positive when a growth developed within 24–36 h; a negative was scored when growth failed to develop within 1 week
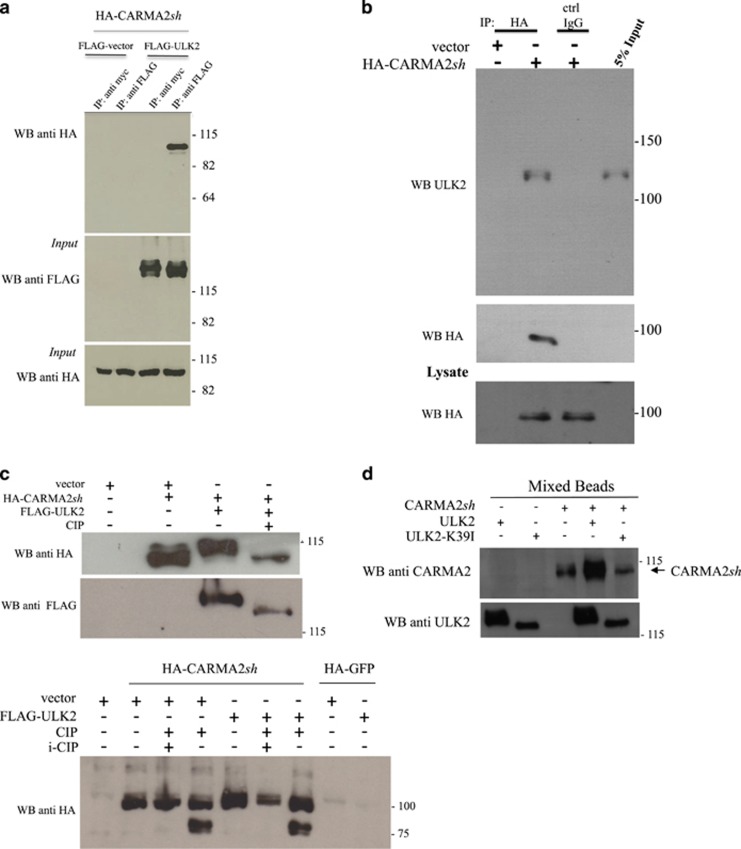
Interaction between CARMA2sh and ULK2 also occurs in mammalian cells, as FLAG-tagged full-length ULK2 coprecipitates with CARMA2sh when lysates prepared from cotransfected HEK293T cells were immunoprecipitated with an anti-FLAG antibody, but not with an anti-myc control antibody (Figure 1a). Also, transfected CARMA2sh coprecipitates endogenous ULK2 in the human keratinocyte HaCaT cell line (Figure 1b).
Figure 1.
ULK2 binds to and phosphorylates CARMA2sh. (a) HEK293T cells were cotransfected with a plasmid encoding for HA-tagged CARMA2sh together with a FLAG-tagged expression vector empty or encoding for ULK2. About 24 h later, lysates were immunoprecipitated with anti-FLAG or anti-myc control antibodies and analyzed for coprecipitating HA-CARMAsh by western blot assay. (b) HaCaT cells were transfected with either an empty plasmid or encoding for HA-tagged CARMA2sh. About 24 h later, cells were lysed and immunoprecipitated with anti-HA or anti-myc control antibodies, and analyzed for coprecipitating endogenous ULK2 protein by western blot assay. (c) HEK293T cells were cotransfected with a plasmid encoding for HA-tagged CARMA2sh together with an expression vector encoding for ULK2. About 24 h later, CARMA2sh expression was analyzed by immunoblot assay probed with anti-HA. Where indicated, the cell lysate was treated with CIP (0.5 U/μg lysate) for 30 min at 37 °C and/or a mixture of phosphatase inhibitors (i-CIP). (d) Lysates from HEK293T separately transfected with the indicated expression plasmids were immunoprecipitated and tested in a mixed beads in vitro kinase assay as described in Material and Methods section. The slower migration in SDS/PAGE of wt ULK2 compared to ULK2K39I is due to ULK2 autophosphorylation (data not shown)
In performing these co-expression experiments, we observed that in SDS-PAGE separations CARMA2sh migrates as a doublet when expressed together with ULK2 in HEK293T cells, suggesting that ULK2 could promote phosphorylation of CARMA2sh (Figure 1c, upper and lower panels). In fact, CARMA2sh doublets disappear when lysates prepared from cotransfected HEK293T cells were treated with calf intestinal phosphatase (CIP) for 30 min immediately before SDS-PAGE separation (Figure 1c, upper panel). For instance, transfected CARMA2sh in SDS/PAGE migrates at an higher molecular mass than that predicted from its amino acid sequence (83.6 kDa predicted versus≈100 kDa observed in SDS/PAGE) (Figure 1c, lower panel, lane 2). When lysates were treated with CIP, a signal appears at ≈80 KDa (lane 4), which corresponds to the predicted molecular mass of CARMA2sh. The ≈80 KDa band disappears again when lysates were treated with a mixture of phosphates inhibitors (lanes 3 and 6), confirming that the ≈100 KDa signal includes phosphorylated forms of CARMA2sh. Of note, co-transfection of ULK2 with CARMA2sh further increases the molecular mass shift of CARMA2sh (lane 5). Overall, these results demonstrate that a consistent fraction of transfected CARMA2sh is present in the cells in a phosphorylated form, and that ULK2 is able to phosphorylate CARMA2sh. Moreover, as shown later in Supplementary Figure 3, CARMA2sh doublets were not observed when CARMA2sh was expressed together with the kinase inactive mutant ULK2K39I.21 Finally, in a mixed beads in vitro kinase assays wt ULK2, but not ULK2K39I, promoted a shift in the SDS/PAGE mobility of CARMA2sh (Figure 1d). Considering altogether these results, we concluded that ULK2 promotes, directly or indirectly, phosphorylation of CARMA2sh.
Next, we tried to map the region of CARMA2sh phosphorylated by ULK2 using deletion mutants of CARMA2sh. The results of these experiments, shown in Supplementary Figure 1, mapped the phosphorylation site of CARMA2sh in the linker region of the protein, within the amino acidic stretch Asp458–Ser500, which contains 10 serine residues and 1 threonine residue. Indeed, a construct encoding for a deleted version of CARMA2sh lacking this region (CARMA2shΔ458-500; predicted molecular mass 79.9 KDa), that we generated and used later in this work, migrates at the expected position in SDS/PAGE separations (Supplementary Figures 3 and 4).
ULK2 represses the NF-κB-inducing activity of wt CARMA2sh, but not that of the non-phosphorylatable or psoriasis-linked CARMA2sh mutants
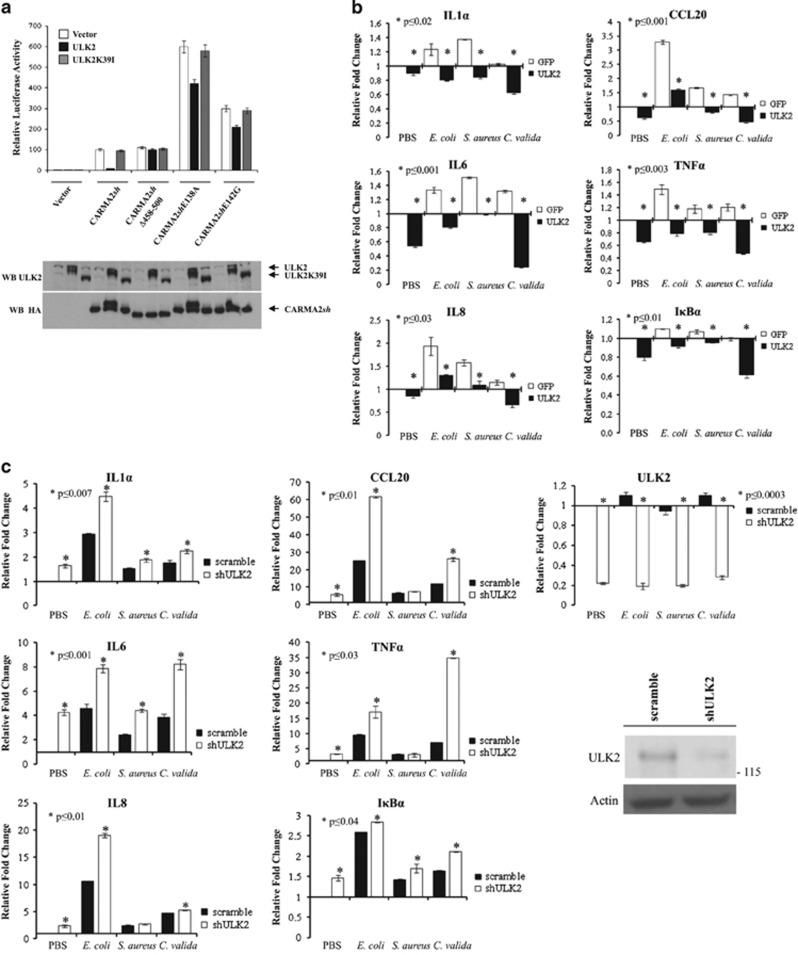
Most of the psoriasis-linked CARMA2sh mutants enhance the transcriptional activity of NF-κB,6, 7, 8, 9, 11 which catalyzes transcription of inflammatory mediators and plays a significant role in the progression of the disease.25 Thus, we tested the effect of ULK2 on the NF-κB-inducing activity of CARMA2sh. Preliminary experiments carried out using an NF-κB-luciferase reporter assay and shown in Figure 2a, indicated that wt ULK2, but not ULK2K39I, dramatically diminishes the NF-κB-inducing activity of CARMA2sh.
Figure 2.
ULK2 represses the NF-κB-inducing activity of CARMA2sh. (a) HEK293T cells were transiently cotransfected with expression vectors encoding for the indicated polypeptides, together with NF-κB-luciferase and β-galactosidase reporter vectors. About 24 h after transfection, cell lysates were prepared and luciferase activity was measured. Data were analyzed by Student's t-test, and a P-value ⩽0.05, indicated with an *, was considered significant. Data shown represent relative luciferase activity normalized against β-galactosidase activity and are representative of at least 10 independent experiments done in triplicate. Lower panel: a fraction of the cell lysate was analyzed by immunoblot assay to monitor protein expression. (b) NHEK were transfected with an expression vector encoding for ULK2 or GFP. About 24 h later, cells were left in PBS or exposed to the indicated heat-killed microorganisms for 16 h, and the expression level of selected NF-κB target genes was monitored by real-time PCR. Graphs show the fold changes respect to the GFP-transfected cells left in PBS. Data were analyzed by Student's t-test, and a P-value ⩽0.05, indicated with an *, was considered significant. Data shown are representative of at least three independent experiments done in triplicate. (c) NHEK cells were infected with lentiviral vectors encoding for shULK2 or a scramble control sequence, and then treated and assayed as in b. Graphs show the fold changes respect to the scramble-infected cells left in PBS. Data shown are representative of at least three independent experiments done in triplicate. Efficacy of ULK2 attenuation by shRNAs was monitored by real-time PCR and immunoblot assay. (d) NHEK were infected with lentiviral vectors encoding for indicated shRNAs or a scramble control sequence and then treated and assayed as in (c). (e) NHEK cells were left in PBS or exposed to heat-killed S. aureus cells for 16 h. Where indicated, the cell lysate was treated with CIP for 30 min at 37 °C and endogenous CARMA2sh expression was analyzed by immunoblot assay
Importantly, in the same assay, the NF-κB-inducing activity of the non-phosphorylatable mutant CARMA2shΔ458-500 was not affected by ULK2 expression, suggesting that CARMA2sh phosphorylation is necessary for ULK2 to operate its negative control on the NF-κB-inducing activity of CARMA2sh. Even more importantly, two of the most recurrent psoriasis-linked mutants, CARMA2shE138A and CARMA2shE142G, which are both stronger activators of NF-κB compared to wt CARMA2sh,6, 7, 8 were only slightly affected by ULK2 expression.
ULK2 represses expression of inflammatory genes in human keratinocytes exposed to PAMPs
Subsequently, we investigated the negative effect of ULK2 on the NF-κB-inducing activity of CARMA2sh in a more physiological experimental system. Given the evidence that CARMA2sh is predominantly expressed in the human skin,7, 10 we decided to use normal human epidermal keratinocytes (NHEK) for a panel of experiments aimed at defining a physiological NF-κB activation pathway dependent on CARMA2sh and inhibited by ULK2. At this point, however, we faced a significant issue. In fact, although it is known that CARMA1 intervenes in the signal transduction pathway that leads to NF-κB activation in B and T lymphocytes following antigen receptor stimulation,12, 13, 14, 15 and that CARMA3 controls NF-κB activation following G-coupled protein receptors triggering in a variety of cell types,12, 13, 14, 15 it is currently not known from which membrane receptor(s) signaling, if any, CARMA2sh triggers NF-κB activation. While testing a number of different stimuli, including several inflammatory cytokines such as TNFα and IL-1β, it has not escaped our notice that a significant repression of inflammatory NF-κB target genes was observed when NHEK-expressing ULK2 were exposed to heat-killed bacterial (Escherichia coli and Staphylococcus aureus) or fungi (Chorizanthe valida) cells (Figure 2b). Heat-killed bacterial and fungi cells display PAMPs and activate NF-κB upon agonistic binding to pattern recognition receptors, including members of the TLRs family expressed on human keratinocytes.26, 27 Consistently with the experiments shown in Figure 2a, short interfering (sh) RNA targeting ULK2 enhanced the expression level of these inflammatory NF-κB target genes in NHEK exposed to the same stimuli (Figure 2c). Thus, in human keratinocytes ULK2 regulates NF-κB activation ensuing from PAMPs recognition, possibly acting on the CBM complex-mediated signaling.
The CARMA2/BCL10/MALT1 complex connects PAMPs recognition to NF-κB activation
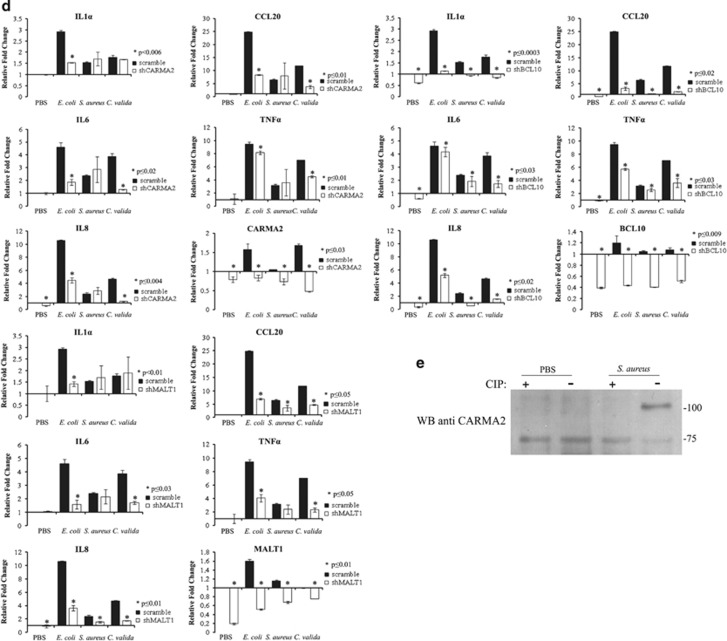
The inhibitory effect on NF-κB activation exerted by ULK2 in NHEK exposed to PAMPs could be due either to an ULK2-mediated suppression of the NF-κB-inducing activity of CARMA2sh, or to an ULK2-sensitive but CARMA2sh-independent mechanisms. To test for this, we used shRNA to individually deplete NHEK of the CBM complex components, id est CARMA2, BCL10 and MALT1. As shown in Figure 2d, abrogation of either CARMA2sh or BCL10 or MALT1 significantly impairs expression of NF-κB target genes in NHEK exposed to PAMPs. Remarkably, NHEK exposure to heat-killed S. aureus promotes a shift in the SDS-PAGE mobility of endogenous CARMA2sh, which was abolished by phosphatase treatment (Figure 2e). These data clearly indicate that in human keratinocytes the CBM complex participates in the signal transduction pathway that connects PAMPs recognition to NF-κB activation. Also, together with the evidences shown before, these experiments indicate that ULK2 negatively regulates this pathway, whereas the two psoriasis-linked CARMA2sh mutants tested escape such a negative control. This observation is particularly remarkable as TLRs activation is considered to play an important role in the pathophysiology of psoriasis.28, 29
ULK2 promotes lysosomal degradation of BCL10
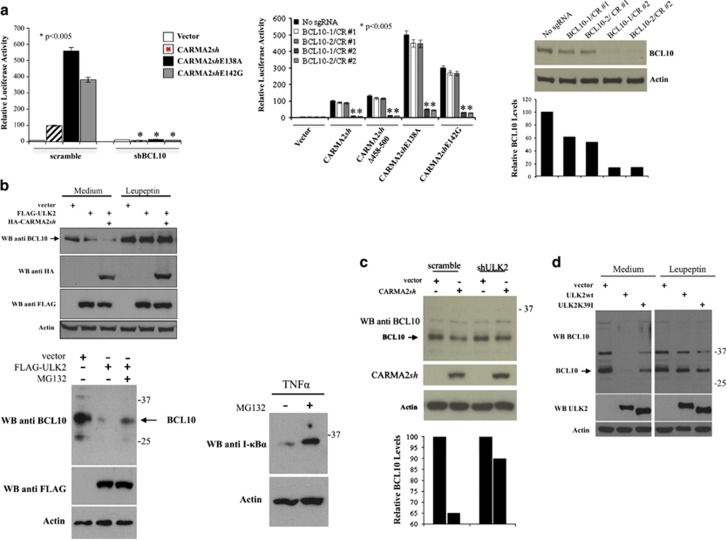
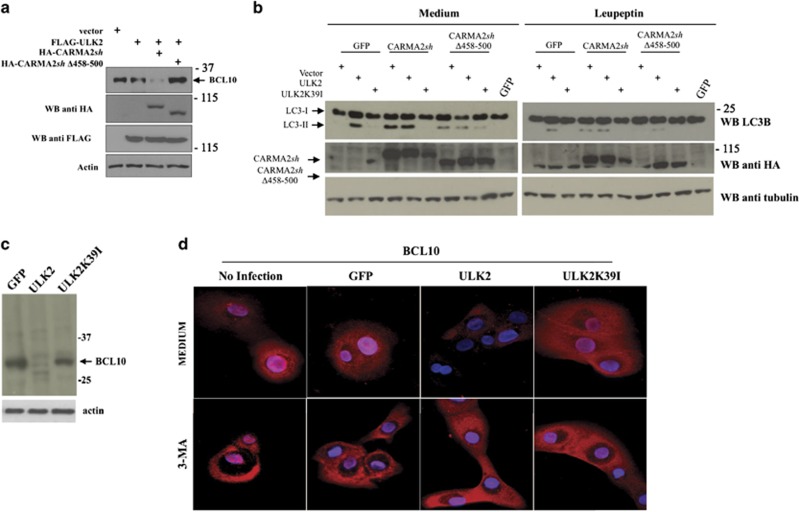
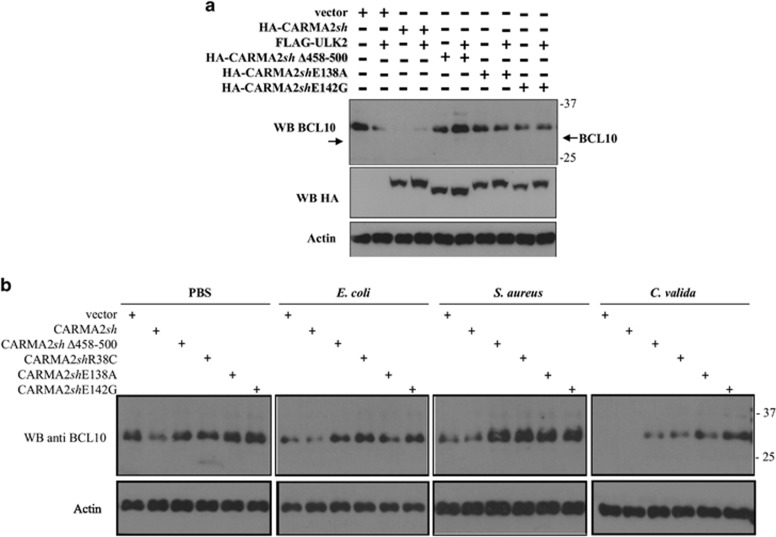
The NF-κB-inducing activity of both wt- and psoriasis-linked CARMA2sh mutants requires BCL10, as assessed by experiments based on shRNA-mediated depletion of BCL10 (Figure 3a, left panel) and disruption of the BCL10 locus through CRISPR/Cas9/sgRNA technology in HEK293T cells (Figure 3a, right panel). Moreover, in the BCL10-targeted cell lines BCL10-1/CR#2 and BCL10-2/CR#2 re-introduction of a plasmid-encoded BCL10 fully restores the NF-κB-inducing activity of CARMA2sh (data not shown). Therefore, we explored the possibility that ULK2 could inhibit the NF-κB-inducing activity of CARMA2sh by regulating the expression level of BCL10. Indeed, as shown in Figure 3b, in HEK293T cells expression of CARMA2sh, which mimics activation of the CBM complex with consequent NF-κB activation, results in a reduction of endogenous BCL10 protein level, which was further accentuated by co-expressed ULK2. BCL10 reduction was rescued by the lysosomal inhibitor leupeptin, but not by the proteasomal inhibitor MG132 (Figure 3b upper and lower panels). Consistently, the lysosomal inhibitor 3-methyladenine (3-MA) rescued the inhibitory effect of ULK2 on the NF-κB-inducing activity of CARMA2sh (Supplementary Figure 2). Reduction of endogenous BCL10 following CARMA2sh expression requires the kinase activity of ULK2, as it was not observed when either shRNA targeting ULK2 or ULK2K39I were used (Figures 3c and d). ULK2-mediated reduction of BCL10 requires phosphorylation of CARMA2sh, as it was not observed when the mutant CARMA2shΔ458-500 was used (Figure 4a).
Figure 3.
ULK2 promotes degradation of BCL10. (a) Left panel: HEK293T cells infected with a lentiviral vector expressing a shRNA targeting BCL10 or a scramble sequence were transiently cotransfected with expression vectors encoding for the indicated polypeptides, together with NF-κB-luciferase and β-galactosidase reporter vectors. About 24 h after transfection, cell lysates were prepared and luciferase activity was measured. Data shown represent relative luciferase activity normalized against β-galactosidase activity and are representative of at least 10 independent experiments done in triplicate. Data were analyzed by Student's t-test, and a P-value ⩽0.05, indicated with an *, was considered significant. Right panel: two independent HEK293T lines (BCL10-1/CR#2 and BCL10-2/CR#2) stably harboring Cas9 and a sgRNA targeting BCL10 were used for the NF-κB luciferase assay described before. The cell lines BCL10-1/CR#1 and BCL10-2/CR#1, harboring an unproductive BCL10 sgRNA, were used as a control. Lower panel: immunoblot and densitometric analysis of BCL10 expression level in BCL10-targeted HEK293T cell lines. (b) HEK293T cells were transiently transfected with the indicated expression vectors and after 24 h treated or not with lysosomal (Leupeptin, 10 μM) or proteasomal (MG132, 10 μM) inhibitors 1 h before lysis. Endogenous BCL10 expression was assessed by immunoblot assay. Lower panel: MG132 efficacy was monitored by assessing proteasomal degradation of IκBα following TNFα stimulation (20 ng/ml for 30 min). (c–d) HEK293T cells expressing the indicated shRNAs and/or polypeptides were assessed for endogenous BCL10 expression by western blot assay
Figure 4.
ULK2 promotes degradation of BCL10. (a) HEK293T cells were transiently transfected with expression vectors encoding for the indicated polypeptides. About 24 h later, cell lysates were prepared and monitored for endogenous BCL10 expression by immunoblot assay. (b) HEK293T cells were transiently transfected with expression vectors encoding for the indicated polypeptides. About 24 h after transfection, cells were left untreated or treated with the lysosome inhibitor leupeptin (10 μM) for 1 h. Cell lysates were then prepared and monitored for conversion of LC3-I to LC3-II by immunoblot assay. (c) NHEK were transfected with an expression vector encoding for wt ULK2 or the kinase-dead mutant ULK2K39I. About 16 h later, cell lysates were prepared and BCL10 expression was determined by immunoblot assay. (d) Immunofluorescence analysis of BCL10 in NHEK infected with lentiviral vectors expressing the indicated polypeptides and left in medium alone or medium supplemented with the autophagy inhibitor 3-MA (5 mM). Data shown are representative of three independent experiments
Together with the experiments done in the presence of lysosomal inhibitors, examination of LC3 conversion from LC3-I to LC3-II by immunoblot assay further pointed to a autophagic route of BCL10 triggered by wt ULK2, but not ULK2K39I, and promoted by wt CARMA2sh but not by CARMA2shΔ458-500 (Figure 4b). Finally, reduction of endogenous BCL10 was also observed in NHEK upon expression of wt ULK2 but not ULK2K39I, and it was rescued by the lysosomal inhibitor 3-MA (Figures 4c and d). Thus, ULK2 negatively regulates the CBM complex signaling, most likely by promoting lysosomal degradation of BCL10.
Psoriasis-linked CARMA2sh mutants fail to promote BCL10 degradation
Next, we investigated for a mechanism that would explain why CARMA2shE138A and CARMA2shE142G mutants elude the ULK2 negative control on NF-κB activation. First of all, we examined whether CARMA2shE138A and CARMA2shE142G mutants are still able to associate to and be phosphorylated by ULK2. The experiments shown in Supplementary Figure 3 indicate that, in transfection experiments, the non-phosphorylatable mutant CARMA2shΔ458-500 and the psoriasis-linked CARMA2shE138A and CARMA2shE142G mutants are all still able to co-immunoprecipitate with ULK2. In addition, as judged by a shift in the SDS/PAGE mobility, both CARMA2shE138A and CARMA2shE142G mutants appear to be phosphorylated by ULK2 (Supplementary Figure 3). Thus, phosphorylation of CARMA2sh by ULK2 is required but not sufficient for ULK2-mediated inhibition of the NF-κB-inducing activity of CARMA2sh. Thus, we explored the possibility that CARMA2shE138A and CARMA2shE142G might fail to promote degradation of BCL10. Indeed, reduction of endogenous BCL10 protein was not observed when the psoriasis-linked mutants CARMA2shE138A or CARMA2shE142G were expressed with ULK2 in NHEK (Figure 5a).
Figure 5.
Psoriasis-linked CARMA2sh mutants fail to promote BCL10 degradation. (a) NHEK cells were transiently transfected with expression vectors encoding for the indicated polypeptides and analyzed for endogenous BCL10 expression by immunoblot assay. (b) NHEK infected with retroviral vectors encoding for the indicated polypeptides were exposed to the indicated stimuli for 4 h and the expression level of BCL10 was determined by immunoblot experiments. Data shown are representative of three independent experiments done in triplicate
We then verified the possibility that the inability of CARMA2shE138A and CARMA2shE142G to promote degradation of BCL10 was somehow associated with their higher capacity to activate NF-κB. For this, we tested an additional psoriasis-linked mutant, CARMA2shR38C, which is incapable to activate NF-κB.6 However, as shown in Supplementary Figure 4, also CARMA2shR38C is unable to support degradation of BCL10.
Moreover, following exposure to PAMPs, BCL10 degradation could be readily observed in mock-transfected NHEK (Figure 5b), or NHEK-expressing wt CARMA2sh, but not in NHEK expressing the mutants CARMA2shΔ458-500, CARMA2shE138A or CARMA2shE142G (Figure 5b). Analysis of BCL10 mRNA expression in NHEK did not reveal any significant change upon ULK2 or CARMA2sh isoforms expression (data not shown), confirming that ULK2-mediated reduction of BCL10 occurs at protein level. Altogether, these data indicate that ULK2-mediated lysosomal degradation of BCL10 represents an intrinsic homeostatic mechanism that restricts NF-κB signaling in keratinocytes, and the psoriasis-linked CARMA2sh mutants we tested failed to promote such a constraining mechanism.
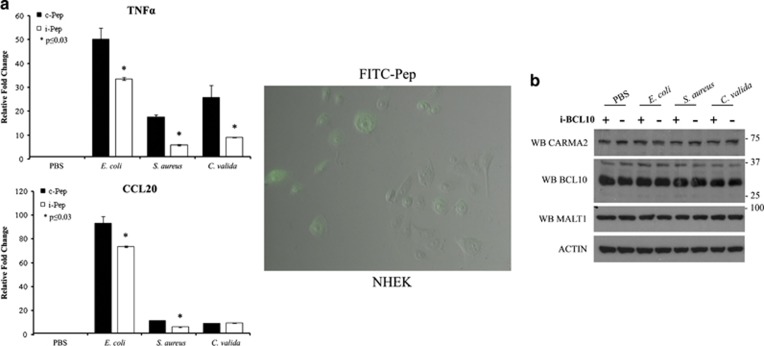
Overall, the data shown above indicate that BCL10 is a key regulator of the NF-κB-inducing activity of wt- and psoriasis-associated CARMA2sh mutants. Thus, we tested whether liposome-assisted delivery of a BCL10 inhibitory peptide30 could reduce NF-κB activation in NHEK exposed to PAMPs. Indeed, the expression level of two monitored NF-κB target genes was partially but significantly diminished when NHEK were treated with a BCL10 inhibitory peptide, but not a control peptide (Figure 6).
Figure 6.
(a) BCL10 inhibitory peptide represses NF-κB signaling upon PAMPs recognition in NHEK. NHEK were liposome-delivered with a BCL10 inhibitory peptide (i-BCL10) or a control scrambled peptide (c-BCL10) and exposed to PAMPs for 24 h. Expression level of the indicated NF-κB responsive genes was determined by real-time PCR. Data shown are representative of six independent experiments done in triplicate. Right panel: FITC-conjugated i-BCL10 peptide was used to monitor delivery efficiency (≅50%). (b) The expression levels of BCL10, MALT1 and CARMA2sh in NHEK left untreated or i-BCL10-delivered were assessed by western blot
Discussion
In this paper we report the identification of ULK2 as a kinase that promotes phosphorylation of CARMA2sh, very likely in the linker region of the protein. This evidence is of particular interest, as it is already known that phosphorylation events occurring in the linker region of CARMA1 control its functional activity. In lymphoid cells, PKC-mediated phosphorylation of the linker region of CARMA1 controls NF-κB activation by triggering a shift from an inactive to an active CARMA1 conformer.31, 32, 33 Thus, a CARMA1 mutant in which Ser552 is mutated fails to mediate TCR-induced NF-κB activation in CARMA1-deficient T cells, whereas deletion of the linker results in constitutive, receptor- and PKC-independent NF-κB activation. As a consequence, constitutive activation of NF-κB driven by mutations in the linker region of CARMA1 is considered to be the molecular basis of some B-cell malignancies.34, 35 We find now that phosphorylation of the linker region in CARMA2sh is somehow also required to trigger the subsequent lysosomal degradation of BCL10, that is, as a signal necessary to terminate CBM complex-mediated activation of NF-κB. Further studies are needed to verify if such a regulatory mechanism applies for CARMA1 as well, as lysosomal degradation of BCL10 has been proposed as an NF-κB signal-off mechanism in activated lymphocytes.17, 18, 19 Our prediction would be that the constitutive NF-κB activity resulting from the linker mutants of CARMA1 also derives from failure to degradate BCL10. In fact, as the NF-κB-inducing activity of CARMA1 requires BCL10, either these mutants do not support degradation of BCL10, or, vice versa, they would be unable to produce constitutive NF-κB activation.
We also show that at least three CARMA2sh mutants associated with genetic psoriasis fail to promote degradation of BCL10, despite being phosphorylated by ULK2. Thus, our data indicate that although phosphorylation of CARMA2sh is a necessary signal to initiate degradation of BCL10, possibly further components must be recruited for selective BCL10 degradation to take place. Much of the information available on ULK2 comes from studies conducted on its close family member ULK1, which represents a central component of the autophagy regulatory complex.23, 36 Although derived from studies on ULK1, these data are perfectly consistent with the evidence shown in our work, pointing to an essential role for ULK2 in triggering lysosomal degradation of BCL10 as a mechanism for quenching NF-κB-activating stimuli conveyed through the CBM complex.
We also demonstrated that a CBM complex comprising CARMA2sh in human keratinocytes transduces NF-κB signaling upon PAMPs recognition on the cell membrane. Evidence in this direction, at least for BCL10 and MALT1, already exist in the literature;20, 21, 22 however, our discovery made in keratinocytes is particularly evocative, either for the historical association between the onset of psoriasis and bacterial infections, either for more recent associations between genetic variants of TLR4, a major player in PAMPs recognition and psoriasis.37, 38–39 Also, this observation is consistent with three recent papers published while this work was in preparation, showing that the proteolytic activity of MALT1 is required for CARMA2sh-induced activation of NF-κB.40, 41, 42
In conclusion, here we have shown that the CBM complex has an essential role in the activation of the NF-κB pathway following human keratinocytes exposure to PAMPs, and that ULK2-mediated degradation of BCL10 represents a mechanism through which this signaling is turned off. Degradation of BCL10 by ULK2 requires phosphorylation of CARMA2sh, and is prevented by several psoriasis-linked CARMA2sh mutations. Thus, the inflammatory phenotype observed in psoriatic skin may result from unrestrained NF-κB signaling due to deficiency of lysosomal BCL10 degradation. Certainly, our data do not exclude the possibility that in addition to the lysosomal pathway also other posttranscriptional mechanisms may contribute to degradation of BCL10. Indeed, as we show in the present work, targeting of BCL10 may represent a novel approach for the treatment of this debilitating disease.
Materials and methods
Two-hybrid screening
The two-hybrid screening was performed using the Matchmaker system (Clontech, Mountain View, CA, USA) as previously described.43 Briefly, yeast strain AH109 GAL4−/− was first transformed with pGBKT7 plasmids carrying the CARMA2sh cDNA bait fused with DBD of GAL4 using lithium acetate/PEG 3000 procedure. Transformant colonies were selected on synthetic dropout plates lacking tryptophan. Expression of bait fusion proteins was assessed by immunoblot analysis. For library screening, yeast AH109 expressing GAL4DBD-CARMA2sh was transformed with a human fetal brain cDNA library cloned in pACT2 vector (Clontech) in fusion with GAL4TAD. About 2 × 106 clones were screened for interaction with GAL4DBD-CARMA2sh using selective growth on minimal medium lacking nutrients whose biosynthesis is mediated by genes under control of the GAL4 transcriptional activity.
Cell culture, plasmids and antibodies
HEK293T and HaCaT cells were obtained from ATCC and cultured in Dulbecco's modified Eagle's medium supplemented with 10% FCS. NHEK were purchased from Lonza (Basel, Switzerland) and cultured according to the provided instructions. HEK293T were transfected by calcium phosphate precipitation; NHEK were transfected using DreamFect Gold Transfection Reagent (OZ Biosciences, Marseille, France) according to the manufacturer's instruction. HaCaT were transfected with Lipofectamine 3000. Retroviral infections were carried out as previously described.44 CARMA2sh and ULK2 mutants were generated through PCR-mediated methods and confirmed by sequencing. Lentiviral vectors expressing shRNAs targeting ULK2 were a gift from Dr. R Shaw (Addgene plasmid #27634, Cambridge, MA, USA). The list of primers and oligos used for this study is shown in the Supplementary Figure 5.
Sources of antisera and monoclonal antibodies were the following: anti-FLAG, anti-β-actin, anti-ULK2, Sigma-Aldrich (St. Louis, MO, USA); anti-CARMA1, Santa Cruz Biotechnology (Dallas, TX, USA). Antisera against BCL10 and CARMA2 were generated in our laboratory and were previously described.11, 45
To disrupt BCL10 gene, a CRISPR/Cas9/single guide (sg) RNA system was designed.46 The sg sequences were cloned into LentiCRISPRv2 containing a Cas9 expression cassette (Addgene) and transduced into HEK293T cells. After puromycin (3 μg/ml) selection, cells stably expressing Cas9/BCL10 sgRNA were isolated. The efficiency of BCL10 knockout was determined by immunoblotting.
Immunoblot analysis and coprecipitation
Cell lysates were made in lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.2, 1% NP40, 2 mM EDTA and a mixture of protease inhibitors). Proteins were separated by SDS-PAGE, transferred onto nitrocellulose membrane and incubated with primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Piscataway, NJ, USA). Blots were developed using the ECL system (Amersham Biosciences). For coimmunoprecipitation experiments, cells were lysed in lysis buffer and immunocomplexes were bound to protein A/G (Roche, Basel, Switzerland), resolved by SDS-PAGE and analyzed by immunoblot assay. All immunoblots were done at least three times using different biological material as sources. Phosphatase Inhibitor Cocktail was purchased from Sigma and used according to the instructions provided.
Luciferase assay
To assess for NF-κB activation state, HEK293T cells were transfected with the indicated plasmidic DNAs together with pNF-κB-luc (Clontech) in 6-well plates. Twenty-four hours after transfection, luciferase activity was determined with Luciferase Assay System (Promega, Madison, WI, USA). Plasmids expressing RSV-β-galactosidase or TK-Renilla were used in transfection mixtures in order to normalize for efficiency of transfection.
Mixed beads in vitro kinase assay
HEK293T cells, separately transfected with plasmids encoding for CARMA2 polypeptides and wild-type or mutant ULK2, were lysed in ice-cold TNTE buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0,3% (vol/vol) Triton X-100, 5 mM EDTA) containing complete protease inhibitor cocktail (Roche), 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 30 nM okadaic acid, 2 mM sodium pyrophosphate. Lysates cleared by centrifugation were incubated with anti-FLAG monoclonal M2 antibody (Sigma Aldrich) for 1 h at room temperature and then washed three times with TNTE buffer. Immunoprecipitates were further washed once with kinase reaction buffer (KRB) (20 mM HEPES, pH 7.5, 20 mM MgCl2, 25 mM β-glycerophosphate, 2 mM DTT, 100 μM sodium orthovanadate and 30 nM okadaic acid) and then mixed and incubated at a final volume of 20 μl in KRB containing 20 μM ATP at 30 °C for 30 min. Reactions were stopped with the addition of 3% SDS sample buffer during heating at 65 °C for 5 min. Mixed beads reaction product were resolved by SDS-PAGE and analyzed by immunoblot assay.
Real-time PCR
Total RNA was isolated from cells or tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The reverse transcriptase reaction was performed using 1 μg of total RNA in a 20 μl reaction and 1 μl of the resulting cDNA was used in the subsequent amplification step along with 300 nM of each primer. The geometric mean values of β-actin and succinyl-CoA synthetase β-subunit fragment were used as normalization factors. The relative transcription level was calculated by using the ΔΔCt method. Real-time PCR reactions were performed in triplicate by using the SYBR Green PCR Master Mix (Qiagen, Hilden, Germany) in a 7900HT system (Applied Biosystems, Foster City, CA, USA).
Acknowledgments
This publication was made possible by a NPRP award (NPRP 7-1189-3-304) from the Qatar National Research Fund (a member of The Qatar Foundation).
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Disclaimer
The statements made herein are solely the responsibility of the authors.
Edited by E Candi
The authors declare no conflict of interest.
Supplementary Material
References
- Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: a comprehensive review. J Autoimmun 2015; 64: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Jin X, Li Y, Jiang H, Tang X, Yang X et al. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet 2014; 46: 45–50. [DOI] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 2012; 44: 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sun L, Yin X, Gao J, Sheng Y, Xu J et al. Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat Commun 2015; 6: 6793–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun 2015; 6: 7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Cao L, Roberson ED, Duan S, Helms CA, Nair RP et al. Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasis. Am J Hum Genet 2012; 90: 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Cao L, Roberson ED, Pierson KC, Yang CF, Joyce CE et al. PSORS2 is due to mutations in CARD14. Am J Hum Genet 2012; 90: 784–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berki DM, Liu L, Choon SE, Burden AD, Griffiths CE, Navarini AA et al. Activating CARD14 mutations are associated with generalized pustular psoriasis but rarely account for familial recurrence in psoriasis vulgaris. J Invest Dermatol 2015; 135: 2964–2970. [DOI] [PubMed] [Google Scholar]
- Fuchs-Telem D, Sarig O, van Steensel MA, Isakov O, Israeli S, Nousbeck J et al. Familial pityriasis rubra pilaris is caused by mutations in CARD14. Am J Hum Genet 2012; 91: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden JL, Lewis SM, Pierson KC, Suárez-Fariñas M, Lentini T, Ortenzio FS et al. CARD14 expression in dermal endothelial cells in psoriasis. PLoS ONE 2014; 9: e111255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudiero I, Zotti T, Ferravante A, Vessichelli M, Vito P, Stilo R. Alternative splicing of CARMA2/CARD14 transcripts generates protein variants with differential effect on NF-κB activation and endoplasmic reticulum stress-induced cell death. J Cell Physiol 2011; 226: 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudiero I, Vito P, Stilo R. The three CARMA sisters: so different, so similar: a portrait of the three CARMA proteins and their involvement in human disorders. J Cell Physiol 2014; 229: 990–997. [DOI] [PubMed] [Google Scholar]
- Blonska M, Lin X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res 2011; 21: 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol 2010; 2: a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell 2001; 104: 33–42. [DOI] [PubMed] [Google Scholar]
- Torres JM, Martinez-Barricarte R, García-Gómez S, Mazariegos MS, Itan Y, Boisson B et al. Inherited BCL10 deficiency impairs hematopoietic and nonhematopoietic immunity. J Clin Invest 2014; 124: 5239–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity 2012; 36: 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Lopez T, Israël A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. Proc Natl Acad Sci USA 2007; 104: 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol Cell Biol 2004; 24: 3860–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dong W, Chen L, Xiang R, Xiao H, De G et al. BCL10 mediates lipopolysaccharide/toll-like receptor-4 signaling through interaction with Pellino2. J Biol Chem 2004; 279: 37436–37444. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Borthakur A, Pant N, Dudeja PK, Tobacman JK. BCL-10 induces lipopolysaccharide induced activation of NF-κB and IL-8 in human intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2007; 293: G429–G437. [DOI] [PubMed] [Google Scholar]
- Fischer KD, Tedford K, Wirth T. New roles for Bcl10 in B-cell development and LPS response. Trends Immunol 2004; 25: 113–116. [DOI] [PubMed] [Google Scholar]
- Alers S, Löffler AS, Wesselborg S, Stork B. The incredible ULKs. Cell Commun Signal 2012; 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 2009; 29: 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldminz AM, Au SC, Kim N, Gottlieb AB, Lizzul PF. NF-κB: an essential transcription factor in psoriasis. J Dermatol Sci 2013; 69: 89–94. [DOI] [PubMed] [Google Scholar]
- Lebre MC, van der Aar AM, van Baarsen L, van Capel TM, Schuitemaker JH, Kapsenberg ML et al. Human keratinocytes express functional toll-like receptor 3, 4, 5, and 9. J Invest Dermatol 2007; 127: 331–341. [DOI] [PubMed] [Google Scholar]
- Miller LS. Toll-like receptors in skin. Adv Dermatol 2008; 24: 71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInturff JE, Modlin RL, Kim J. The role of toll-like receptors in the pathogenesis and treatment of dermatological disease. J Invest Dermatol 2005; 125: 1–8. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, O'Neill LA. The role of the interleukin-1/toll-like receptor superfamily in inflammation and host defence. Microbes Infect 2000; 2: 933–943. [DOI] [PubMed] [Google Scholar]
- Marasco D, Stilo R, Sandomenico A, Monti SM, Tizzano B, de Capua A et al. Generation and functional characterization of a BCL10-inhibitory peptide that represses NF-kappaB activation. Biochem J 2009; 422: 553–561. [DOI] [PubMed] [Google Scholar]
- Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B et al. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB Activation. Immunity 2005; 23: 575–585. [DOI] [PubMed] [Google Scholar]
- Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-García ME, Ovechkina YL et al. Phosphorylation of the CARMA1 linker controls NF-κB activation. Immunity 2005; 23: 561–574. [DOI] [PubMed] [Google Scholar]
- Rueda D, Thome M. Phosphorylation of CARMA1: the link(er) to NF-κB activation. Immunity 2005; 23: 551–553. [DOI] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009; 459: 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 2008; 319: 1676–1679. [DOI] [PubMed] [Google Scholar]
- Gallagher LE, Williamson LE, Chan EY. Advances in autophagy regulatory mechanisms. Cells 2016; 5: E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T, Kanda T, Sato K, Takaishi M, Nakajima K, Yamamoto M et al. Cathepsin K is involved in development of psoriasis-like skin lesions through TLR-dependent Th17 activation. J Immunol 2013; 190: 4805–4811. [DOI] [PubMed] [Google Scholar]
- Wohn C, Ober-Blobaum JL, Haak S, Pantelyushin S, Cheong C, Zahner SP et al. Langerinneg conventional dendritic cells produce IL-23 to drive psoriatic plaque formation in mice. Proc Natl Acad Sci USA 2013; 110: 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RL, Hébert HL, Massey J, Bowes J, Marzo-Ortega H, Ho P et al. Association of toll-like receptor 4 (TLR4) with chronic plaque type psoriasis and psoriatic arthritis. Arch Dermatol Res 2016; 308: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes A, O'Sullivan PA, Breyer F, Ghose A, Cao L, Krappmann D et al. Psoriasis mutations disrupt CARD14 autoinhibition promoting BCL10-MALT1-dependent NF-κB activation. Biochem J 2016; 473: 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Grondona P, Maier T, Brändle M, Schönfeld C, Jäger G et al. MALT1 protease activity controls the expression of inflammatory genes in keratinocytes upon zymosan stimulation. J Invest Dermatol 2016; 136: 788–797. [DOI] [PubMed] [Google Scholar]
- Afonina IS, Van Nuffel E, Baudelet G, Driege Y, Kreike M, Staal J et al. The paracaspase MALT1 mediates CARD14-induced signaling in keratinocytes. EMBO Rep 2016; 17: 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessichelli M, Ferravante A, Zotti T, Reale C, Scudiero I, Picariello G et al. Neuroepithelial transforming gene 1 (Net1) binds to caspase activation and recruitment domain (CARD)- and membrane-associated guanylate kinase-like domain-containing (CARMA) proteins and regulates nuclear factor κB activation. J Biol Chem 2012; 287: 13722–13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiet C, Silvestri E, De Smaele E, Franzoso G, Vito P. c-FLIP efficiently rescues TRAF-2−/− cells from TNF-induced apoptosis. Cell Death Differ 2002; 9: 138–144. [DOI] [PubMed] [Google Scholar]
- Guiet C, Vito P. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate bcl10-induced NF-kappaB activation. J Cell Biol 2000; 148: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014; 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.