Abstract
Background
In non-small-cell lung cancer radiotherapy, radiation pneumonitis ≥ grade 2 (RP2) depends on patients' dosimetric, clinical, biological and genomic characteristics.
Methods
We developed a Bayesian network (BN) approach to explore its potential for interpreting biophysical signaling pathways influencing RP2 from a heterogeneous dataset including single nucleotide polymorphisms, micro RNAs, cytokines, clinical data, and radiation treatment plans before and during the course of radiotherapy. Model building utilized 79 patients (21 with RP2) with complete data, and model testing used 50 additional patients with incomplete data. A developed large-scale Markov blanket approach selected relevant predictors. Resampling by k-fold cross-validation determined the optimal BN structure. Area under the receiver-operating characteristics curve (AUC) measured performance.
Results
Pre- and during-treatment BNs identified biophysical signaling pathways from the patients' relevant variables to RP2 risk. Internal cross-validation for the pre-BN yielded an AUC=0.82 which improved to 0.87 by incorporating during treatment changes. In the testing dataset, the pre- and during AUCs were 0.78 and 0.82, respectively.
Conclusions
Our developed BN approach successfully handled a high number of heterogeneous variables in a small dataset, demonstrating potential for unraveling relevant biophysical features that could enhance prediction of RP2, although the current observations would require further independent validation.
Keywords: Lung cancer, Radiation pneumonitis, Bayesian network analysis, Biophysical interactions
Introduction
Identification of biophysical or pan-Omics models combining clinical, physical, molecular cancer profiling and computational analysis should enable “personalized medicine,” where treatment strategies can be individually tailored based on combinations of diverse existing data resources[1, 2]. However, large scale and diverse radiobiological datasets need to be integrated within a biophysical signaling network of response, interactive manipulation of such network should be supported, and mechanistic radiobiological insights from the analysis would be required. Dose limiting toxicities in the form of radiation pneumonitis (RP) and pulmonary fibrosis are major obstacles in the successful radiation treatment of lung neoplasms. Radiosensitivity of the lung has been shown to depend on multiple factors that include patient's clinical, physical, imaging and molecular characteristics[3–10]. The availability of clinical data, laboratory information, and diagnostic images before and during the course of radiotherapy provides opportunities as well as challenges to build a predictive model for patient specific risks of RP. The purpose of this paper is to develop a systematic methodology based on graphical analysis by Bayesian networks to explore its potential for unraveling biophysical signaling interactions of radiation pneumonitis from a heterogeneous dataset.
Bayesian networks (BNs) are probabilistic graphical models that represent a set of biophysical variables and their conditional dependencies via a directed acyclic graph (DAG) [11]. BNs are also called belief networks, where, in a graphical representation, each node represents a random variable and the edges between the nodes represent probabilistic dependences among the interacting random variables, analogous to using nomograms for representing linear/logistic models in traditional statistical analysis. Thus, BNs are able to explore probabilistic relationships among multiple interacting variables [12]. However, due to limited sample sizes together with large numbers of parameters that need to be optimized in a typical clinical application, an inherent limitation in BN analyses is the need to transform continuous variables into discrete ones, which may lead to loss of information. To mitigate this effect, Hartemink's algorithm, which employs a mutual information approach to retain the most permissible information, can be used for discretization of the data, as it was here. In all, as BNs can effectively probe the interactions of clinical, physical, and molecular data, accommodate hierarchal relationships embedded in radiation response signaling pathways of RP development, and handle missing data, they are promising approaches to support radiation treatment decision making. Previously, a BN was employed to model response in lung cancer including RP [13]. However, that study was limited in scope to 16 selected features that included cytokines and dosimetric variables only. The current study has 200 features, also including genetic SNPs and miRNA markers. Due to the high dimensional nature of this retrospective dataset, different developed methods for feature selection (extended MB) and BN structure learning (Tabu Search) are used to handle large-scale analysis. In addition to internal re-sampling cross-validation using the training data, the current study includes a testing phase, which also assesses the use of the BN for marginalization of missing information.
Materials and methods
Patient Samples
This study involved secondary analysis of NSCLC patients that had been treated on four prospective protocols under IRB approval. The second and third treated patients to standard doses (<74 Gy at 2 Gy per fraction) and the first and fourth were dose escalation studies (total doses up to of 86 Gy over 30 fractions). Radiation dose distributions for total uninvolved lung (left plus right exclusive of gross tumor volume) were computed within the Varian Eclipse treatment planning system using the AAA dose algorithm. All total dose values were converted to their 2 Gy equivalents (EQD2) by the linear-quadric model using locally developed software. Dosimetric features with different parameters were highly correlated with each other in the MB in early screening, and obscured the selection approach to identify important features from other categories for RP2 prediction. Thus, Mean_Lung_Dose computed from EQD2 dose distributions generated with an α/β of 4 Gy was considered to be representative of the dosimetric features [14].
Blood samples were obtained at baseline and after approximately 1/3 and 2/3 of the scheduled radiation doses were completed. Pre-treatment blood samples were analyzed for cytokine levels [15], miRNAs [16], and SNPs [17, 18], which have been identified as candidates from the literature as related to lung cancer or inflammatory disease (Table 1 in Appendix A). The slopes of cytokine changes from before to during treatment were also determined as patients' responses to radiation treatment.
The patients' RP were scored by five grades (CTCAE 3.0) based on clinical assessment and imaging findings [19], and here, RP with grade 2 or above (RP2) was used to represent complication of radiation treatment. The study includes 79 NSCLC patients (with 21 cases of RP2) with complete data (200 features) utilized for biophysical pathway model building and 50 additional patients (with 3 cases of RP2) with incomplete data (missing some blood sample analyses) used for model testing. Input data pre-processing is detailed in Appendix B.
Development of BN approach to identify appropriate biophysical interactions
Step 1. Selection of Relevant Variables for RP Modeling
For feature selection, we used a modified version of the MB approach which represents not only the inner family of inter-related variables as commonly practiced [11] but also their extended family, i.e., next-of-kin variable relationships. The MB of RP2 represents the features most related to RP2, and RP2 is independent from the rest of other features given its MB [20]. However, only including the mostly related features for RP2 prediction may result in loss of extended relationships of higher orders for prediction purposes. Therefore, to minimize this effect, we also extended the MB by one order (next-of-kin MB) for each feature in the original MB. In addition to finding good feature subsets, the MB could also help determine causal relationships among various nodes in a BN [21]. Here, the MBs of RP2 and next-of-kin were found using the HITON algorithm, which is a fast forward selection technique for neighborhood detection designed to exclude irrelevant nodes efficiently based on marginal associations [22]. This algorithm is particularly useful when the number of features is large compared to the number of samples as in the current case.
Step 2. Identifying Appropriate Biophysical Interactions for RP2 Prediction
A BN was employed to integrate the potential variables for RP2 prediction based on the existing data, which is a complex process given the limited sample size. A Tabu Search is a metaheuristic search method employing local search methods used for mathematical optimization [23], and was used in our study to generate a stable BN structure to integrate the selected biophysical variables from bootstrap samples of the original data. Although there are many biophysical interactions related to RP2, and the relationships among potential variables on each interaction are complicated, there existed many known prior biophysical relationships related to RP2 onset that could be exploited to constrain the search space in building the BN structure. These pathways or relationships may have their specific variables from different categories. The known causal influence relationships between them were considered as “biophysical rules” in our BN structure identification, which could be obtained from the literature. An example to identify appropriate biophysical interactions for RP2 prediction is detailed in Appendix C. Therefore, for a selected set of biophysical variables related to RP2, radiobiologically plausible relationships were built among them by adding constraints to the Tabu Search and forcing them to follow the biophysical rules.
Here BN structure learning was intended to find appropriate biophysical relationships to improve RP2 prediction performance by balancing variance reduction and loss of information. While each predictor in the stable BN can contribute to biophysical information for RP2 prediction, it can also cause noise at certain level. In the BN, the dependence between two associated biophysical factors can be described by the strength of the edge/arc between them, which is measured from score gain/loss caused by this edge/arc's removal [24]. In general, the larger strength value represents stronger dependency, and BN structure can be adjusted from selecting different edge strength thresholds. Thus, the properties of edge strength provide an opportunity to discover the relative importance among the predictors in the BN in terms of RP2 prediction. By increasing the threshold of edge strength in a stable BN, an edge with relatively weak strength will disappear. A node without any descendant in the BN is considered as a leaf node, and it has the highest priority to be eliminated for the improvement of RP2 prediction according to the trade-off between variance reduction and loss of information. This is the basic idea of predictor pruning in our BN structure learning. Therefore, the presence of any predictor in the BN is determined by whether it is part of the extended MB, satisfies biophysical rules, and its relative strength within the network to predict RP2.
Cross validation is a technique to assess how a statistical model will generalize to an independent dataset. The Receiver Operating Characteristic (ROC) curve shows the trade-offs between true positive and false positive rates for the diagnostic system, which is widely used in evaluating the prediction power of classification models [25]. The Area Under the ROC Curve (AUC) provides a single measure of the performance of a classifier by assigning 0.5 to a random signal and 1.0 to a perfectly discriminant signal. A binomial smoothing technique was employed here to refine ROC estimates. In our training data, there are 21 RP2 events in 79 patients. To avoid the problem of “inverse cross validation effect” that could appear due to imbalanced event rate [26], a partitioning approach was used [27], [28]. Given the sample size, a 7-fold cross-validation was chosen in this work, where each fold randomly had 3 patients with RP2. Cross validation was employed to guide the Tabu Search and predictor pruning to identify an optimal BN structure with the highest possible RP2 prediction power.
Let N be the total number of the predictors for RP2 prediction, n be the index of these predictors (n=N, N-1,…, 1), Sn be the threshold of the BN's arc strength in a BN with n predictors (0.5 ≤Sn<1). The BN with n predictors and the threshold Sn of arc strength can be denoted as BN(n, Sn), its associated AUC can be described by AUC(n, Sn). Let be the optimal arc strength to maximize AUC(n, Sn) (n=N, N-1,…, 1) with different Sn, the process of obtaining the best BN for RP2 prediction based on R package “bnlearn” is illustrated in Fig. 1.
Fig. 1.
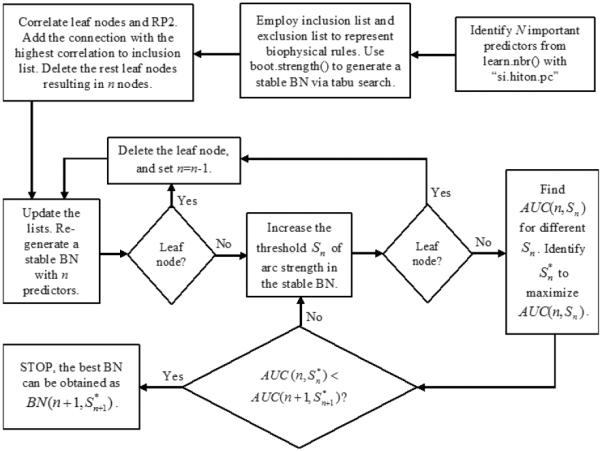
The process of obtaining the best BN
Results
The developed MB of RP2 based on our training data is formed from “Mean_Lung_Dose”, “pre_MCP_1”, “pre_TGF_alpha”, and “pre_eotaxin”. Moreover, each of these variables has its own MB neighborhood as shown in Fig. 2. For example, “V20”, “nos3_Rs1799983”, “Stage”, and “RP2” from the MB of “Mean_Lung_Dose”. As “Mean_Lung_Dose” is closely related to “RP2”, its neighborhood goes back to “RP2” again. In our study, we used extended MB neighborhoods within two layers of RP2 as potential variables of the BN. Our BN building approach considers the whole extended MB (layers#1 and #2), subsequently some predictors from layer#1 of RP2's MB (inner family), such as pre_TGF_alpha, pre_eotaxin, may or may not necessarily be selected as a part of the final pre-treatment BN to predict RP2.
Fig. 2.
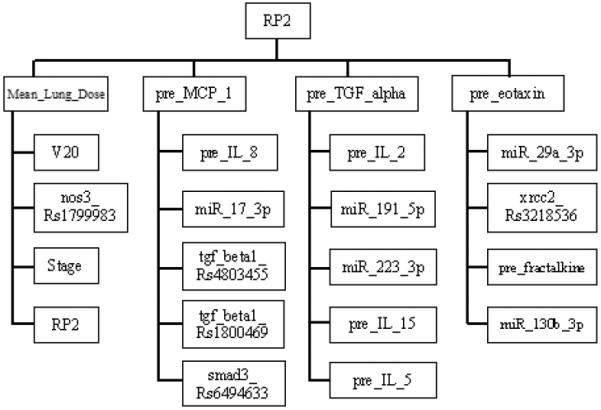
The extended MB neighborhoods of RP2 before radiation treatment, where the upper level shows the inner family of RP2 and the lower levels show the next-of-kin for each of its member.
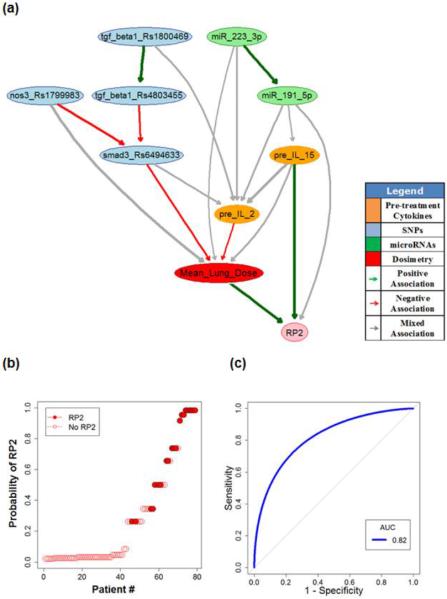
Fig. 3(A) shows a stable pre-treatment BN with edge strength ≥0.65 where nine important biophysical predictors are identified. Their relationships in terms of RP2 prediction are indicated by directed edges, and the thickness of an edge represents the strength of a connection. The green and red lines represent positive and negative influences between the connected predictors, respectively. The gray lines indicate context-dependent mixed influences between the predictors in the network. It is noted that the TGF-β signaling pathway plays an important role in the resulting RP2 BN. Interestingly, a patient's bioprofile seems also to influence (could predict) the radiation dose that s/he receives based on historical cohorts.
Fig. 3.
(A) Pre-treatment BN for RP2 prediction; (B) Patients rank-ordered by probability of RP2 as predicted from the “pre-treatment” BN, where the solid circles represent the patients who got RP2; (C) The internal testing ROC curve of pre-treatment BN based on cross validation.
Fig. 3(B) summarizes the capability of the pre-treatment BN to explain the risk of getting RP2 for the 79 NSCLC patients in the training dataset (AUC of 0.94 with a 95% confidence interval of 0.81–0.98). The corresponding internal testing ROC curve of the pre-treatment BN based on 7-fold cross validation is shown in Fig. 3(C), where the AUC value is 0.82 with a 95% confidence interval of 0.72–0.87. In comparison, a standard model containing only the main dosimetric predictor (Mean_Lung_Dose) had an optimistic AUC=0.67 with a 95% confidence interval of 0.53–0.79 based on 2000 stratified bootstrap replicates. Although these two confidence intervals are overlapping, their means are significantly different according to Delong's test (p-value=0.046). Furthermore, a basic BN model containing only the dosimetric and inflammation related clinical parameters was also generated (Appendix D). The AUC for the ROC of that model, under internal cross validation, is 0.68 with a 95% confidence interval of 0.55–0.80 from 2000 stratified bootstrap replicates. The Delong's test shows that this mean is also significantly different (p-value =0.050).
In the extended model, the slopes of cytokine levels before and during-treatment (SLP) were incorporated as the patients' responses during the course of radiotherapy. The updated extended MB neighborhoods are shown in Fig. 4. The important biophysical predictors during radiation treatment can be found from the large-scale Markov blanket algorithm. In addition, a during-treatment BN with edge strength ≥ 0.65 was developed via BN structure learning as illustrated in Fig. 5(A). While Fig. 5(B) summarizes the ability of the during-treatment BN to explain the 79 patients' RP2 results, Fig. 5(C) shows the internal testing ROC curve of the during-treatment BN based on 7-fold cross validation. The prediction performance of the training dataset is very high (AUC=0.99), and the AUC of the internal testing cross validation is 0.87 with a 95% confidence interval of 0.80–0.91. Potential mechanistic biological interpretation of our BNs is detailed in Appendix E and conditional probability tables (CPTs) for RP2 prediction are given in Appendix F.
Fig. 4.
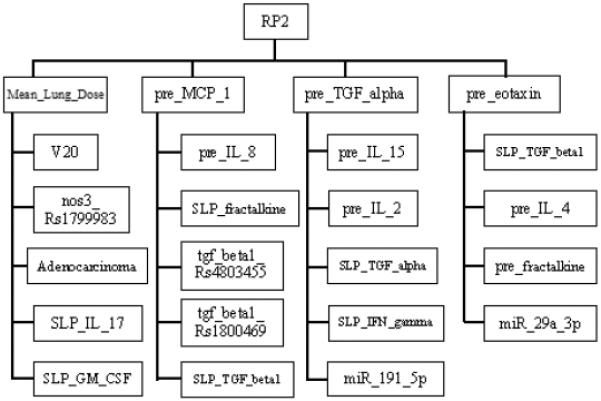
The extended MB neighborhoods of RP2 during radiation treatment, where the upper level shows the inner family of RP2 and the lower levels show the next-of-kin for each of its member.
Fig. 5.
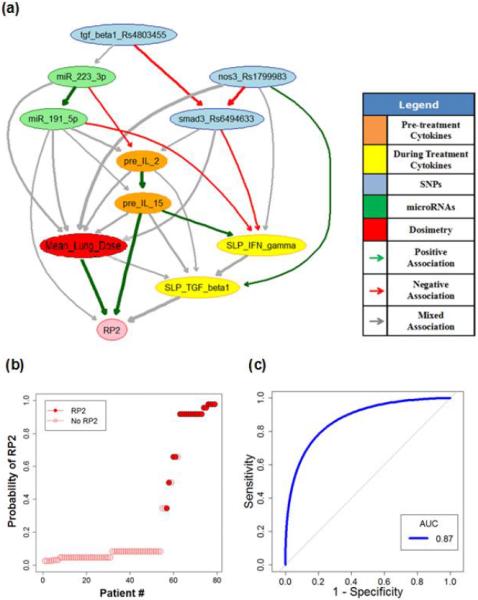
(A) During-treatment BN for RP2 prediction; (B) Patients rank-ordered by probability of RP2 as predicted from the “during-treatment” BN, where the solid circles represent the patients who got RP2; (C) The internal testing ROC curve of during-treatment BN based on cross-validation.
In our study, additional patients from the same original studies randomly carried similar characteristics, but had missing information (primarily some portion of the blood sample analyses). These additional patients (50, with only three cases of RP2) were allocated into a separate cohort used to test the RP2 prediction model developed for the original patients, using the obtained BNs to marginalize the missing data; an important characteristic of BN approaches [29]. A comparison of the characteristics of the training and testing datasets for the selected variables in pre- and during BNs is given in Appendix G. It is noted that the levels of Mean_Lung_Dose in the testing cohort were significantly lower than in the training cohort while SLP_TGF_beta1 was higher; factors that may have contributed to the lower RP2 rate in the testing cohort; dose in particular playing a dominant role. The calibration plots and ROC curves for use of the pre- and during-treatment BNs from the training dataset on the testing dataset are provided in Appendix H. Despite the differences noted above in the two datasets, the AUC values for use of the pre- and during-treatment BNs on the testing dataset were 0.78 and 0.82, respectively, demonstrating some robustness in the BN modeling approach. However, given the relatively small sample size and the limited number of events in the testing cohort, it should be emphasized that further independent analysis would still be required in the future, upon availability of similar datasets, to substantiate the biophysical pathways revealed and to validate the predictive capabilities of the BNs developed here.
Discussion
The main emphasis of this work was to develop a machine learning methodology based on Bayesian network to deal with high number of heterogeneous variables in small datasets to predict outcomes while also revealing associated biophysical pathways. The methodology emphasizes using present knowledge as constraints and resampling based on bootstrapping to avoid overfitting during the construction of the network and cross-validation for internal prediction power validation. Thus, we explored the development of biophysical interaction networks for RP2 in NSCLC patients using pre- and during-treatment information. Upon further external validation, these BNs would appear to allow for prediction of response and personalization of treatment and may add further important insights into RP radiobiology.
Using an extended large-scale Markov blanket algorithm, treatment-relevant SNPs, miRNAs, cytokines, and dosimetric information were identified that appear to be related to RP2 in our dataset. Bayesian network analysis was used to probe the probabilistic relationship among these different factors and their hierarchal relationship with RP2 from genetic variants into changing cytokine levels induced by radiation dose. Our current results generally aligned with known observations made by previous studies [30–35]. However, novel relationships were also generated, which would require further studies to evaluate the importance of these interactions in vitro and in vivo.
Consistent with previous observations, our data indicate a strong correlation between dosimetric information, treatment-relevant cytokines, and the RP2 at the bottom layer of pre-treatment BN. Agrawal et al. showed that the correlation between RP and dosimetric constraints can be added to the classical total lung constraints in order to identify patients likely to develop pulmonary toxicity in response to treatment with chemoradiation [31]. Schaue et al. [32] showed that cytokines can be grouped in a very general way as: pro-inflammatory (IL-1α and β, IL-17), anti-inflammatory (IL-1ra, IL-4, IL-10, TGF-β1), pro-fibrotic (IL-6, TGF-α), chemokines (fractalkine, IL-8, MCP-1, IP-10), immune (IFNϒ, IL-2, IL-4, IL7), and hematopoietic (CSF1, GM-CSF, IL-3, EPO) [32] which is consistent with the groups established through the BN analysis. Moreover, they proposed that these cytokines should be viewed as part of large cross-talking networks that coordinate with other molecular and cellular systems to orchestrate tissue responses as demonstrated in our BN development. The impact of an initial radiation treatment on the radiation outcome can be displayed through changes in cytokine expression. Although pre-treatment cytokines still have a strong influence on RP2, the contribution of cytokines at the mid treatment timepoint is evident. The underlying radiobiology between patients' cytokine expression and their genomic characteritsitcs in terms of RP2 remains elusive. Our data suggest that specific miRNAs might be involved in orchestrating the relationship between genomic characteristics, cytokines and RP2 through their role in regulating the inflammatory response. IL-2 and IL-15 share same cytokine receptors [36], which upon activation engage the Jak1/3-Stat-5 signaling pathway. Such signaling seems to be critical for IL-2 and IL-15 mediated generation and persistence of NK cells, a major producer of IFN-γ[37]. Together, the identification of positive association of IL-2/IL-15 and negative association of smad3_Rs6494633 with IFN-γ during treatment helping us understand the cross-talk between two opposing signaling events, whose imbalance may be critical for RP2 progression.
In this paper, we have shown how the developed methdology based on BN analayses could illustrate the hierachical biophysical influence behind observed radiation outcome, and how it may help predict RP2 under different radiation treatment plans before and during the courses of radiation treatment. It should be noted that the Markov property of BN implies that the probabilty of getting RP2 could be estimated from measuring the nodes that directly connect to RP2 in the BNs (e.g., Mean_Lung_Dose, pre_IL_15, miR_191_5p and SLP_TGF_beta1 in Fig. 5a). Such a “naïve BN” is observed to still perform well with our data (AUC of 0.84). This is contrasted with the de facto predictor of RP2, mean lung dose, which achieves an AUC of only 0.67 by itself. In addition to their potential for RP prediction, the biophysical interactions could be used to explore the in-depth understanding of underlying RP radiobiology, which would be essential to development of targeted intervention. A feature of a BN is conditional probability relationships. Given certain patient characteristics (SNPs, miRNAs, and cytokines information), the network could be expected to predict the dose category (high, medium, low) that the lung is going to receive based on the historical cohorts by following the arrows entering node “Mean_Lung_Dose” in Fig. 3a and 5a. Moreover, we note that mid-treatment information apparently has added value, which could be explained if the initial dose received acts as a probe that would further improve our understanding of the response that the patient is likely to have.
Our developed Bayesian network methodology is a data-driven, response-based approach that appears to provide new potentials for exploring the relationships of biophysical factors leading to radiation-induced RP2. Resulting biophysical Bayesian networks such as those developed here show promise for personalized adaptive radiation treatment planning, although, due to the small sample size and limited number of events in our testing cohort, future validation studies are necessary to confirm the current observations. Additionally, estimated biophysical influences in the current BNs (calculated from the adjacent parent and children nodes) may not reflect the accumulative effect or direct causal interaction of all variables along the network; other intermediate factors (not included in the study) might play a role. The mechanistic biological interpretation of some of the connections is dependent on selecting the most biologically plausible relationships using our extended MB approach and would require further in vitro and in vivo experimental validation. Future work involves enhancing the current Bayesian network analysis, further validation on larger external data, and developing dynamic BNs to guide personlized radiation treatment practice.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health [grant numbers P01 CA059827, R01 CA142840]. The authors wish to thank Paul Stanton, Nan Bi, MD, PhD, and Weili Wang MD, PhD for their work in processing the cytokine, miRNA and SNP data. This work was presented in part at ICTR-PHE 2016, 15–19 February 2016, CICG, Geneva, Switzerland.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El Naqa I. Biomedical informatics and panomics for evidence-based radiation therapy. Wiley Interdisciplinary Reviews-Data Mining and Knowledge Discovery. 2014;4(4):327–340. [Google Scholar]
- 2.Baumann M, Krause M, Overgaard J, et al. Radiation oncology in the era of precision medicine. Nat Rev Cancer. 2016;16(4):234–49. doi: 10.1038/nrc.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Provatopoulou X, Athanasiou E, Gounaris A. Predictive markers of radiation pneumonitis. Anticancer Res. 2008;28(4C):2421–32. [PubMed] [Google Scholar]
- 4.Rodrigues G, Lock M, D'Souza D, et al. Prediction of radiation pneumonitis by dose - volume histogram parameters in lung cancer--a systematic review. Radiother Oncol. 2004;71(2):127–38. doi: 10.1016/j.radonc.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Rancati T, Ceresoli GL, Gagliardi G, et al. Factors predicting radiation pneumonitis in lung cancer patients: a retrospective study. Radiother Oncol. 2003;67(3):275–83. doi: 10.1016/s0167-8140(03)00119-1. [DOI] [PubMed] [Google Scholar]
- 6.Kwaa SLS, Theuwsa JCM, Antoinette Wagenaara A, et al. Evaluation of two dose–volume histogram reduction models for the prediction of radiation pneumonitis. Radiotherapy and Oncology. 1998;48(1):61–69. doi: 10.1016/s0167-8140(98)00020-6. [DOI] [PubMed] [Google Scholar]
- 7.Claude L, Perol D, Ginestet C, et al. A prospective study on radiation pneumonitis following conformal radiation therapy in non-small-cell lung cancer: clinical and dosimetric factors analysis. Radiother Oncol. 2004;71(2):175–81. doi: 10.1016/j.radonc.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Shi A, Zhu G, Wu H, et al. Analysis of clinical and dosimetric factors associated with severe acute radiation pneumonitis in patients with locally advanced non-small cell lung cancer treated with concurrent chemotherapy and intensity-modulated radiotherapy. Radiat Oncol. 2010;5:35. doi: 10.1186/1748-717X-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodemann HP, Bamberg M. Cellular basis of radiation-induced fibrosis. Radiother Oncol. 1995;35(2):83–90. doi: 10.1016/0167-8140(95)01540-w. [DOI] [PubMed] [Google Scholar]
- 10.Ebert N, Baumann M, Troost EG. Radiation-induced lung damage - Clinical risk profiles and predictive imaging on their way to risk-adapted individualized treatment planning? Radiother Oncol. 2015;117(1):1–3. doi: 10.1016/j.radonc.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann Publishers Inc.; San Francisco, CA, USA: 1988. [Google Scholar]
- 12.Gadewadikar J, Kuljaca O, Agyepong K, et al. Exploring Bayesian networks for medical decision support in breast cancer detection. African Journal of Mathematics and Computer Science Research. 2010;3(10):225–231. [Google Scholar]
- 13.Lee S, Ybarra N, Jeyaseelan K, et al. Bayesian network ensemble as a multivariate strategy to predict radiation pneumonitis risk. Medical Physics. 2015;42(5) doi: 10.1118/1.4915284. [DOI] [PubMed] [Google Scholar]
- 14.Bentzen SM, Dische S. Morbidity related to axillary irradiation in the treatment of breast cancer. Acta Oncol. 2000;39(3):337–47. doi: 10.1080/028418600750013113. [DOI] [PubMed] [Google Scholar]
- 15.Fukuyama T, Ichiki Y, Yamada S, et al. Cytokine production of lung cancer cell lines: Correlation between their production and the inflammatory/immunological responses both in vivo and in vitro. Cancer Science. 2007;98(7):1048–1054. doi: 10.1111/j.1349-7006.2007.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo LL, Zhang YS, Zhang LF, et al. MicroRNAs, TGF-beta signaling, and the inflammatory microenvironment in cancer. Tumor Biology. 2016;37(1):115–125. doi: 10.1007/s13277-015-4374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slattery ML, Herrick JS, Lundgreen A, et al. Genetic Variation in the TGF-beta Signaling Pathway and Colon and Rectal Cancer Risk. Cancer Epidemiology Biomarkers & Prevention. 2011;20(1):57–69. doi: 10.1158/1055-9965.EPI-10-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damaraju S, Murray D, Dufour J, et al. Association of DNA repair and steroid metabolism gene polymorphisms with clinical late toxicity in patients treated with conformal radiotherapy for prostate cancer. Clinical Cancer Research. 2006;12(8):2545–2554. doi: 10.1158/1078-0432.CCR-05-2703. [DOI] [PubMed] [Google Scholar]
- 19.Kouloulias V, Zygogianni A, Efstathopoulos E, et al. Suggestion for a New Grading Scale for Radiation Induced Pneumonitis Based on Radiological Findings of Computerized Tomography: Correlation with Clinical and Radiotherapeutic Parameters in Lung Cancer Patients. Asian Pacific Journal of Cancer Prevention. 2013;14:2717–22. doi: 10.7314/apjcp.2013.14.5.2717. [DOI] [PubMed] [Google Scholar]
- 20.Pellet JP, Elisseeff A. Using Markov Blankets for Causal Structure Learning. Journal of Machine Learning Research. 2008;9:1295–1342. [Google Scholar]
- 21.Gevaert O, De Smet F, Timmerman D, et al. Predicting the prognosis of breast cancer by integrating clinical and microarray data with Bayesian networks. Bioinformatics. 2006;22(14):e184–90. doi: 10.1093/bioinformatics/btl230. [DOI] [PubMed] [Google Scholar]
- 22.Aliferis CF, Tsamardinos I, Statnikov A. HITON: a novel Markov Blanket algorithm for optimal variable selection. AMIA Annu Symp Proc. 2003 http://www.ncbi.nlm.nih.gov/pubmed/14728126:21-5. [PMC free article] [PubMed]
- 23.Lokketangen A. Tabu Search - Using the Search Experience to Guide the Search Process - an Introduction with Examples. Ai Communications. 1995;8(2):78–85. [Google Scholar]
- 24.Friedman N, Goldszmidt M, Wyner A. Data analysis with bayesian networks: a bootstrap approach. Proceedings of the Fifteenth conference on Uncertainty in artificial intelligence; Stockholm, Sweden: Morgan Kaufmann Publishers Inc.; 1999. pp. 196–205. [Google Scholar]
- 25.Metz CE. Receiver operating characteristic analysis: a tool for the quantitative evaluation of observer performance and imaging systems. J Am Coll Radiol. 2006;3(6):413–22. doi: 10.1016/j.jacr.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 26.Perlich C, Swirszcz G. On Cross-Validation and Stacking: Building Seemingly Predictive Models on Random Data. ACM SIGKDD Explorations Newsletter. 2010;12(2):11–15. [Google Scholar]
- 27.Parker BJ, Gunter S, Bedo J. Stratification bias in low signal microarray studies. BMC Bioinformatics. 2007;8:326. doi: 10.1186/1471-2105-8-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiller TW, Chen Y, El Naqa I, et al. Modeling radiation-induced lung injury risk with an ensemble of support vector machines. Neurocomputing. 2010;73(10–12):1861–67. [Google Scholar]
- 29.Corani G, Antonucci A, Zaffalon M. Bayesian Networks with Imprecise Probabilities: Theory and Application to Classification. In: Holmes DE, Jain LC, editors. Data Mining: Foundations and Intelligent Paradigms. Springer; Berlin Heidelberg: 2012. pp. 49–93. [Google Scholar]
- 30.Flanders KC, Major CD, Arabshahi A, et al. Interference with transforming growth factor-beta/Smad3 signaling results in accelerated healing of wounds in previously irradiated skin. Am J Pathol. 2003;163(6):2247–57. doi: 10.1016/s0002-9440(10)63582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal S, Kumar S, Lawrence A, et al. Ipsilateral lung dose volume parameters predict radiation pneumonitis in addition to classical dose volume parameters in locally advanced NSCLC treated with combined modality therapy. South Asian J Cancer. 2014;3(1):13–5. doi: 10.4103/2278-330X.126503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaue D, Kachikwu EL, McBride WH. Cytokines in radiobiological responses: a review. Radiat Res. 2012;178(6):505–23. doi: 10.1667/RR3031.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng H, Mertens PR, Gressner AM, et al. IFN-gamma abrogates profibrogenic TGF-beta signaling in liver by targeting expression of inhibitory and receptor Smads. J Hepatol. 2007;46(2):295–303. doi: 10.1016/j.jhep.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Kong FM, Wang S. Nondosimetric risk factors for radiation-induced lung toxicity. Semin Radiat Oncol. 2015;25(2):100–9. doi: 10.1016/j.semradonc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park I, Letterio JJ, Gorham JD. TGF-β1 inhibition of IFN-γ-induced signaling and Th1 gene expression in CD4+ T cells is Smad3 independent but MAP kinase dependent. Molecular Immunology. 2007;44(13):3283–3290. doi: 10.1016/j.molimm.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6(8):595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 37.Carson WE, Giri JG, Lindemann MJ, et al. Interleukin (Il)-15 Is a Novel Cytokine That Activates Human Natural-Killer-Cells Via Components of the Il-2 Receptor. Journal of Experimental Medicine. 1994;180(4):1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.