Abstract
DC-STAMP is a multi-pass transmembrane protein essential for cell-cell fusion of osteoclast precursors during osteoclast (OC) development. DC-STAMP−/− mice have mild osteopetrosis and form mononuclear cells with limited resorption capacity. The identification of an Immunoreceptor Tyrosine-based Inhibitory Motif (ITIM) on the cytoplasmic tail of DC-STAMP suggested a potential signaling function but the absence of known DC-STAMP ligand has hindered examination of downstream signaling pathways. To address this problem, we engineered a light-activatable DC-STAMP chimeric molecule in which light exposure mimics ligand engagement that can be traced by downstream Ca2+ signaling. Deletion of the cytoplasmic ITIM resulted in a significant elevation in the amplitude and duration of intracellular Ca2+ flux. We also found that decreased NFATc1 expression in DC-STAMP−/− cells was restored by DC-STAMP over-expression. Multiple biological phenotypes including cell-cell fusion, bone erosion, cell mobility, DC-STAMP cell surface distribution, and NFATc1 nuclear translocation were altered by deletion of the ITIM and adjacent amino acids. In contrast, mutations on each of tyrosine residues in the ITIM showed no effect on DC-STAMP function. Collectively, our results suggest that ITIM on DC-STAMP is a functional motif that regulates osteoclast differentiation through the NFATc1 / Ca2+ axis.
Introduction
Osteoclasts (OC) are myeloid lineage cells specialized to resorb bone and responsible for pathologic bone loss in inflammatory joint diseases and osteoporosis (Charles and Aliprantis, 2014). Direct involvement of OC in bone erosion has been well documented and recently underscored by the identification of myeloid-derived suppressor cells (MDSC) and inflammatory monocytes in bone pathogenesis (Seeling et al., 2013; Zhang et al., 2015). Following activation by RANKL & M-CSF, circulating osteoclast precursors (OCPs) differentiate into mature OCs with bone resorption activity. Differentiation of OCPs to mature OC is a highly regulated process mediated by temporal and spatial interactions of specialized gene pathways, protein interactions and modifications. (Hobolt-Pedersen et al., 2014; Soe et al., 2015). A critical step in the transformation of monocytes to OC polykaryons is cell-cell fusion.
DC-STAMP is a multi-pass transmembrane protein required for the cells to fuse between 2 lipid bilayers (Yagi et al., 2005). Currently, DC-STAMP is considered a master regulator of osteoclastogenesis (Islam et al., 2014; Zhang et al., 2014). DC-STAMP−/− mice manifest an osteopetrosis phenotype due to the absence of functional multinucleated OC (Yagi et al., 2005). DC-STAMP was recently linked to human disease following the identification of a susceptible mutation on the DC-STAMP cytoplasmic tail in a patient with Paget’s disease (Albagha et al., 2011; Beauregard et al., 2014), and an elevation of DC-STAMP+ cell frequency was reported in psoriatic arthritis patients (Chiu et al., 2012).
In addition to the essential role of DC-STAMP in cell-cell fusion, our previous identification of an Immunoreceptor Tyrosine-based Inhibition Motif (ITIM) on the cytoplasmic tail of DC-STAMP suggests its possible involvement in cell signaling (Chiu et al., 2012). However, the molecular mechanism underlying DC-STAMP-mediated signaling during osteoclastogenesis remains to be elucidated. We proposed a model (Chiu et al., 2012), where the DC-STAMP ITIM- counteracts signaling through Immunoreceptor Tyrosine-based Activation Motif (ITAM)-bearing receptors (Ben Mkaddem et al., 2014; Li et al., 2014); activation signals required for osteoclast differentiation following engagement of RANK by RANKL (Barrow et al., 2011; Humphrey et al., 2005; Nimmerjahn and Ravetch, 2007; Nimmerjahn and Ravetch, 2008; Takayanagi et al., 2002). The integration of these dual signals induces intracellular Ca2+ oscillations (Hwang and Putney, 2011; Kajiya, 2012; Kim et al., 2013; Masuyama et al., 2008), and translocation of NFATc1 from the cytoplasm to the nucleus to turn on genes essential for osteoclast differentiation (Yarilina et al., 2011; Zhao et al., 2010). We previously showed co-precipitation of DC-STAMP and SHIP-1 following exposure of monocytes to an anti-DC-STAMP mAb suggesting a potential signaling role (Chiu et al., 2012).
DC-STAMP knockout (KO) mice were initially established by Yagi et al. (Yagi) These mice harbor the DC-STAMP null mutation and demonstrate an osteopetrosis phenotype due to the inability of DC-STAMP−/− cells to undergo cell-cell fusion and form multinucleated osteoclasts. DC-STAMP−/− cells isolated from the DC-STAMP KO mouse strain are ideal tools to dissect DC-STAMP functions during osteoclastogenesis. Because the endogenous DC-STAMP proteins are not expressed in DC-STAMP−/− cells, these cells allow us to introduce distinct versions of DC-STAMP, either WT or tail-deleted (TD) mutants, and examine the function of DC-STAMP and ITIM regulation at the molecular level by the phenotypes after protein complementation. Considering that the NFATc1/Ca2+ is the major axis of OCgenesis, we sought to determine whether DC-STAMP regulates osteoclast differentiation through NFATc1 and Ca2+. Analysis of downstream DC-STAMP signaling is complicated by the absence of a known DC-STAMP ligand. To address this problem, we engineered photo-activatable and GFP-tagged DC-STAMP molecules. To determine the function of ITIM, we overexpressed the wild-type (WT)- or ITIM-deleted (TD)- versions of chimeric DC-STAMP proteins in DC-STAMP−/− cells and compared the phenotypes related to OCgenesis. In this study, we established several chimeric DC-STAMP fusion proteins to evaluate the effect of tail deletion on DC-STAMP function by examining intracellular Ca2+ signaling, cell-cell fusion, bone erosion, cell surface distribution, and NFATc1 expression. We found that amino acids in and around the ITIM are required for cell fusion but may not require a functional ITIM since mutation of the tyrosine did not disrupt the phenotype. We also found that the ITIM and surrounding amino acids in the tail are required for the expression and nuclear translocation of NFATc1. These data support interaction between NFATc1 and DC-STAMP essential for successful OC differentiation.
Materials & Methods
Animals
DC-STAMP knockout mice were obtained from Dr. Takeshi Miyamoto at Keio University, Japan and were re-derived in the animal facility at the University of Rochester to obtain pathogen-free offspring. All experiments were performed at the University of Rochester Medical Center and were approved (animal protocol # UCAR-2013-015) by the Institutional Animal Care and Use Committee (UCAR) at the University of Rochester Medical Center. The UCAR approved the protocol according to the requirements of animal ethics law and NIH regulations. In the protocol UCAR-2013-015, we took careful considerations to ameliorate suffering including pain relief and euthanasia procedures. All mice were sacrificed with CO2 asphyxiation using a controlled-flow Euthanex apparatus, followed by cervical dislocation, consistent with the new recommendations of the panel on Euthanasia of the American Veterinary Medical Association.
Reagents
OC-promoting media is composed of RANKL and M-CSF. Murine versions of RANKL and M-CSF were purchased from the R&D systems (cat# 462-TEC-010/CF and 416-ML-050/CF). Defined Fetal Bovine Serum was obtained from Hyclone. The fix & perm cell permeabilization reagents (Invitrogen) were added for intracellular staining of cytokines. The concentration of RANKL and M-CSF for cell cultures was 100 ng/ml and 25 ng/ml, respectively. Most of the antibodies were purchased from BD Biosciences unless specified otherwise. Given that anti-DC-STAMP 1A2 antibody is a mouse antibody, to avoid non-specific binding and cross-reactivity on mouse derived cells such as BMM and RAW cells, we performed a complete blocking procedure before IHC staining and flow cytometry analysis. Cells were first incubated with 5% normal mouse sera (NMS, Sigma) followed by mouse FcR blocking (Miltenyi, Cat 130-092-575) for 15 minutes at room temperature. Following addition of NMS and Fc blocker to prevent non-specific binding, cells were stained with the DCSTAMP antibody 1A2 (Millipore, Cat# MABF39-I) followed by APC-conjugated rat-anti-mouse antibody (Cat# M1-14D12, eBioscience).
Cell staining and FACS analysis
For flow cytometry analysis, cells were harvested, washed once with PBS, blocked with 5% normal mouse sera for 10 min at room temperature and stained with antibodies for 20 min. Cells were washed with PBS and fixed in 2% formaldehyde. FACS data were acquired using Canto or LSRII and analyzed using CellQuest (Becton Dickinson) or FlowJo (TreeStar) software. FMO (fluorescence minus one) at each time point is used to define the thresholds of positive and negative events in quadrant analyses.
DNA construct and cloning
The retroviral chimeric plasmid pMX-DCSTAMP-GFP-WT was constructed by inserting a cDNA fragment encoding DC-STAMP into a pMX-IRES-GFP retroviral expression vector (Cell Biolabs, Inc.) via the BamHI and XhoI cloning sites. We used the QuickChange II XL Site-directed Mutagenesis Kit (Stratagene) to delete the ITIM sequence (SFYPKV), from the pMX-DCSTAMP-GFP-WT plasmid and construct the ITIM-deleted mutant plasmid pMX-DCSTAMP-GFP-TD. Two pairs of primers (Y F 5’GACTGACACGAGAATTTTAA3’ & 5’GAGAGGGAGAGAATTGAATAC3’, Y
F 5’GACTGACACGAGAATTTTAA3’ & 5’GAGAGGGAGAGAATTGAATAC3’, Y A 5’GTGTCAGCATCTTTCGCTCCCAGCGTGGAGAGG3’ & 5’CACAGTCGTAGAAAGCGAGGGTCGCACCTCTCC3’) were designed for point mutagenesis. Two photoactivatable (PA) DC-STAMP constructs (WT and TD) were designed with 3 extracellular domains (Figure 3A) replaced with those of rhodopsin with 100% match to the PA-CXCR4-mCherry plasmid (Xu et al., 2014). This design was based on our previous study in which signal generated from the extracellular domain of PA-CXCR4-mCherry plasmid after light activation mimics the signal derived from the endogenous CXCR4 after chemokine activation (Xu et al., 2014). Two PA constructs were ordered from GenScript and their primary sequences were shown in Supplemental Figure S1.
A 5’GTGTCAGCATCTTTCGCTCCCAGCGTGGAGAGG3’ & 5’CACAGTCGTAGAAAGCGAGGGTCGCACCTCTCC3’) were designed for point mutagenesis. Two photoactivatable (PA) DC-STAMP constructs (WT and TD) were designed with 3 extracellular domains (Figure 3A) replaced with those of rhodopsin with 100% match to the PA-CXCR4-mCherry plasmid (Xu et al., 2014). This design was based on our previous study in which signal generated from the extracellular domain of PA-CXCR4-mCherry plasmid after light activation mimics the signal derived from the endogenous CXCR4 after chemokine activation (Xu et al., 2014). Two PA constructs were ordered from GenScript and their primary sequences were shown in Supplemental Figure S1.
Figure 3. DC-STAMP cell surface distribution and OC migration altered by ITIM deletion.
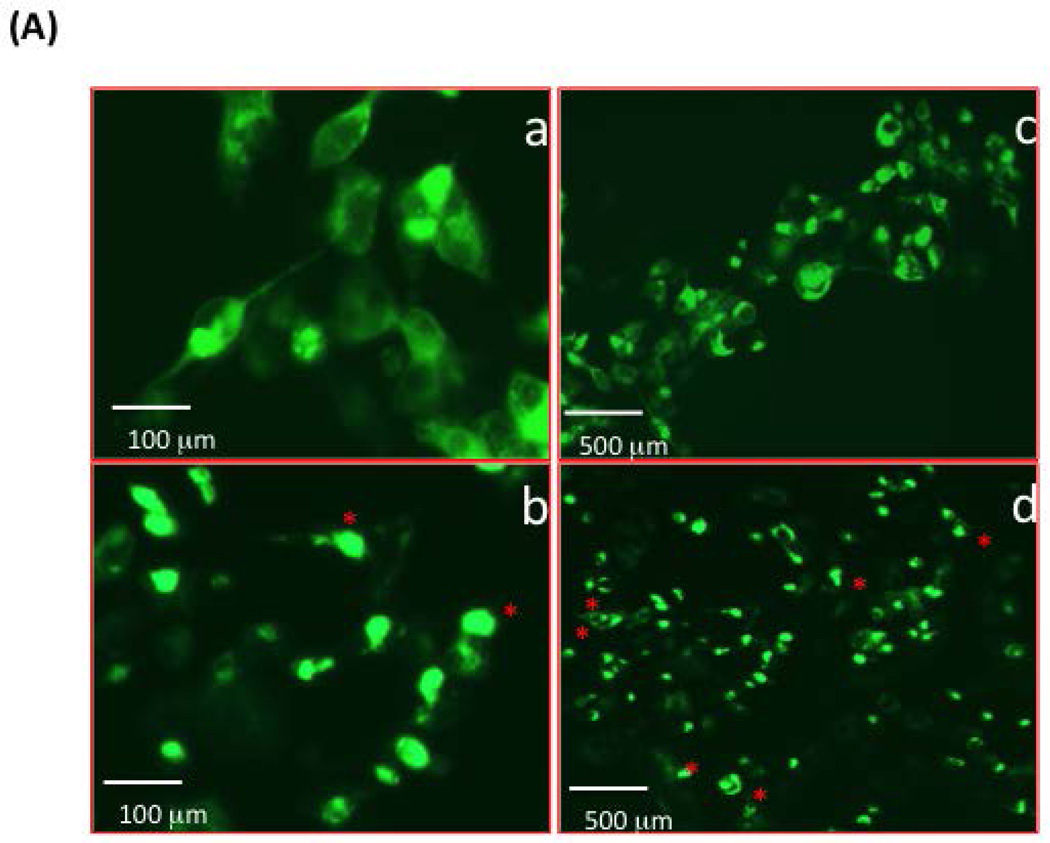
(A) Murine DC-STAMP−/− BMM were infected with GFP-tagged WT- (top row, a & c) or TD-(bottom row, b & d) DC-STAMP. Cellular localization of DC-STAMP was traced by GFP fluorescence. a & b , c & d were photographed under different magnifications as shown by the scale bar; c & d were photographed under the same magnification, TD-DC-STAMP expressing cells demonstrated a more uneven distribution pattern (red asterisks) of DC-STAMP compared to WT-DC-STAMP (d vs. c).
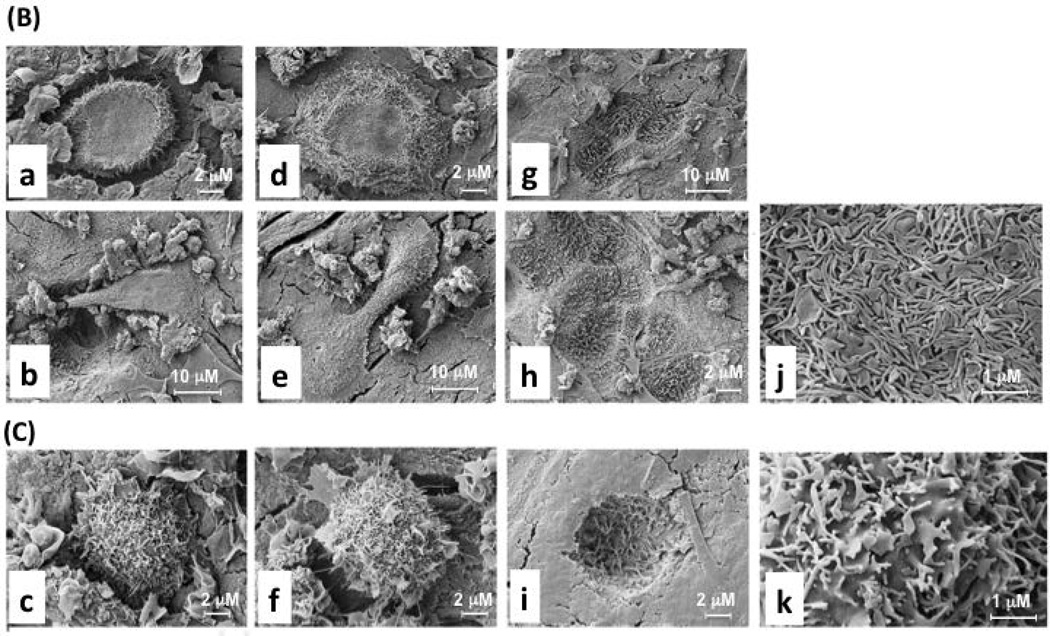
(B) DC-STAMP WT proteins were overexpressed in DC-STAMP−/− cells by retroviral infection, cultured in OC-promoting media (RANKL+M-CSF) on bone wafers, and visualized by scanning electron microscope (SEM).
(C) DC-STAMP TD proteins were overexpressed in DC-STAMP−/− cells following the same experimental conditions as (B). WT DC-STAMP: a, b, d, e, g, h, j; TD DC-STAMP: c, f, i, k. (a)-(f): BMM-derived OC on the surface of bone wafers; (g)-(i): pits eroded by BMM-derived OC; (j)-(k): villous extensions on the cell surface of osteoclasts. Three different scale bars: 1, 2, & 10 µm are shown. The infection efficiency (>70%) by retrovirus on the bone wafer was assessed by GFP expression in a parallel experiment.
Light activation
We used a Nikon Eclipse TE2000-Emicroscope (Nikon) connected with Mosaic microelectromechanical systems (MEMS) Digital Mirror Device (Andor Technology) to apply light illumination to 293T cells in vivo. PA-DC-STAMP-mCherry–expressing 293T cells were generated using the pMX retroviral infection system as described above. The cells were then resuspended in L15 medium (Gibco) containing 2 mg/mL glucose (Sigma) at 4 °C and placed on a mouse ICAM-1–coated Delta T dish (Bioptechs). The cell edge was activated using light (505 nm, 2-µm diameter) in a 30-s light/30-s dark cycle. Time-lapse images were acquired at room temperature or at 37 °C. Image acquisition and analysis were performed using NIS-Element software (Nikon).
Ca2+ assays: set-up, signal conversion, and flux
293T cells (ATCC) were transfected with the pcDNA3.1-PA-DC-STAMP-mCherry plasmid using Lipofectamine 2000 (Invitrogen) on a Delta T culture dish (Biotech). To measure intracellular Ca2+ flux, cells were stained with 2 µg/mL Fluo-4 AM (Molecular Probes) at 37 °C for 30 minutes and incubated at room temperature for an additional 10 min. The cells were re-suspended in Leibovitz’s L15 medium (Gibco) containing 2mg/mL glucose and protected from light. To excite Fluo-4 AM and activate PA-DC-STAMP, the cells were imaged under a GFP HC HiSN Zero Shift Filter set [with an excitation wavelength (450–490 nm), a dichroic mirror (495 nm), and emission filter (500–550 nm)] for 15 min. Time-lapse movies were taken with the Nikon Element software, with 5 seconds frame interval. At the end of the movie recording, 2µM ionomycin (Invitrogen) was added to induce a maximum intracellular calcium release. PA+ transfectants were identified by the red m-Cherry+ signals (Supplemental Figure S3-b & d) from total cells for subsequent analyses.
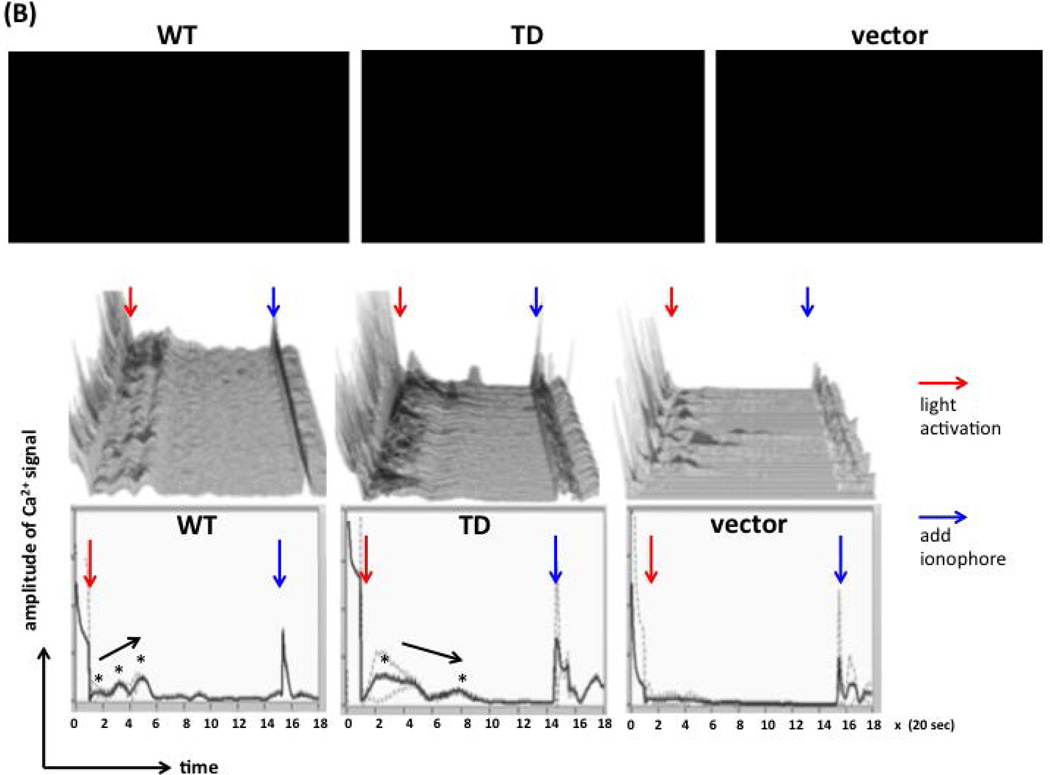
For conversion of Ca2+ signals elicited by each individual cell on 3 videos (top row, Figure 6B), we combined the functions of Amira and Image J for imaging conversion. Briefly, we first manually selected the m-Cherry+ cells, created thresholds to define the edge of m-Cherry+ cells for signal catch in a 0.01 second interval using the Amira software (middle row, Figure 6B), and depicted Ca2+ signals as cumulative curves from single curve of 120 m-Cherry+ cells for each video (bottom row, Figure 6B). The fold changes in Ca2+ signals (Figure 6C), % of cells with spikes (Figure 6D), and duration time of Ca2+ signals (Figure 6E) between WT, TD, and vector-expressing cells were enumerated after the conversion of video signals to digital measurements by the Amira and Image J software.
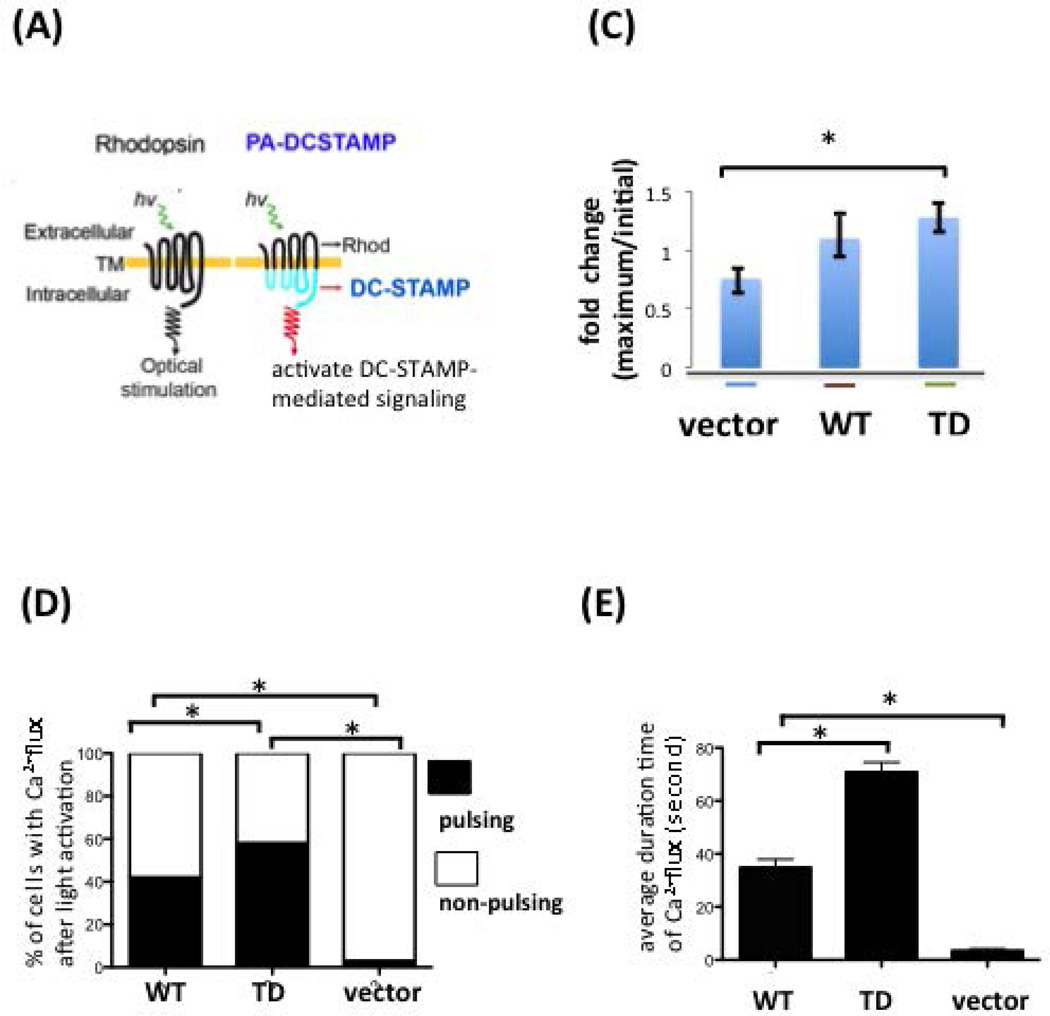
Figure 6. Intracellular Ca2+ flux was altered by ITIM deletion on the DC-STAMP cytoplasmic tail.
(A) The design of the photoactivatable DC-STAMP (PA-DC-STAMP) chimeric construct. Rhod, rhodopsin.
(B) Ca2+ flux assays on WT- or TD-DC-STAMP-overexpressing cells. 293T cells were transiently transfected with WT- or TD-DC-STAMP constructs, labeled with the Fluo-4 Ca2+ dye, and activated with 488-nm light. Cells expressing the PA constructs were selected and identified by the expression of red m-Cherry proteins. Top row: Three videos were recorded for 6 minutes after light activation. Middle row: 120 m-cherry+ cells in each video were selected and the Ca2+ signals, as represented by the green fluorescence, were depicted and converted into curves in a 0.01 second interval using the Amira software. Bottom row: Curves derived from cumulative Ca2+ signals from 120 m-cherry+ cells in each video. The starting and end points of experiments are defined by the light activation (red arrows) and ionophore addition (blue arrows). Click the  button to start each video. Ca2+ pulses are labeled with black asterisks, and the arrows close to the spikes indicate the trend of Ca2+ signal strength.
button to start each video. Ca2+ pulses are labeled with black asterisks, and the arrows close to the spikes indicate the trend of Ca2+ signal strength.
(C) Fold change of intracellular Ca2+ flux before (a) and after (b) light activation on 293T cells transfected with vector, WT-, or –TD-PA-DCSTAMP. A significant difference in Ca2+ flux was detected between the ITIM-deleted PA mutant and vector control (mean +/− SEM; * P ≤ 0.05).
(D) Percentage of WT- and TD- DCSTAMP-expressing 293T cells that showed Ca2+ flux after light activation. * P ≤ 0.05.
(E) Duration time of intracellular Ca2+ signals after light activation in WT-, TD-DCSTAMP, & vector- transfected 293T cells. *P ≤ 0.05.
In vitro OC culture, transfection and retroviral infection
Murine bone marrow cells (4 × 104) were cultured in 96-well plates (α-MEM with 10% FBS and M-CSF (10 ng/ml)) for 2 days, infected with retrovirus for 2 days followed by addition of RANKL (10 ng/ml) for 5 to 7 days. Formation of OC was assessed by counting TRAP+ multinucleated cells with 3 or more nuclei. OC area was assessed using standard stereological methods with an ocular eyepiece grid. NIH imaging software was used to calculate the area of OC. Retrovirus packaging was performed by transfecting plasmids into Plat-E cells (pMY virus). Viral particles were concentrated from cell culture supernatant and used for infection of murine bone marrow macrophage (BMM). Retrovirus packaging was performed by transfecting plasmids into Plat-E cells (pMY virus). Viral particles were concentrated from cell culture supernatant and used for infection of murine bone marrow macrophage (BMM).
Scanning electron microscopy (SEM)
DC-STAMP −/− cells were isolated from the bone marrows of DC-STAMP KO mice, cultured in the OC-promoting media (+M-CSF) for 2 days, and infected with WT or TD DC-STAMP retroviral stock in the presence of RANKL & M-CSF for 14 days on bone wafers. Bone wafers were fixed in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer and post-fixed in buffered 1.0% osmium tetroxide, dehydrated in a graded series of ethanol to 100%, transitioned into a series of ethanol/HMDS to 100% HMDS and dried overnight. For imaging, the wafers were sputter coated with gold and imaged using a Zeiss-Auriga Field Emission Scanning Electron Microscope with a Gatan digital camera. The NIH Image J software was used to quantify the bone erosion areas on EM.
Bone erosion assay
BM cells isolated from DC-STAMP KO mice were infected with retrovirus expressing WT or TD DC-STAMP. They were cultured with M-CSF on bone slices to generate OCPs and RANKL was added 2 days later. Cells were fixed and stained for TRAP activity for OC enumeration. Three wells of OC were counted for each sample to obtain an average OC count/per million of cultured cells. Bone slices were stained with 0.5% toluidine blue to visualize resorption pits as previously described (Chiu et al., 2010).
NFATc1 nuclear translocation
WT or TD DC-STAMP-GFP virus-infected BMM were cultured with RANKL and M-CSF to generate osteoclasts in 8-chamber slides. After mature osteoclasts were observed, cells were fixed with 10% neutral buffered formalin and permeabilized using 0.1% Triton X-100. Immunofluorescent staining was performed using mouse anti-NFATc1 antibody (7A6, Santa Cruz, Biotechnology, Santa Cruz, CA, USA) followed by DyLight 594-conjugated goat anti-mouse IgG (Thermo Scientific Pierce Biotech). The nuclear location was labeled with DAPI (Vectashield Mounting Media, H-1200, Vector Laboratory, Burlingame, CA). Subcellular localization of DyLight 594-labeled NFATc1 was observed using confocal microscopy.
Statistical analysis
All results shown in this study were repeated 3 times for each condition. Data were analyzed by the Prism 6 software and presented as mean ± SD. Statistical analyses were performed with StatView statistical software (SAS Institute, Cary, NC, USA). Differences between two groups were compared using an unpaired Student’s t test, whereas more than two groups were compared using one-way analysis of variance (ANOVA) between groups, and p values <0.05 were considered to be statistically significant.
Results
The ITIM on DC-STAMP is essential for cell-cell fusion and normal bone erosion activity
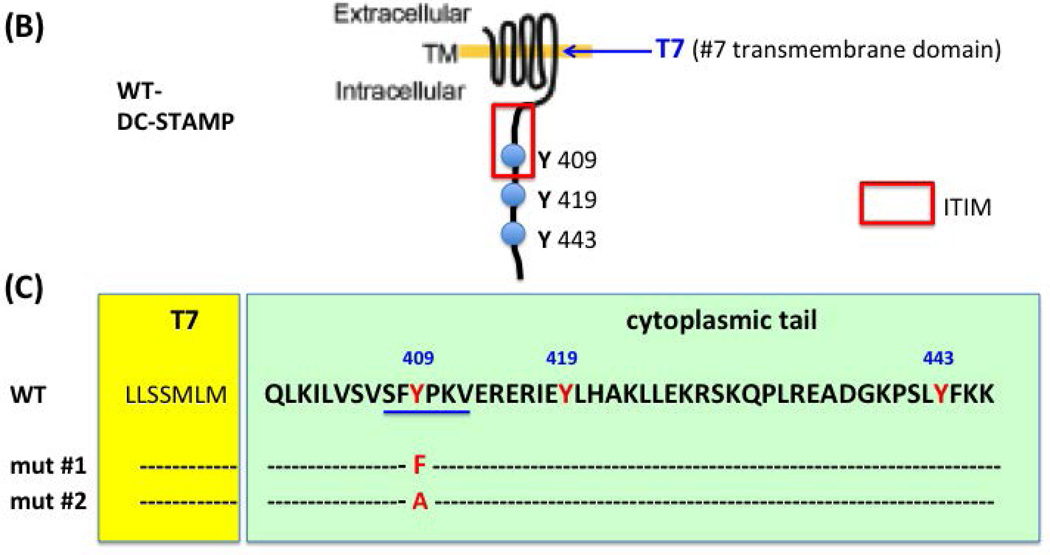
DC-STAMP−/− cells failed to generate multi-nucleated OC, even in the presence of RANKL and M-CSF (Yagi et al., 2005). To examine the function of the ITIM, we established three mutated versions of DC-STAMP (Figure 1C). The first, a 6 amino acid-deletion in the ITIM (TD), the second, a tyrosine to phenylalanine (Y409 to F409) mutation, and the third, a tyrosine to alanine (Y409 to A409) point mutation (Figure 1A-1C). These DC-STAMP mutants were designed to determine if the intact ITIM, the tyrosine residue within the ITIM or amino acids in the cytoplasmic tail are essential for the function of DC-STAMP. We introduced these 3 DC-STAMP constructs into DC-STAMP−/− cells and examined OC formation, cell-cell fusion, bone erosion, and DC-STAMP cell surface expression compared to transfection with WT DC-STAMP constructs.
Figure 1. Comparison of human and murine sequences of DC-STAMP. Susceptible (L400 to F400) point mutations in Paget's disease and ITIM are present in the DC-STAMP cytoplasmic tail.
(A) The transmembrane (T) regions of DC-STAMP are labeled with T1-T7. The point mutation (L to F) identified in Paget’s disease is labeled with a red arrow; ITIM labeled with a red bracket, 3 tyrosine residues close to ITIM are labeled in red, and deleted protein sequences (TD) in GFP and PA chimeric constructs are underlined.
(B) Three tyrosine residues (in red) are close to ITIM. . Red rectangle: ITIM; Blue: 6 amino-acid deletions in tail-deleted mutant (TD).
(C) Two point mutations (mut#1: Y409 F409; mut#2: Y409
F409; mut#2: Y409 A409) were introduced in the ITIM.
A409) were introduced in the ITIM.
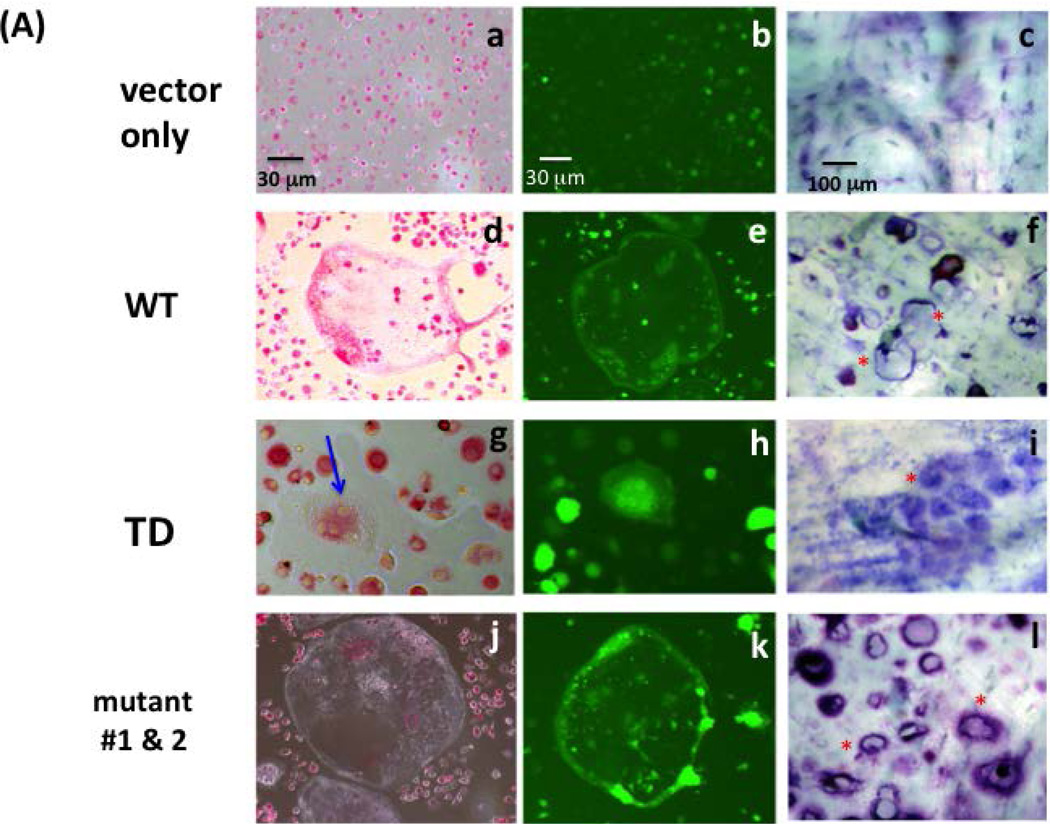
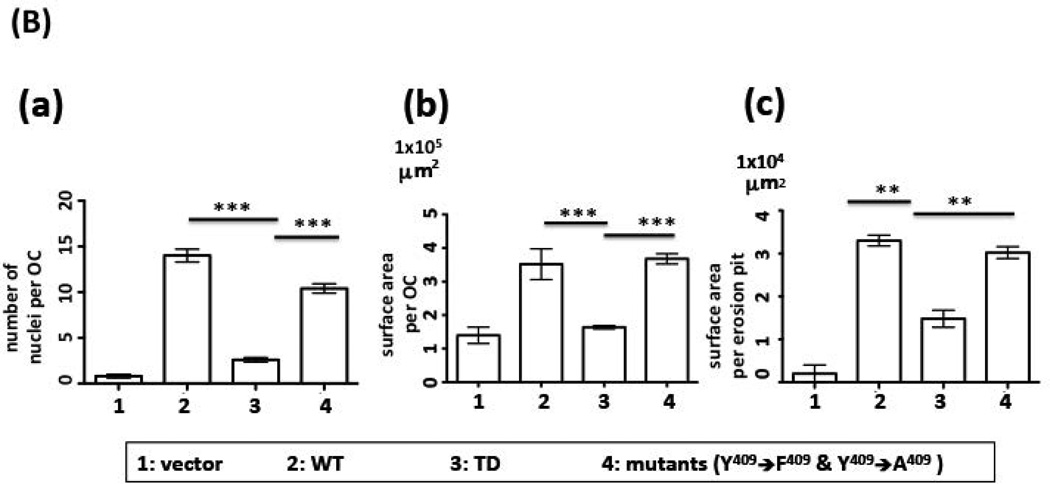
As shown in Figure 2, the WT version of DC-STAMP-GFP rescued the cell-cell fusion deficiency in DC-STAMP−/− cells as evidenced by the generation of multi-nucleated OC (Figure 2A-d). The green fluorescence of GFP confirmed the expression of the DC-STAMP construct (Figure 2A-e). In contrast, ITIM-deleted (TD) DC-STAMP failed to rescue the OC-forming deficiency of DC-STAMP−/− cells. The only multi-nucleated cells found in TD complementation contained <4 nuclei (blue) and most cells were mononuclear. The frequency of cells with 4 nuclei in TD-infected BMM was low (4.0 ± 0.8 per 106 GFP+ cells). Of interest, the (Y to F, Y409F) mutant complemented cell-cell fusion deficiency of DC-STAMP−/− cells to a level similar to that of WT DC-STAMP (Figure 2A-j vs. 2A-d). In addition, a punctate aggregated DC-STAMP surface distribution was detected (Figure 2A-k vs. 2A-e). To test bone erosion activity, a parallel set of WT, TD, and (Y409F) infected BMM cells were examined in a bone wafer assay. Consistent with the ability of WT and (Y409F) but not TD mutant to complement the cell-cell fusion deficiency of DC-STAMP−/− cells (Figure 2A-b, e, h, k), extensive bone resorption was only detected in cells infected with WT and the (Y409F) mutant (Figure 2A-f & 2A-l), but not in TD-overexpressing cells (Figure 2A-i). Cells infected with vector were added as control (Figure 2A, a-c). The complementation activity of WT, TD and (Y409F) to cell-cell fusion deficiency on DC-STAMP−/− cells was evaluated by the # of nuclei/cell, the area of OC, and the area of formed pits (Figure 2B, a-c).
Figure 2. Deletion of ITIM on DC-STAMP alters cell-cell fusion, cell volume and bone resorption.
(A) DC-STAMP−/− cells were infected with GFP-tagged DC-STAMP constructs: wild-type (WT, d-f), ITIM-deleted (TD, g-i), tyrosine mutants (Y409 F409 or
F409 or  A409, j-l), or vector (a-c). After viral infection, cells were cultured in OC-promoting media (RANKL and M-CSF). 1st column: TRAP staining (a,d,g,j); 2nd column: GFP fluorescent images (b,e,h,k); 3rd column: bone wafer assay (c,f,i,l). Red asterisks identify selected erosion pits.
A409, j-l), or vector (a-c). After viral infection, cells were cultured in OC-promoting media (RANKL and M-CSF). 1st column: TRAP staining (a,d,g,j); 2nd column: GFP fluorescent images (b,e,h,k); 3rd column: bone wafer assay (c,f,i,l). Red asterisks identify selected erosion pits.
(B) Summary of results shown in (A). (a) Number of nuclei per OC, (b) surface area per OC, (c) surface area per erosion pit. (***P=0.001; **P=0.01).
ITIM deletion on DC-STAMP alters morphology and mobility of osteoclasts
The relative cell surface distribution of DC-STAMP, CD47, CD9, and syncytin-1 are critical for optimal cell-cell fusion (Hobolt-Pedersen et al., 2014; Soe et al., 2015). Altered cell-cell fusion and bone erosion activity associated with the ITIM deletion (Figure 2A) suggests this motif may affect the cell surface distribution of DC-STAMP. To address this possibility, we compared DC-STAMP distribution on WT- or TD-infected 293T (Figure 3A-a, b) and mouse BMM (Figure 3A-c, d). Cells transfected with the ITIM deletion on DC-STAMP demonstrated an uneven and punctate cell surface distribution of DC-STAMP, which was especially prominent in 293T cells (Figure 3A-b, red asterisks).
To complete bone erosion, osteoclasts extend their ruffled borders with villous extensions, form a tight seal with bone, acidify the extracellular area to solubilize the mineral and release enzymes which dissolve the matrix (Henriksen et al., 2011; Henriksen et al., 2006). Given that bone erosion activity is greatly impaired by ITIM deletion (Figure 2A), we examined the effect of ITIM deletion on osteoclast morphology and bone erosion activity by scanning electron microscopy (SEM). SEM is commonly applied to evaluate two characteristics of an active osteoclast, ruffled extending borders and a tight attachment of osteoclast to the bone surface (Rifkin and Gay, 1992).
The shape, gliding mobility, flagellum-like cytoplasmic extensions, and bone erosion activity in WT- and TD-infected DC-STAMP−/− cells were examined by SEM after 14-day incubation on bone wafers (Figure 3B: WT; Figure 3C). Major differences in cell shape, gliding and resorption activity, and cell surface microvilli structure were noted in WT and TD DC-STAMP-expressing osteoclasts. First, DC-STAMPWT-expressing osteoclasts were characterized by a thin, extended morphology and attached to the bone surface tightly (Figure 3B-a, b, d & e), whereas DC-STAMP-TD -expressing cells were rounded and more compact (Figure 3C-c & f). Of note, many of rounded DC-STAMP-TD -expressing osteoclasts lacked membrane ruffles or microvilli. Second, DC-STAMPWT-expressing osteoclasts excavated more resorption pits, a larger resorption area (Figure 3B-g & h, >300 µM2 / pit) and adjacent regions of scalloped bone (>75% of resorption pits), suggesting a higher gliding activity, characteristic of a mobile multinucleated OC. In contrast, single pits without adjacent areas of resorption were observed on bone wafers incubated with DC-STAMP-TD -expressing osteoclasts (Figure 3C-i), suggesting that DC-STAMP-TD cells are not mobile. We were unable to quantify the depth of the pits, however, with this technique. The images of resorption pits also revealed a difference in the membrane ruffles and microvilli between DC-STAMPWT (Figure 3B-g & h) and DC-STAMP-TD (Figure 3C-i)-expressing osteoclasts. Under a higher magnification, microvilli on DC-STAMPWT cells were denser compared to the flattened ruffles on DC-STAMP-TD (Figure 3B-j vs. 3C-k). The SEM results were consistent with the observation from the bone wafer assay in which a larger erosion area was detected in WT- compared to TD- DC-STAMP-overexpressing cells (Figure 2A-f vs. 2A-i),
ITIM-deleted DC-STAMP is stably expressed on the cell surface
The fact that multiple defects including cell-cell fusion, bone erosion, and cell mobility were observed in ITIM deleted constructs (Figure 2 & 3) raised the possibility that deletion of ITIM affects the stability of DC-STAMP protein and its cell surface expression. To assess if the punctate cellular structures of TD DC-STAMP represent accumulation of membrane vesicle-like structures or misfolding TD proteins wrapped within lysosome in apoptotic cells, we examined the co-expression of GFP and DC-STAMP on WT and TD DC-STAMP by flow cytometry (Figure S4(A)). Co-expression of GFP and DC-STAMP were detected in both WT- and TD-expressing cells (Figure S4-a & S4-b). Intriguingly, under a defined GFP mean fluorescence intensity (MFI), two cell subsets (red arrows in Figure S4-a & S4-b) can be clearly distinguished by MFI of DC-STAMP, suggesting a heterogenous cell population in TD-overexpressing cells. The overlay histogram (Figure S4, A-c) showed that the cell surface expression level of DC-STAMP is higher in DC-STAMPTD than DC-STAMPWT (Figure S4(A), blue vs. green lines).
ITIM deletion on DC-STAMP impairs NFATc1 nuclear translocation
NFATc1 is a master regulator that regulates NFkB-induced OC differentiation (Kim et al., 2008; Negishi-Koga and Takayanagi, 2009). De-phosphorylation of NFATc1 by calcineurin results in a conformational change that exposes a nuclear localization signal of NFATc1 and results in NFATc1 nuclear import (Yarilina et al., 2011; Zhao et al., 2010). NFATc1 translocation from cytoplasm to the nucleus is a crucial step that determines the activation of transcriptional regulators involved in bone homeostasis (Yamashita et al., 2007; Yarilina et al., 2011). Given that targeted inhibition of DC-STAMP by miRNA, miR-7b, was associated with suppression of NFATc1 (Dou et al., 2014), as well as decreased cell-cell fusion, bone erosion, and cell mobility, characteristics observed in ITIM deletion constructs (Figure 2 & 3), we examined the effect of ITIM deletion on NFATc1 cellular localization with confocal microscopy and western blot analysis.
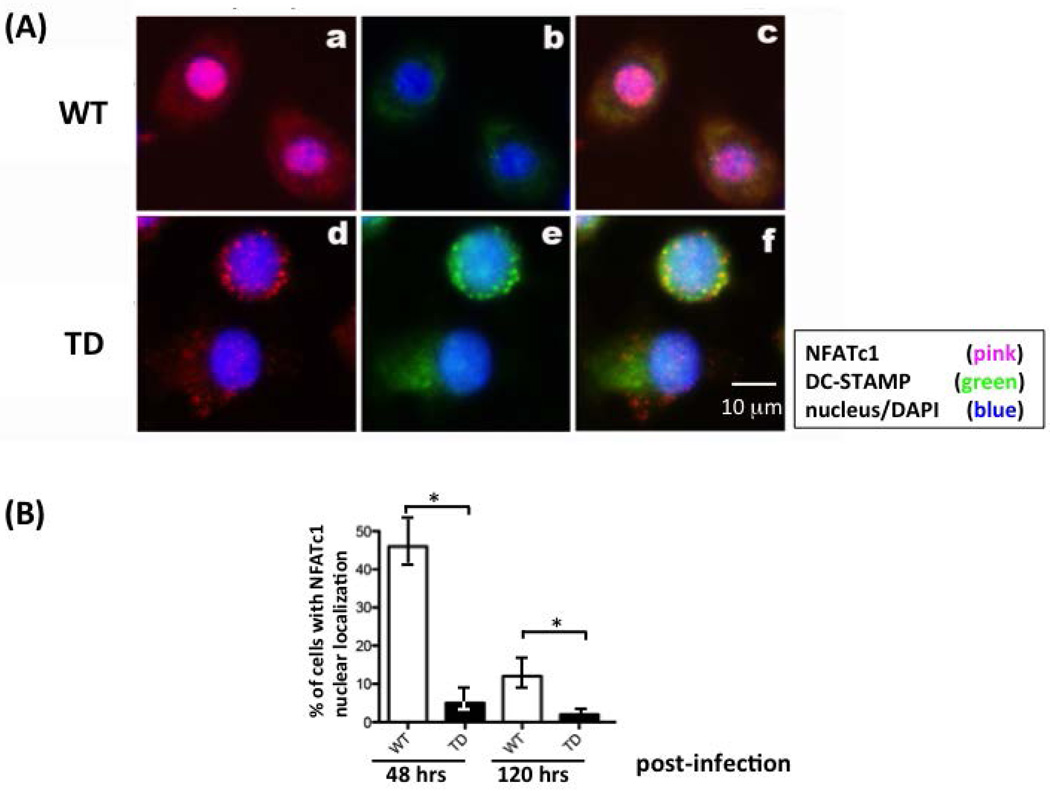
Cellular localization of NFATc1 was examined on DC-STAMP−/− cells that overexpress DC-STAMPWT or DC-STAMPTD proteins after retroviral infection. NFATc1 was present in the nuclei of DC-STAMPWT-expressing cells (Figure 4(A)a-c, S4(B)a & b, & S4(C)a to d,), but remained in the cytoplasm of DC-STAMPTD-expressing cells (Figure 4(A)d-f, S4(B)c & d, & S4(C)e to h,). These data are consistent with the concept that amino acids in the DC-STAMP cytoplasmic tail are involved, either directly or indirectly, in the nuclear translocation of NFATc1.
Figure 4. Nuclear translocation of NFATc1 is decreased in cells infected with DC-STAMP-TD constructs.
(A) DC-STAMP−/− BMM were transiently infected with GFP-tagged WT (top row) or TD (bottom row) DC-STAMP. a & d: NFATc1/nucleus overlay; b & e: DC-STAMP/nucleus overlay; c & f: overlay of NFATc1/DC-STAMP/nucleus.
(B) Percentage of cells with NFATc1 nuclear localization. The percentage of cells with NFATc1 nuclear translocation was calculated by examining 100 GFP+ cells at two time points post-infection (48 & 120 hours). Figure S4 depicts WT- and TD-DC-STAMP-expressing cells in a lower magnification.
Knockout of DC-STAMP affects NFATc1 expression at the protein level
It has been previously shown that knock down of DC-STAMP does not affect the expression of NFATc1 at the transcriptional level (Yagi et al., 2005). To confirm this finding, we examined NFATc1 gene expression in DC-STAMP−/− macrophages by RT-PCR and the level was significantly reduced compared to DC-STAMP+/+ cells (data not shown). Intriguingly, a significant decrease in NFATc1 proteins was also detected in DC-STAMP−/− BMMs by Western blot analysis compared to WT BMM (Figure 5A), suggesting that ablation of DC-STAMP affects NFATc1 protein expression either through regulation of translation efficiency or protein degradation.
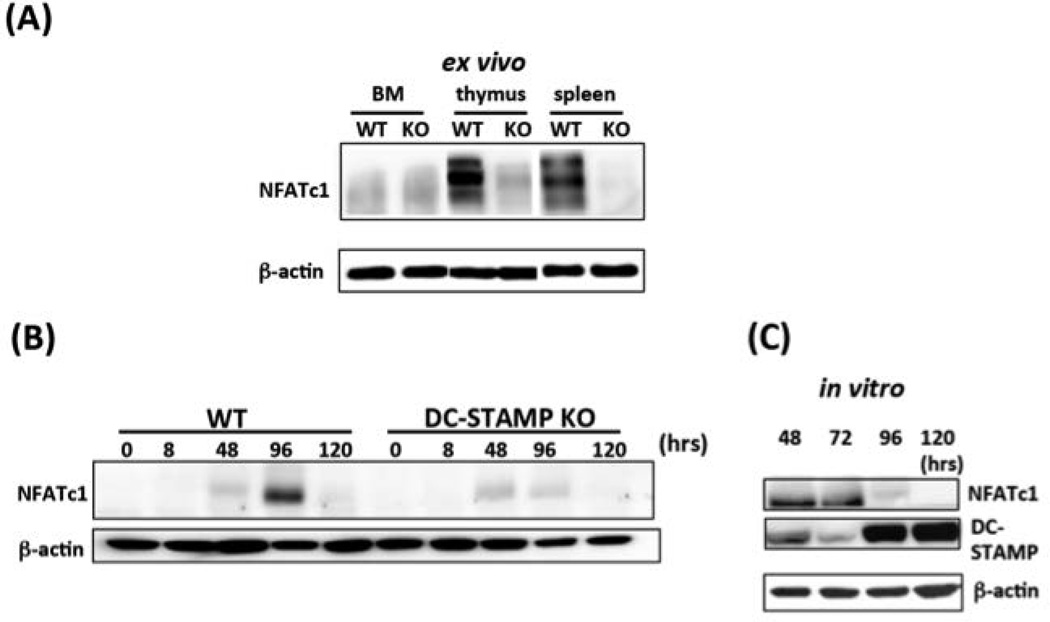
Figure 5. Relationship between DC-STAMP and NFATc1 protein expression.
(A) Ex vivo expression of DC-STAMP. DC-STAMP−/− cells demonstrate reduced expression of NFATc1 protein in bone marrow, thymus, and spleen. Western blots depict proteins isolated from DC-STAMP +/+ (WT) & DC-STAMP −/− (KO) littermates.
(B) Expression of DC-STAMP in WT and DC-STAMP−/− cells cultured with RANKL and M-CSF. NFATc1 protein expression was induced when DC-STAMP+/+ and DC-STAMP−/− BMMs were cultured in the presence of RANKL+M-CSF. The maximal NFATc1 expression in +/+ and −/− cells was noted at 96 and 48 hours post-infection, respectively. The expression of NFATc1 on DC-STAMP+/+ (left) and DC-STAMP−/− (right) cells after 8, 48, 96 and 120- hours in the presence of RANKL+M-CSF is shown.
(C) Reduced NFATc1 protein level in DC-STAMP−/− cells is restored by DC-STAMP overexpression. Reduced NFATc1 protein expression in DC-STAMP−/− cells (Fig 2B) was restored following retroviral infection with WT DC-STAMP construct. Total proteins were isolated at 4 time points post-infection (48, 72, 96, 120 hours) and subjected to western blot analysis.
Next, we addressed whether overexpression of WT DC-STAMP rescues expression of NFATc1 protein. NFATc1 proteins were monitored by Western blotting in DC-STAMP−/− cells infected with WT or TD DC-STAMP at 8, 48, 96 and 120 hours, (Figure 5B, WT: lanes 1–5; TD: 6–10). The expression of NFATc1 was detected at 48 hours in enriched WT bone marrow macrophages (Figure 5B, lane 3) and reached a peak at 96-hr (Figure 5B, lane 4). In contrast, maximal NFATc1 expression was detected in DC-STAMP−/− cells at 48 hours (Figure 5B, lane 8) and declined at 96 hours post culture (Figure 5B, lane 9). These data suggest that the overall decrease of NFATc1 by DC-STAMP knockout can be rescued by DC-STAMP overexpression, which prompted us to further investigate the interaction between NFATc1 and DC-STAMP proteins.
As shown in Figure 5C, we quantified both NFATc1 and DC-STAMP protein after DC-STAMP−/− cells were infected with wild-type DC-STAMP retrovirus (construct #3 in the supplemental Figure S1), and cultured in vitro for 48, 72, 96, and 120 hrs. Expression of NFATc1 was first noted after 48-hr (Figure 5C, top panel, lane 1), but gradually declined after 96-hr post-infection (Figure 5C, top panel, lanes 2–4). The expression level of DC-STAMP steadily increased between 48–120 hours post-infection (Figure 5C, center panel, lanes 1–4). These observations confirm that DC-STAMP can rescue decreased NFATc1 expression observed in DC-STAMP−/− cells (Figure 5C, top panel, lanes 1 & 2) at early time points (48–72 post-infection) before the maximal level of DC-STAMP proteins was detected (Figure 5C, middle panel, lanes 3 & 4, for 96- & 120 hour post-infection). The maximal expression of DC-STAMP and NFATc1 proteins peaked at different time points (Figure 5C, NFATc1 peaks at 48 and 72 hours, whereas DC-STAMP peaked at 96 and 120 hours). These data indicate some type of reciprocal regulation between DC-STAMP and NFATc1 during osteoclastogenesis. Additional studies are in progress to examine this this interaction in more detail.
Deletion of ITIM on DC-STAMP alters the patterns of intracellular Ca2+ signals
As shown above, ITIM deletion on DC-STAMP impairs NFATc1 nuclear translocation (Figure 4 & S4-(B) & (C)) and affects NFATc1 expression at the protein level (Figure 5). These observations, together with the fact that NFATc1/Ca2+ signaling is an essential axis of osteoclast differentiation (Negishi-Koga and Takayanagi, 2009; Zhao et al., 2010), raised the possibility that ITIM deletion might also disturb intracellular Ca2+ signaling. The absence of a known DC-STAMP ligand, however, precludes detailed analysis of downstream signaling of this molecule.
To overcome this barrier, we employed an optogenetic approach previously established for the study of the Photo-Activatable-chemokine C-X-C motif receptor 4 (PA-CXCR4), a protein that shares a high structure similarity with DC-STAMP (Xu et al., 2014). We engineered two photo-activatable DC-STAMP chimeric molecules, PA-DCSTAMPWT and PA-DCSTAMPTD (Figure 6A, S1: constructs #5–8), in which the extracellular and transmembrane domains of DC-STAMP were replaced with rhodopsin (Supplemental Figure S2) (Xu et al., 2014). Transfected cells can be easily distinguished from non-transfected cells by the expression of red m-Cherry proteins tagged to the C’-termini of PA-DCSTAMPWT and PA-DCSTAMPTD molecules (Supplemental Figure S3-b). Overlay of the green [Ca2+] signals with red PA-DC-STAMP-positive transfectants enabled us to identify cells that express chimeric PA-DC-STAMP proteins with high [Ca2+] signals at the time of imaging (Supplemental Figure S3-d).
To determine whether intracellular [Ca2+] flux is affected by ITIM deletion on DC-STAMP, we transiently transfected 293T cells with PA-DCSTAMPWT and PA-DCSTAMPTD, stimulated cells with 488 nm light, and monitored [Ca2+] signals by video imaging. As shown in the videos of Figure 6B (top panel), light activation was sufficient to induce prominent downstream [Ca2+] signals in PA-DC-STAMP-expressing cells (WT and TD) but not in vector alone control. We combined the automatic coding function of Amira and ImageJ to randomly select 120 red m-cherry+ cells from each video, and converted their Ca2+ signals into digital measurements with 1/100 sec interval from the beginning to the end of the video recording (light activation and ionophore addition, red and blue arrows in Figure 6B middle panel, respectively). The Ca2+ signal from each cell is represented with a single line (middle panel of Figure 6B). In addition, we also added up the Ca2+ signals from these 120 randomly selected cells and depicted them in a single cumulative curve (bottom panel of Figure 6B). Of note, no spikes were detected in cells transfected with vector control. Collectively, these results suggested that PA-DCSTAMPWT and PA-DCSTAMPTD (1) are functional molecules that respond to light stimulation; with distinct patterns. After light activation, PA-DCSTAMPWT showed significant flux as evidenced by 3 distinct spikes (black asterisks, far left, bottom panel of Figure 6B) with an increasing trend. In contrast, only 2 distinct spikes were found in the PA-DCSTAMPTD–expressing cells with a trend of decreasing Ca2+ magnitude (black asterisks, middle in the bottom panel of Figure 6B).
We further calculated the magnitude of Ca2+ intensity (Figure 6C), percentage of cells with Ca2+ spikes (Figure 6D), and the duration time of detectable Ca2+ signals after light activation (Figure 6E). The magnitude of Ca2+ intensity of DCSTAMPWT and PA-DCSTAMPTD after light activation was calculated by the ratio of maximal to minimum Ca2+ digital signals (Figure 6C). A significantly higher percentage of cells with intracellular Ca2+ signals was found in TD transfectants (Figure 6D), suggesting that ITIM deletion either promotes the uptake of Fluo-4 AM Ca2+ indicator or inhibits the extracellular release of Fluo-4 AM Ca2+ indicator from the Ca2+ pump. We also measured the duration of the Ca2+ signal in 100 WT- and TD-DC-STAMP cells, respectively. TD-DC-STAMP transfectants showed sustained and prolonged Ca2+ signaling compared to WT-DC-STAMP cells that was significantly different (Figure 6E). Taken together, our results suggest that the ITIM on the cytoplasmic tail of DC-STAMP is essential for induction of intracellular Ca2+ flux. Deletion of ITIM on DC-STAMP was associated with altered Ca2+ flux resulting in an elevated and sustained intracellular Ca2+ levels.
Discussion
Formation of multinucleated osteoclasts requires several rounds of cell-cell fusion between single nucleated OCPs (Helming and Gordon, 2009). Cell-cell fusion does not occur randomly but takes place in a selective manner that is determined by the maturation stage and cellular organization of fusion factors and molecules (Hobolt-Pedersen et al., 2014; Soe et al., 2011). DC-STAMP is considered a major regulator of osteoclastogenesis (Islam et al., 2014; Yagi et al., 2005; Yagi et al., 2006) among several critical osteoclastogenic proteins (Islam et al., 2014; Lee et al., 2006; Zhang et al., 2014), because it is required for the formation of multinucleated OC (Chiu et al., 2012; Zhang et al., 2014). However, the molecular mechanisms underlying DC-STAMP-mediated osteoclastogenesis remains largely unknown. Previous identification the ITIM on the cytoplasmic tail of DC-STAMP (Chiu et al., 2012) suggested its role in signaling, which prompted us to propose a model in which ITIM serves as an inhibitory signal to regulate osteoclastogenesis through Ca2+ oscillation (Chiu et al., 2012). Unfortunately, formally testing this model proved difficult because the ligand of DC-STAMP is not known. We addressed this technical hurdle by employing an optogenetic approach. We constructed a chimeric DC-STAMP molecule by replacing the extracellular domain with rhodopsin to mimic endogenous ligand engagement with light activation. We further examined the role of ITIM on signaling and OC function by comparing multiple WT and ITIM-deleted (TD) DC-STAMP constructs in the DC-STAMP−/− knockout background. Results shown in this study suggest that ITIM is a bona fide signaling motif that regulates cell-cell fusion and other biological functions related to osteoclastogenesis through the Ca2+/ NFATc1 axis.
The RNA and protein levels of NFATc1 are regulated by OC-related genes such as Tgif2, c-fos, NF-kβ (Krzeszinski et al., 2014; Matsuo et al., 2004; Yamashita et al., 2007). Although it is still unknown whether similar regulation occurs between DC-STAMP and NFATc1, several lines of evidence suggest that there is a mutual regulation between DC-STAMP and NFATc1. First, constitutive overexpression of NFATc1 in BMMs induces osteoclast fusion even in the absence of RANKL (Takayanagi et al., 2002), suggesting that NFATc1 can override RANKL deprivation and is independent from the RANKL-driven OCgenesis pathway. Second, overexpression of NFATc1 induces cell-cell fusion via up-regulation of DC-STAMP (Kim et al., 2008). Third, overexpression of DC-STAMP rescues cell-cell fusion deficiency of pre-OC despite reduced NFATc1 activity (Kim et al., 2008). Fourth, NFATc1 binds to the promoter of DC-STAMP, which determines the initiation of transcription of DC-STAMP (Yagi et al., 2007). Lastly, DC-STAMP-specific miRNA, miR-7b, suppresses NFATc1 signaling during osteoclastogenesis (Dou et al., 2014). These findings, together with our data that NFATc1 nuclear translocation and protein expression are altered by DC-STAMP (Figure 4 & 5), suggest a reciprocal regulation between DC-STAMP and NFATc1 at the levels of gene and protein expression. Inhibition of DC-STAMP either by DC-STAMP knockout (Figure 5A) or miRNA suppression (Dou et al., 2014) was associated with decreased osteoclastogenesis and NFATc1 suppression. Intriguingly, DC-STAMP knockdown was associated with a decrease in NFATc1 protein (Figure 5A). but not mRNA (Yagi et al., 2005), In addition, we showed that the expression levels of NFATc1 and DC-STAMP proteins peaked at different time points after re-introduction and overexpression of DC-STAMP in DC-STAMP−/− cells (Figure 5C). Given that binding of RANKL to its receptor RANK initiates a signaling cascade that promotes Ca2+ mobilization and induces c-Fos followed by NFATc1 expression (Yamashita et al., 2007), results shown in this study are consistent with previous studies which suggest DC-STAMP regulates osteoclastogenesis thru the NFATc1- Ca2+ axis (Asagiri and Takayanagi, 2007; Yarilina et al., 2011).
We chose to use PA chimeric proteins to mimic ligand engagement by light activation in 293T cells instead of murine bone marrow macrophage-based on the following considerations: (1) 293 T cells is a cell line commonly used for testing protein biochemical property (Schindl et al., 2012) and is also accepted for characterizing biological functions of osteoclasts (Ong et al., 2013). The initial characterization of DC-STAMP was also performed on the 293T cell line (Hartgers et al., 2000); (2) PA-DCSTAMP chimeric proteins (WT or TD) can only be introduced into murine bone marrow by retroviral infection, which takes at least 2–3 days for the expression of transduced proteins and thus it’s technically challenging to determine the optimum point for light activation. In contrast, 293 T cells are easily transiently transfected with PA-DC-STAMP and the expression of transduced proteins can be detected within 24 hours and confirmed by the expression of red m-Cherry protein fused to the C’-terminus of DC-STAMP. Results from 293T cells, however, does reflect the effect of ITIM-deletion on osteoclastogenesis after light activation because 293T cells don’t form osteoclasts. We are now infecting murine bone marrow macrophage (BMM) with WT or TD of PA-DC-STAMP constructs and optimizing the optogenetic parameters to address this critical question.
Initiation of osteoclastogenesis requires a dual-signal activation, one from RANKL engagement and a second costimulatory signal from ITAM-containing receptors/adaptors (Dou et al., 2014; Li et al., 2014). ITAM-mediated signals depend on the phosphorylation and dephosphorylation on the tyrosine residues surrounding the ITAM by Syk or zeta-associated protein kDa tyrosine kinases (Humphrey et al., 2005). Thus, alternation and/or replacement of tyrosine residues close to ITAM causes either the loss function of ITAM in vitro (Li et al., 2014) or switch of ITAM function from an activation to an inhibitory configuration (Ben Mkaddem et al., 2014). Of clinical relevance, replacement of tyrosine residues surrounding or on the ITAM dramatically changes the outcomes from susceptibility to resistance in regard of arthritis pathogenesis in an animal model (Ben Mkaddem et al., 2014). These results, together with the counteracting function between ITAM- and ITIM-bearing receptors (Crotti et al., 2015; Ivashkiv, 2009; Koga et al., 2004; Takayanagi, 2007), led us to propose a model in which DC-STAMP, an ITIM-bearing transmembrane protein, regulates the function of ITIM-ITAM network in osteoclastogenesis (Chiu et al., 2012). We showed a physical interaction between DC-STAMP and SHP-1, an SH2-containing tyrosine-specific protein phosphatase, by co-immunoprecipitation (co-IP), supporting the involvement of DC-STAMP in signaling, through the tyrosine phosphorylation/dephosphorylation (Chiu et al., 2012). To further test this model, we established several DC-STAMP tail mutants on either GFP-tagged or photoactivatable vector backbones (Figure 1B, supplemental figure S1 & S2) and examined the effect of these mutations on DC-STAMP function in DC-STAMP−/− cells. Our results showed that deletion of 6 or 16 amino acids surrounding the ITIM in GFP-tagged (blue underline in Figure 1A) or photoactivatable DC-STAMP chimera (red underline in Supplemental Figure S2) inhibited DC-STAMP function in regard to multinucleation and bone resorption (Figure 2A-g, h, i). On the contrary, point mutations on the tyrosine residue of ITIM (Y409F and Y409A) did not affect DC-STAMP function (Figure 2A-j,k,l). Collectively, these results suggest that an intact ITIM sequence is required for complementing OC-forming deficiency in DC-STAMP−/− cells, whereas Y409 phosphorylation or/and structure of cytoplasmic tail might not play a critical role in DC-STAMP function as previously proposed (Chiu et al., 2012).
In summary, results shown in this study provide observations on the function of ITIM on the cytoplasmic tail of DC-STAMP (Chiu et al., 2012). Our results suggest that an intact ITIM is essential for DC-STAMP function, not only cell-cell fusion, but also for optimal bone resorption, cell mobility, DC-STAMP cell surface distribution, NFATc1 nuclear translocation, and calcium flux. Additional studies are underway to elucidate the molecular mechanisms underlying the role of ITIM in the regulation of these biological aspects during osteoclastogenesis. The finding of a point mutation on the cytoplasmic tail of DC-STAMP in patients with Paget’s disease (Beauregard et al., 2014) coupled with a report that murine arthritis severity can be attenuated by switching the function of ITAM from activation to inhibition via tyrosine replacement (Ben Mkaddem et al., 2014), raises the possibility that modulating ITIM activity in DC-STAMP by ITIM-specific targeting biologics such as siRNA or CRISPR has therapeutic potential in inflammatory arthritis and metabolic bone disorders.
Supplementary Material
Acknowledgments
We would like to thank Dr. Takeshi Miyamoto at Keio University in Japan for providing us the DC-STAMP KO mouse strain and staff at University of Rochester animal facility for animal care. We would like to thank Dr. Carrie Dykes for her help on editing of our manuscript. This study is supported by funds from the National Institutes of Health (NIH) AR0169000, 5UL1TR000042-09, and P30AR069655.
List of abbreviations
- ITIM
Immunoreceptor Tyrosine-based Inhibitory Motif
- RANKL
Receptor Activator of NF-κB Ligand
- M-CSF
macrophage colony-stimulating factor
- Ca
calcium
- NFATc1
nuclear factor of activated T cells, cytoplasmic 1
- DC-STAMP
dendritic cell-specific transmembrane protein
- RT-PCR
reverse-transcriptase polymerase chain reaction
Footnotes
Competing interests
The authors have no conflicts for this manuscript.
Authors' contributions
YGC: experimental design, molecular cloning, photoactivation, data analysis, & manuscript preparation
ES: experimental design, data analysis & manuscript preparation
DL: molecular cloning, photoactivation, retroviral infection
YX: photoactivation
TS: confocal microscopy JL: western blot
BW, TW, WH: molecular cloning
KB: electron microscopy
RW: video conversion
LA: molecular cloning
MK: photoactivation
CTR: experimental design, data analysis, & manuscript preparation
References
- Albagha OM, Wani SE, Visconti MR, Alonso N, Goodman K, Brandi ML, Cundy T, Chung PY, Dargie R, Devogelaer JP, Falchetti A, Fraser WD, Gennari L, Gianfrancesco F, Hooper MJ, Van Hul W, Isaia G, Nicholson GC, Nuti R, Papapoulos S, Montes Jdel P, Ratajczak T, Rea SL, Rendina D, Gonzalez-Sarmiento R, Di Stefano M, Ward LC, Walsh JP, Ralston SH Genetic Determinants of Paget's Disease C. Genome-wide association identifies three new susceptibility loci for Paget's disease of bone. Nat Genet. 2011;43(7):685–689. doi: 10.1038/ng.845. [DOI] [PubMed] [Google Scholar]
- Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40(2):251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Barrow AD, Raynal N, Andersen TL, Slatter DA, Bihan D, Pugh N, Cella M, Kim T, Rho J, Negishi-Koga T, Delaisse JM, Takayanagi H, Lorenzo J, Colonna M, Farndale RW, Choi Y, Trowsdale J. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J Clin Invest. 2011;121(9):3505–3516. doi: 10.1172/JCI45913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard M, Gagnon E, Guay-Belanger S, Morissette J, Brown JP, Michou L. Identification of rare genetic variants in novel loci associated with Paget's disease of bone. Hum Genet. 2014;133(6):755–768. doi: 10.1007/s00439-013-1409-x. [DOI] [PubMed] [Google Scholar]
- Ben Mkaddem S, Hayem G, Jonsson F, Rossato E, Boedec E, Boussetta T, El Benna J, Launay P, Goujon JM, Benhamou M, Bruhns P, Monteiro RC. Shifting FcgammaRIIA-ITAM from activation to inhibitory configuration ameliorates arthritis. J Clin Invest. 2014;124(9):3945–3959. doi: 10.1172/JCI74572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, Aliprantis AO. Osteoclasts: more than 'bone eaters'. Trends Mol Med. 2014;20(8):449–459. doi: 10.1016/j.molmed.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YG, Shao T, Feng C, Mensah KA, Thullen M, Schwarz EM, Ritchlin CT. CD16 (FcRgammaIII) as a potential marker of osteoclast precursors in psoriatic arthritis. Arthritis Research & Therapy. 2010;12(1):R14. doi: 10.1186/ar2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Mensah KA, Schwarz EM, Ju Y, Takahata M, Feng C, McMahon LA, Hicks DG, Panepento B, Keng PC, Ritchlin CT. Regulation of human osteoclast development by dendritic cell-specific transmembrane protein (DC-STAMP) J Bone Miner Res. 2012;27(1):79–92. doi: 10.1002/jbmr.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti TN, Dharmapatni AA, Alias E, Haynes DR. Osteoimmunology: Major and Costimulatory Pathway Expression Associated with Chronic Inflammatory Induced Bone Loss. J Immunol Res. 2015;2015:281287. doi: 10.1155/2015/281287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou C, Zhang C, Kang F, Yang X, Jiang H, Bai Y, Xiang J, Xu J, Dong S. MiR-7b directly targets DC-STAMP causing suppression of NFATc1 and c-Fos signaling during osteoclast fusion and differentiation. Biochim Biophys Acta. 2014;1839(11):1084–1096. doi: 10.1016/j.bbagrm.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Hartgers FC, Vissers JL, Looman MW, van Zoelen C, Huffine C, Figdor CG, Adema GJ. DC-STAMP, a novel multimembrane-spanning molecule preferentially expressed by dendritic cells. Eur J Immunol. 2000;30(12):3585–3590. doi: 10.1002/1521-4141(200012)30:12<3585::AID-IMMU3585>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19(10):514–522. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Henriksen K, Bollerslev J, Everts V, Karsdal MA. Osteoclast activity and subtypes as a function of physiology and pathology--implications for future treatments of osteoporosis. Endocr Rev. 2011;32(1):31–63. doi: 10.1210/er.2010-0006. [DOI] [PubMed] [Google Scholar]
- Henriksen K, Sorensen MG, Nielsen RH, Gram J, Schaller S, Dziegiel MH, Everts V, Bollerslev J, Karsdal MA. Degradation of the organic phase of bone by osteoclasts: a secondary role for lysosomal acidification. J Bone Miner Res. 2006;21(1):58–66. doi: 10.1359/JBMR.050905. [DOI] [PubMed] [Google Scholar]
- Hobolt-Pedersen AS, Delaisse JM, Soe K. Osteoclast fusion is based on heterogeneity between fusion partners. Calcif Tissue Int. 2014;95(1):73–82. doi: 10.1007/s00223-014-9864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey MB, Lanier LL, Nakamura MC. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- Hwang SY, Putney JW., Jr Calcium signaling in osteoclasts. Biochim Biophys Acta. 2011;1813(5):979–983. doi: 10.1016/j.bbamcr.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R, Bae HS, Yoon WJ, Woo KM, Baek JH, Kim HH, Uchida T, Ryoo HM. Pin1 regulates osteoclast fusion through suppression of the master regulator of cell fusion DC-STAMP. J Cell Physiol. 2014;229(12):2166–2174. doi: 10.1002/jcp.24679. [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nat Immunol. 2009;10(4):340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiya H. Calcium signaling in osteoclast differentiation and bone resorption. Adv Exp Med Biol. 2012;740:917–932. doi: 10.1007/978-94-007-2888-2_41. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim T, Jeong BC, Cho IT, Han D, Takegahara N, Negishi-Koga T, Takayanagi H, Lee JH, Sul JY, Prasad V, Lee SH, Choi Y. Tmem64 modulates calcium signaling during RANKL-mediated osteoclast differentiation. Cell Metab. 2013;17(2):249–260. doi: 10.1016/j.cmet.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lee SH, Ha Kim J, Choi Y, Kim N. NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and the dendritic cell-specific transmembrane protein (DC-STAMP) Mol Endocrinol. 2008;22(1):176–185. doi: 10.1210/me.2007-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Inui M, Inoue K, Kim S, Suematsu A, Kobayashi E, Iwata T, Ohnishi H, Matozaki T, Kodama T, Taniguchi T, Takayanagi H, Takai T. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428(6984):758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512(7515):431–435. doi: 10.1038/nature13375. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, Choi Y. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12(12):1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- Li S, Miller CH, Giannopoulou E, Hu X, Ivashkiv LB, Zhao B. RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. J Clin Invest. 2014;124(11):5057–5073. doi: 10.1172/JCI71882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama R, Vriens J, Voets T, Karashima Y, Owsianik G, Vennekens R, Lieben L, Torrekens S, Moermans K, Vanden Bosch A, Bouillon R, Nilius B, Carmeliet G. TRPV4-mediated calcium influx regulates terminal differentiation of osteoclasts. Cell Metab. 2008;8(3):257–265. doi: 10.1016/j.cmet.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZ, Bachler MA, Amano H, Aburatani H, Ishikawa H, Wagner EF. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279(25):26475–26480. doi: 10.1074/jbc.M313973200. [DOI] [PubMed] [Google Scholar]
- Negishi-Koga T, Takayanagi H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol Rev. 2009;231(1):241–256. doi: 10.1111/j.1600-065X.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fc-receptors as regulators of immunity. Adv Immunol. 2007;96:179–204. doi: 10.1016/S0065-2776(07)96005-8. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Ong EC, Nesin V, Long CL, Bai CX, Guz JL, Ivanov IP, Abramowitz J, Birnbaumer L, Humphrey MB, Tsiokas L. A TRPC1 protein-dependent pathway regulates osteoclast formation and function. J Biol Chem. 2013;288(31):22219–22232. doi: 10.1074/jbc.M113.459826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin BR, Gay CV. Biology and physiology of the osteoclast. Boca Raton: CRC Press; 1992. [Google Scholar]
- Schindl R, Fritsch R, Jardin I, Frischauf I, Kahr H, Muik M, Riedl MC, Groschner K, Romanin C. Canonical transient receptor potential (TRPC) 1 acts as a negative regulator for vanilloid TRPV6-mediated Ca2+ influx. J Biol Chem. 2012;287(42):35612–35620. doi: 10.1074/jbc.M112.400952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeling M, Hillenhoff U, David JP, Schett G, Tuckermann J, Lux A, Nimmerjahn F. Inflammatory monocytes and Fcgamma receptor IV on osteoclasts are critical for bone destruction during inflammatory arthritis in mice. Proc Natl Acad Sci U S A. 2013;110(26):10729–10734. doi: 10.1073/pnas.1301001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe K, Andersen TL, Hobolt-Pedersen AS, Bjerregaard B, Larsson LI, Delaisse JM. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone. 2011;48(4):837–846. doi: 10.1016/j.bone.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Soe K, Hobolt-Pedersen AS, Delaisse JM. The elementary fusion modalities of osteoclasts. Bone. 2015;73C:181–189. doi: 10.1016/j.bone.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7(4):292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Xu Y, Hyun YM, Lim K, Lee H, Cummings RJ, Gerber SA, Bae S, Cho TY, Lord EM, Kim M. Optogenetic control of chemokine receptor signal and T-cell migration. Proc Natl Acad Sci U S A. 2014;111(17):6371–6376. doi: 10.1073/pnas.1319296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, Morita K, Ninomiya K, Suzuki T, Miyamoto K, Oike Y, Takeya M, Toyama Y, Suda T. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202(3):345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M, Miyamoto T, Toyama Y, Suda T. Role of DC-STAMP in cellular fusion of osteoclasts and macrophage giant cells. J Bone Miner Metab. 2006;24(5):355–358. doi: 10.1007/s00774-006-0697-9. [DOI] [PubMed] [Google Scholar]
- Yagi M, Ninomiya K, Fujita N, Suzuki T, Iwasaki R, Morita K, Hosogane N, Matsuo K, Toyama Y, Suda T, Miyamoto T. Induction of DC-STAMP by alternative activation and downstream signaling mechanisms. J Bone Miner Res. 2007;22(7):992–1001. doi: 10.1359/jbmr.070401. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Yao Z, Li F, Zhang Q, Badell IR, Schwarz EM, Takeshita S, Wagner EF, Noda M, Matsuo K, Xing L, Boyce BF. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J Biol Chem. 2007;282(25):18245–18253. doi: 10.1074/jbc.M610701200. [DOI] [PubMed] [Google Scholar]
- Yarilina A, Xu K, Chen J, Ivashkiv LB. TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proc Natl Acad Sci U S A. 2011;108(4):1573–1578. doi: 10.1073/pnas.1010030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dou C, Xu J, Dong S. DC-STAMP, the Key Fusion-Mediating Molecule in Osteoclastogenesis. J Cell Physiol. 2014 doi: 10.1002/jcp.24553. [DOI] [PubMed] [Google Scholar]
- Zhang H, Huang Y, Wang S, Fu R, Guo C, Wang H, Zhao J, Gaskin F, Chen J, Yang N, Fu SM. Myeloid-derived suppressor cells contribute to bone erosion in collagen-induced arthritis by differentiating to osteoclasts. J Autoimmun. 2015;65:82–89. doi: 10.1016/j.jaut.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang X, Liu Y, He A, Jia R. NFATc1: functions in osteoclasts. Int J Biochem Cell Biol. 2010;42(5):576–579. doi: 10.1016/j.biocel.2009.12.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.