Abstract
The herpes simplex virus type 1 capsid is a protective shell that acts as a container for the genetic material of the virus. After assembly of the capsid, the viral DNA is translocated into the capsid interior through a channel formed by the portal. The portal is composed of a dodecamer of UL6 molecules which form a ring-like structure found at a single vertex within the icosahedron. Formation of portal-containing capsids minimally requires the four structural proteins (VP5, VP19C, VP23, and UL6) and a scaffolding protein (UL26.5). Recently, an interaction between UL26.5 and the portal has been identified, suggesting the scaffold functions by delivering the portal to the growing capsid shell. The aim of this study was to identify regions within UL26.5 required for its interaction with the portal. A specific region was identified by mutational analysis. Deletion of scaffold amino acids (aa) 143 to 151 was found to be sufficient to inhibit formation of the scaffold-portal complex as assayed in vitro. The aa 143 to 151 contain the sequence YYPGE, which is highly conserved among alphaherpesviruses. Although it did not bind to the portal, the Δ143-151 mutant was found to retain the ability to support assembly of morphologically normal capsids in vitro. Such capsids, however, did not contain the portal. The results suggest assembly of portal-containing capsids requires formation of a scaffold-portal complex in which intermolecular contact is dependent on scaffold aa 143 to 151.
Methods of electron cryomicroscopy and three-dimensional image reconstruction have been employed to determine the structure of the herpes simplex virus (HSV-1) capsid at 8.5 Å resolution (29, 34). The capsid has been found to be an icosahedral protein shell approximately 125 nm in diameter, 15 nm thick, and composed primarily of VP5 (UL19). The structural components include 162 capsomers (150 hexons and 12 pentons) that are arranged on a T=16 icosahedral lattice. Hexons form the edges and faces of the capsid and are each composed of six VP5 molecules. In contrast, one penton is found at each of the 12 vertices. Eleven of the pentons are VP5 pentamers, while the last vertex is occupied by the portal complex. Adjacent capsomers are linked together in groups of three by triplexes, heterotrimeric complexes composed of one molecule of VP19C and two molecules of VP23.
The portal complex is composed of 12 UL6 molecules, which form a ring (23) similar in structure and dimensions to the portals found in the capsids of double-stranded DNA bacteriophages, such as P22 (5), φ29 (9, 30), and SPP1 (26). Like bacteriophage portals, the HSV-1 portal functions as a channel through which the viral DNA enters the preformed capsid (12, 17). The mechanism for introduction of DNA into the HSV-1 capsid is also thought to resemble the mechanism found in bacteriophages, with the portal and the terminase subunits (UL15 and UL28 in HSV-1 [1, 2, 33]) forming a packaging “machine” (4, 7).
In addition to the structural proteins, HSV-1 capsid assembly requires the action of a scaffolding protein. Two such proteins have been identified, UL26 and UL26.5. Of these, UL26.5 is found to be the predominant species present in infected cells (25). UL26.5 interacts directly with VP5 and is thought to drive capsid formation by bringing the major capsid molecules together through a mechanism of scaffold self-interaction (12). The regions of UL26.5 required for interaction with VP5 (13, 31), as well as the regions required for intermolecular self-interaction (27, 28), have been identified and shown to be critical for capsid formation. Although the scaffold plays a central role in capsid assembly, no scaffolding protein is found within the mature capsid or the virion. As the procapsid matures, the VP5 binding region (the C-terminal 25 amino acids) of the scaffold is removed by the viral protease (UL26), and the released scaffold is lost from the capsid interior (35).
Recently, an interaction between UL26.5 and the portal has been demonstrated by in vitro experiments with purified proteins (24). In addition, evidence suggests that the HSV-1 scaffold-portal interaction plays an important role in recruitment of the portal into the capsid as it is formed. Experiments have shown that the drug WAY-150138 blocks assembly of the portal into HSV-1 capsids, presumably by interfering with the ability of scaffold to interact with the portal (20). To further characterize the role of the scaffolding protein in capsid assembly, we have begun to map the specific regions within the scaffold that are required for interaction with the portal. Deletions in the 329-amino-acid (aa) UL26.5 protein were created and tested for their ability to bind the portal.
MATERIALS AND METHODS
Construction of UL26.5 mutants.
The pAC373 baculovirus transfer vector was used for all the mutant constructs and has been described previously (21). Mutations in the UL26.5 gene were created by PCR of the HSV-1 (KOS) genomic DNA and cloned into the BamHI and KpnI sites of the pAC373 backbone.
Forward primers for the creation of N-terminal truncations contained a BglII site at the 5′ end followed by an ATG in frame with the codon sequence immediately downstream of the deletion site. Reverse primers contained the sequence complementary to the C terminus of UL26.5 with a KpnI site located downstream of the coding sequence. PCR amplification resulted in removal of codons upstream from the deletion site and insertion of an ATG in frame with the downstream codons.
C-terminal truncations were created by using a similar PCR strategy, inserting a premature stop codon, and removing the region downstream of the truncation point. Forward primers contained a BglII site at the 5′ end, followed by the original ATG and several downstream codons. Reverse primers contained sequence complementary to the region immediately 5′ of the termination site, followed by a stop codon and a KpnI restriction site.
Internal deletions within the UL26.5 gene were created using a two-step PCR procedure (11). Step one involved two independent PCRs, utilizing either a forward or reverse primer that contained the sequence of the regions flanking the deletion site. This resulted in the amplification of two fragments of UL26.5, a portion of UL26.5 upstream of the deletion site and a second fragment containing the downstream sequence. Both fragments contained the deletion. The fragments were gel purified and used as the template for the second PCR with forward and reverse primers from the full-length wild-type (wt) UL26.5. This second PCR step combined the sequences, resulting in the amplification of a single UL26.5 product containing the desired mutation.
The Thermal Ace DNA polymerase kit (Invitrogen, Carlsbad, Calif.) was used for PCR amplification. The PCR protocol consisted of an initial incubation at 98°C for 3 min, followed by 30 amplification cycles (50 s at 98°C, 30 s at 60°C, and 60 s at 72°C). All PCR products were run on a 1% agarose gel, verified for the appropriate size, and purified from the agarose using the QIAquick gel extraction kit (QIAGEN, Valencia, Calif.). The PCR fragments were inserted into the pAC373 backbone using standard cloning protocols (11). All clones were confirmed by DNA sequencing. Large quantities of each clone were purified by using the HiSpeed plasmid midi kit (QIAGEN).
Recombinant baculovirus.
Baculoviruses containing the UL26.5 mutants were created using the BaculoGold kit (BD Biosciences, San Diego, Calif.) and Spodoptera frugiperda (Sf9) cells maintained in Graces insect medium supplemented with 10% fetal calf serum (Invitrogen) and 1% penicillin-streptomycin (Invitrogen), using the provided protocol. A T25 flask contained 4 × 106 Sf9 cells which were cotransfected with 5 μg of the pAC transfer vector containing the mutant UL26.5 gene and 0.5 μg of linearized baculovirus DNA, using the transfection reagents and protocol provided with the BaculoGold kit. After completion of transfection, the cells were incubated at 28°C for 4 days. The culture supernatant was harvested and clarified by centrifugation at 1,500 rpm for 5 min in a Sorvall RT7 centrifuge (RTH-250 rotor). One milliliter of the clarified supernatant was used to infect a T150 flask (Corning, Cambridge, Mass.) containing 3 × 107 Sf9 cells. Infected cells were incubated at 28°C for 72 h. The supernatant was harvested, stored at 4°C, and used as stock for infections. The pellet was tested for protein expression via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis utilizing a rabbit polyclonal antibody (antibody NC3,4) specific for the UL26.5 protein, generously provided by Roselyn Eisenberg and Gary Cohen (6).
Protein expression and purification.
The production of insect cells expressing the UL26.5, UL19, triplex, and UL6 genes has been described previously (24). All infected insect cells were stored as 1-ml aliquots consisting of 300 μl of cells in 700 μl of phosphate-buffered saline (PBS) supplemented with 100 μl of protease inhibitor (Roche Complete protease inhibitor cocktail tablets; one tablet dissolved in 5 ml of PBS) at −80°C. Extraction of pre-VP22a (UL26.5), VP5 (UL19), and triplex (UL18 and UL38) from Sf9 cells was carried out by cell lysis followed by ammonium sulfate precipitation as previously described (22). The same method was used for purification of the UL26.5 truncation mutants.
Portals composed of UL6 protein were purified as previously described (23). Briefly, inclusion bodies containing UL6 protein were isolated after treatment of infected Sf9 cells with Triton X-100 and DNase. Inclusion bodies were then resuspended in TNE (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 1.0 mM EDTA) containing 1 M arginine, a treatment that solubilizes the UL6. Portals were further purified by sedimentation in sucrose gradients containing 1 M arginine. Peak fractions containing UL6 protein were identified by dot blotting and stored at 4°C. In such preparations the UL6 concentrations were ∼0.5 mg/ml, as determined by Coomassie staining of an SDS-12% polyacrylamide gel.
Purification and expression of the UL26.5 internal deletions involved a small-scale infection and purification similar to the larger-scale method described above. A T150 flask containing ∼2 × 107 Sf9 cells (60 to 70% confluent) was infected with 1 ml of stock virus, and cells were incubated at 28°C. At 72 h postinfection, the cells were manually dislodged from the bottom of the flask by gently tapping the side until cells were suspended in solution. Medium was harvested and centrifuged at 1,500 rpm in a Sorvall RT7 centrifuge for 5 min at room temperature to pellet the cells. The supernatant was discarded, and the cell pellet was resuspended in 500 μl of PBS containing protease inhibitor. The suspension was stored at −80°C for at least 24 h. The packed cells were lysed by one cycle of freezing (−80°C) and thawing and subjected to 5 min of centrifugation at 16,000 × g at 4°C in a bench-top microcentrifuge. The supernatant was removed, added to 0.2 volumes of saturated ammonium sulfate, and incubated on ice for 30 min. The resulting precipitate, which contained UL26.5 or UL26.5 mutant, was collected by 15 min of centrifugation at 16,000 × g at 4°C in a bench-top microcentrifuge, and the supernatant was discarded. The pellet was resuspended in 100 μl of PBS containing protease inhibitor and stored at 4°C, resulting in a ∼2-mg/ml concentration (0.2 mg) of purified scaffolding protein, as determined by Coomassie staining of an SDS-12% polyacrylamide gel.
UL6 interaction assay.
To examine the interaction of UL6 with UL26.5, 2 μl of purified UL6 (0.5 mg/ml) was added to 30 μl of purified UL26.5 or the UL26.5 mutant in PBS. The mixture was incubated at room temperature for 15 min and then centrifuged for 10 min at 16,000 × g to remove any UL6 not bound to UL26.5. A 25-μl aliquot of the supernatant was added to a fresh tube, taking care not to include any material from the pellet. Supernatant was then subjected to either agarose gel electrophoresis (internal deletion mutants) or SDS-PAGE (truncation mutants). Agarose gel analysis was used for comparisons where all mutants formed scaffold particles (see below), while SDS-PAGE was used in comparisons where mutant scaffolding proteins differed in their ability to form scaffold particles (the truncation mutants).
Agarose gel electrophoresis was performed with samples prepared by gently mixing 5 μl of Ficoll (15% Ficoll with 0.25% bromophenol blue dye) with 25 μl of reaction supernatant (see above). The 30-μl samples were loaded onto a 1% agarose gel (prepared in 20 mM Tris-phosphate [pH 7.5]) and run on a BRL Horizon 58 electrophoresis unit at 110 V until the dye front ran off the end of the gel (∼1.5 h). The gel was then electrophoretically transferred onto an Immobilon-P polyvinylidine difluoride transfer membrane (Millipore, Billerica, Mass.) using a Hoefer TE series Transphor electrophoresis unit overnight at 50 mA in transfer buffer (20% methanol, 25 mM Tris, and 200 mM glycine). To identify the protein bands, the membrane was stained in 0.1% Ponceau S and 1% acetic acid for 10 min and washed with water until the protein bands were distinguishable from background. The stained blot was recorded by densitometric scanning, and the membrane was stained with UL6-specific monoclonal antibody 1C9 as previously described (24).
SDS-PAGE analysis of the scaffold-UL6 reaction supernatant was performed on 50-μl samples prepared by mixing 25 μl of reaction mix (described above) with 25 μl of sample buffer (0.3 M dithiothreitol, 50 mM EDTA, 2.0% SDS, 65 mM Tris [pH 6.8], 5% sucrose, and 0.1% bromophenol blue and supplemented with protease inhibitor) and boiling for 2 min. The sample was then divided in half, and each half was run on a 12% polyacrylamide gel in a Hoefer PAGE unit at 110 V until the dye front ran off the bottom of the gel (∼2 h). One gel was stained with Coomassie, while the second was transferred onto an Immobilon-P membrane and probed for UL6 under the conditions described for the agarose gel (see above). Both the Coomassie-stained gel and the Western blot were recorded, and the relative amounts of each protein were determined by densitometric analysis. The Coomassie-stained bands that corresponded to each truncation mutant were identified by comparison with baculovirus-infected Sf9 cell lysate containing a different truncation mutant construct, and they were confirmed by comparison of its relative position, in the gel, to that of wt scaffold.
To express the portal binding activity achieved for each sample, the amount of UL6 retained in solution (determined by UL6 Western blotting) was divided by the amount of scaffold, or scaffold mutant, in the sample (determined by Coomassie staining). Each experiment contained dilutions of wt scaffold as a positive control and a standard for portal binding. Each mutant scaffold's ability to bind UL6 protein was normalized to the wt scaffold binding activity and expressed as the percentage of wt scaffold binding [(mutant binding ability)/(wt binding ability) × 100], allowing for comparison of results between experiments.
In vitro capsid assembly.
Proteins required for capsid assembly (UL26.5, VP5, and triplex) were purified as described above. A 13.5-μg portion of UL26.5, 30 μg of VP5, and 0.9 μg of triplex were mixed in a 0.5-ml Eppendorf tube, and PBS was added to make the total volume 100 μl. A 10-μl aliquot of assembly mix (400 mM EDTA and 50 mM dithiothreitol in PBS) and 3.3 μl of protease inhibitor were added. Finally, 4 μl (2 μg) of purified UL6 was added and the reaction mix was incubated at 36°C overnight. Formation of capsids was confirmed by electron microscopy (see below). Large debris was removed by a brief spin (30 s) at 8,000 × g in a bench-top microcentrifuge. The clarified supernatant was transferred to a fresh 600-μl tube (Beckman Ultra-Clear; 5- by 41-mm tubes) and diluted to a volume of 570 μl with TNE. The sample was underlaid with 30 μl of a 35% (wt/vol) sucrose cushion, and capsids were pelleted by centrifugation for 30 min at 22,500 rpm in a Beckman SW55Ti rotor at 22°C containing adapters. The capsid pellet was resuspended in 50 μl of TNE containing protease inhibitor and layered onto a 600-μl 20-to-50% (wt/vol) sucrose (in TNE) gradient with a 50-μl 65% sucrose cushion and spun at 22,500 rpm for 40 min at 22°C in a Beckman SW55Ti rotor. The gradient was then fractionated into 15 equal portions, and the peak fractions, containing capsids, were identified by Ponceau S staining of a dot blot. Capsids from these fractions were then analyzed by SDS-PAGE, electron microscopy, and agarose gel electrophoresis as described above.
Scaffold particle formation assay.
To determine the oligomeric state of UL26.5 and UL26.5 mutants, ∼5 μg of purified protein was added to a 5-ml continuous 10-to-30% (wt/vol) sucrose gradient in PBS and centrifuged in a Beckman SW55Ti rotor at 35,000 rpm for 2 h at 4°C. Fifteen fractions were collected from each gradient, and 20 μl of each fraction was run on an SDS-PAGE gel. The ability of the scaffold to enter the sucrose gradient was used as the test for scaffold particle formation. Truncation mutants that were unable to form scaffold particles remained at the top of the gradient.
Electron microscopy.
Electron microscopic analysis of in vitro-assembled capsids was performed on both the raw assembly mix, to confirm the formation of capsids prior to purification, and on capsids purified by sucrose gradient centrifugation. In both cases, samples were diluted, absorbed onto glow-discharged carbon-Formvar-coated copper grids, and stained with 1% uranyl acetate. Grids were examined and photographed in a Philips 400T electron microscope operated at 80 keV. Photographs were digitized by scanning on a flatbed scanner, and images were analyzed with Photoshop 5.0.
RESULTS
Identification of portal binding sites in UL26.5.
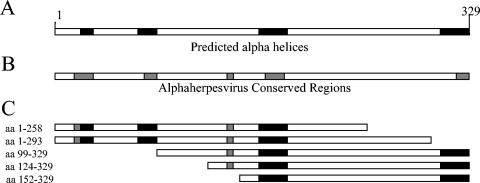
To identify specific sites within the scaffold that are required for interaction with the portal, five UL26.5 truncation mutants were produced (Fig. 1). These truncations effectively separated the protein into three primary sections: the N-terminal and C-terminal regions isolated by deletion and an undisturbed central core (aa 152 to 258). Attempts at making C-terminal truncations that spanned the core resulted in proteins that were highly insoluble and impossible to test for portal binding activity. By determining the ability of each soluble truncation mutant to bind the portal, the location of domains required for interaction could be identified.
FIG. 1.
Schematic drawing showing the scaffolding protein (UL26.5; A and B) and the truncation mutant constructs (C). Selected regions of either the N terminus or C terminus were removed by PCR manipulation and cloned into a baculovirus transfer vector for expression in insect cells as described in Materials and Methods. Truncations were designed to isolate regions which contain sequence homology with other alphaherpesviruses (B) or that are predicted to form α-helices (A).
Three N-terminal truncation mutants were created by engineering start codons into UL26.5 at amino acid positions 98, 123, and 151 and removing the codons upstream of the truncation sites. Similarly, two C-terminal truncations were constructed by the insertion of premature stop codons at positions 259 and 294. A C-terminal truncation at position 219 resulted in a product that was insoluble and difficult to test for portal binding activity (data not shown). The truncation mutants created are summarized in Fig. 1. The mutant constructs were expressed in Sf9 cells through the use of a baculovirus expression system, and the proteins were partially purified by ammonium sulfate precipitation. Although the levels of protein expressed by each of the mutant constructs were similar to what has been achieved with wt scaffold, the purified mutant scaffolds appeared to lack the purity and stability of the wt preparations. The truncation mutants showed an increased presence of background protein and a heightened sensitivity to proteolysis. To overcome this difficulty, the recombinant proteins were used within 24 h of purification, limiting the effect of the increased proteolysis.
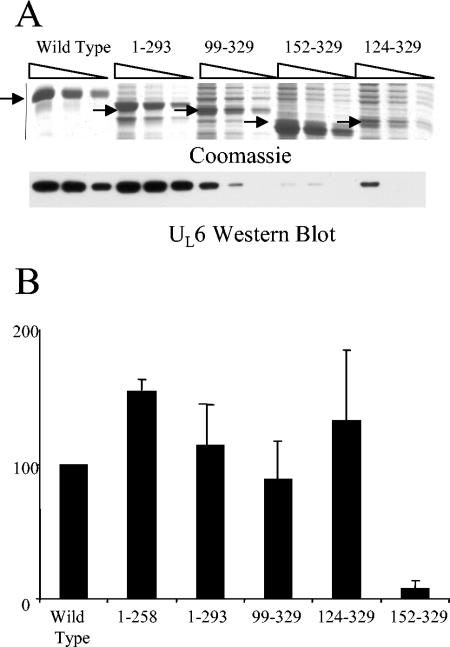
The purified truncation mutants were tested for the ability to interact with purified portal by measuring their ability to retain UL6 protein in solution. Because purified UL6 is only soluble in 1 M arginine, dilution of the arginine to less than 0.1 M causes the portal to precipitate. However, when the arginine is diluted in the presence of a protein that is capable of interaction, a protein-portal complex is formed, retaining the UL6 in solution. Samples containing possible mutant scaffold-portal complexes were split in half and subjected to separate SDS-PAGE analyses. One polyacrylamide gel was stained with Coomassie, and the second was transferred to Immobilon-P membrane and probed for UL6 protein. Dilutions of wt scaffold tested in this manner showed a direct relationship between scaffold dose and amount of portal retained in solution (Fig. 2A, compare the amount of portal retained in lanes 1, 2, and 3 to the corresponding amount of scaffold). In contrast, no UL6 was retained in solution with irrelevant proteins, including bovine serum albumin and DNase I (data not shown). This establishes the ability to retain UL6 in solution as a system for demonstrating portal binding. Figure 2A shows an experiment in which four truncation mutants were tested for portal interaction. For each mutant, a range of input concentrations was tested to ensure results were within the linear range for Western immunoblotting quantification and to eliminate the possibility of UL6 saturation. Figure 2B shows a graph of the portal binding activity for the scaffold truncation mutants.
FIG. 2.
Portal interaction with the scaffold and scaffold truncation mutants. The ability to retain the portal in solution was used as a measure of portal binding activity as described in Materials and Methods. (A) Samples containing the portal mixed with each individual truncation mutant were prepared and analyzed by Coomassie staining of an SDS-PAGE gel (top) and an anti-UL6 Western immunoblot (bottom). The relative amounts of scaffold and UL6 present in each sample were determined by densitometric scanning of the scaffold bands observed in the Coomassie gel (darkest-staining bands) and the UL6 identified by the Western immunoblotting. Additional bands in the mutant tracks corresponded to contaminating proteins present in the scaffold protein purifications. The wt scaffold was used as a positive control (lanes 1 to 3). (B) Relative UL6 binding activity for each of the truncation mutants, expressed as a percentage of wt binding activity. The results shown represent the average of at least three independent experiments for each truncation mutant. Note that scaffold 152-329 was the only mutant that failed to bind portal at or near the level of the wt.
The truncation mutant scaffold 152-329 was found to be significantly impaired in portal binding, showing only 5% of wt binding activity (Fig. 2B). In contrast, all the other truncation mutants showed binding activity similar to that of the wt, including the scaffold 124-329 mutant. These results identify a site between aa 124 and 152 that is necessary for scaffold interaction with the portal (Fig. 3A).
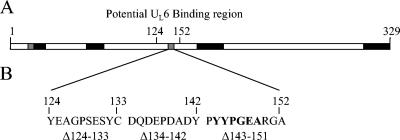
FIG. 3.
Schematic representation of the scaffolding protein (A) and the internal deletion mutants (B). Truncation mutant analysis identified a potential UL6 interaction domain located between aa 124 and 152. Three internal deletion mutants that span this region were created, and the regions deleted are shown in panel B. Note the presence of a region of alphaherpesvirus sequence homology (bold) in the region deleted from scaffold Δ143-151.
UL6 interaction site.
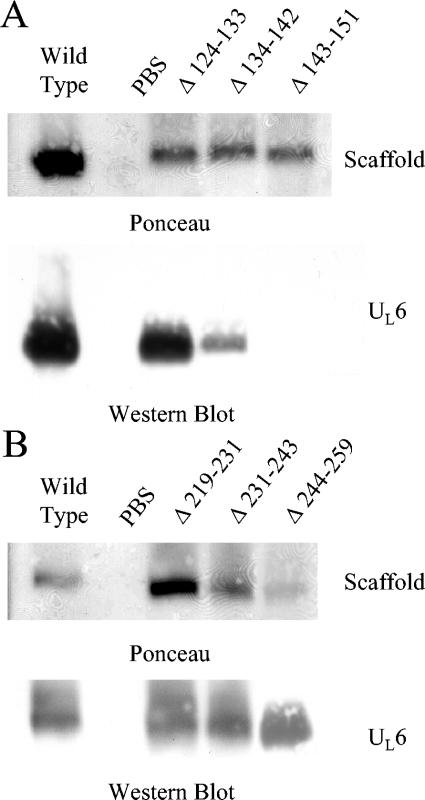
To further isolate the location of a scaffold region required for portal interaction, internal deletions spanning the proposed binding site were constructed. Three scaffold mutants containing deletions of 8 to 12 aa each were created, and together these spanned the region implicated by the truncation mutants (Fig. 3). Using an agarose gel electrophoresis method, each mutant was tested for the ability to form a complex with the UL6 protein (Fig. 4A). Whereas the Δ124-133 mutant retained full portal binding activity, the Δ134-142 mutant was partially inhibited and binding by the Δ143-151 mutant was not detected. The results suggest an important role for aa 143 to 151 in portal binding.
FIG. 4.
Agarose gel analysis of scaffold internal deletion mutants. Internal deletions were tested for the ability to form a complex with the portal. Samples containing the individual deletion mutants were mixed with the portal, separated by agarose gel electrophoresis, and transferred to an Immobilon-P membrane. The locations of the scaffold bands were determined by Ponceau S staining (top panels), and the presence of the portal at this location was determined by anti-UL6 immunoblotting (bottom panels). (A) Scaffold mutants that span the region identified (aa 124 to 151) by the truncation mutant analysis. (B) Internal deletion mutants that span aa 219 to 259 of the scaffold were also tested for the ability to interact with the portal. Note that scaffold Δ143-151 failed to interact with the portal.
To determine if region 219-259, which could not be tested by truncation mutant analysis, is required for interaction with the portal, three additional internal deletion mutants that span this region were created. These internal deletion mutants were tested for the ability to interact with the portal (Fig. 4B), and all three were found to retain the ability to interact, indicating that aa 219 to 259 are not required for portal interaction.
Capsid assembly with scaffold Δ143-151.
In addition to identifying a specific region of scaffold required for interaction with the portal, the scaffold Δ143-151 mutant provided an opportunity to further test the importance of scaffold-portal complex formation in assembly of capsids. If the scaffold-portal complex were critical for incorporation of the portal into the nascent capsid structure, the scaffold Δ143-151 mutant should fail to assemble portal-containing capsids. To test this hypothesis, the ability of scaffold Δ143-151 to assemble portal-containing capsids was examined using an in vitro capsid assembly system (22). Purified VP5, triplex, and either wt UL26.5 or scaffold Δ143-151 were mixed, and UL6 protein was added. The assembly reaction mixture was incubated overnight to allow capsids to form, and the morphology of the resulting capsids was analyzed by electron microscopy. Both the wt and mutant scaffolds were found to be capable of forming intact capsids, with no obvious morphological differences (Fig. 5).
FIG. 5.
Electron microscopic analysis of capsids produced by wt scaffold (top) or the scaffold mutant Δ143-151 (bottom). Capsids were produced in an in vitro assembly reaction and purified by sucrose gradient sedimentation. The peak fractions were analyzed by negative-stain electron microscopy. Note that capsids produced with Δ143-151 are morphologically indistinguishable from capsids produced with the wt scaffold.
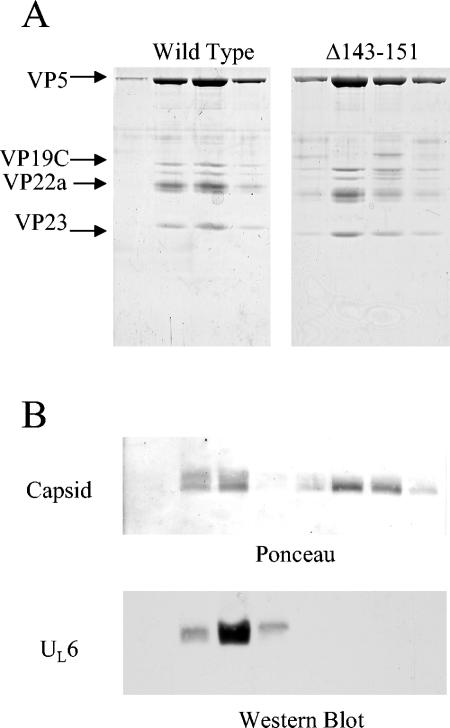
The remaining capsids for each of the preparations were purified by sucrose gradient sedimentation, and fractions were collected. The four fractions containing the peak protein concentrations for both the wt and Δ143-151 capsid preparations were subjected to SDS-PAGE to determine their protein compositions (Fig. 6A). Bands corresponding to the major structural components of the capsid were identified. Similar concentrations of VP5 were observed in both preparations, suggesting that similar amounts of capsids were produced by the wt and Δ143-151 scaffolds. The four fractions for both preparations were further analyzed by agarose gel electrophoresis and transferred to an Immobilon-P membrane. The location of the capsid band was visualized by Ponceau staining and was found to coincide with the location of the UL6 protein as determined by Western immunoblotting (Fig. 6B). The capsids produced with the wt scaffold showed a significant amount of UL6 associated with the capsid band, while the capsids produced by scaffold Δ143-151 showed no detectable portal incorporation. These observations indicate that deletion of the portal binding domain of scaffold prevents portal incorporation into the capsid without affecting assembly of capsids lacking portals.
FIG. 6.
Analysis of capsids assembled in vitro with the scaffold Δ143-151 mutant. Capsids were formed with either the wt or Δ143-151 scaffold proteins, using an in vitro assembly reaction. The resulting capsids were purified by sucrose gradient sedimentation as described in Materials and Methods. Protein compositions of the four peak fractions for both capsid samples were determined by SDS-PAGE analysis (A), and the amount of UL6 associated with the capsids was determined by agarose gel electrophoresis (B). The agarose gel was transferred to an Immobilon-P membrane, and the locations of the capsid bands were identified by Ponceau S staining (B, top). Colocalization of the UL6 with the capsid was observed by anti-UL6 Western immunoblotting (B, bottom). Note the lack of UL6 colocalization with capsids produced by the Δ143-151 mutant scaffold.
Scaffold particles and UL6 interaction.
The intermolecular self-association characteristics of UL26.5 cause the scaffold to form into large scaffold particles ∼550 Å in diameter and consisting of several hundred UL26.5 molecules (15, 21). Because the scaffold particles have been observed to interact directly with the portal (24), the importance of scaffold particle formation for portal interaction was assayed for each truncation mutant. A correlation between the ability to form scaffold particles and the ability to interact with the portal would suggest that the formation of scaffold particles may be important for portal interaction.
To determine which truncation mutants are capable of forming scaffold particles, the purified proteins were subjected to sucrose gradient sedimentation (Fig. 7). Scaffold particles enter such gradients, forming a distinct band, while small particles remain at the top. The C-terminal truncation mutant scaffold 1-293 was found to be capable of forming scaffold particles but, compared to the wt, the 1-293 particles formed a broader band within the gradient, suggesting the particles were not as homogenous in size as the wt particles. None of the N-terminal truncation mutants retained this ability. Consistent with observations made in scaffold intermolecular self-association studies (28), deletion of the N-terminal 99 aa was found to be sufficient to inhibit scaffold particle formation. Because the N-terminal truncation mutant scaffold 99-329 and scaffold 124-329 both showed significant portal binding activity, it must be the case that portal binding occurs regardless of whether the scaffold mutant forms small or large oligomers.
FIG. 7.
Sucrose gradient analysis of the scaffolding protein and the scaffold truncation mutants. Truncation mutants were subjected to sucrose gradient sedimentation, and the protein composition for each gradient fraction was determined by SDS-PAGE. Formation of a distinct band within the sucrose gradient provided evidence for scaffold particle formation. Note that only the wt scaffold and scaffold 1-293 were able to form scaffold particles. The abilities of the truncation mutants to interact with the UL6 protein were compared to the abilities to form scaffold particles, as shown in the two right-most columns.
DISCUSSION
Although the mechanism for incorporation of a portal into the HSV-1 capsid has not been established, the process must involve three distinct functions: (i) the portal must be recruited to the capsid assembly site, (ii) the portal must be assimilated into the capsid, and (iii) portal incorporation must be restricted to a single vertex. A basic requirement for each of the above functions is the ability of the portal to be recognized by other proteins involved in assembly. Recognition likely occurs through a specific interaction between the portal and one of the other assembly proteins. While all of the capsid proteins are possible candidates for this role, the scaffold has been implicated by its ability to interact with purified portals (24). Identification of specific sites within the scaffolding protein would support the view that the scaffold-portal interaction is specific, and it would reinforce the idea that the scaffold-portal complex is involved with the assembly of the portal into the nascent capsid.
Site of UL6 interaction.
By deleting large sections of the scaffolding protein, we have identified a single site within the scaffold that is required for portal interaction. Further analysis by internal deletions produced a mutant with 9 aa deleted (position 143 to 151) that was sufficient to prevent interaction with the portal complex. We conclude, therefore, that the scaffold-portal interaction is dependent upon this specific site. It is possible, however, that the site identified is not the only site required for interaction. Other required sites may be present within the scaffold, located within regions (e.g., the central core) that were not tested by deletion analysis.
The creation of large C-terminal truncations resulted in highly insoluble products, making it difficult to test them for portal interaction (i.e., aa 219 to 259). To overcome this problem, several smaller internal deletions were created and tested for the ability to interact with the portal. The results demonstrated that no sites required for portal interaction were located within aa 219 to 259 (Fig. 4B). The only amino acid residues that were not tested for the ability to interact with the portal, by deletion analysis, were located in the central core (aa 152 to 219). Although it cannot be ruled out that the core contains an additional site required for interaction, the complete loss of interaction ability observed with the deletion of aa 143 to 151 argues against the existence of a second site.
Interaction site sequence analysis.
Previous amino acid sequence alignment of HSV-1 UL26.5 with other alphaherpesvirus scaffolding proteins has suggested five regions of high homology, four of which are located at positions that have a high probability of forming α-helices (Fig. 1) (27, 28). Three presumptive helical regions have been shown to be required for the formation of large scaffold particles, implicating them in the self-association characteristics of the scaffold (28), and one, at the C terminus, is involved in interaction with the major capsid protein (13, 31). However, one of the regions (aa 144 to 148) containing the conserved sequence YYPGE is not expected to form any known secondary structure, is not required for UL26.5 self-association, and has no known impact on capsid formation. Interestingly, this region is located within the 9-aa region required for portal binding (Fig. 8). The presence of sequence conservation in this region suggests the possibility of a common motif required for portal interaction among the alphaherpesviruses.
FIG. 8.
Sequence comparison of the HSV-1 scaffold region involved in UL6 interaction with similar regions in 10 other alphaherpesviruses. The comparison revealed a site of sequence homology within the putative UL6 binding domain, corresponding to aa 144 to 148 of the HSV-1 scaffold. CHV, cercopithecine herpesvirus; BHV, bovine herpesvirus; SMHV, squirrel monkey herpesvirus; GHV, gallid herpesvirus; EHV, equine herpesvirus; VZV, varicella-zoster virus.
Capsid formation.
In vitro formation of portal-containing capsids minimally requires four components, the major capsid, triplex, portal, and scaffolding proteins. Capsids produced in this manner are indistinguishable from in vivo capsids by electron microscopic analysis and show similar protein composition and structural characteristics (24). Assembly reactions using scaffold Δ143-151 retained the ability to form capsids which were morphologically indistinguishable from capsids made with the wt scaffold (Fig. 5). The mutant scaffold did not show any impairment of assembly, producing a similar number of capsids, with similar protein composition to the wt (Fig. 6A). When the portal protein was included in the assembly reaction, no portal was found associated with the capsid structure (Fig. 6B). Thus, deletion of the scaffold site responsible for interaction with the portal does not affect formation of capsids in vitro but has a specific effect on incorporation of the portal.
The above results are consistent with the hypothesis that the scaffold-portal complex is involved in assembly of portal-containing capsids and are reminiscent of observations made in bacteriophage systems. For instance, experiments performed with phage P22 have identified scaffold mutations which assemble only portal-negative capsids (3, 8). This is suggestive of a direct interaction between the scaffold and portal proteins and demonstrates a specific role for the scaffolding protein in portal acquisition. In phage φ29, an interaction between the scaffolding protein and the portal has also been identified biochemically, and observations suggest the interaction plays an important role in the assembly of morphologically normal capsids (10). Such observations support the view that the formation of a scaffold-portal complex is required for assembly of portal-containing capsids.
Inclusion of the portal into the capsid structure likely occurs either by a mechanism of assembly initiation or during the stage of nascent capsid growth. Experiments with P22 have shown that the addition of the portal to preformed procapsids does not result in portal-containing capsids (16), arguing against the possibility that the portal is acquired at the completion of assembly. Initiation of capsid assembly by the portal would provide a mechanism for preventing the incorporation of multiple portals, since only the initiating portal would be incorporated (19). The formation of a scaffold-portal complex could provide the nucleation event triggering the start of assembly (32).
However, several observations argue against a role for the portal in initiation. While capsid assembly in phage T4 requires the presence of the portal (14), HSV-1 and P22 are capable of assembling morphologically normal capsids in the absence of portal protein (3, 22). It must be, therefore, that the scaffold-portal complex is required only for formation of portal-containing capsids. Portal-negative capsids must be formed in a different way. Because the kinetics of P22 assembly is unaffected by addition of the portal (3), assembly of portal-negative and -positive capsids likely uses related pathways. When the portal is absent it is possible that a different scaffold complex may act as the initiator, taking the place of the scaffold-portal complex. Alternatively, the portal may be incorporated into the capsid structure after initiation, during addition of the major capsid protein to the growing structure. A mechanism could be envisioned in which the scaffold-portal complex delivers the portal to the growing structure, and the incorporation of the portal prevents the recruitment of additional portals in an as-yet-uncharacterized manner (18).
The present study demonstrates that deletion of a 9-aa region within the scaffold is sufficient to prevent interaction of UL26.5 with the portal. This scaffold mutant retains the ability to form capsids in vitro, but it is no longer capable of producing capsids containing a portal. Our results are consistent with the observations made with the drug WAY-150138, demonstrating the necessity for the formation of a scaffold-portal complex in the assembly of portal-containing capsids (20). While the mechanism of portal delivery remains unclear, our results establish that the scaffold is a critical component in the portal recognition and delivery events of the assembly process. Future experiments will be directed at identifying how the scaffold-portal complex incorporates a single portal into the capsid.
Acknowledgments
We thank Katherine Larsen and Oneida Mason for excellent technical assistance.
This work was supported by National Institutes of Health award AI41644.
REFERENCES
- 1.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., A. P. Poon, J. Rovnak, and B. Roizman. 1994. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazinet, C., and J. King. 1988. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 202:77-86. [DOI] [PubMed] [Google Scholar]
- 4.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cingolani, G., S. D. Moore, P. E. Prevelige, Jr., and J. E. Johnson. 2002. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J. Struct. Biol. 139:46-54. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 8.Greene, B., and J. King. 1996. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology 225:82-96. [DOI] [PubMed] [Google Scholar]
- 9.Guasch, A., J. Pous, B. Ibarra, F. X. Gomis-Ruth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa, and M. Coll. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 315:663-676. [DOI] [PubMed] [Google Scholar]
- 10.Guo, P. X., S. Erickson, W. Xu, N. Olson, T. S. Baker, and D. Anderson. 1991. Regulation of the phage phi 29 prohead shape and size by the portal vertex. Virology 183:366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, N.Y.
- 12.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 13.Hong, Z., M. Beaudet-Miller, J. Durkin, R. Zhang, and A. D. Kwong. 1996. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J. Virol. 70:533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellenberger, E. 1990. Form determination of the heads of bacteriophages. Eur. J. Biochem. 190:233-248. [DOI] [PubMed] [Google Scholar]
- 15.McClelland, D. A., J. D. Aitken, D. Bhella, D. McNab, J. Mitchell, S. M. Kelly, N. C. Price, and F. J. Rixon. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore, S. D., and P. E. Prevelige, Jr. 2002. Bacteriophage p22 portal vertex formation in vivo. J. Mol. Biol. 315:975-994. [DOI] [PubMed] [Google Scholar]
- 17.Moore, S. D., and P. E. Prevelige, Jr. 2002. DNA packaging: a new class of molecular motors. Curr. Biol. 12:R96-R98. [DOI] [PubMed] [Google Scholar]
- 18.Moore, S. D., and P. E. Prevelige, Jr. 2002. A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J. Virol. 76:10245-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murialdo, H., and A. Becker. 1978. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol. Rev. 42:529-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., and J. C. Brown. 2002. Inhibition of herpes simplex virus replication by WAY-150138: assembly of capsids depleted of the portal and terminase proteins involved in DNA encapsidation. J. Virol. 76:10084-10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., F. L. Homa, D. R. Thomsen, Z. Ye, and J. C. Brown. 1994. Cell-free assembly of the herpes simplex virus capsid. J. Virol. 68:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 26.Orlova, E. V., B. Gowen, A. Droge, A. Stiege, F. Weise, R. Lurz, M. van Heel, and P. Tavares. 2003. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 22:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier, A., F. Do, J. J. Brisebois, L. Lagace, and M. G. Cordingley. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J. Virol. 71:5197-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston, V. G., and I. M. McDougall. 2002. Regions of the herpes simplex virus scaffolding protein that are important for intermolecular self-interaction. J. Virol. 76:673-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrag, J. D., B. V. Prasad, F. J. Rixon, and W. Chiu. 1989. Three-dimensional structure of the HSV1 nucleocapsid. Cell 56:651-660. [DOI] [PubMed] [Google Scholar]
- 30.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomsen, D. R., W. W. Newcomb, J. C. Brown, and F. L. Homa. 1995. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 69:3690-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Driel, R., and E. Couture. 1978. Assembly of the scaffolding core of bacteriophage T4 proheads. J. Mol. Biol. 123:713-719. [DOI] [PubMed] [Google Scholar]
- 33.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, Z. H., S. J. Macnab, J. Jakana, L. R. Scott, W. Chiu, and F. J. Rixon. 1998. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc. Natl. Acad. Sci. USA 95:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]