Abstract
Sedoheptulose‐7‐phosphate isomerase, GmhA, is the first enzyme in the biosynthesis of nucleotide‐activated‐glycero‐manno‐heptoses and an attractive, yet underexploited, target for development of broad‐spectrum antibiotics. We demonstrated that GmhA homologs in Neisseria gonorrhoeae and N. meningitidis (hereafter called GmhAGC and GmhANM, respectively) were interchangeable proteins essential for lipooligosaccharide (LOS) synthesis, and their depletion had adverse effects on neisserial viability. In contrast, the Escherichia coli ortholog failed to complement GmhAGC depletion. Furthermore, we showed that GmhAGC is a cytoplasmic enzyme with induced expression at mid‐logarithmic phase, upon iron deprivation and anaerobiosis, and conserved in contemporary gonococcal clinical isolates including the 2016 WHO reference strains. The untagged GmhAGC crystallized as a tetramer in the closed conformation with four zinc ions in the active site, supporting that this is most likely the catalytically active conformation of the enzyme. Finally, site‐directed mutagenesis studies showed that the active site residues E65 and H183 were important for LOS synthesis but not for GmhAGC function in bacterial viability. Our studies bring insights into the importance and mechanism of action of GmhA and may ultimately facilitate targeting the enzyme with small molecule inhibitors.
Keywords: crystal structure, drug target, Neisseria gonorrhoeae, sedoheptulose‐7‐phosphate isomerase GmhA
1. Introduction
The World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) emphasized an urgent need for the development of antimicrobials with novel modes of action against antibiotic‐resistant threats with severe consequences for public health, including infections caused by drug‐resistant Neisseria gonorrhoeae (Centers for Disease Control and Prevention, 2013a, 2013b; World Health Organization, 2012, 2015). Gonorrhea is highly prevalent throughout the world, and if untreated or inadequately treated, often leads to serious repercussions on reproductive health including ectopic pregnancy, pelvic inflammatory disease, and infertility (Centers For Disease Control And Prevention, 2013b, Low, Unemo, Skov Jensen, Breuer, & Stephenson, 2014; World Health Organization, 2011). In the absence of a protective gonorrhea vaccine, antibiotics remain the sole therapeutic intervention. However, the well‐documented ability of gonococci to acquire antibiotic resistance continues to threaten available treatment options (Unemo, 2015; Unemo & Shafer, 2014). To meet the needs raised by WHO and CDC, our laboratory focuses on identification and validation of new molecular targets for the development of gonorrhea treatments (Bonventre, Zielke, Korotkov, & Sikora, 2016; Zielke, Wierzbicki, Baarda, & Sikora, 2015; Zielke, Wierzbicki, Weber, Gafken, & Sikora, 2014; Zielke et al., 2016).
Targeting the first enzymes in the nucleotide‐activated‐glycero‐manno‐heptose pathways is a relatively unexplored field, although it appears to be an alternative approach to the discovery of broad‐spectrum antibacterial drugs (Bauer, Stevens, & Hansen, 1998; Darby, Ananth, Tan, & Hinnebusch, 2005; Tamaki, Sato, & Matsuhashi, 1971; Valvano, Messner, & Kosma, 2002). Nucleotide‐activated‐glycero‐manno‐heptoses, while absent in eukaryotic cells, are widely present in bacteria and are crucial components of the lipopolysaccharides (LPS), lipooligosaccharides (LOS), capsules, O‐antigens, and glycan moieties of bacterial cell surface (S‐layer) glycoproteins (Valvano et al., 2002). In particular, one such potential drug target of significant interest is sedoheptulose‐7‐phosphate isomerase, GmhA, annotated previously as TfrA (Havekes, Lugtenberg, & Hoekstra, 1976) and LpcA (Brooke & Valvano, 1996a). GmhA is conserved in many Gram‐negative and some Gram‐positive bacteria and is responsible for catalyzing isomerization of the D‐sedoheptulose 7‐phosphate into D‐glycero‐α,β‐D‐manno‐heptose‐7‐phosphate (Eidels & Osborn, 1974), which is the first and common step for parallel biosynthetic pathways leading to generation of GDP‐D‐glycero‐α‐D‐manno‐heptose (D,D‐heptose) and ADP‐L‐glycero‐β‐D‐manno‐heptose [L,D‐heptose; reviewed in Ref: (Valvano et al., 2002)]. D,D‐heptose is required for glycosylation of flagella and capsular polysaccharides, and for the assembly of disaccharide repeating units (D,D‐heptose linked to L‐rhamnose) composing the S‐layer glycan in the Gram‐positive Eubacterium, Aneurinibacillus thermoaerophilus DSM 10155 (Eidels & Osborn, 1974; Kosma, Wugeditsch, Christian, Zayni, & Messner, 1995; Wugeditsch et al., 1999). L,D‐heptose is used for glycosylation of capsular polysaccharides (Valvano et al., 2002) and as a primary building block of LPS/LOS core oligosaccharide (Brooke & Valvano, 1996a; Eidels & Osborn, 1971). In addition, a large family of bacterial autotransporter heptosyltransferases (BAHTs) utilizes L,D‐heptose as a sugar donor to modify serine residues on their substrate autotransporters, which has a significant impact on the virulence of Gram‐negative pathogens (Lu, Li, & Shao, 2015). The L,D‐heptose is synthesized in sequential reactions catalyzed in order by GmhA‐HldE(HldA)‐GmhB‐HldE(HldC)‐HldD [reviewed in Refs: (Raetz & Whitfield, 2002; Valvano et al., 2002)]. Usually one or more L,D‐heptose molecules and two 2‐keto‐3‐deoxy‐D‐manno‐oct‐2‐ulosonic acid (KDO) residues form the inner portion (lipid A proximal) of the LPS/LOS core oligosaccharide, which is typically more conserved than the structurally diverse outer core (Raetz & Whitfield, 2002). Similarly, gonococcal LOS contain two basal heptose molecules, designated Hep I and Hep II, forming elongation centers α, β, and χ (Gibson et al., 1989; John, Griffiss, Apicella, Mandrell, & Gibson, 1991; Yamasaki, Bacon, Nasholds, Schneider, & Griffiss, 1991). The individual chains can be decorated with structures that mimic human carbohydrate epitopes (α chain linked to Hep I; Apicella & Mandrell, 1989); a single glucose, lactose, or glucose with additional sugars (β chain extending from Hep II; Gibson et al., 1989; Yamasaki et al., 1994); and GlcNac, GlcNac acetate, or occasionally galactose (χ chain; Mcleod Griffiss, Brandt, Saunders, & Zollinger, 2000). In addition, phosphate or phosphoethanolamine groups may be attached to the heptose residues (Preston, Mandrell, Gibson, & Apicella, 1996; Raetz & Whitfield, 2002). The phosphoric residues participate in the ionic interactions between LPS/LOS and outer membrane proteins, as well as divalent cations, providing a barrier against entry of detergents, antibiotics, and hydrophobic compounds (Preston et al., 1996; Raetz & Whitfield, 2002). The N. gonorrhoeae heptose‐monophosphate was recently linked with the clinical and epidemiological synergy of gonorrhea and HIV (Malott et al., 2013). At the molecular level, this interplay involves the unique ability of gonococci to efficiently liberate phosphorylated L,D‐heptose into the extracellular milieu, which elicits an immune response and induces HIV‐1 expression and viral production in cluster of differentiation 4‐positive (CD4+) T cells (Malott et al., 2013).
Mutations in genes encoding GmhA in different bacterial species examined to date resulted in pleiotropic effects including production of truncated LPS/LOS composed of lipid A and KDO residues, increased susceptibility to antibiotics and detergents, impaired biofilm formation, and attenuated virulence (Aballay, Drenkard, Hilbun, & Ausubel, 2003; Bauer et al., 1998; Brooke & Valvano, 1996b; Darby et al., 2005). In addition, lack of HldA, which acts immediately downstream from GmhA in the L,D‐heptose biosynthetic pathway, rendered gonococci unable to induce HIV‐1 expression (Malott et al., 2013). Therefore, we propose GmhA in N. gonorrhoeae, GmhAGC, as a molecular target for the development of new antigonococcal drugs. Herein, we performed characterization of GmhAGC at the molecular, functional, and structural levels to facilitate the future targeting of this enzyme with small molecule inhibitors.
2. Experimental Procedures
2.1. Bacterial strains and growth conditions
Strains of bacteria used in this study are listed in Table 1. Neisseria gonorrhoeae and N. meningitidis were cultured either on gonococcal base solid medium (GCB, Difco), or in gonococcal base liquid (GCBL) medium supplemented with Kellogg's supplement I and II in ratios 1:100 and 1:1,000, respectively (Spence, Wright, & Clark, 2008). GCBL was additionally supplemented with sodium bicarbonate at a final concentration of 0.042%. In vitro host‐relevant growth conditions (iron deprivation, presence of normal human serum, anoxia) were procured as described previously (Zielke et al., 2016). Neisseria were cultured on solid media for 18–22 hr at 37°C in the presence of 5% atmospheric CO2. For N. gonorrhoeae, piliated or nonpiliated variants were passaged onto GCB and incubated for additional 18–22 hr. Colonies with piliated morphology were used for DNA transformation, while nonpiliated variants were used in all other experiments. Escherichia coli strains were grown either on Luria–Bertani agar (LBA, Difco) or cultured in Luria–Bertani broth (LB, Difco) at 37°C.
Table 1.
Bacterial strains used in this study
| Bacterial strains | Reference |
|---|---|
| N. gonorrhoeae | |
| FA1090 | (Connell et al., 1988) |
| MS11 | (Meyer, Mlawer, & So, 1982) |
| 1291 | (Apicella, Breen, & Gagliardi, 1978) |
| F62 | (Sparling, 1966) |
| FA1090 ∆gmhA GC/P lac :: gmhA GC | This study |
| FA1090 ∆gmhA GC/P lac :: gmhA GC E65A | This study |
| FA1090 ∆gmhA GC/P lac :: gmhA GC H183A | This study |
| FA1090 ∆gmhA GC/P lac :: gmhA NM | This study |
| Baltimore collection 1991–1994:LGB1, LG14, LG2, LGB26, LG20 | (Garvin et al., 2008; Zielke et al., 2014) |
| Clinical isolates from Public Health–Seattle & King County Sexually Transmitted Disease Clinic:UW01, UW02, UW03, UW04, UW05, UW06, UW07, UW08, UW09, UW10, UW11, UW12, UW13 | (Zielke et al., 2016) |
| 2016 WHO reference strains:F, G, K, L, M, N, O, P, W, X, Y, Z, U, V | (Unemo et al., 2016) |
| N. meningitidis | |
| MC58 | (McGuinness et al., 1991) |
| MC58 ∆gmhA NM/P lac :: gmhA NM | This study |
| MC58 ∆gmhA NM/P lac :: gmhA GC | This study |
| E. coli | |
| MC1061 | (Casadaban & Cohen, 1980) |
| BL21(DE3) | (Studier & Moffatt, 1986) |
Antibiotics were used on selected bacteria in the following concentrations: for N. gonorrhoeae: kanamycin 40 μg/ml, erythromycin 0.5 μg/ml; for N. meningitidis: kanamycin 80 μg/ml, erythromycin 2 μg/ml; for E. coli: kanamycin 50 μg/ml, erythromycin 250 μg/ml.
2.2. Genetic manipulations, site‐directed mutagenesis, and transcomplementation
Plasmids and primers used in this study are listed in Table S1. Oligonucleotides were designed based on the genomic sequence of N. gonorrhoeae FA1090 (NC_002946), N. meningitidis MC58 (NC_003112), and E. coli BL21(DE3) (NC_012892) using SnapGene software version 2.8 (GSL Biotech LLC) and synthesized by IDT Technologies. Genomic DNA of gonococcal strains, N. meningitidis MC58, and E. coli BL21(DE3) was isolated with the Wizard Genomic DNA Purification Kit (Promega). PCR products and plasmid DNA were purified using QIAprep Spin Miniprep Kit (QIAGEN). PCR reactions were performed using chromosomal or plasmid DNA as template, appropriate oligonucleotides, and Q5® High‐Fidelity DNA Polymerase (NEB). E. coli MC1061 was used as the host during the molecular cloning and site‐directed mutagenesis procedures. All created constructs and suppressor mutations in ∆gmhA GC/P lac ::gmhA GC were verified by Sanger Sequencing at the Center for Genomic Research and Biocomputing at Oregon State University. Transformation of N. gonorrhoeae and N. meningitidis was performed as described previously (Alexander, Richardson, & Stojiljkovic, 2004; Zielke et al., 2014).
The recombinant GmhAGC (rGmhAGC) containing N‐terminal‐6 × His‐tag followed by the tobacco etch virus (TEV) protease recognition site was obtained by amplifying the DNA region of the ngo1986 lacking stop codon with primers rNGO1986‐F and rNGO1986‐R and cloning the obtained PCR product (615 bp) into NcoI/HindIII sites of pRSF‐NT (Table S1).
The conditional GmhAGC mutant, FA1090 ∆gmhA GC/P lac ::gmhA GC, was constructed using a strategy as described by (Zielke et al., 2016) by placing an additional copy of ngo1986 under the control of the isopropyl β‐D‐1‐thiogalactopyranoside (IPTG)‐inducible promoter, Plac, within an intergenic region located between lctP and aspC in the FA1090 chromosome (Mehr, Long, Serkin, & Seifert, 2000) and a subsequent in‐frame replacement of the ngo1986 in its native chromosomal locus with the nonpolar kanamycin resistance cassette. Specifically, the ngo1986 containing its native ribosomal binding site (RBS) was amplified with primers NGO1986‐RBS‐F and NGO1986‐RBS‐R. The resulting 628 bp PCR product was digested with FseI and inserted into ScaI/FseI‐treated pGCC4, yielding pGCC4‐GmhAGC. After transformation of FA1090 with pGCC4‐GmhAGC, gonococci were selected on GCB with 0.5 μg/ml erythromycin and verified by PCR with primers pGCC4‐Ver‐F and pGCC4‐Ver‐R.
The constructs for deletion of gmhA GC were obtained by amplification of the 552 bp upstream DNA region and 571 bp downstream from ngo1986 with primers NGO1986‐UP‐F/NGO1986‐UP‐R and NGO1986‐Down‐F/NGO1986‐Down‐R, respectively. The upstream fragment was digested with EcoRI/KpnI and cloned into similarly cleaved pUC18K, yielding pUC18K‐GmhAGC‐Up. Next, the downstream fragment was inserted into the BamHI/HindIII‐cleaved pUC18K‐GmhAGC‐Up. The resulting pUC18K‐GmhAGC was linearized with HindIII and used to transform FA1090 carrying the second copy of GmhA on the chromosome, FA1090P lac ::gmhA GC. Clones were selected on a solid medium supplemented with kanamycin and 0.1 mmol/L IPTG, and verified by PCR with NGO1986‐Ver‐F and NGO1986‐Ver‐R primers. Subsequently, selected gonococci were further verified by immunoblotting analyses using anti‐GmhAGC antisera.
To generate GmhAGCE65A and GmhAGCH183A, the template for site‐directed mutagenesis (pUC18‐GmhAGC) was constructed by amplifying ngo1986 with its native RBS using primers NGO1986‐RBS‐F and NGO1986‐RBS‐R and cloning into SmaI‐cleaved pUC18. Site‐directed mutagenesis was performed using the template; primer pairs E65A‐F/E65A‐R or H183A‐F/H183A‐R, respectively, and Q5 Site‐Directed Mutagenesis Kit (NEB), according to the manufacturer's manual. Subsequently, the mutated variants of GmhAGC were amplified with NGO1986‐RBS‐F and NGO1986‐RBS‐R, cleaved with FseI, cloned into ScaI/FseI‐digested pGCC4, introduced into the FA1090 chromosome, and the native ngo1986 was deleted as described above.
For transcomplementation studies, GmhA homologs from N. meningitidis MC58 (nmb2090) and E. coli BL21(DE3) (ECBD_3400) were amplified with primer pairs NGO1986‐RBS‐F/NGO1986‐RBS‐R and ECBD3400‐RBS‐F, respectively. The resulting PCR products were digested with FseI and introduced into ScaI/FseI‐treated pGCC4. Introduction into the FA1090 chromosome and deletion of GmhA were performed as outlined above.
The cytoplasmic marker control for subfractionation experiments, the zwf gene (ngo0715) encoding glucose‐6‐phosphate‐1‐dehydrogenase was amplified using primers RAZ548 and RAZ549. The PCR product was cleaved with NcoI and HindIII and cloned into similarly digested pRSF‐NT to obtain plasmid pRSF‐NT‐Zwf.
2.3. Proteome subfractionation procedures
Colonies of N. gonorrhoeae FA1090 were collected from GCB, suspended in 500 ml of GCBL to OD600 of 0.1, and cultured with aeration at 37°C until OD600 of ~0.8. Bacterial cells were separated from the suspension by centrifugation (10 min, 6,000g, 4°C). The crude cell envelopes were purified using a sodium carbonate extraction procedure while naturally released membrane vesicles (MVs) and soluble proteins (SS) were fractionated from culture supernatants by ultracentrifugation as described previously (Zielke et al., 2016).
2.4. GmhAGC depletion studies
Colonies of FA1090 ∆gmhA GC/P lac :: gmhA GC were collected with a cotton swab from GCB supplemented with 10 μmol/L IPTG and suspended in GCBL to OD600 of 0.1. After two washes in prewarmed GCBL, bacterial suspension was split and incubated with shaking (220 rpm) with or without IPTG for 3 hr at 37°C. Cultures were back‐diluted to OD600 of 0.1 in fresh GCBL, as described above, and cultured for additional 6 hr. At specific time points indicated in the text, OD600 measurements were taken, samples for western blotting and LOS isolation were withdrawn, and cultures were serially diluted followed by plating on GCB for enumeration of colony‐forming units (CFUs). Experiments were performed on three separate occasions and mean values and SEM are presented.
2.5. Isolation of LOS and silver staining
LOS was isolated from N. gonorrhoeae and N. meningitidis based on the method described previously (Hitchcock & Brown, 1983). Bacteria were either collected from GCB or GCBL, as specified in the text, suspended in 1.5 ml of GCBL to OD600 of 0.2 and spun down for 1.5 min at 15,000g. Pelleted cells were lysed by addition of 50 μl of lysis buffer (2% SDS, 4% β‐mercaptoethanol, 10% glycerol, 1mol/L Tris‐HCl pH 6.8, and 0.01% bromophenol blue) and incubation at 100°C for 10 min. Samples were allowed to cool down to room temperature and proteins were digested by addition of 25 μg proteinase K in 10 μl of lysis buffer and incubated for 1 hr at 60°C. Isolated LOS was resolved on 16.5% Mini‐PROTEAN® Tris‐Tricine Gel (Bio‐Rad) and visualized by a silver staining procedure (Tsai & Frasch, 1982).
2.6. Fitness assessment
Colonies of different neisserial strains, as indicated in the text, were collected from GCB and reconstituted in GCBL to OD600 of 0.1. Bacterial cultures were incubated in the absence of IPTG for 3 hr at 37°C with aeration and subsequently back‐diluted to OD600 of 0.2, serially diluted, and plated on GCB with or without IPTG for CFUs scoring. Experiments were repeated in three biological replicates and mean values with corresponding SEM are presented.
2.7. Purification of the rGmhAGC and rZwf and production of polyclonal rabbit antibodies
E. coli BL21(DE3) strain carrying either pRSF‐NT‐GmhAGC or pRSF‐NT‐Zwf was used as heterologous host for overproduction and purification of rGmhAGC and recombinant Zwf (rZwf), respectively. Overnight cultures were back‐diluted in 2.0 L of LB supplemented with kanamycin and incubated with aeration at 37°C until the optical density (OD600) reached ~0.5. Overproduction of rGmhAGC and rZwf was induced with 0.1 and 1 mmol/L IPTG, respectively, and cultures were incubated for additional 3 hr at 37°C. Cells were harvested by centrifugation (6,000g, 10 min, 4°C). Pelleted bacteria carrying pRSF‐NT‐GmhAGC were suspended in lysis buffer (20 mmol/L Tris‐HCl pH 7.0, 1 mol/L NaCl, 10 mmol/L imidazole, 5% glycerol) supplemented with a Pierce Protease Inhibitor Mini Tablet (Thermo Scientific) and lysed by passaging six times through a French pressure cell at 12,000 psi. Cell debris and unbroken cells were separated from soluble protein fraction by centrifugation at 16,000g for 30 min at 4°C. The supernatant was passed through 0.22 μm membrane filter (VWR International) and applied onto Bio‐Scale MiniProfinity IMAC cartridges (Bio‐Rad) on the NGC Scout Chromatography system (Bio‐Rad). Loosely bound proteins were removed with 10 column volumes of wash buffer (20 mmol/L Tris pH 8.0, 500 mmol/L NaCl, 40 mmol/L imidazole) and elutions were conducted with a 40–250 mmol/L imidazole gradient. Fractions containing rGmhAGC were combined and a PD‐10 column (GE Healthcare) was used to exchange the buffer to 20 mmol/L HEPES pH 7.5, 100 mmol/L NaCl, 5% glycerol. Subsequently, the N‐terminal‐6 × His‐tag of the rGmhAGC was removed by TEV protease. Cleavage reaction was prepared by mixing rGmhAGC with TEV in 20:1 (w:w ratio) in 500 μl of cleavage buffer (0.5 mol/L Tris‐HCl pH 8.0, 5 mmol/L EDTA), 10 μl of 0.5 mol/L DTT, and 500 μl of Ni‐NTA agarose (Qiagen) equilibrated with cleavage buffer. Following overnight incubation at room temperature, cleavage mixture was loaded onto a 5 ml polypropylene column (Thermo Scientific) and supernatant containing 6 × His‐tag‐free rGmhAGC was collected. To remove residual TEV protease that coeluted with rGmhAGC, the mixture was incubated again with 500 μl of Ni‐NTA agarose (Qiagen) equilibrated with cleavage buffer for 1 hr with rotation at 4°C, and rGmhAGC was eluted. The purified rGmhAGC was applied to a PD‐10 column (GE Healthcare) for a buffer exchange into 20 mmol/L HEPES pH 7.5, 100 mmol/L NaCl, and 5% glycerol.
Purification of rZwf was accomplished using the same procedures as described above with the following modifications. E. coli cells were resuspended in lysis buffer (20 mmol/L Tris‐HCl pH 8.0, 10 mmol/L imidazole, 450 mmol/L NaCl). Cells were lysed using French Press and rZwf was purified using 5 ml Bio‐Scale Mini Nuvia IMAC Ni‐Charged column (Bio‐Rad) connected to a NGC Chromatography System (Bio‐Rad). Bound peptides were eluted using elution buffer (20 mmol/L Tris‐HCl pH 8.0, 250 mmol/L imidazole, 450 mmol/L NaCl). Fractions containing proteins were pooled, EDTA and DTT were added to final concentrations of 0.5 mmol/L and 1 mmol/L, respectively, and the His‐tag was removed by overnight incubation with TEV protease (ratio 1:100 w/w). After cleavage, the proteins were separated using Hi Load 16/600 Superdex 75 pg column (GE Healthcare) and buffer containing 20 mmol/L Tris pH 8, 150 mmol/L NaCl connected to the NGC Chromatography System. Fractions containing Zwf were pooled together and concentrated using 10 kDa Vivaspin 20 concentrators (GE Healthcare). Glycerol was added to a final concentration of 10% and the protein was aliquoted and stored at −80°C.
The polyclonal rabbit anti‐GmhAGC and anti‐Zwf antibodies were generated using 6 × His‐tag‐free rGmhAGC and rZwf, respectively. Standard 13‐week antibody production protocols were applied, utilizing four New Zealand White rabbits, animal handling was performed according to the Animal Protocol #1 approved by IACUC, in a certified animal facility (USDA 93‐R‐283) and the NIH Animal Welfare Assurance Program (#A4182‐01) at the Pacific Immunology Corporation. Anti‐GmhAGC and anti‐Zwf antisera were used at 1:10,000.
2.8. Size exclusion chromatography
The NGC Scout Chromatography system (Bio‐Rad) employing a HiLoad 16/600 Superdex 75 pg column (GE Healthcare Life Sciences) was used to separate purified rGmhAGC based on the molecular size. Buffer for the chromatography (20 mmol/L Tris‐HCl pH 8.0, 500 mmol/L NaCl) was applied at 1 ml/min flow rate. Gel Filtration Standard (Bio‐Rad) was used to determine the size of the separated proteins.
2.9. Crystallization and structure determination of N. gonorrhoeae GmhAGC
The screening for initial crystallization conditions was performed using JCSG Core Suites I‐IV (Qiagen) (Newman et al., 2005). The optimized crystals were grown using 0.1 mol/L Tris‐HCl pH 8.5, 0.2 mol/L magnesium chloride, 30% PEG4000. Crystals were transferred to a cryoprotectant solution supplemented with 20% glycerol and flash‐frozen in liquid nitrogen. The diffraction data were collected from a single crystal at the beamline 22‐ID, Southeast Regional Collaborative Access Team (SER‐CAT) at the Advanced Photon Source, Argonne National Laboratory. Data were integrated and scaled using XDS and XSCALE (Kabsch, 2010a). The structure was solved by molecular replacement using Phaser (McCoy et al., 2007) and structure of Pseudomonas aeruginosa GmhA (PDB 3BJZ) as a search model (Taylor et al., 2008). The electron density modification was performed using Parrot (Cowtan, 2010) followed by automated model rebuilding using Buccaneer (Cowtan, 2006). The model was completed by manual rebuilding in Coot (Emsley, Lohkamp, Scott, & Cowtan, 2010) and was refined using REFMAC5 (Murshudov et al., 2011). The structure was validated using Coot and the MolProbity server (Chen et al., 2010). The structural figures were generated using PyMol (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).
2.10. SDS‐PAGE and immunoblotting
Whole cell lysates of Neisseria strains indicated in the text were either collected from solid media after 22 hr of aerobic or 48 hr of anaerobic growth, or harvested from liquid media following procedures described (Zielke et al., 2014, 2016). E. coli cells were collected from LB agar plates after overnight incubation. Samples were normalized based on OD600 values or based on total protein concentration, and SDS‐PAGE, staining with Coomassie brilliant blue G‐250, and immunoblotting analyses were performed exactly as described previously (Zielke et al., 2014, 2016).
2.11. Statistical analysis
GraphPad Prism's build‐in t‐test was used for determination of statistically significant differences between obtained experimental results. A confidence level of 95% was used for all analyses.
2.12. Accession numbers
The coordinates and structure factors were deposited to the Protein Data Bank with accession code 5I01.
3. Results
3.1. Chromosomal location and purification of GmhAGC
Genes encoding the enzymes of L,D‐heptose biosynthesis pathway are scattered throughout the chromosome in the majority of bacteria including N. gonorrhoeae (Valvano et al., 2002). The GmhA homolog in N. gonorrhoeae strain FA1090 is encoded by ngo1986, which is located between ngo1985, coding for an outer membrane lipoprotein (Zielke et al., 2014) and ngo1987 encoding a putative endonuclease (YraN). Comparison of the gmhA location using BioCyc Pathway/Genome Database Collection (http://biocyc.org/) showed that this genetic arrangement is conserved among the various deposited Neisseria species and isolates (n = 70) with only a few exceptions, including N. bacilliformis ATCC BAA‐1200, N. sp. oral taxon 020 F0370, N. shayeganii 871, and N. subflava NJ9703.
GmhAGC consists of 197 amino acids with residues 37–197 comprising a sugar isomerase domain (SIS) (Bateman, 1999) that is shared between all ketose/aldose isomerases (Golinelli‐Pimpaneau, Le Goffic, & Badet, 1989). At the amino acid level, GmhAGC shows 43–57% identity with crystallized orthologous proteins (Table 3) and contains the key conserved residues observed in all GmhA homologs, including three serine residues and a threonine that presumably interact with the phosphate group of sedoheptulose 7‐phosphate or D‐glycero‐D‐manno‐heptose 7‐phosphate (Harmer, 2010; Valvano et al., 2002).
Table 3.
Structural homologs of N. gonorrhoeae GmhAGC
| Protein | Organism | r.m.s.d.a | Sequence identitya (%) | Ligands | PDB ID | Reference |
|---|---|---|---|---|---|---|
| GmhA | P. aeruginosa | 0.5 | 57 | D‐glycero‐α‐D‐manno‐heptopyranose‐7‐phosphate | 1X92 | (Taylor et al., 2008) |
| DiaA | E. coli | 0.7 | 52 | apo | 2YVA | (Keyamura et al., 2007) |
| DiaA | E. coli | 0.7 | 52 | apo | 4U6N | unpublished |
| GmhA | F. tularensis | 0.7 | 45 | apo | 3TRJ | (Chaudhury et al., 2013) |
| GmhA | C. psychrerythraea | 0.7 | 48 | apo | 5BY2 | (Do et al., 2015) |
| GmhA | C. jejuni | 0.7 | 54 | apo | 1TK9 | (Seetharaman et al., 2006) |
| GmhA | P. aeruginosa | 0.8 | 56 | apo | 3BJZ | (Taylor et al., 2008) |
| GmhA | B. pseudomallei | 0.8 | 43 | Zn2+ | 2X3Y | (Harmer, 2010) |
| GmhA | B. pseudomallei | 0.9 | 43 | Zn2+, D‐glycero‐α‐D‐manno‐heptopyranose‐7‐phosphate | 2XBL | (Harmer, 2010) |
| GmhA | E. coli | 1.4 | 45 | apo | 2I2W | (Taylor et al., 2008) |
| GmhA | E. coli | 1.1 | 45 | D‐sedoheptulose‐7‐phosphate | 2I22 | |
| GmhA | V. cholerae | 1.1 | 43 | apo | 1X94 | (Seetharaman et al., 2006) |
As reported by the Dali server (Holm & Rosenstrom, 2010) for superposition of monomers. A mean value is listed for the structures containing more than one protein subunit in the asymmetric unit.
To characterize GmhAGC, we first purified recombinant protein with an N‐terminal 6 × His‐tag followed by the tobacco etch virus (TEV) protease cleavage site and prepared untagged protein. Size exclusion chromatography indicated that the native GmhAGC forms tetrameric structures in the solution (Fig. S1A). The denatured protein migrated on SDS‐PAGE according to the predicted molecular mass of 21.093 kDa (Fig. S1B). Untagged GmhAGC was subsequently used in crystallization and to obtain polyclonal rabbit antisera.
3.2. N. gonorrhoeae deprived of GmhAGC displays defect in LOS synthesis and growth cessation
The genetic inactivation of gmhA in E. coli, Haemophilus influenzae, H. ducreyi, Salmonella enterica, and Yersinia pestis resulted in altered outer membrane permeability and decreased virulence in animal infection models, however, the mutants did not display drastic growth defects in vitro (Aballay et al., 2003; Bauer et al., 1998; Brooke & Valvano, 1996b; Darby et al., 2005). Therefore, we were surprised by the failure of our numerous attempts to generate a clean deletion of ngo1986 in N. gonorrhoeae FA1090. As an alternative strategy, an additional copy of gmhA GC was placed under the control of the IPTG‐inducible P lac promoter and introduced into the chromosome of wild‐type FA1090, followed by an allelic replacement of ngo1986 with the kanamycin resistance cassette. The obtained ∆gmhA GC/P lac ::gmhA GC mutant grew scarcely after passage on solid media without IPTG, whereas abundant colonies were observed in the presence of the inducer (Figure 1a). The depletion of GmhAGC was corroborated by probing whole cell lysates of these gonococci with anti‐GmhAGC antisera (Figure 1b). Visualization of LOS species by Tricine‐SDS‐PAGE coupled with silver staining showed, as expected, that bacteria deprived of GmhAGC carried a truncated version of LOS that migrated faster in comparison with the LOS species isolated from wild‐type and ∆gmhA GC/P lac ::gmhA GC grown in the presence of IPTG (Figure 1b).
Figure 1.
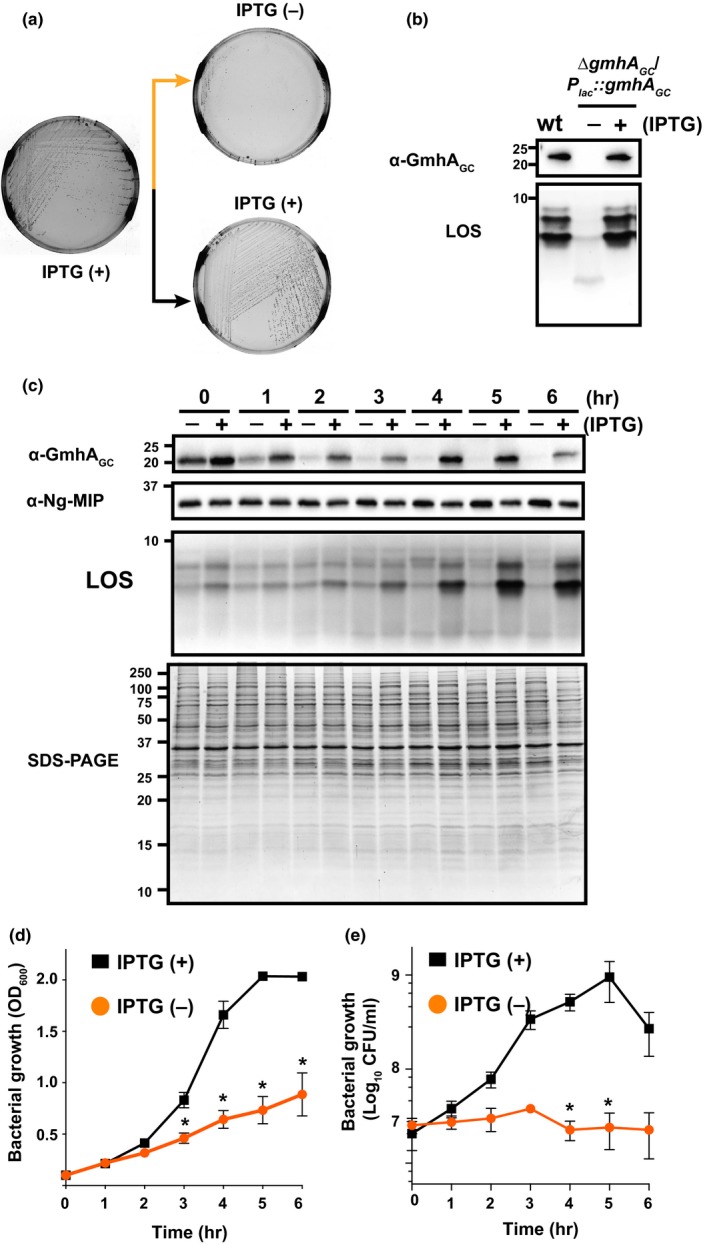
GmhAGC is pivotal for N. gonorrhoeae growth and lipooligosaccharide (LOS) synthesis. (a) The N. gonorrhoeae FA1090 gmh AGC conditional knockout strain, ∆gmh AGC/P lac ::gmh AGC, was streaked out from frozen glycerol stock on solid media supplemented with 20 μmol/L isopropyl β‐D‐1‐thiogalactopyranoside (IPTG). After 18 hr incubation at 37°C in the presence of 5% atmospheric CO 2, the colonies were passaged onto plates either with (+) or without IPTG (−). (b) Whole cell lysates of FA1090 wild‐type and isogenic ∆gmh AGC/P lac ::gmh AGC harvested from plates with (+) or without 20 μmol/L IPTG (−) were either probed with polyclonal rabbit antisera or subjected to LOS extraction using proteinase K followed by silver staining. (c–e) The FA1090 ∆gmh AGC/P lac ::gmh AGC cells were collected from solid media supplemented with 20 μmol/L IPTG, suspended to OD 600 of 0.1, washed twice, divided, and cultured either in the presence or absence of IPTG for 3 hr. At time point designated as 0 hr, corresponding cultures were back‐diluted to the same density (OD 600 of 0.1) into fresh media with or without IPTG and incubated for additional 6 hr. Samples of bacterial cultures were collected every hour for GmhAGC and Ng‐MIP immunoblotting analysis, LOS, and whole cell protein profiles (c), monitoring of bacterial proliferation by measurement of density of the cultures at OD 600, (d) and spotting serially diluted bacteria on solid media with IPTG for CFU scoring (e). Whole cell lysates were matched by the same OD 600 units. As loading control, samples separated by SDS‐PAGE were either probed with anti‐Ng‐MIP antibodies or stained with Coomassie brilliant blue G‐250. The migration of molecular mass marker (kDa) is indicated on the left. All experiments were performed in three biological replicates. Means and SEM are presented on graphs; *p < .05
Subsequently, time course experiments under standard laboratory growth conditions were conducted to examine the correlation between depletion of GmhAGC, LOS formation, and N. gonorrhoeae fitness. An initial suspension of nonpiliated colonies of ∆gmhA GC/P lac ::gmhA GC collected from solid media supplemented with IPTG was divided and cultured in liquid media in the presence or absence of IPTG [(+) or (−), respectively]. After 3 hr, to ensure GmhAGC depletion, the corresponding cultures were diluted to OD600 of 0.1 in media with or without IPTG. From this point onward (shown as 0 hr in Figure 1c–e), samples were withdrawn every hour to assess GmhAGC levels and LOS formation (Figure 1c), culture density (Figure 1d), and colony forming units (CFUs; Figure 1e). Immunoblotting with anti‐GmhAGC antisera showed that in the absence of IPTG, GmhAGC levels were slightly decreased at 0 hr and complete GmhAGC depletion was achieved three hours later (Figure 1c). Also, it was noted that despite the presence of inducer, GmhAGC had a peculiar expression pattern; decreased amounts of protein in comparison with these observed at 0 hr were detected at 1 hr and in early‐log phase (corresponding to 2–3 hr data points). Furthermore, increased GmhAGC levels were found in late‐log phase (4, 5 hr) followed by a decline in stationary phase (6 hr; Figure 1c–e). In contrast, no major differences were noticed in the overall protein profiles and the level of Ng‐MIP (Figure 1c), which is ubiquitously expressed throughout phases of gonococci growth (Zielke et al., 2016). As expected based on data presented in Figure 1b, the wild‐type LOS species diminished over time and were barely detectable upon GmhAGC diminution (Figure 1c). Finally, analysis of culture density and number of viable gonococci showed that in media lacking IPTG, bacterial growth was greatly reduced concomitant to depletion of GmhAGC (Figure 1c–e), whereas in the presence of IPTG, the generation time of the ∆gmhA GC/P lac ::gmhA GC mutant was 61.2 min (Figure 1d–e), a value similar to the parental wild‐type strain (56.5 min; Figure 2b). In addition, under nonpermissive conditions, we observed that the OD600 of the mutant culture continued to increase (Figure 1d) while the number of viable bacteria remained at 107 CFUs/ml (Figure 1e). These results suggested that suppressors were arising in the mutant population but had not reached the point where they overcome the persistent population by the end of the 6‐hr growth curve. We tested this possibility by passaging ∆gmhA GC/P lac ::gmhA GC on solid media without and with IPTG. After the third passage, ample bacterial colonies were obtained with several mutations within the P lac , allowing expression of GmhAGC and LOS synthesis (Fig. S2).
Figure 2.
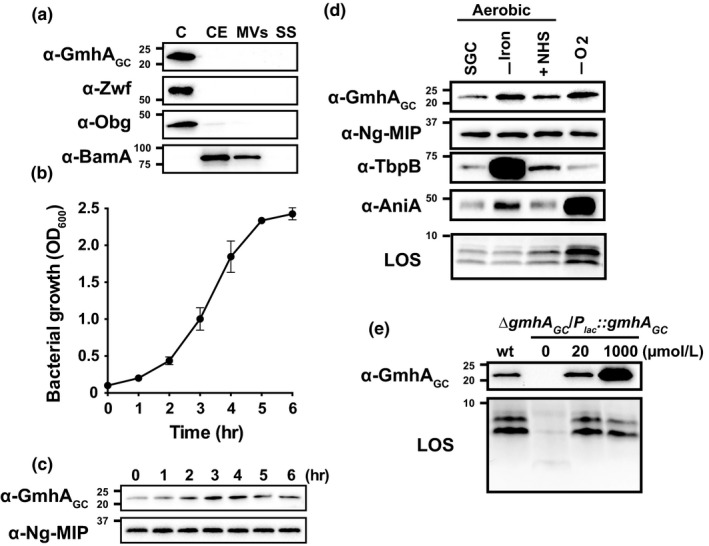
Assessment of GmhAGC subcellular localization and expression patterns. (a) N. gonorrhoeae wild‐type FA1090 cells were harvested during mid‐exponential phase and subjected to proteome extraction to separate cytoplasmic/periplasmic proteins (C), cell envelopes (CE), naturally released membrane vesicles (MVs), and soluble proteins in culture supernatants (SS). Individual fractions were loaded based on the total amount of protein (μg) and probed with antisera as indicated on the left. (b–c) Wild‐type N. gonorrhoeae FA1090 was cultured in liquid media and at indicated time points, OD 600 measurements were taken (b) and samples were withdrawn and processed for immunoblotting analysis of GmhAGC and Ng‐MIP (c). (d) Quantities of GmhAGC and lipooligosaccharide (LOS) in wild‐type FA1090 during in vitro conditions relevant to different infection sites [standard growth under aerobic conditions (SGC), iron deprivation (‐Iron), presence of normal human serum (+NHS), and anaerobiosis (‐O2)], were assessed by probing the whole cell lysates with anti‐GmhAGC antibodies and silver staining of proteinase K‐extracted LOS, respectively. Immunoblotting experiments with antisera against Ng‐MIP, TbpB, and AniA were used as markers for ubiquitous expression, iron‐limiting, and anaerobic conditions, respectively. (e) Effect of overexpression of GmhAGC on LOS amounts was examined by harvesting wild‐type FA1090 and isogenic ∆gmh AGC/P lac ::gmh AGC grown on solid media with different isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) concentrations (0, 20, or 1,000 μmol/L). Whole cell lysates were either probed with anti‐GmhAGC antisera or treated with proteinase K and LOS was visualized by silver staining. Experiments were performed in biological triplicates and representative immunoblots and silver‐stained gels are shown. Mean values and corresponding SEM are presented on the graph. Migration of a molecular mass marker (kDa) is indicated on the left
Together, these findings demonstrated that GmhAGC is pivotal for LOS synthesis and optimal growth of N. gonorrhoeae.
3.3. GmhAGC subcellular localization and expression patterns
Preliminary results and absence of signal peptide suggested that GmhA of E. coli is a cytoplasmic enzyme (Brooke & Valvano, 1996a). However, subcellular localization of GmhA and expression patterns were never assessed. To examine the localization of GmhAGC, N. gonorrhoeae FA1090 was harvested at mid‐logarithmic phase of growth and subjected to subfractionation procedures. Equal amounts of extracted proteome fractions including cytoplasm, cell envelopes, naturally released membrane vesicles, and soluble proteins in culture supernatants were separated by SDS‐PAGE and probed with polyclonal antibodies (Figure 2a). GmhAGC was found solely in the cytoplasmic protein fraction, similarly to the cytoplasmic enzyme glucose‐6‐phosphate 1‐dehydrogenase, Zwf. As expected, the GTPase Obg (ObgGC), which primarily associates with 50S ribosomal subunits and partly with the peripheral inner membrane proteome (Papanastasiou et al., 2013; Zielke et al., 2015), was detected mainly in the cytoplasm and minute amounts were also found in the cell envelope fraction; whereas, antisera against the outer membrane protein marker BamA (Zielke et al., 2016) cross‐reacted with fractions containing cell envelopes and membrane vesicles.
The expression of GmhAGC was subsequently examined in wild‐type FA1090 during routine aerobic growth in liquid media (Figure 2b–c) and on solid media under conditions that more closely mimic clinical infection, such as iron deprivation, exposure to human serum, and anoxia (Figure 2d). Antibodies against Ng‐MIP, TbpB, and AniA were used as markers for ubiquitous expression, iron‐limiting conditions, and anaerobiosis, respectively (Cornelissen, 2008; Zielke et al., 2014, 2016). Immunoblotting analyses of whole cell lysates showed that expression of GmhAGC peaked during mid‐exponential phase (3 and 4 hr, Figure 2b–c). Increased GmhAGC levels were also detected during iron deprivation and anaerobic growth in comparison with standard laboratory conditions (Figure 2d). As expected, expression of Ng‐MIP remained constant throughout different phases of gonococcal growth and was unchanged under all tested conditions (Zielke et al., 2015, 2016), while TbpB and AniA were the most highly upregulated during iron deprivation and anaerobic growth, respectively (Figure 2d).
The increase in GmhAGC expression did not influence LOS migration patterns in wild‐type gonococci, albeit increased amounts of total LOS were observed in bacteria cultured in the presence of normal human serum and anaerobically (Figure 2d). To further assess the possible correlation between expression of GmhAGC and LOS levels, ∆gmhA GC/P lac ::gmhA GC was cultured in increasing concentrations of IPTG. The vast overexpression of GmhAGC achieved with 1,000 μmol/L IPTG did not have adverse effect on bacterial growth (data not shown) and had no effect on the LOS quantities (Figure 2e).
Cumulatively, these experiments demonstrated that GmhAGC is a cytoplasmic enzyme with augmented expression during mid‐logarithmic phase, iron depletion and anaerobiosis, and that overproduction of GmhAGC alone does not alter gonococcal LOS abundance.
3.4. Conservation of gmhA among Neisseria
Analysis of gmhA conservation showed that the gene (locus NGO1986, NMB2090, NMC2070) is present in all of the 39,182 Neisseria spp. genomes deposited into the PubMLST database (http://pubmlst.org/neisseria/ as of July, 20, 2016) and that there are 340 alleles and 323 single nucleotide polymorphic sites (Fig. S4).
Expression of GmhAGC among 36 different N. gonorrhoeae strains isolated from patients at different times and geographic locations, including the 2016 WHO reference strains (Unemo et al., 2016), was also assessed by immunoblotting. Whole cell lysates were resolved by SDS‐PAGE and probed with either polyclonal anti‐GmhAGC antisera or Zwf antibodies, or stained with Coomassie brilliant blue G‐250 as a loading control. Antisera against GmhAGC cross‐reacted with all clinical isolates of N. gonorrhoeae, but no cross‐reactivity was detected for the E. coli GmhA homolog (Figure 3). In addition, there were noticeable differences in GmhAGC protein abundance between the strains, while Zwf was uniformly expressed and there were no discrepancies in regard to samples normalization and loading (Figure 3 and Fig. S3).
Figure 3.
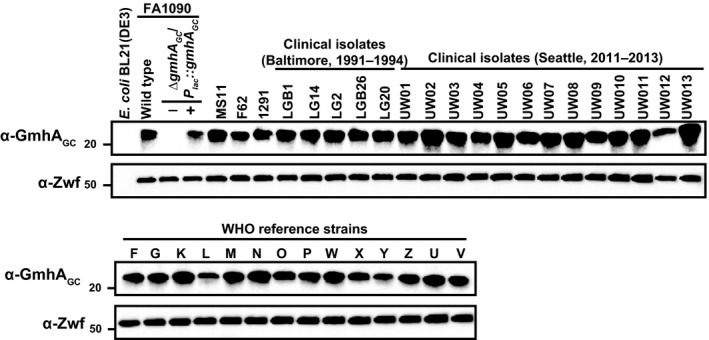
GmhAGC expression in a panel of N. gonorrhoeae isolates. Whole cell lysates of E. coli, wild‐type FA1090, ∆gmh AGC/P lac ::gmh AGC grown either with (+) or without (−) isopropyl β‐D‐1‐thiogalactopyranoside (IPTG), and 36 additional strains of N. gonorrhoeae were resolved on a 10–20% Tris‐Glycine gel and probed with anti‐GmhAGC antibodies. Immunoblotting with anti‐Zwf antisera was used as a loading control. Migration of a molecular mass marker (kDa) is indicated on the left
3.5. Hindering GmhAGC isomerase activity does not influence N. gonorrhoeae growth
The crucial side chains for GmhA enzymatic activity appeared to be E65 and H180 in the E. coli ortholog as analyzed by in vitro kinetic assays, LOS synthesis, and novobiocin sensitivity studies. It has been proposed that these two residues act as the base and the acid, respectively, to promote the isomerization reaction of D‐sedoheptulose 7‐phosphate into D‐glycero‐α,β‐D‐manno‐heptose‐7‐phosphate (Taylor et al., 2008). Therefore, to determine whether the observed decrease in N. gonorrhoeae survival upon GmhAGC depletion is a consequence of abolished LOS synthesis, site‐directed mutagenesis of corresponding residues (E65 and H183 in GmhAGC) was employed. The obtained GmhAGC E65A and H183A variants were placed under the IPTG‐inducible promoter and introduced into the chromosome of wild‐type FA1090, followed by allelic exchange of ngo1986 with the kanamycin resistance cassette. Both mutated proteins were stably produced in N. gonorrhoeae; however, the H183A variant was present at higher levels than the wild‐type GmhAGC (Figure 4a). Silver staining analysis of LOS revealed that bacteria expressing either E65A or H183A constructs produced truncated LOS, regardless of the presence of IPTG (Figure 4a). Furthermore, under nonpermissive conditions (without IPTG), the ∆gmhA GC/P lac ::gmhA GCE65A and ∆gmhA GC/P lac ::gmA GCH183A, similar to the ∆gmhA GC/P lac ::gmhA GC, had decreased viability, which was demonstrated by a 3214‐, and 8653‐fold decline, respectively, in CFUs in comparison with wild‐type gonococci (Figure 4b). In contrast, the expression of either mutated version of GmhAGC rescued bacterial viability to the wild‐type level.
Figure 4.
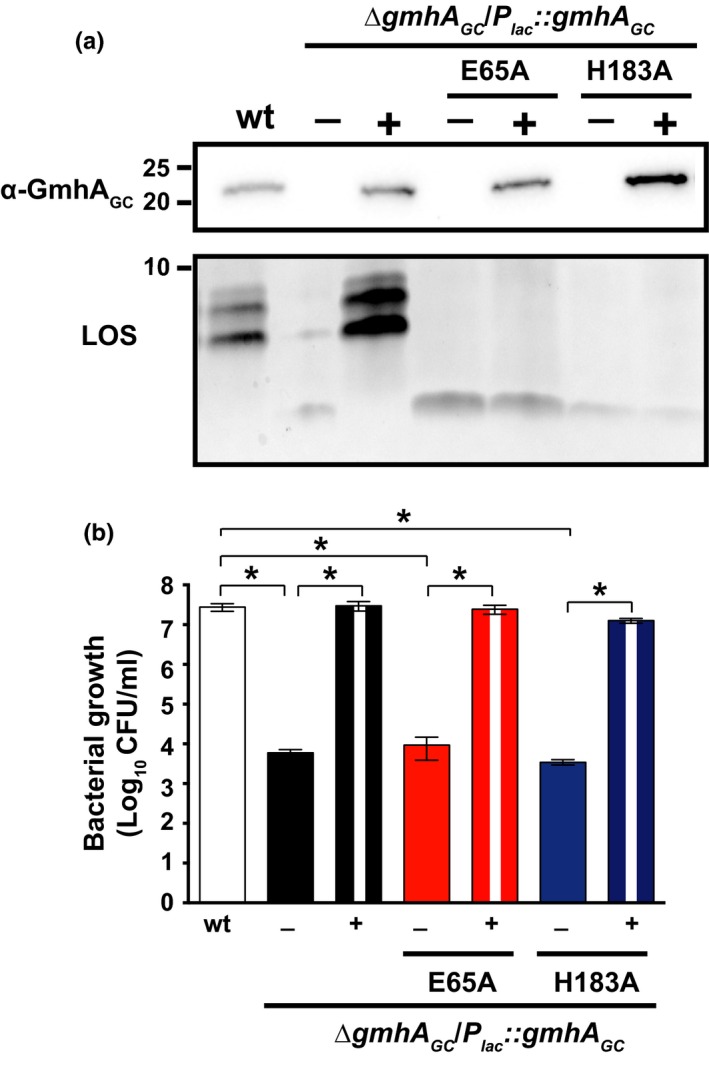
Hindering GmhAGC isomerase activity does not influence N. gonorrhoeae growth. (a) Wild‐type FA1090 and isogenic conditional mutants carrying either native gmh AGC (∆gmh AGC/P lac ::gmh AGC) or mutated variants of GmhAGC (∆gmh AGC/P lac ::gmh AGCE65A or ∆gmh AGC/P lac ::gmh AGCH183A) were collected from solid media with (+) and without (−) isopropyl β‐D‐1‐thiogalactopyranoside (IPTG). Expression of individual GmhAGC variants and lipooligosaccharide (LOS) patterns were examined in whole cell extracts by immunoblotting and silver staining, respectively. Samples were matched by equivalent OD 600 units. Migration of a molecular mass marker (kDa) is indicated on the left. (b) Wild‐type FA1090 and conditional mutants ∆gmh AGC/P lac ::gmh AGC, ∆gmh AGC/P lac ::gmh AGCE65A, and ∆gmh AGC/P lac ::gmh AGCH183A were collected from solid media supplemented with 20 μmol/L IPTG, suspended in liquid media to OD 600 of 0.1, cultured for 3 hr, back‐diluted to equal OD 600 of 0.2, serially diluted, and spotted on solid media in the presence (+) and absence (−) of IPTG. CFUs were scored. The data show averages of CFUs with corresponding SEM of at least three separate experiments; *p < .05
These in vivo studies demonstrated the importance of residues E65 and H183 in GmhAGC activity in the production of full‐length LOS and suggested that abolition of LOS synthesis is disconnected from the GmhAGC‐dependent effect on N. gonorrhoeae viability.
3.6. Transcomplementation studies of GmhA
GmhA homologs of H. influenzae and H. ducreyi restored the synthesis of full‐length LPS in the E. coli ∆gmhA mutant (Bauer et al., 1998; Brooke & Valvano, 1996a). This suggested that GmhA proteins can function interchangeably. However, failure of multiple attempts to remove ngo1986 in FA1090 carrying the E. coli gmhA gene cloned under the Plac promoter and integrated into the chromosome ruled out the possibility of a functional interspecies complementation. The E. coli GmhA (GmhAEC) shares 74% and 75% amino acid identity with H. influenzae and H. ducreyi proteins, respectively, while only 50% identity exists between GmhAEC and GmhAGC (Table 3). In contrast, N. meningitidis GmhA (GmhANM) shows 98% identity to GmhAGC, and anti‐GmhAGC antisera readily recognized GmhANM (Figure 5a). Therefore, we decided to use an analogous strategy to create N. gonorrhoeae and N. meningitidis ∆gmhA strains expressing either endogenous or nonendogenous GmhA. Immunoblotting experiments demonstrated that under permissive conditions, both proteins were stably expressed in each host, while without IPTG, neither GmhAGC nor GmhANM were detected (Figure 5a). Functional interchangeability of GmhA between N. gonorrhoeae and N. meningitidis was then evaluated by analysis of LOS patterns. In FA1090, ∆gmhA GC/P lac ::gmhA NM expression of GmhANM resulted in LOS migrating exactly as LOS species extracted from the wild‐type gonococci (Figure 5a). Likewise, expression of GmhAGC restored LOS synthesis in the N. meningitidis MC58 ∆gmhA NM/P lac ::gmhA GC. Furthermore, as expected from our studies in N. gonorrhoeae (Figure 1), depletion of GmhANM in N. meningitidis MC58 ∆gmhA NM/P lac ::gmhA NM had adverse effects on bacterial viability and resulted in a 300‐ and 235.7‐fold decrease in CFUs in comparison with wild‐type and ∆gmhA NM/P lac ::gmhA NM cultured in the presence of IPTG, respectively (Figure 5b). There was no statistically significant difference in the number of CFUs between the wild‐type N. gonorrhoeae and its isogenic ∆gmhA GC mutant expressing either GmhAGC or GmhANM (Figure 5b). Similarly, both GmhANM and GmhAGC fully complemented the lack of GmhANM in N. meningitidis.
Figure 5.
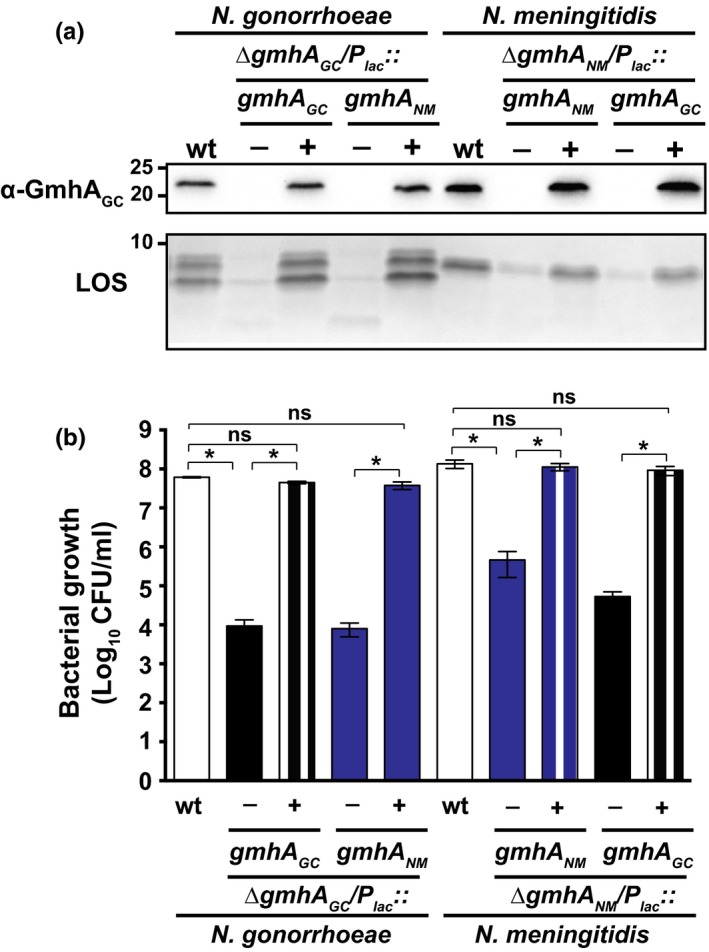
Homologs of GmhA from N. gonorrhoeae and N. meningitidis function interchangeably. (a) N. gonorrhoeae FA1090 wild‐type and isogenic conditional gmhA mutants bearing either endogenous (∆gmh AGC/P lac ::gmh AGC) or N. meningitidis‐derived gmh ANM (∆gmh AGC/P lac ::gmh ANM), as well as N. meningitidis MC58 wild‐type and conditional gmhA mutants carrying gmhA alleles (∆gmh ANM/P lac ::gmh ANM or ∆gmh ANM/P lac ::gmh AGC) were harvested from GCB after 22 hr of incubation either with (+) or without (−) isopropyl β‐D‐1‐thiogalactopyranoside (IPTG). Whole cell lysates were probed with anti‐GmhAGC antibodies or treated with proteinase K and lipooligosaccharide (LOS) was visualized by silver staining. Migration of a molecular mass marker (kDa) is indicated on the left. (b) Wild‐type and mutant strains of N. gonorrhoeae and N. meningitidis, as described above, were collected from GCB supplemented with 20 μmol/L IPTG and 15 μmol/L IPTG, respectively. Bacteria were suspended in gonococcal base liquid (GCBL) to OD 600 of 0.1, cultured for 3 hr, back‐diluted to equal OD 600 of 0.2, serially diluted, and plated on GCB with (+) and without (−) IPTG. CFUs were counted following 22 hr of incubation. Experiments were performed in three independent replicates. Mean values and SEM are presented; *p < .05
Together, the transcomplementation studies showed that the N. gonorrhoeae and N. meningitidis GmhA can function interchangeably. Expression of either of the homologs restored both viability and LOS synthesis. Additionally, we concluded that E. coli GmhAEC is not able to complement GmhAGC functions, as we were not able to generate a viable N. gonorrhoeae ∆gmhA GC/P lac ::gmhA EC mutant.
3.7. The structure of N. gonorrhoeae GmhA
To gain insights into the function of GmhAGC and to facilitate the future targeting of this enzyme with small molecule inhibitors, we obtained recombinant protein for structural studies (Fig. S1). The structure of N. gonorrhoeae GmhA was determined by molecular replacement and was refined to 2.37 Å resolution with R work 0.207, R free 0.267, and excellent stereochemical parameters (Table 2). Four monomers of GmhAGC were present in the asymmetric unit (Figure 6a). The tetrameric architecture is consistent with the results of size exclusion chromatography (Fig. S1A), and with the previously determined structures of GmhA homologs from other bacteria (Table 3). The interface area of the GmhAGC tetramer is extensive and buries 14,160 Å2 of surface area as calculated by the PISA server (Krissinel & Henrick, 2007). Four subunits of GmhAGC adopt highly similar structures with root mean square deviation (r.m.s.d.) 0.1–0.2 Å between subunits. Residues 69–74 (chains A and B) and 69–75 (chains C and D) are disordered in the structure. These residues form a loop in the vicinity of the active site and could become ordered upon substrate binding. A similar disorder was observed in the homologous region of E. coli GmhA (Taylor et al., 2008).
Table 2.
Data collection and refinement statistics
| N. gonorrhoeae GmhAGC (PDB 5I01) | |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.0000 |
| Space group | P21212 |
| Cell dimensions: | |
| a, b, c (Å) | 114.36, 130.15, 47.15 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 85.91–2.37 (2.43–2.37)a |
| R sym | 0.134 (1.032) |
| CC1/2 b | 99.6 (62.4) |
| I/σI | 9.3 (1.7) |
| Completeness (%) | 95.8 (97.8) |
| Multiplicity | 3.7 (3.6) |
| Refinement | |
| Resolution (Å) | 85.91–2.37 |
| No. reflections (total/free) | 28191/1405 |
| R work/R free | 0.207/0.267 |
| Number of atoms: | |
| Protein | 5411 |
| Ligand/ion | 4 |
| Water | 101 |
| B‐factors: | |
| Protein | 37.6 |
| Ligand/ion | 54.9 |
| Water | 32.7 |
| All atoms | 37.2 |
| Wilson B | 41.2 |
| R.m.s. deviations: | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.224 |
| Ramachandran distributionc (%): | |
| Favored | 97.8 |
| Outliers | 0 |
Values in parentheses are for the highest resolution shell.
CC1/2 correlation coefficient as defined in (Karplus & Diederichs, 2012) and calculated by XSCALE (Kabsch, 2010b).
Calculated using the MolProbity server (http://molprobity.biochem.duke.edu) (Chen et al., 2010).
Figure 6.
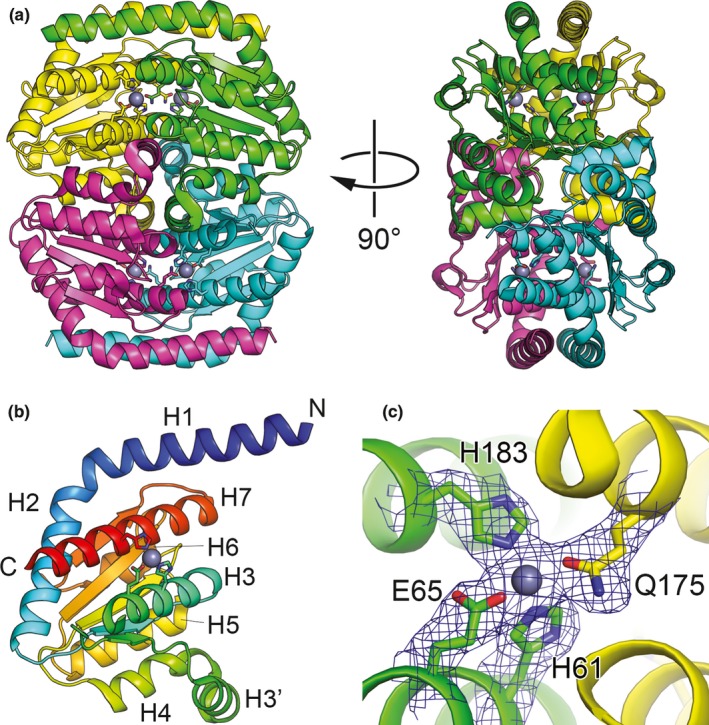
The structure of N. gonorrhoeae GmhAGC. (a) The ribbon representation of GmhAGC tetramer with monomers colored in green, yellow, cyan, and magenta. Zn2+ ions are shown as gray spheres. (b) The structure of GmhAGC monomer is colored in rainbow colors from blue (N‐terminus) to red (C‐terminus). The α‐helices are labeled according to Seetharaman et al., 2006. (c) A close‐up view of the Zn2+‐binding site of GmhAGC. The coordinating side chains are shown in stick representation. The σA‐weighted 2FO–FC electron density map countered at 1.0 σ is displayed as blue mesh
Each monomer of GmhAGC is composed of a central five‐stranded parallel β‐sheet flanked by four α‐helices from each side (Figure 6b). Four zinc ions are present in the GmhAGC tetramer. The zinc ions are coordinated by the side chains of residues H61, E65, and H183 of one monomer, and the side chain of Q175 of the neighboring monomer (Figure 6c). Therefore, the zinc‐binding sites of GmhAGC are identical to the zinc‐binding sites of B. pseudomallei GmhA (Harmer, 2010).
Overall, the structure of GmhAGC is similar to the previously determined structures of GmhA from other bacteria, as well as E. coli DiaA (Table 3). The monomer structure of GmhAGC could be superimposed to the homologous structures with r.m.s.d. of 0.5–1.4 Å. The tetramer structure of GmhAGC forms a “closed” conformation, similar to the GmhA structures of P. aeruginosa, V. cholerae, B. pseudomallei, and C. psychrerythraea (Do et al., 2015; Harmer, 2010; Seetharaman et al., 2006; Taylor et al., 2008).
4. Discussion
GmhA is a conserved sedoheptulose‐7‐phosphate isomerase involved in the first biosynthesis step of the L,D‐heptose component of the LPS/LOS. Deprivation of heptoses results in pleiotropic phenotypes including synthesis of LPS/LOS molecules composed only of lipid A and KDO residues, increased susceptibility to antimicrobial agents, defects in plasmid F conjugations and P1 bacteriophage transduction, and decreased virulence and biofilm formation (Bauer et al., 1998; Brooke & Valvano, 1996a; Earl et al., 2015; Havekes et al., 1976; Malott et al., 2013; Tamaki et al., 1971). In N. gonorrhoeae, heptoses are also crucial for LOS function in the evasion of the host immune system and induction of HIV expression (Malott et al., 2013; Preston et al., 1996). Variability in LOS molecules on the gonococcal surface is driven by phase variable enzymes responsible for decorating the sugar moiety of the outer core oligosaccharide (Apicella et al., 1987; Danaher et al., 1995). N. gonorrhoeae scavenges sialic acid from the human body and modifies the lacto‐N‐neotetraose attached to the HepI of the LOS core oligosaccharide, which provides protection from killing by classical and alternative complement pathways (Elkins et al., 1992; Ram et al., 1998). Thus, inhibition of GmhA could aid in treatment of infections caused by different pathogenic bacteria including N. gonorrhoeae.
Accordingly, in this work, we presented initial characterization of GmhAGC at the functional and structural levels. Our experiments demonstrated that gonococci and meningococci depleted in GmhA produced overall significantly less LOS molecules that migrated faster in comparison with the LOS‐derived from wild‐type bacteria (Figures 1b and 5a). In contrast to other orthologs, however, diminution of the GmhA cellular pool had severe consequences on neisserial growth (Figures 1a, d–e and 5b). This protein is likely not essential for bacterial viability, as GmhA‐depleted neisserial cells still arose on solid media under nonpermissive conditions (Figure 1a, d–e). Corroborating this observation, gmhA GC was not found among 827 gonococcal core essential genes identified using a high‐density Tn5 transposon library (Remmele et al., 2014). In addition, expression of GmhAGC variants carrying substitutions in the catalytic residues coordinating zinc ions, E65, and H183 (Figure 6c), led to synthesis of truncated LOS while retaining ample N. gonorrhoeae viability (Figure 4). These findings suggested that GmhAGC may be involved in additional physiological function(s) in Neisseria. Not surprisingly, a complete functional transcomplementation was achieved between GhmAGC and GmhANM (Figure 5a–b), whereas multiple attempts to generate a viable strain of N. gonorrhoeae expressing only GmhAEC were unsuccessful (data not shown). Based on these findings, we concluded that GmhA plays a critical role in LOS synthesis and is a fundamental growth factor for both N. gonorrhoeae and N. meningitidis.
Profiling of GmhAGC expression showed that amounts of GmhAGC increased during exponential growth of wild‐type N. gonorrhoeae (Figure 2c). Surprisingly, the isogenic conditional ∆gmhA GC/P lac ::gmhA GC mutant cultured in the presence of IPTG had elevated levels of GmhAGC during the late logarithmic phase (Figure 1c–e), suggesting that expression of GmhAGC is regulated at the posttranscriptional (e.g., small noncoding RNAs, RNA‐binding proteins, RNases, and thermos‐switches) or posttranslational levels. Higher amounts of GmhAGC were also noted upon exposure of N. gonorrhoeae to environmental cues relevant to infection; iron deprivation and anaerobiosis (Figure 2d). Transcriptomic studies, however, did not identify GmhAGC as an iron‐regulated gene (Ducey, Carson, Orvis, Stintzi, & Dyer, 2005; Jackson et al., 2010). The observed upregulation of GhmAGC under anaerobic conditions was in agreement with deep sequencing analysis of the N. gonorrhoeae anaerobic stimulon, where gmhA GC was found to be 4.2‐ and 6.1‐fold upregulated in two biological experiments (Isabella & Clark, 2011). Higher levels of enzymes acting downstream from GmhA including HldA (NGO0402) and GmhB (NGO2070) were identified in anaerobically versus aerobically cultured gonococci in our recent high‐throughput proteomic studies (Zielke et al., 2016), suggesting that the L,D‐heptose biosynthesis pathway is upregulated during anoxia. Accordingly, increased amounts of LOS were detected in anaerobically grown gonococci (Figure 2d), while vast overexpression of GhmAGC alone had no effect on LOS levels (Figure 2e). GmhAGC expression was also studied in a diverse collection of gonococcal isolates, including the 2016 WHO reference strains (Unemo et al., 2016) containing multidrug‐resistant N. gonorrhoeae (Figure 3). Potential involvement of GmhA in antibiotic resistance via increased synthesis of LPS was suggested recently in Salmonella typhimurium DT104B multiresistant strain with additional fluoroquinolone resistance (Correia et al., 2016). Notwithstanding this suggestion, the amounts of GmhAGC varied widely between the WHO strains K, L, V, W, X, Y, and Z, which display overall high levels of ciprofloxacin resistance (MIC>32 mg/L) and decreased susceptibility to gemifloxacin and moxifloxacin (Unemo et al., 2016). Additionally, strain L, which has high MICs for all quinolones (MICs in mg/L for ciprofloxacin, gemifloxacin, and moxifloxacin of >32, 8, and 16, respectively), showed significantly lower GmhAGC levels in comparison with WHO isolates F, O, P, and U, which are all ciprofloxacin sensitive (MIC of 0.004 mg/L) and are significantly more sensitive to gemifloxacin and moxifloxacin (MICs ranging from 0.004 to 0.016 mg/L).
Finally, to better understand the pivotal function of GmhAGC, we have determined the three‐dimensional structure of the untagged enzyme (Figure 6). Comparison of our GmhAGC crystal structure with the structures of homologs from other bacteria did not reveal significant differences in the general organization of the structure or within the catalytic site of the enzyme. Zinc ions were not added to any of the protein purification steps or crystallization buffers, yet similar to Burkholderia pseudomallei GmhA (Harmer, 2010), GmhAGC held zinc ions in the active site (Figure 6c), providing further support that GmhA is a metalloenzyme. Zinc is likely retained from the cell of the heterologous host during protein overproduction and natively bound to the active site of the enzyme. Zinc binding likely drives the “closed” conformation of GmhAGC, which was also observed in structures of B. pseudomallei, P. aeruginosa, and V. cholerae GmhA (Table 3), thus providing further support that the “closed” conformation is catalytically relevant. In addition, site‐directed mutagenesis coupled with in vivo studies confirmed the pivotal function of GmhAGC E65 and H183 in the isomerization of the sedoheptulose‐7‐phosphate (Figures 4 and 6).
In conclusion, understanding the function and structure of individual GmhA proteins will facilitate drug discovery approaches focused on targeting this protein with small molecule inhibitors. In particular, our work demonstrated the crucial function of GmhA in neisserial growth and LOS synthesis, positive regulation of expression by host‐relevant environmental stimuli, and conservation among different isolates, as well as provided further support for the mode of action of GmhA. These findings underscore the significance of GmhAGC as a target for antigonorrhea therapeutics. Future work involving determining the interacting partner(s) of GmhAGC and analysis of global changes at the proteome and metabolome levels are required to elucidate the whole scope of physiological function(s) of GmhAGC in N. gonorrhoeae.
Conflict of Interest
The authors have declared that no conflict of interest exist.
Supporting information
Acknowledgments
We thank staff members of Southeast Regional Collaborative Access Team (SER‐CAT) at the Advanced Photon Source, Argonne National Laboratory, for assistance during data collection. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE‐AC02‐06CH11357. We thank Benjamin I. Baarda for critical reading of the manuscript. We acknowledge the Protein Core at the Center for Molecular Medicine, University of Kentucky, that is supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant number P30GM110787. Funding for this work was provided to AES by grant R01‐AI117235 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and was partially supported by an Institutional Development Award (IDeA) from the NIGMS/NIH under grant number P20GM103486 to KVK.
Wierzbicki, I.H. , Zielke, R.A. , Korotkov, K.V. and Sikora, A.E. Functional and structural studies on the Neisseria gonorrhoeae GmhA, the first enzyme in the glycero‐manno‐heptose biosynthesis pathways, demonstrate a critical role in lipooligosaccharide synthesis and gonococcal viability. MicrobiologyOpen. 2017;6:e00432 https://doi.org/10.1002/mbo3.432
References
- Aballay, A. , Drenkard, E. , Hilbun, L. R. , & Ausubel, F. M. (2003). Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Current Biology, 13, 47–52. [DOI] [PubMed] [Google Scholar]
- Alexander, H. L. , Richardson, A. R. , & Stojiljkovic, I. (2004). Natural transformation and phase variation modulation in Neisseria meningitidis. Molecular Microbiology, 52, 771–783. [DOI] [PubMed] [Google Scholar]
- Apicella, M. A. , Breen, J. F. , & Gagliardi, N. C. (1978). Degradation of the polysaccharide component of gonococcal lipopolysaccharide by gonococcal and meningococcal sonic extracts. Infection and Immunity, 20, 228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicella, M. A. , & Mandrell, R. E. (1989). Molecular mimicry as a factor in the pathogenesis of human neisserial infections: In vitro and in vivo modification of the lipooligosaccharide of Neisseria gonorrhoeae by N‐acetylneuraminic acid. The Pediatric Infectious Disease Journal, 8, 901–902. [DOI] [PubMed] [Google Scholar]
- Apicella, M. A. , Shero, M. , Jarvis, G. A. , Griffiss, J. M. , Mandrell, R. E. , & Schneider, H. (1987). Phenotypic variation in epitope expression of the Neisseria gonorrhoeae lipooligosaccharide. Infection and Immunity, 55, 1755–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A. (1999). The SIS domain: A phosphosugar‐binding domain. Trends in Biochemical Sciences, 24, 94–95. [DOI] [PubMed] [Google Scholar]
- Bauer, B. A. , Stevens, M. K. , & Hansen, E. J. (1998). Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infection and Immunity, 66, 4290–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre, J. A. , Zielke, R. A. , Korotkov, K. V. , & Sikora, A. E. (2016). Targeting an Essential GTPase Obg for the Development of Broad‐Spectrum Antibiotics. PLoS ONE, 11, e0148222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke, J. S. , & Valvano, M. A. (1996a). Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. Journal of Biological Chemistry, 271, 3608–3614. [DOI] [PubMed] [Google Scholar]
- Brooke, J. S. , & Valvano, M. A. (1996b). Molecular cloning of the Haemophilus influenzae gmhA (lpcA) gene encoding a phosphoheptose isomerase required for lipooligosaccharide biosynthesis. Journal of Bacteriology, 178, 3339–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban, M. J. , & Cohen, S. N. (1980). Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. Journal of Molecular Biology, 138, 179–207. [DOI] [PubMed] [Google Scholar]
- Centers For Disease Control And Prevention . (2013a). Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, US Department of Health and Human Services [Online].
- Centers For Disease Control And Prevention . (2013b). Grand Rounds: the growing threat of multidrug‐resistant gonorrhea. MMWR Morb Mortal Wkly Rep [Online], 62. Retrieved from: http://www.ncbi.nlm.nih.gov/pubmed/23407126 [Accessed Feb 15].
- Chaudhury, S. , Abdulhameed, M. D. , Singh, N. , Tawa, G. J. , D'haeseleer, P. M. , Zemla, A. T. , … Wallqvist, A . (2013). Rapid countermeasure discovery against Francisella tularensis based on a metabolic network reconstruction. PLoS ONE, 8, e63369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, V. B. , Arendall, W. B., 3RD , Headd, J. J. , Keedy, D. A. , Immormino, R. M. , Kapral, G. J. , … Richardson, D. C . (2010). MolProbity: All‐atom structure validation for macromolecular crystallography. Acta Crystallographica. Section D, Biological Crystallography, 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell, T. D. , Black, W. J. , Kawula, T. H. , Barritt, D. S. , Dempsey, J. A. , Kverneland, K. JR. , … Cannon, J. G . (1988). Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Molecular Microbiology, 2, 227–236. [DOI] [PubMed] [Google Scholar]
- Cornelissen, C. N. (2008). Identification and characterization of gonococcal iron transport systems as potential vaccine antigens. Future Microbiology, 3, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia, S. , Hebraud, M. , Chafsey, I. , Chambon, C. , Viala, D. , Torres, C. , … Igrejas, G . (2016). Impacts of experimentally induced and clinically acquired quinolone resistance on the membrane and intracellular subproteomes of Salmonella Typhimurium DT104B. Journal of Proteomics, 145, 46–59. [DOI] [PubMed] [Google Scholar]
- Cowtan, K. (2006). The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallographica. Section D, Biological Crystallography, 62, 1002–1011. [DOI] [PubMed] [Google Scholar]
- Cowtan, K. (2010). Recent developments in classical density modification. Acta Crystallographica. Section D, Biological Crystallography, 66, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaher, R. J. , Levin, J. C. , Arking, D. , Burch, C. L. , Sandlin, R. , & Stein, D. C. (1995). Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. Journal of Bacteriology, 177, 7275–7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby, C. , Ananth, S. L. , Tan, L. , & Hinnebusch, B. J. (2005). Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infection and Immunity, 73, 7236–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, H. , Yun, J. S. , Lee, C. W. , Choi, Y. J. , Kim, H. Y. , Kim, Y. J. , … Lee, J. H. (2015). Crystal Structure and Comparative Sequence Analysis of GmhA from Colwellia psychrerythraea Strain 34H Provides Insight into Functional Similarity with DiaA. Molecules and Cells, 38, 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducey, T. F. , Carson, M. B. , Orvis, J. , Stintzi, A. P. , & Dyer, D. W. (2005). Identification of the iron‐responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. Journal of Bacteriology, 187, 4865–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl, S. C. , Rogers, M. T. , Keen, J. , Bland, D. M. , Houppert, A. S. , Miller, C. , … Marketon, M. M. (2015). Resistance to Innate Immunity Contributes to Colonization of the Insect Gut by Yersinia pestis. PLoS ONE, 10, e0133318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidels, L. , & Osborn, M. J. (1971). Lipopolysaccharide and aldoheptose biosynthesis in transketolase mutants of Salmonella typhimurium . Proceedings of the National Academy of Sciences U S A, 68, 1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidels, L. , & Osborn, M. J. (1974). Phosphoheptose isomerase, first enzyme in the biosynthesis of aldoheptose in Salmonella typhimurium. Journal of Biological Chemistry, 249, 5642–5648. [PubMed] [Google Scholar]
- Elkins, C. , Carbonetti, N. H. , Varela, V. A. , Stirewalt, D. , Klapper, D. G. , & Sparling, P. F. (1992). Antibodies to N‐terminal peptides of gonococcal porin are bactericidal when gonococcal lipopolysaccharide is not sialylated. Molecular Microbiology, 6, 2617–2628. [DOI] [PubMed] [Google Scholar]
- Emsley, P. , Lohkamp, B. , Scott, W. G. , & Cowtan, K. (2010). Features and development of Coot. Acta Crystallographica. Section D, Biological Crystallography, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin, L. E. , Bash, M. C. , Keys, C. , Warner, D. M. , Ram, S. , Shafer, W. M. , & Jerse, A. E. (2008). Phenotypic and genotypic analyses of Neisseria gonorrhoeae isolates that express frequently recovered PorB PIA variable region types suggest that certain P1a porin sequences confer a selective advantage for urogenital tract infection. Infection and Immunity, 76, 3700–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, B. W. , Webb, J. W. , Yamasaki, R. , Fisher, S. J. , Burlingame, A. L. , Mandrell, R. E. , … Griffiss, J. M. (1989). Structure and heterogeneity of the oligosaccharides from the lipopolysaccharides of a pyocin‐resistant Neisseria gonorrhoeae . Proceedings of the National Academy of Sciences U S A, 86, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinelli‐Pimpaneau, B. , Le Goffic, F. , & Badet, B . (1989). Glucosamine‐6‐phosphate from Escherichia coli: Mechanism of the reaction at the fructose‐6‐phosphate binding site. Journal of the American Chemical Society, 111, 3029–3034. [Google Scholar]
- Harmer, N. J. (2010). The structure of sedoheptulose‐7‐phosphate isomerase from Burkholderia pseudomallei reveals a zinc binding site at the heart of the active site. Journal of Molecular Biology, 400, 379–392. [DOI] [PubMed] [Google Scholar]
- Havekes, L. M. , Lugtenberg, B. J. , & Hoekstra, W. P. (1976). Conjugation deficient E. coli K12 F‐ mutants with heptose‐less lipopolysaccharide. Molecular and General Genetics, 146, 43–50. [DOI] [PubMed] [Google Scholar]
- Hitchcock, P. J. , & Brown, T. M. (1983). Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver‐stained polyacrylamide gels. Journal of Bacteriology, 154, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. , & Rosenstrom, P. (2010). Dali server: Conservation mapping in 3D. Nucleic Acids Research, 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella, V. M. , & Clark, V. L. (2011). Deep sequencing‐based analysis of the anaerobic stimulon in Neisseria gonorrhoeae. BMC Genomics, 12, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, L. A. , Ducey, T. F. , Day, M. W. , Zaitshik, J. B. , Orvis, J. , & Dyer, D. W. (2010). Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. Journal of Bacteriology, 192, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, C. M. , Griffiss, J. M. , Apicella, M. A. , Mandrell, R. E. , & Gibson, B. W. (1991). The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. Journal of Biological Chemistry, 266, 19303–19311. [PubMed] [Google Scholar]
- Kabsch, W. (2010a). Integration, scaling, space‐group assignment and post‐refinement. Acta Crystallographica. Section D, Biological Crystallography, 66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W . (2010b). Xds. Acta Crystallographica Section D: Biological Crystallography, 66, 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, P. A. , & Diederichs, K. (2012). Linking crystallographic model and data quality. Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyamura, K. , Fujikawa, N. , Ishida, T. , Ozaki, S. , Su'etsugu, M. , Fujimitsu, K. , … Katayama, T . (2007). The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA‐specific initiation complexes. Genes & Development, 21, 2083–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma, P. , Wugeditsch, T. , Christian, R. , Zayni, S. , & Messner, P. (1995). Glycan structure of a heptose‐containing S‐layer glycoprotein of Bacillus thermoaerophilus . Glycobiology, 5, 791–796. [DOI] [PubMed] [Google Scholar]
- Krissinel, E. , & Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology, 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Low, N. , Unemo, M. , Skov Jensen, J. , Breuer, J. , & Stephenson, J. M . (2014). Molecular diagnostics for gonorrhoea: Implications for antimicrobial resistance and the threat of untreatable gonorrhoea. PLoS Medicine, 11, e1001598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q. , Li, S. , & Shao, F. (2015). Sweet Talk: Protein Glycosylation in Bacterial Interaction With the Host. Trends in Microbiology, 23, 630–641. [DOI] [PubMed] [Google Scholar]
- Malott, R. J. , Keller, B. O. , Gaudet, R. G. , Mccaw, S. E. , Lai, C. C. , Dobson‐Belaire, W. N. , … Gray‐Owen, S. D . (2013). Neisseria gonorrhoeae‐derived heptose elicits an innate immune response and drives HIV‐1 expression. Proceedings of the National Academy of Sciences U S A, 110, 10234–10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A. J. , Grosse‐Kunstleve, R. W. , Adams, P. D. , Winn, M. D. , Storoni, L. C. , & Read, R. J. (2007). Phaser crystallographic software. Journal of Applied Crystallography, 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness, B. T. , Clarke, I. N. , Lambden, P. R. , Barlow, A. K. , Poolman, J. T. , Jones, D. M. , & Heckels, J. E. (1991). Point mutation in meningococcal por A gene associated with increased endemic disease. Lancet, 337, 514–517. [DOI] [PubMed] [Google Scholar]
- Mcleod Griffiss, J. , Brandt, B. L. , Saunders, N. B. , & Zollinger, W. (2000). Structural relationships and sialylation among meningococcal L1, L8, and L3,7 lipooligosaccharide serotypes. Journal of Biological Chemistry, 275, 9716–9724. [DOI] [PubMed] [Google Scholar]
- Mehr, I. J. , Long, C. D. , Serkin, C. D. , & Seifert, H. S. (2000). A homologue of the recombination‐dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics, 154, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, T. F. , Mlawer, N. , & So, M. (1982). Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell, 30, 45–52. [DOI] [PubMed] [Google Scholar]
- Murshudov, G. N. , Skubak, P. , Lebedev, A. A. , Pannu, N. S. , Steiner, R. A. , Nicholls, R. A. , … Vagin, A. A. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallographica. Section D, Biological Crystallography, 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, J. , Egan, D. , Walter, T. S. , Meged, R. , Berry, I. , Ben Jelloul, M. , … Perrakis, A. (2005). Towards rationalization of crystallization screening for small‐ to medium‐sized academic laboratories: The PACT/JCSG+ strategy. Acta Crystallographica. Section D, Biological Crystallography, 61, 1426–1431. [DOI] [PubMed] [Google Scholar]
- Papanastasiou, M. , Orfanoudaki, G. , Koukaki, M. , Kountourakis, N. , Sardis, M. F. , Aivaliotis, M. , … Economou, A. (2013). The Escherichia coli peripheral inner membrane proteome. Molecular & Cellular Proteomics: MCP, 12, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, A. , Mandrell, R. E. , Gibson, B. W. , & Apicella, M. A. (1996). The lipooligosaccharides of pathogenic gram‐negative bacteria. Critical Reviews in Microbiology, 22, 139–180. [DOI] [PubMed] [Google Scholar]
- Raetz, C. R. , & Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annual Review of Biochemistry, 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram, S. , McQuillen, D. P. , Gulati, S. , Elkins, C. , Pangburn, M. K. , & Rice, P. A. (1998). Binding of complement factor H to loop 5 of porin protein 1A: A molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae . Journal of Experimental Medicine, 188, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele, C. W. , Xian, Y. , Albrecht, M. , Faulstich, M. , Fraunholz, M. , Heinrichs, E. , … Rudel, T . (2014). Transcriptional landscape and essential genes of Neisseria gonorrhoeae , Nucleic Acids Res, 42, 10579–10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman, J. , Rajashankar, K. R. , Solorzano, V. , Kniewel, R. , Lima, C. D. , Bonanno, J. B. , … Swaminathan, S. (2006). Crystal structures of two putative phosphoheptose isomerases. Proteins, 63, 1092–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling, P. F. (1966). Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. Journal of Bacteriology, 92, 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J. M. , Wright, L. , & Clark, V. L. (2008). Laboratory maintenance of Neisseria gonorrhoeae . Current Protocols in Microbiology, Chapter 4, Unit 4A 1. [DOI] [PubMed] [Google Scholar]
- Studier, F. W. , & Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high‐level expression of cloned genes. Journal of Molecular Biology, 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Tamaki, S. , Sato, T. , & Matsuhashi, M. (1971). Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K‐12. Journal of Bacteriology, 105, 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, P. L. , Blakely, K. M. , de Leon, G. P. , Walker, J. R. , McArthur, F. , Evdokimova, E. , … Junop, M. S. (2008). Structure and function of sedoheptulose‐7‐phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. Journal of Biological Chemistry, 283, 2835–2845. [DOI] [PubMed] [Google Scholar]
- Tsai, C. M. , & Frasch, C. E. (1982). A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Analytical Biochemistry, 119, 115–119. [DOI] [PubMed] [Google Scholar]
- Unemo, M. (2015). Current and future antimicrobial treatment of gonorrhoea ‐ the rapidly evolving Neisseria gonorrhoeae continues to challenge. BMC Infectious Diseases, 15, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M. , Golparian, D. , Sanchez‐Buso, L. , Grad, Y. , Jacobsson, S. , Ohnishi, M. , … Harris, S. R. (2016). The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: Phenotypic, genetic and reference genome characterization. Journal of Antimicrobial Chemotherapy, 71, 3096–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo, M. , & Shafer, W. M. (2014). Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: Past, evolution, and future. Clinical Microbiology Reviews, 27, 587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvano, M. A. , Messner, P. , & Kosma, P. (2002). Novel pathways for biosynthesis of nucleotide‐activated glycero‐manno‐heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology, 148, 1979–1989. [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2011). Prevalence and incidence of selected sexually transmitted infections, Chlamydia trachomatis, Neisseria gonorrhoeae, syphilis and Trichomonas vaginalis. Methods and results used by WHO to generate 2005 estimates., 36.
- World Health Organization . (2012). Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. Retrieved from: http://whqlibdoc.who.int/publications/2012/9789241503501_eng.pdf.
- World Health Organization . (2015). Antimicrobial resistance draft global action plan on antimicrobial resistance.
- Wugeditsch, T. , Zachara, N. E. , Puchberger, M. , Kosma, P. , Gooley, A. A. , & Messner, P. (1999). Structural heterogeneity in the core oligosaccharide of the S‐layer glycoprotein from Aneurinibacillus thermoaerophilus DSM 10155. Glycobiology, 9, 787–795. [DOI] [PubMed] [Google Scholar]
- Yamasaki, R. , Bacon, B. E. , Nasholds, W. , Schneider, H. , & Griffiss, J. M. (1991). Structural determination of oligosaccharides derived from lipooligosaccharide of Neisseria gonorrhoeae F62 by chemical, enzymatic, and two‐dimensional NMR methods. Biochemistry, 30, 10566–10575. [DOI] [PubMed] [Google Scholar]
- Yamasaki, R. , Kerwood, D. E. , Schneider, H. , Quinn, K. P. , Griffiss, J. M. , & Mandrell, R. E. (1994). The structure of lipooligosaccharide produced by Neisseria gonorrhoeae, strain 15253, isolated from a patient with disseminated infection. Evidence for a new glycosylation pathway of the gonococcal lipooligosaccharide. Journal of Biological Chemistry, 269, 30345–30351. [PubMed] [Google Scholar]
- Zielke, R. A. , wierzbicki, I. H. , Baarda, B. I. , Gafken, P. R. , Soge, O. O. , Holmes, K. K. , … Sikora, A. E. (2016). Proteomics‐driven antigen discovery for development of vaccines against gonorrhea. Molecular & Cellular Proteomics: MCP, 5, 2338–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke, R. A. , Wierzbicki, I. H. , Baarda, B. I. , & Sikora, A. E. (2015). The Neisseria gonorrhoeae Obg protein is an essential ribosome‐associated GTPase and a potential drug target. BMC Microbiology, 15, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke, R. A. , Wierzbicki, I. H. , Weber, J. V. , Gafken, P. R. , & Sikora, A. E. (2014). Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Molecular & Cellular Proteomics: MCP, 13, 1299–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


