Abstract
The eye lens crystallins represent an ideal target for studying the effects of aging on protein structure. Herein we examine separately the water-soluble (WS) and water-insoluble (WI) crystallin fractions and identify sites of isomerization and epimerization. Both collision-induced dissociation and radical-directed dissociation are needed for detection of these non-mass-shifting post-translational modifications. Isomerization levels differ significantly between the WS and the WI fractions from sheep, pig, and cow eye lenses. Residues that are most susceptible to isomerization are identified site-specifically and are found to reside in structurally disordered regions. However, isomerization in structured domains, although less common, often yields more dramatic effects on solubility. Numerous isomerization hotspots were also identified and occur in regions with aspartic acid and serine repeats. For example, 128KMEIVDDDVPSLW140 in βB3 crystallin contains three sequential aspartic acid residues and is isomerized heavily in the WI fractions, while it is not modified at all in the WS fractions. Potential causes for enhanced isomerization at sites with acidic residue repeats are presented. The importance of acidic residue repeats extends beyond the lens, as they are found in many other long-lived proteins associated with disease.
Keywords: photodissociation, long-lived proteins, isomerization, epimerization
Introduction
In most cells, proteostasis involves continuous regulation of protein synthesis and degradation, but the eye lens is a notable exception.1 During the final stages of maturation, lens cells eject all organelles, preventing normal protein turnover.2 The lens therefore provides a unique opportunity to observe the effects of aging in relation to damage accumulation in long-lived proteins, which are increasingly a subject of interest.3 The α-crystallins are important multimeric chaperones that help prevent protein aggregation. They constitute approximately 35–40% of the total soluble mass of the lens and are composed of a 3:1 ratio of αA to αB.4 While αA is found almost exclusively in the lens, αB has been found throughout the body including heart, glia, muscle, kidney, lung, and skin cells.5 Gene knockout studies revealed that a mouse lens can develop normally without αB but not without αA, which led to prompt cataract formation.6,7 Nevertheless, the chaperone activity of heteropolymers formed from a 3:1 ratio of αA to αB is higher than either homopolymer, suggesting that the combination of both proteins is optimal.8 Beta and gamma crystallins are the two other major subgroups of crystallins in the lens. They act in conjunction with the α-crystallins to maintain lens structure and transparency and to achieve a suitable refractive index.9,10
To behave properly as chaperones, α-crystallin monomers must interact with each other to form polydisperse oligomers. αA and αB form dimers that then assemble into larger complexes ranging from 15 to 50 monomers, with the subunits dynamically intermixing and exchanging.11 Consequently, obtaining crystal structures of the α-crystallins has been daunting. Removing the N- and C-terminal extensions allows for determination of partial crystal structures.12,13 When crystallins do not interact with each other properly, they become water-insoluble and begin to aggregate. The influence of many post-translational modifications (PTMs) on this process has been investigated, including deamidation, oxidation, disulfide formation, and truncation.14,15 Importantly, all of these PTMs lead to mass shifts that are easily detectable by mass spectrometry (MS).
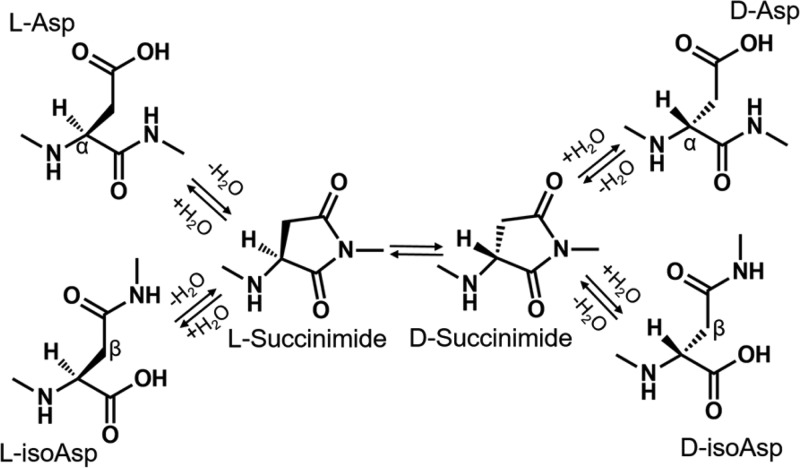
More subtle modifications that also increase with age, such as isomerization and epimerization, do not lead to easily detectable mass shifts and have been significantly less studied.16,17 Methods for detecting isomerization in peptides (meaning either inversion of the chiral center of an amino acid to produce an epimer or conversion of aspartic acid to isoaspartic acid) include stereoselective enzymatic digestion,18 ion mobility,19 and MS.20,21 Previous studies have shown that aspartic acid and serine are the two amino acids most susceptible to spontaneous isomerization.22 Deamidation of asparagine residues generates aspartic acid, and this transformation can also be accompanied by isomerization. The mechanism by which l-Asp converts into l-isoAsp, d-isoAsp, and d-Asp has been studied previously and is known to proceed via nonenzymatic formation of a succinimide ring, which can yield four isomers, as shown in Scheme 1. l-Succinimide can be converted to d-succinimide via keto–enol tautomerism and then hydrolyzed to form d-Asp or d-isoAsp.23 The mechanism by which l-Ser epimerizes to d-Ser in proteins is not well-established, although proposals have been made.24,25
Scheme 1. Mechanism of Spontaneous Aspartic Acid Isomerization via a Succinimide Intermediate.
Alpha and beta carbons are labeled to highlight the difference between l-asp and l-isoasp, respectively.
Although isomerization might appear to be a “benign” PTM, studies have shown it can cause major perturbations in protein structure. Noguchi and coworkers successfully crystallized hen egg-white lysozyme with an isoaspartic acid substitution at Asp101 that caused backbone deflection of nearly 90° relative to the native structure.26 Crystal structures of a modified ribonuclease revealed that isoAsp-32 induces conversion of an α-helix to a U-shaped loop.27 Isomerization not only perturbs 3D structure but also can affect physical properties such as solubility and bioactivity. For example, isomerization of Asp92 in immunoglobulin γ2 (IgG2) leads to deactivation of the antigen-binding region.28
Large-scale (i.e., proteomic) identification of single amino acid isomerism within peptides is challenging because there is no change in mass. However, differences in fragment intensities following MS/MS analysis can be used to detect isomers. This method was first applied to stereoisomers by Tao and coworkers, who reported differences as a ratio of relative abundances of a pair of fragment ions differing most between l- and d- enantiomers (Rchiral = RD/RL).29 It has been shown that radical-directed dissociation30 (RDD) yields the best chiral discrimination for analysis of peptide epimers.31 Previous work on the detection of isomerization and epimerization in the sheep lens using tandem LC–MS revealed novel sites of isomerization in the crystallins.32
The present study focuses on changes in isomerization and epimerization for αA, αB, and βB3 crystallin between water-soluble (WS) and water-insoluble (WI) protein fractions from cow, sheep, and pig eye lenses. To determine which regions of these proteins are most susceptible to isomerization, enzymatic digestion into peptides was followed by LC–MS/MS analysis using both collision-induced dissociation (CID) and RDD. Importantly, we found several regions in the well-ordered crystallin domain that are disproportionately isomerized in the WI fractions. Specific isomerization “hotspots” were identified and correspond to regions with serine or aspartic acid repeats. Potential explanations for the isomerization of these sites are offered, and comparison with other long-lived and problematic proteins reveals that these sequence motifs are common.
Experimental Methods
Protein Extraction and Digestion
Cow, pig, and sheep eye lenses were acquired as discarded tissue from Corona Cattle, Inc. (Corona, CA). The approximate ages for each of the animals were 12–18 months for the cow, 5–6 months for the pig, and 6–8 months for the sheep. The lenses were separated and washed with distilled H2O. Whole lenses were then homogenized in 50 mM Tris-HCl pH 7.8. The supernatant was separated from the precipitate following centrifugation at 15,100g for 20 min at 4 °C. The supernatant (WS) was purified by dialysis against water. The precipitate (WI) was solubilized in 6 M urea and purified by dialysis against 6 M urea. For WS digestion, the protein was dissolved in 50 mM NH4HCO3 buffer, pH 7.8; disulfide bonds were then reduced with 100 mM dithiothreitol (DTT) at 95 °C for 5 min. After returning to room temperature, reduced cysteine residues were capped using 100 mM iodoacetamide in the dark for 20 min. Finally, the proteins were digested with trypsin overnight at 37 °C using a 50:1 protein to enzyme ratio. For the WI digestion, the proteins were dissolved using 6 M 50 mM Tris-HCl, pH 8.0. Disulfide bonds were cleaved using 200 mM DTT in Tris-HCl, pH 8.0 at 37 °C for 20 min. Following this, 200 mM iodoacetamide in Tris-HCl, pH 8.0 was added and the mixture was incubated in the dark for 1 h. To consume unreacted iodoacetamide, 200 mM DTT was added and incubated for 1 h in the dark. Next, the urea concentration was diluted to <0.6 M using 50 mM Tris-HCl, 1 mM CaCl2, pH 7.6. The proteins were digested using trypsin with a 50:1 protein to enzyme ratio for 16 h at 37 °C. For the iodobenzoic acid modification, the digested peptides were desalted and cleaned using a peptide trap (Michrom Bioresource). Approximately 5 nmoles of the digestion mixture, 15 μL of 15 mM 4-iodobenzoic acid NHS-activated ester in dioxane, and 5 μL of borate buffer (pH 8.6) were combined and incubated for 1 h at 37 °C. Important: Dimethyl sulfoxide should not be substituted for dioxane in this step because it can cause aspartic acid isomerization. The modification side products at arginine and tyrosine side chains were removed by incubating the reaction mixture in 1 M hydroxylamine, pH 8.5. The same procedure was used for the synthetic peptide standards. These procedures have been previously determined not to yield any detectable isomerization/epimerization in control experiments.32
Calculation of R Values
To quantify isomer identification, an Rchiral value approach, originally reported by Tao et al., was utilized.29 In this paper, Risomer represents the ratios of the relative intensities of a pair of fragments that varies the most between two isomers (RA/RB). If Risomer = 1, then there the two tandem MS spectra are indistinguishable and the species are likely not isomers. If Risomer > 1, then a larger number indicates a higher probability that two unique molecules are represented. To confidently identify each of these isomers by MS/MS, we use a threshold that was determined by performing a t test on the Risomer values obtained by performing CID and RDD on a mixture of synthetic peptides separated by LC–MS. Using 99% confidence intervals, the Risomer threshold for CID is >1.9 and for RDD it is >2.4.32
Additional detailed methods are available in the Supporting Information (SI) Methods.
Results and Discussion
General Observations
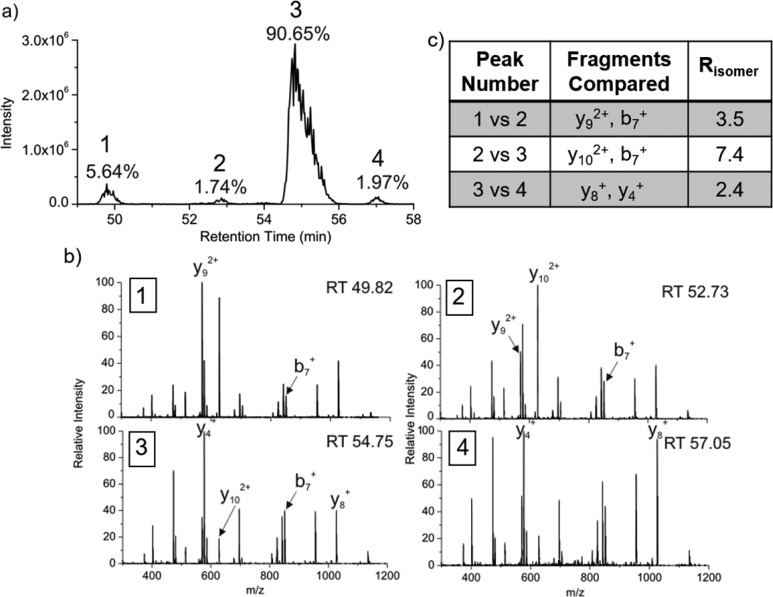
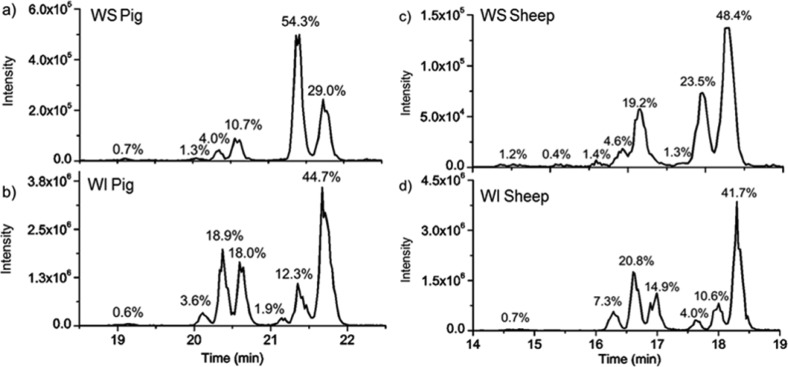
We have previously described a modified bottom-up strategy for detecting sites of isomerization, including epimerization, in proteins.32 In brief, proteolytically digested peptides are separated by reverse-phase column chromatography and analyzed by MS with minimal time exclusion windows to favor repeated analysis of the same m/z. For example, the N-terminal fragment of αA crystallin from the WI protein fraction of sheep lens, acetyl-MDIAIQHPWFK, elutes at four different times, each separated by ∼2 min, as shown in Figure 1a. To establish that these peaks represent isomers, repeated MS/MS analysis of each eluting peak is required (both CID and RDD are used in separate runs).32Figure 1b shows CID spectra from each of the LC peaks in Figure 1a. Although the spectra are similar, suggesting that they originate from the same peptide sequence, the relative intensities of certain peaks change noticeably. For example, the intensities of y92+, b7+, and y102+ ions vary considerably between LC peaks 1 and 2. These differences can be quantified into Risomer values,32 which are provided in Figure 1c. For CID analysis, Risomer values above 1.9 represent statistically significant differences, meaning that all four peaks in Figure 1a are different isomers of Acetyl-MDIAIQHPWFK. Mass spectra alone do not provide information about which isomer corresponds to the native l-Asp peptide, but in this case, the largest peak constitutes >90% of the total relative abundance and likely represents the unmodified l-Asp isomer. If the WI fraction is highly isomerized, in which case identification of the l-Asp isomer becomes ambiguous, then comparison with the less isomerized WS digest or an authentic standard is used to identify the l-Asp isomer.
Figure 1.
(a) LC chromatogram for the separation of the four isomers of Ac-MDIAIQHPWFK in the WI sheep lens digest. The percentages represent the calculated peak area. (b) CID spectra from each of the LC peaks. Labeled fragments are those used to determine Risomer values. (c) Table of the Risomer values including which fragments were compared.
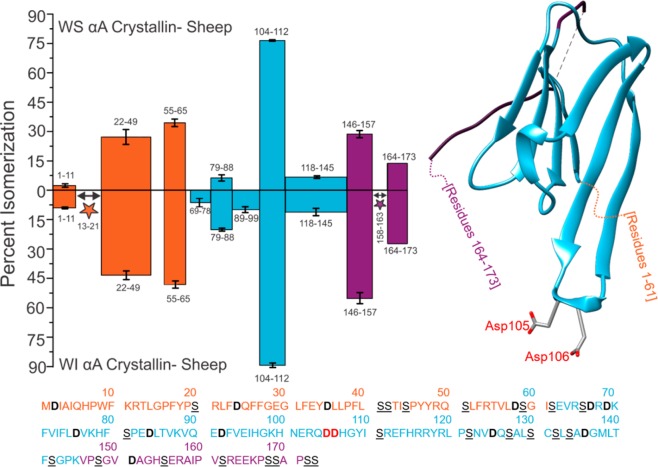
The degree of peptide isomerization from the WS and WI protein fractions of sheep αA crystallin is summarized in Figure 2. All isomers were confirmed by comparison of either CID or RDD MS/MS spectra, as described above. For example, the two isomers of 158AIPVSR163 were indistinguishable by CID but could be confidently detected using RDD (see SI Figure S1). Additionally, some isomers coelute, preventing quantitation of the relative abundance of each form. Isomerization of these peptides is denoted with stars. The first downward bar on the left side of Figure 2a (1–11) represents the data from Figure 1. This peptide, acetyl-MDIAIQHPWK, was found to be 9% isomerized, with error bars representing standard deviations from three technical replicates. The color coding of the bars corresponds to the three distinct structural regions present in α-crystallins, the N-terminal disordered region, the crystallin-ordered region, and the disordered C-terminal extension.33 These regions are illustrated relative to a partial crystal structure on the right side of Figure 2.13 The full protein sequence is provided at the bottom of Figure 2, color coded and with residues of interest marked for easy location. The trends in Figure 2 illustrate that the average amount of isomerization is significantly higher in the N and C termini than in the well-ordered Crystallin domain for both the WS and WI fractions. These dynamic regions play important roles in the assembly of α-crystallins into higher-order structures,34 but this flexibility may also enable isomerization by facilitating more frequent access to the pathways outlined in Scheme 1.35
Figure 2.
Percent isomerization of water-soluble (WS) αA Sheep versus water-insoluble (WI) αA Sheep. Orange, disordered N-terminus; blue, structured α-crystallin domain; purple, disordered C-terminus. Three separate digests were performed; error bars represent standard deviations. Number ranges represent peptide sequences. Peptide 164–173 does not contain error bars because it only appeared baseline-resolved in one digest. The full protein sequence is given below the plot, with aspartic acid residues in bolded/black and serine residues in underlined/black. Asp105 and Asp106 are in bold red text in the amino acid sequence and are shown explicitly in the crystal structure (PDB 3L1F) to highlight an important region of isomerization. Stars indicate isomerized regions where isomerization was identified, but quantitation was not possible due to incomplete chromatographic separation.
Interestingly, the degree of isomerization in the disordered regions, although slightly more abundant in the WI fraction, is similar in both the WS and WI fractions. This suggests that these modifications do not significantly drive aggregation and loss of solubility, as previously reported.36 Differences in degree of isomerization are more notable within the crystallin region, where isomerization is significantly more abundant in the WI fraction (except for the dramatically modified peptide 104–112 that will be discussed further below). Cumulatively, these results imply that modifications to flexible regions may be more facile but also less consequential in terms of altered functionality.
Results from an identical analysis of αB are shown in Figure 3. Fewer peptides are isomerized in αB, and the average degree of isomerization is less in both the WS and WI fractions compared with αA. The flexible N-terminal domain is highly isomerized, similar to αA. Interestingly, the C-terminal extension is not isomerized, which contrasts with αA and is largely due to the fact that the C-terminal extension of αB lacks any aspartic acid residues and contains only a single serine. The degree of isomerization observed in the WS versus WI fractions also varies more dramatically compared with αA, with greater isomerization being observed in the WI fraction for all peptides. αA and αB share similar functionality and freely intermix to form higher order structures, yet comparison of Figures 2 and 3 illustrates significant differences in propensity and effects of isomerization. The sequence alignment of the two proteins is <60% (Figure S2), but the tertiary structures of αA and αB are very similar, as illustrated by the crystal structure of truncated bovine αB on the right side of Figure 3.13 Despite the similarity, sequence variation appears to have a significant effect on the ability of αB to accommodate isomerization and retain solubility, in agreement with previous observations.37 Another interesting difference between the two α-crystallins is the overall abundance of acidic amino acids. Aspartic acid in αA comprises 8.8% of residues and glutamic acid contributes another 5.8%, whereas αB contains 6.3% aspartic acid and 8.0% glutamic acid. Although isomerization of glutamic acid is possible, the formation of the glutarimide intermediate is much slower compared with the succinimide equivalent in aspartic acid.38 Therefore, even though both proteins contain a similar percentage of acidic residues, αA contains more residues prone to isomerization, and these residues often reside in regions where isomerization is facile.
Figure 3.
Isomerization of αB from sheep, which differs significantly from what was observed for αA (see Figure 2a, formatting is identical). Asp109 in the amino acid sequence is in red bold text and shown explicitly in the crystal structure (PDB 3L1G) to highlight an important site of isomerization.
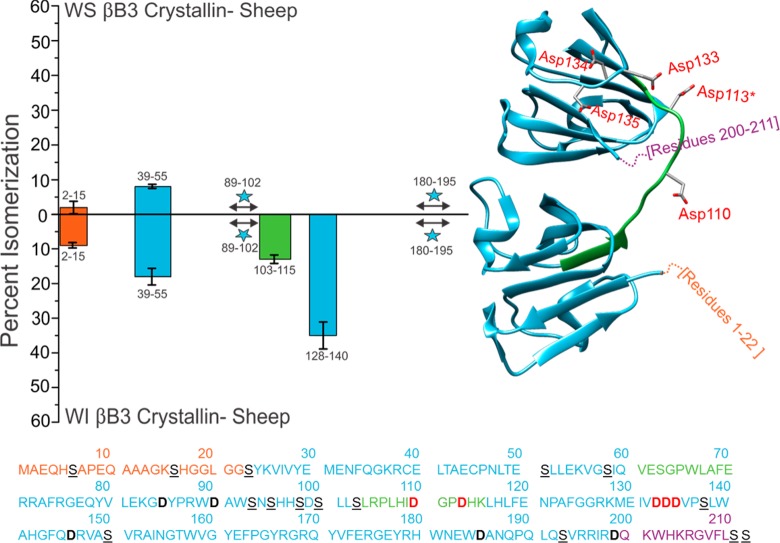
The same analysis that was performed on the α crystallins was also conducted on one of the β crystallins. Results for βB3 crystallin are shown in Figure 4. βB3 is composed of 211 amino acids, with two large structured domains connected by a flexible linker. The disordered N-terminal and C-terminal regions are much smaller than those observed in the α-crystallins. The total amount of isomerization detected in the WS and WI fractions of βB3 is lower than that in αA and αB, which is likely due to the greater fraction of highly structured regions. Similar to αB, βB3 is characterized by significant disparity between the degree of isomerization in the WS versus WI fractions, with greater isomerization being observed in the latter. This suggests again that structural perturbations in βB3 are consequential and lead to rapid loss of solubility. The isomerized versions of several peptides, including 103SLRPLHIDGPDHK115 and 128KMEIVDDDVPSLW140, are only detectable in the WI fractions (see Figure S3). 103SLRPLHIDGPDHK115 is part of the structurally critical connecting peptide that joins the two domains of the monomer, denoted by the green bar in Figure 4. 128KMEIVDDDVPSLW140 contains aspartic acid residues that form ion pairs during assembly into higher order oligomers.39 Sequence alignment for human, pig, cow, and sheep βB3 shows that each contains this aspartic acid repeat motif (Figure S4). The results in Figure 4 suggest that isomerization of either of these peptides leads to dramatic loss of solubility and, by extension, function.
Figure 4.
Percent isomerization of WS versus WI βB3 Sheep. Formatting is identical to Figure 2, except for the linker of the structured domains, shown in green. Asp110, Asp113, Asp133, Asp134, and Asp135 are all in red bold text in the amino acid sequence and are shown explicitly in the human crystal structure (PDB 3QK3) to highlight important regions of isomerization. His113 from human βB3 was mutated to Asp113*.
The same analysis was carried out on pig and cow lenses, and the results are summarized with the sheep results in Figure 5. The percent isomerization per protein was determined by taking the sum of the percent of isomerization per peptide and dividing it by the total number of peptides, including a value of zero for those that were not isomerized. Peptides where the degree of isomerization could not be determined were omitted for all samples of the same protein. For example, tryptic digestion of 173 residue αA yielded 12 peptides that cover 92% of the sequence. Contributions from these same peptides were used to calculate the numbers for each of the WS and WI αA digests for each animal. The data were then normalized to the αA digest that was most isomerized, which was the WI sheep digest. The same approach was applied to αB and βB3. Although there is some variation between species, in general, similar results were obtained for all three organisms. The degree of isomerization is, on average, significantly greater in the WI fractions for all species. Specific data from the WI sheep fraction are shown in Table S1.
Figure 5.
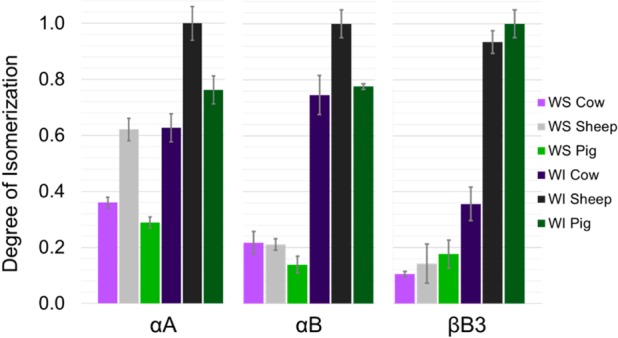
Total normalized degree of isomerization among the three major crystallin proteins detected.
Multiple Aspartic Acid Effect
Several of the peptides in Figures 2–4 appear to exhibit an unusual degree of isomerization relative to their peers. For example, 104QDDHGYISR112 from αA is not only is the most isomerized site in the protein but also is significantly more isomerized than any other peptide in the structured α-crystallin domain. Quantitatively, the average amount of isomerization per peptide in the structured domain of αA is 17.9% for the WS digest and 27.3% in the WI digest. At 76.7 and 89.3% isomerization in the WS and WI fractions, 104QDDHGYISR112 is isomerized ∼4× the average rate in the ordered crystallin region. The analogous sequence in αB, 108QDEHGFISR116, is also the most isomerized peptide extracted from the WI fraction, although full quantitative comparisons cannot be made due to lack of separation of some isomers for other peptides in the ordered region (see Figure S5 for details). Finally, 128KMEIVDDDVPSLW140 is the most isomerized peptide in the WI fraction from βB3. Each of these peptides shares a common feature, sequential repeats of acidic residues. In two cases, multiple sequential aspartic acid residues are present. These observations suggest that sequential acidic residues represent sites of greater propensity for isomerization.
There are at least two possible explanations that could account for increased isomerization at acid residue repeats. First, if there are multiple aspartic acid residues, then isomerization at multiple sites is possible. Structural perturbation engendered by the first modification could easily lead to increased local backbone flexibility, facilitating isomerization at additional sites. Indeed, examination of the elution profiles for these peptides reveals an abundance of isomers, indicating modification at more than one residue. For example, the elution profiles for the WS and WI fractions from pig for 104pQDDHGYSIR112 reveal the presence of seven isomers; see Figure 6. The lowercase “p” at the N-terminus of the sequence denotes pyroglutamate, which is a common product during trypsin digestion. MS2 experiments confirm that each peak in Figure 6 represents a different isomer. Comparison with synthetically prepared all l-Asp isomer was used to confirm the peak corresponding to the canonical peptide. Risomer values (4.8, 5.5, 3.7, 4.6, 4.5, 1.2, and 3.4) relative to the synthetic version confirm that the sixth peak in the digest is the all l-Asp isomer (see Figure S6 for details). The WS/WI degree of isomerization for 104pQDDHGYSIR112 from sheep is shown in Figure 6c,d. A similar elution profile is seen between WI pig and sheep; however, the amount of l-Asp in the WS pig lens is nearly twice as great as it is in WS sheep.
Figure 6.
(a) WS chromatogram and (b) WI chromatogram for 104pQDDHGYSIR112 from pig. (c) WS Sheep. (d) WI Sheep. Lower case “p” indicates the pyroglutamate that forms during tryptic digestion of N-terminal glutamine residues. Isomers were confirmed by MS/MS analysis and are labeled with percent abundance.
Another factor contributing to isomerization at sites with multiple acidic residues, which may be more important than increased backbone flexibility, is inhibited repair by protein isoaspartyl methyltransferase (PIMT). PIMT is the only known repair enzyme that targets protein damage caused by aging. It is present and active in bovine lenses and reverts l-isoAsp back to l-Asp.40,41d-Asp is also repaired, although much less efficiently, but d-isoAsp is not a substrate.42 Importantly, sequence effects for substrate recognition have been identified and revealed that PIMT has substantially lower affinity for isoAsp if the n+1, n+2, or n+3 residues are negatively charged or if the n+1 site is proline.43 Therefore, the sequence regions with multiple acidic residues that we have observed to be highly modified are unlikely to be repaired by PIMT, and our results confirm the importance of this enzyme in the repair of long-lived proteins.
Acidic residue repeat sites may therefore be prone to sequential isomerization and difficult to repair, a potent combination with important consequences for protein aging. Indeed, examination of other long-lived proteins associated with age-related diseases reveals the presence of numerous acidic acid repeat sites that would similarly be susceptible to isomerization. For example, amyloid precursor protein associated with Alzheimer’s disease contains 27 acidic residue repeat sites that would be poor substrates for PIMT. Table 1 lists long-lived proteins that contain aspartic acid repeats, are aggregation-prone, and are associated with diseases. It is clear that the results obtained here may be relevant on a much broader scale for any system composed of long-lived proteins.
Table 1. Aggregation-Prone Proteins Associated with Various Diseases.
| protein | tau-protein P10636-1 | amyloid precursor protein (APP) P05067-1 | amyloid-beta (1–42) | alpha-synuclein P37840-1 | TAR TDP-43 Q13148-1 | Huntingtin P42858-1 | alpha-A crystallin P02489 | alpha-B crystallin P02511 | beta-B3 crystallin P26998-1 |
|---|---|---|---|---|---|---|---|---|---|
| disease | PDa | ADb | AD | PD/AD | ALSc | HDd | Cate | Cat | Cat |
| length | 758 | 770 | 42 | 140 | 414 | 3142 | 173 | 175 | 211 |
| % Asp | 5.8 | 6.8 | 7.1 | 4.3 | 5.3 | 4.4 | 8.8 | 6.3 | 6.2 |
| “DD” or “DDD” | “DD” × 1 | “DD” × 4 and “DDD” × 1 | none | none | “DD” × 2 | “DD” × 5 and “DDD” × 1 | “DD” × 2 | none | “DDD” × 1 |
| acidic repeatsf | 16 | 27 | 1 | 1 | 9 | 44 | 4 | 3 | 2 |
| % Ser | 10.6 | 4.5 | 4.8 | 2.9 | 9.9 | 9.7 | 10.4 | 9.7 | 8.1 |
Parkinson’s disease.
Alzheimer’s disease.
Amyotrophic lateral sclerosis.
Huntington’s disease.
Cataracts.
Number of Asp residues with an acidic residue in the n+1, n+2, or n+3 position.
We have also identified isomerized regions with multiple serine residues, such as 164EEKPSSAPSS173. In the WS and WI fraction of the sheep lens this peptide elutes in six different peaks (Figure S7), and comparison of resulting CID spectra confirms that they represent different isomers. It is interesting to note that the total amount of isomerization in the WI sheep fraction for 164EEKPSSAPSS173 is much lower than what was observed for 104QDDHGYISR112, 27.3% compared with 90.4%, respectively. It is possible that isomerization of aspartic acid may have a larger influence on local structure than epimerization of serine, inducing isomerization of nearby residues more efficiently. Alternatively, there is no known repair enzyme for epimerization of serine, which may suggest that the failure of PIMT to repair isomerization may be the most important factor influencing isomerization at acidic residue repeats. However, the number of peptides with unambiguous epimerization at serine, that is, peptides that are isomerized and do not contain aspartic acid, is relatively small. Therefore, more data will need to be acquired before strong conclusions about serine epimerization can be drawn.
Conclusions
Peptide epimerization and isomerization are difficult to detect and remain among the least studied PTMs despite their potentially important role in numerous diseases. We have evaluated these modifications in crystallin proteins from the eye lenses of several organisms, and our results suggest that isomerization and epimerization lead to reduced protein solubility in many cases. Although there is variation between species, several common themes emerge from analysis of the data. For example, isomerization occurs more readily in regions of proteins that are disordered. This finding suggests that many other long-lived proteins with known disordered regions, such as α-synuclein, tau, and β-amyloid, may also exhibit pathology related to isomerization or epimerization. Although less prone to modification, isomerization within well-structured regions of proteins leads to more drastic changes in behavior, including unchecked aggregation. It was previously known that serine and aspartic acid are the most easily isomerized among the natural amino acids, but we have further demonstrated that when multiple acidic residues are close in sequence, the propensity for modification is enhanced further. We postulate that increased flexibility following an initial modification and failure of PIMT to repair damaged residues contribute to the dramatic isomerization of sites with multiple acidic residues. The degree of isomerization at these sites also confirms the importance of PIMT for long-term maintenance of protein structure. Proteins associated with Parkinson’s disease, Alzheimer’s disease, amyotrophic lateral sclerosis, and Huntington’s disease all contain regions with multiple aspartic acid or serine residues in close proximity, suggesting that these proteins may also exhibit hotspots of isomerization.
Both αA and αB exhibit similar structural and functional behavior, yet αA is found only in the eye lens. The isomerization behavior of αA and αB are quite distinct, with αA being more prone to isomerization, but with αB suffering greater loss of solubility following isomerization. The ability of αA to sustain isomerization without loss of solubility may make it uniquely suited to the zero-turnover environment of the eye lens. It is clear that further, detailed examination of isomerization and epimerization in other tissues is needed and will expand our understanding of the role of long-lived proteins in aging-related diseases.
Acknowledgments
We are grateful for funding from the NIH, NIGMS grant R01GM107099.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.7b00073.
SI Methods. An expanded description of experimental methods. Figure S1: LC chromatograms and resulting CID and RDD spectra from 158AIPVSR163 from the WI sheep digest. Figure S2: Sequence alignment between αA and αB Crystallin from sheep using the Clustal Omega matrix. Figure S3: LC chromatograms and resulting CID spectra from 128KMEIVDDDVPSLW140 from the WS and WI pig digests. Table S1: Risomer values for the l-Asp synthetic version of 128KMEIVDDDVPSLW140 versus each peak from the WI pig digest. Figure S4: Sequence alignment of human, pig, cow and sheep βB3 proteins using Clustal Omega. Table S2: Tryptic peptides found to be isomerized in the three major proteins from water-insoluble sheep digest. Figure S5: LC chromatogram and resulting CID spectra from 108pQDEHGFISR116 from the WS pig digests. Table S3: Risomer values for each of the synthetic 108pQDEHGFISR116 versus the digest peaks from the WS pig digest. Figure S6: LC chromatogram and resulting CID spectra from 104pQDDHGYSIR112 from the WI pig digest. Table S4: Risomer values for the l-Asp synthetic version of 104pQDDHGYSIR112 versus each of the peaks in the LC. Figure S7. Comparison of the degree of isomerization in 164EEKPSSAPSS173between WS and WI sheep LC chromatograms. (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Toyama B. H.; Hetzer M. W. Protein homeostasis: live long, won’t prosper. Nat. Rev. Mol. Cell Biol. 2012, 14 (1), 55–61. 10.1038/nrm3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. Lens organelle degradation. Exp. Eye Res. 2002, 74 (1), 1–6. 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- Toyama B. H.; Savas J. N.; Park S. K.; Harris M. S.; Ingolia N. T.; Yates J. R.; Hetzer M. W. Identification of Long-Lived Proteins Reveals Exceptional Stability of Essential Cellular Structures. Cell 2013, 154 (5), 971–982. 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz J.; Bova M. P.; Ding L. L.; Haley D. a; Stewart P. L. Lens alpha-Crystallin: function and structure. Eye 1999, 13 (3b), 403–408. 10.1038/eye.1999.114. [DOI] [PubMed] [Google Scholar]
- Oertel M. F.; May C. A.; Bloemendal H.; Lutjen-Drecoll E. Alpha-B-Crystallin expression in tissues derived from different species in different age groups. Ophthalmologica 2000, 214 (1), 13–23. 10.1159/000027469. [DOI] [PubMed] [Google Scholar]
- Brady J. P.; Garland D.; Duglas-Tabor Y.; Robison W. G.; Groome A.; Wawrousek E. F. Targeted disruption of the mouse alpha A-Crystallin gene induces cataract and cytoplasmic inclusion bodies containing the small heat shock protein alpha B-Crystallin. Proc. Natl. Acad. Sci. U. S. A. 1997, 94 (3), 884–889. 10.1073/pnas.94.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J. P.; Garland D. L.; Green D. E.; Tamm E. R.; Giblin F. J.; Wawrousek E. F. alphaB-Crystallin in lens development and muscle integrity: A gene knockout approach. Investig. Ophthalmol. Vis. Sci. 2001, 42 (12), 2924–2934. [PubMed] [Google Scholar]
- Srinivas P.; Narahari A.; Petrash J. M.; Swamy M. J.; Reddy G. B. Importance of eye lens α-Crystallin heteropolymer with 3:1 αA to αB ratio: Stability, aggregation, and modifications. IUBMB Life 2010, 62 (9), 693–702. 10.1002/iub.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbers G.A.; Hoekman W. A.; Bloemendal H.; Jong W. W.; Kleinschmidt T.; Braunitzer G. Homology between the primary structures of the major bovine beta-Crystallin chains. Eur. J. Biochem. 1984, 139 (3), 467–479. 10.1111/j.1432-1033.1984.tb08029.x. [DOI] [PubMed] [Google Scholar]
- Mahendiran K.; Elie C.; Nebel J.-C.; Ryan A.; Pierscionek B. K. Primary sequence contribution to the optical function of the eye lens. Sci. Rep. 2014, 4, 5195. 10.1038/srep05195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. J.; Lioe H.; Robinson C. V.; Kay L. E.; Benesch J. L. P. α-B-Crystallin Polydispersity Is a Consequence of Unbiased Quaternary Dynamics. J. Mol. Biol. 2011, 413 (2), 297–309. 10.1016/j.jmb.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Bagnéris C.; Bateman O. A.; Naylor C. E.; Cronin N.; Boelens W. C.; Keep N. H.; Slingsby C. Crystal Structures of α-Crystallin Domain Dimers of αB-Crystallin and Hsp20. J. Mol. Biol. 2009, 392 (5), 1242–1252. 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- Laganowsky A.; Benesch J. L. P.; Landau M.; Ding L.; Sawaya M. R.; Cascio D.; Huang Q.; Robinson C. V.; Horwitz J.; Eisenberg D. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010, 19, 1031–1043. 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson S. R.; Hasan A.; Smith D. L.; Smith J. B. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 2000, 71 (2), 195–207. 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- Lund A. L.; Smith J. B.; Smith D. L. Modifications of the water-insoluble human lens alpha-crystallins. Exp. Eye Res. 1996, 63 (6), 661–672. 10.1006/exer.1996.0160. [DOI] [PubMed] [Google Scholar]
- Hooi M. Y. S.; Raftery M. J.; Truscott R. J. W. Accelerated aging of Asp 58 in alphaA Crystallin and human cataract formation. Exp. Eye Res. 2013, 106, 34–39. 10.1016/j.exer.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Masters P. M.; Bada J. L.; Zigler J. S. Aspartic acid racemization in heavy molecular weight crystallins and water insoluble protein from normal human lenses and cataracts. Proc. Natl. Acad. Sci. U. S. A. 1978, 75 (3), 1204–1208. 10.1073/pnas.75.3.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N.; Takata T.; Fujii N.; Aki K. Isomerization of aspartyl residues in crystallins and its influence upon cataract. Biochim. Biophys. Acta, Gen. Subj. 2016, 1860 (1), 183–191. 10.1016/j.bbagen.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Jia C.; Lietz C. B.; Yu Q.; Li L. Site-Specific Characterization of d-Amino Acid Containing Peptide Epimers by Ion Mobility Spectrometry. Anal. Chem. 2014, 86 (6), 2972–2981. 10.1021/ac4033824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor P. B.; Cournoyer J. J.; Pitteri S. J.; Chrisman P. A.; McLuckey S. A. Differentiation of aspartic and isoaspartic acids using electron transfer dissociation. J. Am. Soc. Mass Spectrom. 2006, 17 (1), 15–19. 10.1016/j.jasms.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Cournoyer J. J.; Pittman J. L.; Ivleva V. B.; Fallows E.; Waskell L.; Costello C. E.; O’Connor P. B. Deamidation: Differentiation of Aspartyl from Isoaspartyl Products in Peptides by Electron Capture Dissociation. Protein Sci. 2005, 14 (2), 452–463. 10.1110/ps.041062905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooi M. Y. S.; Truscott R. J. W. Racemisation and human cataract. D-Ser, D-Asp/Asn and d-Thr are higher in the lifelong proteins of cataract lenses than in age-matched normal lenses. Age 2011, 33 (2), 131–141. 10.1007/s11357-010-9171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi O.; Kirikoshi R.; Manabe N. Racemization of the succinimide intermediate formed in proteins and peptides: A computational study of the mechanism catalyzed by dihydrogen phosphate ion. Int. J. Mol. Sci. 2016, 17 (10), 1698. 10.3390/ijms17101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi B.; Collins M.; Bergström E.; Dowle A.; Penkman K.; Thomas-Oates J.; Wilson J. New Experimental Evidence for In-Chain Amino Acid Racemization of Serine in a Model Peptide. Anal. Chem. 2013, 85 (12), 5835–5842. 10.1021/ac4005869. [DOI] [PubMed] [Google Scholar]
- Takahashi O. Two-water-assisted racemization of the succinimide intermediate formed in proteins. A computational model study. Health 2013, 5 (12), 2018–2021. 10.4236/health.2013.512273. [DOI] [Google Scholar]
- Noguchi S.; Miyawaki K.; Satow Y. Succinimide and isoaspartate residues in the crystal structures of hen egg-white lysozyme complexed with tri-N-acetylchitotriose. J. Mol. Biol. 1998, 278 (1), 231–238. 10.1006/jmbi.1998.1674. [DOI] [PubMed] [Google Scholar]
- Noguchi S. Structural changes induced by the deamidation and isomerization of asparagine revealed by the crystal Structure of Ustilago sphaerogena ribonuclease U2B. Biopolymers 2010, 93 (11), 1003–1010. 10.1002/bip.21514. [DOI] [PubMed] [Google Scholar]
- Rehder D. S.; Chelius D.; McAuley A.; Dillon T. M.; Xiao G.; Crouse-Zeineddini J.; Vardanyan L.; Perico N.; Mukku V.; Brems D. N.; et al. Isomerization of a Single Aspartyl Residue of Anti-Epidermal Growth Factor Receptor Immunoglobulin γ2 Antibody Highlights the Role Avidity Plays in Antibody Activity. Biochemistry 2008, 47 (8), 2518–2530. 10.1021/bi7018223. [DOI] [PubMed] [Google Scholar]
- Tao W. A.; Zhang D.; Nikolaev E. N.; Cooks R. G. Copper(II)-assisted enantiomeric analysis of D,L-amino acids using the kinetic method: Chiral recognition and quantification in the gas phase. J. Am. Chem. Soc. 2000, 122 (43), 10598–10609. 10.1021/ja000127o. [DOI] [Google Scholar]
- Sun Q. Y.; Nelson H.; Ly T.; Stoltz B. M.; Julian R. R. Side Chain Chemistry Mediates Backbone Fragmentation in Hydrogen Deficient Peptide Radicals. J. Proteome Res. 2009, 8 (2), 958–966. 10.1021/pr800592t. [DOI] [PubMed] [Google Scholar]
- Tao Y.; Quebbemann N. R.; Julian R. R. Discriminating d-amino acid-containing peptide epimers by radical-directed dissociation mass spectrometry. Anal. Chem. 2012, 84 (15), 6814–6820. 10.1021/ac3013434. [DOI] [PubMed] [Google Scholar]
- Tao Y.; Julian R. R. Identification of amino acid epimerization and isomerization in Crystallin proteins by tandem LC-MS. Anal. Chem. 2014, 86 (19), 9733–9741. 10.1021/ac502296c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. K.; Kim R.; Kim S. H. Crystal structure of a small heat-shock protein. Nature 1998, 394 (6693), 595–599. 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Kumar L. V. S.; Rao C. M. Domain swapping in human alphaA and alphaB crystallins affects oligomerization and enhances chaperone-like activity. J. Biol. Chem. 2000, 275 (29), 22009–22013. 10.1074/jbc.M003307200. [DOI] [PubMed] [Google Scholar]
- Clarke S. Propensity for spontaneous succinimide formation from aspartyl and asparaginyl residues in cellular proteins. Int. J. Pept. Protein Res. 1987, 30 (6), 808–821. 10.1111/j.1399-3011.1987.tb03390.x. [DOI] [PubMed] [Google Scholar]
- Takata T.; Fujii N. Isomerization of Asp residues plays an important role in αA-Crystallin dissociation. FEBS J. 2016, 283 (5), 850–859. 10.1111/febs.13635. [DOI] [PubMed] [Google Scholar]
- Sakaue H.; Takata T.; Fujii N.; Sasaki H.; Fujii N. Alpha B- and βA3-Crystallins Containing D-Aspartic Acids Exist in a Monomeric State. Biochim. Biophys. Acta, Proteins Proteomics 2015, 1854 (1), 1–9. 10.1016/j.bbapap.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Robinson N. E.; Robinson Z. W.; Robinson B. R.; Robinson A. L.; Robinson J. A.; Robinson M. L.; Robinson A. B. Structure-dependent nonenzymatic deamidation of glutaminyl and asparaginyl pentapeptides. J. Pept. Res. 2004, 63 (5), 426–436. 10.1111/j.1399-3011.2004.00151.x. [DOI] [PubMed] [Google Scholar]
- Slingsby C.; Wistow G. J.; Clark A. R. Evolution of crystallins for a role in the vertebrate eye lens. Protein Sci. 2013, 22 (4), 367–380. 10.1002/pro.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden P. N.; Horwitz J.; Clarke S. Protein carboxyl methyltransferase from cow eye lens. Biochem. Biophys. Res. Commun. 1983, 113 (2), 418–424. 10.1016/0006-291X(83)91742-4. [DOI] [PubMed] [Google Scholar]
- McFadden P. N.; Clarke S. Chemical conversion of aspartyl peptides to isoaspartyl peptides. A method for generating new methyl-accepting substrates for the erythrocyte D-aspartyl/L-isoaspartyl protein methyltransferase. J. Biol. Chem. 1986, 261 (25), 11503–11511. [PubMed] [Google Scholar]
- Lowenson J. D.; Clarke S. Recognition of D-aspartyl residues in polypeptides by the erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis. J. Biol. Chem. 1992, 267 (9), 5985–5995. [PubMed] [Google Scholar]
- Lowenson J. D.; Clarke S. Structural elements affecting the recognition of L-isoaspartyl residues by the L-isoaspartyl/D-aspartyl protein methyltransferase. Implications for the repair hypothesis*. J. Biol. Chem. 1991, 266 (29), 19396–19406. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.