Abstract
In the budding yeast Saccharomyces cerevisiae, the cell division cycle and sporulation are mutually exclusive cell fates; glucose, which stimulates the cell division cycle, is a potent inhibitor of sporulation. Addition of moderate concentrations of glucose (0.5%) to sporulation medium did not inhibit transcription of two key activators of sporulation, IME1 and IME2, but did increase levels of Sic1p, a cyclin-dependent kinase inhibitor, resulting in a block to meiotic DNA replication. The effects of glucose on Sic1p levels and DNA replication required Grr1p, a component of the SCFGrr1p ubiquitin ligase. Sic1p is negatively regulated by Ime2p kinase, and several observations indicate that glucose inhibits meiotic DNA replication through SCFGrr1p-mediated destruction of this kinase. First, Ime2p was destabilized in the presence of glucose, and this turnover required Grr1p, a second component of SCFGrr1p, Cdc53p, and an SCFGrr1p-associated E2 enzyme, Cdc34p. Second, Ime2p-ubiquitin conjugates were detected under conditions of rapid Ime2p turnover, and conjugation of Ime2p to ubiquitin required GRR1. Third, a mutant form of Ime2p (Ime2ΔPEST), in which a putative Grr1p-interacting sequence was deleted, was more stable than wild-type Ime2p. Finally, expression of the IME2ΔPEST allele bypassed the block to meiotic DNA replication caused by 0.5% glucose. In addition, Grr1p is required for later events in sporulation independently of its role in Ime2p turnover.
Sporulation in the yeast Saccharomyces cerevisiae is regulated by three principal nutritional signals: nonfermentable carbon sources and starvation for essential growth nutrients promote sporulation, whereas glucose inhibits sporulation. Sporulation in yeast transforms a diploid cell into four haploid spores (reviewed in reference 30) and consists of two sequential processes—meiosis and spore formation. Nutritional signals regulate both the initiation of meiosis and later stages of sporulation (reviewed in references 22 and 26).
Glucose regulates initiation of meiosis through several mechanisms. At one level, glucose inhibits transcription of IME1. This gene encodes a “master regulator” transcription factor, which activates transcription of most early meiotic genes. At a second level, glucose inhibits assembly of Ime1p and other transcription factors into an active complex (33, 41, 50, 53). At a third level, glucose may inhibit the activity Ime2p kinase, which is encoded by an early meiotic gene. Ime2p kinase is required at multiple stages throughout sporulation, suggesting that Ime2p drives meiosis in the same way that cyclin/Cdk drives the cell division cycle (reviewed in reference 18). Although nutritional controls on Ime2p kinase have not been identified, this kinase is regulated by phosphorylation (3), association with other proteins (12), and stability (7).
Glucose inhibits sporulation through several different signal transduction pathways. Both the protein kinase A pathway and the glucose-repression pathway mediate glucose controls on meiosis as well as many other cellular processes (reviewed in references 9 and 49). These pathways regulate both initiation of meiosis and later stages of sporulation (21, 34). A third pathway, the glucose induction pathway, may also regulate sporulation since a key regulator of this pathway, Grr1p, is required to prevent sporulation during late stages of growth (38).
Grr1p activates the glucose induction pathway by targeting destruction of the Mth1p transcription factor, leading to the activation of hexose transporter genes (14, 25, 35). Grr1p is also required in an array of other cellular processes, including amino acid import (4), pseudohyphal differentiation (6), and G1 cyclin expression (reviewed in reference 48). Grr1p is the F-box protein component of the SCFGrr1p ubiquitin ligase complex, also termed an E3 (reviewed in reference 51). SCFGrr1p targets specific proteins for ubiquitination, recognizing a conserved PEST domain in these proteins. The target protein must also be phosphorylated before it can be recognized by the SCF. Once ubiquitinated, these targeted proteins, e.g., Cln2p and Gic2p, are degraded by the proteosome.
In this study, we present evidence that SCFGrr1p promotes ubiquitination and turnover of Ime2p in response to moderate amounts of glucose, a condition commonly encountered during late stages of growth. We propose that destruction of Ime2p under these conditions stabilizes Sic1p, a cyclin-dependent kinase inhibitor, resulting in a block to meiotic DNA replication. In addition, GRR1 has separate roles in regulating late meiotic events.
MATERIALS AND METHODS
Yeast strains and plasmids.
All yeast strains used in this study (Table 1) are isogenic to W303 or SH777, a derivative of W303. Because auxotrophic markers affect the transition from mitosis to meiosis, we compared strains with identical auxotrophies. For each mutant, at least two independent isolates were constructed and analyzed.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SH777 | MATa/MATα ade2/ade2 can1:ADE2:CAN1/can1:ADE2:CAN1 his3-11,15/his3-11,15 leu2-3,112/ leu2-3,112 lys2 (3′Δ):HIS3:lys2(5′Δ)/LYS2 trp1-1/trp1-3′Δ ura3-1/ura3-1 | 32 |
| SH1740 | SH777 except grr1Δ::TRP1/grr1Δ::TRP1 | This study |
| SH1970 | SH777 except TRP1/TRP1 LEU2/LEU2 | 38 |
| SH1752 | SH777 except grr1Δ::TRP1/grr1Δ::TRP1 LEU2/LEU2 | 38 |
| SH2778 | SH777 except trp1/TRP1 | This study |
| SH2716 | MATα ade2 can1:ADE2:CAN1 his3-11,15 leu2-3,112 trp1-3′Δ ura3-1 grr1::URA3 w/GAL1-IME2::6XHA-TRP1 | S. Irniger |
| SH2811 | SH1970 containing pS652 (tetO-IME2-6XHA-URA3) | This study |
| SH2813 | SH1752 containing pS652 (tetO-IME2-6XHA-URA3) | This study |
| SH2877 | SH1970 containing pS669 [tetO-IME2(pestΔ)-6XHA-URA3] | This study |
| SH3219 | SH1740 containing pS714 (tetO-IME2-6XHA-LEU2) and pTer62 (cup1-mycUb.Ko-URA3) | This study |
| SH3204 | SH2778 containing pRS315 (LEU2) and pTer62 (Cup1-mycUb.Ko-URA3) | This study |
| SH2362 | SH1970 containing pRS316 (URA3) | This study |
| SH3195 | SH2778 containing pS714 (tetO-IME2-6XHA-LEU2) and pTer62 (Cup1-mycUb.Ko-URA3) | This study |
| W303 | MATa/MATα ade2/ade2 can1/can1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 | |
| MTY1537 | W303 except cdc34-2/cdc34-2 | M. Tyers |
| MTY1538 | W303 except cdc53-1/cdc53-1 | M. Tyers |
| MTY1831 | MATaade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sic1Δ | M. Tyers |
| SH3179 | MTY1537 with pS652 (tetO-IME2-6XHA-URA3) | This study |
| SH3181 | MTY1538 with pS652 (tetO-IME2-6XHA-URA3) | This study |
| SH3183 | W303 with pS652 (tetO-IME2-6XHA-URA3) | This study |
| SH2898 | SH777 except grr1Δ::TRP1/GRR1 LEU2/leu2-3,112 | This study |
| SH2896 | SH777 except grr1Δ::TRP1/grr1Δ::TRP1 LEU2/leu2-3,112 | This study |
pS652, which contains the tetO-IME2-6XHA gene, was constructed by PCR amplification of the IME2-6XHA gene from yeast strain SH2716, digestion of the fragment with BglII and SbfI, and cloning of the resulting BglII-SbfI fragment into the BamHI and PstI sites of pCM188 (Euroscarf). pS714, a LEU2 derivative of pS652, was generated by digesting pS652 with StuI, which cleaves the plasmid in URA3, and cotransforming this linearized plasmid with a PCR fragment containing the LEU2 gene flanked by 40 bp of homology to the 5′ or 3′ end of the URA3 open reading frame. pS669, a derivative of pS652 with the IME2 PEST sequence precisely deleted, was constructed by cotransforming yeast with pS652 (linearized at BstEII) and a PCR fragment containing the IME2ΔPEST allele. This IME2ΔPEST fragment was constructed by fusion PCR between a 1.5-kb fragment homologous to the region immediately 5′ to the deletion and a 650-bp fragment homologous to the region immediately 3′ to the deletion. The 650-bp fragment also contained 40 bp at its 5′ end that was homologous to the sequence immediately 5′ to the deletion, allowing fusion of the two fragments. pS588, used to prepare the CLN2 probe, was constructed by cloning a 1.6-kb EcoRI/HindIII fragment from pCM214 (10) into the EcoRI and HindIII sites of pRS306 (46). All plasmids constructed for this study were verified by DNA sequencing. pRS315, pTER62, and pCM188 have been described elsewhere (1, 16, 46).
Growth and sporulation conditions.
Cultures were inoculated at a concentration of 5 × 104 cells/ml in 10 to 100 ml of growth medium. Mid-log-phase cultures were harvested after 12 to 16 h at a cell concentration of 0.5 × 107 to 1 × 107 cells/ml, and late-log phase cultures were harvested after 36 h at a cell concentration of 5 × 107 to 7 × 107 cells/ml. To prepare sporulation cultures, growth cultures were harvested, washed three times, and then resuspended in sporulation medium. All cultures were incubated at 30°C with constant shaking.
Growth medium was synthetic complete (SC) medium (pH 4.6) containing glucose as the sole carbon source (40). For strains containing plasmids, the medium lacked leucine, uracil, or tryptophan to maintain selection for the plasmid. Sporulation medium (Sp) was 2% potassium acetate (pH 7.0) supplemented with amino acids to balance auxotrophies and 0.17% yeast nitrogen base (lacking ammonium sulfate and amino acids). Sp+0.5 and Sp+2 media contained the same components as Sp medium with the addition of 0.5 and 2.0% glucose, respectively.
In experiments that measured the stability of tetO-IME2-6XHA, cultures were grown to mid-log phase in SC medium lacking tetracycline and then transferred to Sp or Sp+0.5 medium containing 2 μg of tetracycline/ml to repress transcription from the tetO promoter. In experiments with temperature-sensitive strains, cultures were grown to mid-log phase at 25°C, shifted to 37°C for 1 h, and then transferred to Sp+0.5 medium containing 2 μg of tetracycline/ml. In experiments to detect conjugation of ubiquitin to Ime2p, cultures were grown to mid-log phase in SC medium containing 2 μg of tetracycline/ml and then transferred to Sp+0.5 medium lacking tetracycline and containing 150 μM CuSO4 for 6 h to induce both myc-Ub.KO and Ime2p-6XHA.
Assays for DNA replication, recombination, nuclear divisions, spore formation, and spore viability.
Fluorescence-activated cell sorter (FACS) analysis of late-log-phase cultures was performed as described previously (32). To avoid complications in FACS analysis caused by branched or elongated grr1Δ cells, meiotic DNA replication was monitored by lysing cells, staining DNA in the extracts with diaminobenzoic acid, and measuring DNA content with a spectrofluorimeter as described elsewhere (27).
DNA recombination between LEU2 and MAT was monitored by random spore analysis as follows. Diploid strains (SH2896 and SH2898) heterozygous at the LEU2 locus (LEU2/leu2-3,112) were incubated for 72 h in sporulation medium and then plated on medium lacking leucine and arginine and containing canavanine, which selects for haploids (32). The colonies that formed on this medium were assayed for mating type as described elsewhere (40). Map distance was calculated as 100 cM multiplied by the frequency of Leu+ Canr colonies that displayed a change in coupling between LEU2 and MAT.
Meiotic chromosome segregation was assayed by staining nuclei with 4′,6′-diamidino-2-phenylindole (DAPI) and visualizing nuclei by fluorescence microscopy (20). Spore formation was assayed by light microscopy. In both assays, at least 300 cells were counted for each determination. Spore viability was determined by dissecting tetrads and determining the ability of each spore to form a colony. For these studies, 20 to 30 tetrads were dissected in each of three independent trials.
All values for the frequency of DNA replication, recombination, nuclear divisions, and spore formation are the means of at least three independent experiments. Data in the text are represented as the mean ± standard error. In graphs, the error bars represent the standard error; where error bars are not shown, they are less than the width of the symbol.
RNA and protein assays.
Yeast RNA was isolated by vortexing 2 × 108 to 4 × 108 yeast cells with glass beads and phenol (13). Transcript levels were measured by S1 nuclease protection (32). Templates for the IME1, IME2, and DED1 riboprobes used in S1 nuclease analysis have been described previously (21, 38); the CLN2 riboprobe was prepared using T3 RNA polymerase and pS588 cleaved with XmnI.
For analysis of protein expression levels, extracts were prepared by glass bead-trichloroacetic acid lysis (17). Ime2-6XHAp was detected by Western blotting using monoclonal antibody to the hemagglutinin (HA) epitope (12CA5; Covance), and Sic1p was detected by rabbit polyclonal antibodies (gift of M. Tyers, University of Toronto). Cdc28p was detected using anti-Cdc2 (PSTAIRE) antibody (Santa Cruz Biotechnology). Immunoprecipitation was performed using an ethanol-glass bead lysis method (31); the immunoprecipitation buffer contained 20 μl of protein inhibitor cocktail for yeast (Sigma)/ml, 20 μg of leupeptin/ml, 20 μg of pepstatin A/ml, and 1 mM phenylmethylsulfonyl fluoride. Ime2p-6XHA was immunoprecipitated with rabbit polyclonal anti-HA (Covance), and ubiquitin-conjugated proteins were detected with monoclonal anti-myc antibody (Santa Cruz Biotechnology). Protein bands from Western blot assays were visualized with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) and a fluorescence detection system (ECLplus; Amersham). Bands from RNA or protein analysis were quantitated with a STORM PhosporImager.
RESULTS
Addition of 0.5% glucose to sporulation medium inhibits meiotic DNA replication without inhibiting IME1 and IME2 transcription.
In addition to glucose inhibition of IME1 and IME2 transcription, glucose may also regulate meiotic initiation through additional targets. For example, in late-log-phase cultures, IME1 and IME2 are expressed to moderate levels, but meiotic DNA replication and later stages of meiosis are blocked (21). These cultures contain moderate amounts of glucose as well as nonfermentable carbon sources such as ethanol. Late-log-growth cultures containing less glucose initiate meiosis at higher frequencies than cultures containing more glucose (38).
To identify mechanisms by which glucose inhibits meiotic initiation after IME1 and IME2 have been transcribed, we transferred late-log-phase glucose cultures to sporulation medium lacking glucose (Sp), sporulation medium containing a moderate amount of added glucose (Sp+0.5), or sporulation medium containing a higher amount of added glucose (Sp+2). As an initial experiment, we measured spore formation after 24, 48, and 72 h (Fig. 1A). We found that addition of either the moderate or high concentration of glucose to sporulation medium almost completely blocked spore formation even after 72 h.
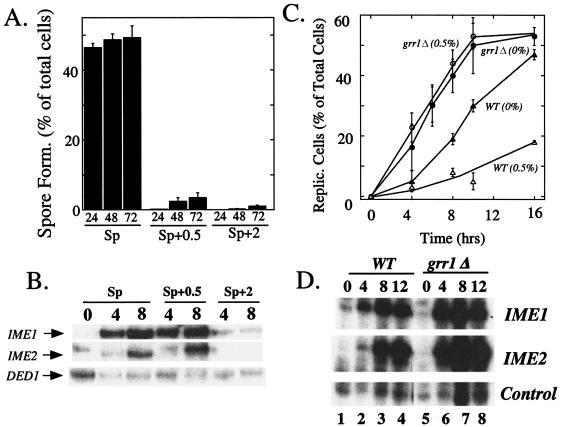
FIG. 1.
Inhibition of meiotic DNA replication by moderate concentrations of glucose requires GRR1. (A) Late-log-phase cultures of a wild-type strain (SH1970) were transferred to Sp, Sp+0.5, or Sp+2. Spore formation was measured at the indicated times after transfer. (B) Late-log-phase cultures of SH1970 were transferred as described for panel A, and expression levels of IME1, IME2, and a control (DED1) transcript were measured at the indicated times by S1 analysis. (C) Late-log-phase cultures of a wild type (SH1970) and a grr1Δ mutant (SH1742) were transferred to Sp or Sp+0.5 medium, and DNA replication was measured at the indicated times after transfer. (D) Wild-type and grr1Δ late-log-phase cultures were transferred to Sp medium, and IME1, IME2, and control (DED1) transcript levels were analyzed at the indicated times as described for panel B.
We next examined the induction of IME1 and IME2 in Sp, Sp+0.5, and Sp+2 media. Whereas IME1 and IME2 induction was almost completely blocked in Sp+2 medium, induction of these genes was delayed only slightly if at all in Sp+0.5 medium (Fig. 1B). Thus, addition of 0.5% glucose to sporulation medium was not sufficient to inhibit IME1 and IME2 transcription but was sufficient to block sporulation at some later stage.
We next compared the timing and efficiency of the first cellular event of sporulation, meiotic DNA replication, in Sp and Sp+0. 5 medium. To measure meiotic DNA replication, cultures were grown to late log phase, transferred to Sp or Sp+0.5 medium, and then assayed for DNA content at various times after the transfer. At the time of transfer, >85% of cells were in G1 phase, as measured by FACS analysis (K. Purnapatre and S. Honigberg, unpublished data). After transfer, meiotic replication occurred efficiently in Sp medium but occurred with much lower efficiency in Sp+0.5 medium (Fig. 1C). Thus, moderate concentrations of glucose inhibit sporulation at some stage after IME2 transcription but before initiation of meiotic DNA replication.
GRR1 is required for glucose to inhibit DNA replication.
Because grr1Δ mutants are defective in repressing sporulation during late stages of growth (38) and Grr1p is required in the glucose induction pathway, inhibition of meiotic DNA replication by moderate concentrations of glucose might require Grr1p. To test this hypothesis, a grr1Δ mutant was grown to late-log phase and then transferred to either Sp or Sp+0.5 medium. DNA replication was monitored at various times after transfer, as described above for the wild type. In contrast to the results described above for the wild type, the timing of meiotic DNA replication in the grr1Δ mutant was approximately the same in Sp+0.5 medium as in Sp medium (Fig. 1C). Thus, inhibition of meiotic DNA replication by 0.5% glucose required GRR1.
Interestingly, even in the absence of glucose, meiotic DNA replication occurred approximately 4 h earlier in the grr1Δ mutant than in the wild type (Fig. 1C). Consistent with this result, IME1 and IME2 transcript appeared several hours earlier in this mutant than in the wild type (Fig. 1D). The reason for early initiation of meiosis in this mutant is not known, but one possibility is that the grr1Δ mutant adapts from glucose growth medium to sporulation medium more rapidly than the wild type. In addition, at later times in sporulation medium, IME2 transcript accumulates to higher levels in the grr1Δ mutant than in the wild type (Fig. 1D). As discussed below, this accumulation may result from increased Ime2p stability.
Grr1p is required for Sic1p stabilization in Sp+0.5 medium.
The cyclin-dependent kinase inhibitor Sic1p binds and inhibits Cdc28p/Clb5,6p, preventing the onset of DNA replication until Sic1p is destroyed (11, 44). To determine whether glucose affects Sic1p levels, cultures were grown to mid-log phase and then transferred to Sp or Sp+0.5 medium. Samples were removed at the indicated times, and the level of Sic1p and a control protein (Cdc28p) were measured by Western blotting. As a control, we verified that the Sic1p band was absent in a sic1Δ strain (M. Gray and S. Honigberg, unpublished data).
In Sp medium, Sic1p levels were moderate at the time of transfer, remained low or decreased slightly after 2 or 4 h, and increased slightly by 6 h, at which time meiotic replication had initiated in most cells (Fig. 2A, lanes 1 to 4, and B). In contrast, in Sp+0.5 medium, Sic1p levels increased continuously from 0 to 6 h and reached levels approximately fivefold higher than in Sp medium (Fig. 2A, lanes 5 to 7, and B). Thus, addition of 0.5% glucose to sporulation medium led to increased Sic1p levels, a likely cause of the block to meiotic replication in this medium.
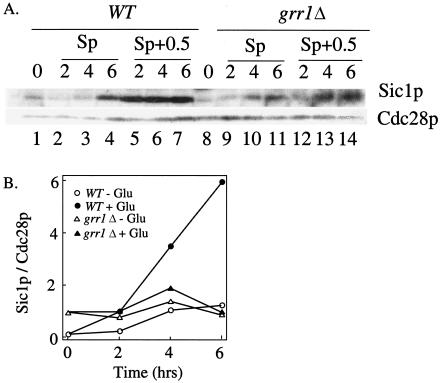
FIG. 2.
Sic1p accumulates in wild-type but not grr1Δ Sp+0.5 cultures. Wild-type and grr1Δ cultures were transferred to Sp or Sp+0.5 medium. After transfer, samples were removed from these cultures at the indicated times, and the levels of Sic1p and a control protein (Cdc28p) in the samples were measured. (A) Western blot analysis; (B) quantitation of Western blot assay in a trial independent from the data shown in panel A.
We next asked whether the increase in Sic1p caused by glucose depends on Grr1p. The grr1Δ mutant was assayed for Sic1p levels as described above for the wild type. In Sp medium, the pattern of Ime2p expression was similar to that in the wild type. However, in Sp+0.5 medium, Sic1p levels in this mutant failed to increase significantly (Fig. 2A, lanes 8 to 14, and B). Thus, Grr1p is required for the increase in Sic1p levels caused by addition of 0.5% glucose to sporulation medium.
Ime2p-6XHA is stabilized in a grr1Δ mutant.
Although Grr1p and Sic1p both regulate meiotic DNA replication, it is unlikely that Sic1p is targeted for destruction by SCFGrr1p. For one thing, the grr1Δ mutant displays decreased, rather than increased, Sic1p levels. Furthermore, during the cell cycle, Sic1p is targeted for destruction not by SCFGrr1p but by SCFCdc4p. In order for SCFCdc4p to target Sic1p for destruction, the latter protein must be specifically phosphorylated (36), and in meiosis this phosphorylation requires Ime2p kinase (11). These findings suggest the hypothesis that SCFGrr1p stabilizes Sic1p by targeting Ime2p for destruction.
To test whether Ime2p is destabilized in the presence of glucose, we compared the stability of an epitope-tagged version of Ime2p, Ime2p-6XHA, in Sp and Sp+0.5 media. To separate transcriptional control of IME2 from posttranscriptional control on Ime2p, IME2-6XHA was expressed from the tetO promoter, which is repressed by tetracycline and induced in its absence (2). The tetO-IME2-6XHA gene was first induced in growth medium lacking tetracycline, and then cultures were transferred to Sp or Sp+0.5 medium containing tetracycline to shut off transcription of this gene. Indeed, IME2-6XHA transcript levels decreased rapidly after transfer to either Sp medium (Purnapatre and Honigberg, unpublished) or Sp+0.5 medium (Fig. 3A, lanes 1 to 6).
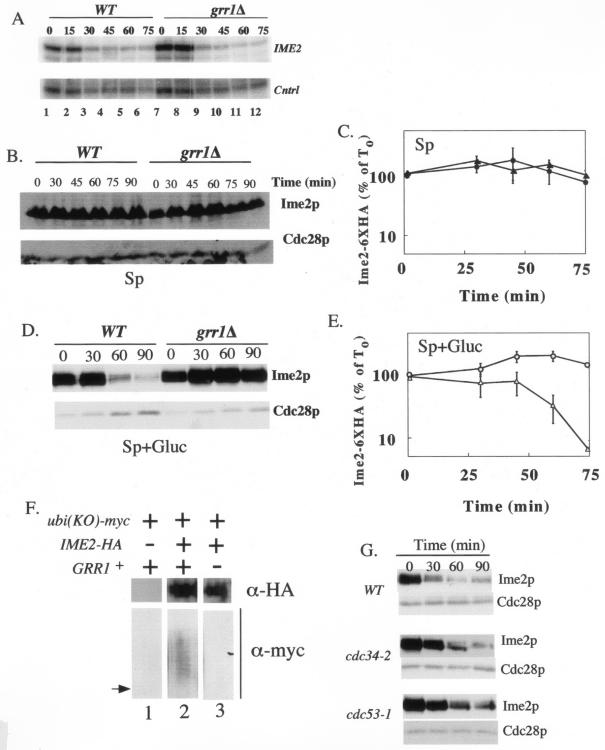
FIG. 3.
Ime2p-6XHA is stabilized in the grr1Δ mutant. (A) Shutoff of transcription from the tetO promoter occurs normally in the grr1Δ mutant. Wild-type (WT) or grr1Δ strains expressing the tetO-IME2-6XHA allele (SH2811 and 2813, respectively) were transferred to sporulation medium containing tetracycline, samples were removed at the indicated times (0 to 75 min), and IME1, IME2, and a control transcript (Cntrl) were analyzed as described for Fig. 1. (B) Ime2p-6XHA levels in Sp medium. For the cultures shown in panel A, samples were removed at the indicated times (0 to 90 min) and Ime2p-6XHA levels and Cdc28p were analyzed by Western blotting. (C) Graph representing Ime2p-6XHA expression relative to Cdc28p expression in wild-type (triangles) and grr1Δ (circles) strain from three independent trials of the experiment shown in panel B. (D) Ime2p-6XHA stability in Sp+0.5 medium. Cultures were grown and analyzed as described for panel B. (E) Graph representing Ime2p-6XHA expression relative to Cdc28p expression in wild-type (triangles) and grr1Δ (circles) strains for three independent trials of the experiment shown in panel D. Error bars in panels C and E represent the standard errors of the means of three independent experiments. (F) Strains expressing both Ime2p-6XHA and myc-Ubi(KO) were incubated in Sp+0.5 medium for 6 h, immunoprecipitated with anti-HA antibodies, and analyzed for both Ime2p-6XHA and myc-Ubi(KO)-conjugated proteins by Western blotting. The arrow points to the expected position of unmodified Ime2p-6XHA on the anti-Myc blot based on comparison of this blot with the anti-HA blot. (G) Ime2p-6XHA stability in Sp+0.5, analyzed as described in the legend for panel B, for wild-type (WT), cdc34-2, and cdc53-1 strains at 37°C (SH3183, SH3179, and SH3181, respectively).
At various times after cultures were transferred to Sp or Sp+0.5 medium, we measured the level of Ime2p-6XHA protein by Western blotting. Ime2p-6HA was relatively stable in Sp medium (Fig. 3B, lanes 1 to 6, and C) over the time period examined (0 to 90 min), but it was turned over rapidly in Sp+0.5 medium (Fig. 3D, lanes 1 to 6, and E). Thus, addition of glucose to sporulation medium increases the rate of Ime2p-HA turnover.
We next asked whether turnover of Ime2p-HA in Sp+0.5 was dependent on GRR1. We repeated the experiments described in the previous paragraph in a grr1Δ mutant. Transcription shutoff in this mutant was equivalent to shutoff in the wild type (Fig. 3A). In Sp medium, the stability of Ime2p-HA was approximately the same in the grr1Δ mutant as in the wild type. In contrast, in Sp+0.5 medium Ime2p-HA was much more stable in the grr1Δ mutant than in the wild type. Thus, turnover of Ime2p-HA in Sp+0.5 medium depends on GRR1.
Ubiquitination of Ime2p-6XHA in Sp+0.5 medium requires Grr1p.
Because Grr1p is required for Ime2p-6XHA turnover, we tested whether Grr1p was also required to ubiquitinate Ime2p. To determine whether Ime2p-6XHA was ubiquitinated, we utilized a plasmid bearing a Myc-tagged version of ubiquitin (mUb.Ko) under the control of the CUP1 promoter (1). The Ub.Ko allele contains mutations that change all five lysines in ubiquitin to arginine, thus preventing the formation of multi-Ub chains. Cultures containing the CUP1-mUb.Ko plasmid and/or the tetO-IME2-6XHA plasmid were grown to mid-log phase and then transferred to Sp+0.5 medium for 6 h. Extracts were prepared from these cultures, Ime2p-6XHA was immunoprecipitated, and the immunoprecipitate was analyzed by Western blotting for Ime2p-6XHA and for ubiquitin-conjugated proteins. From strains expressing both mUb.Ko and Ime2p-6XHA, a ladder of protein bands was detectable with antibody to mUb.Ko (Fig. 3F, lane 2). This pattern of bands indicated recovery of a protein that is ubiquitinated at multiple residues. As expected, the fastest-mobility band in the ladder migrated slightly slower than unmodified Ime2p-6XHA. This ladder of bands was absent in a strain which lacked the Ime2p-6XHA plasmid (Fig. 3F, lane 1). These results indicate that in Sp+0.5 medium, Ime2p becomes conjugated to ubiquitin at multiple residues.
To determine whether conjugation of ubiquitin to Ime2p requires Grr1p, the above experiments were repeated in a grr1Δ mutant. Ime2p was immunoprecipitated from the grr1Δ mutant at least as efficiently as from the wild type, but no ladder of ubiquitinated products was detected (Fig. 3F, lane 3). These results indicate that Grr1p is required for conjugation of Ime2p to ubiquitin.
Ime2p instability requires the SCFGrr1p component, Cdc53p, and the associated E2, Cdc34p.
To verify the role of SCFGrr1p in Ime2p turnover, we determined whether rapid destruction of Ime2p in Sp+0.5 medium requires a second component of this SCF, Cdc53p. At the same time, we determined whether the ubiquitin-conjugating enzyme (E2) that associates with SCFGrr1p, Cdc34p, is also required for Ime2p turnover. CDC53 and CDC34 are essential genes, so we employed temperature-sensitive cdc53-1 and cdc34-2 mutants. Both mutants are defective in ubiquitination of the SCFGrr1p substrates, Cln2p and Gic2p, at 37°C but grow almost normally at a lower temperature (25°C) (23, 52).
The tetO-IME2-6XHA plasmid was expressed in both mutants and in a wild-type control strain. These strains were grown at the permissive temperature and then shifted to Sp+0.5 medium at 37°C. Under this condition, Ime2p-6XHA was more stable in the mutants than in the control, indicating that Cdc53p and Cdc34p, like Grr1p, are involved in glucose-stimulated turnover of Ime2p (Fig. 3G). However, Ime2p-6XHA was not completely stabilized in either mutant at 37°C; indeed, Ime2p-6XHA was not completely stabilized at 37°C even in a grr1Δ mutant (data not shown). Incomplete stabilization of Ime2p-6XHA in the temperature-sensitive mutants could reflect residual SCFGrr1p activity at the restrictive temperature or thermal instability of Ime2p-6XHA. Interestingly, Cln2p-HA is also only partially stabilized in cdc53-1 and cdc34-2 mutants (52). In any case, the delayed turnover of Ime2p-6XHA in cdc53-1 and cdc34-2 mutants is consistent with SCFGrr1p being required for Ime2p degradation in Sp+0.5 medium.
An allele of IME2 lacking the C-terminal PEST sequence is stabilized in Sp+0.5 medium.
SCFGrr1p recognizes a PEST sequence at the C terminus of Cln2p (5). A putative PEST sequence (HSIPDVGTDSTISDSIDETELSK) can also be found near the C terminus of Ime2p (Fig. 4A). PEST sequences contain at least one proline and several serines, threonines, glutamates, and aspartates and are flanked by positively charged amino acids. The C-terminal domain of IME2, including the putative PEST sequence, has been implicated as a negative regulatory domain (29), consistent with a role of this domain in Ime2p stability.
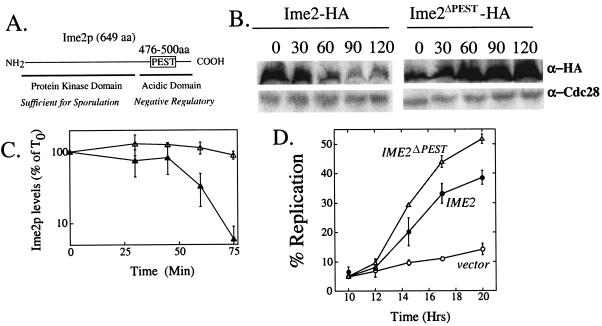
FIG. 4.
Expression of Ime2pPESTΔ in Sp+0.5 medium. (A) Diagram of Ime2p, highlighting a putative PEST sequence. The proposed negative regulatory acidic domain and kinase domain of Ime2p are indicated below the line. (B) Western blot analysis of Ime2p and Ime2pPESTD. Mid-log cultures containing Ime2p-6XHA or Ime2pPESTΔ-6XHA were transferred to Sp+0.5 medium containing tetracycline (SH2811 and SH2877, respectively). Samples were removed at the indicated times (0 to 120 min), and blots were probed with anti-HA and anti-Cdc28 antibodies. (C) Graph representing Ime2p-6XHA (filled triangles) and Ime2pPESTΔ-6XHA (open triangles) levels relative to Cdc28p in Sp+0.5 for repeated trials of the experiment shown in panel B. Error bars represent the standard errors of the means of three independent experiments. (D) Meiotic DNA replication in Sp+0.5 medium in wild-type strains expressing Ime2pPESTΔ-6XHA (SH2877; open triangles), Ime2p-6XHA (SH2811; filled circles), or the vector, pRS316 (SH2362; open circles).
To determine whether the Ime2p-PEST sequence was required for Ime2p destruction, we generated a tagged allele of IME2 on a plasmid with the putative PEST sequence precisely deleted (tetO-IME2ΔPEST-6XHA). The stability of Ime2pΔPEST-6XHA was compared to that of Ime2p-6XHA under the conditions where the latter protein was found to be unstable (Sp+0.5). We found that Ime2pΔPEST-6XHA was much more stable than Ime2p-6XHA in Sp+0.5 medium (Fig. 4B and C). Thus, the IME2 PEST domain is required for glucose-dependent turnover of Ime2p, consistent with this regulation being mediated through Grr1p. In the mutant, Ime2pΔPEST-6XHA levels actually increase two- to fivefold (Fig. 4A), implying that Ime2p synthesis continues even after the addition of tetracycline to the medium.
Expression of the IME2ΔPEST-6XHA allele bypasses inhibition of replication by glucose.
As a direct test of the hypothesis that glucose represses meiotic DNA replication by stimulating Ime2p turnover, we asked whether expression of the stabilized allele of IME2, IME2ΔPEST, would bypass inhibition of meiotic replication by glucose. We measured the timing of meiotic DNA replication in Sp+0.5 medium in a strain containing tetO-IME2ΔPEST, a strain containing tetO-IME2, and a strain bearing only the tetO vector. Under these conditions, DNA replication in the IME2ΔPEST strain occurred much more rapidly and more efficiently than did the control strain (Fig. 4D). Overexpression of the wild-type tetO-IME2 allele was less effective than IME2ΔPEST at bypassing the replication block (Fig. 4D), consistent with the idea that glucose represses meiotic replication by stimulating Ime2p turnover.
Spore packaging is defective in the grr1Δ mutant.
Because meiotic DNA replication is resistant to glucose in the grr1Δ mutant, spore formation might also be expected to be resistant to glucose. However, we found that spore formation was sensitive to glucose in this mutant. When wild-type mid-log-phase cultures were transferred to Sp+0.5 or Sp medium, the efficiency of spore formation was twofold lower in Sp+0.5 than in Sp medium. For unknown reasons, addition of glucose to sporulation medium inhibited spore formation more effectively in cultures transferred from late log phase (Fig. 1A) than in those transferred from mid-log phase (Table 2). In contrast to the wild type, mid-log-phase grr1Δ cultures transferred to sporulation medium displayed 10-fold-lower spore formation in Sp+0.5 medium than in Sp medium (Table 2). Thus, while meiotic DNA replication is resistant to glucose in the grr1Δ mutant, spore formation in this mutant is actually more sensitive to glucose.
TABLE 2.
Spore formation, recombination, and spore viabilitya
| Assay |
WT
|
grr1Δ
|
||
|---|---|---|---|---|
| Sp | Sp + glucose | Sp | Sp + glucose | |
| Spore formation | 52 ± 3 | 25 ± 5 | 44 ± 3 | 4.3 ± 0.9 |
| Dyads, total asci (%) | 34 ± 2 | 49 ± 7 | 64 ± 3 | 93 ± 4 |
| LEU2-MAT recomb. (cM) | 33 ± 2 | 30 ± 2 | 38 ± 1 | 34 ± 3 |
| Spore viability (%) | 93 ± 1 | 96 ± 2 | 71 ± 3 | 44 ± 8 |
Mid-log-phase cultures were transferred to Sp or Sp plus glucose medium, incubated for 72 h, and then assayed for spore formation, the fraction of dyads among total asci, recombination (recomb.) in the LEU2-MAT interval, and spore viability. Recombination was assayed in SH2898 (grr1Δ/grr1Δ) or SH2896 (grr1Δ/GRR1); all other experiments were assayed in SH1752 (grr1Δ/grr1Δ) or SH1970 (WT). Values are means ± standard errors. WT, wild type.
We also observed that late-log-phase grr1Δ cultures have a reduced ability to form spores relative to mid-log-phase grr1Δ cultures. When mid-log-phase cultures were transferred to Sp medium, the grr1Δ mutant formed spores almost as efficiently as the wild type (Table 2). In contrast, when late-log-phase cultures were transferred to Sp medium, the grr1Δ mutant formed asci much less efficiently than the wild type (only 3.3% ± 1.6% asci from the mutant compared to 57% ± 2% from the wild type). Because IME1 and IME2 transcription and meiotic replication occur efficiently after transfer of either mid- or late-log-phase cultures to Sp medium (Fig. 1) (Gray and Honigberg, unpublished), the defect in spore formation from late-log-phase cultures was likely specific to the later stages of sporulation. As reported previously (6), late-log-phase grr1Δ cultures contained many pseudohyphal cells, i.e., cells containing elongated or branched buds. Because pseudohyphal differentiation may interfere with spore formation (43), we used mid-log-phase cultures to characterize late stages of sporulation in the grr1Δ mutant.
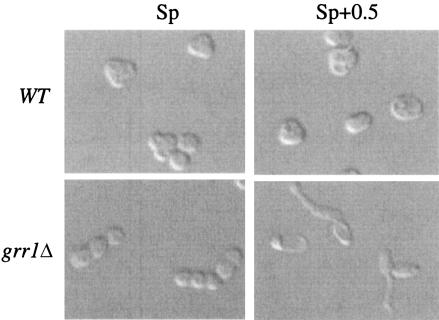
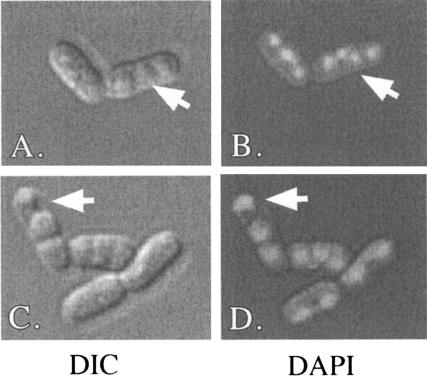
Even though the grr1Δ mutant formed spores efficiently when cells were transferred from mid-log-phase cultures to Sp medium, the asci formed from these cultures were morphologically distinct from wild-type asci. Specifically, grr1Δ spores were usually packaged in a linear arrangement, whereas wild-type spores were packaged in a tetrahedral arrangement (Fig. 5). In addition, the frequency of two-spore and three-spore asci was much higher in the grr1Δ mutant than in the wild type (Table 2). Using fluorescence microscopy, we determined that 30% of grr1Δ asci contained at least one unpackaged nucleus, and these types of asci were rare in the wild type. In at least some of these asci, small structures that may have been anucleate spores were observed (Fig. 6). Thus, one cause of the decreased number of spores per asci in the grr1Δ mutant is the failure to efficiently package nuclei into spores. The cause of the linear morphology of grr1Δ asci is not known but could be related to the cell polarity defect in grr1Δ mutants (8, 23, 24).
FIG. 5.
Cell morphology in the wild type (WT, SH1970) and the grr1Δ mutant (SH1752) in Sp and Sp+0.5 medium. Mid-log cultures were transferred to Sp or Sp+0.5. After 72 h, images were captured using Nomarski optics on a Nikon TE2000-U with a ColorView imaging system and analySIS software (SIS).
FIG. 6.
Spore packaging in the grr1Δ mutant. Mid-log-phase cultures of a grr1Δ mutant (SH1752) were transferred to Sp. After 72 h, nuclei were stained with DAPI and images were captured using Nomarski (left side) and fluorescence (right side) optics, with Nomarski optics on a Nikon TE2000-U with a ColorView imaging system and analySIS software (SIS). Arrows indicate unpackaged nuclei. DIC, differential interference contrast.
Late stages of meiosis are sensitive to glucose in the grr1Δ mutant.
To further characterize defects in sporulation observed in the grr1Δ mutant, we measured several cellular events that occur after meiotic DNA replication but prior to spore formation. For each of these events, we compared the efficiency in the mutant to that in the wild type in both Sp and Sp+0.5 medium.
Meiotic DNA recombination occurs immediately after meiotic DNA replication. To determine if grr1Δ mutants are defective in meiotic recombination, we measured recombination in the LEU2-MAT interval on chromosome III (see Materials and Methods). In both Sp and Sp+0.5 media, the frequency of recombination in this interval was approximately the same in the grr1Δ mutant as the wild type (Table 2). The frequency of recombination observed in these experiments was very close to the expected frequency based on map distance (35 cM). Thus, GRR1 is not required for crossover recombination in meiosis.
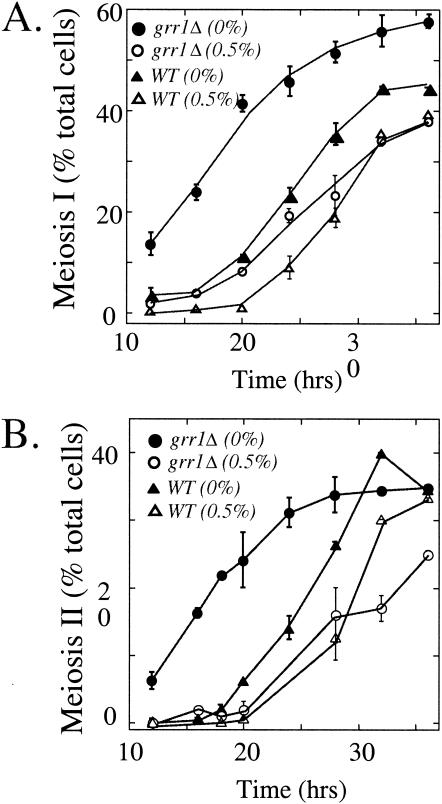
After completion of meiotic DNA recombination, meiotic cells undergo two cycles of chromosome segregation. We monitored meiotic chromosome segregation over time in the grr1Δ mutant and the wild-type control by the appearance of binucleate and tetranucleate cells. In the wild type, both cycles of chromosome segregation were delayed and somewhat less efficient in Sp+0.5 medium than in Sp medium (Fig. 7). In the grr1Δ mutant, the meiotic divisions occurred more rapidly than in the wild-type in Sp medium, consistent with the earlier timing of IME1 induction, IME2 induction, and meiotic DNA replication in this mutant. In contrast, the meiotic divisions were substantially delayed in this mutant in Sp+0.5 medium (Fig. 7). Comparing the grr1Δ mutant to the wild type, addition of glucose delayed chromosome segregation by 12 to 16 h in the mutant but only by 5 to 6 h in the wild type. Thus, in Sp+0.5 medium the grr1Δ mutant initiates meiosis more rapidly than the wild type but completes chromosome segregation less efficiently.
FIG. 7.
Meiotic divisions in the grr1Δ mutant. (A) Mid-log-phase cultures of the wild type (WT) and the grr1Δ mutant (SH1970 and SH1752, respectively) were transferred to Sp or Sp+0.5 medium. Samples were removed from these cultures at the indicated times (0 to 36 h) and analyzed for the meiosis I nuclear division (fraction of cells in the culture that are in binucleate or later stages of meiosis). (B) The same samples as shown in panel A were assayed for meiosis II nuclear division (fraction of cells in the culture that are in tetranucleate or later stages of meiosis).
Defects in either chromosome segregation or spore formation, such as were observed in the grr1Δ mutant, can lead to decreased spore viability. To measure spore viability, we dissected asci of both the grr1Δ mutant and the wild type after incubation for 72 h in either Sp or Sp+0.5 medium. In the wild type, spore viability was >90% in either medium (Table 2). In contrast, in the grr1Δ mutant, spore viability was significantly less than the wild type in Sp medium and even lower in Sp+0.5 medium. Thus, the grr1Δ mutant is defective in producing viable spores, and this defect is exacerbated by moderate concentrations of glucose.
Expression of IME2ΔPEST does not cause defects in spore formation.
Because Ime2p is stabilized in the grr1Δ mutant, we asked whether this stabilization caused the observed defects in spore formation. To test this idea, a wild-type strain containing a plasmid bearing IME2ΔPEST was grown to mid-log phase and then transferred to Sp+0.5 medium (SH2877). After 72 h, we compared the frequency of spore formation in this strain to that in a control strain bearing an empty vector (SH2362). We found that expression of IME2ΔPEST increased rather than decreased the level of spore formation (30% ± 4% of cells formed spores in the tetO-IME2ΔPEST strain, compared to only 13% ± 3% of cells from the control strain). This result is consistent with earlier studies showing that Ime2p is required at late stages of meiosis as well as for the initiation of meiosis (3). Thus, it is unlikely that glucose inhibits late stages of meiosis in the grr1Δ mutant by stabilizing Ime2p.
DISCUSSION
The principal conclusions from this study are the following: (i) adding moderate concentrations of glucose to sporulation medium destabilizes Ime2p, stabilizes Sic1p, and inhibits meiotic DNA replication; and (ii) these three effects of glucose require the SCFGrr1p ubiquitin ligase.
Several lines of evidence indicate that the SCFGrr1p complex directly causes ubiquitination and destruction of Ime2p. First, Ime2p is less stable in Sp+0.5 medium than in Sp medium and is stabilized in the former medium in grr1Δ, cdc53-1, and cdc34-2 mutants. Second, Ime2p is ubiquitinated in Sp+0.5 medium, and this ubiquitination requires Grr1p. Third, a region of Ime2p containing a putative PEST sequence, a domain found in other SCFGrr1p substrates, is necessary for Ime2p destruction. Fourth, strains containing an IME2 allele that lacks this PEST sequence bypass the block to meiotic replication in Sp+0.5 medium.
Degradation of ectopically expressed Ime2p during rapid growth in glucose medium does not depend on Cdc34p (7), the E2 that associates with SCFGrr1p; thus, it is likely that Ime2p is degraded during rapid growth through an SCFGrr1p-independent pathway, in contrast to the SCFGrr1p-dependent pathway operable in Sp+0.5 medium. In this regard, we note that Ime2p-6XHA was only partially stabilized in the cdc34-2 and cdc53-1 mutants in Sp+0.5 medium, possibly because these mutants retain residual Cdc34p or Cdc53p activity at the restrictive temperature. Because destruction of SCFGrr1p targets such as Cln2p, Gic2p, and Mth1p requires phosphorylation of these targets, it is likely that glucose also regulates Ime2p stability by phosphorylation, although the kinase that mediates this step is not known. Nitrogen and glucose may regulate Ime2p through independent pathways, since Gpa2p, the alpha-subunit of a trimeric G-protein, binds Ime2p in vitro and is required for nitrogen to inhibit Ime2p kinase in vivo (12).
Surprisingly, the grr1Δ mutant is less sensitive to glucose than the wild type during early stages of sporulation and more sensitive during late stages. For example, both meiotic chromosome segregation and spore formation were more sensitive to glucose in the grr1Δ mutant than in the wild type. This increased glucose sensitivity does not result from stabilization of Ime2p, because expression of the stabilized form of Ime2p, Ime2ΔPESTp, does not increase glucose sensitivity. Wild-type meiotic cells become resistant to glucose as the nuclear divisions initiate (commitment to meiosis) (15, 19, 28). The continued sensitivity to glucose in late stages of sporulation suggests that the grr1Δ mutant may be defective in commitment to meiosis.
Turnover of Ime2p when glucose is present inhibits meiotic DNA replication (at least in part) by stabilizing Sic1p. Sic1p inhibits Clb5,6p/Cdc28p kinase activity, which is necessary for replication (44), and so for replication to ensue Sic1p must be destroyed and this destruction requires Ime2p (11). Indeed, addition of glucose to sporulation medium led to increased Sic1p levels, and this increase depended on Grr1p. Interestingly, an analogous pathway of kinase turnover and Sic1p stabilization occurs in the cell division cycle. In the cell cycle, SCFGrr1p targets Cln1p and Cln2p cyclins for destruction, and these cyclins (together with Cdc28p kinase) phosphorylate Sic1p, targeting it for ubiquitination by SCFCdc4p and subsequent destruction. Thus, DNA replication is regulated by SCFGrr1p in both meiosis and mitosis, but the substrate for this ubiquitin ligase is different in the two programs. Ime2p has additional roles in meiosis beyond regulating Sic1p (3, 37, 42), so Ime2p turnover likely prevents progression of meiosis through additional mechanisms. Indeed, we observed an accumulation of IME2 transcript in the grr1Δ mutant. Because Ime2p kinase activates the Ndt80p transcription factor (3, 45, 47), which may activate IME2 transcription during late stages of meiosis (22), it is possible that stabilization of Ime2p may lead to increased levels of IME2 transcript via an IME2-NDT80 positive feedback loop.
As described in the introduction, glucose inhibits sporulation through several signaling pathways and targets. One potential advantage of these multiple layers of control may be to permit specialized responses to different glucose concentrations. As described in this study, addition of high concentrations of glucose to sporulation medium completely blocks IME1 and IME2 transcription, whereas addition of lower concentrations of glucose allows transcription of these genes without allowing meiotic DNA replication. The latter condition approximates the conditions present during late stages of growth in glucose. During these stages, cells express moderate levels of IME1 but do not initiate meiotic DNA replication (21). We propose that moderate concentrations of glucose present during late stages of growth block meiotic DNA replication via the Grr1p/Ime2p/Sic1p pathway without preventing moderate expression of IME1 and IME2. This moderate expression of IME1 and IME2 during late stages of growth may explain why these cultures can initiate meiosis much faster than mid-log cultures (39).
Acknowledgments
We are grateful to Michael Ellison (University of Alberta), Jon Gerton (Stowers Institute), F. Irniger (Georg-August University), and Mike Tyers (University of Toronto) for yeast strains, plasmids, and antibodies. We thank M. Tyers, S. Irniger, Mark Hochstrasser (Yale University), Antony Cooper (University of Missouri—Kansas City), and Kathryn Hill (University of Missouri—Kansas City) for helpful discussions.
This work was supported by grant R01-GM58017 by the National Institutes of Health and a grant from the University of Missouri Research Board.
REFERENCES
- 1.Arnason, T., and M. J. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase, Ime2. Genes Dev. 17:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard, F., and B. Andre. 2001. Ubiquitin and the SCFGrr1 ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496:81-85. [DOI] [PubMed] [Google Scholar]
- 5.Berset, C., P. Griac, R. Tempel, J. La Rue, C. Wittenberg, and S. Lanker. 2002. Transferable domain in the G1 cyclin Cln2 sufficient to switch degradation of Sic1 from the E3 ubiquitin ligase SCFCdc4 to SCFGrr1. Mol. Cell. Biol. 22:4463-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blacketer, M. J., P. Madaule, and A. M. Myers. 1995. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics 140:1259-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolte, M., P. Steigemann, G. H. Braus, and S. Irniger. 2002. Inhibition of APC-mediated proteolysis by the meiosis-specific protein kinase Ime2. Proc. Natl. Acad. Sci. USA 99:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, J. L., M. Jaquenoud, M. P. Gulli, J. Chant, and M. Peter. 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11:2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 10.Colomina, N., E. Gari, C. Gallego, E. Herrero, and M. Aldea. 1999. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 18:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 12.Donzeau, M., and W. Bandlow. 1999. The yeast trimeric guanine nucleotide-binding protein alpha subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol. Cell. Biol. 19:6110-6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elder, R. T., E. Y. Loh, and R. W. Davis. 1983. RNA from yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc. Natl. Acad. Sci. USA 80:2432-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flick, K. M., N. Spielewoy, T. I. Kalashnikova, M. Guaderrama, Q. Zhu, H. C. Chang, and C. Wittenberg. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 14:3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesan, A. T., H. Holter, and C. Roberts. 1958. Some observations on sporulation in Saccharomyces. C. R. Trav. Lab. Carlsberg (Chim.) 13:1-6. [PubMed] [Google Scholar]
- 16.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 17.Hann, B. C., and P. Walter. 1991. The signal recognition particle in S. cerevisiae. Cell 67:131-144. [DOI] [PubMed] [Google Scholar]
- 18.Honigberg, S. M. 2004. Ime2p and Cdc28p: co-pilots driving meiotic development. J. Cell Biochem. 92:1025-1033. [DOI] [PubMed] [Google Scholar]
- 19.Honigberg, S. M., C. Conicella, and R. E. Esposito. 1992. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics 130:703-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honigberg, S. M., and R. E. Esposito. 1994. Reversal of cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth. Proc. Natl. Acad. Sci. USA 91:6559-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honigberg, S. M., and R. H. Lee. 1998. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4548-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honigberg, S. M., and K. Purnapatre. 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116:2137-2147. [DOI] [PubMed] [Google Scholar]
- 23.Jaquenoud, M., M. P. Gulli, K. Peter, and M. Peter. 1998. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 17:5360-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaquenoud, M., and M. Peter. 2000. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p). Mol. Cell. Biol. 20:6244-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniak, A., Z. Xue, D. Macool, J. H. Kim, and M. Johnston. 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 27.Kassir, Y., and G. Simchen. 1991. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 194:94-110. [DOI] [PubMed] [Google Scholar]
- 28.Kirsop, B. H. 1954. Studies in yeast sporulation. I. Some factors influencing sporulation. J. Inst. Brew. 60:393. [Google Scholar]
- 29.Kominami, K., Y. Sakata, M. Sakai, and I. Yamashita. 1993. Protein kinase activity associated with the IME2 gene product, a meiotic inducer in the yeast Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 57:1731-1735. [DOI] [PubMed] [Google Scholar]
- 30.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Laney, J. D., and M. Hochstrasser. 2002. Assaying protein ubiquitination in Saccharomyces cerevisiae. Methods Enzymol. 351:248-257. [DOI] [PubMed] [Google Scholar]
- 32.Lee, R. H., and S. M. Honigberg. 1996. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol. Cell. Biol. 16:3222-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malathi, K., Y. Xiao, and A. P. Mitchell. 1999. Catalytic roles of yeast GSK3β/shaggy homolog Rim11p in meiotic activation. Genetics 153:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuura, A., M. Treinin, H. Mitsuzawa, Y. Kassir, I. Uno, and G. Simchen. 1990. The adenylate cyclase/protein kinase cascade regulates entry into meiosis in Saccharomyces cerevisiae through the gene IME1. EMBO J. 9:3225-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriya, H., and M. Johnston. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 101:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 37.Ofir, Y., S. Sagee, N. Guttmann-Raviv, L. Pnueli, and Y. Kassir. 2004. The role and regulation of the preRC component Cdc6 in the initiation of premeiotic DNA replication. Mol. Biol. Cell 15:2230-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purnapatre, K., and S. M. Honigberg. 2002. Meiotic differentiation during colony maturation in Saccharomyces cerevisiae. Curr. Genetics 42:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Purnapatre, K., S. Piccarillo, B. L. Schneider, and S. M. Honigberg. 2002. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells 7:675-691. [DOI] [PubMed] [Google Scholar]
- 40.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Rubin-Bejerano, I., S. Mandel, K. Robzyk, and Y. Kassir. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16:2518-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindler, K., K. R. Benjamin, A. Martin, A. Boglioli, I. Herskowitz, and E. Winter. 2003. The Cdk-activating kinase Cak1p promotes meiotic S phase through Ime2p. Mol. Cell. Biol. 23:8718-8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroder, M., J. S. Chang, and R. J. Kaufman. 2000. The unfolded protein response represses nitrogen starvation-induced developmental differentiation in yeast. Genes Dev. 14:2962-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwob, E., T. Bohm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
- 45.Shubassi, G., N. Luca, J. Pak, and J. Segall. 2003. Activity of phosphoforms and truncated versions of Ndt80, a checkpoint-regulated sporulation-specific transcription factor of Saccharomyces cerevisiae. Mol. Genet. Genomics 270:324-336. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sopko, R., S. Raithatha, and D. Stuart. 2002. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22:7024-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spruck, C. H., and H. M. Strohmaier. 2002. Seek and destroy: SCF ubiquitin ligases in mammalian cell cycle control. Cell Cycle 1:250-254. [PubMed] [Google Scholar]
- 49.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 50.Vidan, S., and A. P. Mitchell. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17:2688-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems, A. R., T. Goh, L. Taylor, I. Chernushevich, A. Shevchenko, and M. Tyers. 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R. Soc. London B 354:1533-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453-463. [DOI] [PubMed] [Google Scholar]
- 53.Xiao, Y., and A. P. Mitchell. 2000. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol. Cell. Biol. 20:5447-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]