Abstract
Cells contain powerful RNA decay machinery to eliminate unneeded RNA from the cell, and this process is an important and regulated part of controlling gene expression. However, certain structured RNAs have been found that can robustly resist degradation and extend the lifetime of an RNA. In this review, we present three RNA structures that use a specific three-dimensional fold to provide protection from RNA degradation, and discuss how the recently-solved structures of these RNAs explain their function. Specifically, we describe the Xrn1-resistant RNAs from arthropod-borne flaviviruses, exosome-resistant long non-coding RNAs associated with lung cancer metastasis and found in Kaposi’s Sarcoma-associated herpesvirus, and tRNA-like sequences occurring in certain plant viruses. These three structures reveal three different mechanisms to protect RNAs from decay and suggest RNA structure-based nuclease resistance may be a widespread mechanism of regulation.
Keywords: RNA structure, Xrn1, exosome, RNA decay, MALAT1, tRNA-like sequence, small flavivirus RNA (sfRNA), RNA triple-helix
INTRODUCTION
Cells have many ways to precisely control the pathways of RNA decay as part of their overall strategy to regulate RNA levels. The pathway used to eliminate most messenger RNAs (mRNAs) in eukaryotes initiates by shortening of the mRNA’s 3′ poly-A tail, followed by either degradation by the 3′→5′ exonuclease complex (exosome) or enzymatic decapping of the mRNA’s 5′ end and degradation by the 5′→3′ exonuclease Xrn1 [1,2](Figure 1). Related RNA degradation pathways regulate mRNA levels and provide mRNA quality control, including deadenylation-independent, endonuclease-mediated, nonsense mediated, no-go, and non-stop decay pathways [3–9]. The rate of decay of specific mRNAs can be controlled by several processes [10–13], including ones that use RNA structure. In the simplest case, a thermostable RNA secondary structure at the 5′ or 3′ end of an RNA might slow the degradation machinery, but in more specific cases it is now clear that some RNAs form folded three-dimensional conformations that use more refined mechanisms to confound or evade the decay machinery. Here we discuss three recently-solved viral RNA structures that use different strategies to actively evade the powerful decay machinery. In one case, an analogous RNA structure has been found in cells, suggesting that continued characterization of degradation machine-evading viral RNAs may lead to the discovery of cellular versions, perhaps with broad biological significance.
Figure 1.
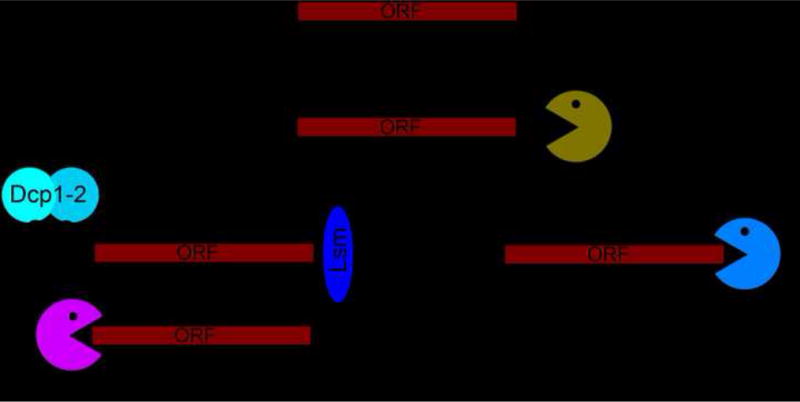
The primary path of mRNA degradation in eukaryotes. Shortening of the poly-A tail is followed by decapping by the Dcp1/Dcp2 complex and then either 5′→3′ decay by Xrn1 or 3′→5′ decay by the exosome complex.
Blocking degradation from the 5′ direction: Flaviviral Xrn1-resistant structures
The first RNA structure discussed here is found in arthropod-borne flaviviruses (FVs), a world-wide health threat [14,15]. These FVs have genomes comprising (+)sense single-stranded RNA that contain conserved 5′ and 3′ untranslated regions (UTRs) [16]. FV infection results in accumulation of both the genomic RNA and subgenomic flaviviral RNAs (sfRNAs); the latter are associated with infection-induced cytopathicity and pathogenicity [17]. sfRNAs interact with many cellular proteins, provide interferon resistance, alter mRNA degradation rates, and effect miRNA-dependent pathways [18–31]. The mechanism of sfRNA production depends on partial degradation of the FV genomic RNA by Xrn1. The enzyme loads and progresses in a 5′→3′ direction until it halts at defined locations in the genome’s 3′ UTR; the protected RNAs are sfRNAs [17]. Thus, this is an example of a virus not only confounding the decay machinery, but exploiting it to the viruses’ advantage (Figure 2a). This ability to resist Xrn1 is surprising as Xrn1 can degrade highly structured RNAs [32,33]. Initial characterization of the 3′ UTRs of diverse FVs suggested that Xrn1 resistance occurs at conserved RNA “stem-loop” (SL) secondary structures in the 3′ UTR [17,34–36]. Subsequent biophysical and biochemical characterization of isolated SL structures (dubbed Xrn1-resistant RNAs or xrRNAs) showed them to be compactly folded RNAs whose structure and function depends on conserved nucleotides [37].
Figure 2.
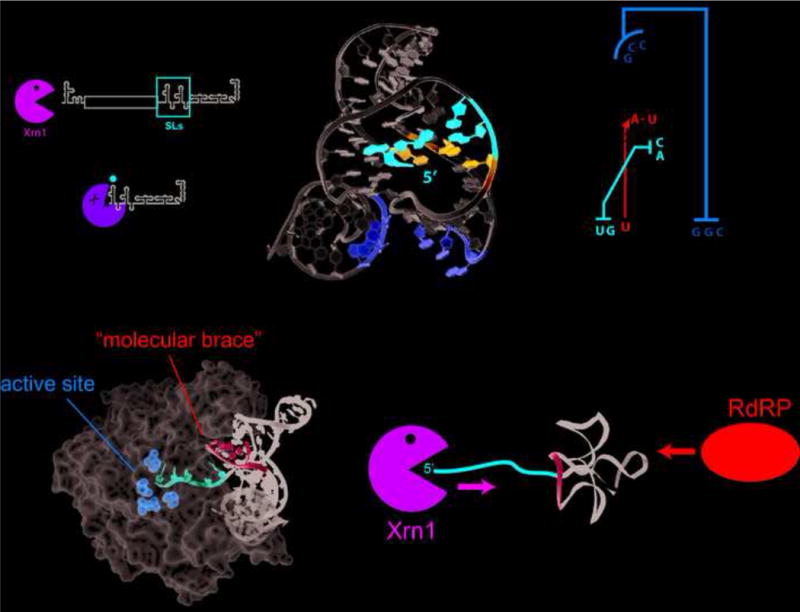
Structure of Xrn1-resistant RNAs from mosquito-borne FVs. (a) These FV genomes contain conserved 5′ and 3′ untranslated regions (UTRs) with a single open reading frame (ORF) encoding multiple viral proteins. Xrn1 digestion of the genomic RNA leads to stalling of Xrn1 at conserved stem-loop (SL) structures in the 3′ UTR, generating subgenomic flaviviral RNAs (sfRNAs). (b) Structure (left) and secondary structure diagram (right) of an xrRNA from Murray Valley encephalitis (MVE) virus (PDB: 4PQV). The 5′ end of the RNA is inserted through a ring-like element formed by the three-helix junction of this RNA, facilitated by conserved long-range base pairs (red) and a base-triple interaction (blue). This is further stabilized by a pseudoknot interaction (yellow) that was formed in the crystal as a domain-exchanged crystal contact between separate molecules. (c) The three-dimensional structure was modeled onto the structure of the Drosophila melanogaster Xrn1 protein bound to a DNA substrate analog (PDB: 2Y35). The model suggests that the xrRNA provides a molecular brace (cyan) allowing only 5 nts of single-stranded RNA (red) into the active site of the enzyme consistent with the previously mapped halt site of Xrn1[37]. (d) The molecular brace is positioned to prevent 5′→3′ degradation by Xrn1, however it does not shelter the 3′ end which can be readily denatured by enzymes acting in a 3′→5′ manner, such as the viral RdRP which must replicate the viral genome.
Insight into this mechanism of Xrn1 resistance was provided by the structure of an xrRNA from Murray Valley encephalitis (MVE) virus solved by x-ray crystallography [38](Figure 2b). The xrRNA’s three-dimensional fold is centered on a three-helix junction that forms an unusual ring-like architecture in which the 5′ end of the RNA passes from one side of the structure to the other. This fold depends on a second, unpredicted pseudoknot formed by base-pairing between the 5′ end of the resistant RNA and nucleotides in the three-way junction. Interestingly, the crystal structure captured the xrRNA in a state where the predicted pseudoknot was not formed, suggesting this element may be conformationally dynamic. Modeling of the xrRNA structure onto the structure of D. melanogaster Xrn1 [39] suggested that the ring-like structure of the xrRNA contacts the enzyme surface around the entrance to the active site, perhaps acting like a molecular brace (Figure 2c). This may shelter the next base-pair to be unwound from the domains of Xrn1 that provide helicase-like activity. Thus, the structure suggests Xrn1 resistance is conferred in part by a mechanical unfolding problem the enzyme cannot resolve. The structure also suggests how the viral RNA-dependent RNA polymerase (RdRP) traveling in the 3′→5′ direction can readily traverse this structure: the molecular brace will only be encountered by processive molecules (such as Xrn1) seeking to unwind the structure from the 5′ side (Figure 2d).
sfRNAs affect several pathways in infected cells [18,21,24,25,27–31], one of these appears to be directly related to the Xrn1 resistance that forms sfRNA. Data suggest that after halting, Xrn1 remains bound to the resistant structure and this Xrn1 sequestration dysregulates cellular mRNA decay [21,40]. Evidence for this includes cell-based examination of the alteration of mRNA levels as a result of sfRNA production and demonstration of the ability of xrRNAs to protect other RNAs from Xrn1 in trans [21]. Thus, there may be specific contacts formed between the xrRNA and Xrn1 that go beyond the “mechanical brace” model proposed above. The molecular details of the sfRNA-Xrn1 interaction remain unexplored but may provide insight into how a single folded RNA can globally alter RNA degradation patterns [41].
Preventing access to the 3′ end: KSHV PAN and MALAT1 RNAs
Unlike the FV xrRNAs that block Xrn1’s 5′→3′ exonuclease activity, other structured RNAs inhibit 3′→5′ degradation by the exosome. The two discussed here are the Kaposi’s sarcoma–associated herpesvirus (KSHV) polyadenylated expression and nuclear retention element (PAN ENE) and a similar cellular RNA element found on the 3′ end of the processed metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) RNA (Figure 3a). Like the FV xrRNAs these are both RNAs with direct importance to human health. The PAN ENE RNA is a highly abundant long non-coding RNA expressed in the nucleus during the lytic phase of KSHV infection whereas the MALAT1 RNA was identified as a highly stable nuclear marker in different lung cancers by subtractive hybridization [42,43]. These RNAs have exceptionally long half-lives and significant sequence homology, and functional studies suggested both sequester the 3′ A-rich or poly(A) sequence from the exosome within in a triple helix structure [44–46]
Figure 3.
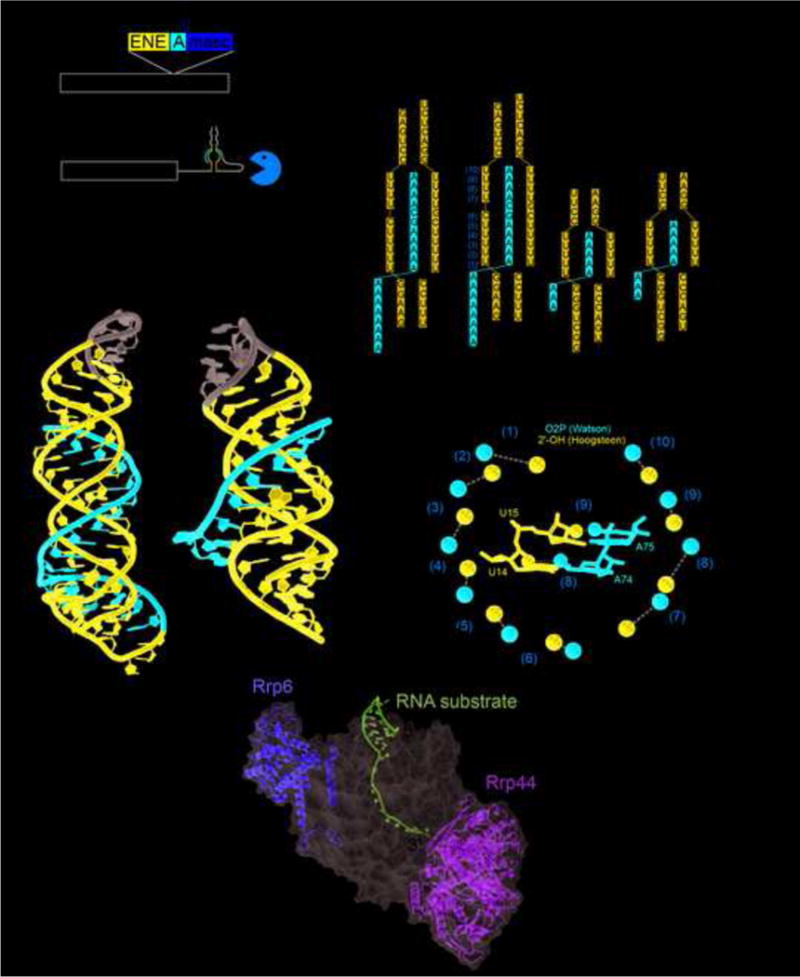
MALAT1 and PAN ENE crystal structures. (a) Schematic of the human MALAT1 transcript and the product after processing by RNase P (cut site indicated by arrowhead at nucleotide 8356). The exosome is able to degrade the unprocessed RNA, but the exosome is significantly hindered once the MALAT1 structure is at the 3′ end. (b) The crystal structure of MALAT1 (PDB: 4PLX) and PAN ENE (PDB: 3P22). The Watson strand with the 3′ end is in red, the Crick and Hoogsteen strands are in blue, and the engineered tetraloops are in gray. (c) The secondary structure of the conserved sequences and constructs crystallized for MALAT1 and PAN ENE. Leontis and Westhof symbols indicate non-WC base pairs. (d) Crystal structure of the 12-subunit exosome; a bound structured RNA with the 3′ end extending into the exosome is shown in magenta. Nucleases Rrp6 and Rrp44 are yellow and green, and the rest of the exosome is represented as a semi-transparent gray surface. (PDB: 5C0X). (e) The OP2 and 2′ OH of the Watson and Hoogsteen strands are shown as red or blue spheres respectively. The distances between them are given and orange numbering corresponds to the secondary structure in panel (b). The grey arrow depicts moving along the Watson strand from the 5′ to the 3′ direction and the location of the G-C pair in the middle that provides the “reset.” In the middle the same coloring is used to highlight interactions of the U14 and A74 (pair (8)) and U15 and A75 (pair (9)).
Structures of both of these RNA elements solved by x-ray crystallography demonstrate how they resist 3′→5′ decay by sequestering the 3′ end from the unwinding and endonuclease activities of the exosome [45,46] (Figure 3b). In both structures, an A-rich 3′ end engages both sides of a U-rich internal loop to form a series of U·A-U base triples. Specifically, the A-rich tail forms A-U Watson-Crick base-pairs with one side of the U-rich internal loop, and the other side of the U-rich loop forms U·A Hoogsteen interactions with these Watson-Crick pairs (Figure 3c). Mutations that disrupt these interactions render the RNA unable to resist 3′→5′ degradation [45–49]. Hence, by engaging the A-rich 3′ tail in stable RNA structure, the PAN ENE and MALAT1 achieve structural sequestration that presumably prevents efficient loading and progression of the exosome, which requires ~30 nucleotides of single-stranded RNA [50,51] (Figure 3d). Despite a very similar structural strategy, differences between PAN and MALAT1 show how an RNA structure can be adjusted to achieve different behaviors. The PAN RNA allows for sequestration almost anywhere along a long 3′ poly(A) tail, thus the tail can overhang the structure [45]. As a result, PAN RNA exhibits biphasic decay kinetics with rapid degradation of the overhanging tail then slow decay through the RNA triple-helix [46,47,52]. In contrast, MALAT1 has a specific sequence and length resulting in the 3′ end forming a blunt ended triple-helix without an overhang and a correspondingly slow single decay rate [46]. This poly(A) sequestration strategy may be widespread, as potentially similar elements have been found in cellular MEN β and Sno-lncRNAs [53] and the Plautia stali intestine virus genomic RNA [49].
The MALAT1 RNA also reveals a potentially fundamental discovery about RNA triple helices. Processed MALAT1 does not have a continuous poly(A) tail, it has the sequence AAAAAGCAAAA at its 3′ end. All nine adenosines are in U·A-U triples, there is a C+·G-C interaction for the guanine, and a Watson-Crick G-C base-pair for the cytosine. Why the G-C base-pair in the middle? The structure reveals that the distance between the 2′ OH of the Hoogsteen strand and the O2P of the Watson strand decreases with each step along the 3′ tail (Figure 3e). Modeling revealed a steric clash between these two atoms if a seventh base triple were present, likely limiting the maximum length of such a triple helix [46]. The G-C base-pair “resets” the helical register, allowing for the start of another triple-helix segment [46]. This understanding of basic RNA structure will be valuable in predicting the behavior and structures of other RNAs with potentially long runs of base triples, including those that use this as a 3′ end protection strategy.
Mimicry, conformational plasticity, and chemical modification: the TYMV TLS RNA
The third structure we discuss is the tRNA-like structure (TLS) found at the extreme 3′ end of the turnip yellow mosaic virus (TYMV) (for review: [54]). The TYMV TLS is a substrate for tRNA-modifying enzymes including cellular amino-acyl-tRNA synthetases (aaRS) that add an amino acid to the viral genome’s 3′ terminus (a valine for TYMV) [55,56], allowing binding of eukaryotic elongation factor 1a (eEF1A). TLS RNAs play multiple roles during infection including enhancing translation and doubling the half-life of the viral RNA to a similar degree as a poly-A tail but less than the PAN or MALAT1 RNAs [57]. The TLS’s ability to enhance stability (and its other functions), depends on 3′ aminoacylation [54]. Overall, the TYMV TLS is a multifunctional RNA that appears to have structural plasticity built in: it must mimic a tRNA, but must also readily unfold to serve as a template for replication.
The global conformation of the TYMV TLS matches the classic tRNA L-shaped structure [58,59] (Figure 4a,b). One “face” of the structure mimics a tRNA to interact with the aaRS and with eIF1a [59,60] (Figure 4c). However, the other face of the TLS and many of the interactions between elements analogous to a tRNA’s D loop, T loop, and V loop, are very different from a tRNA’s [58,59] (Figure 4b). Whereas a tRNA’s structure is held together by a large number of tertiary contacts, the TLS relies on a more limited set; this likely is what enables the RNA to readily unfold to allow replication.
Figure 4.
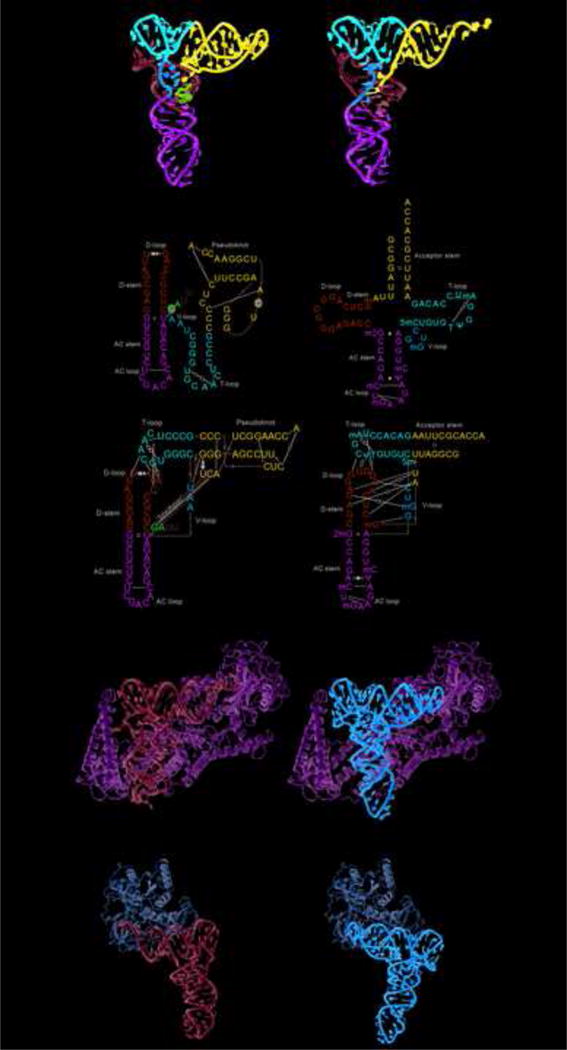
Structure of the TYMV TLS and tRNA. (a) The crystal structures of the TYMV TLS (PDB: 4P5J) and tRNAPhe (PDB: 1EHZ) with analogous structural elements labeled. The AC loops and stems are green, the D-loops and D-stems are cyan, the T-loop is red, and the V-loop is orange. The pseudoknot of the TLS and acceptor stem of the tRNA are blue and the linchpin region of the TLS is magenta. (b) Secondary structures, depicted several ways, of the TLS and tRNA using the coloring of panel (a). Leontis and Westhof symbols indicate non-WC base pairs. The secondary structures on the top compare the TLS to the standard tRNA cloverleaf depiction, those on the bottom emphasize the interactions that stabilize each fold. (c) Left: Structure of a valine amino-acyl-tRNA synthetase (ValRS) (light green) bound to cognate tRNA (cyan) (PDB: 1GAX). Right The model of the TYMV TLS (orange) bound to ValRS (light green) made by aligning the TLS to the tRNA’s T-loop and stem regions. (d) Left: Structure of EF-Tu (bacterial homolog of eEF1A) bound to a tRNA with the protein in light brown and the tRNA in cyan (PDB: 1TTT). Left: Model of the TYMV TLS bound to the protein made by aligning the TLS with the tRNA’s by the T-loop and stem regions.
There are several possible mechanisms by which the TYMV TLS RNA structure may increase the lifetime of the RNA to which it is affixed. The presence of a stable and complex RNA structure on the 3′ end of the RNA may inhibit loading of the exosome, aminoacylation may create a chemically invalid substrate for the exosome, or eIF1a binding may block access by the decay machinery. The TLS has been shown to bind to the ribosome in vitro, if this occurs in cells it could also sequester the 3′ end from exonuclease activity [59]. Because it looks like a tRNA, the TLS could also be subject to the rapid tRNA decay pathway (RTD), which uses Met22, Rat1, and Xrn1 to degrade hypomodified tRNA, such as tRNAVal lacking 7mG and 5mC modifications in V loop and T stem [61]. The TLS could avoid this because its 5′ end is in a pseudoknot structure and it has a structurally divergent V loop. Overall, the TYMV TLS’s ability to enhance the stability of the viral RNA may be due to a combination of these effects; understanding how these are integrated with the TLS’ other functions promises to give new insight into how structurally plastic and multifunctional RNAs might control mRNA decay.
Conclusions
The three structures discussed here reveal different putative strategies for evading the 5′→3′ and 3′→5′ degradation machinery: 1) A mechanical unfolding problem that stymies Xrn1, 2) Sequestration of the poly(A) tail to hide from the exosome, and 3) A likely combination of chemical alteration, structural stability, and protein binding. The diversity of structures and strategies underscores the rich repertoire available to alter the decay fate of an mRNA and also suggests there are more RNA three-dimensional strategies yet to be discovered. In addition, although the MALAT1 RNA is an example of a cellular RNA that is using a strategy also found in viruses (PAN RNA), RNAs operating analogously to the TYMV TLS and FV xrRNAs have not been found in cellular RNAs. The search for such elements is a prime area for ongoing inquiry as is the search for more diverse structured RNAs that manipulate and regulate the powerful RNA degradation machinery.
HIGHLIGHTS.
RNAs can inhibit RNA degrading enzymes using three-dimensional folded structures.
Many of these RNAs are viral, teaching lessons about analogous cellular RNAs.
Recent structures reveal how these RNAs confound or evade degradation machinery.
Acknowledgments
The authors wish to thank members of the Kieft Lab for helpful discussions. Viral RNA structural studies in the Kieft Lab are supported by National Insitutes of Health grants GM081346 and GM097333 to JSK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 3.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harigaya Y, Parker R. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA. 2010;1:132–141. doi: 10.1002/wrna.17. [DOI] [PubMed] [Google Scholar]
- 6.Klauer AA, van Hoof A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip Rev RNA. 2012;3:649–660. doi: 10.1002/wrna.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 9.Stevens A, Wang Y, Bremer K, Zhang J, Hoepfner R, Antoniou M, Schoenberg DR, Maquat LE. Beta -Globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc Natl Acad Sci U S A. 2002;99:12741–12746. doi: 10.1073/pnas.192442399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau PW, MacRae IJ. The molecular machines that mediate microRNA maturation. J Cell Mol Med. 2009;13:54–60. doi: 10.1111/j.1582-4934.2008.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2013;4:423–435. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2012;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 15.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 17••.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. Demonstrates that sfRNAs are linked to cytopathicity and pathogenicity during West Nile Virus infection. They identify that this cytopathic effect relies on Xrn1-resistant RNA elements. [DOI] [PubMed] [Google Scholar]
- 18.Clarke BD, Roby JA, Slonchak A, Khromykh AA. Functional non-coding RNAs derived from the flavivirus 3′ untranslated region. Virus Res. 2015;206:53–61. doi: 10.1016/j.virusres.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 19.Bidet K, Garcia-Blanco MA. Flaviviral RNAs: weapons and targets in the war between virus and host. Biochem J. 2014;462:215–230. doi: 10.1042/BJ20140456. [DOI] [PubMed] [Google Scholar]
- 20.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011;8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. The authors demonstrate that sfRNAs inhibit the activity of Xrn1 on other transcripts, suggesting that sfRNAs may form long-lived inhibitory complexes with the Xrn1 enzyme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon SL, Wilusz J. Cytoplasmic viruses: rage against the (cellular RNA decay) machine. PLoS Pathog. 2013;9:e1003762. doi: 10.1371/journal.ppat.1003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emara MM, Liu H, Davis WG, Brinton MA. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J Virol. 2008;82:10657–10670. doi: 10.1128/JVI.00991-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015 doi: 10.1126/science.aab3369. This manuscript underscored the importance of the flaviviral sfRNA as a determinant of Dengue virus fitness. They showed the sfRNA binds to cellular protein TRIM25 as part of its strategy to inhibit the inate immune response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Liu H, Zou J, Zhang B, Yuan Z. Dengue virus subgenomic RNA induces apoptosis through the Bcl-2-mediated PI3k/Akt signaling pathway. Virology. 2014;448:15–25. doi: 10.1016/j.virol.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang RY, Hsu TW, Chen YL, Liu SF, Tsai YJ, Lin YT, Chen YS, Fan YH. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet Microbiol. 2013;166:11–21. doi: 10.1016/j.vetmic.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 29.Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan YH, Nadar M, Chen CC, Weng CC, Lin YT, Chang RY. Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans. Virol J. 2011;8:492. doi: 10.1186/1743-422X-8-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Roby JA, Pijlman GP, Wilusz J, Khromykh AA. Noncoding Subgenomic Flavivirus RNA: Multiple Functions in West Nile Virus Pathogenesis and Modulation of Host Responses. Viruses-Basel. 2014;6:404–427. doi: 10.3390/v6020404. This review provides an excellent introduction to, and overview o, the formation and function of sfRNAs during flaviviral infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5′–>3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta. 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5′–>3′ exoribonuclease Xrn1. Nat Struct Mol Biol. 2011;18:270–276. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Yue L, Li X, Yu X, Zhao H, Jiang Z, Qin E, Qin C. Identification and characterization of small sub-genomic RNAs in dengue 1–4 virus-infected cell cultures and tissues. Biochem Biophys Res Commun. 2010;391:1099–1103. doi: 10.1016/j.bbrc.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 37••.Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife. 2014;3:e01892. doi: 10.7554/eLife.01892. The manuscript established that Xrn1 resistance by structured RNAs in flavivirus is due to a compactly folded RNA, with the fold centered around a three-way junction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. This paper presents the high-resolution structure of an xrRNA from Murray Valley Encephalitis Virus, detailing how the three-dimensional fold of the RNA accomplishes Xrn1 resisitance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinek M, Coyle SM, Doudna JA. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charley PA, Wilusz J. Sponging of cellular proteins by viral RNAs. Curr Opin Virol. 2014;9:14–18. doi: 10.1016/j.coviro.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Villordo SM, Filomatori CV, Sanchez-Vargas I, Blair CD, Gamarnik AV. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog. 2015;11:e1004604. doi: 10.1371/journal.ppat.1004604. The manuscript revealed that cycling of Dengue virus between human and mostuito hosts is associated with a build-up of mutations within the structures that act to inhibit Xrn1 and create the viral sfRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 44.Conrad NK, Steitz JA. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. This manuscript presented the crystal structure of the PAN element from Kaposi’s sarcoma-associated herpesvirus, revelaing the formation of triple helix that protects the 3′ end from decay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. The mansucript presents the crystal structure of the MALAT1 ENE, revlealing how it forms a triple helix to sequester the 2′ end from degradation. This paper also revealed a potentially fundamental feature of RNA triple helices: the need to “reset.”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci U S A. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tycowski KT, Shu MD, Borah S, Shi M, Steitz JA. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Makino DL, Baumgartner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- 51.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 52.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 53.Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dreher TW. Viral tRNAs and tRNA-like structures. Wiley Interdiscip Rev RNA. 2010;1:402–414. doi: 10.1002/wrna.42. [DOI] [PubMed] [Google Scholar]
- 55.Yot P, Pinck M, Haenni AL, Duranton HM, Chapeville F. Valine-specific tRNA-like structure in turnip yellow mosaic virus RNA. Proc Natl Acad Sci U S A. 1970;67:1345–1352. doi: 10.1073/pnas.67.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinck M, Yot P, Chapeville F, Duranton HM. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature. 1970;226:954–956. doi: 10.1038/226954a0. [DOI] [PubMed] [Google Scholar]
- 57.Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 58.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59••.Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature. 2014;511:366–369. doi: 10.1038/nature13378. This paper presented the high-resolution structure of the tRNA-like structure from the Turnip Yellow Mosaic Viurs, revealing its two-faced architecture and novel folding patterns. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 61.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]


