Abstract
Influenza virus infections represent a worldwide public health and economic problem due to the significant morbidity and mortality caused by seasonal epidemics and pandemics. Sensitive and convenient methodologies for detection of influenza viruses are essential for further disease control. Loop-mediated isothermal amplification (LAMP) is the most commonly used method of nucleic acid isothermal amplification. However, with regard to multiplex LAMP, differentiating the ladder-like LAMP products derived from multiple targets is still challenging today. The requirement of specialized instruments has further hindered the on-site application of multiplex LAMP. We have developed an integrated assay coupling multiplex reverse transcription LAMP with cascade invasive reaction using nanoparticles (mRT-LAMP-CIRN) as a sensor for the detection of three subtypes of influenza viruses: A/H1N1pdm09, A/H3 and influenza B. The analytic sensitivities of the mRT-LAMP-CIRN assay were 101 copies of RNA for both A/H1N1pdm09 and A/H3, and 102 copies of RNA for influenza B. This assay demonstrated highly specific detection of target viruses and could differentiate them from other genetically or clinically related viruses. Clinical specimen analysis showed the mRT-LAMP-CIRN assay had an overall sensitivity and specificity of 98.3% and 100%, respectively. In summary, the mRT-LAMP-CIRN assay is highly sensitive and specific, and can be used as a cost-saving and instrument-free method for the detection of influenza viruses, especially for on-site use.
Keywords: multiplex, LAMP, gold nanoparticles, on-site detection, influenza virus
Introduction
Influenza viruses pose an ongoing threat to human health principally through their ability to cause acute respiratory diseases worldwide.1,2 Influenza virus evolves rapidly by reassortment and genetic drift and is one of the most common causes of morbidity and mortality, leading to a significant economic impact every year.3,4 According to the World Health Organization (WHO), seasonal influenza infections result in ~3–5 million severe cases and 300,000–500,000 deaths around the world yearly.5 A rapid and sensitive methodology for the early detection of seasonal influenza viruses is of paramount importance to facilitate clinical care, infection control, as well as epidemiologic investigations. Among the diagnostic tools, nucleic acid-based tests are more rapid and sensitive than traditional techniques such as serological assays and influenza virus isolation. At present, real-time reverse transcription polymerase chain reaction (RT-PCR) is a powerful molecular diagnostic method for influenza virus infections.6–9 In contrast to PCR-based target gene detection, which requires bulky and expensive equipments and highly skilled technicians, recent research has focused on the development of isothermal amplification methods for simple and user-friendly pathogen detection. As the reaction is conducted under isothermal conditions, the thermal cycler is not required, which makes these methods more suitable for use in resource-limited regions or for field use.
As the most commonly used method of isothermal amplification, loop-mediated isothermal amplification (LAMP) allows amplification of DNA with high sensitivity and specificity.10,11 LAMP can also be used to detect RNA template by simply adding a reverse transcriptase enzyme under identical reaction conditions as LAMP.10,12 To date, RT-LAMP methods have been developed to detect various RNA viruses, including influenza viruses.13–17 Multiplex detection represents the development trend of molecular diagnostic methods due to the properties of cost- and time-saving. Although previous studies have described several multiplex LAMP methods, how to differentiate the ladder-like LAMP products derived from multiple targets is still challenging. These methods used either end-point (pyrosequencing,18 gel electrophoresis,19 etc) or real-time (annealing curve,20 fluorescence technology,21,22 etc) analysis to differentiate the multiple target sequences. The main drawback of these strategies is the requirement of complicated and specialized instruments, which hinders the on-site use of multiplex LAMP.
To take full advantage of multiplex LAMP, the product identification methodology should be simple and cost-effective. Hence, the colorimetric detection would be ideal due to its simplicity, user-friendliness, and ease to interpret the result. Utilization of oligonucleotide probe-modified gold nanoparticles (AuNPs), the aggregation of which can cause notable changes in optical property,23–25 seems to satisfy this condition. In the present study, we developed a multiplex RT-LAMP (mRT-LAMP) assay for simultaneous amplification of three subtypes of influenza viruses (influenza A/H1N1pdm09, A/H3 and influenza B), and the RT-LAMP amplicons were further identified by cascade invasive reaction26,27 using oligonucleotide probe-modified AuNPs as a sensor. The results can be observed and determined easily based on color changes by naked eyes. mRT-LAMP coupled with cascade invasive reaction using nanoparticles as a sensor (mRT-LAMP-CIRN) provides us a sensitive, specific, cost-saving and instrument-free diagnostic tool for the identification of influenza viruses, especially in resource-limited settings.
Materials and methods
Viral isolates and clinical specimens
Three influenza virus strains A/Jiangsu/2/2009(H1N1), A/Nanjing/1/2009(H3N2) and B/Jiangsu/01/2015 isolated from patient specimens at Jiangsu Provincial Center for Disease Control and Prevention were used for the development of mRT-LAMP-CIRN assay. Eight clinically or genetically related viruses were used as control to evaluate the specificity of the assay, including seasonal influenza virus A/H1N1, avian influenza viruses A/H5N1, A/H7N9 and A/H9N2, respiratory syncytial viruses types A and B, and parainfluenza viruses types 1 and 2. A total of 94 nasopharyngeal swab specimens collected from influenza-like cases, including 58 known influenza-positive specimens (35 A/H1N1pdm09, 14 A/H3N2, and nine influenza B), were used to validate the mRT-LAMP-CIRN assay. All the isolates and specimens were kept at −80°C until use. Written informed consent regarding the use of clinical specimens for investigational purposes was obtained from all the patients involved. This study was approved by the Ethics Committee of the Jiangsu Provincial Center for Disease Control and Prevention. All the experiments were carried out in accordance with the ethical guidelines for medical and health research involving human subjects.
Isolation of viral RNA
Viral RNA was extracted from 200 μL of specimens with a High Pure viral RNA kit (Roche Diagnostics, Manheim, Germany) according to the manufacturer’s instructions. Purified RNA was eluted with 50 μL of elution buffer (pH =8.0) and kept at −80°C until further analysis.
Preparation of RNA standards
The hemagglutinin (HA) gene, matrix protein 1 (M1) gene and nonstructural protein 1 (NS1) gene were amplified from the influenza viruses A/Jiangsu/2/2009(H1N1), A/Nanjing/1/2009(H3N2) and B/Jiangsu/01/2015 with primers containing T7 promoter sequence on reverse sides. RNA was transcribed directly from purified PCR amplicons containing the full-length region of each target for mRT-LAMP by the use of T7 RNA polymerase (TaKaRa Biotechnology Co. Ltd., Dalian, People’s Republic of China). The synthetic RNA transcripts were purified and quantified by reading absorbance at A260 nm,28 and then ten-fold diluted as RNA standards ranging from 104 to 100 RNA copies/μL.
Design of primers and probes
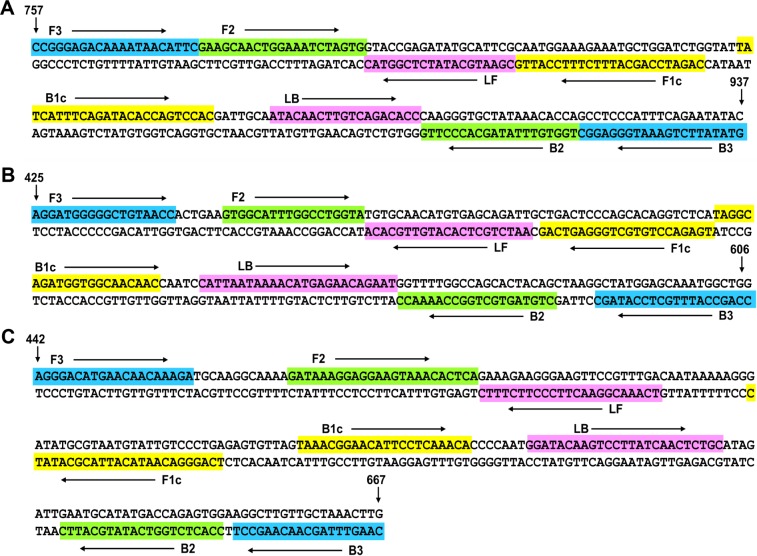
mRT-LAMP primers were designed based on the conserved regions of HA gene, M1 gene and NS1 gene for influenza A/H1N1pdm09, A/H3 and influenza B, respectively. RT-LAMP primers were designed using a PrimerExplorer version 4 program on the Eiken website (http://primerexplorer.jp/e/). A set of six primers, including two outer primers (F3/B3), two inner primers (FIP/BIP), and two loop primers (LF/LB), were selected for each of the three gene targets. The specificity and feasibility of the selected primers were subsequently checked by Basic Local Alignment Search Tool search against National Center for Biotechnology Information database. The sequence information of the selected final mRT-LAMP primers containing degenerate bases is shown in Table 1, and the details of the primers are shown in Figure 1.
Table 1.
Primers used for multiplex RT-LAMP assay
| Target | Name | Type | Sequence (5′-3′) | Length (nt) |
|---|---|---|---|---|
| A/H1N1pdm09, (HA gene) | 09H1-F3 | Forward outer | CCGGGAGACAAAATAACATTC | 21 |
| 09H1-B3 | Reverse outer | GTATATTCTGAAATGGGAGGC | 21 | |
| 09H1-FIB | Forward inner | CAGATCCAGCATTTCTTTCCATTGGAAG CAACTGGAAATCTAGTG(F1C+F2) |
45 | |
| 09H1-BIP | Reverse inner | TATCATTTCAGATACACCAGTCCACTGG TGTTTATAGCACCCTTG(B1C+B2) |
45 | |
| 09H1-LF | Forward loop | CGAATGCATATCTCGGYAC | 19 | |
| 09H1-LB | Reverse loop | ATACAACTTGTCARACACC | 19 | |
| A/H3, (M1 gene) | H3-F3 | Forward outer | AGGATGGGGGCTGTAACC | 18 |
| H3-B3 | Reverse outer | CCAGCCATTTGCTCCATAGC | 20 | |
| H3-FIB | Forward inner | TGAGACCTGTGCTGGGAGTCAGGTG GCMTTTGGCCTGGTA |
40 | |
| H3-BIP | Reverse inner | TAGGCARATGGTGGCAACAACCTGT AGTGCTGGCCARAACC |
41 | |
| H3-LF | Forward loop | AATCTGYTCACATGTTGCACA | 21 | |
| H3-LB | Reverse loop | CATTAATAARACATGAGAACAGAAT | 25 | |
| Influenza B, (NS1 gene) | FluB-F3 | Forward outer | AGGGACATGAACAACAAAGA | 20 |
| FluB-B3 | Reverse outer | CAAGTTTAGCAACAAGCCT | 19 | |
| FluB-FIB | Forward inner | TMARGGACAATACATTACGCATATC GATAAAGGAGGAAGTAAACACTCA |
49 | |
| FluB-BIP | Reverse inner | TAAAYGGAACATTCCTCAAACACCACTC TGGTCATAGGCATTC |
43 | |
| FluB-LF | Forward loop | TYAAACGGAACTTCCCTTCTTTC | 23 | |
| FluB-LB | Reverse loop | GGATACAAGTCCTTATYAACTCTGC | 25 |
Notes: The polymorphic sites of the primers are underlined; Y represents degenerate C or T; R represents degenerate A or G; M represents degenerate A or C.
Abbreviations: RT-LAMP, reverse-transcription loop-mediated isothermal amplification; HA, hemagglutinin; M1, matrix protein 1; NS1, nonstructural protein 1; nt, nucleotide.
Figure 1.
Schematic showing location of multiplex RT-LAMP primer binding sites.
Notes: (A) HA gene of influenza A/H1N1pdm09. Assay spans region from nucleotide 757–937 with reference to the HA gene sequence of the H1N1pdm09 virus strain A/Jiangsu/2/2009(H1N1). (B) M1 gene of influenza A/H3N2. Assay spans region from nucleotide 425–606 with reference to the M1 gene sequence of the H3N2 virus strain A/Nanjing/1/2009(H3N2). (C) NS1 gene of influenza B. Assay spans region from nucleotide 442–667 with reference to the NS1 gene sequence of the influenza B virus strain B/Jiangsu/01/2015.
Abbreviations: HA, hemagglutinin; M1, matrix protein 1; NS1, nonstructural protein 1; RT-LAMP, reverse-transcription loop-mediated isothermal amplification.
Using the Universal Invader Design Software version 1.2.4 (Third Wave Technologies, Inc., Madison, WI, USA), three sets of cascade invasive reaction upstream and downstream probes were designed based on the sequences of mRT-LAMP amplicons. The sequence information of the selected upstream, downstream and hairpin probes used in secondary invasive reaction, as well as two types of oligonucleotide probes (nanoparticle probes) used for modification of AuNPs is shown in Table 2. All the primers and probes were synthesized and modified by TaKaRa Biotechnology Co. Ltd.
Table 2.
Probes used for cascade invasive reaction and modification of AuNPs
| Target | Name | Type | Sequence (5′-3′) | Modification |
|---|---|---|---|---|
| A/H1N1pdm09 | 09H1-Up | Up | ATCTGGTATTATCATTTCAGATACACCAGTT | |
| 09H1-Dp | Dp | CGCGCCGAGGCCACGATTGCAATACAAC | ||
| A/H3 | H3-Up | Up | GTCTCATAGGCAGATGGTGGCT | |
| H3-Dp | Dp | CGCGCCGAGGAACAACCAATCCATTAATAAAAC | ||
| Influenza B | FluB-Up | Up | TGTATTGTCCTTGAGAGTGTTGGTAAAT | |
| FluB-Dp | Dp | CGCGCCGAGGCGGAACATTCCTCAAACA | ||
| Universal | Hp | Hp | GTCTTGTGGTACTGCACTCGTCTCGGTTTTCCGA GACGAGTCCTCGGCGCGATCGTGATGAACCAT |
|
| 3′-AuNP | nanoparticle probe | GCAGTACCACAAGACAAAAAAAAAA | 3′-SH C6 | |
| 5′-AuNP | nanoparticle probe | AAAAAAAAAAATGGTTCATCACGAT | 5′-SH C6 |
Abbreviations: AuNPs, gold nanoparticles; Up, upstream probe; Dp, downstream probe; Hp, hairpin probe.
Preparation and modification of AuNPs
AuNPs were synthesized by reducing tetrachloroauric acid with trisodium citrate, and then modified with oligonucleotide probes according to the protocol described previously.29 Two types of nanoparticle probes (Table 2), which were complementary to the hairpin probe, were used to modify the two batches of AuNPs separately. Transmission electron microscopy (TEM) (JEM 2100F; JEOL Co. Ltd., Tokyo, Japan) was used to measure the sizes of oligonucleotide probe-modified AuNPs, which resulted in an average diameter of 13 nm. The ultraviolet (UV) spectrum and TEM images of oligonucleotide probe-modified AuNPs are described in detail in the Supplementary material.
mRT-LAMP reaction
mRT-LAMP reaction was carried out using the RNA Amplification Kit (Eiken China Co., Ltd., Shanghai, People’s Republic of China) in a final volume of 20 μL containing 10 μL of Reaction Mix, three sets of primers for influenza A/H1N1pdm09, A/H3 and influenza B (0.15 μM each of F3 and B3 primers, 1.2 μM each of FIP and BIP primers and 0.6 μM each of LF and LB primers), 0.8 μL of Enzyme Mix and 1 μL of RNA template. Distilled water was used as no-template control. Twenty-five microliters of mineral oil (Sigma-Aldrich, St Louis, MO, USA) was added to the top of the reaction mixture. mRT-LAMP reactions were run at different temperatures including 60°C, 63°C and 65°C using a real-time turbidimeter (LA320C; Teramecs, Tokyo, Japan) in order to determine the optimal reaction temperature for obtaining the highest amplification efficiency.
Cascade invasive reaction and AuNPs hybridization
Three target-specific cascade invasive reactions for each mRT-LAMP reaction were performed in a volume of 20 μL containing 1× reaction buffer (10 mM 3-(N-morpholino) propanesulfonic acid, pH 7.5, 0.05% Tween-20, 0.05% Nonidet P40 and 7.5 mM MgCl2); 0.2 μM each of upstream and downstream probes; 0.2 μM hairpin probe; 100 ng Archaeoglobus fulgidus flap endonuclease (AfuFEN) enzyme, which was prepared in our lab as described previously;30 and 5 μL diluted RT-LAMP amplification products (30 μL deionized water was added to 20 μL products). The mixtures were then incubated at 85°C for 1 min followed by 63°C for 20 min. After cascade invasive reaction, 3 μL of each previously prepared oligonucleotide probe-modified AuNPs (30 nM) and 5 μL of NaCl (500 mM) were together added into each reaction tube. Hybridization was performed at 55°C for 30 min. Then, the result was observed with naked eyes directly or after brief centrifugation (1,000× g for 30 s) of the products. The negative reaction mixture became colorless because of the aggregation of AuNPs, while the positive reaction mixture remained red.
Analytical sensitivity and specificity of mRT-LAMP-CIRN
To evaluate the analytical sensitivity of the mRT-LAMP-CIRN assay, synthetic RNA standard for each target was tenfold serially diluted (ranging from 104 to 100 RNA copies/μL) and was used as mRT-LAMP-CIRN template. The specificity of the assay was assessed by analyzing the RNA extracts from influenza viruses A/Jiangsu/2/2009(H1N1), A/Nanjing/1/2009(H3N2) and B/Jiangsu/01/2015, and from each control virus mentioned above separately. RNA extracts from the three influenza virus strains were also tested in different combinations in a single reaction.
Evaluation of the mRT-LAMP-CIRN assay with clinical specimens
To evaluate the performance characteristics of the mRT-LAMP-CIRN assay, 94 clinical nasopharyngeal swab specimens collected from influenza-like cases were analyzed by mRT-LAMP-CIRN assay with the parallel analysis by real-time RT-PCR where primers and probes were all recommended by the WHO.31,32 Real-time RT-PCR was performed using the SuperScript® III Platinum One-Step qRT-PCR System (Thermo Fisher Scientific, Carlsbad, CA, USA). The cycling conditions were: 50°C for 20 min, 95°C for 2 min, followed by 40 cycles in 15 s at 94°C and 1 min at 60°C.
Results
Principle of mRT-LAMP-CIRN assay
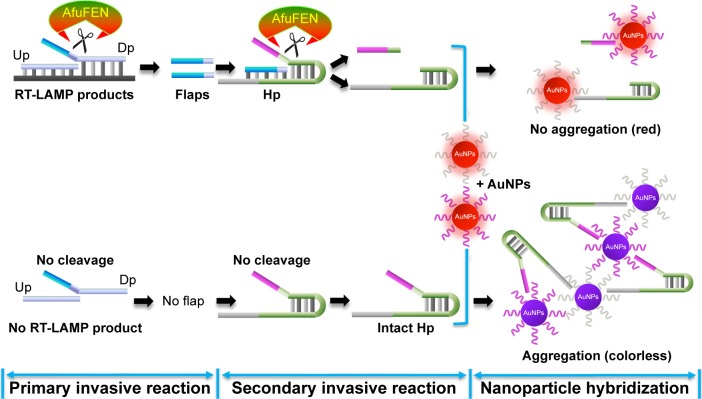
The overall process of the mRT-LAMP-CIRN assay includes RT-LAMP amplification of viral nucleic acids, signal amplification by cascade invasive reaction and optical detection using oligonucleotide probe-modified AuNPs. A schematic of the principles of cascade invasive reaction and signal detection with AuNPs is shown in Figure 2. Briefly, the upstream and downstream probes hybridize with RT-LAMP products, forming a one base overlapping structure at the 3′ end of the upstream probe, which can be recognized and cleaved by AfuFEN enzyme. Then the cleaved 5′ flap of the downstream probe hybridizes with the hairpin probe, which can drive a secondary invasive reaction. To visibly detect whether the hairpin probe is cleaved, two types of oligonucleotide probe-modified AuNPs, which can hybridize with the two ends of hairpin probe, are added. If the target of RT-LAMP product is present, the cleaved hairpin probe cannot trigger the aggregation of oligonucleotide probe-modified AuNPs, leading to red color of the reaction. Conversely, if the target of RT-LAMP product is absent and the hairpin probe is intact without cleavage, leading to the aggregation of AuNPs, then the reaction becomes colorless.
Figure 2.
The principles of cascade invasive reaction and signal detection with AuNPs for detecting multiplex RT-LAMP products.
Notes: The system contains three steps: primary invasive reaction, secondary invasive reaction and nanoparticle hybridization. Primary invasive reaction: the Up, Dp and RT-LAMP amplicon form a one-base overlapping structure at the 3′end of the Up. AfuFEN enzyme specifically recognizes the structure and cleaves the 5′ flap of the Dp. Secondary invasive reaction: the cleaved 5′ flap from the target-specific primary invasive reaction hybridizes with the Hp and drives a secondary invasive reaction. Then the Hp is cleaved by AfuFEN. Nanoparticle hybridization: if the target of RT-LAMP product is present, the cleaved Hp cannot trigger the aggregation of AuNPs, resulting in red-colored dispersions of AuNPs. Conversely, if the target of RT-LAMP product is absent, the Hp remains intact without DNA cleavage, yielding nanoparticle aggregation-induced precipitation, and then the reaction mixture becomes colorless.
Abbreviations: Up, upstream probe; Dp, downstream probe; Hp, hairpin probe; AfuFEN, Archaeoglobus fulgidus flap endonuclease; AuNPs, gold nanoparticles; RT-LAMP, reverse-transcription loop-mediated isothermal amplification.
Optimization of the mRT-LAMP-CIRN assay
The real-time turbidimetry device enables observation of LAMP primer kinetics. To optimize the mRT-LAMP conditions, at the commencement of this study, we used a real-time turbidimeter (LA320C; Teramecs, Tokyo, Japan) to determine the optimal primer sequences, primer concentrations, reaction temperature, as well as amplification time. The optimal primer sequences and concentrations, determined using various concentrations of synthetic RNA standards as templates, are shown in Table 1 and described in Materials and methods. The details of the optimization of reaction temperature and amplification time are described in the Supplementary material.
Analytical sensitivity of the mRT-LAMP-CIRN
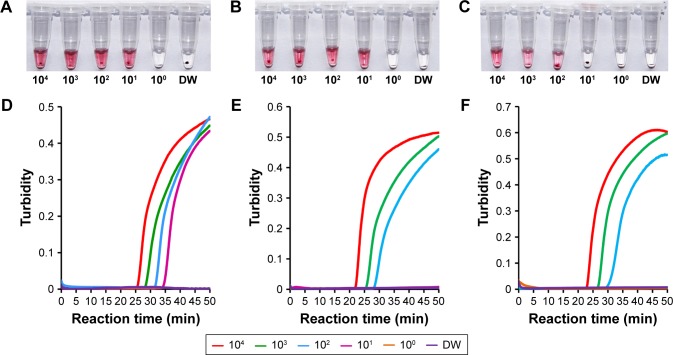
The analytical sensitivity of the mRT-LAMP-CIRN assay was evaluated by testing synthetic RNA standards ranging from 104 to 100 RNA copies/μL. As shown in Figure 3A–C, the mRT-LAMP-CIRN assay could detect 101 copies of synthetic RNA for both A/H1N1pdm09 and A/H3, and 102 copies of synthetic RNA for influenza B. Identical results were obtained on three separate days indicating that the mRT-LAMP-CIRN assay was both robust and reproducible. The mRT-LAMP reactions were also monitored by a real-time turbidimeter. As shown in Figure 3D and F, the detection limits for A/H1N1pdm09 and influenza B were 101 and 102 copies of synthetic RNA, respectively; these were equivalent to those obtained by mRT-LAMP-CIRN (Figure 3A and C). Nevertheless, the detection limit for A/H3 obtained by real-time turbidity detection was an order of magnitude lower than that obtained by mRT-LAMP-CIRN (Figure 3B and E), indicating that for RT-LAMP product analysis, the sensitivity of cascade invasive reaction using AuNPs as a sensor was higher than real-time turbidity detection.
Figure 3.
Sensitivity test results of the mRT-LAMP-CIRN assay for detecting influenza A/H1N1pdm09, A/H3 and influenza B.
Notes: Ten-fold serial dilutions of A/H1N1pdm09 (A), A/H3 (B) and influenza B (C) RNA transcripts (ranging from 104 to 100 RNA copies) were detected by the mRT-LAMP-CIRN assay. The multiplex RT-LAMP amplification reactions for A/H1N1pdm09 (D), A/H3 (E) and influenza B (F) RNA transcripts were also real-time monitored by a turbidimeter and the corresponding curves of concentrations of templates were marked in the figure.
Abbreviations: mRT-LAMP-CIRN, multiplex reverse-transcription loop-mediated isothermal amplification coupled with cascade invasive reaction using nanoparticles as a sensor; DW, distilled water.
Analytical specificity of the mRT-LAMP-CIRN
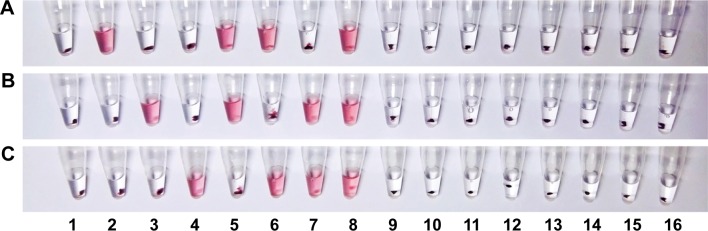
To evaluate the analytical specificity of the mRT-LAMP-CIRN assay, RNA extracts from influenza viruses A/Jiangsu/2/2009(H1N1), A/Nanjing/1/2009(H3N2) and B/Jiangsu/01/2015, and from various control viruses that could cause similar symptoms were tested separately. RNA extracts from the three influenza virus strains were also tested in different combinations in a single reaction. As shown in Figure 4, for single target detection, the positive reactions with red color in the reaction tubes were only observed in the extracts of influenza A/H1N1pdm09, A/H3N2 and influenza B viruses, whereas all of the control viruses, including the seasonal influenza virus A/H1N1, avian influenza viruses A/H5N1, A/H7N9 and A/H9N2, respiratory syncytial viruses types A and B, and parainfluenza viruses types 1 and 2 were negative. For multiple target detection, the mRT-LAMP-CIRN assay could differentiate two or three targets in a single reaction successfully (Figure 4). These results demonstrated the high analytical specificity of the mRT-LAMP-CIRN assay.
Figure 4.
Specificity test results of the mRT-LAMP-CIRN assay for detection of influenza viruses.
Notes: The analytical specificity of the mRT-LAMP-CIRN assay was assessed by testing the RNA extracts from influenza A/H1N1pdm09, A/H3N2 and influenza B, and from various control viruses separately. RNA extracts from the three influenza virus strains were also tested in different combinations in a single reaction. (A) The cascade invasive reactions were performed with 09H1-Up, 09H1-Dp and Hp. (B) The cascade invasive reactions were performed with H3-Up, H3-Dp and Hp. (C) The cascade invasive reactions were performed with FluB-Up, FluB-Dp and Hp. 1: no-template control; 2: A/H1N1pdm09; 3: A/H3N2; 4: influenza B; 5: A/H1N1pdm09 and A/H3N2; 6: A/H1N1pdm09 and influenza B; 7: A/H3N2 and influenza B; 8: A/H1N1pdm09, A/H3N2 and influenza B; 9: A/H1N1; 10: A/H5N1; 11: A/H7N9; 12: A/H9N2; 13: respiratory syncytial viruses type A; 14: respiratory syncytial viruses type B; 15: parainfluenza viruses types 1; 16: parainfluenza viruses types 2.
Abbreviations: mRT-LAMP-CIRN, multiplex reverse-transcription loop-mediated isothermal amplification coupled with cascade invasive reaction using nanoparticles as a sensor; Up, upstream probe; Dp, downstream probe; Hp, hairpin probe.
Clinical performance
To evaluate the clinical performance characteristics of the mRT-LAMP-CIRN assay, a total of 94 clinical specimens collected from influenza-like cases were analyzed by mRT-LAMP-CIRN assay and by real-time RT-PCR in parallel. As shown in Table 3, out of 58 specimens that were proven positive for influenza viruses by real-time RT-PCR, mRT-LAMP-CIRN detected 34/35 influenza A/H1N1pdm09, 14/14 influenza A/H3 and 9/9 influenza B. Compared to real-time RT-PCR, the sensitivity and specificity of the mRT-LAMP-CIRN assay for detecting influenza A/H1N1pdm09 were 97.1% (34/35) and 100% (59/59), respectively. For detecting influenza A/H3, the sensitivity and specificity of the assay were 100% (14/14) and 100% (80/80), and for influenza B, the sensitivity and specificity were also 100% (9/9) and 100% (85/85), respectively. In summary, the mRT-LAMP-CIRN assay had an overall sensitivity and specificity of 98.3% (57/58) and 100% (36/36), respectively.
Table 3.
Clinical performance of mRT-LAMP-CIRN compared with real-time RT-PCR for detecting influenza A/H1N1pdm09, A/H3 and influenza B
| mRT-LAMP-CIRN | Real-time RT-PCR
|
Performance characteristics
|
||
|---|---|---|---|---|
| Positive | Negative | Sensitivity, % | Specificity, % | |
| A/H1N1pdm09 | ||||
| Positive | 34 | 0 | 97.1 | 100 |
| Negative | 1 | 59 | ||
| A/H3 | ||||
| Positive | 14 | 0 | 100 | 100 |
| Negative | 0 | 80 | ||
| Influenza B | ||||
| Positive | 9 | 0 | 100 | 100 |
| Negative | 0 | 85 | ||
| Combined A/H1N1pdm09, A/H3 and FluB | ||||
| Positive | 57 | 0 | 98.3 | 100 |
| Negative | 1 | 36 | ||
Abbreviations: mRT-LAMP-CIRN, multiplex reverse-transcription loop-mediated isothermal amplification coupled with cascade invasive reaction using nanoparticles as a sensor; RT-PCR, reverse-transcription polymerase chain reaction.
Discussion
Since many respiratory viruses, including influenza viruses can cause similar symptoms, it is impossible for clinicians to distinguish one virus from another; therefore, they rely on the laboratory tests to identify the etiologic agent. Considering the morbidity and mortality caused by influenza viruses every year, sensitive and convenient laboratory methods to identify influenza virus subtypes are urgently needed. These are important for initial clinical treatment, avoidance of antibiotic misuse, as well as prevention of influenza virus transmission. In this study, we developed a sensitive mRT-LAMP-CIRN assay for the specific detection of three subtypes of influenza viruses, including influenza A/H1N1pdm09, A/H3 and influenza B. Because no specialized instrument is required, and the read-out can be easily analyzed by visual inspection by operators, the mRT-LAMP-CIRN assay described in this study is more suitable for use in low-equipment setting laboratory and on-site testing.
Several isothermal amplification methods have been developed that can pave the way for molecular diagnosis of infectious pathogens in simplified formats. LAMP is a promising method that has been developed to detect various pathogens.11,13–17 LAMP amplicons can be determined by agarose gel electrophoresis,33 lateral flow dipstick,14 turbidity or fluorescence measurement,34,35 or even visual inspection of white precipitates or color changes.36,37 Due to the properties of cost- and time-saving, multiplex detection represents the development trend of bioassays. Numerous multiplex molecular diagnostic methods such as multiplex PCR/RT-PCR have been developed.38 However, for multiplex LAMP, the differentiation of the ladder-like amplicons derived from multiple targets is still challenging today. To achieve multiplex LAMP detection, methods based on either endpoint18,19 or real-time analysis20–22 have been developed to identify the amplicons derived from multiple targets. Nevertheless, all these strategies require specialized and complicated instruments, which hinders the on-site application of multiplex LAMP assay. Invasive reaction is reported to be a highly sensitive and specific DNA detection method that amplifies the target-DNA-specific signal instead of the DNA template.26,27 By using invasive reaction probes that were specific for multiplex LAMP amplicons, the ladder-like LAMP products from different targets were successfully differentiated and identified. By taking advantage of cascade invasive reaction and the remarkable optical property of AuNPs in different aggregated states, the mRT-LAMP-CIRN assay developed in this study presents us with an alternative molecular method for sensitive multiplex LAMP product detection without the need for any complicated instruments. Because the mRT-LAMP-CIRN assay amplifies three targets simultaneously in a single tube, the cost for amplification of each target is reduced to one-third of the traditional singleplex LAMP. As the AfuFEN enzyme and AuNPs were prepared in our lab, the total cost of mRT-LAMP-CIRN assay for detecting one target is about two-thirds of traditional LAMP. Moreover, this pilot study may provide a strategy for establishing multiplex LAMP to much more than three plex, so that the cost for detecting each target would be reduced further.
The high analytical sensitivity of LAMP has been reported previously.39 Several possible factors may contribute to this fact. For example, the LAMP reaction is less affected by the presence of inhibitory substances in biological materials, less sensitive to various salts, and can tolerate the inhibitory effect of large concentrations of templates.40 However, previous studies have shown that due to the mutual interference of multiple primers, the sensitivities of multiplex nucleic acid detection tests tend to decrease. In this study, by coupling cascade invasive reaction with mRT-LAMP, one RT-LAMP amplicon could yield large numbers of cleaved hairpin probes (Figure 2); thus, the analytical sensitivity of mRT-LAMP might be increased by this signal amplification step. Indeed, the sensitivity for A/H3 obtained by mRT-LAMP-CIRN was ten times higher than that obtained by real-time turbidity detection. However, for 100 copy of A/H1N1pdm09 template and 101 copies of influenza B templates (one order of magnitude lower than their detection limits), the mRT-LAMP amplicons might be too limited, or no amplification occurred. So, they also could not be detected by cascade invasive reaction using nanoparticles as a sensor despite the signal amplification effect.
Because LAMP offers highly efficient amplification of target DNA, once the reaction tube is opened after LAMP reaction, there is a risk of aerosol contamination.41 To overcome this problem, in this study, mineral oil was added to the top of the LAMP reaction mixture to ensure that the amplicons did not disseminate into the air when the reaction tubes were opened. To further reduce the chances of contamination, protocols similar to those followed for PCR were adopted in this study. For example, the sample treatment, solution preparation and amplicon identification were carried out in separate areas.
The amplification mechanism of LAMP confers on the mRT-LAMP-CIRN assay inherent specificity because eight regions of each target sequence must be recognized by the four sequence-specific primers and two loop primers for amplification to occur. In addition, the invasive reaction is a specific DNA detection method because the mRT-LAMP amplicon must contain a sequence to which the invasive reaction upstream and downstream probes will hybridize in order to produce a positive result. By combining the specificities of both LAMP and cascade invasive reaction, the mRT-LAMP-CIRN assay developed in this study should provide higher specificity in theory. Our results showed that the mRT-LAMP-CIRN assay had 100% analytical specificity for identification of influenza A/H1N1pdm09, A/H3N2 and influenza B viruses, and there was no cross-reaction with other genetically or clinically related control viruses tested in this study.
As real-time RT-PCR has been widely accepted as the standard method for detecting influenza virus RNA, during clinical specimen detection, we compared the new assay described in this study with real-time RT-PCR. The results showed that, compared to real-time RT-PCR, the mRT-LAMP-CIRN assay had an overall sensitivity and specificity of 98.3% and 100%, respectively. Out of 35 specimens that were positive for influenza A/H1N1pdm09 as detected by real-time RT-PCR, 34 were determined to be positive by mRT-LAMP-CIRN. The difference in number detected may be due to the fact that eight regions of the target sequence must be recognized by the six primers in mRT-LAMP reaction, while only three regions of the target sequence need to be recognized in real-time RT-PCR reaction. Because of the frequent mutation of influenza virus genome, there might be a base variation in this specimen that affected the detection effect of mRT-LAMP-CIRN but not real-time RT-PCR. Further testing on additional clinical specimens is still needed to evaluate the clinical performance characteristics of this mRT-LAMP-CIRN assay.
In conclusion, a highly sensitive and specific mRT-LAMP-CIRN assay has been developed for identification of influenza viruses, including influenza A/H1N1pdm09, A/H3 and influenza B. Cost savings and no need for any complicated instruments make this assay more suitable for on-site use and low-equipment setting laboratory use.
Supplementary materials
UV spectrum and TEM images of oligonucleotide probe-modified AuNPs
The normalized optical absorption spectrum for the gold nanoparticles (AuNPs) after modification with oligonucleotide probes is shown in Figure S1A. We found that the oligonucleotide probe-modified AuNPs exhibited a ultraviolet (UV) peak at 530 nm. The Transmission electron microscopy (TEM) images of the free and aggregated AuNPs are shown in Figure S1B and C.
Optimization of the mRT-LAMP-CIRN assay
RNA extracts from three influenza viral isolates: A/Jiangsu/2/2009(H1N1), A/Nanjing/1/2009(H3N2) and B/Jiangsu/01/2015 were used as templates to optimize the multiplex reverse-transcription loop-mediated isothermal amplification (mRT-LAMP) conditions. The mRT-LAMP reactions were run at three different temperatures including 60°C, 63°C, and 65°C to determine the optimal reaction temperature for obtaining the highest amplification efficiency. At 63°C, the shortest amplification time and best amplification efficiency were obtained for all the three targets (Figure S2), and thus 63°C was chosen as the optimal temperature for all subsequent multiplex RT-LAMP reactions. According to the amplification curves obtained by real-time turbidimetry device, 50 min was chosen as the standard assay time because almost all the reactions had reached the plateau phase of amplification and longer reaction time yielded similar results (Figure S2; Figure 3).
Characterization of oligonucleotide probe-modified AuNPs.
Notes: (A) The normalized optical absorption spectrum for the AuNPs after modification with oligonucleotide probes. The TEM images of the free (B) and aggregated (C) AuNPs. Scale bar: 50 nm.
Abbreviations: AuNPs, gold nanoparticles; TEM, transmission electron microscopy.
Optimization of the mRT-LAMP-CIRN assay.
Notes: RNA extracts from A/Jiangsu/2/2009(H1N1) (A), A/Nanjing/1/2009(H3N2) (B) and B/Jiangsu/01/2015 (C) were used as templates to optimize the mRT-LAMP conditions. Distilled water was used as no-template control (D). The mRT-LAMP reactions were run at 60°C, 63°C, and 65°C for 60 min to determine the optimal reaction temperature and amplification time.
Abbreviation: mRT-LAMP-CIRN, multiplex reverse-transcription loop-mediated isothermal amplification coupled with cascade invasive reaction using nanoparticles as a sensor.
Acknowledgments
The study was supported in part by the Important National Science & Technology Specific Projects during the 25-year plan period (2013ZX10004103–006), Jiangsu Province Science & Technology Demonstration Project for Emerging Infectious Diseases Control and Prevention (No BE2015714), the National Natural Science Foundation of China (81501785, 81601732, 61401217), the Natural Science Foundation of Jiangsu Province (BK20141030, BK20161583, BK20140900), the key discipline of Jiangsu Province (ZDXKA2016008) and the Young Medical Talent Project of Jiangsu Province (QNRC2016537).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Simonsen L, Spreeuwenberg P, Lustig R, et al. Global mortality estimates for the 2009 Influenza Pandemic from the GLaMOR project: a modeling study. PLoS Med. 2013;10(11):e1001558. doi: 10.1371/journal.pmed.1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9(8):590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richard M, Fouchier RA. Influenza A virus transmission via respiratory aerosols or droplets as it relates to pandemic potential. FEMS Microbiol Rev. 2016;40(1):68–85. doi: 10.1093/femsre/fuv039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molinari NA, Ortega-Sanchez IR, Messonnier ML, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25(27):5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 5.Girard MP, Cherian T, Pervikov Y, Kieny MP. A review of vaccine research and development: human acute respiratory infections. Vaccine. 2005;23(50):5708–5724. doi: 10.1016/j.vaccine.2005.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito M, Nukuzuma S, Sugie M, et al. Detection of pandemic influenza A (H1N1) 2009 virus RNA by real-time reverse transcription polymerase chain reaction. Pediatr Int. 2012;54(6):959–962. doi: 10.1111/j.1442-200X.2012.03720.x. [DOI] [PubMed] [Google Scholar]
- 7.Suwannakarn K, Payungporn S, Chieochansin T, et al. Typing (A/B) and subtyping (H1/H3/H5) of influenza A viruses by multiplex real-time RT-PCR assays. J Virol Methods. 2008;152(1–2):25–31. doi: 10.1016/j.jviromet.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Choi JH, Kim MS, Lee JY, et al. Development and evaluation of multiplex real-time RT-PCR assays for seasonal, pandemic A/H1pdm09 and avian A/H5 influenza viruses detection. J Microbiol. 2013;51(2):252–257. doi: 10.1007/s12275-013-2452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munro SB, Kuypers J, Jerome KR. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J Clin Microbiol. 2013;51(4):1124–1129. doi: 10.1128/JCM.03113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parida M, Sannarangaiah S, Dash PK, Rao PV, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18(6):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiting SH, Champoux JJ. Properties of strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase: mechanistic implications. J Mol Biol. 1998;278(3):559–577. doi: 10.1006/jmbi.1998.1720. [DOI] [PubMed] [Google Scholar]
- 13.Huang XY, Hu XN, Ma H, et al. Detection of new bunyavirus RNA by reverse transcription-loop-mediated isothermal amplification. J Clin Microbiol. 2014;52(2):531–535. doi: 10.1128/JCM.01813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Y, Wu B, Qi X, et al. Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PLoS One. 2013;8(8):e69941. doi: 10.1371/journal.pone.0069941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Zhu JP, Zhang Q, et al. Detection of enterovirus 71 using reverse transcription loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2012;179(2):330–334. doi: 10.1016/j.jviromet.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Shigemoto N, Fukuda S, Takao S, et al. Rapid detection of novel influenza A virus and seasonal influenza A (H1N1, H3N2) viruses by reverse transcription-loop-mediated isothermal amplification (RT-LAMP) Kansenshogaku Zasshi. 2010;84(4):431–436. doi: 10.11150/kansenshogakuzasshi.84.431. Japanese. [DOI] [PubMed] [Google Scholar]
- 17.Curtis KA, Rudolph DL, Owen SM. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2008;151(2):264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Liang C, Chu Y, Cheng S, et al. Multiplex loop-mediated isothermal amplification detection by sequence-based barcodes coupled with nicking endonuclease-mediated pyrosequencing. Anal Chem. 2012;84(8):3758–3763. doi: 10.1021/ac3003825. [DOI] [PubMed] [Google Scholar]
- 19.Iseki H, Alhassan A, Ohta N, et al. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J Microbiol Methods. 2007;71(3):281–287. doi: 10.1016/j.mimet.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Mahony J, Chong S, Bulir D, Ruyter A, Mwawasi K, Waltho D. Multiplex loop-mediated isothermal amplification (M-LAMP) assay for the detection of influenza A/H1, A/H3 and influenza B can provide a specimen-to-result diagnosis in 40 min with single genome copy sensitivity. J Clin Virol. 2013;58(1):127–131. doi: 10.1016/j.jcv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Tanner NA, Zhang Y, Evans TC., Jr Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques. 2012;53(2):81–89. doi: 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Wang Y, Lan R, et al. Multiple endonuclease restriction real-time loop-mediated isothermal amplification: a novel analytically rapid, sensitive, multiplex loop-mediated isothermal amplification detection technique. J Mol Diagn. 2015;17(4):392–401. doi: 10.1016/j.jmoldx.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Xu W, Xue X, Li T, Zeng H, Liu X. Ultrasensitive and selective colorimetric DNA detection by nicking endonuclease assisted nanoparticle amplification. Angew Chem Int Ed Engl. 2009;48(37):6849–6852. doi: 10.1002/anie.200901772. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, Xie X, Li D, Yang Z, Li T, Liu X. Ultrasensitive colorimetric DNA detection using a combination of rolling circle amplification and nicking endonuclease-assisted nanoparticle amplification (NEANA) Small. 2012;8(12):1846–1850. doi: 10.1002/smll.201200263. [DOI] [PubMed] [Google Scholar]
- 25.Zou B, Cao X, Wu H, et al. Sensitive and specific colorimetric DNA detection by invasive reaction coupled with nicking endonuclease-assisted nanoparticles amplification. Biosens Bioelectron. 2015;66:50–54. doi: 10.1016/j.bios.2014.10.077. [DOI] [PubMed] [Google Scholar]
- 26.Lyamichev VI, Kaiser MW, Lyamicheva NE, et al. Experimental and theoretical analysis of the invasive signal amplification reaction. Biochemistry. 2000;39(31):9523–9532. doi: 10.1021/bi0007829. [DOI] [PubMed] [Google Scholar]
- 27.Hall JG, Eis PS, Law SM, et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc Natl Acad Sci U S A. 2000;97(15):8272–8277. doi: 10.1073/pnas.140225597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fronhoffs S, Totzke G, Stier S, et al. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol Cell Probes. 2002;16(2):99–110. doi: 10.1006/mcpr.2002.0405. [DOI] [PubMed] [Google Scholar]
- 29.Hill HD, Mirkin CA. The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat Protoc. 2006;1(1):324–336. doi: 10.1038/nprot.2006.51. [DOI] [PubMed] [Google Scholar]
- 30.Song Q, Qi X, Jia H, et al. Invader assisted enzyme-linked immunosorbent assay for colorimetric detection of disease biomarkers using oligonucleotide probe-modified gold nanoparticles. J Biomed Nanotechnol. 2016;12(4):831–839. doi: 10.1166/jbn.2016.2257. [DOI] [PubMed] [Google Scholar]
- 31. WHO.int [homepage on the Internet] CDC protocol of realtime RTPCR for influenza A(H1N1) [Accessed January 6, 2017]. Available from: http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf.
- 32. WHO.int [homepage on the Internet] WHO information for molecular diagnosis of influenza virus in humans – update. [Accessed January 6, 2017]. Available from: http://www.who.int/influenza/resources/documents/molecular_diagnosis_influenza_virus_humans_update_201108.pdf.
- 33.Sun J, Li X, Zeng H, et al. Development and evaluation of loop-mediated isothermal amplification (LAMP) for the rapid diagnosis of Penicillium marneffei in archived tissue samples. FEMS Immunol Med Microbiol. 2010;58(3):381–388. doi: 10.1111/j.1574-695X.2009.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori Y, Kitao M, Tomita N, Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59(2):145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Nyan DC, Swinson KL. A novel multiplex isothermal amplification method for rapid detection and identification of viruses. Sci Rep. 2015;5:17925. doi: 10.1038/srep17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XJ, Sun Y, Liu L, Belak S, Qiu HJ. Validation of a loop-mediated isothermal amplification assay for visualised detection of wild-type classical swine fever virus. J Virol Methods. 2010;167(1):74–78. doi: 10.1016/j.jviromet.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Krause JC, Panning M, Hengel H, Henneke P. The role of multiplex PCR in respiratory tract infections in children. Deutsches Arztebl Int. 2014;111(38):639–645. doi: 10.3238/arztebl.2014.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Shen X, Lu J, Zhang L. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) system for rapid detection of HDV genotype 1. Lett Appl Microbiol. 2013;56(3):229–235. doi: 10.1111/lam.12039. [DOI] [PubMed] [Google Scholar]
- 40.Kaneko H, Kawana T, Fukushima E, Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Mao Z, Qiu Y, Zheng L, Chen J, Yang J. Development of a visual loop-mediated isothermal amplification method for rapid detection of the bacterial pathogen Pseudomonas putida of the large yellow croaker (Pseudosciaena crocea) J Microbiol Methods. 2012;89(3):179–184. doi: 10.1016/j.mimet.2012.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of oligonucleotide probe-modified AuNPs.
Notes: (A) The normalized optical absorption spectrum for the AuNPs after modification with oligonucleotide probes. The TEM images of the free (B) and aggregated (C) AuNPs. Scale bar: 50 nm.
Abbreviations: AuNPs, gold nanoparticles; TEM, transmission electron microscopy.
Optimization of the mRT-LAMP-CIRN assay.
Notes: RNA extracts from A/Jiangsu/2/2009(H1N1) (A), A/Nanjing/1/2009(H3N2) (B) and B/Jiangsu/01/2015 (C) were used as templates to optimize the mRT-LAMP conditions. Distilled water was used as no-template control (D). The mRT-LAMP reactions were run at 60°C, 63°C, and 65°C for 60 min to determine the optimal reaction temperature and amplification time.
Abbreviation: mRT-LAMP-CIRN, multiplex reverse-transcription loop-mediated isothermal amplification coupled with cascade invasive reaction using nanoparticles as a sensor.