ABSTRACT
Whether obesity accelerates or suppresses autophagy in adipose tissue is still debatable. To clarify dysregulation of autophagy and its role in pathologies of obese adipose tissue, we focused on lysosomal function, protease maturation and activity, both in vivo and in vitro. First, we showed that autophagosome formation was accelerated, but autophagic clearance was impaired in obese adipose tissue. We also found protein and activity levels of CTSL (cathepsin L) were suppressed in obese adipose tissue, while the activity of CTSB (cathepsin B) was significantly enhanced. Moreover, cellular senescence and inflammasomes were activated in obese adipose tissue. In 3T3L1 adipocytes, downregulation of CTSL deteriorated autophagic clearance, upregulated expression of CTSB, promoted cellular senescence and activated inflammasomes. Upregulation of CTSB promoted additional activation of inflammasomes. Therefore, we suggest lysosomal dysfunction observed in obese adipose tissue leads to lower autophagic clearance, resulting in autophagosome accumulation. Simultaneously, lysosomal abnormalities, including deteriorated CTSL function and compensatory activation of CTSB, caused cellular senescence and inflammasome activation. Our findings strongly suggest lysosomal dysfunction is involved in early pathologies of obese adipose tissue.
KEYWORDS: adipose tissue, autophagy, cathepsin, inflammasome, lysosome, obesity, senescence
Introduction
Defined as increased adiposity, obesity is closely related with insulin resistance, hyperglycemia and hyperlipidemia, and often progresses to metabolic diseases,1,2 In obese individuals, energy intake exceeds energy consumption, and excess energy accumulates in the form of triacylglycerides (TG), predominantly in white adipose tissue (WAT). As an organ, WAT not only stores energy, but also participates in endocrine activity by secreting adipose tissue-derived cytokines, known as adipokines. As obesity progresses, WAT secretes less anti-inflammatory adipokines, such as adiponectin, and more proinflammatory adipokines, such as IL6 (interleukin 6) and TNF (tumor necrosis factor), which contributes to the development of insulin resistance and type 2 diabetes.3,4 Obese WAT appears to undergo chronic and low-grade inflammatory conditions with infiltration of M1 macrophages.5 More recently, activation of NLRP3 (NLR family, pyrin domain containing 3), an inflammasome consisting of NLRP3, PYCARD/ASC (PYD and CARD domain containing) and CASP1 (caspase 1) cysteine protease, reportedly contributes to onset of inflammation in WAT both in diet-induced and genetically obese mice.6,7 In response to stress signaling, such as reactive oxygen species (ROS) or TNF, the NLRP3 inflammasome interacts with 45-kDa pro-CASP1, leading to its cleavage into 2 active forms: p20 and p10. Activation of CASP1 results in processing of IL1B (interleukin 1 β) and IL18 (interleukin 18), which promotes inflammation.6-8 It is widely accepted that obese WAT exhibits a senescence-like phenotype including upregulation of TP53, (tumor protein p53; in the nomenclature for TP53/TRP53/p53, note that the mouse nomenclature is TRP53, “transformation related protein 53,” whereas the rat nomenclature is TP53, but we use TP53 hereafter to refer to both the human and murine genes/proteins for simplicity) and its downstream target, CDKN1A/p21.9-11 This phenotype, which shows positivity for senescence-associated GLB1/β-galactosidase (SA-GLB1) and increased production of proinflammatory cytokines including TNF and CCL2 (C-C motif chemokine ligand 2), is involved in insulin resistance and diabetes in a TP53-dependent manner.11
Autophagy is a cellular catabolic process occurring via lysosomal clearance that can be divided into 3 main types: macroautophagy, microautophagy and chaperone-mediated autophagy. In the present study, we focused on macroautophagy (hereafter referred to as autophagy). In this process, damaged organelles and proteins are sequestered inside double-membrane vesicles, autophagosomes, which fuse with acidic lysosomes to form autolysosomes that degrade the intramembranous contents. Thus, autophagy plays an important role in cell, tissue and organism homeostasis, and has been implicated in a diverse range of pathologies including cancer, neurodegeneration and diabetes.12-14 Mice with systemic deletion of the autophagy-related genes Atg5 or Atg7 exhibit accumulation of mitochondrial mass and increased β-oxidation in WAT, as well as enhanced insulin sensitivity with a reduction in plasma LEP (leptin) levels. Moreover, these mice show resistance to high fat diet (HFD)-induced obesity. Collectively, these findings indicate that autophagy regulates characteristics of both adipocytes and adipose tissue.15,16 However, it remains unclear whether autophagy is activated or suppressed in obese WAT because of technical difficulties analyzing autophagy function in vivo.17 Ost and colleagues report increases of autophagosomes in obese WAT obtained from diabetic patients and autophagic flux analyzed using an MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) turnover assay with both rapamycin and chloroquine.18 In contrast, we and another group report impairment of autophagic flux in WAT of obese mice, which results in accumulation of autophagosomes.19,20
As lysosomal destabilization and CTSB activation occur in WAT during early development of obesity, leading to adipocyte cell death and macrophage infiltration,21 we focused on lysosomal impairment to clarify discrepancies among previous reports. As an acid organelle involved in various cellular functions including autophagy,22 lysosomes contain more than 50 hydrolytic enzymes, such as proteases, lipases and nucleases, that are critical for autophagic degradation. CTSB, CTSL and CTSD (cathepsin D) are the most abundant lysosomal proteases.23 We show here that lysosomal dysfunction, particularly functional derangement of CTSB and CTSL, causes early pathologies in obese adipose tissue including autophagosome accumulation, enhanced cellular senescence and activated inflammasomes.
Results
Autophagic flux in obese WAT
To examine whether autophagic flux is activated or suppressed in obese WAT, we analyzed expression levels of autophagy-related proteins in WAT of obese mice. Along with body weight and WAT mass, conversion of LC3-I to LC3-II and expression levels of SQSTM1/p62 (sequestosome 1) protein were significantly increased in obese WAT (Fig. 1A to C). In contrast, ATG5 and BECN1/Beclin 1, which also participate in the autophagy machinery, were unchanged in obese WAT (Fig. 1A, D and E). The amount of LC3-II is considered to generally represent both the number of autophagosomes24 and SQSTM1 protein selectively degraded by autophagy.25 Thus, while our findings imply alteration of autophagy in WAT of HFD mice, it is difficult to confirm whether autophagy is accelerated or suppressed because LC3-II upregulation indicates both enhancement of autophagic clearance and accumulation of autophagosomes.26
Figure 1.
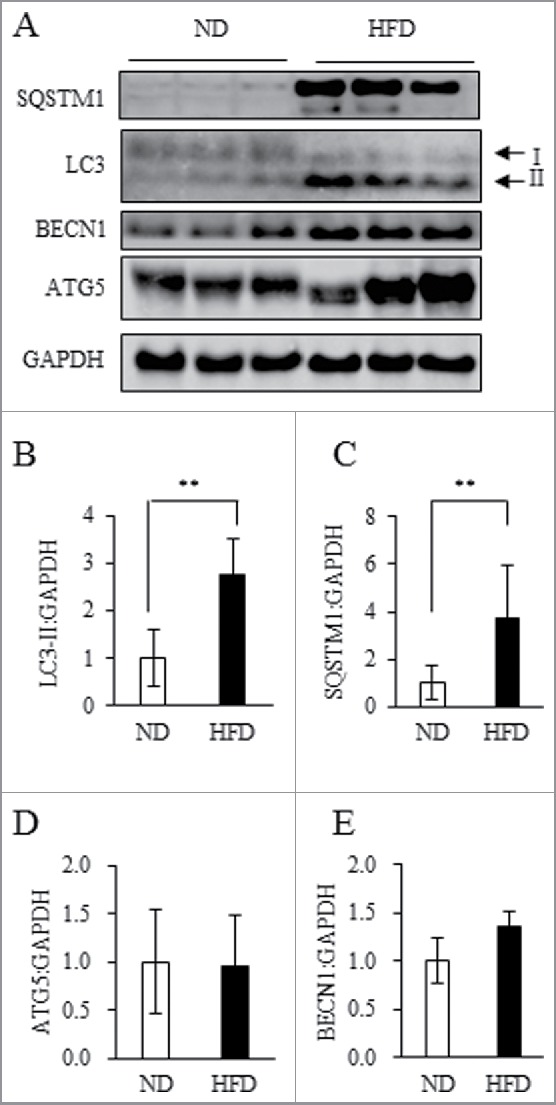
HFD treatment induced obesity and upregulated expression of certain autophagy-related proteins in WAT. (A to E) Total protein extracted from WAT of ND mice or 18HFD mice analyzed by western blot using anti-SQSTM1, LC3, BECN1, ATG5 and GAPDH antibodies (A) with quantitative data shown (B to E). Representative images and the quantitative data (ND: n = 13, HFD: n = 9) were shown. Intensity of GAPDH was used as a loading control. Values indicate mean ± SD (ND: n = 13, HFD: n = 9). Differences between values were analyzed by the Student t test. Statistical significance shown as *P < 0.05, **P < 0.01.
To analyze autophagic flux more accurately, an LC3-II turnover assay has recently become more widely used.26 First, we applied the LC3-II turnover assay in ex vivo WAT, as previously reported.27,28 In this assay, WAT explants were incubated with or without chloroquine, an inhibitor of lysosomal acidification and autophagic clearance. Compared with normal diet (ND) mice, chloroquine significantly increased expression levels of both LC3-II and SQSTM1 in WAT of HFD mice (Fig. 2A to C), also as previously reported.27,28 A SQSTM1 turnover assay with or without addition of rapamycin, an inhibitor of MTORC1 (mechanistic target of rapamycin complex 1) activity and autophagy activator, resulted in an observed reduction of SQSTM1 expression in WAT of ND mice, but not HFD mice. Moreover, rapamycin treatment increased LC3-II in obese WAT (Fig. 2A, D and E). Overall, these ex vivo analyses suggest autophagosome formation was accelerated in obese WAT, however, autophagic clearance was likely impaired.
Figure 2.
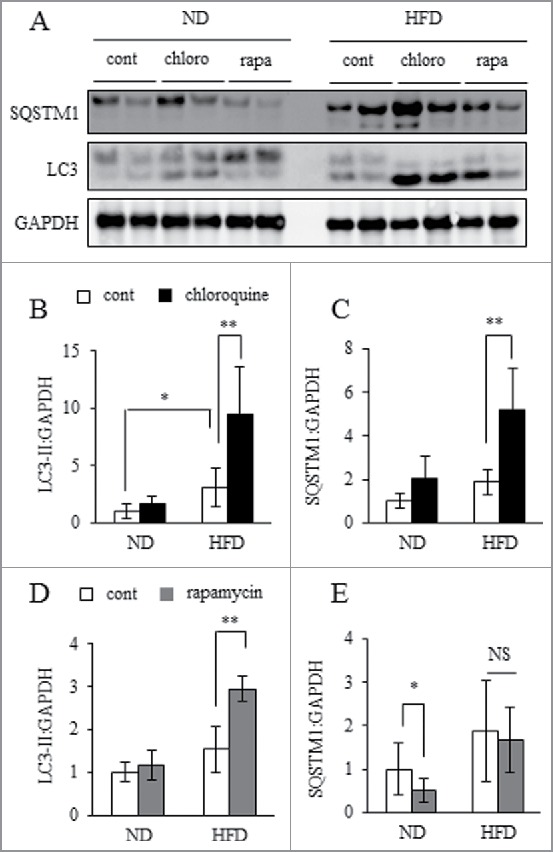
Autophagosome formation was accelerating but lysosomal clearance was impaired. WAT explants from ND or 30HFD mice were incubated in the presence or absence of 10 mM chloroquine (chloro) or 500 nM rapamycin (rapa) for 24 h, and then samples were collected and assayed. Total protein extracted from WAT was analyzed by western blot using anti-SQSTM1, LC3 and GAPDH antibodies (A) with quantitative data shown (B to E). Representative images and the quantitative data (n = 4) were shown. Intensity of GAPDH was used as a loading control. Values indicate mean ± SD (n = 6). Differences between values were analyzed by the Student t test. Statistical significance shown as *P < 0.05, **P < 0.01.
Lysosomal function in obese WAT
To assess the contribution of various molecular cathepsin species to autophagy in adipocytes, we performed experiments using 2 cathepsin inhibitors: E64-d, which selectively inhibits CTSL and CTSB, and pepstatin A, a selective inhibitor of CTSD and CTSE (cathepsin E). While E64-d treatment increased LC3-II and SQSTM1 expression, pepstatin A did not, indicating CTSL and/or CTSB play an important role in autophagy for adipocytes (Fig. S1). Therefore, we focused on CTSL and CTSB in WAT and 3T3L1 adipocytes. Cathepsins are synthesized in the endoplasmic reticulum (ER), processed post-translationally and shuttled to the lysosome, where they are processed proteolytically into procathepsins for cathepsin maturation under acidic conditions.29 Compared with ND mice, pro-CTSL expression was upregulated in WAT of HFD mice; however, mature CTSL expression was significantly downregulated (Fig. S2A to C). Similarly, enzymatic activity of CTSL was significantly decreased in obese WAT (Fig. S2D); in contrast, expression of both pro- and mature forms of CTSB was upregulated in obese WAT (Fig. S2A, F and G). Enzymatic activity and Ctsb mRNA expression were equally upregulated in obese WAT (Fig. S2H and I). Ctsl mRNA expression was upregulated in obese WAT in a similar manner (Fig. S2E).
In addition, to clarify whether obesity itself affects downregulation of CTSL activity, we analyzed the WAT of LEP-deficient, genetically obese mice (ob/ob) fed a ND or HFD (Fig. S3A to C). As protein levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in ob/ob mice were markedly upregulated, as shown in Figure S4A and B, individual protein levels were standardized to total protein level for evaluation. Similar to the results observed for HFD mice, conversion of LC3-I to LC3-II and expression levels of SQSTM1 protein were significantly increased in ob/ob mice, particularly in ob/ob HFD mice (Fig. S4C to E). The mature form of CTSL protein and its activity were significantly downregulated, while Ctsl mRNA levels were increased (Fig. S4C and G to I). The mature form of CTSB protein was unchanged, which was inconsistent with results obtained in HFD mice (Fig. S2G); however, Ctsb mRNA levels and its protein activity were significantly increased (Fig. S4C and J to L). These results suggest that mature CTSL protein and its activity were suppressed, while the activity of CTSB was significantly enhanced in WAT derived from both HFD-induced and genetically obese mice.
To further elucidate mechanisms underlying dysregulation of cathepsins in obese WAT, alterations in cathepsin expression and enzymatic activity induced by HFD were analyzed over a time-course (Fig. S5A to C). Pro-CTSL protein expression was significantly increased in the 4HFD group (HFD intake for 4 wk); moreover, CTSL activity declined significantly this group (Fig. 3A, B and D). In the 8HFD group, mature CTSL protein expression was significantly decreased (Fig. 3A and C). In contrast, pro- and mature CTSB expression was significantly and slightly increased in 4HFD animals (Fig. 3A, F and G), respectively, and activity was significantly increased in the 8HFD group (Fig. 3H). Both Ctsl and Ctsb mRNA expressions were significantly increased in 4HFD, 8HFD and 18HFD animals (Fig. 3E and I). As it has recently been reported that TFEB (transcription factor EB) transcriptionally regulates autophagy and lysosomal biogenesis, and is involved in cellular lipid metabolism,30,31 we predicted that upregulation of Ctsl and Ctsb mRNA was induced through TFEB. Upon investigating TFEB protein expression in obese WAT, we found it was not upregulated in 4HFD, 8HFD or 18HFD animals, and was significantly decreased in 8HFD (Fig. S6A and B). This indicates upregulation of Ctsl and Ctsb mRNA expression is unlikely to be dependent on TFEB. We also examined alterations in levels of Lc3b and Sqstm1 mRNA transcripts in obese WAT. The results showed that Lc3b mRNA expression was decreased in 8HFD and 18HFD mice, while Sqstm1 mRNA expression was significantly increased in 4HFD, 8HFD and 18HFD animals (Fig. 3K and M). In obese WAT, however, the increased ratio of SQSTM1 protein markedly exaggerated that of Sqstm1 mRNA, particularly in 8HFD and 18HFD groups (Fig. 3L and M), suggesting increased LC3-II and SQSTM1 was not caused by significant upregulation of transcription. Interestingly, downregulation of CTSL enzymatic activity appeared to precede upregulation of CTSB expression based on these findings. Therefore, we considered the enzymatic activity of CTSL to be primarily suppressed in obese WAT, whereby compensatory activation of CTSB expression subsequently occurred in a transcription- and/or post-transcription-dependent manner. Such deviation within cathepsin regulatory mechanisms may lead to impaired autophagic clearance.
Figure 3.
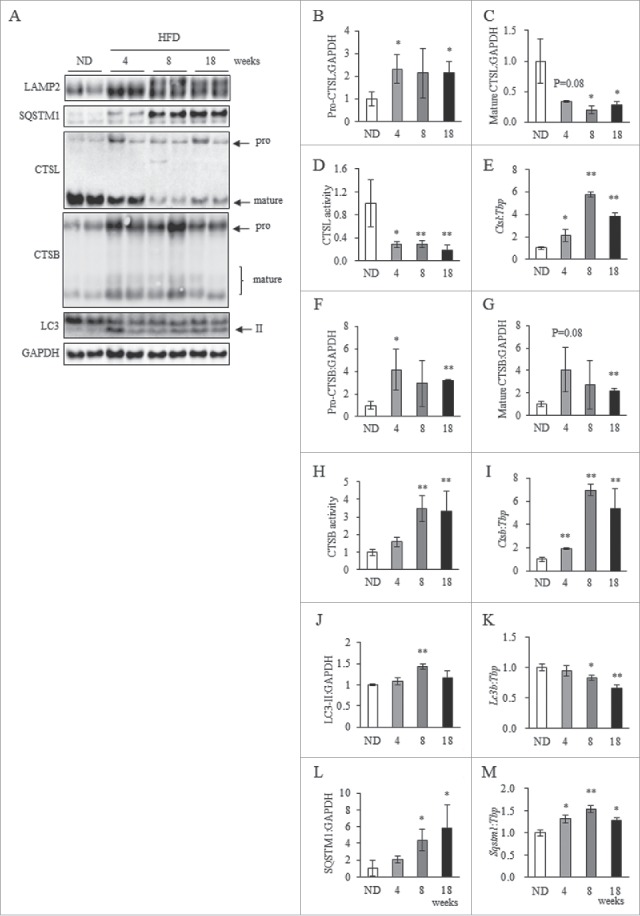
Dysregulation of cathepsins was observed in HFD-induced obese WAT. (A) Total protein extracted from WAT of ND, 4HFD, 8HFD and 18HFD mice analyzed by western blot using anti-LAMP2, CTSL, CTSB, SQSTM1, LC3 and GAPDH antibodies (A) with quantitative data shown (B, C, F, G, J and L). Representative images and the quantitative data (n = 4) were shown. Intensity of GAPDH was used as a loading control (n = 4). Enzymatic assay of CTSL (D) and CTSB (H) in WAT of ND, 4HFD, 8HFD and 18HFD mice, as indicated, analyzed by selective substrate. mRNA expression of Ctsl (E), Ctsb (I), Lc3b (K) and Sqstm1 (M) in WAT of ND, 4HFD, 8HFD and 18HFD mice, as indicated, analyzed by real-time RT-PCR (n = 4). Data were normalized against Tbp (n = 4). Values indicate mean ± SD. Differences between values were analyzed by the Student t test with Bonferroni correction *P < 0.05, **P < 0.01.
Effect of CTSL inhibition on CTSB expression in 3T3L1 adipocytes
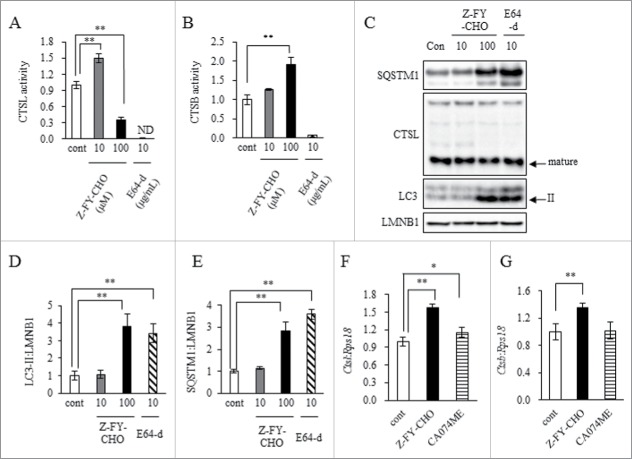
To establish whether CTSL inhibition causes an increase in CTSB expression, we treated 3T3L1 adipocytes with a CTSL-selective inhibitor (Z-FY-CHO), CTSB-selective inhibitor (CA074ME), or a CTSL and CTSB inhibitor (E64-d). Ten micromolar Z-FY-CHO treatment increased CTSL activity, but 100 μM treatment resulted in a marked decrease (Fig. 4A). Both LC3-II and SQSTM1 protein levels increased significantly with 100 μM Z-FY-CHO treatment, indicating accumulation of autophagosomes occurs with CTSL suppression (Fig. 4C, D and E). Autophagosome accumulation induced by 10 μg/mL E64-d treatment produced similar results. Surprisingly, Z-FY-CHO treatment increased CTSB activity, indicating CTSL suppression caused compensatory activation of CTSB (Fig. 4B). Z-FY-CHO treatment also increased Ctsl and Ctsb mRNA expression (Fig. 4F and G). In contrast, CA074ME treatment did not markedly increase expression of these mRNAs (Fig. 4F and G).
Figure 4.
CTSL inhibition impaired autophagic flux in 3T3L1 adipocytes and enhanced CTSB activity. 3T3L1 preadipocytes were differentiated into adipocytes at day 11 and treated with 10 to 100 μM CTSL inhibitor, Z-FY-CHO, or 10 μg/mL cathepsin inhibitor, E64-d. Adipocytes were harvested and analyzed. Enzymatic assay of CTSL (A) and CTSB (B) analyzed by selective substrate. Total cell lysates analyzed by western blot using anti-SQSTM1, CTSL, LC3 and LMNB1 antibodies (C) with quantitative data shown (D and E). Representative images and the quantitative data (n = 4) were shown. Intensity of LMNB1 was used as a loading control (n = 4). 3T3L1 preadipocytes were differentiated into adipocytes at d 11 and treated with 100 μM Z-FY-CHO or 10 μM CTSB inhibitor, CA074ME. Adipocytes were harvested and analyzed. mRNA expression of Ctsl (F) and Ctsb (G) were analyzed by real-time RT-PCR (n = 4). Data were normalized against Rps18 (n = 4). Values indicate mean ± SD. Differences between values were analyzed by Tukey-Kramer method with *P < 0.05, **P < 0.01.
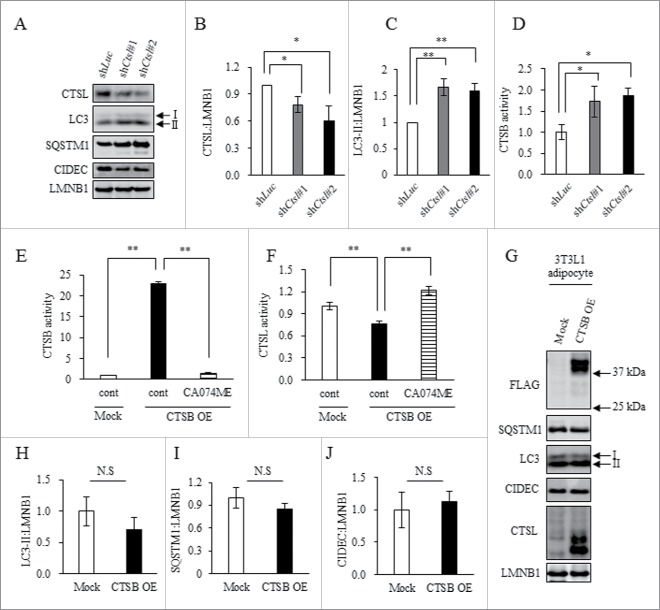
Expression of CIDEC/Fsp27 (cell death inducing DFFA like effector c), a marker for lipid droplet and adipocyte differentiation, and size of lipid droplets decreased significantly with knockdown of CTSL in 3T3L1 adipocytes (Fig. S7A and B); whereas, expression of LC3-II protein significantly increased (Fig. 5A to C). Surprisingly, under the same conditions, CTSB activity was accelerated (Fig. 5D). These findings indicate downregulation of CTSL suppressed adipocyte differentiation while inducing autophagosome accumulation and compensatory activation of CTSB.
Figure 5.
CTSL knockdown promoted complementary activation of CTSB, and CTSB overexpression did not affect autophagic clearance. (A to D) 3T3L1 shLuc (control), as well as shCtsl#1 and shCtsl#2 preadipocytes were differentiated at Day 8 and adipocytes were collected. (A to C) Total cell lysates were analyzed by western blotting using anti-CTSL, LC3, SQSTM1, CIDEC and LMNB1 antibodies (A) with quantitative data shown (B and C). Representative images and the quantitative data (n = 4) were shown. Intensity of LMNB1 was used as a loading control (n = 4). (D) Enzymatic assay of CTSB. Values indicate mean ± SD (n = 4). Differences between values were analyzed by the Student t test. Statistical significance was shown as *P<0.05, **P<0.01. (E to J) 3T3L1 (Mock) or 3T3L1 CTSB (OE) preadipocytes were differentiated into adipocytes at d 8 and treated with 10 μM CA074ME for 24 h. Enzymatic assay of CTSB (E) and CTSL (F) were analyzed by selective substrate. Values indicate mean ± SD. Differences between values were analyzed by Tukey-Kramer method with *P < 0.05, **P < 0.01. (G to J) Total cell lysates analyzed by western blot using anti-FLAG, SQSTM1, LC3, CIDEC, CTSL and LMNB1 antibodies (G) with quantitative data shown (H to J). Representative images and the quantitative data (n = 4) were shown. Intensity of LMNB1 was used as a loading control. Values indicate mean ± SD. Differences between values were analyzed by the Student t test with *P < 0.05, **P < 0.01.
To determine if compensatory activation of CTSB affects autophagy in adipocytes, we analyzed CTSB-overexpressing 3T3L1 adipocytes. CA074ME treatment suppressed increased activity of CTSB in CTSB-overexpressing 3T3L1 adipocytes (Fig. 5E). Interestingly, CTSB upregulation inhibited CTSL activity and CA074ME treatment ameliorated this reduction in enzymatic activity (Fig. 5F). However, CTSB activation did not alter expression of LC3-II, SQSTM1 or CIDEC (Fig. 5G to J), suggesting compensatory activation of CTSB does not contribute to alteration of autophagy in adipocytes.
Impact of cathepsin derangement on senescence-like changes and inflammasomes in adipocytes
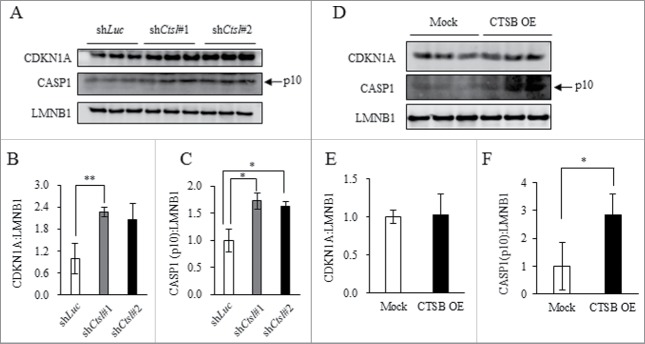
In human neuroblastoma, CTSL inhibition reportedly causes cellular senescence.32 Senescence-like changes observed in obese WAT occur in a TP53-dependent manner.11 Moreover, it has been shown that CTSB is responsible for activation of inflammasomes via lysosomal permeabilization33,34 which leads to inflammation in obese WAT.6,7 Therefore, we investigated whether cathepsin derangement in obese WAT is involved in either senescence-like changes or inflammasome activation. In CTSL-knockdown or CTSB-overexpressing 3T3L1 adipocytes, expression of markers for senescence (TP53 and CDKN1A) and inflammasome activation (cleaved CASP1, p10 subunit) were examined by western blotting. Our results indicated CTSL knockdown significantly increased expression of both CDKN1A (Fig. 6A and B) and cleaved CASP1 (Fig. 6A and C). Treatment with a CTSL inhibitor significantly increased expression of CDKN1A and cleaved CASP1 in a dose-dependent manner (Fig. S8A to C). CTSB overexpression did not enhance expression of CDKN1A (Fig. 6D and E), but increased cleaved CASP1 (Fig. 6D and F). Expression of TP53 was not detected in either CTSL-knockdown or CTSB-overexpressing 3T3L1 adipocytes. Collectively, these findings suggest the possibility that dysregulation of CTSL and CTSB induces senescence-like features and inflammasome activation in 3T3L1 adipocytes.
Figure 6.
CTSL knockdown promoted senescence-like changes, and complementary activation of CTSB enhanced inflammasome activity. (A to E) 3T3L1 shLuc (control), as well as shCtsl#1 and shCtsl#2 preadipocytes were differentiated at Day 8 and adipocytes were collected. Total cell lysates analyzed by western blot using anti-CDKN1A, CASP1 and LMNB1 antibodies (A) with quantitative data shown (B and C). Representative images and the quantitative data (n = 4) were shown. Intensity of LMNB1 was used as a loading control. (D to F) 3T3L1 (Mock) or 3T3L1 CTSB (OE) preadipocytes were differentiated into adipocytes at Day 8. Total cell lysates analyzed by western blot using anti-CDKN1A, CASP1 and LMNB1 antibodies (D) with quantitative data shown (E and F). Representative images and the quantitative data (n = 4) were shown. Intensity of LMNB1 was used as a loading control. Values indicate mean ± SD (n = 4). Differences between values were analyzed by the Student t test. Statistical significance shown as *P < 0.05, **P < 0.01.
Cellular senescence and inflammasome activation in obese WAT
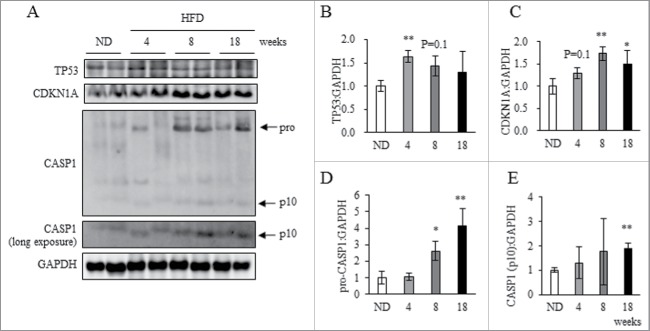
Finally, we investigated senescence-like features and inflammasome activation in obese WAT. In a similar and time-dependent manner to downregulation of CTSL in WAT (Fig. 3), expression levels of TP53 and CDKN1A protein were significantly increased in both 4HFD and 8HFD groups (Fig. 7A to C), and Cdkn1a mRNA levels were significantly increased in the 4HFD, 8HFD and 18HFD groups (Fig. S9). Moreover, expression of pro-CASP1 (45 kDa) and cleaved CASP1 was significantly increased in 8HFD and 18HFD animals (Fig. 7A, D and E).
Figure 7.
Cellular senescence and inflammasome activation in obese WAT. Total protein extracted from WAT of ND, 4HFD, 8HFD and 18HFD mice analyzed by western blot using anti-TP53, CDKN1A, CASP1, and GAPDH antibodies (A) with quantitative data shown (B to E). Representative images and the quantitative data (n = 4) were shown. Intensity of GAPDH was used as a loading control. Values indicate mean ± SD (n = 4). Differences between values were analyzed by the Student t test with Bonferroni correction. Statistical significance shown as *P < 0.05, **P < 0.01.
Discussion
Alteration of autophagic machinery in obese WAT
Several studies have demonstrated that obesity affects autophagic machinery. For example, obesity suppresses autophagy via downregulation of ATG7 in the liver, where autophagic dysfunction contributes to deterioration of ER stress responses and insulin resistance.35 Although, autophagic flux in β cells appears to be enhanced as an adaptive response against HFD-induced insulin resistance.36 Yoshizaki et al. conclude that autophagy in WAT is suppressed in diet-induced obese mice.19 In contrast, Jansen et al. report that autophagy activity is enhanced in WAT of obese individuals.28 Generally, in vivo autophagy analysis has been performed by morphological observation using electron microscopy. However, this method is insufficient as autophagosomes increase whenever autophagic flux is either promoted or suppressed.17 Here, we applied ex vivo analysis of autophagic flux in obese WAT. A careful evaluation of autophagy is required because a considerable amount of stress is placed on WAT explants during performance of ex vivo LC3-II or SQSTM1 turnover assays. Moreover, SQSTM1 protein is involved in various cellular functions by interacting with many proteins such as KEAP1 and NFE2L2/NRF2, as well as TRAF6 and NFKB1.25,37 Therefore, it is important to investigate and carefully evaluate alterations of autophagy in obese WAT. Here we show that in WAT of HFD-induced obese mice, autophagosome formation was accelerated but autophagic clearance was impaired due to lysosomal dysfunction, leading to autophagosome accumulation. In fact, treatment with chloroquine or E64-d dramatically increased protein expression of both LC3-II and SQSTM1, suggesting lysosomal impairment enhances autophagosome formation.38 Moreover, knockdown of ATP6V0C (ATPase H+ transporting V0 subunit c), a V-ATPase subunit that regulates lysosomal acidification, increases autophagosome accumulation both at basal levels and at low bafilomycin A1 (BAF) concentrations.39 These results support our data indicating lysosomal dysfunction promotes autophagosome formation and accumulation in obese WAT.
Functional impairment of lysosomes observed in obese WAT involves dysregulation of both CTSL and CTSB
CTSL and CTSB, both of which are ubiquitously expressed, are a cysteine endopeptidase and exopeptidase, respectively. While CTSB-deficient mice do not exhibit a characteristic phenotype,40 CTSL-deficient mice exhibit skin abnormalities and impaired bone development.41 Double CTSL- and CTSB-deficient mice have a lifespan of 2 to 4 wk and exhibit early-onset postnatal brain atrophy.42 However, no previous report has indicated whether these deficiencies affect WAT function. CTSL is synthesized as pro-CTSL (39 kDa), and then shuttled to the endosome and then lysosome, where it is processed into mature CTSL (30 kDa) in an acidic environment.29 CTSB, which is synthesized as pro-CTSB (44 kDa) and then subsequently processed into mature CTSB (33 kDa),29,43 is far more stable than CTSL under neutral conditions.44 In this study, we found mature CTSL expression and its activity were significantly reduced in WAT of both HFD-induced obese mice and ob/ob mice. In contrast, mRNA and pro-CTSL protein expression increased significantly, suggesting its compensatory transcriptional activation occurs with impairment of CTSL maturation in obese WAT. Generally, cathepsin maturation is regulated by lysosomal acidic pH, other proteases like CTSD, and endogenous cathepsin inhibitors including cystatins, thyropins and serpins.45 Oxidative stress induces lysosomal instability and membrane permeabilization.46 Moreover, in trabecular meshwork cells, oxidative stress decreases both cathepsin activity and lysosomal acidity, resulting in decreased autophagic flux.47 When HFD mice were treated with an antioxidant, N-acetylcysteine, CTSL maturation was significantly restored; although, CTSL enzymatic activity was not (data not shown). In autolysosomes derived from livers of ob/ob mice, lysosomal acidity is significantly reduced and enzymatic activities of both CTSB and CTSL are suppressed.48 Furthermore, a mutation in PSEN1 (presenilin 1), a pathogenic gene associated with familial Alzheimer disease, causes lysosomal alkalization following reduction of CTSD maturation. These cells also exhibit accumulation of both LC3-II and SQSTM1 proteins.49 In addition, the MHC class II-associated p41 invariant chain fragment selectively inhibits CTSL activity.50 Therefore, it is important to consider the possibility that both lysosomal acidifying agents and endogenous CTSL inhibitors (such as PSEN1 and p41 invariant chain fragment, respectively) have predominant roles in CTSL dysfunction in obese WAT.
Based on in vitro experiments using cathepsin inhibitors and genetic knockdown, our findings also suggest CTSL inhibition induces compensatory transcriptional upregulation and enzymatic activation of CTSB in 3T3L1 adipocytes and obese WAT. In the brain tissue of mice with Niemann–Pick type C disease, expression of mature protein forms for both CTSB and CTSD are upregulated with accumulation of autophagosomes and ubiquitinated proteins.51 Hannaford et al. report upregulated expression levels of both Ctsl and Ctsb mRNA, pro- and mature proteins in obese WAT, particularly within stromal vascular fractions.52 Our findings about CTSB in WAT of HFD mice are similar, with the exception of our result indicating that mature CTSL protein is downregulated. While we do not know the reason for this discrepancy, we confirmed a reduction in CTSL enzymatic activity was present in WAT of both HFD and ob/ob mice. Based on our data, enzymatic activity of CTSL may be suppressed by oxidative stress, lysosomal pH acidifying factors and/or endogenous CTSL inhibitors, as previously mentioned. At this point, compensatory upregulation of mature CTSB proteins and its enzymatic activation occurs in WAT of HFD mice. In contrast, only an induction of CTSB activity was detected in ob/ob mice. In general, maturation of cathepsin is regulated by other cathepsins.45 Our data, therefore, suggests that enhanced compensatory upregulation or enzymatic activation of CTSB promotes CTSL cleavage, resulting in further suppression of CTSL enzymatic activity. Furthermore, the mechanism of these compensatory reactions may differ among underlying causes of obesity, for example, HFD-induced or genetic. We could confirm here that CTSL dysfunction directly caused accumulation of autophagosomes and compensatory activation of CTSB only in vitro using Ctsl knockdown 3T3-L1 cells. Therefore, further examination is necessary to demonstrate a direct link between CTSL dysfunction and the subsequent responses in vivo.
Activation of cellular senescence and inflammasomes via lysosomal dysfunction in obese WAT
Senescence-associated phenotypes, including activation of SA-GLB1, are reportedly promoted in a TP53-dependent manner in obese WAT.11 We show here that inhibition of CTSL by either chemical inhibitor or genetic knockdown increases expression of CDKN1A in differentiated 3T3L1 adipocytes. In obese WAT, maturation of CTSL was impaired, its enzymatic activity was lowered, and expression of TP53 was increased. Generally, the TP53-CDKN1A and CDKN2A/p16 pathways regulate cellular senescence independently;53 however, senescence regulated by the TP53-CDKN1A pathway is more dominant.54 Thus, we concluded that impairment of CTSL maturation and/or reduction of its enzymatic activity promotes cellular senescence in obese WAT. Moreover, dysregulation of CTSL induces compensatory upregulation of CTSB expression, leading to activation of inflammasomes. In fact, expression of IL1 (interleukin 1), IL6, and CCL2 was upregulated when autophagosomes were accumulating in obese WAT.19 Similarly, we found expression levels of both TNF and SERPINE1 (serpin family E member 1) increased in differentiated 3T3L1 adipocytes when lysosomal activity was inhibited with BAF (data not shown). Therefore, we suggest lysosomal dysfunction triggers upregulation of inflammatory cytokines via inflammasome activation in obese WAT.
In conclusion, we demonstrated impairment of lysosomal clearance in obese WAT, resulting in autophagosome accumulation. This effect is attributable to lysosomal dysfunction, as represented by CTSL inhibition and compensatory activation of CTSB. Lysosomal dysfunction also induces senescence-like changes and inflammasome activation in obese WAT, leading to disturbances in adipocyte homeostasis. Our results provide novel insight into early pathologies of obese WAT. To clarify valuable therapeutic strategies for obesity, further analyses concerning lysosomal dysfunction and dysregulation of CTSL and CTSB are required.
Materials and methods
Animals and diets
Experiments using mice were conducted in accordance with provisions of the Ethics Review Committee for Animal Experimentation at the Tokyo University of Science. Male C57BL/6 mice (3 wk old) were purchased from CREA Japan and maintained in temperature-controlled, specific pathogen-free conditions with 12-h light/dark cycles within the animal facility at Tokyo University of Science. In the first experiment, male C57BL/6 mice at 4 wk of age were randomly divided into 2 groups: normal diet (ND; Nosan) and high fat diet (HFD; CREA, HFD32). After 18 or 30 wk, mice were killed and epididymal WAT was collected. In the second experiment, male C57BL/6 mice (4 wk of age) were randomly divided into 4 groups: ND, 4HFD, 8HFD and 18HFD. The last 3 groups were provided HFD for 4, 8 or 18 wk before euthanasia, respectively; whereas, ND mice were provided ND throughout their lives. After 22 wk, all 4 groups of mice were killed and epididymal WAT was collected. In the third experiment, male C57BL/6 mice (5 wk of age) and ob/ob mice (5 wk of age) were purchased from Charles River Laboratories Japan (Yokohama, Japan). At 6 wk of age, ob/ob mice were randomly divided into 2 groups: ND and HFD. C57BL/6 mice were provided ND throughout their lives, as control. At 10 wk of age, all 3 groups of mice were killed and epididymal WAT was collected.
Ex vivo experiment
Ex vivo analyses were performed as described previously.27 Briefly, WAT explants from ND mice or 30-wk-old HFD mice were minced into small tissue fragments and preincubated for 1 h in Dulbecco's modified Eagle's medium (Wako, 041–29775) supplemented with 10% fetal bovine serum (Bovogen Biologicals, SFBS-F) and 1% penicillin/streptomycin (Sigma-Aldrich, P0781) in a humidified incubator (37°C, 5% CO2). After preincubation, samples were incubated for an additional 24 h in the same medium with or without 10 μM chloroquine (Wako, 038–17971) or 500 nM rapamycin (LC Laboratories, R5000). Samples were then collected and western blotting was performed.
Reagents
CTSL inhibitor (Z-FY-CHO; sc-3132) was purchased from Santa Cruz Biotechnology. E64-d (4321-v), pepstatin A1 (4397-v), CA074 (4322-v) and CA074ME (4323-v) were purchased from Peptide Institute.
Cell culture and differentiation
3T3-L1 preadipocytes were purchased from RIKEN Bioresource Center (Ibaraki, Japan) and were maintained in Dulbecco's modified Eagle's medium (low glucose) (Wako, 041–29775) with 10% fetal bovine serum and 1% penicillin/streptomycin (Sigma).55,56 Differentiation of 3T3-L1 preadipocytes to adipocytes was performed as previously reported by our laboratory.55,56
Vector construction
pMXs-neo-mU6 vector was constructed as previously reported.55,56 We designed a mouse Ctsl shRNA expression vector based on target sequences for effective CTSL protein knockdown, also as previously reported.56 Oligonucleotides for shCtsl and shRNA for the firefly luciferase (Photinus pyralis) control (shLuc) were chemically synthesized (Operon Biotechnology), with sequences listed in Table S1. For our Ctsb-overexpression vector, Ctsb cDNA was amplified from a Ctsb vector (Addgene, 11249; deposited by H. Choe) by PCR using PrimeSTAR® HS DNA polymerase (Takara, R010A) and then subcloned into Xho1- and Not1-digested pMXs-AMNN-Puro vector.54 Oligonucleotide primers for Ctsb were chemically synthesized as follows: 5′-GGGCTCGAGCACCATGTGGCAGCTCTGGGCC-3′ (Xho1) and 5′-GCCGCGGCCGCTTACTTATCGTCGTCATCCTTGTAATCGATCTTTTCCCAGTACTGATCGGTG-3′ (Not1), with restriction enzyme sites indicated in italics and antisense of FLAG-tag coding sequences underlined.
CTSL knockdown and CTSB overexpression in 3T3L1
For CTSL knockdown, mock (shLuc) and 2 CTSL-knockdown 3T3L1 cell lines (shCtsl#1 and shCtsl#2) were generated using a retroviral system with pMXs-neo-mU6 vector, as previously reported.55,56 For CTSB overexpression, mock and CTSB-overexpressing 3T3L1 cell lines (CTSB [OE]) were generated using a retroviral system with pMXs-Puro-AMNN vector. Preadipocyte cell lines were differentiated into mature adipocytes as previously reported.20,55,56 Day 8 to 12 3T3L1 adipocytes, which were fully differentiated with accumulated TG, were used in this study.
Western blot analysis
Western blotting with Immunostar® LD chemiluminescent substrates (Wako, 290–69904) was performed. Signals were detected with an LAS-3000 image analyzer (Fujifilm, Tokyo, Japan) and analyzed using Multigauge software (Fujifilm) as described previously.20,55,56 LC3 (PM036), SQSTM1 (PM045), ATG5 (M153–3), BECN1 (PD017) and LMNB1/Lamin B1 (PM064) were purchased from MBL. LAMP2 (ab13524), CTSB (ab58802), CTSL (ab133641), TFEB (ab2636) and CIDEC (ab16760) were purchased from Abcam. CDKN1A (sc-397) and CASP1 (sc-514) were purchased from Santa Cruz Biotechnology. FLAG M2 (F1804) and ACTB/β-actin (A1978) were purchased from Sigma-Aldrich. TP53 (OP03) was purchased from Millipore. GAPDH (010–25521) was purchased from WAKO.
Cathepsin activity assay
Cathepsin activity was assayed fluorometrically with Z-Arg-Arg-MCA (Peptide Institute, 3123-v) for CTSB and Z-Phe-Arg-MCA for CTSL (Peptide Institute, 3095-v), as previously reported.57 Briefly, WAT or cell pellets were resuspended in lysis buffer (352 mM KH2PO4, 48 mM Na2HPO4, 4 mM EDTA, pH 6.0) and incubated on ice for 60 min before centrifugation for 10 min at 2,100 ×g. Supernatant was collected and protein concentrations were determined with a BCA kit (Pierce, 23225). Supernatant was then added to the reaction buffer (4 mM DTT in lysis buffer) as an assay buffer.
CTSB activity. A total of 100 μL assay buffer (containing 1 μg of protein) was mixed with 100 μL substrate buffer (10 μM Z-Arg-Arg-AMC diluted in 0.1% Brij 35; Sigma, B4184) and incubated at 37°C for 30 min. Fluorescence was measured using a Wallac ARVO MX/Light 1420 Multilabel/Luminescence Counter (PerkinElmer, Waltham, MA, USA) with an excitation/emission of 360/460 nm.
CTSL activity. A total of 100 μL assay buffer (containing 10 μg of protein) was mixed with 100 μL substrate buffer (10 μM Z-Phe-Arg-AMC and 10 μM CA074) and incubated at 37°C for 30 min. Fluorescence was measured as described above.
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed using a CFX Connect™ Real-Time PCR System (Bio-Rad, Hercules, CA, USA) with SYBR® Premix ExTaq™II (Takara, RR820B), as described previously.20,55,56 The following primers were used: Ctsl (forward) 5′-TCG GTG ACA TGA CCA ATG AG-3′, (reverse) 5′-CAC ACA ACC CTT TTC TCT CCA G-3′; Ctsb (forward) 5′-CAT GAC AAG CCT TCC TTC CAC-3′, (reverse) 5′-ATT GTT CCC GTG CAT CAA AG-3′; Lc3b (forward) 5′-CCA GTG ATT ATA GAG CGA TAC AAG G-3′, (reverse) 5′-AAG AAG GCT TGG TTA GCA TTG AG-3′; Sqstm1 (forward) 5′-GAA GCT GAA ACA TGG ACA CTT TG-3′, (reverse) 5′-CAT TGG GAT CTT CTG GTG GAG-3′; Cdkn1a (forward) 5′-AGT ACT TCC TCT GCC CTG CTG-3′, (reverse) 5′-GCG CTT GGA GTG ATA GAA ATC TG-3′; Lamp1 (forward) 5′-TCA GCA TCT CCA ACC ATT CAC-3′, (reverse) 5′-TGAACACACTCTTCCACAGACC-3′; Tbp (forward) 5′-CAG TAC AGC AAT CAA CAT CTC AGC-3′, (reverse) 5′-CAA GTT TAC AGC CAA GAT TCA CG-3′; Rps18 (forward) 5′-TGC GAG TAC TCA ACA CCA ACA T-3′, (reverse) 5′-CTT TCC TCA ACA CCA CAT GAG C-3′.
Statistical analyses
Statistical analyses were performed using the Tukey–Kramer test (with R software) or the Student t test (with or without Bonferroni correction). Data are presented as mean ± standard deviation (SD). P values < 0.05 were considered to be statistically significant.
Supplementary Material
Abbreviations
- ACTB/β-actin
actin, β
- ATG
autophagy related
- BECN1
Beclin 1
- CASP
caspase
- CCL2
C-C motif chemokine ligand 2
- CIDEC
cell death-inducing DFFA-like effector c
- CTSB
cathepsin B
- CTSD
cathepsin D
- CTSL
cathepsin L
- EDTA
ethylenediaminetetraacetic acid
- ER
endoplasmic reticulum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HFD
high fat diet
- IL
interleukin
- LAMP
lysosomal associated membrane protein
- LEP
leptin
- LMNB1
Lamin B1
- MAP1LC3A/B/LC3A/B
microtubule associated protein 1 light chain 3 α/ β
- MTORC1
mechanistic target of rapamycin complex 1
- ND
normal diet
- PCR
polymerase chain reaction
- PYCARD/ASC
PYD and CARD domain containing
- ROS
reactive oxygen species
- RT-PCR
reverse transcription polymerase chain reaction
- SA-GLB1/SA β-gal
senescence-associated galactosidase β 1
- SERPINE1
serpin family E member 1
- shRNA
small hairpin RNA
- SQSTM1
sequestosome 1
- TFEB
transcription factor EB
- TG
triacylglycerides
- TNF
tumor necrosis factor
- TP53
tumor protein 53 (note that the mouse nomenclature is TRP53 and rat nomenclature is TP53, but we use TP53 to refer to both human and murine genes/proteins for simplicity)
- WAT
white adipose tissue
Disclosure of potential conflicts of interest
We certify that there is no conflict of interest of any type regarding the material discussed in the manuscript.
Acknowledgments
We are deeply grateful to all members of the Laboratory of Molecular Pathology and Metabolic Disease of Faculty of Pharmaceutical Sciences, Tokyo University of Science (TUS) for their cooperation. The authors also thank Drs. Fumio Fukai, Takuya Iyoda, and Naoko Otani (TUS) for their help.
Funding
This work was supported by Grants-in-Aid for Scientific Research (C) (No. 19590396) and for Challenging Exploratory Research (No. 26670193) from the Japan Society for the Promotion of Science, and by the MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2014–2018.
References
- [1].Mokdad AH, Ford ES, Bowman BA, Dietz WH, Vinicor F, Bales VS, Marks JS. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 2003; 289(1):76-9; PMID:12503980; http://dx.doi.org/ 10.1001/jama.289.1.76 [DOI] [PubMed] [Google Scholar]
- [2].Yusuf S, Hawken S, Ounpuu S, Bautista L, Franzosi MG, Commerford P, Lang CC, Rumboldt Z, Onen CL, Lisheng L, et al.. INTERHEART Study Investigators. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet 2005; 366(9497):1640-9; PMID:16271645; http://dx.doi.org/ 10.1016/S0140-6736(05)67663-5 [DOI] [PubMed] [Google Scholar]
- [3].Gnacińska M, Małgorzewicz S, Stojek M, Łysiak-Szydłowska W, Sworczak K. Role of adipokines in complications related to obesity: a review. Adv Med Sci 2009; 54(2):150-7; PMID:19875356 [DOI] [PubMed] [Google Scholar]
- [4].Ouchi N, Parker JL, Lugus JJ, Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol 2011; 11(2):85-97; PMID:21252989; http://dx.doi.org/ 10.1038/nri2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 2005; 46(11):2347-5; PMID:16150820; http://dx.doi.org/ 10.1194/jlr.M500294-JLR200 [DOI] [PubMed] [Google Scholar]
- [6].Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 2011; 17(2):179-88; PMID:21217695; http://dx.doi.org/ 10.1038/nm.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stienstra R, van Diepen JA, Tack CJ, Zaki MH, van de Veerdonk FL, Perera D, Neale GA, Hooiveld GJ, Hijmans A, Vroegrijk I, et al.. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc Natl Acad Sci U S A 2011; 108(37):15324-9; PMID:21876127; http://dx.doi.org/ 10.1073/pnas.1100255108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature 2012; 481(7381):278-86; PMID:22258606; http://dx.doi.org/ 10.1038/nature10759 [DOI] [PubMed] [Google Scholar]
- [9].Yahagi N, Shimano H, Matsuzaka T, Najima Y, Sekiya M, Nakagawa Y, Ide T, Tomita S, Okazaki H, Tamura Y, et al.. p53 Activation in adipocytes of obese mice. J Biol Chem 2003; 278(28):25395-400; PMID:12734185; http://dx.doi.org/ 10.1074/jbc.M302364200 [DOI] [PubMed] [Google Scholar]
- [10].Inoue N, Shimano H, Nakakuki M, Matsuzaka T, Nakagawa Y, Yamamoto T, Sato R, Takahashi A, Sone H, Yahagi N, et al.. Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor. Mol Cell Biol 2005; 25(20):8938-47; PMID:16199872; http://dx.doi.org/ 10.1128/MCB.25.20.8938-8947.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Minamino T, Orimo M, Shimizu I, Kunieda T, Yokoyama M, Ito T, Nojima A, Nabetani A, Oike Y, Matsubara H, et al.. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med 2009; 15(9):1082-7; PMID:19718037; http://dx.doi.org/ 10.1038/nm.2014 [DOI] [PubMed] [Google Scholar]
- [12].Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell 2008; 132:27-42; PMID:18191218; http://dx.doi.org/ 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451(7182):1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; http://dx.doi.org/ 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- [15].Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 2009; 119:3329-39; PMID:19855132; http://dx.doi.org/ 10.1172/JCI35541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA 2009; 106:19860-5; PMID:19910529; http://dx.doi.org/ 10.1073/pnas.0906048106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Perry CN, Kyoi S, Hariharan N, Takagi H, Sadoshima J, Gottlieb RA. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol 2009; 453:325-42; PMID:19216914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ost A, Svensson K, Ruishalme I, Brännmark C, Franck N, Krook H, Sandström P, Kjolhede P, Strålfors P. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med 2010; 16:235-46; PMID:20386866; http://dx.doi.org/ 10.2119/molmed.2010.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yoshizaki T, Kusunoki C, Kondo M, Yasuda M, Kume S, Morino K, Sekine O, Ugi S, Uzu T, Nishio Y, et al.. Autophagy regulates inflammation in adipocytes. Biochem Biophys Res Commun 2012; 417:352-7; PMID:22155234; http://dx.doi.org/ 10.1016/j.bbrc.2011.11.114 [DOI] [PubMed] [Google Scholar]
- [20].Mikami K, Okita N, Tokunaga Y, Ichikawa T, Okazaki T, Takemoto K, Nagai W, Matsushima S, Higami Y. Autophagosomes accumulate in differentiated and hypertrophic adipocytes in a p53-independent manner. Biochem Biophys Res Commun 2012; 427:758-63; PMID:23041188; http://dx.doi.org/ 10.1016/j.bbrc.2012.09.134 [DOI] [PubMed] [Google Scholar]
- [21].Gornicka A, Fettig J, Eguchi A, Berk MP, Thapaliya S, Dixon LJ, Feldstein AE. Adipocyte hypertrophy is associated with lysosomal permeability both in vivo and in vitro: role in adipose tissue inflammation. Am J Physiol Endocrinol Metab 2012; 303(5):E597-E606; PMID:22739104; http://dx.doi.org/ 10.1152/ajpendo.00022.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013; 14(5):283-96; PMID:23609508; http://dx.doi.org/ 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta 2012; 1824(1):44-50; PMID:21640203; http://dx.doi.org/ 10.1016/j.bbapap.2011.05.013 [DOI] [PubMed] [Google Scholar]
- [24].Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005; 1(2):84-91; PMID:16874052; http://dx.doi.org/ 10.4161/auto.1.2.1697 [DOI] [PubMed] [Google Scholar]
- [25].Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett 2010; 584(7):1374-8; PMID:20153326; http://dx.doi.org/ 10.1016/j.febslet.2010.02.017 [DOI] [PubMed] [Google Scholar]
- [26].Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/ 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kovsan J, Blüher M, Tarnovscki T, Klöting N, Kirshtein B, Madar L, Shai I, Golan R, Harman-Boehm I, Schön MR, et al.. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab 2011; 96:E268-77; PMID:21047928; http://dx.doi.org/ 10.1210/jc.2010-1681 [DOI] [PubMed] [Google Scholar]
- [28].Jansen HJ, van Essen P, Koenen T, Joosten LA, Netea MG, Tack CJ, Stienstra R. Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology 2012; 153(12):5866-74; PMID:23117929; http://dx.doi.org/ 10.1210/en.2012-1625 [DOI] [PubMed] [Google Scholar]
- [29].Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest 2010; 120(10):3421-31; PMID:20921628; http://dx.doi.org/ 10.1172/JCI42918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332(6036):1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al.. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013; 15(6):647-58; PMID:23604321; http://dx.doi.org/ 10.1038/ncb2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zheng X, Chu F, Mirkin BL, Sudha T, Mousa SA, Rebbaa A. Role of the proteolytic hierarchy between Cathepsin L, cathepsin D and caspase-3 in regulation of cellular susceptibility to apoptosis and autophagy. Biochim Biophys Acta 2008; 1783(12):2294-300; PMID:18775751; http://dx.doi.org/ 10.1016/j.bbamcr.2008.07.027 [DOI] [PubMed] [Google Scholar]
- [33].Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 2008; 9(8):847-56; PMID:18604214; http://dx.doi.org/ 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 2008; 9(8):857-65; PMID:18604209; http://dx.doi.org/ 10.1038/ni.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab 2010; 11(6):467-78; PMID:20519119; http://dx.doi.org/ 10.1016/j.cmet.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al.. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 2008; 8(4):325-32; PMID:18840363; http://dx.doi.org/ 10.1016/j.cmet.2008.08.009 [DOI] [PubMed] [Google Scholar]
- [37].Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009; 137(6):1001-4; PMID:19524504; http://dx.doi.org/ 10.1016/j.cell.2009.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li M, Khambu B, Zhang H, Kang JH, Chen X, Chen D, Vollmer L, Liu PQ, Vogt A, Yin XM. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem 2013; 288(50):35769-80; PMID:24174532; http://dx.doi.org/ 10.1074/jbc.M113.511212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mangieri LR, Mader BJ, Thomas CE, Taylor CA, Luker AM, Tse TE, Huisingh C, Shacka JJ. ATP6V0C knockdown in neuroblastoma cells alters autophagy-lysosome pathway function and metabolism of proteins that accumulate in neurodegenerative disease. PLoS One 2014; 9(4):e93257; PMID:24695574; http://dx.doi.org/ 10.1371/journal.pone.0093257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Deussing J, Roth W, Saftig P, Peters C, Ploegh HL, Villadangos JA. Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc Natl Acad Sci USA 1998; 95(8):4516-21; PMID:9539769; http://dx.doi.org/ 10.1073/pnas.95.8.4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Potts W, Bowyer J, Jones H, Tucker D, Freemont AJ, Millest A, Martin C, Vernon W, Neerunjun D, Slynn G, et al.. CTSL-deficient mice exhibit abnormal skin and bone development and show increased resistance to osteoporosis following ovariectomy. Int J Exp Pathol 2004; 85(2):85-96; PMID:15154914; http://dx.doi.org/ 10.1111/j.0959-9673.2004.00373.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, Olsen BR. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci USA 2002; 99(12):7883-8; PMID:12048238; http://dx.doi.org/ 10.1073/pnas.112632299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mach L, Stüwe K, Hagen A, Ballaun C, Glössl J. Proteolytic processing and glycosylation of Cathepsin B. The role of the primary structure of the latent precursor and of the carbohydrate moiety for cell-type-specific molecular forms of the enzyme. Biochem J 1992; 282(Pt 2):577-82; PMID:1312333; http://dx.doi.org/ 10.1042/bj2820577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Turk B, Bieth JG, Björk I, Dolenc I, Turk D, Cimerman N, Kos J, Colic A, Stoka V, Turk V. Regulation of the activity of lysosomal cysteine proteinases by pH-induced inactivation and/or endogenous protein inhibitors, cystatins. Biol Chem Hoppe Seyler 1995; 376(4):225-30; PMID:7626231; http://dx.doi.org/ 10.1515/bchm3.1995.376.4.225 [DOI] [PubMed] [Google Scholar]
- [45].Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 2012; 1824(1):68-88; PMID:22024571; http://dx.doi.org/ 10.1016/j.bbapap.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene 2008; 27(50):6434-51; PMID:18955971; http://dx.doi.org/ 10.1038/onc.2008.310 [DOI] [PubMed] [Google Scholar]
- [47].Porter K, Nallathambi J, Lin Y, Liton PB. Lysosomal basification and decreased autophagic flux in oxidatively stressed trabecular meshwork cells: implications for glaucoma pathogenesis. Autophagy 2013; 9(4):581-94; PMID:23360789; http://dx.doi.org/ 10.4161/auto.23568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Inami Y, Yamashina S, Izumi K, Ueno T, Tanida I, Ikejima K, Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun 2011; 412(4):618-25; PMID:21856284; http://dx.doi.org/ 10.1016/j.bbrc.2011.08.012 [DOI] [PubMed] [Google Scholar]
- [49].Coffey EE, Beckel JM, Laties AM, Mitchell CH. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer's disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 2014; 263:111-24; PMID:24418614; http://dx.doi.org/ 10.1016/j.neuroscience.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bevec T, Stoka V, Pungercic G, Dolenc I, Turk V. Major histocompatibility complex class II-associated p41 invariant chain fragment is a strong inhibitor of lysosomal CTSL. J Exp Med 1996; 183(4):1331-8; PMID:8666891; http://dx.doi.org/ 10.1084/jem.183.4.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Liao G, Yao Y, Liu J, Yu Z, Cheung S, Xie A, Liang X, Bi X. Cholesterol accumulation is associated with lysosomal dysfunction and autophagic stress in Npc1 −/− mouse brain. Am J Pathol 2007; 171(3):962-75; PMID:17631520; http://dx.doi.org/ 10.2353/ajpath.2007.070052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hannaford J, Guo H, Chen X. Involvement of cathepsins B and L in inflammation and cholesterol trafficking protein NPC2 secretion in macrophages. Obesity (Silver Spring) 2013; 21(8):1586-95; PMID:23666609; http://dx.doi.org/ 10.1002/oby.20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8(9):729-40; PMID:17667954; http://dx.doi.org/ 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- [54].Smogorzewska A, de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO 2002; 21(16):4338-48; http://dx.doi.org/ 10.1093/emboj/cdf433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Matsushima S, Okita N, Oku M, Nagai W, Kobayashi M, Higami Y. An Mdm2 antagonist, Nutlin-3a, induces p53-dependent and proteasome-mediated poly(ADP-ribose) polymerase1 degradation in mouse fibroblasts. Biochem Biophys Res Commun 2011; 407(3):557-61; PMID:21419099; http://dx.doi.org/ 10.1016/j.bbrc.2011.03.061 [DOI] [PubMed] [Google Scholar]
- [56].Okita N, Ishikawa N, Mizunoe Y, Oku M, Nagai W, Suzuki Y, Matsushima S, Mikami K, Okado H, Sasaki T, et al.. Inhibitory effect of p53 on mitochondrial content and function during adipogenesis. Biochem Biophys Res Commun 2014; 446(1):91-7; PMID:24565844; http://dx.doi.org/ 10.1016/j.bbrc.2014.02.059 [DOI] [PubMed] [Google Scholar]
- [57].Barrett AJ, Kirschke H. CTSB, cathepsin H, and CTSL. Methods Enzymol 1981; 80 Pt C:535-61; PMID:7043200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.