Abstract
The mechanism of nonflagellar swimming of marine unicellular cyanobacteria remains poorly understood. SwmA is an abundant cell surface-associated 130-kDa glycoprotein that is required for the generation of thrust in Synechococcus sp. strain WH8102. Ultrastructural comparisons of wild-type cells to a mutant strain in which the gene encoding SwmA has been insertionally inactivated reveal that the mutant lacks a layer external to the outer membrane. Cryofixation and freeze-substitution are required for the preservation of this external layer. Freeze fracturing and etching reveal that this additional layer is an S-layer. How the S-layer might function in motility remains elusive; however, this work describes an ultrastructural component required for this unique type of swimming. In addition, the work presented here describes the envelope structure of a model swimming cyanobacterium.
The mechanism of swimming motility in marine Synechococcus, in the apparent absence of readily visible locomotor structures, remains a mystery 19 years after its discovery (33). The swimming behavior of marine Synechococcus resembles that of flagellated bacteria in several respects. Swimming Synechococcus strains are observed to rotate about their longitudinal axis as they translocate at speeds of up to 25 μm/s. Moreover, cells that become fortuitously attached to a microscope slide or coverslip rotate about the point of attachment (34). Nevertheless, numerous attempts at visualizing an appendage or organelle, including transmission electron microscopy (TEM) of negatively stained cells, high-intensity dark-field microscopy, and shearing experiments, did not reveal any (33, 34).
The rotational behavior of swimming and attached cells suggests that the cell surface or some component thereof rotates and hence may function in swimming. One component of the cell surface that is required for swimming has been identified. SwmA is a 130-kDa cell surface glycoprotein that is required for swimming (5). It is not an integral outer membrane (OM) protein but can be removed from cells by treatment with EDTA (5). Cells in which swmA has been insertionally inactivated, which hence do not express the SwmA protein, are nonmotile yet, when fortuitously attached to a microscope slide or coverslip, are still observed to rotate about their point of attachment. Thus, SwmA is somehow required for the generation of thrust but not torque (5). In order to understand how SwmA functions in the generation of thrust, we sought to determine its localization in cells of Synechococcus. Preliminary TEM experiments analyzing immunolabeled thin sections of motile Synechococcus cells indicated that SwmA is associated with the outer membrane (6; B. Brahamsha, unpublished results). To understand better the localization of SwmA, further TEM analyses of thin sections as well as of whole cells were undertaken.
Synechococcus sp. strain S1A1, in which the swmA gene is insertionally inactivated, continues to express all of the other major and minor cell surface polypeptides found in the wild type and lacks only the SwmA protein (5). We compared the ultrastructure of strain S1A1 to that of the wild-type strain, WH8102. By the use of cryofixation combined with freeze-substitution, methods known for preserving delicate structures that may be destroyed by standard fixation procedures (12, 16), we show here that strain S1A1 lacks an outer cell layer. Furthermore, by use of the freeze-etching technique, we show that this additional layer is a surface layer (S-layer). The use of these techniques allowed us to detect an ultrastructural component important for this novel type of swimming motility.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Motile Synechococcus sp. strain WH8102 was used along with a nonmotile mutant strain, S1A1, in which the swmA gene had been insertionally inactivated. The inactivation was carried out as described in reference 5, using an internal fragment of the swmA gene (nucleotides 84507 to 84926 of the Synechococcus sp. strain WH8102 genome) (22) cloned into the EcoRI site of the suicide vector pMUT100. Both strains were grown in SN medium (32) prepared with local seawater as described previously (5). Cultures of S1A1 contained 15 μg of kanamycin ml−1 to select for and maintain the inserted plasmid inactivating swmA. Fifty-milliliter cultures were maintained in 125-ml flasks under constant illumination (10 microeinsteins · m−2 · s−1) without shaking at 25°C.
Chemical fixation.
Cells were fixed and prepared according to techniques described by Beveridge et al. (3) with some modifications. Cells were fixed in SN medium containing 2% (wt/vol) osmium tetroxide for 1.5 h at 4°C, rinsed three times with SN medium, and fixed with 2% glutaraldehyde (EM grade; Sigma) for 1.5 h at 4°C, followed by three rinses in phosphate-buffered saline. Following fixation, cells were enrobed in 1.5% low-melting-point agarose (FMC Bioproducts, Rockland, Maine) as described by Rippka et al. (25) with some modifications. The cell-containing agarose was cut into 1-mm cubes and washed with deionized water two times for 5 min each time. The cubes were stained with 1% (wt/vol) uranyl acetate for 30 min at room temperature and then rinsed with deionized water three times for 5 min each time. The cubes were then dehydrated in a graded series of ethanol and acetone washes. Once dehydrated, the agarose cubes were embedded in Durcupan resin and polymerized in a vacuum oven at 60°C for a minimum of 24 h.
Cryofixation and freeze-substitution.
In addition to conventional fixation, samples were prepared for TEM by cryofixation and freeze-substitution. Cells were centrifuged at 7,000 × g at room temperature for 10 min to form a loose pellet. A small amount of this pellet was then sandwiched between 300-mesh Gilder Fine Bar Beehive copper grids (Ted Pella, Inc.) and rapidly plunged into liquid propane maintained at −188°C. The grids, along with adhered cells, were incubated at −87°C in anhydrous acetone containing 2% (wt/vol) osmium tetroxide for 48 h. The grids were then slowly (1°C min−1) and incrementally (with a pause at −20°C for 2 h and another at −4°C for 1 h) brought up to room temperature. Grids were rinsed in anhydrous acetone and stained in anhydrous acetone containing 2% (wt/vol) uranyl acetate for 1 h at −4°C. The grids were then dehydrated and embedded as described above. Thin sections (∼80 nm) were cut on a Reichert-Jung Ultracut-E ultramicrotome and mounted on Formvar- and carbon-coated copper grids. Grids were poststained in 1% (wt/vol) uranyl acetate and then stained in Sato lead (27). Sections were then visualized and recorded using a JEOL 100CX transmission electron microscope at an acceleration voltage of 80 kV.
Freeze fracturing and etching.
Samples were processed as previously described by Heuser and Salpeter (13-15). Briefly, cells were concentrated by centrifugation at 7,000 × g at room temperature for 10 min to form a loose pellet and then immediately quick-frozen by slamming onto a supercooled copper block. Samples were then fractured and freeze-etched to various degrees in a Balzer's vacuum evaporator at −100°C. Replication with a 2-nm-thick layer of platinum, rotary-deposited from 18° above the horizontal, was achieved by standard evaporation techniques. Samples were backed with 4 to 6 nm of pure carbon, rotary-deposited from 75° above the horizontal. Replicas were visualized and recorded using a JEOL TEM microscope at an acceleration voltage of 100 kV.
RESULTS
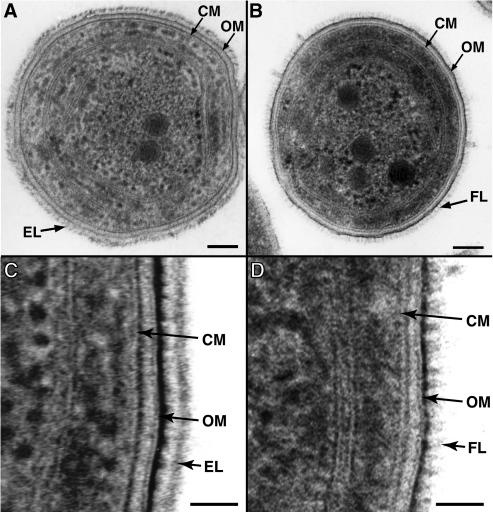
Thin sections of wild-type Synechococcus sp. strain WH8102 prepared by cryosubstitution revealed a multilayered gram-negative cell envelope profile characteristic of cyanobacteria. The cytoplasmic membrane (CM) is closely associated with a 6-nm-wide layer of darkly staining peptidoglycan (Fig. 1 A and C). This close association of peptidoglycan with the CM has been observed in other freeze-substituted preparations of cyanobacteria (16) and some other gram-negative bacteria (12). A double-tracked 9-nm-wide OM is then separated from the peptidoglycan and CM by a 9-nm-wide lightly stained periplasmic space. The OM is asymmetrically stained, with the outer leaflet being more densely stained and thicker than the inner leaflet, as has been seen in various other cryopreserved bacteria (11, 12). Immediately external to the outer membrane there are a light-staining region 12 nm wide and then a dark-staining layer approximately 11 nm wide. In some cross sections, regions of this dark-staining external layer (EL) exhibit a distinct periodic arrangement (Fig. 1A). Cross sections show that this EL extends around the entire cell circumference. The location and periodic nature of this layer observed in thin sections suggest that this EL may be an S-layer. Such surface structures are better visualized by freeze fracturing and are described in more detail below.
FIG. 1.
Thin-section transmission electron micrographs of Synechococcus sp. strains WH8102 (A, C) and S1A1 (B, D) prepared by cryofixation and freeze-substitution. The external layer (EL) is observed to extend around the whole wild-type WH8102 cell and exhibits distinct periodicity (A). In strain S1A1 a fibrillar layer (FL), which also extends around the whole cell, is observed. Higher-magnification micrographs show detail of the cell envelope structures of wild-type (C) and S1A1 mutant (D) cells. CM, cytoplasmic membrane; OM, outer membrane. Bars, 100 (A, B) and 30 nm (C, D).
In contrast, this EL is missing in the swmA mutant (Fig. 1B and D). Cryosubstituted thin sections of strain S1A1 exhibit the typical multilayered cell envelope observed in wild-type cells. Strain S1A1 cells also display staining of material external to the outer membrane, although its appearance is distinct from that of the EL of the wild type. These nonmotile cells possess a layer of diffuse-staining material with a periodic fibril-like appearance. This fibrillar layer (FL) appears as short, 18-nm-long fibrils of various thicknesses extending perpendicular from the external surface of the outer membrane (Fig. 1B and D). We have examined several hundred cells of both the wild type and the mutant, and the results shown here are representative. Cross sections show that this layer also extends around the entire cell surface (Fig. 1B and D). Cells prepared by conventional chemical fixation failed to show these differences (data not shown), thus highlighting the importance of quick-freeze fixation for preserving such structures.
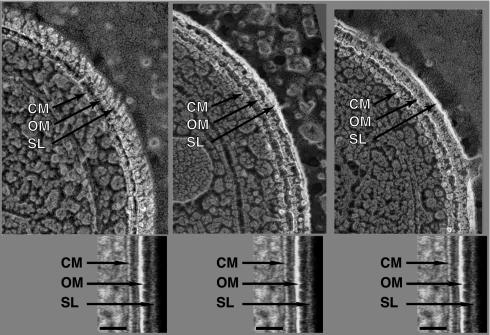
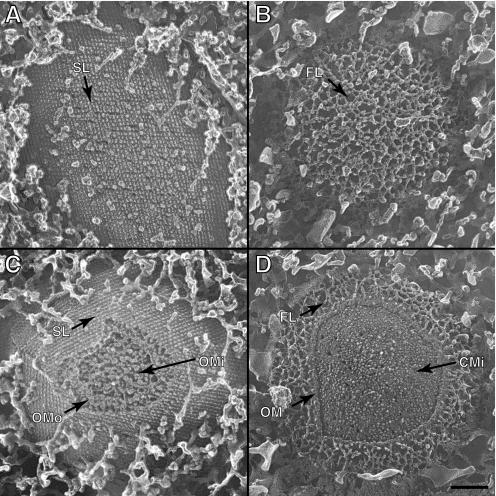
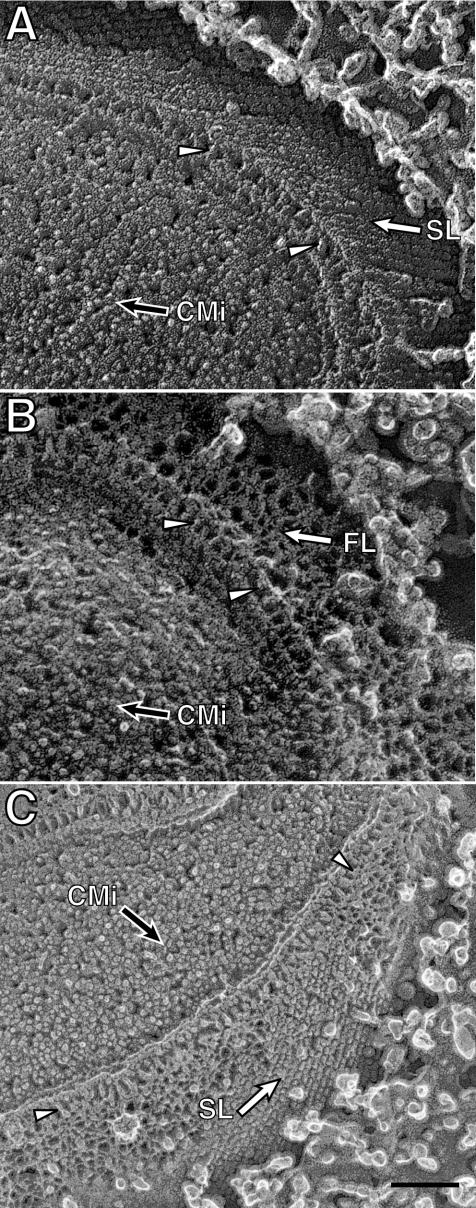
Intact cells of the wild type and the swmA mutant were also prepared by quick-freezing, fracturing, and in some cases, etching. Fractures producing clean cross sections across the cell envelope exhibit a multilayered cell envelope profile identical to that observed in thin-section preparations. An alignment of images from thin-section and freeze-fractured preparations illustrates the similarity of the structures observed by these two methods (Fig. 2). Oblique fractures providing surface views of the outermost layer of wild-type cells clearly exhibit the paracrystalline array of an S-layer (Fig. 3A). The S-layer appears as a regular array of hemispheres on the surface of the cell. These hemispheres have center-to-center spacing of 12 nm and yield an oblique 110° lattice. Cross-sectional fractures of wild-type cells reveal the S-layer as an 8-nm-thick outermost layer. The S-layer is separated from the outer membrane by a 14-nm space in which there are apparent columnar connections between the continuous S-layer and the outer membrane surface (Fig. 4A). Additionally, fractures removing the S-layer reveal a layer of fibrillar material beneath the S-layer in wild-type cells (Fig. 4C). These aerial views of fractured wild-type cells expose fibrillar material that is quite similar in appearance to the fibrillar material seen in surface views of the mutant (Fig. 3B and D and 4B). These observations are summarized in the model shown in Fig. 5.
FIG. 2.
Comparison of Synechococcus sp. strain WH8102 cells prepared by freeze fracturing and etching (upper panels) and cryofixation and freeze-substitution (lower panels). Both cryopreservation techniques yield similar cell envelope profiles. SL, S-layer. Bars, 30 nm.
FIG. 3.
Transmission electron micrographs of Synechococcus sp. strains WH8102 (A, C) and S1A1 (B, D) prepared by freeze fracturing and etching. Aerial views display the paracrystalline lattice structure of an S-layer on wild-type cells (A, C), while S1A1 cells (B, D) lack the structured surface layer and display irregular organization of the outermost material on these cells. Partial fractures of WH8102 cells (C) and S1A1 cells (D) reveal layers of cell envelope covered by the outermost layers. SL, S-layer; OMo, outer membrane outer leaflet; OMi, outer membrane inner leaflet; CMi, cytoplasmic membrane inner leaflet. Bar, 100 nm (all panels).
FIG. 4.
Freeze fractured and etched preparations of Synechococcus sp. strain WH8102 (A, C) and S1A1 (B) cells. Etching has uncovered the outermost surfaces of the cells (open arrows), while partial cross-fractures reveal the cytoplasmic membrane inner leaflet face (CMi, filled arrows) and cell envelope layers. Fracturing has revealed fibril-like structures (arrowheads) immediately external to the outer membrane in both strains. Panel C shows a cell in which the S-layer (open arrow) has been fractured away, revealing the material immediately underneath. The fracture plane then crossed several cell envelope layers to reveal the cytoplasmic membrane inner leaflet face. SL, S-layer; CMi, cytoplasmic membrane inner leaflet. Bar, 100 nm (all panels).
FIG. 5.
Model of Synechococcus sp. strain WH8102 cell envelope structure.
The swmA mutant prepared by the quick-freezing and deep-etching method displays all of the same cell envelope structures as the wild type, with the exception of the missing S-layer. The outermost surface of S1A1 lacked regularity and the lattice-like appearance of the outer surface as seen in wild-type cells (Fig. 3B and D). Cross-sectional fractures reveal fibril-like structures arrayed perpendicular to the cell envelope (Fig. 4B). This FL extends 19 nm from the cell surface. Just as seen in thin sections, fractured cells of S1A1 lack the outermost S-layer. The outermost FL appears irregularly arranged in surface views (Fig. 3).
DISCUSSION
The mechanism of swimming motility in marine Synechococcus remains elusive due in large part to the lack of readily visible propulsive organelles. In order to address this question, we are making use of swimming motility mutants (5; J. McCarren and B. Brahamsha, unpublished data). The results presented here show, for the first time, an ultrastructural difference between a wild-type Synechococcus strain that is capable of swimming and a mutant that is not. Wild-type cells exhibit a cell envelope characterized by the presence of a crystalline S-layer, which is lacking in a mutant in which a single gene, swmA, has been insertionally inactivated. Polar effects of the insertion are unlikely since swmA does not appear to be part of an operon (5, 22), and 5′ and 3′ neighboring genes are encoded on the opposite strand.
Several observations suggest that the S-layer is composed of SwmA. First, swmA mutants lack the S-layer. Furthermore, as shown previously (5), a comparison of the outer membrane protein profiles of the wild type and the swmA mutant revealed that the only difference between these strains is the loss of SwmA. In addition, SwmA is one of the most abundant proteins in motile cells (5), which is a characteristic shared by other S-layer proteins (20) and is consistent with the hypothesis that this protein forms a layer covering the entire cell surface. Also, divalent cations, specifically Ca2+, have been shown to be important for the integrity, attachment, and recrystallization of S-layers in various bacteria (9, 10, 21, 37), and treatment with divalent cation chelators causes some S-layers to dissociate from the cell surface. Treatment of Synechococcus sp. strain WH8102 with the chelator EDTA removes the outer membrane and solubilizes SwmA (5). Finally, while S-layer proteins are highly divergent, sharing essentially no homology in amino acid comparisons (9), SwmA, like other S-layer proteins, has numerous glycine- and aspartate-rich repeats that are thought to function in calcium ion bridging to the outer membrane (5).
S-layers are not uncommon in cyanobacteria (16, 17, 30). Additionally, fibrillar structures similar in size and arrangement to the ones we report here, spanning the region between the S-layer and outer membrane, have been observed in TEM analyses of both a coastal marine strain (23) and a freshwater strain of Synechococcus (28), neither of which exhibits swimming motility. Samuel et al. (26) have also used rapid-freezing and fracturing methods to examine cells of Synechococcus sp. strain WH8113, a motile strain closely related to strain WH8102 (31), and their results are similar to the ones reported here in several respects. Strain WH8113 was shown to have an S-layer arranged in a similar 110° rhomboid organization with 12-nm spacing separating individual units. In addition, however, these authors visualized short fibrils (5 nm wide and 150 nm long), or spicules, covering the surface of the cell. They proposed that these spicules, which were reported to span the entire cell envelope, may be able to transmit motion in the cytoplasmic or outer membrane to the surrounding medium. Wolgemuth et al. (36) further refined this model by postulating the presence of a mechanochemical filament located in the cytoplasmic membrane, which would be attached to these spicules and would cause them to move by bending. These are plausible models; however, it is clear from the results presented here that the presence of these spicules is not a characteristic of all swimming Synechococcus strains. Using methods like the ones used by Samuel et al. (26), we have never observed such spicules on the surface of WH8102 cells. The absence of spicules in the swimming strain WH8102 suggests that such structures are not required for motility in all swimming Synechococcus strains. Ehlers et al. (8) have proposed a model in which localized contractions and expansions of the OM generate surface waves of sufficient amplitude and frequency to propel Synechococcus at the observed speeds. This model does not require spicules and hence remains a possibility. The challenge to identify the mechanism of wave generation required by such a model remains.
How then does the S-layer of swimming Synechococcus strains function in motility? While S-layers are also ubiquitous in archaea and common in many bacteria (29), in only a few cases has the function of an S-layer been determined (4). S-layers are known to be sites of attachment for exoenzymes (7, 19). Perhaps in swimming Synechococcus strains, the S-layer serves as the site of attachment or the site for interaction of yet-to-be-discovered motility components. S-layers have also been shown to perform a shape-maintaining role in archaea (2). Conceivably, the S-layer may function to maintain cell shape or cell envelope integrity in the face of continuous disruption associated with the surface wave generation mechanism proposed by Ehlers et al. (8).
In several strains of Oscillatoria, a filamentous gliding cyanobacterium, an array of helically arranged fibers is present between the peptidoglycan and the outer membrane (1), and one model to explain the gliding motility of these organisms invokes the generation of surface waves produced by the contraction of such an array. While we have not observed such an array in Synechococcus sp. strain WH8102, there may be other ways of generating surface waves, such as localized swelling of the cell surface (18, 24). Could the S-layer be involved in such swelling? The source of energy for swimming of Synechococcus is the sodium motive force (35), suggesting that the “motor” responsible for converting chemical into mechanical energy is located in the cytoplasmic membrane. How is this energy transduced to the cell surface? While they are unable to generate thrust, and hence unable to translocate, swmA mutants can generate torque, since attached cells can still rotate about a point of attachment (5, 6). What is the mechanism of torque generation? How is it coupled to thrust? In order to answer these questions, it will be necessary to identify the other components of the motility machinery as well as to identify the proteins with which SwmA interacts.
The complete sequence of the genome of Synechococcus sp. strain WH8102 has recently been obtained (22). An examination of this genome provides further indication of the uniqueness of the mechanism of swimming of Synechococcus strains, as no homologs for proteins associated with other forms of prokaryotic motility were found (22). Recently, through transposon mutagenesis, three clusters of genes (swmA is in one of them) that are required for swimming were identified (22; McCarren and Brahamsha, unpublished). An analysis of these mutants, using both biochemical approaches and the ultrastructural methods described here, should provide further insights into what remains a fascinating mystery.
Acknowledgments
We thank Brian Palenik for critically reading the manuscript.
This work was supported by grants NSF MCB97-27759 and DOE DE-FG03-01ER63148. Work conducted in the laboratory of J. Heuser was supported by USPHS grant GM29647. Work performed at the National Center for Microscopy and Imaging Research at UCSD was supported by grant RR04050 to Mark Ellisman from the National Center of Research Resources of the National Institutes of Health.
REFERENCES
- 1.Adams, D. G., D. Ashworth, and B. Nelmes. 1999. Fibrillar array in the cell wall of a gliding filamentous cyanobacterium. J. Bacteriol. 181:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister, W., and G. Lembcke. 1992. Structural features of archaebacterial cell envelopes. J. Bioenerg. Biomembr. 24:567-575. [DOI] [PubMed] [Google Scholar]
- 3.Beveridge, T. J., T. J. Popkin, and R. M. Cole. 1994. Electron microscopy, p. 42-71. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 4.Beveridge, T. J., P. H. Pouwels, M. Sara, A. Kotiranta, K. Lounatmaa, K. Kari, E. Kerosuo, M. Haapasalo, E. M. Egelseer, I. Schocher, U. B. Sleytr, L. Morelli, M. L. Callegari, J. F. Nomellini, W. H. Bingle, J. Smit, E. Leibovitz, M. Lemaire, I. Miras, S. Salamitou, P. Beguin, H. Ohayon, P. Gounon, M. Matuschek, K. Sahm, H. Bahl, R. Grogono-Thomas, J. Dworkin, M. J. Blaser, R. M. Woodland, D. G. Newell, M. Kessel, and S. F. Koval. 1997. Functions of S-layers. FEMS Microbiol. Rev. 20:99-149. [DOI] [PubMed] [Google Scholar]
- 5.Brahamsha, B. 1996. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. USA 93:6504-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahamsha, B. 1999. Non-flagellar swimming in marine Synechococcus. J. Mol. Microbiol. Biotechnol. 1:59-62. [PubMed] [Google Scholar]
- 7.Egelseer, E., I. Schocher, M. Sara, and U. B. Sleytr. 1995. The S-layer from Bacillus stearothermophilus DSM 2358 functions as an adhesion site for a high-molecular-weight amylase. J. Bacteriol. 177:1444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehlers, K. M., A. D. T. Samuel, H. C. Berg, and R. Montgomery. 1996. Do cyanobacteria swim using traveling surface waves? Proc. Natl. Acad. Sci. USA 93:8340-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 10.Garduno, R., B. Phipps, and W. Kay. 1995. Physical and functional S-layer reconstitution in Aeromonas salmonicida. J. Bacteriol. 177:2684-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, L. L., and T. J. Beveridge. 1990. Evaluation of freeze-substitution and conventional embedding protocols for routine electron microscopic processing of eubacteria. J. Bacteriol. 172:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, L. L., R. Harris, W. Villiger, and T. J. Beveridge. 1991. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J. Bacteriol. 173:1623-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heuser, J. 1981. Preparing biological samples for stereomicroscopy by the quick-freeze, deep-etch, rotary-replication technique. Methods Cell Biol. 22:97-122. [DOI] [PubMed] [Google Scholar]
- 14.Heuser, J. 1980. The quick-freeze, deep-etch method of preparing samples for high-resolution, 3-D microscopy. Trends Biochem. Sci. 6:64-68. [Google Scholar]
- 15.Heuser, J., and S. R. Salpeter. 1979. Organization of acetylcholine receptors in quick-frozen, deep-etched, and rotary-replicated Torpedo postsynaptic membrane. J. Cell Biol. 82:150-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiczyk, E., and W. Baumeister. 1995. Envelope structure of four gliding filamentous cyanobacteria. J. Bacteriol. 177:2387-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoiczyk, E., and A. Hansel. 2000. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J. Bacteriol. 182:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch, A. L. 1990. The sacculus contraction/expansion model for gliding motility. J. Theor. Biol. 142:95-112. [Google Scholar]
- 19.Matuschek, M., G. Burchhardt, K. Sahm, and H. Bahl. 1994. Pullulanase of Thermoanaerobacterium thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene, composite structure of the enzyme, and a common model for its attachment to the cell surface. J. Bacteriol. 176:3295-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messner, P., and U. Sleytr. 1992. Crystalline bacterial cell-surface layers, p. 213-273. In A. H. Rose (ed.), Advances in microbial physiology, vol. 33. Academic Press, San Diego, Calif. [DOI] [PubMed] [Google Scholar]
- 21.Nomellini, J., S. Kupcu, U. Sleytr, and J. Smit. 1997. Factors controlling in vitro recrystallization of the Caulobacter crescentus paracrystalline S-layer. J. Bacteriol. 179:6349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 23.Perkins, F., L. Haas, D. Phillips, and K. Webb. 1981. Ultrastructure of a marine Synechococcus possessing spinae. Can. J. Microbiol. 27:318-329. [DOI] [PubMed] [Google Scholar]
- 24.Pitta, T. P., E. E. Sherwood, A. M. Kobel, and H. C. Berg. 1997. Calcium is required for swimming by the nonflagellated cyanobacterium Synechococcus strain WH8113. J. Bacteriol. 179:2524-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rippka, R., J. B. Waterbury, and G. Cohen-Bazire. 1974. A cyanobacterium which lacks thylakoids. Arch. Microbiol. 100:419-436. [Google Scholar]
- 26.Samuel, A. D. T., J. D. Peterson, and T. S. Reese. 24 April 2001, posting date. Envelope structure of Synechococcus sp. WH8113, a nonflagellated swimming cyanobacterium. BMC Microbiol. 1:4. [Online.] http://www.biomedcentral.com/1471-2180/1/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato, T. 1967. A modified method for lead staining. J. Electron Microsc. 16:133-141. [PubMed] [Google Scholar]
- 28.Schultze-Lam, S., and T. J. Beveridge. 1994. Physicochemical characteristics of the mineral-forming S-layer from the cyanobacterium Synechococcus strain GL24. Can. J. Microbiol. 40:216-223. [Google Scholar]
- 29.Sleytr, U. B., P. Messner, D. Pum, and M. Sara. 1996. Crystalline bacterial cell surface proteins. Academic Press, Austin, Tex.
- 30.Smarda, J., D. Smajs, J. Komrska, and V. Krzyzanek. 2002. S-layers on cell walls of cyanobacteria. Micron 33:257-277. [DOI] [PubMed] [Google Scholar]
- 31.Toledo, G., B. Palenik, and B. Brahamsha. 1999. Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl. Environ. Microbiol. 65:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterbury, J. B., and J. M. Willey. 1988. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 167:100-105. [Google Scholar]
- 33.Waterbury, J. B., J. M. Willey, D. G. Franks, F. W. Valois, and S. W. Watson. 1985. A cyanobacterium capable of swimming motility. Science 230:74-76. [DOI] [PubMed] [Google Scholar]
- 34.Willey, J. M. 1988. Characterization of swimming motility in a marine cyanobacterium. Ph.D. thesis. Woods Hole Oceanographic Institution and Massachusetts Institution of Technology, Cambridge, Mass.
- 35.Willey, J. M., J. B. Waterbury, and E. P. Greenberg. 1987. Sodium-coupled motility in a swimming cyanobacterium. J. Bacteriol. 169:3429-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolgemuth, C. W., O. Igoshin, and G. Oster. 2003. The motility of mollicutes. Biophys. J. 85:828-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, L., Z. Pei, S. Fujimoto, and M. Blaser. 1992. Reattachment of surface array proteins to Campylobacter fetus cells. J. Bacteriol. 174:1258-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]