ABSTRACT
Most species of filamentous cyanobacteria are capable of gliding motility, likely via a conserved type IV pilus-like system that may also secrete a motility-associated polysaccharide. In a subset of these organisms, motility is achieved only after the transient differentiation of hormogonia, which are specialized filaments that enter a nongrowth state dedicated to motility. Despite the fundamental importance of hormogonia to the life cycles of many filamentous cyanobacteria, the molecular regulation of hormogonium development is largely undefined. To systematically identify genes essential for hormogonium development and motility in the model heterocyst-forming filamentous cyanobacterium Nostoc punctiforme, a forward genetic screen was employed. The first gene identified using this screen, designated ogtA, encodes a putative O-linked β-N-acetylglucosamine transferase (OGT). The deletion of ogtA abolished motility, while ectopic expression of ogtA induced hormogonium development even under hormogonium-repressing conditions. Transcription of ogtA is rapidly upregulated (1 h) following hormogonium induction, and an OgtA-GFPuv fusion protein localized to the cytoplasm. In developing hormogonia, accumulation of PilA but not HmpD is dependent on ogtA. Reverse transcription-quantitative PCR (RT-qPCR) analysis indicated equivalent levels of pilA transcript in the wild-type and ΔogtA mutant strains, while a reporter construct consisting of the intergenic region in the 5′ direction of pilA fused to gfp produced lower levels of fluorescence in the ΔogtA mutant strain than in the wild type. The production of hormogonium polysaccharide in the ΔogtA mutant strain is reduced compared to that in the wild type but comparable to that in a pilA deletion strain. Collectively, these results imply that O-GlcNAc protein modification regulates the accumulation of PilA via a posttranscriptional mechanism in developing hormogonia.
IMPORTANCE Filamentous cyanobacteria are among the most developmentally complex prokaryotes. Species such as Nostoc punctiforme develop an array of cell types, including nitrogen-fixing heterocysts, spore-like akinetes, and motile hormogonia, that function in dispersal as well as the establishment of nitrogen-fixing symbioses with plants and fungi. These symbioses are major contributors to global nitrogen fixation. Despite the fundamental importance of hormogonia to the life cycle of filamentous cyanobacteria and the establishment of symbioses, the molecular regulation of hormogonium development is largely undefined. We employed a genetic screen to identify genes essential for hormogonium development and motility in Nostoc punctiforme. The first gene identified using this screen encodes a eukaryotic-like O-linked β-N-acetylglucosamine transferase that is required for accumulation of PilA in hormogonia.
KEYWORDS: gliding motility, hormogonia, Nostoc punctiforme, O-linked β-N-acetylglucosamine transferase, type IV pili
INTRODUCTION
Most species of filamentous cyanobacteria are capable of gliding motility, likely via a conserved type IV pilus-like system that may also secrete a motility-associated polysaccharide (1–3). In a subset of these organisms, motility is achieved only after the transient differentiation of hormogonia, specialized filaments that enter a nongrowth state dedicated to motility (4). In both hormogonium-forming and non-hormogonium-forming strains, motile filaments perform phototaxis to seek out favorable light environments (5) and can establish supracellular structures that arise from the movement of individual filaments within conglomerates (2, 6). In certain heterocyst-forming species, such as Nostoc punctiforme, hormogonia serve as the infective unit for establishing symbioses with plants and fungi, after which nitrogen-fixing heterocysts provide the eukaryotic partner with a source of fixed nitrogen (2, 7, 8). These symbioses are major contributors to global nitrogen fixation (9).
Despite the fundamental importance of hormogonia to the life cycle of many filamentous cyanobacteria and the establishment of these ecologically important symbioses, the molecular regulation of hormogonium development is largely undefined. What is currently known about hormogonium development comes primarily from studies on the model filamentous cyanobacterium N. punctiforme. In wild-type N. punctiforme strains, a constitutive level of hormogonia develops under standard growth conditions, but a number of factors can positively or negatively affect hormogonium differentiation, including putative autogenic repressors and signals from symbiotic plant partners (10–14).
Recent studies have shown that developing hormogonia undergo an extensive program of differential gene transcription (1, 15–17) and have implicated a chemotaxis system encoded by the hmp locus as being essential for hormogonium development (1, 2). Comparative transcriptomics revealed that slightly less than half of the hormogonium-specific changes in gene expression are dependent on a functional hmp locus (1). The deletion of hmp genes does not affect the expression of the putative major pilin PilA or its export out of the cell, implying that expression and assembly of the type IV pilus (T4P) system are independent of the hmp locus (2). In contrast, both the production of hormogonium polysaccharide (HPS) and increased transcription of the HPS synthesis-associated gene cluster, hpsE to hpsK (hpsE–K), are dependent on a functional hmp locus (1, 2). These results imply the existence of at least two independent genetic circuits during hormogonium development, an hmp-independent circuit that includes the genes encoding the T4P-like motor complex, and an hmp-dependent circuit that includes the gene products involved in the synthesis of the hormogonium polysaccharide.
To systematically identify additional components of the hormogonium gene regulatory network, a forward genetic screen was employed. The first gene identified using this screen, designated ogtA, encodes a putative O-linked β-N-acetylglucosamine transferase (OGT). In eukaryotes, OGTs consist of an N-terminal series of tetratricopeptide (TPR) repeats, followed by a C-terminal glycosyl transferase 41 (GT41) domain, and are involved in cell signaling and development by modulating target protein activity via the reversible addition of O-GlcNAc to serine and threonine residues (18, 19). While structurally similar putative OGTs are also widespread among prokaryotes, their function is largely unknown (20–22). A recent study found that inactivation of a gene encoding a putative OGT in the cyanobacterium Synechococcus elongatus PCC 7942 resulted in a pleiotropic phenotype, and it provided biochemical evidence of OGT enzymatic activity for this gene product (23). Here, we provide a detailed characterization of the phenotype for an N. punctiforme ΔogtA mutant strain, the results of which implicate a role for O-GlcNAc protein modification in hormogonium development.
RESULTS
Forward genetic screen to identify genes essential for hormogonium development and motility.
To identify uncharacterized components of the hormogonium motility apparatus, as well as begin to define the gene regulatory network governing hormogonium development, a forward genetic screen using transposon mutagenesis was employed. Wild-type N. punctiforme constitutively produces hormogonia, evidenced by rapid colony spreading on plates. This robust motility makes the wild-type strain ideal for studying hormogonium development and motility. However, in this instance, the rapid colony spreading would result in confluent colonies on agar plates, potentially obscuring any nonmotile mutants. Therefore, the laboratory-derived spontaneous mutant strain UCD153 was used instead (15). Strain UCD153 retains the ability to differentiate motile hormogonia when induced by the removal of combined nitrogen or the addition of plant exudates, but it does not constitutively differentiate hormogonia and displays less robust motility. Exconjugate libraries of strain UCD153 carrying random transposon insertions were generated and screened for nonmotile colonies, identified by the absence of colony spreading after transfer from Allan and Arnon medium diluted 4-fold (AA/4) to BG-110 0.8% Noble agar (Fig. 1). Transposon insertion sites were identified for isolates that failed to show any signs of motility following several rounds of transfer to fresh BG-110 medium. To date, a total of 263 nonmotile strains have been identified from a library of 18,082 transposon mutants. Sequenced insertion sites include several genes experimentally demonstrated to be essential for hormogonium development and motility, including those in the pil (pilB, 5 isolates; pilN, 1 isolate; pilQ, 1 isolate), hmp (hmpC, 1 isolate; hmpD, 3 isolates; hmpE, 3 isolates), and hps (hpsA, 1 isolate; hpsF, 1 isolate) gene loci, providing proof of principle for the experimental design (1–3).
FIG 1.
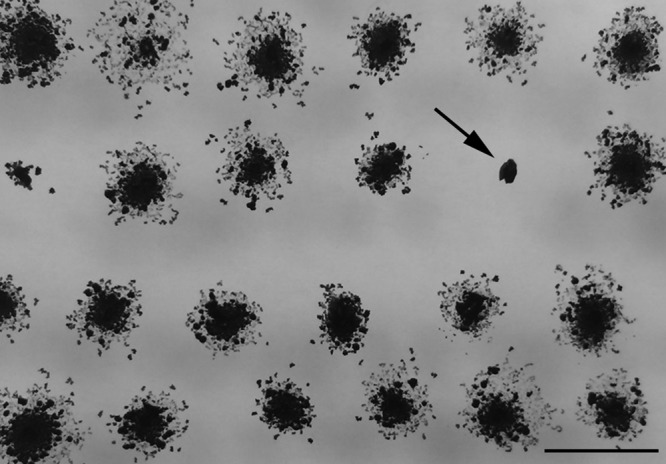
Identification of nonmotile transposon mutants. A photograph of a motility screening plate 2 weeks following inoculation from selection plates is shown. The arrow indicates a nonmotile colony. Bar = 1 cm.
Putative OGT is essential for normal hormogonium development and motility.
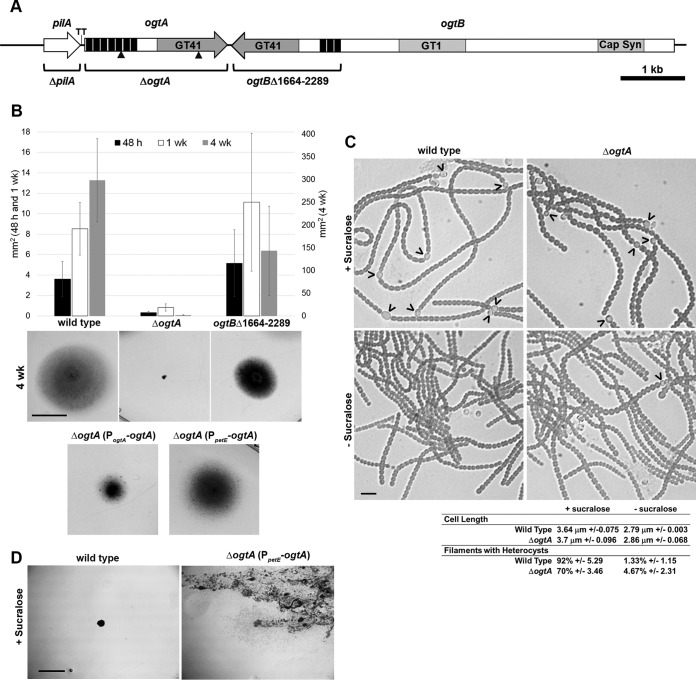
Two nonmotile isolates, TNM103 and TNM428, harbored transposon insertions in an open reading frame, herein designated ogtA (locus tag Npun_F0677), which encodes a putative O-linked β-N-acetylglucosamine transferase (OGT) (Fig. 2A). The putative OGT encoded by ogtA has a series of 7 TPR repeats at the N terminus (24), followed by a C-terminal GT41 domain, similar to eukaryotic OGTs (TPR, pfam13414; GT41, pfam13844) (NCBI BLASTP). To confirm that ogtA was essential for motility, an in-frame deletion strain was constructed in the wild-type genetic background. Following induction of hormogonia from liquid cultures by the supplementation and subsequent removal of sucralose, a potent inhibitor of hormogonium development (25), the ΔogtA mutant strain failed to show any signs of motility in both plate (Fig. 2B) and time-lapse motility assays (SMOV 1 + 2) but could be induced to form filaments with hormogonium morphology, including a reduction in cell length and the absence of heterocysts, similar to the wild-type strain (Fig. 2C). The ogtA gene is directly downstream of pilA, which has been experimentally demonstrated to be essential for motility (3), raising the possibility that removal of ogtA had a cis-acting effect on the upstream pilA gene or that a mutation in pilA was inadvertently introduced during construction of the ΔogtA mutant strain. To confirm that the nonmotile phenotype was due to the deletion of ogtA rather than the disruption of pilA, a shuttle vector harboring ogtA and the 119-bp 5′-intergenic region was introduced into the ΔogtA mutant strain to test for complementation. Reintroduction of ogtA in trans on this shuttle vector restored motility, although not to the same level as the wild-type strain (Fig. 2B). This result confirms that the nonmotile phenotype of the ΔogtA mutant strain is primarily due to the loss of ogtA rather than an inadvertent effect on the upstream pilA gene, or the gene immediately downstream of ogtA (see below). A shuttle vector expressing ogtA from the petE promoter was also introduced into the ΔogtA mutant strain to determine the effect of ectopic overexpression. Expression of genes from the petE promoter on a multicopy replicative shuttle vector has been shown to produce higher levels of gene expression than that from native chromosomal loci (2, 26, 27). Ectopic expression of ogtA restored motility in the ΔogtA mutant strain (Fig. 2B) and promoted the formation of motile hormogonia even in the presence of sucralose (Fig. 2D), but it reduced growth and resulted in the loss of viability upon repeated subculture in liquid medium. Because hormogonia enter a nongrowth state, prolonged persistence of or rapid reentry into the hormogonium state following dedifferentiation would limit cell growth and lead to the extinction of the culture, which could explain the deleterious effect of ogtA overexpression. In our hands, we have found that the expression of several different genes from the petE promoter in Nostoc punctiforme, when present on a multicopy plasmid, is leaky; thus, attempts to omit copper from the growth medium did not limit hormogonium differentiation or correct for the growth defect in the complemented strain. It is unclear if the leaky expression from the petE promoter under these conditions is due to inadvertent introduction of copper via copper-contaminated labware and reagents, or if the removal of copper is insufficient to reduce expression from the petE promoter in N. punctiforme, at least in the context of a multicopy replicative shuttle vector, in contrast to regulation of PpetE in the closely related filamentous cyanobacterium Anabaena sp. strain PCC 7120 (26).
FIG 2.
Phenotypic characterization of the ΔogtA and ogtBΔ1664–2289 mutant strains. (A) Schematic diagram of the genomic region harboring pilA, ogtA, and ogtB. Triangles indicate the location of transposon insertions. Deleted regions are indicated for each deletion strain. GT41, glycosyl transferase 41; GT1, glycosyl transferase 1; Cap Syn, capsular polysaccharide export domain; black bars, TPR repeats; TT, transcription terminator. (B) Quantification (average of 3 biological replicates; error bars = 1 standard deviation [SD]) and representative photographs (4-week images; bar = 1 cm) of results from plate motility assays. (C) Light micrographs of and quantitative data for hormogonium characteristics for the wild-type and ΔogtA mutant strains before and after induction of hormogonia by the removal of sucralose. Heterocysts attached to filaments are indicated by carets; ± values indicate 1 SD. Bar = 10 μm. (D) Photographs depicting serial transfer of colonies of the wild-type strain or attempts to streak for isolation of the ΔogtA mutant strain expressing ogtA from the petE promoter (PpetE-ogtA) on medium containing sucralose. Bar = 5 mm. The presence of diffuse cell material is indicative of the spreading of colonies due to the differentiation of motile hormogonia.
A second open reading frame encoding a putative OGT, herein designated ogtB (locus tag Npun_R0678), is located immediately downstream and in the opposite orientation of ogtA (Fig. 2A). ogtB encodes a multidomain protein which includes, from the N to C terminus, a capsular polysaccharide export domain (pfam05159), a group 1 glycosyl transferase (pfam00534), and an OGT domain (TPR, pfam13414; GT41, pfam13844) (NCBI BLASTP) (Fig. 2A). In most other filamentous cyanobacteria, the homologous OGT is encoded separately from the gene encoding the corresponding capsular polysaccharide export and group 1 glycosyl transferase domains (IMG Gene Ortholog Neighborhoods) (28). To determine if the OGT domain encoded by ogtB is also essential for functional hormogonia, a mutant with an in-frame deletion of the OGT-encoding portion of ogtB (corresponding to amino acid residues 1664 to 2289) was constructed. The resulting strain, the ogtBΔ1664–2289 mutant, formed functional hormogonia and was indistinguishable from the wild-type strain in plate colony-spreading assays up to 1 week after transfer to AA/4 0.5% Noble agar medium (Fig. 2B). However, by 4 weeks posttransfer, wild-type colonies covered a substantially greater area than that of the ogtBΔ1664–2289 mutant strain, while the cell density for this strain appeared greater than that of the wild type (Fig. 2B). These results imply that the OGT domain encoded by ogtB is dispensable for hormogonium development and motility but may exert a subtle effect on the basal rate of hormogonium differentiation.
Expression and protein localization of ogtA.
Previous studies using DNA microarrays to measure global transcription reported that the expression of ogtA increases rapidly upon hormogonium induction (1, 15–17). To confirm this, RT-qPCR was used to measure the transcription of ogtA and the upstream pilA gene over the course of hormogonium development. Consistent with previous studies, the expression of ogtA was induced in developing hormogonia, with transcript abundance increasing 10-fold at 1 h postinduction and remaining elevated throughout the 24-h time course (Fig. 3A). This expression profile differs from that of the upstream pilA gene (Fig. 3A), indicating that pilA and ogtA are most likely not cotranscribed but rather are under the control of separate promoters. Transcript mapping based on RNA sequencing (RNA-seq) analysis of RNA extracted 0 and 18 h post-hormogonium induction did not reveal the presence of any transcripts antisense to the ogtA coding region, making it unlikely that the phenotype of the ΔogtA mutant strain was due to disruption of an anti-sense-encoded small regulatory RNA (Fig. 3B). The RNA-seq data also provide evidence that ogtA and pilA are transcribed independently. At 18 h postinduction, the abundance of pilA mRNA (11,750 reads/kb/million mapped reads) was substantially higher than that of ogtA (321 reads/kb/million mapped reads), and the read coverage for both coding regions was substantially higher than that of the 119-bp intergenic region between pilA and ogtA. Consistent with the transcript mapping, the position of a predicted rho-independent transcriptional terminator (29) corresponds well with a drastic decrease in read coverage on the 3′ end of pilA.
FIG 3.
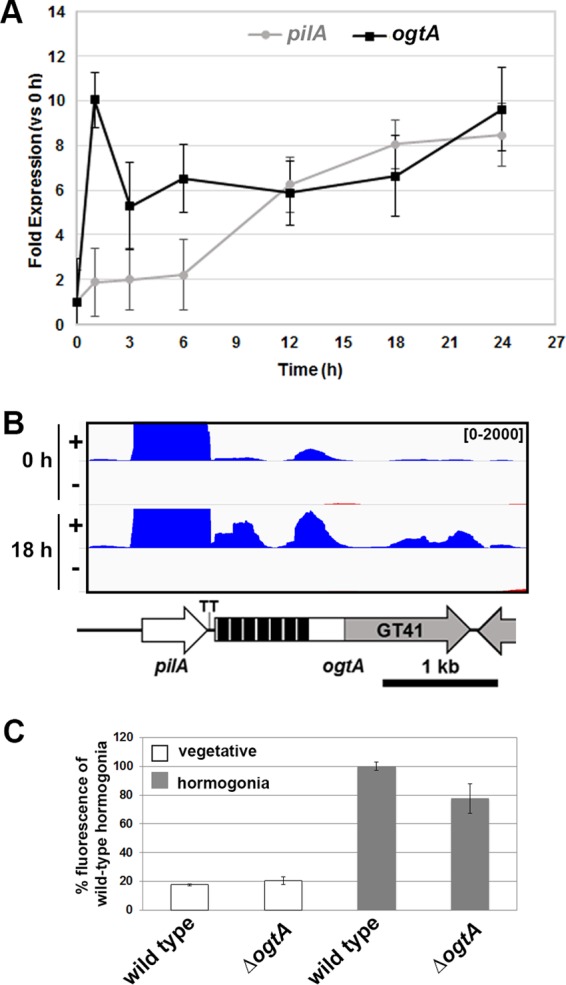
Transcription of ogtA and pilA. (A) qPCR analysis of ogtA and pilA expression over the time course of hormogonium development in the wild-type strain (n = 3; error bars = 1 SD). (B) Transcript map displaying the read coverage from RNA-seq data for both the positive and negative strands (as indicated) for the genomic region containing pilA and ogtA at 0 and 18 h post-hormogonium induction. Value range, 0 to 2,000 reads per bp. Details on the gene map are given in the legend to Fig. 2. (C) Quantification of sfgfp-aav expression from an ogtA promoter region-gfp reporter construct (PogtA-sfgfp-aav in pBK111) in vegetative filaments or hormogonia (24 h postinduction) of the wild-type or ΔogtA mutant strain. Values are expressed as a percentage of the fluorescence intensity for wild-type hormogonia 24 h postinduction (n = 3; error bars = 1 SD).
The activity of the putative ogtA promoter was further assessed by introduction of a shuttle vector containing the 119-bp intergenic region between pilA and ogtA upstream of gfp (PogtA-sfgfp-aav in pBK111) into the wild-type strain. In the resulting strain, hormogonia produced substantially higher fluorescence than vegetative filaments, confirming the presence of a hormogonium-specific promoter directly upstream of ogtA (Fig. 3C; see also Fig. S1 in the supplemental material). To determine if the ogtA promoter is subject to either negative or positive autoregulation, the shuttle vector containing PogtA-sfgfp-aav was introduced into the ΔogtA mutant strain. The resulting strain produced levels of fluorescence comparable to those of the wild-type strain in both vegetative and hormogonium filaments, indicating that ogtA is not subject to autoregulation.
Many of the proteins known to be essential for hormogonium development and motility, including Pil proteins and the chemotaxis protein-like Hmp proteins, localize to rings adjacent to the cell junctions in motile hormogonia (2, 3). Initially, to determine the subcellular localization of OgtA, the chromosomal allele of ogtA was replaced with a gfpuv-tagged variant. The resulting strain differentiated motile hormogonia, but OgtA-GFPuv was undetectable by either fluorescence microscopy or immunoblot analysis with anti-green fluorescent protein (anti-GFP) antibodies. These results imply that either ogtA is expressed at low levels in the cell or the GFPuv tag is unstable or fails to fold properly. The results from RNA-seq analysis of ogtA transcript abundance (see above and Fig. 3B) support the idea of ogtA being expressed at low levels in the cell. Subsequently, a shuttle vector expressing ogtA-gfpuv under the control of the petE promoter was introduced into the ΔogtA mutant strain to increase the cellular levels of OgtA-GFPuv. Neomycin-resistant (Neor) exconjugant colonies that arose on selection plates following introduction of this shuttle vector grew slowly and lost viability immediately upon subculture. However, direct examination of these Neor colonies revealed the presence of motile hormogonia (SMOV 3), with detectable levels of fluorescence from the OgtA-GFPuv fusion protein (Fig. S2). The fluorescent signal was dispersed throughout the cytoplasm of cells in motile hormogonia, with no obvious bias in localization to the cell poles or cell periphery.
Accumulation of PilA in hormogonia is dependent on ogtA.
To further define the role of ogtA in hormogonium development and motility, production of the type IV pilus system component PilA, the Hmp chemotaxis-like system protein HmpD, and hormogonium polysaccharide (HPS) was assessed via immunological or lectin-based assays. Immunoblot analysis demonstrated that the wild-type and ΔogtA mutant strains accumulated similar levels of HmpD following hormogonium induction (Fig. 4A). HmpD was undetectable in the ΔhmpD mutant strain, which served as a negative control. In contrast, the level of PilA was drastically reduced in the ΔogtA mutant strain but accumulated to high levels in hormogonia of the wild-type or ΔhmpD mutant strain (Fig. 4A), as previously reported (2). Consistent with previous reports, a lower-molecular-weight isoform of PilA was detected for the ΔhmpD mutant strain than for the wild type (2).
FIG 4.
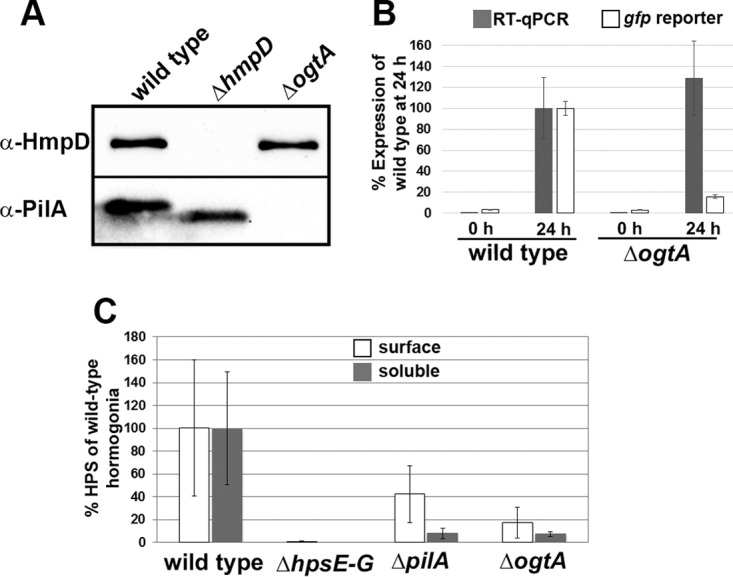
Expression of PilA, HmpD, and HPS in the ΔogtA mutant strain. (A) Western blot analysis of HmpD and PilA in hormogonia of the wild-type, ΔhmpD mutant, and ΔogtA mutant strains. (B) Quantification of pilA expression by using RT-qPCR and a pilA promoter region-gfp reporter construct (PpilA-sfgfp-aav in pBK109) in the wild-type or ΔogtA mutant strain 0 and 24 h post-hormogonium induction. Values are expressed as a percentage of the expression level in wild-type hormogonia 24 h postinduction (n = 3; error bars = 1 SD). (C) Quantification of the cell surface and soluble fractions of HPS in the wild-type, ΔhpsE–D mutant, ΔpilA mutant, and ΔogtA mutant strains. Values are expressed as a percentage of the fluorescence (surface fraction) or chemiluminescent (soluble fraction) intensity for wild-type hormogonia 24 h postinduction (n = 3; error bars = 1 SD).
The decreased accumulation of PilA in the ΔogtA mutant strain implies that ogtA is required for transcription of pilA in developing hormogonia, or for a subsequent step in the synthesis of PilA, such as efficient translation of pilA mRNA or stability of PilA protein following synthesis. To determine the role of transcriptional regulation, pilA transcript abundance in the wild-type and ΔogtA mutant strains 0 h and 24 h following hormogonium induction was quantified using RT-qPCR (Fig. 4B). The wild-type and ΔogtA mutant strains induced transcription of pilA to comparable levels following hormogonium induction, implying that the effect of ogtA on PilA levels is posttranscriptional. The activity of the pilA promoter was also assessed indirectly in the wild-type and ΔogtA mutant strains by introduction of a shuttle vector containing the 655-bp intergenic region between pilA and the Npun_F0675 locus upstream of gfp (PpilA-sfgfp-aav in pBK109). In the resulting strains, wild-type hormogonia accumulated substantially higher levels of fluorescence than hormogonia of the ΔogtA mutant strain, although both strains accumulated more fluorescence in hormogonia than in vegetative filaments (Fig. 4B and S1B). Collectively, these results imply that activation of pilA transcription is not disrupted by the loss of ogtA, but ogtA may regulate pilA expression posttranscriptionally.
In comparison to the wild-type stain, the ΔogtA mutant strain produced substantially smaller amounts of both soluble and surface-associated HPS (Fig. 4C and S3A and B). Because the production of normal levels of HPS is dependent on pilA (3) and the ΔogtA mutant strain fails to accumulate PilA, the amount of polysaccharide produced by the ΔogtA mutant strain was also compared to that produced by a ΔpilA mutant strain. The production levels of HPS in the ogtA and pilA deletion strains were indistinguishable, and both strains produced more HPS than the ΔhpsE–G mutant strain, which failed to produce any detectable HPS (Fig. 4C and S3A and B).
DISCUSSION
The evidence presented here supports a role for O-GlcNAc protein modification in regulating hormogonium development in N. punctiforme. First, deletion of ogtA, which encodes a putative OGT, abolishes hormogonium motility, while overexpression leads to aberrant hormogonium development even under normally repressive conditions. Second, the expression of ogtA is induced early in hormogonium development (Fig. 3A), well before expression of the genes thought to comprise the gliding motor, such as those in the pil and hps loci (1). This implies a role in development via modification of a regulatory protein, rather than one directly involved in motility. The localization of OgtA to the cytoplasm, rather than the cell junctions where the motor complex is thought to reside, is also consistent with a regulatory role in hormogonium development, as opposed to a functional role in the gliding motor. Third, the accumulation of at least one protein known to be required for motility, PilA, is dependent on ogtA. Analysis of data generated from a previous study on the conservation of predicted proteins of N. punctiforme across 121 different cyanobacterial species identified homologs of OgtA (>20% sequence identity) encoded in the genomes of 47 of 51 filamentous cyanobacteria but only 31 of 70 unicellular strains (3). These data support a conserved role for O-GlcNAc protein modification in the generation of motile filaments in both hormogonium-forming and non-hormogonium-forming filamentous cyanobacteria.
Although we do not provide biochemical evidence for O-GlcNAcylation activity of the putative OGT encoded by ogtA, such evidence has been reported for the OGT of Synechococcus elongatus PCC 7942 (seOGT) (23). Given that the catalytic GT41 domain of N. punctiforme OgtA (amino acids 374 to 729 of OgtA) shares 29% sequence identity with seOGT (NCBI BLASTP) and maintains conserved histidine and lysine residues (H280 and K445 of seOGT, corresponding to H386 and K554 of N. punctiforme OgtA) essential for seOGT function (23), it seems likely that ogtA encodes a functional OGT. Several attempts were made to detect the presence of O-GlcNAcylated proteins in developing hormogonia using either the anti-O-GlcNAc monoclonal antibody CTD110.6 or succinylated wheat germ agglutinin (sWGA). Reactive proteins of various intensities were detected by each method, but in both cases, we were unable to detect any definitive difference in reactive proteins between the wild-type and a ΔogtA ogtBΔ1664–2289 double-deletion strain, where all putative OGT-encoding genes were deleted (Fig. S4). Thus, the identity of the O-GlcNAcylated proteins, if any, is currently unknown.
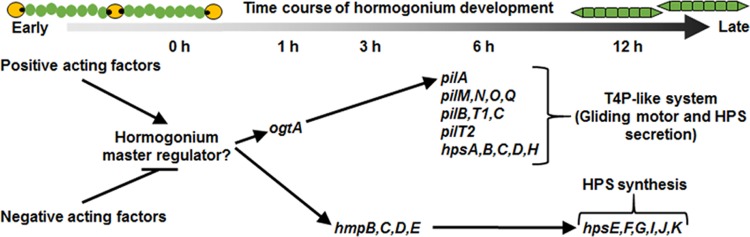
Based on the results presented here, as well as those from previous studies, we have incorporated ogtA into a model of the gene regulatory network promoting hormogonium development (Fig. 5). The first step in the pathway involves the integration of various positive and negative acting factors that ultimately control activation of a hypothetical master regulator of hormogonium development. This master regulator in turn activates at least two independent genetic circuits.
FIG 5.
A model of the gene regulatory network promoting hormogonium development, depicting genes or gene clusters essential for functional hormogonia. Arrows indicate positive interactions between genes or their mRNA or protein products, and lines with bars indicate negative interactions. Timing of events, based on gene expression (1), is depicted above. Based on the evidence presented here, regulation of pilA by ogtA is posttranscriptional, and it is currently unknown if ogtA regulates other components of the type 4 pilus-like system in addition to pilA.
The first genetic circuit is an hmp-dependent circuit, where increased expression of the hmp locus in turn leads to transcriptional activation of genes involved in HPS synthesis. The fact that the ΔogtA mutant strain accumulates wild-type levels of HmpD and at least some HPS supports this model. The decreased production of HPS in the ΔogtA mutant strain could be attributed to either a direct effect of ogtA on expression of the genes involved in HPS synthesis or an indirect effect due to loss of PilA in the ΔogtA mutant strain. Given that PilA is known to be essential for secretion of wild-type levels of HPS, and that the ΔogtA and ΔpilA mutant strains produce similar levels of HPS, we favor an indirect relationship between ogtA and HPS production but cannot rule out a direct interaction. The second genetic circuit activated by the hypothetical hormogonium master regulator is an ogtA-dependent circuit, where expression of ogtA in turn promotes increased accumulation of PilA. This is supported by the loss of PilA upon removal of ogtA.
The mechanism by which ogtA modulates PilA levels is currently unknown but appears to be independent of pilA transcription given the equivalent induction of pilA transcripts in the wild-type and ΔogtA mutant strains, as assessed by RT-qPCR. This implies that ogtA affects posttranscriptional regulation of pilA mRNA, posttranslational regulation of PilA protein levels, or both. The reduced fluorescent signal from the pilA promoter region-gfp reporter in the ΔogtA mutant strain compared to the wild-type strain provides limited evidence in favor of a posttranscriptional mechanism for regulation of PilA levels, possibly via a regulatory element within the 5′ untranslated region of pilA mRNA, which would be present on transcripts derived from the pilA promoter region-gfp reporter construct. In the unicellular cyanobacterium Synechocystis sp. strain PCC 6803, there is evidence that a protein with homology to the RNA-binding protein Hfq interacts with the pilus extension ATPase PilB and may regulate the expression of some type IV pilus genes via a posttranscriptional mechanism (30, 31). It is possible that a similar mechanism controls PilA levels in N. punctiforme and is modulated by ogtA. Alternatively, it is possible that protein stability and turnover of PilA are modulated by OgtA, perhaps directly via O-GlcNAcylation of PilA acting to stabilize the protein. This appears to be the case in Streptococcus gordonii, where stability of the cell surface adhesin GspB requires glycosylation with N-acetylglucosamine and glucose (32). In this case, the failure to detect O-GlcNAc-PilA by immunological techniques may be due to the addition of subsequent sugars to the GlcNAc residue, which could occlude binding of antibodies or lectins, or the presence of cross-reactive proteins with a similar molecular weight that obscure the detection of modified PilA. Additional experimentation is required to definitively determine the mode by which ogtA regulates PilA levels.
At this point, it is also unclear if ogtA affects accumulation of other proteins involved in hormogonium development and motility, such as additional components of the type IV pilus system, or if it is limited to controlling the accumulation of PilA. We anticipate that the forward genetic screen employed here will facilitate the identification of other genes essential for hormogonium development and motility, eventually leading to a more complete model of the underlying gene regulatory network promoting hormogonium development.
MATERIALS AND METHODS
Strains and culture conditions.
For a detailed description of the plasmids, strains, and oligonucleotides used in this study, refer to Tables S1 and S2 in the supplemental material. N. punctiforme ATCC 29133 and its derivatives were cultured in Allan and Arnon medium diluted 4-fold (AA/4), without supplementation of fixed nitrogen, as previously described (15), with the exception that 5 and 10 mM sucralose was added to liquid and solid media, respectively, to inhibit hormogonium formation (25). For hormogonium induction, the equivalent of 30 μg · ml−1 chlorophyll a (Chl a) of cell material from cultures at a Chl a concentration of 10 to 20 μg · ml−1 was harvested at 2,000 × g for 3 min, washed two times with AA/4, and resuspended in 2 ml of fresh AA/4. For selective growth, the medium was supplemented with 50 μg · ml−1 neomycin. Escherichia coli cultures were grown in lysogeny broth (LB) for liquid cultures or LB supplemented with 1.5% (wt/vol) agar for plates. Selective growth medium was supplemented with 50 μg · ml−1 kanamycin, 50 μg · ml−1 ampicillin, and 10 μg · ml−1 chloramphenicol.
Plasmid and strain construction.
The primers used for plasmid construction are indicated in Tables S1 and S2. All constructs were sequenced to ensure sequence fidelity.
For in-frame deletion of genes or gene segments, approximately 900 bp of flanking DNA on either side of the gene and the first and last three or four codons of each gene were amplified via overlap extension PCR and cloned into pRL278 (33) as BamHI-SacI fragments using restriction sites introduced on the primers.
For replacement of the chromosomal allele of ogtA with a C-terminal gfpuv-tagged variant, approximately 900 bp of DNA downstream of the stop codon was amplified via PCR and cloned into pSCR569 (27), which contains gfpuv, as a SpeI-SacI fragment using restriction sites introduced on the primers. Approximately 900 bp of DNA upstream of the stop codon was then amplified via PCR and cloned into this plasmid as a BamHI-SmaI fragment using restriction sites introduced on the primers, resulting in plasmid pDDR360.
To construct a mobilizable shuttle vector containing ogtA and its putative promoter region, the ogtA coding and 119-bp 5′ intergenic regions were amplified via PCR and subsequently cloned into pAM504 (34) as a BamHI-SacI fragment using restriction sites introduced on the primers to create pBK112.
To construct a mobilizable shuttle vector expressing ogtA from the petE promoter, the ogtA coding region was amplified via PCR and subsequently cloned into pDDR155 (2) as a BamHI-SacI fragment, replacing the hmpA-gfp coding region, using restriction sites introduced on the primers to create pDDR387.
To construct a mobilizable shuttle vector expressing ogtA-gfpuv from the petE promoter, the ogtA coding region was amplified via PCR and subsequently cloned into pDDR155 (2) as a BamHI-SmaI fragment, replacing the hmpA coding region, using restriction sites introduced on the primers to create pDDR388.
The C terminus of GFP can be appended with 3 amino acid degradation tags, producing GFP variants with a short half-life, which is desirable for in situ studies of gene expression (35, 36). To construct a mobilizable shuttle vector containing the superfolder variant of gfp with an AAV-degradation tag (sfgfp-aav) under the control of the pilA or ogtA promoter region, the sfgfp coding region was amplified from pDDR338 (3) using a primer that introduced codons corresponding to the amino acids AAV to the 3′ end of sfgfp. The sfgfp-aav gene was then fused to the intergenic regions upstream of pilA and ogtA via overlap extension PCR and subsequently cloned into pAM504 (34) as BamHI-SacI fragments using restriction sites introduced on the primers to create pBK109 and pBK111, respectively.
Gene deletions and allelic replacements were performed as previously described (1), with the exception that N. punctiforme cultures used for mating with E. coli were grown in medium supplemented with 5 mM sucralose, which inhibited hormogonium development and enhanced the efficiency of recovering single recombinants following conjugal transfer of suicide vectors. Gene deletion or allelic substitution strains were constructed by introduction of appropriate suicide vectors into wild-type N. punctiforme, with the exception of strain UOP122, the ΔogtA ogtBΔ1664–2289 double-deletion mutant strain, which was created by the introduction of plasmid pBK100 into strain UOP106, the ΔogtA mutant strain. It should be noted that the deletion of ogtA removed 744 bp of the 899-bp region used for homologous recombination between pBK100 and the genomic region on the 3′ end of ogtB. Despite the shortened region for homologous recombination, we were able to successfully recover double recombinant ogtBΔ1664–2289 deletion mutants from the ΔogtA genetic background.
Generation of transposon mutant libraries and screening for nonmotile transposon mutants.
Plasmid pRL1063a (37) was introduced into strain UCD 153 (15) via conjugation, according to previously established conjugation protocols (1). After several weeks of incubation on selection plates (AA/4 supplemented with neomycin), Neor exconjugant colonies were transferred to motility screening plates (BG-110, 0.8% [wt/vol] Noble agar, supplemented with neomycin). Colonies were manually picked from the selection plates using an inoculating needle and plunged into the agar on the motility screening plates, as this produced the most robust motility. Nonmotile colonies were subsequently transferred to fresh motility screening plates several times, and colonies that continually displayed no signs of motility upon repeated subculture were transferred to liquid cultures to produce enough biomass for subsequent identification of the transposon insertion site.
Identification of transposon insertion sites.
Cell material equivalent to 30 μg of Chl a was harvested from liquid cultures by centrifugation at 2,000 × g for 3 min and washed once with 1 ml of TES (10 mM Tris, 25 mM EDTA, 500 mM NaCl [pH 8.0]), and chromosomal DNA was extracted using the QIAamp minikit (Qiagen), according to the manufacturer's protocol. Approximately 500 ng of chromosomal DNA was digested with HpaII (New England BioLabs) for 1 h at 37°C, followed by heat inactivation at 80°C for 20 min. The heat-inactivated restriction digest was self-ligated with T4 DNA ligase (New England BioLabs) overnight at 16°C and then used as the template for inverse PCR using the primers Tn5-seq-F and Tn5-seq-R. PCR products were separated on a 2% agarose gel, and the bands excised and purified using the QIAEX II gel extraction kit and sequenced (Quintara Biosciences) using the nested primer Tn5-seq-F-nest.
Motility assays.
Both plate and time-lapse motility assays were performed as previously described (3), with the exception that a Bio-Rad Molecular Imager ChemiDoc XRS+ was used for imaging of plate motility assays at 4 weeks postinduction.
qPCR.
Transcripts of ogtA, pilA, and rnpB were amplified using primer sets qNpun_F0677_F2 and qNpun_F0677_R2, qpilA-F2 and qpilA-R2, and qNpun_R018_F1 and qNpun_R018_R1, respectively, using a StepOnePlus real-time PCR system (Applied Biosystems) and SensiFAST SYBR No-ROX kit (Bioline), according to the specifications of the manufacturers. Quantification of pilA and ogtA transcripts from the wild-type strain during the time course of hormogonium development (Fig. 3A) was performed using 8 ng of cDNA, synthesized as previously described (27) from previously generated total RNA extracted at 0, 1, 3, 6, 12, 18, and 24 h following induction of hormogonia (1). For quantification of pilA and ogtA transcripts in the wild-type and ΔogtA mutant strains at 0 h and 24 h postinduction (Fig. 4B), total RNA was extracted for each of 3 biological replicates from each strain at each time point, according to previously published methods (15), and 500 ng of total RNA was used to synthesize cDNA with the ProtoScript first-strand cDNA synthesis kit and random hexamer primers (New England BioLabs, Inc.), according to the manufacturers' protocols, after which 1 μl of cDNA was used as the template for quantitative PCR (qPCR). For all experiments, transcript abundance of ogtA and pilA was normalized relative to rnpB using the 2−ΔΔCT method (38), as the averages of two technical replicates from each of three biological replicates.
RNA-seq.
For transcript mapping using RNA-seq, library preparation and sequencing were carried out at the University of California Berkeley QB3 Vincent J. Coates Genomics Sequencing Laboratory using 10 μg of total RNA, previously extracted 18 h following hormogonium induction (1). rRNA was removed using the Ribo-Zero rRNA removal kit (bacteria) (Illumina, Inc.). Directional cDNA libraries were synthesized from the rRNA-depleted samples, sheared to a library size of 200 bp, appended with adapters, and sequenced in a single lane of an Illumina HiSeq 2000 flow cell, multiplexed with 4 other libraries, generating 50-bp paired-end reads. Analysis of sequencing data was performed using the software package Rockhopper (39) (with default parameters), and transcript maps were generated using Integrated Genome Viewer (40). A total of 44,445,052 and 36,321,509 reads mapped to the N. punctiforme genome for the 0- and 18-h time points, respectively, with less than 0.1% mapping to rRNA.
Generation of anti-HmpD polyclonal antibodies.
To avoid raising antibodies that may cross-react with other methyl-accepting chemotaxis proteins encoded in the N. punctiforme genome, only the first 499 amino acids of HmpD, which contain the unique sensing domain, but not the highly conserved transmembrane and signaling domains, were used. For purification of HmpD amino acids 1 to 499 (HmpD1–499), 500 ml of LB supplemented with the appropriate antibiotics was inoculated with a 1:25 dilution of an overnight culture of E. coli BL21(DE3) harboring pDDR116 and incubated at 37°C with shaking at 225 rpm until an optical density at 600 nm (OD600) of 0.6 was reached. The culture was induced for protein expression by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to 1 mM and incubated for an additional 4 h. Cells were harvested by centrifugation at 5,000 × g and the protein purified under native conditions, according to the manufacturer's protocol (QIAexpressionist; Qiagen). The eluent was dialyzed against phosphate-buffered saline (PBS) to remove imidazole, and purified protein at 2 mg · ml−1 was used to generate rabbit polyclonal antisera (UC Davis School of Veterinary Medicine, Comparative Pathology Lab).
Immunological or lectin-based assays.
For immunoblot analyses, total cellular protein was extracted as previously described (3). Detection of PilA was performed as previously described (2), while detection of HmpD followed the same protocol (using a 1:10,000 dilution of primary antibody), with the exception that a 10% SDS-PAGE gel was used. Detection of O-GlcNAcylated proteins using the anti-O-GlcNAc monoclonal antibody CTD110.6 (Enzo Life Sciences, Inc.) in conjunction with goat anti-mouse IgG–horseradish-conjugated peroxidase (HRP) (sc2005; Santa Cruz Biotechnology), or biotinylated succinylated wheat germ agglutinin (sWGA) (Vector Laboratories) in conjunction with the avidin-HRP conjugate Vectastain ABC reagent, was performed as previously described (2, 41). Detection and quantification of soluble hormogonium polysaccharide in the culture medium by lectin blotting with biotinylated Ulex europaeus agglutinin (UEA) were performed as previously described (3). The detection of cell-associated hormogonium polysaccharide by staining with fluorescein-UEA, followed by confocal fluorescence microscopy, was performed as previously described (3). Quantification of cell-associated hormogonium polysaccharide from fluorescence micrographs was performed using ImageJ (NIH) as follows: for each of 3 biological replicates, the sum of the pixel intensities from both UEA-fluorescein-derived fluorescence as well as cellular autofluorescence was calculated for each of 4 images, and the amount of cell-associated hormogonium polysaccharide was expressed as the average of the ratio of UEA-fluorescein-derived fluorescence/autofluorescence. The incorporation of cellular autofluorescence was required to normalize the value from each image to account for differences in the total amount of cell material imaged.
Microscopy.
Light microscopy of filaments and fluorescence microscopy of cellular autofluorescence, fluorescently labeled hormogonium polysaccharide, and GFP variants were acquired with a Leica DMIRE2 inverted fluorescence microscope using MetaMorph software (Molecular Devices) and a Yokogawa CSU-X1 spinning disc confocal scanner unit with a QuantEM:5125C camera. Excitation and emission were as follows: 491 nm excitation and 525 (±25) nm emission for fluorescently labeled UEA-fluorescein and sfGFP-aav, 405 nm excitation and 525 (±25) nm emission for GFPuv-labeled proteins, and 561 nm excitation and 605 (±25.5) nm emission for cellular autofluorescence. For determination of the average cell length and percentage of filaments with heterocysts, 50 filaments each from 3 biological replicates were scored for the presence of a heterocyst and the average cell length from a contiguous 5-vegetative-cell interval using ImageJ (NIH). For quantification of fluorescence intensities derived from pilA or ogtA promoter-driven expression of sfgfp-aav, the mean pixel intensity was measured for each of 30 individual filaments from each of three biological replicates using ImageJ (NIH).
Accession number(s).
RNA-seq data can be accessed at NCBI GEO (accession no. GSE94020).
Supplementary Material
ACKNOWLEDGMENTS
We thank John C. Meeks for assistance with production of the anti-HmpD polyclonal antibody, which was supported by grant IOS 0822008 from the U.S. National Science Foundation, as well as his helpful comments on the manuscript. We also thank the Lin-Cereghino lab for use of and assistance with their vacuum blot module for lectin blot analysis, Karen Lundy and Shana McDevitt of the University of California Berkeley QB3 Vincent J. Coates Genomics Sequencing Laboratory for technical assistance with generation of RNA-seq data, and Carrie Kozina, Rajneet Padda, Sabreen Chahal, and the microbiology (BIOL 145) laboratory course students for purification of chromosomal DNA from nonmotile transposon mutants of N. punctiforme.
This work was supported by a Pacific Fund grant awarded to D.D.R. from the University of the Pacific.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00075-17.
REFERENCES
- 1.Risser DD, Meeks JC. 2013. Comparative transcriptomics with a motility-deficient mutant leads to identification of a novel polysaccharide secretion system in Nostoc punctiforme. Mol Microbiol 87:884–893. doi: 10.1111/mmi.12138. [DOI] [PubMed] [Google Scholar]
- 2.Risser DD, Chew WG, Meeks JC. 2014. Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme. Mol Microbiol 92:222–233. doi: 10.1111/mmi.12552. [DOI] [PubMed] [Google Scholar]
- 3.Khayatan B, Meeks JC, Risser DD. 2015. Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria. Mol Microbiol 98:1021–1036. doi: 10.1111/mmi.13205. [DOI] [PubMed] [Google Scholar]
- 4.Rippka R, Castenholz RW, Herdman M. 2001. Oxygenic photosynthetic bacteria. Subsection IV, p 562–589. In Boone DR, Castenholz RW, Garrity GM (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 1 Springer, New York, NY. [Google Scholar]
- 5.Castenholz RW. 1982. Motility and taxis, p 413–419. In Carr NG, Whitton BA (ed), The biology of cyanobacteria. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 6.Shepard RN, Sumner DY. 2010. Undirected motility of filamentous cyanobacteria produces reticulate mats. Geobiology 8:179–190. doi: 10.1111/j.1472-4669.2010.00235.x. [DOI] [PubMed] [Google Scholar]
- 7.Meeks JC. 2006. Molecular mechanisms in the nitrogen-fixing Nostoc-bryophyte symbiosis. Prog Mol Subcell Biol 41:165–196. doi: 10.1007/3-540-28221-1_9. [DOI] [PubMed] [Google Scholar]
- 8.Wong FC, Meeks JC. 2002. Establishment of a functional symbiosis between the cyanobacterium Nostoc punctiforme and the bryophyte Anthoceros punctatus requires genes involved in nitrogen control and initiation of heterocyst differentiation. Microbiology 148:315–323. doi: 10.1099/00221287-148-1-315. [DOI] [PubMed] [Google Scholar]
- 9.Elbert W, Webber B, Burrows S, Steinkamp J, Budel B, Andreae M, Poschl U. 2012. Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat Geosci 5:459–462. doi: 10.1038/ngeo1486. [DOI] [Google Scholar]
- 10.Liaimer A, Jenke-Kodama H, Ishida K, Hinrichs K, Stangeland J, Hertweck C, Dittmann E. 2011. A polyketide interferes with cellular differentiation in the symbiotic cyanobacterium Nostoc punctiforme. Environ Microbiol Rep 3:550–558. doi: 10.1111/j.1758-2229.2011.00258.x. [DOI] [PubMed] [Google Scholar]
- 11.Campbell EL, Wong FC, Meeks JC. 2003. DNA binding properties of the HrmR protein of Nostoc punctiforme responsible for transcriptional regulation of genes involved in the differentiation of hormogonia. Mol Microbiol 47:573–582. doi: 10.1046/j.1365-2958.2003.03320.x. [DOI] [PubMed] [Google Scholar]
- 12.Cohen MF, Meeks JC. 1997. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol Plant Microbe Interact 10:280–289. doi: 10.1094/MPMI.1997.10.2.280. [DOI] [PubMed] [Google Scholar]
- 13.Campbell EL, Meeks JC. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl Environ Microbiol 55:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liaimer A, Helfrich EJN, Hinrichs K, Guljamow A, Ishida K, Hertweck C, Dittmann E. 2015. Nostopeptolide plays a governing role during cellular differentiation of the symbiotic cyanobacterium Nostoc punctiforme. Proc Natl Acad Sci U S A 112:1862–1867. doi: 10.1073/pnas.1419543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol 189:5247–5256. doi: 10.1128/JB.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell EL, Christman H, Meeks JC. 2008. DNA microarray comparisons of plant factor- and nitrogen deprivation-induced Hormogonia reveal decision-making transcriptional regulation patterns in Nostoc punctiforme. J Bacteriol 190:7382–7391. doi: 10.1128/JB.00990-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christman HD, Campbell EL, Meeks JC. 2011. Global transcription profiles of the nitrogen stress response resulting in heterocyst or hormogonium development in Nostoc punctiforme. J Bacteriol 193:6874–6886. doi: 10.1128/JB.05999-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart GW, Housley MP, Slawson C. 2007. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 19.Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. 2012. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olszewski NE, West CM, Sassi SO, Hartweck LM. 2010. O-GlcNAc protein modification in plants: evolution and function. Biochim Biophys Acta 1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke AJ, Hurtado-Guerrero R, Pathak S, Schüttelkopf AW, Borodkin V, Shepherd SM, Ibrahim AFM, van Aalten DMF. 2008. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J 27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ. 2008. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol 15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 23.Sokol KA, Olszewski NE. 2014. The putative eukaryote-like O-GlcNAc transferase of the cyanobacterium Synechococcus elongatus PCC 7942 hydrolyzes UDP-GlcNAc and is involved in multiple cellular processes. J Bacteriol 197:354–361. doi: 10.1128/JB.01948-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karpenahalli MR, Lupas AN, Soding J. 2007. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8:1–8. doi: 10.1186/1471-2105-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Splitt S, Risser DD. 2015. The non-metabolizable sucrose analog sucralose is a potent inhibitor of hormogonium differentiation in the filamentous cyanobacterium Nostoc punctiforme. Arch Microbiol 198:137–147. doi: 10.1007/s00203-015-1171-7. [DOI] [PubMed] [Google Scholar]
- 26.Buikema WJ, Haselkorn R. 2000. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc Natl Acad Sci U S A 98:2729–2734. doi: 10.1073/pnas.051624898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risser DD, Wong FC, Meeks JC. 2012. Biased inheritance of the protein PatN frees vegetative cells to initiate patterned heterocyst differentiation. Proc Natl Acad Sci U S A 109:15342–15347. doi: 10.1073/pnas.1207530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. 2000. Prediction of transcription terminators in bacterial genomes. J Mol Biol 301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 30.Dienst D, Dühring U, Mollenkopf HJ, Vogel J, Golecki J, Hess WR, Wilde A. 2008. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 154:3134–3143. [DOI] [PubMed] [Google Scholar]
- 31.Schuergers N, Ruppert U, Watanabe S, Nürnberg DJ, Lochnit G, Dienst D, Mullineaux CW, Wilde A. 2014. Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol Microbiol 92:840–852. doi: 10.1111/mmi.12595. [DOI] [PubMed] [Google Scholar]
- 32.Takamatsu D, Bensing BA, Sullam PM. 2004. Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J Bacteriol 186:7100–7111. doi: 10.1128/JB.186.21.7100-7111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai YP, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei TF, Ramasubramanian TS, Golden JW. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol 176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen JB, Sternberg C, Poulsen LK, Bjørn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64:2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H, Camsund D, Lindblad P, Heidorn T. 2010. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res 38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolk CP, Cai Y, Panoff JM. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci U S A 88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- 39.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zachara NE, Vosseller K, Hart GW. 2011. Detection and analysis of proteins modified by O-linked N-acetylglucosamine. Curr Protoc Protein Sci 66:12.8.1–12.8.33. doi: 10.1002/0471140864.ps1208s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.