ABSTRACT
Host-associated bacteria, such as Escherichia coli, often encounter various host-related stresses, such as nutritional deprivation, oxidative stress, and temperature shifts. There is growing interest in searching for small endogenous proteins that mediate stress responses. Here, we characterized the small C-tail-anchored inner membrane protein ElaB in E. coli. ElaB belongs to a class of tail-anchored inner membrane proteins with a C-terminal transmembrane domain but lacking an N-terminal signal sequence for membrane targeting. Proteins from this family have been shown to play vital roles, such as in membrane trafficking and apoptosis, in eukaryotes; however, their role in prokaryotes is largely unexplored. Here, we found that the transcription of elaB is induced in the stationary phase in E. coli and stationary-phase sigma factor RpoS regulates elaB transcription by binding to the promoter of elaB. Moreover, ElaB protects cells against oxidative stress and heat shock stress. However, unlike membrane peptide toxins TisB and GhoT, ElaB does not lead to cell death, and the deletion of elaB greatly increases persister cell formation. Therefore, we demonstrate that disruption of C-tail-anchored inner membrane proteins can reduce stress resistance; it can also lead to deleterious effects, such as increased persistence, in E. coli.
IMPORTANCE Escherichia coli synthesizes dozens of poorly understood small membrane proteins containing a predicted transmembrane domain. In this study, we characterized the function of the C-tail-anchored inner membrane protein ElaB in E. coli. ElaB increases resistance to oxidative stress and heat stress, while inactivation of ElaB leads to high persister cell formation. We also demonstrated that the transcription of elaB is under the direct regulation of stationary-phase sigma factor RpoS. Thus, our study reveals that small inner membrane proteins may have important cellular roles during the stress response.
KEYWORDS: C-tail-anchored membrane protein, oxidative stress, heat shock, persistence
INTRODUCTION
Membrane proteins interact with, or are part of, biological membranes, and they separate the cytoplasm from the extracellular environment in all living cells. These membrane proteins are prevalent as they are encoded by 20 to 30% of all genes in most genomes (1, 2). For Escherichia coli, the inner membrane is a phospholipid bilayer, and integral inner membrane proteins are mostly α-helical (3). The inner membrane proteins include transporters, channels, receptors, enzymes, and structural membrane-anchoring domains for myriad tasks, including energy transduction and cell adhesion (4).
Most proteins are inserted into the membrane by the well-conserved Sec pathway, consisting of the membrane-spanning translocase SecYEG in bacteria. Many accessory proteins, including the signal recognition particle (SRP) and its cognate membrane receptor FtsY, aid in protein targeting and insertion (5). To be tagged to the membrane via the Sec pathway, a protein usually has an N-terminal signal sequence for recognition by the SRP. Translation of the N-terminal signal peptide that target proteins to the SRP pathway is required for directing mRNAs to the membrane (6). In E. coli, YidC can be specifically cross-linked to the transmembrane domain of newly synthesized peptides during their membrane insertion via translocons (complexes of proteins associated with the translocation of polypeptides across membranes) (7, 8). Several lines of evidence have revealed that in E. coli, as well as in mammalian cells, ribosomes translating inner membrane proteins interact cotranslationally with translocons in the membrane, and this interaction is required for the proper insertion of nascent polypeptides into the membrane (6, 9).
C-tail-anchored inner membrane proteins, a class of proteins characterized by their lack of N-terminal signal sequences, were first identified in eukaryotes, and they play critical roles in membrane traffic, apoptosis, and protein translocation in eukaryotes (10–12). Tail-anchored inner membrane proteins have been found in Streptomyces coelicolor, and they are capable of targeting proteins to the membrane in the absence of an N-terminal signal sequence; the C-terminal transmembrane domain is sufficient for membrane targeting (13). By considering the distance of the transmembrane domain from the C terminus (less than 30 residues) and also the lack of an N-terminal signal peptide, 12 proteins were identified as C-terminal-anchored proteins in E. coli (14). These 12 proteins include those known to be associated with the inner face of the cytoplasmic membrane, such as the flagella assembly protein FlK, the TraL protein involved in F pilus formation, and enzymes with hydrophobic substrates that are expected to be favored by membrane anchorage (15, 16). Only three of the 12 C-tail-anchored proteins, namely, YgaM and paralogs ElaB and YqjD, have not been characterized (14). Recently, these three proteins were shown to be associated with stationary-phase ribosomes (17). Complexes in the inner membrane of E. coli are often involved in key processes, such as energy generation, cell division, signal transduction, and transport (18). Targeting small stress resistance proteins is an emerging area for treating bacterial infections (19–22); however, there are risks in targeting YgaM, ElaB, and YqjD as their function is not known.
In this study, we focused on the physiological role of ElaB when cells encounter external stress. The elaABCD genes in the E. coli genome were previously named without a phenotypic explanation. ElaC was renamed Rbn and has both endoribonuclease and exoribonuclease activities, while ElaD, previously misannotated as a putative sulfatase/phosphatase, is an efficient and specific deubiquitinating enzyme (23–25). It was reported that ElaB is mainly expressed in the stationary phase (17), and we demonstrate here that elaB transcription is regulated by the stationary-phase sigma factor RpoS. We also provide evidence that ElaB protects cells against heat shock and oxidative stress. However, unexpectedly, disruption of ElaB greatly increases persister cell formation. Thus, ElaB represents a new transmembrane protein that participates in various stress responses.
RESULTS
ElaB is an inner membrane protein with a conserved C-terminal transmembrane domain.
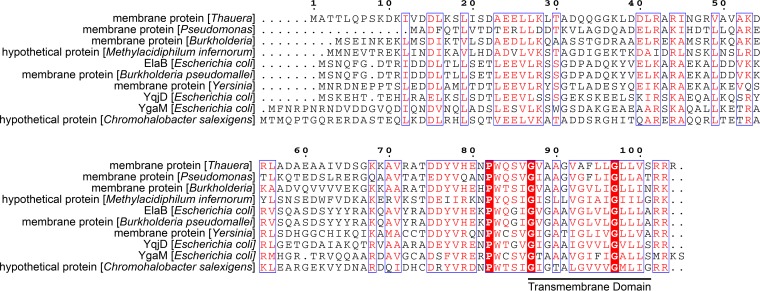
ElaB is a small protein of 101 amino acids. It has one transmembrane domain at the C terminus, and this domain is conserved in several bacterial species, including opportunistic pathogens (Fig. 1). By contrast, the N termini of these proteins show much less conservation in terms of length and amino acid composition. Two proteins in E. coli, YqjD and YgaM, have C-terminal transmembrane domains that are the same as in ElaB. Moreover, the transmembrane domain is very close to the end of the C terminus and is followed by two to four residues containing one to three arginines (Fig. 1).
FIG 1.
Conserved C-terminal transmembrane domain in ElaB. Alignment of amino acid sequences among ElaB and other hypothetical membrane proteins from different bacterial species was performed based on the Dense Alignment Surface method (http://www.sbc.su.se/~miklos/DAS/). The predicted transmembrane motif is underlined.
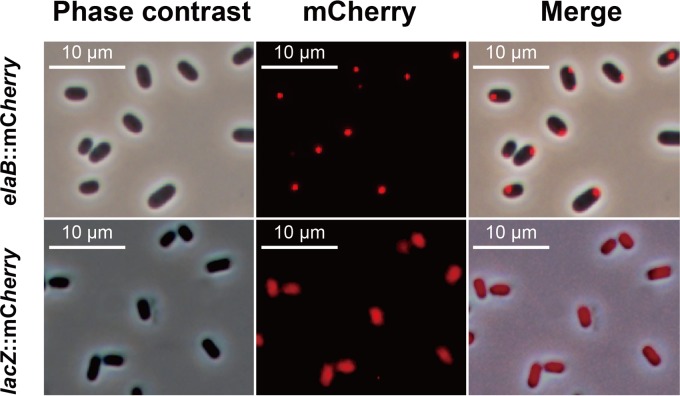
ElaB localization in E. coli was first checked by fusing the red fluorescent protein gene mCherry to the C terminus of the elaB gene in the chromosome of BW25113 wild-type cells to express the fused protein ElaB-mCherry. As expected, ElaB-mCherry was localized to the cell poles (Fig. 2, upper panel). To exclude potential effects of C-terminal mCherry on localization, we also fused the mCherry gene to the C terminus of the lacZ gene in the chromosome of BW25113 wild-type cells, and the fused protein LacZ-mCherry was localized to the cytoplasm (Fig. 2, lower panel). To further check the ElaB localization, ElaB with a green fluorescent protein (GFP) tagged at the C terminus was produced using pCA24N-elaB-gfp. As expected, GFP-fused ElaB was also localized to the cell poles (see Fig. S1 in the supplemental material). Localization of GFP-fused membrane proteins at the cell poles was also observed with the previously characterized inner membrane proteins YqjD (26) and YidC (27), suggesting that ElaB is anchored in the membrane.
FIG 2.
ElaB is an inner membrane-anchored protein. Coexpression of the mCherry fusion protein ElaB or LacZ in BW25113 at a turbidity at 600 nm of 1.0. mCherry fluorescence (middle panels) is overlaid with the phase-contrast images (left panels) in the same field to obtain merged images (right panels). Three independent cultures were used, and only one representative is shown here.
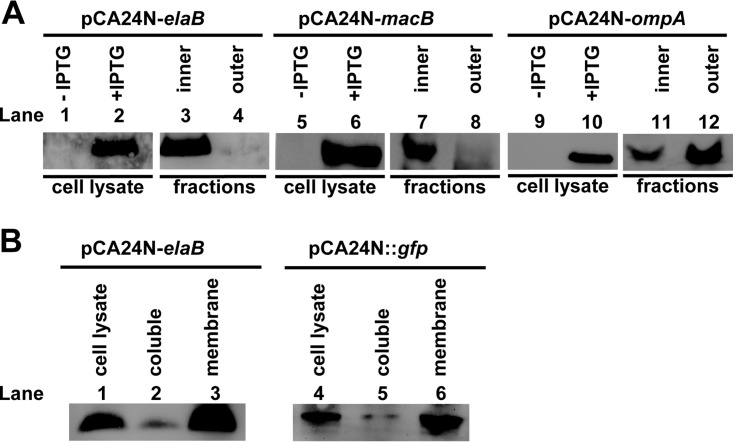
To further determine if ElaB resides in the inner membrane or the outer membrane, the membrane proteins of E. coli were fractionated by a sucrose cushion protocol, which takes advantage of the fact that the inner membrane of E. coli has a lower density than the outer membrane (17, 28). Different cell fractions were obtained from E. coli producing ElaB with an N-terminal His tag using plasmid pCA24N-elaB. Western blotting was conducted to detect the His-tagged ElaB in the membrane fractions using an antibody to the His tag. His-ElaB was present only in the inner membrane fraction but not in the outer membrane fraction (Fig. 3A, lanes 1 to 4). We also used inner membrane protein MacB (29) and outer membrane protein OmpA (30) as positive controls for the membrane protein localization assay. As expected, MacB was found only in the inner membrane fraction (Fig. 3A, lanes 5 to 8), while OmpA was largely present in the outer membrane fraction with a small proportion in the inner membrane fraction due to its high abundance (31) (Fig. 3A, lanes 9 to 12).
FIG 3.
ElaB localization by Western blotting. (A) Cell lysates and fractions containing the inner and outer membrane proteins were obtained as described in Materials and Methods. The His tag antibody was used to determine ElaB levels when elaB was overexpressed using pCA24N-elaB. Cells producing inner membrane protein MacB and outer membrane protein OmpA were also used as positive controls. (B) Expression of ElaB via pCA24N and chromosomal ElaB-GFP and its levels in soluble fractions and membrane fractions were determined. The GFP antibody was used to determine ElaB levels when gfp and elaB were coexpressed in the chromosome.
A previous study showed that YqjD and ElaB are ribosome-associated proteins and that YqjD is also anchored in the inner membrane (17). To see whether the ribosome-associated protein ElaB is also present in the soluble fraction, we quantified the levels of GFP-fused ElaB in the soluble and membrane fractions. More than 90% of the ElaB expressed by pCA24N-elaB in the whole-cell lysate was recovered in the membrane fraction, with a small proportion present in the soluble fraction (Fig. 3B, lanes 1 to 3). Similar results were obtained using GFP-fused ElaB integrated in the chromosome of the BW25113 wild type (Fig. 3B, lanes 4 to 6). In E. coli, ribosomes translating membrane proteins interact cotranslationally with translocons in the membrane, a process that is essential for proper insertion of nascent polypeptides into the membrane (9). Here, we demonstrate that ElaB, which shares a conserved transmembrane motif with YqjD, is also an inner membrane protein. Unlike with YqjD, the production of ElaB did not inhibit cell growth or lead to cell lysis (data not shown).
elaB is regulated by RpoS.
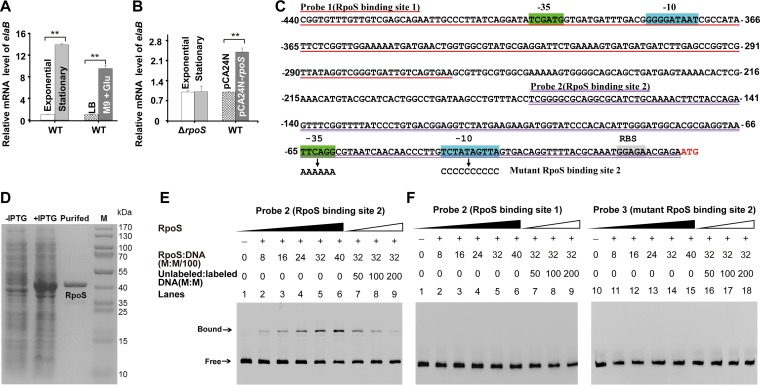
To determine when ElaB is induced, we first examined elaB expression during different growth conditions. The transcription of elaB was upregulated 13.9- ± 0.2-fold when cells entered the stationary phase, and transcription of elaB was also upregulated 9.5- ± 0.6-fold when cells were growing in nutrient-limited minimal medium (M9 minimal medium with 0.4% glucose) compared with that in LB medium during the exponential growing phase (Fig. 4A). By contrast, the expression of the neighboring upstream gene elaA did not change during these growth conditions (data not shown).
FIG 4.
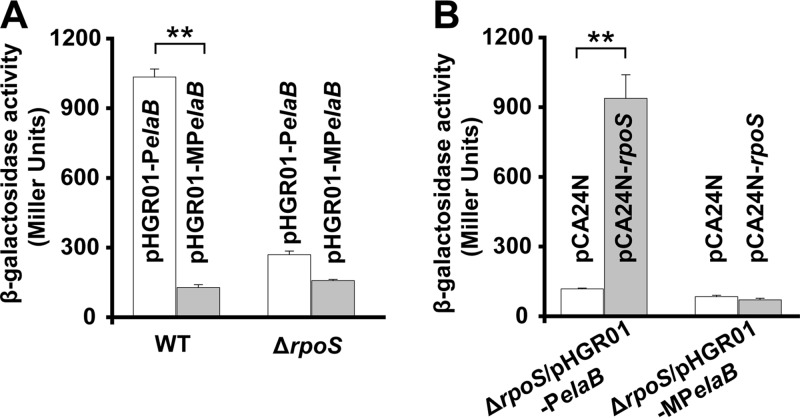
ElaB is induced during the stationary phase, and elaB is regulated by RpoS. (A) Cells grown to the exponential phase (optical density at 600 nm [OD600], 0.7) and stationary phase (OD600, 6.0) were harvested, and total RNA was isolated. The transcriptional level of elaB was determined using qRT-PCR and calculated using 2−ΔΔCT. The expression of elaB was normalized to that of rrsG. The relative expression of elaB in the stationary phase was compared to that in the exponential phase, and the relative expression in M9-glucose medium was normalized to that in LB medium. (B) elaB expression is induced by RpoS. The ΔrpoS cells grown to the exponential phase (OD600, 0.7) and stationary phase (OD600, 6.0) were harvested, and the transcriptional levels of elaB were determined as described for panel A. The cells harboring pCA24N-rpoS were induced with 1 mM IPTG for 1 h at an OD600 of 1.0, the cells were harvested, and total RNA was isolated. The expression of elaB was determined, and vector pCA24N was used as a negative control. Three independent cultures for each strain were used for panels A and B, and the data are shown as means ± standard deviations. **, P < 0.01. (C) The promoter region of elaB and the sequences of the two probes each containing one putative RpoS binding site. The numbers indicate the locations relative to the start codon of elaB. Two predicted binding sites of RpoS are marked. −10 and −35 regions and the ribosome binding site (RBS) are highlighted. The start codon of elaB is shown in red letters. The arrows point to the mutated sequences of the −10 and −35 regions in RpoS binding site 2. (D) Purification of RpoS. A total of 20 μg protein was loaded in each lane. M indicates a protein marker. (E) RpoS binds to probe 2 in a concentration-dependent manner with the addition of RNA core polymerase (lanes 1 to 6). The addition of an excess amount of unlabeled probe reduced the binding of RpoS to the labeled probe in a concentration-dependent manner (lanes 7 to 9). (F) RpoS did not bind to probe 1 (lanes 1 to 9) or the mutant RpoS binding site 2 (probe 3, lanes 10 to 18) under the same conditions.
To test whether expression of elaB is under the direct control of the stationary sigma factor RpoS (σ38), elaB expression was first tested in an rpoS deletion mutant. As expected, there was no induction of elaB in the absence of RpoS in the stationary phase (Fig. 4B). Moreover, the transcription of elaB was upregulated 2.4- ± 0.1-fold by overexpressing rpoS via pCA24N-rpoS in minimal medium during the exponential growing phase (Fig. 4B). Next, Virtual Footprint (32) and FGENESB (Softberry) programs were applied to predict potential RpoS binding sites in the elaB promoter. Two RpoS binding sites were identified at 65 nucleotides (nt) and 401 nt upstream of the start codon of elaB (Fig. 4C). The predicted binding sites were also aligned with previously identified consensus sequences for the RpoS regulon (33–39), and the −10 region in the putative RpoS binding site 2 near the elaB start codon showed high similarity to the previously proposed −10 region consensus sequence of TGN0–2CYATAMT (Y stands for C or T, M stands for A or C, and N0–2 stands for up to 2 of any nucleotide) (36) or CTATA(C/A)T (38). Moreover, the −35 region is 18 nucleotides from the −10 region in the putative RpoS binding site 2, which is regarded as a functional location (17 ± 2 nucleotides) (39).
To determine how RpoS regulates elaB transcription, we conducted an electrophoretic mobility shift assay (EMSA) with two DNA probes amplified from the promoter of elaB (probe 1 containing the putative RpoS binding site 1 and probe 2 containing the putative RpoS binding site 2) using purified RpoS (Fig. 3D) in the presence of E. coli core RNA polymerase. As shown in Fig. 4E and F, RpoS bound and shifted only the DNA fragment containing the RpoS binding site close to elaB start codon (probe 2) in a dose-dependent manner, but did not bind or shift the DNA fragments containing the RpoS binding site 1 (probe 1) or the mutant RpoS binding site 2 (probe 3) (Fig. 4C).
To further confirm the regulation of RpoS in vivo, we conducted promoter activity assays by fusing the promoter of elaB with the lacZ gene in pHGR01 to construct the lacZ reporter plasmid pHGR01-PelaB. As expected, the BW25113 wild-type cells harboring pHGR01-PelaB showed significantly higher β-galactosidase activity (1,034.2 ± 34.2 Miller units [MU]) than the ΔrpoS cells (268.9 ± 15.9 MU) (Fig. 5A). Moreover, we performed site-directed mutagenesis to change the conserved RpoS binding site from TTCAGG (−35 region)…TCTATAGTTA (−10 region) to AAAAAA (−35 region)…CCCCCCCCCC (−10 region) (Fig. 4C) in pHGR01-PelaB to construct a mutant lacZ reporter plasmid, pHGR01-MPelaB. Both the wild-type and ΔrpoS hosts carrying pHGR01-MPelaB had similar β-galactosidase activities, suggesting that RpoS no longer regulates elaB transcription once the conserved binding site is altered (Fig. 5A). To further confirm the regulation of RpoS on elaB, we conducted the complementary experiment of the promoter activity assay using pCA24N-rpoS to overexpress rpoS in the ΔrpoS host cells. As expected, using pHGR01-PelaB as the reporter plasmid, ΔrpoS cells overexpressing rpoS via pCA24N-rpoS showed an 8.0- ± 0.9-fold increase in the promoter activity compared with that of the empty plasmid pCA24N (937.6 ± 101.5 MU for ΔrpoS/pCA24N-rpoS cells versus 117.3 ± 4.0 MU for ΔrpoS/pCA24N cells). However, when pHGR01-MPelaB was used as the reporter plasmid, the activity of the mutated promoter of elaB did not change with overexpression of rpoS in the ΔrpoS cells (70.4 ± 6.7 MU for ΔrpoS/pCA24N-rpoS cells versus 83.7 ± 6.2 MU for ΔrpoS/pCA24N cells) (Fig. 5B). Collectively, these results demonstrate that elaB is induced in the stationary phase and that the transcription of elaB is under the direct control of stationary-phase sigma factor RpoS.
FIG 5.
Promoter activity of elaB is induced by RpoS. (A) Wild-type BW25113 (WT) and ΔrpoS cells harboring pHGR01-PelaB and pHGR01-MPelaB in the stationary phase were collected, and β-galactosidase activities were tested. (B) ΔrpoS/pHGR01-PelaB and ΔrpoS/pHGR01-MPelaB cells expressing RpoS were induced with 0.5 mM IPTG for 2 h at an OD600 of 1.0, and β-galactosidase activities were tested. The pCA24N vector was used as a negative control. Three independent cultures for each strain were used. **, P < 0.01.
ElaB increases survival during heat and oxidative stress.
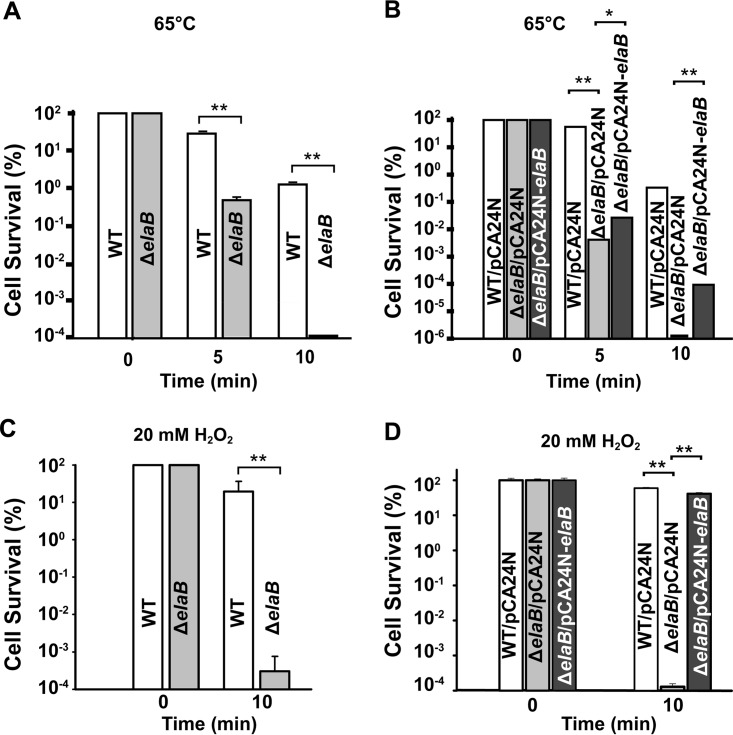
To determine the physiological role of ElaB, we tested whether elaB contributes to survival under stress conditions using the elaB deletion mutant. To test whether ElaB contributes to survival under heat shock, cell viability was determined with and without elaB at 65°C for 10 min. Survival was greatly reduced (3.3 ± 0.1) × 105-fold when elaB was deleted (Fig. 6A), and complementation of the elaB mutation increased survival upon heat shock (Fig. 6B). Similarly, cell survival after treatment with 20 mM H2O2 for 10 min was reduced approximately 3.6 × 104-fold when elaB was deleted (Fig. 6C), and complementation of the elaB mutation increased cell viability 1.2 × 105-fold to levels similar to those of the wild type (Fig. 6D). Collectively, these results show that ElaB increases oxidative and heat stress resistance.
FIG 6.
ElaB increases survival during heat and oxidative stress. (A) Wild-type BW25113 (WT) and ΔelaB cells were grown until the turbidity reached 1.0 and were treated with heat stress (65°C for 5 min and 10 min); cell survival (%) was then tested. (B) Overnight cultures were diluted until the turbidity reached 0.5, and then 1 mM IPTG was added for 2 h. The OD600 was adjusted to 1.0, and cell survival was determined as described for panel A. (C) Wild-type and ΔelaB cells were treated with 10 mM H2O2 for the indicated times, and then cell survival was assayed. (D) Cells were collected as described for panel B, and cell survival was determined after treatment with 10 mM H2O2. Each assay was performed with three independent cultures, and one standard deviation is shown. *, P < 0.05; **, P < 0.01.
ElaB decreases persister cell formation.
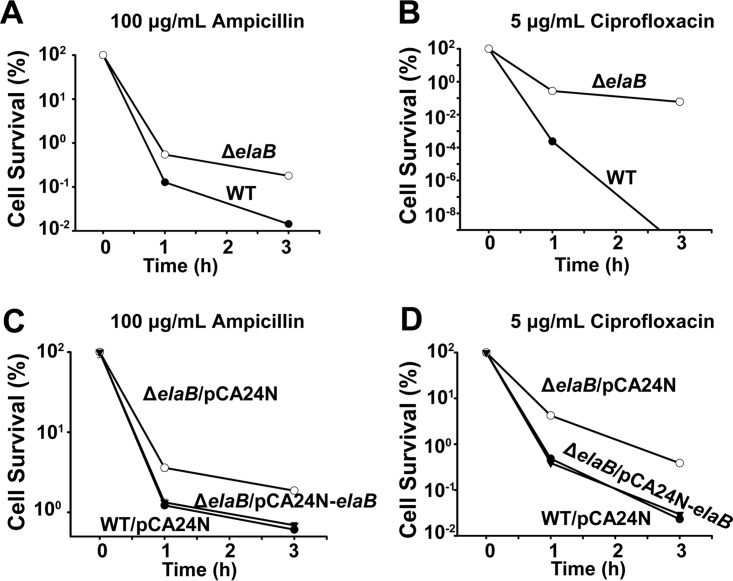
Inner membrane proteins can mediate antibiotic resistance as well as persister cell formation either by damaging the cell membrane or by affecting membrane permeability (40–44). Persister cells are phenotypic variants of regular cells, play a major role in the high antibiotic resistance of bacterial biofilms, and are likely responsible for the recalcitrance of chronic infections to antibiotics (45–47). To investigate the role of ElaB in persister cell formation, we first measured the MICs of 10 antibiotics commonly used for bacterial infections for the ΔelaB strain and the wild-type strain. As shown in Table 1, changes in MIC values for the 10 antibiotics between the two strains were within 2-fold. Next, we performed the persister cell assay using high concentrations (>10× MIC) of ampicillin and ciprofloxacin. The numbers of persister cells formed in the ΔelaB strain were 4.3- ± 0.5-fold higher after 1 h and 12.6- ± 0.5-fold higher after 3 h when treated with 100 μg/ml ampicillin (Fig. 7A). Critically, persister cell formation in the ΔelaB strain was (1.15 ± 0.1) × 103-fold higher than in the wild-type cells when treated with 5 μg/ml ciprofloxacin for 1 h. After 3 h, no persister cells were detected in the wild-type strain, while the proportion of persister cells in the ΔelaB strain was 0.06% ± 0.01% (Fig. 7B). Similar to what was reported earlier (48), ciprofloxacin appears to be more effective in killing regular nongrowing cells than ampicillin. Additionally, complementation of elaB via pCA24N-elaB reduced the persister cell formation compared with that from ΔelaB cells harboring an empty pCA24N plasmid (Fig. 7CD). Collectively, these results suggest that the inactivation of ElaB greatly increases persister cell formation.
TABLE 1.
MICs of 10 antibiotics for wild-type BW25113 and the ΔelaB mutant
| Antibiotic | MIC (μg/ml) for: |
|
|---|---|---|
| BW25113 | ΔelaB mutant | |
| Ampicillin | 4 | 4 |
| Polymyxin B | 0.5 | 0.5 |
| Cefoxitin | 8 | 4 |
| Ceftazidime | 0.5 | <0.25 |
| Imipenem | <0.125 | <0.125 |
| Cefotaxime | <0.25 | <0.25 |
| Cefepime | <0.5 | <0.5 |
| Ciprofloxacin | <0.125 | <0.125 |
| Gentamicin | <0.5 | <0.5 |
| Tetracycline | 2 | 2 |
FIG 7.
ElaB decreases persistence. (A and B) Survival of wild-type BW25113 (WT) and ΔelaB cells was determined with 100 μg/ml ampicillin and 5 μg/ml ciprofloxacin at the indicated time points. Overnight cultures were diluted and cultured until the turbidity reached 1.0 before the addition of ampicillin or ciprofloxacin. (C and D). Cell survival upon expression of elaB via pCA24N-elaB in the wild-type and ΔelaB cells was determined with 100 μg/ml ampicillin and 5 μg/ml ciprofloxacin at the indicated time points. Overnight cultures were diluted and cultured until the turbidity reached 0.5, and then 1 mM IPTG was added for 2 h. The turbidity was then adjusted to 1.0, the cells were treated with antibiotics, and persistence was determined at the indicated time points. Empty plasmid pCA24N was used as a negative control. Three independent cultures of each strain were evaluated for panels A to D.
DISCUSSION
Bacterial membranes play essential roles in the responses to various stresses. Here, we identified a new small inner membrane protein with a C-terminal transmembrane domain in E. coli, ElaB, which has a role in multiple stress responses. We found that (i) expression of elaB is upregulated during the stationary phase and it is positively regulated by RpoS in a concentration-dependent manner by the binding of RpoS to the elaB promoter, (ii) ElaB protects cells against oxidative stress and heat shock, and (iii) ElaB decreases persister cell formation. C-tail-anchored inner membrane proteins were first identified in eukaryotes and have been shown to play critical roles in apoptosis and other vital processes (10–12). In Streptomyces coelicolor, they may also play an important role (49). Here, we provide evidence that the C-tail-anchored inner membrane protein ElaB in E. coli participates in important cellular processes such as the responses to heat stress and oxidative stress.
Sigma factor RpoS is a general stress response regulator, and in E. coli, environmental stresses such as oxidative stress stimulate the expression of the rpoS gene and thus turn on the expression of the RpoS-controlled regulon (22, 50, 51). The transcription of genes recognized by RpoS through specific sequences in the promoter enables the activation of a “general stress response,” thus protecting cells from harmful conditions (39). In this study, we showed that the previously uncharacterized ElaB is under the direct control of RpoS, and ElaB protects cells against oxidative stress. Thus, under stress conditions where RpoS is upregulated (22), elaB transcription should also be induced, illustrating another example of the multifactorial regulation of RpoS. In addition, the upregulation of elaB by RpoS was also previously detected by other groups using DNA microarray studies with stress conditions (50, 52, 53). The deletion of rpoS, as well as the genes that RpoS controls, increases E. coli persister formation dramatically to the extent that nearly the whole population becomes persistent (54). Moreover, biofilms of the rpoS mutant were much more resistant to being killed by tobramycin than were wild-type P. aeruginosa biofilms (55). However, the underlying mechanism is unclear. Here, we found that ElaB, activated by RpoS, greatly decreased the tolerance to high concentrations of ciprofloxacin by promoting the formation of more persister cells. Two other identified C-terminus-anchored membrane proteins, YqjD and YgaM, are also induced in the stationary phase in E. coli (17). However, the physiological functions of YgaM and YqjD are largely unknown except that YqjD has been suggested to inhibit cell growth by inactivating the translational properties of ribosomes (17). We previously found that the deletion of the acid stress-related genes (e.g., gadB) that are activated by RpoS (52) or its transcriptional activator gadX greatly increased persister cell formation (54). Nevertheless, these results suggest that RpoS and genes under the direct control of RpoS are potentially important for the general stress response, but the underlying mechanism of how these genes contribute to the reduced tolerance to high concentrations of antibiotic while maintaining enhanced resistance to nonantibiotic environmental stresses needs further investigation. Thus, the disruption of C-tail-anchored inner membrane proteins can reduce stress resistance; it can also lead to deleterious effects, such as increased persistence in E. coli.
Our BLAST search indicates that ElaB, as well as YqjD and YgaM, lack N-terminal signal sequences. The high diversity of the N-terminal domains of these proteins, which exhibit no universally conserved sequence characteristics, argues for a membrane-targeting mechanism that depends primarily, if not entirely, on the C-terminal domains. Although the “twin-arginine repeat,” or TAT, pathway is involved in the secretion of folded proteins (56) and ElaB has two arginines at the end of the C terminus, it lacks the characteristic Z-R-R-ϕ-X-X sequence (where Z is a polar residue, X-X are hydrophobic residues, and ϕ is any residue) recognized by the TAT system (Fig. 1). Hence, it is unlikely that ElaB is secreted by the TAT system. In the absence of their N-terminal signal sequences, the C-terminal transmembrane domains of these C-tail-anchored proteins identified in S. coelicolor are sufficient for membrane targeting (13). However, the underlying targeting pathway has not been elucidated. The identification of this new targeting pathway suggests it may be an important target for antimicrobial agents.
Two small hydrophobic polypeptides, TisB (41) and GhoT (40, 42, 57), are toxin components of toxin-antitoxin systems. Neither of these is under the regulation of RpoS, but they are found to increase persister cell formation by reducing cell metabolism to create dormancy. GhoT is a small membrane protein with two transmembrane domains (residues 7 to 27 and 37 to 57) (40). Additionally, TisB (41) and GhoT damage cell envelopes and lead to cell death when overproduced, but overproduction of ElaB did not cause growth inhibition, indicating that different inner membrane proteins and transmembrane proteins may function differently in the presence of general stress. By using in vivo fluorescence imaging and next-generation sequencing, bacterial persister cells, in addition to dormancy (58), employ an “active defense” to pump antibiotics out and reduce intracellular drug concentrations through enhanced efflux activity (43). Since ElaB does not seem to affect the metabolic state of the cell, it is possible that the function of ElaB is involved with efflux pumps either by enhanced pumping of the antibiotic out of the cell or by limiting the uptake of the antibiotic. Although the C-terminal transmembrane domains of ElaB and YqjD are highly conserved, the functions of ElaB and YqjD seem different. YqjD inhibits cell growth by binding to the 30S subunit in the 70S and 100S ribosomes at the N-terminal region (17). However, the N-terminal regions of these two proteins are not highly conserved, and overproduction of ElaB does not lead to growth inhibition or cell lysis. Future studies are needed to elucidate the biochemical properties of these C-tailed inner membrane proteins and how they interact with efflux pumps or other cellular components in prokaryotes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 2. Luria-Bertani (LB) broth (59) and M9 minimal medium with 0.4% glucose (60) were used as indicated. The Keio collection (61) and the ASKA library (62) were used for deleting and overexpressing single genes. Chloramphenicol (30 μg/ml) was used for maintaining pCA24N-based plasmids, and kanamycin (50 μg/ml) was used for preculturing the isogenic knockout mutants.
TABLE 2.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli K-12 BW25113 | ||
| Wild type | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 rph-1 | 61 |
| ΔelaB mutant | ΔelaB ΔKmr | 61 |
| ΔrpoS mutant | ΔrpoS ΔKmr | 61 |
| elaB::mCherry mutant | mCherry was fused before the elaB stop codon in the wild-type strain | This study |
| lacZ::mCherry mutant | mCherry was fused before the lacZ stop codon in the wild-type strain | This study |
| elaB::gfp mutant | gfp was fused before the elaB stop codon in the wild-type strain | This study |
| Plasmids | ||
| pCA24N | Cmr lacIq | 62 |
| pCA24N-elaB | Cmr lacIq PT5-lac::elaB | 62 |
| pCA24N-gfp | Cmr lacIq, with gfp gene | 62 |
| pCA24N-elaB-gfp | Cmr lacIq PT5-lac::elaB, with gfp gene | 62 |
| pCA24N-ompA | Cmr lacIq PT5-lac::ompA | 62 |
| pCA24N-macB | Cmr lacIq PT5-lac::macB | 62 |
| pCA24N-rpoS | Cmr lacIq PT5-lac::rpoS | 62 |
| pET28b-rpoS | Kmr lacIq, pET28b PT7-lac::rpoS with C-terminal His tag | This study |
| pHGR01 | Kmr, R6K ori, promoterless-lacZ reporter vector | 71 |
| pHGR01-PelaB | Fused elaB promoter with lacZ in pHGR01 | This study |
| pHGR01-MPelaB | RpoS binding site in elaB promoter was mutated | This study |
Cmr and Kmr indicate chloramphenicol and kanamycin resistance, respectively. P and MP indicate promoter and mutant promoter, respectively.
Construction of plasmids.
For the purification of sigma factor RpoS, the coding region of rpoS was amplified using BW25113 genomic DNA as the template with the primer pair listed in Table S1 in the supplemental material. PCR products were purified using a gel extraction kit (Qiagen, Hilden, Germany), were digested with XbaI and HindIII, and were purified with a PCR product purification kit (Qiagen). The purified PCR products were ligated into the pET28b plasmid and transferred into E. coli BL21. The correct construct was verified by DNA sequencing using primer M13.
For the promoter activity assay, a 300-nt fragment from −300 to −1 relative to the elaB translational start site was amplified by PCR with primers pHGR01-PelaB-f and pHGR01-PelaB-r (Table S1). The PCR product was purified and digested with EcoRI and HindIII and then was ligated into pHGR01. The correct vector pHGR01-PelaB was verified by DNA sequencing using the primer pair pHGR01-f and pHGR01-r listed in Table S1. To perform site mutagenesis of the RpoS binding site in the elaB promoter in pHGR01-PelaB (pHGR01-MPelaB), from TTCAGG (−35)…TCTATAGTTA (−10) to AAAAAA (−35)…CCCCCCCCCC (−10), three rounds of PCR were performed. The first-round PCR was amplified with pHGR01-PelaB-f and M-pHGR01-PelaB-r2 (Table S1) using the wild-type genomic DNA as the template, the purified PCR product was then used as the template and the secondary-round PCR was amplified with pHGR01-PelaB-f and M-pHGR01-PelaB-r3 (Table S1). The purified products from the secondary PCR were further used as the template and amplified with pHGR01-PelaB-f and pHGR01-PelaB-r. The last-round PCR products were further cloned into pHGR01 to make pHGR01-MPelaB, and the correct mutation in the elaB promoter region in pHGR01-MPelaB was validated by DNA sequencing using primers pHGR01-f and pHGR01-r listed in Table S1.
Generation of mCherry and gfp fused strains.
To generate elaB::mCherry, the one-step inactivation method (63) was applied to fuse mCherry, which encodes a red fluorescent protein, before the stop codon of elaB to generate the protein ElaB-mCherry. The coding region of mCherry without its start codon was amplified by PCR using pmCherry-N1 (Clontech, Mountain View, CA) as the template with primers mCherry-f and mCherry-r. The primer pair mCherry-KM-f and KM-r was used to amplify the kanamycin resistance (Kmr) cassette, bordered by FLP recombination target (FRT) sites, from plasmid pKD4. The PCR products served as the templates in overlapping extension PCR with primers mCherry-f and KM-r to generate a DNA fragment carrying mCherry and Kmr cassettes flanked by ∼60-nt regions up- and downstream of the elaB stop codon. The PCR products were purified using a gel extraction kit (Qiagen), and purified fragments were electroporated into BW25113/pKD46 competent cells. The strain of BW25113 elaB::mCherry was confirmed by PCR followed by DNA sequencing using primers of conf-f and conf-r. The same procedures were performed to fuse mCherry before the stop codon of the lacZ gene, as well as for the construction of the elaB::gfp strain.
Protein localization.
For localization of ElaB using the mCherry-ElaB fusion, strains were cultured to a turbidity at 600 nm of 1.0, were washed with 0.85% NaCl, and were imaged with fluorescence microscopy (Zeiss Axiophot) using an oil immersion objective (100×). For localization of ElaB by GFP fusion, overnight cultures of BW25113 harboring pCA24N-elaB-gfp or pCA24N-gfp were inoculated into LB broth with chloramphenicol (30 μg/ml) to a turbidity at 600 nm of 0.1, and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce ElaB-GFP expression for 2 h before imaging.
Fractionation of membrane proteins.
To study the localization of ElaB, proteins from the inner and outer membranes were obtained by differential centrifugation as described previously with modification (17, 28). IPTG (0.5 mM) was added to induce ElaB expression using pCA24N-elaB for 3 h. Cell pellets were resuspended in TE buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA), and 1 mg/ml lysozyme (Sigma-Aldrich) was added to lyse the cells. Cell debris was removed by centrifugation at 3,200 × g at 4°C for 10 min. The supernatant was further centrifuged at 4°C at 500,000 × g for 1.5 h, and the pellets (membrane protein factions) were resuspended in 2 ml of TE buffer. An aliquot (1/10 of the suspension) was then loaded on a 20-ml sucrose step gradient prepared with 10 ml of 70% (wt/vol) sucrose in TE buffer layered with 10 ml of 53% (wt/vol) sucrose in TE buffer to form a gradient and subjected to centrifugation at 4°C at 400,000 × g for 5 h. The upper band (inner membrane proteins) and lower band (outer membrane proteins) were collected from the top of the gradient. After removing sucrose using a filter (Amicon Ultracel-3K; Millipore), the membrane fractions were solubilized in sodium dodecyl sulfate (SDS) sample buffer and were used for Western blotting. Cells harboring pCA24N-ompA and pCA24N-macB prepared under the same conditions were used as controls.
Western blot analysis.
A total of 2.5 μg protein was loaded and run using Tricine-SDS-PAGE as described previously (64). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA), and Western blotting was performed with primary antibodies raised against the His tag (Cell Signaling Technology, Danvers, MA) or GFP (Abmart, Shanghai, China) and horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (Bio-Rad, Richmond, CA).
qRT-PCR.
Total RNA was isolated (65) using an RNA isolation kit (Invitrogen, Carlsbad, CA). DNase was applied during the RNA isolation process to avoid contamination by DNA. A total of 50 ng total RNA was used for quantitative real-time PCR (qRT-PCR) using the Power SYBR Green RNA-to-CT 1-step kit and the StepOne real-time PCR system (Applied Biosystems). Primers were annealed at 60°C, and rrsG (66) was used for normalizing the data. The relative expression levels for the induction or repression of elaB under different conditions were calculated as described previously (67).
Electrophoretic mobility shift assay.
EMSAs were performed as previously described (33, 68). Briefly, DNA fragments were amplified using the primer pairs shown in Table S1. PCR products were gel purified with a QIAquick gel extraction kit (Qiagen) and labeled with the Pierce biotin 3′ end DNA labeling kit (Thermo Scientific, Rockford, IL). For the binding reactions, the E. coli RNA core polymerase (NEB, Ipswich, MA, USA) was mixed 1:2 (M:M) with RpoS at room temperature for 5 min to form the holoenzyme before the addition of biotin-labeled DNA probes (0.05 pmol). The binding reactions were performed with the nonspecific competitor DNA [poly(dI-dC)] and Nonidet P-40 in buffer containing 10 mM HEPES (pH 7.3), 20 mM KCl, 1 mM MgCl2, and 5% glycerol at 25°C for 3 h. The final mixtures were run on a 6% DNA retardation gel (Invitrogen), were transferred to a nylon membrane, and were UV cross-linked. Chemiluminescence was performed with the LightShift chemiluminescent EMSA kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's protocol.
β-Galactosidase activity assay.
BW25113 wild-type and ΔrpoS cells harboring pHGR01-PelaB or pHGR01-MPelaB were cultured to a turbidity at 600 nm of 6.0, and 800 μl cultures were diluted with 4 ml PM2 (70 mM Na2HPO4·12H2O, 30 mM NaH2PO4·H2O, 1 mM MgSO4 and 0.2 mM MnSO4, pH 7.0) (69). Next, 30 μl of toluene and 35 μl of a 0.1% SDS solution were added to 2.5 ml of bacterial suspension to permeabilize cells. The mixtures were vortexed for 10 s and incubated at 37°C for 45 min in a water bath for the evaporation of toluene. For the enzymatic reactions, 250 μl of permeabilized cells was added to PM2 supplemented with β-mercaptoethanol (final concentration, 100 mM) to a final volume of 1 ml. The reactions were started by adding 250 μl of 4 mg/ml o-nitrophenol-galactoside in PM2. Next, 500 μl of 1 M Na2CO3 was added to stop the reactions and the turbidity at 420 nm was measured. The β-galactosidase activity (Miller units) was calculated as described previously (70). For rpoS complementation studies, cells carrying pCA24N-rpoS were cultured to a turbidity at 600 nm of 1.0, and the expression of RpoS was induced with 0.5 mM IPTG for 2 h; β-galactosidase activity was measured as above.
Survival assay.
Overnight cultures were diluted to a turbidity at 600 nm of 0.05 in LB broth and incubated at 37°C with shaking at 250 rpm until the cultures reached a turbidity of 1.0. Next, 1 ml was collected for a cell viability assay to measure the initial population. For strains using pCA24N-based plasmids, overnight cultures were diluted to a turbidity of 0.05 and grown to a turbidity of 0.5. To induce gene expression for 2 h, 1 mM IPTG was added and the turbidity was adjusted to 1.0 (66). For the oxidative stress assay, 1 ml cultures were treated with or without 20 mM H2O2 for 10 min, and cells were serially diluted in 0.85% NaCl solution and applied as 10 μl drops on LB agar. For the persistence assay, 100 μg/ml ampicillin or 5.0 μg/ml ciprofloxacin was added to cultures with a turbidity of 1.0 followed by incubation for 3 h; cell survival was determined by a drop assay. For heat shock stress, cultures at a turbidity of 1.0 were transferred from 37°C to 65°C using a water shaking incubator for 5 min and 10 min.
Determination of MICs.
MICs were determined using Biofosun (Shanghai, China) drug-sensitive test plates. In brief, overnight cultures were diluted to turbidity of 0.05 in LB medium and incubated at 37°C until the turbidity reached 0.1. Cultures were diluted 1:100 in fresh LB medium, and 100 μl of the cell suspension inoculated into the wells which were treated with different concentrations of antibiotics. The cells were then cultured without shaking for 20 h at 37°C. The MICs were determined as the lowest antibiotic concentrations at which no visible growth occurred.
Statistical analysis.
Statistical analysis was performed using SPSS (16.0 version). The differences of the corresponding values among different conditions were tested by one-way analysis of variance (ANOVA) followed by a least significant difference test. A probability level (P value) of less than 0.05 was regarded as statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the Keio and ASKA strains provided by the Genome Analysis Project in Japan.
This work was supported by the National Science Foundation of China (grant no. 31290233, 31625001, and 31500025), by the National Science Foundation of Guangdong Province (2015A030310405), by the China Postdoctoral Science Foundation funded project (2013M542217 and 2014T70830), and by the Army Research Office (W911NF-14-1-0279). X.W. is a recipient of the 1000-Youth Elite Program (the Recruitment Program of Global Experts in China).
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00057-17.
REFERENCES
- 1.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 2.Liszewski K. 2015. Dissecting the structure of membrane proteins. Genet Eng News 35:17. [Google Scholar]
- 3.Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 4.Saier MH Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The transporter classification database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen K, Shan SO. 2010. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci U S A 107:7698–7703. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffitt JR, Pandey S, Boettiger AN, Wang SY, Zhuang XW. 2016. Spatial organization shapes the turnover of a bacterial transcriptome. eLife 5:e13065. doi: 10.7554/eLife.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kol S, Nouwen N, Driessen AJ. 2008. Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J Biol Chem 283:31269–31273. doi: 10.1074/jbc.R800029200. [DOI] [PubMed] [Google Scholar]
- 8.Johnson AE, van Waes MA. 1999. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol 15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 9.Herskovits AA, Bibi E. 2000. Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc Natl Acad Sci U S A 97:4621–4626. doi: 10.1073/pnas.080077197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalbfleisch T, Cambon A, Wattenberg BW. 2007. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic 8:1687–1694. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- 11.Pedrazzini E. 2009. Tail-anchored proteins in plants. J Plant Biol 52:88–101. doi: 10.1007/s12374-009-9014-1. [DOI] [Google Scholar]
- 12.Kriechbaumer V, Shaw R, Mukherjee J, Bowsher CG, Harrison AM, Abell BM. 2009. Subcellular distribution of tail-anchored proteins in arabidopsis. Traffic 10:1753–1764. doi: 10.1111/j.1600-0854.2009.00991.x. [DOI] [PubMed] [Google Scholar]
- 13.Craney A, Tahlan K, Andrews D, Nodwell J. 2011. Bacterial transmembrane proteins that lack N-terminal signal sequences. PLoS One 6:e19421. doi: 10.1371/journal.pone.0019421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borgese N, Righi M. 2010. Remote origins of tail-anchored proteins. Traffic 11:877–885. doi: 10.1111/j.1600-0854.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- 15.Karlinsey JE, Pease AJ, Winkler ME, Bailey JL, Hughes KT. 1997. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J Bacteriol 179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost LS, Paranchych W, Willetts NS. 1984. DNA sequence of the F traALE region that includes the gene for F pilin. J Bacteriol 160:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida H, Maki Y, Furuike S, Sakai A, Ueta M, Wada A. 2012. YqjD is an inner membrane protein associated with stationary-phase ribosomes in Escherichia coli. J Bacteriol 194:4178–4183. doi: 10.1128/JB.00396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luirink J, Yu Z, Wagner S, de Gier JW. 2012. Biogenesis of inner membrane proteins in Escherichia coli. Biochim Biophys Acta 1817:965–976. doi: 10.1016/j.bbabio.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev 55:561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jara LM, Cortes P, Bou G, Barbe J, Aranda J. 2015. Differential roles of antimicrobials in the acquisition of drug resistance through activation of the SOS response in Acinetobacter baumannii. Antimicrob Agents Chemother 59:4318–4320. doi: 10.1128/AAC.04918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaniol V, Bernhard S, Aebi C. 2015. Moraxella catarrhalis AcrAB-OprM efflux pump contributes to antimicrobial resistance and is enhanced during cold shock response. Antimicrob Agents Chemother 59:1886–1894. doi: 10.1128/AAC.03727-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta T, Deutscher MP. 2009. Catalytic properties of RNase BN/RNase Z from Escherichia coli: RNase BN is both an exo- and endonuclease. J Biol Chem 284:15425–15431. doi: 10.1074/jbc.M109.005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asha PK, Blouin RT, Zaniewski R, Deutscher MP. 1983. Ribonuclease BN: identification and partial characterization of a new tRNA processing enzyme. Proc Natl Acad Sci U S A 80:3301–3304. doi: 10.1073/pnas.80.11.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catic A, Misaghi S, Korbel GA, Ploegh HL. 2007. ElaD, a deubiquitinating protease expressed by E. coli. PLoS One 2:e381. doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Young KD. 2012. Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol Microbiol 84:276–295. doi: 10.1111/j.1365-2958.2012.08021.x. [DOI] [PubMed] [Google Scholar]
- 27.Urbanus ML, Fröderberg L, Drew D, Björk P, de Gier J-WL, Brunner J, Oudega B, Luirink J. 2002. Targeting, insertion, and localization of Escherichia coli YidC. J Biol Chem 277:12718–12723. doi: 10.1074/jbc.M200311200. [DOI] [PubMed] [Google Scholar]
- 28.Castanie-Cornet MP, Cam K, Jacq A. 2006. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J Bacteriol 188:4264–4270. doi: 10.1128/JB.00004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daley DO, Rapp M, Granseth E, Melén K, Drew D, von Heijne G. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 30.Freudl R, Schwarz H, Stierhof Y, Gamon K, Hindennach I, Henning U. 1986. An outer membrane protein (OmpA) of Escherichia coli K-12 undergoes a conformational change during export. J Biol Chem 261:11355–11361. [PubMed] [Google Scholar]
- 31.Fontaine F, Fuchs RT, Storz G. 2011. Membrane localization of small proteins in Escherichia coli. J Biol Chem 286:32464–32474. doi: 10.1074/jbc.M111.245696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Gralla JD. 2001. Sigma38 (rpoS) RNA polymerase promoter engagement via −10 region on nucleotides. J Biol Chem 276:30064–30071. doi: 10.1074/jbc.M102886200. [DOI] [PubMed] [Google Scholar]
- 34.Peano C, Wolf J, Demol J, Rossi E, Petiti L, De Bellis G, Geiselmann J, Egli T, Lacour S, Landini P. 2015. Characterization of the Escherichia coli sigma(S) core regulon by chromatin immunoprecipitation-sequencing (ChIP-seq) analysis. Sci Rep 5:10469. doi: 10.1038/srep10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciag A, Peano C, Pietrelli A, Egli T, De Bellis G, Landini P. 2011. In vitro transcription profiling of the sigmaS subunit of bacterial RNA polymerase: re-definition of the sigmaS regulon and identification of sigmaS-specific promoter sequence elements. Nucleic Acids Res 39:5338–5355. doi: 10.1093/nar/gkr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacour S, Landini P. 2004. sigma(S)-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigma(S)-dependent genes and identification of their promoter sequences. J Bacteriol 186:7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Typas A, Hengge R. 2006. Role of the spacer between the −35 and −10 regions in sigmaS promoter selectivity in Escherichia coli. Mol Microbiol 59:1037–1051. doi: 10.1111/j.1365-2958.2005.04998.x. [DOI] [PubMed] [Google Scholar]
- 38.Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, Gourse RL. 2001. Promoter recognition and discrimination by EsigmaS RNA polymerase. Mol Microbiol 42:939–954. doi: 10.1046/j.1365-2958.2001.02703.x. [DOI] [PubMed] [Google Scholar]
- 39.Landini P, Egli T, Wolf J, Lacour S. 2014. sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ Microbiol Rep 6:1–13. doi: 10.1111/1758-2229.12112. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Lord DM, Cheng H-Y, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. 2012. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol 8:858–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng HY, Soo VW, Islam S, McAnulty MJ, Benedik MJ, Wood TK. 2014. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ Microbiol 16:1741–1754. doi: 10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 43.Pu YY, Zhao ZL, Li YX, Zou J, Ma Q, Zhao YN, Ke YH, Zhu Y, Chen HY, Baker MAB, Ge H, Sun YJ, Xie XS, Bai F. 2016. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerdes K, Semsey S. 2016. Pumping persisters. Nature 534:41–42. doi: 10.1038/nature18442. [DOI] [PubMed] [Google Scholar]
- 45.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.del Pozo JL, Patel R. 2007. The challenge of treating biofilm-associated bacterial infections. Clin Pharmacol Ther 82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 47.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 48.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 49.Caraney A. 2012. PhD dissertation. Small molecule interrogation of Streptomyces coelicolor growth, development and secondary metabolism. McMaster University, Hamilton, Ontario, Canada. [Google Scholar]
- 50.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hengge R. 2009. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res Microbiol 160:667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics 272:580–591. doi: 10.1007/s00438-004-1089-2. [DOI] [PubMed] [Google Scholar]
- 53.Ito A, May T, Kawata K, Okabe S. 2008. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol Bioeng 99:1462–1471. doi: 10.1002/bit.21695. [DOI] [PubMed] [Google Scholar]
- 54.Hong SH, Wang XX, O'Connor HF, Benedik MJ, Wood TK. 2012. Bacterial persistence increases as environmental fitness decreases. Microb Biotechnol 5:509–522. doi: 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teltzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 56.Lee PA, Tullman-Ercek D, Georgiou G. 2006. The bacterial twin-arginine translocation pathway. Annu Rev Microbiol 60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Lord DM, Hong SH, Peti W, Benedik MJ, Page R, Wood TK. 2013. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ Microbiol 15:1734–1744. doi: 10.1111/1462-2920.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A 107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maniatis T, Sambrook J, Fritsch EF. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 60.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 63.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schägger H. 2006. Tricine-SDS-PAGE. Nat Protoc 1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 65.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. 2004. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol 64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, Wood TK. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol 7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y, Quiroga C, Chen Q, McAnulty MJ, Benedik MJ, Wood TK, Wang X. 2014. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res 42:6448–6462. doi: 10.1093/nar/gku279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao K, Liu M, Burgess RR. 2005. The global transcriptional response of Escherichia coli to induced sigma 32 protein involves sigma 32 regulon activation followed by inactivation and degradation of sigma 32 in vivo. J Biol Chem 280:17758–17768. doi: 10.1074/jbc.M500393200. [DOI] [PubMed] [Google Scholar]
- 69.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frias JE, Flores E. 2015. Induction of the nitrate assimilation nira operon and protein-protein interactions in the maturation of nitrate and nitrite reductases in the Cyanobacterium Anabaena sp. strain PCC. 7120. J Bacteriol 197:2442–2452. doi: 10.1128/JB.00198-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu HH, Jin M, Ju LL, Mao YT, Gao HC. 2014. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ Microbiol 16:3181–3195. doi: 10.1111/1462-2920.12457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.