Abstract
Lycorine potently inhibits flaviviruses in cell culture. At 1.2-μM concentration, lycorine reduced viral titers of West Nile (WNV), dengue, and yellow fever viruses by 102- to 104-fold. However, the compound did not inhibit an alphavirus (Western equine encephalitis virus) or a rhabdovirus (vesicular stomatitis virus), indicating a selective antiviral spectrum. The compound exerts its antiviral activity mainly through suppression of viral RNA replication. A Val → Met substitution at the 9th amino acid position of the viral 2K peptide (spanning the endoplasmic reticulum membrane between NS4A and NS4B proteins) confers WNV resistance to lycorine, through enhancement of viral RNA replication. Initial chemistry synthesis demonstrated that modifications of the two hydroxyl groups of lycorine can increase the compound's potency, while reducing its cytotoxicity. Taken together, the results have established lycorine as a flavivirus inhibitor for antiviral development. The lycorine-resistance results demonstrate a direct role of the 2K peptide in flavivirus RNA synthesis.
Keywords: Flavivirus replication, Antiviral, West Nile virus, Flavivirus 2K peptide, Viral resistance
Introduction
The family Flaviviridae consists of three genera, the flaviviruses, the pestiviruses, and the hepatitis C viruses. Many members of the genus Flavivirus are arthropod-borne human pathogens, including four serotypes of dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV) (Gubler et al., 2007). More than 50 million, 200,000, and 50,000 human cases were reported annually for DENV, YFV, and JEV infections, respectively (Gubler et al., 2007). Since the initial outbreak of WNV in New York in 1999, the virus has caused thousands of human cases of infections each year and has spread throughout North America (Kramer et al., 2007). No effective antiviral therapy has been approved for the treatment of flavivirus infections. Human vaccines are currently available only for JEV, YFV, and TBEV. It has been well recognized that development of a vaccine for DENV is particularly challenging, because of the need to simultaneously immunize against all four DENV serotypes. Therefore, development of therapeutics is the priority for intervention in flavivirus infections.
The flavivirus genome is a plus-sense, single-stranded RNA of about 11,000 nucleotides (Lindenbach et al., 2007). The genomic RNA consists of a 5′ untranslated region (UTR), a single open reading frame (ORF), and a 3′UTR. The single ORF encodes a long polyprotein that is co-translationally and post-translationally processed by viral and host proteases into ten mature viral proteins. The N-terminus of the polyprotein contains three structural proteins: capsid (C), premembrane (prM/M), and envelope (E). The C-terminus of the polyprotein contains seven nonstructural (NS) proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. Complete cleavage of the polyprotein generates a 2K peptide between NS4A and NS4B. The 2K peptide spans the membrane of endoplasmic reticulum (ER). Two viral proteins have enzymatic activities. NS3 functions as a protease (with NS2B as a cofactor), a nucleotide triphosphatase, an RNA triphosphatase, and a helicase (Falgout et al., 1993, Li et al., 1999, Warrener et al., 1993, Wengler and Wengler, 1991). NS5 acts as a methyltransferase (MTase) and an RNA-dependent-RNA polymerase (RdRp) (Ackermann and Padmanabhan, 2001, Egloff et al., 2002, Guyatt et al., 2001, Ray et al., 2006, Tan et al., 1996). Besides functioning in replication, flaviviral NS proteins play roles in virion assembly (Jones et al., 2005, Kummerer and Rice, 2002, Leung et al., 2001, Liu et al., 2003) and evasion of host innate immune responses (Best et al., 2005, Guo et al., 2005, Liu et al., 2005, Munoz-Jordan et al., 2005, Munoz-Jordan et al., 2003). Upon viral entry and nucleocapsid uncoating, the genomic RNA is translated into proteins, which are translocated across the ER membrane to form the replication complexes (Lindenbach et al., 2007). The molecular details of individual NS proteins and their roles in flavivirus replication remain to be characterized.
Lycorine is an alkaloid compound found in several plants, such as daffodil (Narcissus pseudonarcissus) and bush lily (Clivia miniata). A number of biological activities have been reported for lycorine, including inhibition of protein and DNA synthesis (Chattopadhyay et al., 1984), cell growth and division (De Leo et al., 1973), and anti-leukemia effect (Liu et al., 2004, Liu et al., 2007). In addition, the compound has been shown to inhibit poliovirus (Ieven et al., 1982), Severe Acute Respiratory Syndrome-associated coronavirus (SARS-CoV) (Li et al., 2005), herpes simplex virus (type 1) (Renard-Nozaki et al., 1989), and vaccinia virus (Zhou et al., 2003).
In this paper, we report that lycorine inhibits flaviviruses with a selective antiviral spectrum. Mode-of-action analysis indicates that lycorine inhibits flaviviruses mainly through suppression of viral RNA synthesis. Structural modifications of the lycorine compound increased its potency while decreasing its cytotoxicity, indicating the potential for the compound's therapeutic development. Furthermore, we found that a single-amino acid substitution in WNV 2K peptide confers resistance to lycorine, partially through enhancement of viral RNA synthesis, thus revealing a direct role of the 2K peptide in flavivirus RNA replication.
Results
Identification of lycorine as an inhibitor of WNV and DENV-1
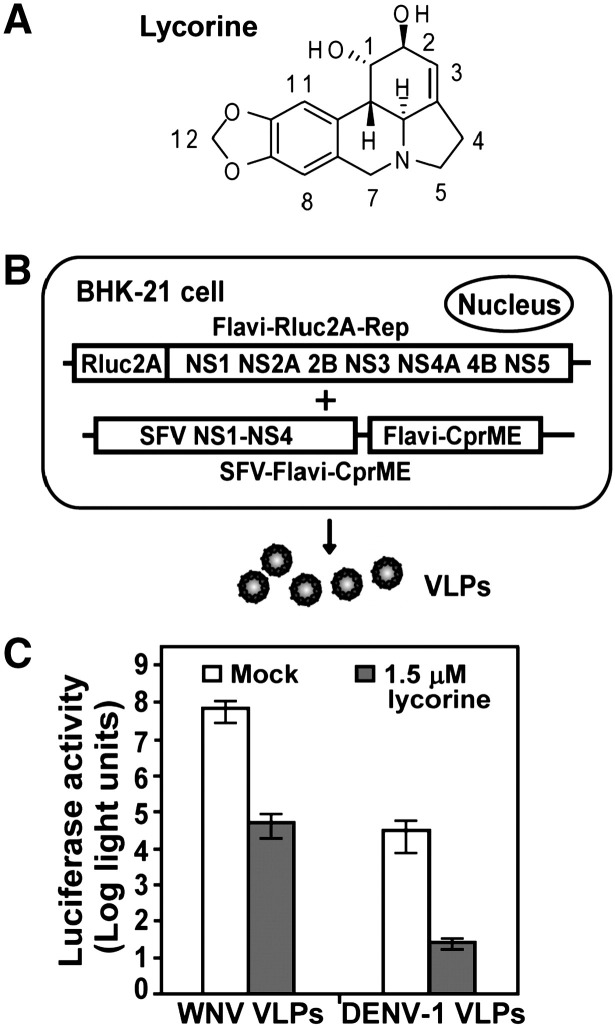
Lycorine (Fig. 1A) has been reported to have antiviral activities (See Introduction). To test whether lycorine inhibits flaviviruses, we initially screened the compound using a viral-like particle (VLP)-based infection assay. As depicted in Fig. 1B, VLPs of WNV and DENV-1 were prepared by trans-supply viral structural proteins (CprME; through an alphavirus Semliki Forest virus [SFV] expression vector) to package corresponding replicon RNAs containing a luciferase reporter (Rluc2A-Rep). The titers of the VLPs were estimated to be 2.5 × 106 and 2.4 × 103 FFU/ml for WNV and DENV-1, respectively. Vero cells were infected with 1 FFU/cell of WNV VLP or with 0.01 FFU/cell of DENV-1 VLP (due to the low titer of DENV-1 VLP). The infected cells were treated with 1.5 μM lycorine or were mock-treated with 1% DMSO. At 24 h post-infection (p.i.), lycorine reduced the luciferase signals by 1400- and 1200-fold in the WNV and DENV-1 VLP-infected cells, respectively (Fig. 1C). Higher concentrations of lycorine were also tested; however, due to cytotoxicity, these results are not presented (see next section). The results indicate that lycorine inhibits WNV and DENV-1.
Fig. 1.
Identification of lycorine as an inhibitor of WNV and DENV-1 VLP infections. (A) Structure of lycorine. The carbon positions of the lycorine molecule are numbered. (B) Production of flavivirus VLPs. Flavivirus VLPs were prepared by sequential transfection of BHK-21 cells with a luciferase-reporting replicon (Flavi-Rluc2A-Rep) and an SFV vector expressing flavivirus structural proteins (SFV-Flavi-CprME). See Materials and methods for details. (C) Inhibition of WNV and DENV-1 VLP infections by lycorine. Vero cells were infected with WNV (1 FFU/cell) or DENV-1 (0.01 FFU/cell) VLPs in the presence of 1.5 μM lycorine. Luciferase activities were measured at 24 h post-infection. Average results of three independent experiments are shown.
Inhibition of an epidemic strain of WNV at noncytotoxic concentrations
We performed an MTT assay to determine the cytotoxicity of lycorine in order to exclude the possibility that the above-described antiviral activity was due to compound-mediated cytotoxicity (Fig. 2A). Vero cells were incubated with various concentrations of lycorine for 48 h; cell viability was indicated by cellular metabolism of MTT tetrazolium salt. The CC50 (50% cytotoxic concentration) value of the compound was estimated to be 24 μM. At 1.5 μM, cell viability was > 90%. Therefore, all following experiments were performed at < 1.5 μM of lycorine.
Fig. 2.
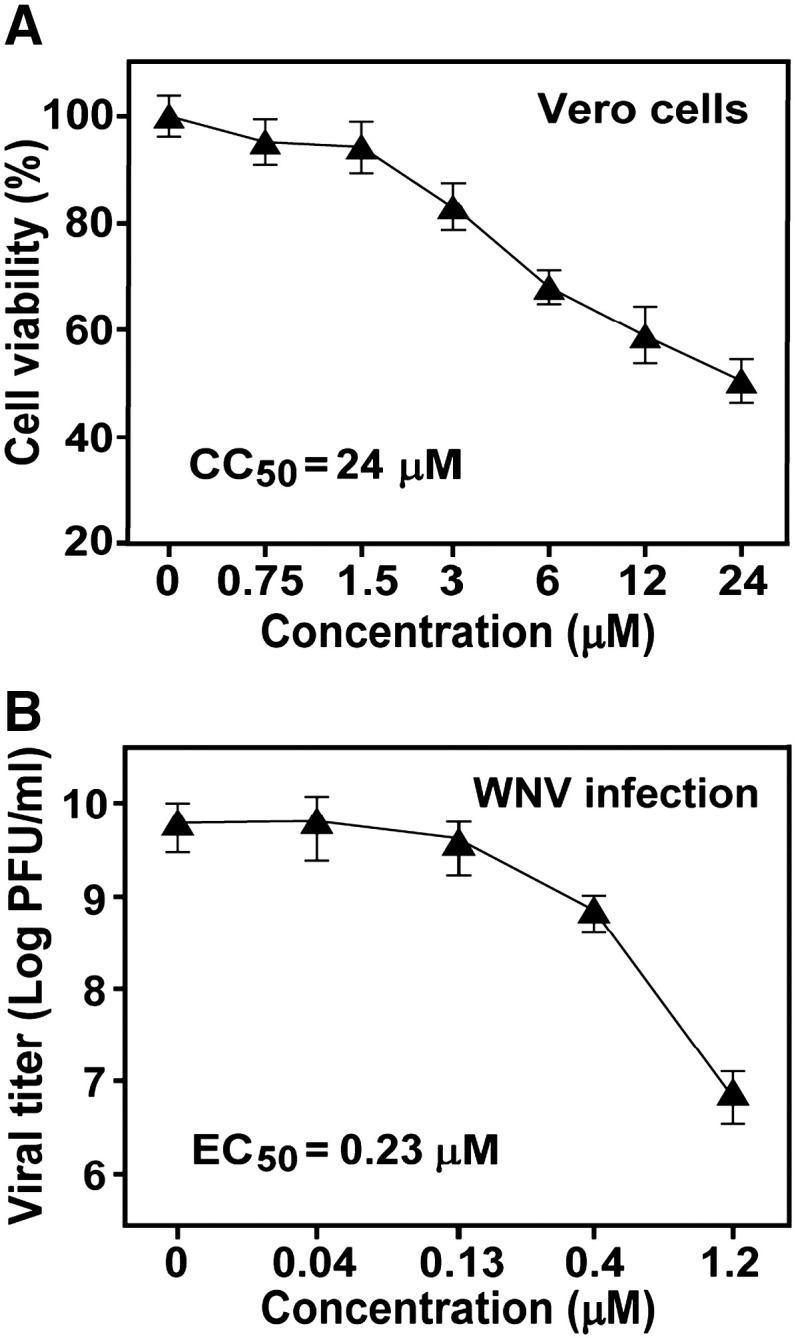
Cytotoxicity and potency of lycorine against an epidemic strain of WNV. (A) Cytotoxicity of lycorine in Vero cells. Cytotoxicity was examined by incubation of Vero cells with the indicated concentrations of lycorine. Cell viability was measured by an MTT assay, and is presented as a percentage of colorimetric absorbance derived from the compound-treated cells compared with that from the mock-treated cells (with 1% DMSO). Average results from three experiments are shown. (B) Inhibition of WNV infection in cell culture. Vero cells were infected with an epidemic strain of WNV (0.1 MOI). The infected cells were immediately treated with lycorine at the indicated concentrations. Viral titers in culture fluids at 42 h p.i. were determined by plaque assays.
Next, we assayed the antiviral activity of lycorine using an authentic WNV infection assay. Vero cells were infected with an epidemic strain of WNV (0.1 MOI). The infections were treated with various concentrations of the compound, and assayed for viral yields in culture medium at 42 h p.i. (Fig. 2B). The compound suppressed viral titer in a dose-responsive manner. At 1.2 μM, the compound reduced the viral titer by 910-fold. The EC50 (50% effective concentration) value was estimated to be 0.23 μM. The results demonstrate that lycorine inhibits WNV at noncytotoxic concentrations.
Selective antiviral spectrum
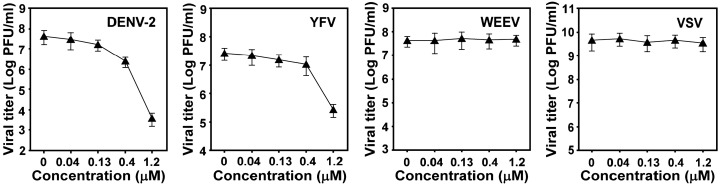
To examine the antiviral spectrum of lycorine, we performed viral titer reduction assays using other flaviviruses (DENV-2 and YFV), a plus-strand RNA alphavirus (Western equine encephalitis virus, WEEV), and a negative-strand RNA rhabdovirus (vesicular stomatitis virus, VSV). Lycorine inhibited both DENV-2 and YFV (Fig. 3 ). At 1.2 μM, the compound reduced viral titers of DENV-2 and YFV by 1.1 × 104- and 98-fold, respectively. In contrast, the compound suppressed titers of neither WEEV nor VSV at the tested concentrations. The latter results further suggest that lycorine at 1.2 μM is not cytotoxic. Overall, the results indicate that the antiviral spectrum of the compound is selective for flaviviruses.
Fig. 3.
Antiviral spectrum of lycorine. Viral titer reduction assays were performed for DENV-2, YFV, WEEV, and VSV in the presence of various concentrations of lycorine. See details in Materials and methods.
Inhibitory step(s) of viral infection
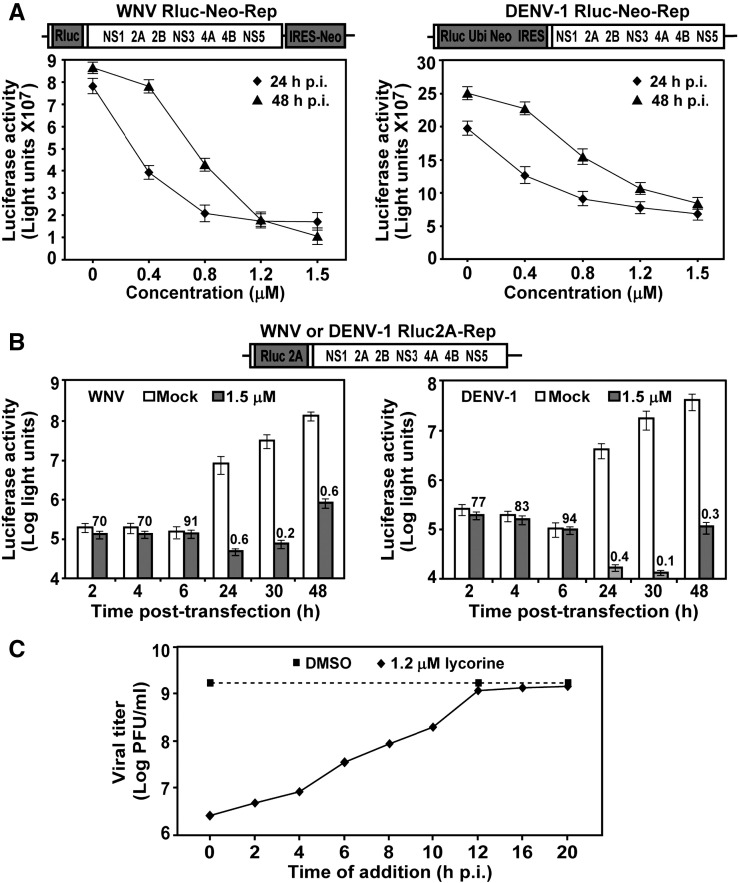
Given that lycorine inhibits VLP-mediated infection (Fig. 1C), the compound could block steps of viral entry and/or replication. To pinpoint the step(s) of compound inhibition, we tested the compound in the Vero cell lines containing persistently replicating replicon of WNV or DENV (Rluc-Neo-Rep; Fig. 4A). Incubation of the Rluc-Neo-Rep cells with 1.5-μM lycorine for 24 h or 48 h led to reduced luciferase activities, suggesting that the compound inhibits viral translation and/or RNA synthesis.
Fig. 4.
Mechanism of lycorine-mediated inhibition of flaviviruses. (A) Antiviral activities in Vero cells containing Rluc-Neo-Rep of WNV (left) and DENV-1 (right). Vero cells containing a WNV or DENV-1 replicon (Rluc-Neo-Rep) were treated with lycorine at the indicated concentrations, and were measured for luciferase activities at 24 or 48 h post-treatment. (B) Analysis of lycorine using transient replicons of WNV (left) and DENV-1 (right). Luciferase replicons (Rluc2A-Rep) of WNV or DENV-1 were electroporated into BHK-21 cells. The transfected cells were immediately incubated with 1.5 μM lycorine, and were measured for luciferase activities at the indicated time points post-transfection. Numbers above the lycorine-treated datum points indicate percentages of luciferase signals from the compound-treated transfection compared with those from the mock-treated transfection (set to 100%). Error bars indicate the standard deviations from three independent experiments. (C) Time-of-addition analysis of lycorine in WNV infection. Vero cells were infected with WNV at an MOI of 10 at 4°C for 1 h. The infected cells were washed three times with PBS. Lycorine (1.2 μM) was then added to the cells at the indicated time points post-infection. The supernatants were assayed for viral titers at 20 h post-infection. As controls, 1% DMSO was added to the infected cells at 0, 12 and 20 h p.i. for estimation of its effect on viral production.
We then used a transient replicon system (Rluc2A-Rep; Fig. 4B) to differentiate between inhibition of viral translation and inhibition of RNA synthesis. Previous studies showed that transfection of BHK-21 cells with the Rluc2A-Rep of WNV or DENV-1 produces two luciferase peaks, one at 1 to 8 h post-transfection (p.t.), and another at > 20 h p.t.; these respectively represent viral translation and RNA replication (Lo et al., 2003, Puig-Basagoiti et al., 2006). As shown in Fig. 4B, whereas lycorine suppressed early luciferase signals (at 2, 4, and 6 h p.t.) by < 30%, it reduced late luciferase signals (at 24, 30 and 48 h p.t.) by > 99%. Taken together, the results suggest that lycorine weakly inhibits viral translation, but strongly suppresses RNA synthesis.
Time-of-addition analysis
A time-of-addition experiment was performed to further elucidate the mode-of-action of lycorine (Fig. 4C). Vero cells were synchronously infected with WNV (Puig-Basagoiti et al., 2006). Lycorine (1.2 μM) was added to the infected cells at various time points post-infection. Viral titers in the culture medium were determined at 20 h post-infection. As controls, 1% DMSO was added to infected cells at 0, 12, or 20 h p.i., for estimation of its effect on viral yield. The results showed that the inhibitory effect of lycorine on viral titer gradually diminished when the compound was added at time points up to 10 h p.i.; the compound completely lost its antiviral activity when added after 10 h post-infection. These results agree with the transient replicon results, which indicated that lycorine weakly inhibits viral translation, but strongly suppresses RNA synthesis.
Lycorine does not inhibit WNV protease, NTPase, MTase, or RdRp activities
To identify potential antiviral target(s), we directly tested lycorine in our previously established enzyme assays, using recombinant proteins of WNV, including protease (with NS2B), NTPase, MTase, and RdRp (Ray et al., 2006, Wong et al., 2003). None of the enzyme activities were suppressed by the compound at concentrations up to 100 μM (data not shown). The results suggest that the compound does not directly target the enzyme functions of the viral NS3 or NS5 proteins.
Selection and characterization of lycorine-resistant WNV
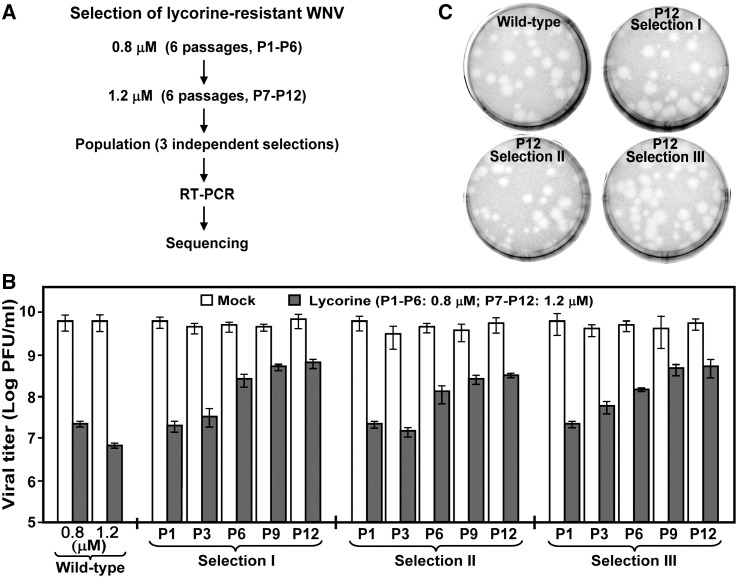
As an alternative means by which to identify antiviral target(s), we selected lycorine-resistant WNV by culturing wild-type (WT) virus in the presence of increasing concentrations of the compound (Fig. 5A). Three independent selections (I-III) were performed. For each selection, a total of 12 passages were carried out, with the first 6 passages (P1-P6) selected at 0.8 μM lycorine and the last 6 passages (P7-P12) selected at 1.2 μM lycorine. Viruses from each passage were assayed for their resistance, through comparison of viral titers from mock-treated infection with viral titers from lycorine-treated infection (harvested at 42 h p.i.). Fig. 5B shows representative data from such resistance assays for P1, P3, P6, P9, and P12. Viral resistance gradually improved from P1 to P10; no further improvement was observed from P10 to P12 (Fig. 5B and data not shown). We therefore terminated the selections at P12. Notably, the P12 viruses did not show complete resistance to lycorine. At 1.2 μM, the compound reduced viral titers of P12 virus-infected cells by approximately 10-fold. In contrast, the compound suppressed viral titers of the WT virus-infected cells by about 1000-fold at the same concentration (Fig. 5B). The results indicated that the resistance of the P12 viruses increased by approximately 100-fold over that of the WT. Similar results were obtained for all three independent selections.
Fig. 5.
Selection and characterization of lycorine-resistant WNV. (A) Scheme for selection of lycorine-resistant WNV. Three independent selections were performed. P1 through P6 were selected at 0.8 μM lycorine; P7 through P12 were selected at 1.2 μM. (B) Resistance profile during selection. Viruses from each of the 12 passages were monitored for their resistance. Vero cells were infected with viruses at an MOI of 0.1 in the presence of 0.8 μM lycorine (P1-P6), 1.2 μM lycorine (P7-P12), or 1% DMSO (as a negative control). At 42 h p.i., viral titers in culture fluids were quantified by plaque assays. Resistance is quantified by comparison of the viral titers from the lycorine-treated infections with the viral titers from the mock-treated infections. Results for representative passages from the three selections are shown. (C) Plaque morphologies of WT and lycorine-resistant viruses. Plaque assays for WT and P12 viruses were performed on Vero cells in the absence of lycorine.
Plaque morphologies of the WT and lycorine-resistant P12 viruses were compared in agar containing no lycorine (Fig. 5C). Plaques derived from the P12 viruses were slightly more heterogeneous in size than the plaques derived from the WT virus, indicating that the P12 viruses were composed of quasispecies. However, the majority of the plaques from the P12 viruses are similar in size to the plaques derived from the WT virus. Collectively, the results demonstrated that WNVs partially resistant to lycorine can be reproducibly selected in cell culture.
Sequencing of lycorine-resistant WNV
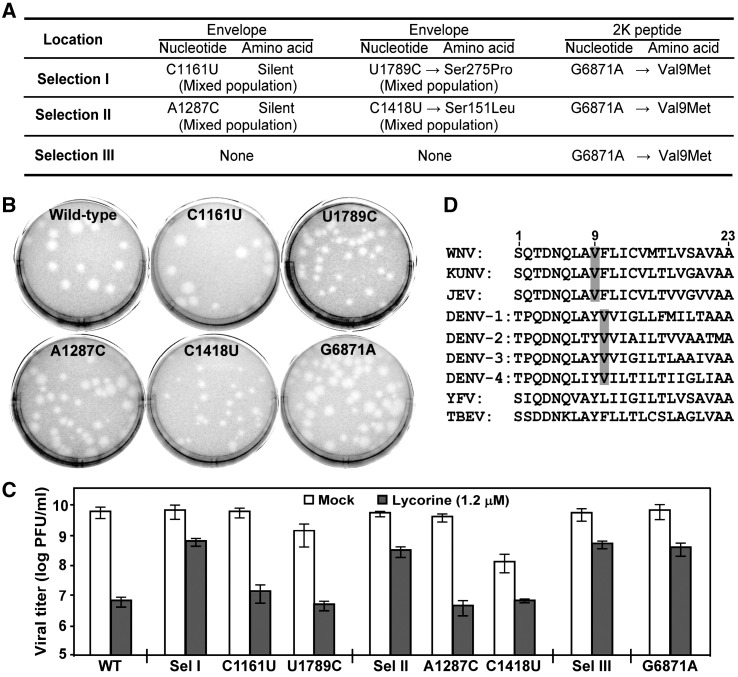
The complete genomes of P12 viruses from the three independent selections were sequenced, to identify potential resistance mutation(s). We performed population sequencing by directly sequencing the RT-PCR products amplified from the RNA extracted from the P12 culture supernatants (Fig. 5A). All selections had accumulated a consensus nucleotide change from G to A at position 6871 (G6871A), resulting in an amino acid substitution from Val to Met at position 9 (Val9Met) in the 2K peptide. No other mutation was detected from Selection III. For Selections I and II, two additional, but distinct, nucleotide mutations were recovered in the E gene: one mutation was silent, while the other one led to an amino-acid change (Fig. 6A). Sequencing chromatograms indicated that the E mutations existed in mixed populations, as represented by both WT and mutant nucleotide peaks at the specific positions (data not shown). As a negative control, viruses passaged in 1% DMSO did not give rise to any mutations.
Fig. 6.
Identification of a single-amino acid change in the 2K peptide as a resistance determinant. (A) Summary of mutations identified from the three selections. Locations of the nucleotide and/or amino acid changes are indicated. Mixed populations (containing both the WT-nucleotide and the indicated mutant-nucleotide) were found in the E gene from selections I and II. (B) Plaque morphologies of WT and recombinant C1161U, U1789C, A1287C, C1418U, and G6871A viruses. Plaques were developed in the absence of lycorine. (C) Resistance analyses of WT virus, three independently selected P12 viruses (from Sel, I, II, and III), and recombinant C1161U, U1789C, A1287C, C1418U, and G6871A viruses. The resistance assays were performed as described in the legend to Fig. 5B. (D) Alignment of amino-acid sequences of flavivirus 2K peptide. The 23-amino acid sequences of 2K peptide are aligned for nine flaviviruses. The conserved Val residues in viruses from JEV- and DENV-serocomplexes are shaded in grey. The 2K peptide sequences of WNV, KUNV, JEV, DENV-1, DENV-2, DENV-3, DENV-4, YFV and TBEV are derived from GenBank accession numbers AF404756, D00246, AF315119, U88535, M29095, M93130, AY947539, X03700, and AF069066, respectively.
Identification of a single-amino acid mutation in the 2K peptide as a resistance determinant
We prepared two panels of recombinant viruses, to identify the lycorine-resistance determinant. The first panel of viruses contained the mutations in the E gene. Each of the four mutations in the E region was individually engineered into an infectious cDNA clone of WNV. Transfection of BHK-21 cells with the genome-length RNAs resulted in four mutant viruses (Fig. 6B): C1161U and U1789C (derived from Selection I), and A1287C and C1418U (derived from Selection II). The four mutant viruses exhibited different plaque morphologies: the two viruses containing the silent mutations (C1161U and A1287C) yielded plaques similar to those of the WT virus, whereas the mutant viruses containing amino acid changes in the E protein (C1418U and U1789C) generated smaller plaques than those of the WT virus. Resistance assays showed that, after treatment of infected cells (0.1 MOI) with lycorine (1.2 μM lycorine for 42 h), none of the four mutants yielded viral titers that were significantly higher than the titers of the WT virus (Fig. 6C). The results indicate that the E mutations are not responsible for resistance. Notably, the smaller plaques (Fig. 6B) and the lower titers from mock-treated infections for mutant viruses U1789C and C1418U (Fig. 6C) suggest that the amino acid changes in the E gene negatively affect viral replication.
The second panel of viruses was prepared to examine the mutation G6817A in the 2K peptide. The plaque morphology of G6871A virus was similar to that of the WT virus (Fig. 6B). Remarkably, the G6871A virus showed a resistance level close to those of the P12 viruses from all three selections (Fig. 6C). Sequencing of the G6871A mutant virus indicated that the engineered mutation was retained without extra changes; furthermore, the G6871A mutation was retained after passaging the mutant virus in Vero cells for five rounds (total 10 days; data not shown). These results demonstrate that the G6871A mutation in the 2K peptide is the major determinant for lycorine resistance. Sequence alignment (Fig. 6D) indicates that the mutated Val at amino-acid position 9 of the 2K peptide (total of 23 amino acids) is conserved among the members of the JEV-serocomplex; and a Val at position 10 is conserved among the members of the DENV-serocomplex; whereas no Val is found at a similar position in the 2K peptides from members of the YFV- and TBEV-serocomplexes.
Replication kinetics of WT and 2K-mutant viruses in the presence and absence of lycorine
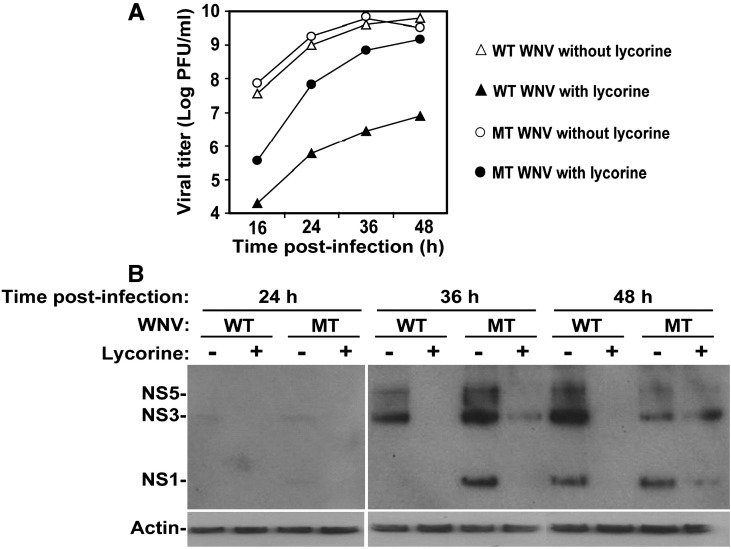
We further characterized the G6871A mutation by comparing the replication kinetics between the WT and mutant (MT) viruses in the presence and absence of lycorine inhibitor. Vero cells were synchronizely infected with the WT and the G6871A MT viruses, and treated with or without 1.2 μM lycorine. As expected, the compound inhibited WT virus more dramatically than it did on the MT virus (Fig. 7A). Western blotting analysis showed that, at 16 h p.i., no viral proteins could be detected in cells infected with either WT or mutant viruses (data not shown). For the MT virus, the expression levels of viral NS1, NS3, and NS5 increased from 24 to 36 h p.i., but decreased at 48 h p.i.; the decrease in protein expression at 48 h p.i. was due to cytopathic effect (i.e., cell lysis). At each time point, lycorine treatment suppressed viral protein expression. These results confirm the critical role of the G6871A mutation in lycorine resistance.
Fig. 7.
Replication kinetics of WT and 2K-mutant viruses in the presence and absence of lycorine. Vero cells, in a 12-well plate, were infected with WNV (WT and G6871A MT) at an MOI of 5, incubated at 4°C for 1 h, washed three times with PBS, and incubated at 37°C with medium containing 1.2-μM lycorine or with medium containing 1% DMSO. At 16, 24, 36, and 48 h p.i., viral titers in supernatants were determined by plaque assays (A). The infected cells were washed twice with PBS, lysed with 250 μl of lysis buffer, and frozen at-80°C. The cell lysates (10 μl) were analyzed by western blotting (B). Four monoclonal antibodies against NS1 (1:1000 dilution; purchased from Sigma), NS3 (1:4000 dilution; in-house generated), NS5 (1:4000 dilution; in-house generated), or β-actin (1:1000 dilution; purchased from Chemicon) were mixed and used as primary antibodies. Horse radish peroxidase (HRP)-labeled anti-mouse IgG (1:4000 dilution) was used as a secondary antibody. β-actin was used as a loading control. One representative of two experiments is shown.
Enhancement of RNA replication by the 2K peptide mutation
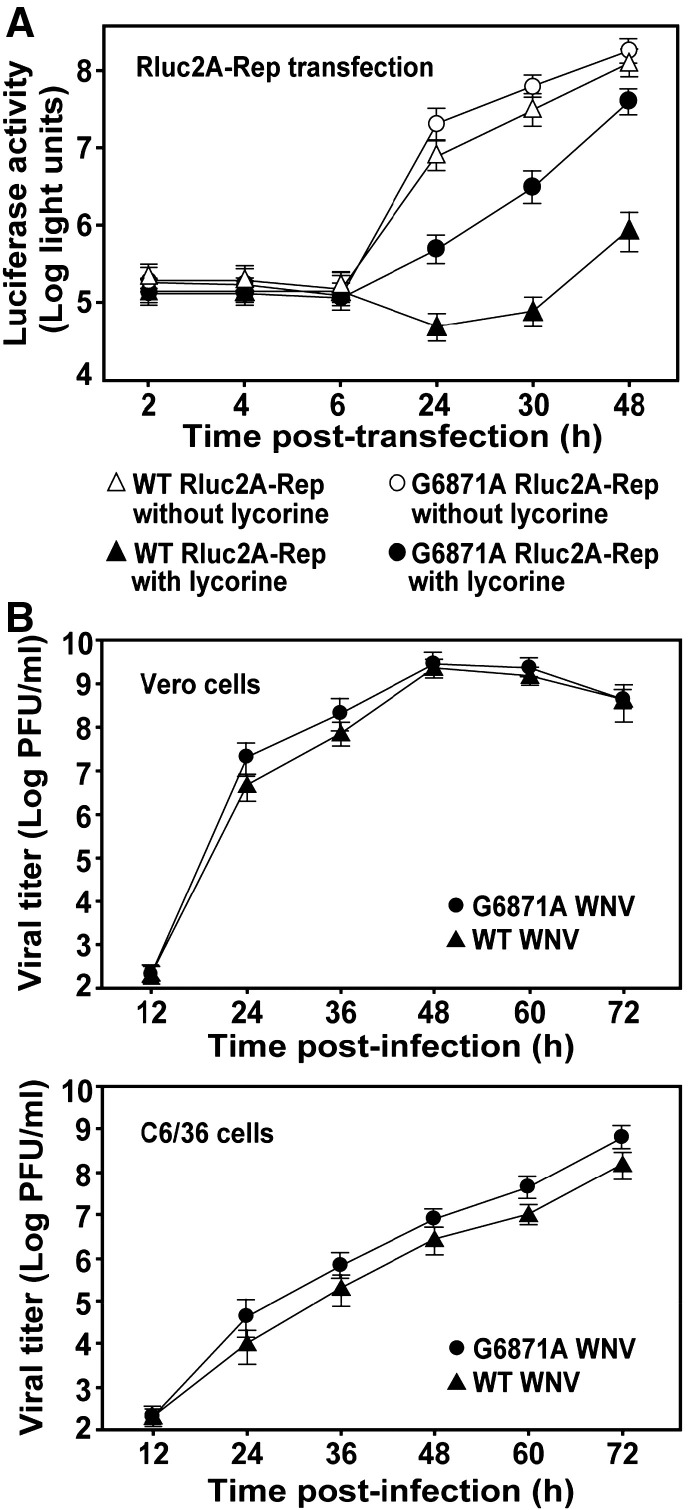
To further validate the contribution of the G6871A change to resistance, we engineered the mutation into a reporting replicon (Rluc2A-Rep) of WNV. In the absence of lycorine, the mutant and WT replicons yielded equal levels of luciferase activities at 2, 4, and 6 h p.t.; in contrast, the mutant replicon exhibited luciferase signals 2.5-, 2-, and 1.4-fold higher than the WT replicon at 24, 30, and 48 h p.t., respectively (Fig. 8A). These results demonstrate that the 2K peptide mutation does not affect viral translation, but it instead enhances RNA synthesis, especially at the early replication stage. In the presence of inhibitor (1.5 μM), the luciferase signals from the WT and mutant replicons were reduced. The reporting signals at 2, 4, and 6 h p.t. were slightly suppressed (< 30%) for both replicons; however, the luciferase activities at 24, 30, and 48 h p.t. from the WT replicon were more dramatically suppressed by the compound than were the activities from the mutant replicon. These results again demonstrate that the single-amino acid change in the 2K peptide is responsible for lycorine resistance.
Fig. 8.
Enhancement of viral replication through mutation of the 2K peptide. (A) Resistance and replication analyses using WNV replicon. The effect of the G6871A mutation in the 2K peptide on resistance to lycorine was quantified using a transient replicon (Rluc2A-Rep) assay. Equal amounts (10 μg) of WT and G6871A mutant replicon RNAs were electroporated into BHK-21 cells. The transfected cells were immediately treated with lycorine (1.5 μM) or without lycorine (1% DMSO as controls). Luciferase activities were measured at the indicated time points post-transfection. Average results from three independent experiments are presented. (B) Growth kinetics of WT and 2K peptide G6871A mutant viruses. Vero and C6/36 cells were infected with the WT and the 2K peptide mutant viruses at an MOI of 0.05. After 1-h incubation, the cells were washed three times with PBS and the medium was replenished. Viral titers in culture fluids were quantified at the indicated time points using plaque assays.
To further examine the effect of the G6871A mutation in the 2K peptide on viral replication, we compared the growth kinetics of the WT and mutant viruses. Vero and mosquito C6/36 cells were each infected with WT or mutant virus (0.05 MOI) and were then monitored for viral yields. In Vero cells, the mutant virus produced titers 4.2- and 2.7-fold higher than those of the WT virus at 24 and 36 h p.i., respectively; the difference in viral titer was reduced to < 1.4-fold after 48 h p.i. (Fig. 8B). In C6/36 cells, the mutant virus generated titers 2.8-to 4.4-fold higher than those of the WT virus at all tested time points. For both Vero and C6/36 cells, similar titers of the WT and mutant viruses were detected at 12 h p.i.; these low viral titers were most likely derived from the input viruses during inoculation, and are not substantially composed of progeny viruses. These results, together with the transient replicon data (Fig. 8A), strongly indicate that the 2K peptide mutation enhances viral replication. It should be noted that the replication enhancement was not evident when the plaque sizes were compared between the WT and mutant viruses (Fig. 6B).
Improvement of antiviral profile of lycorine compounds
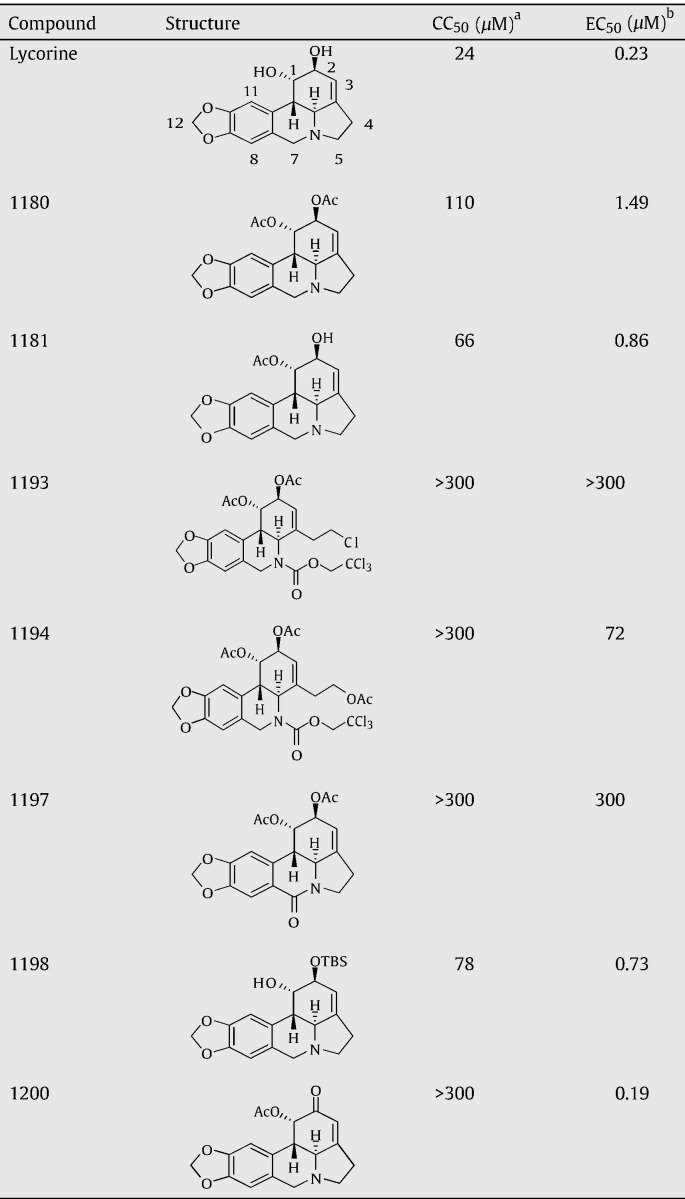
We initiated chemistry studies by synthesizing seven lycorine analogues (Table 1 ). The analogues were each examined for cytotoxicity and antiviral activity. Three regions of the parental lycorine compound were modified. (i) One or two hydroxyl groups at the C1 and C2 positions of lycorine were substituted with other chemical groups. All seven analogues that contained these modifications showed CC50 values higher than the value for parental lycorine. Among them, compound 1200, in which the hydroxyl group at the C1-position is acetylated and the C2 hydroxyl group is oxidized to form a carbonyl group, slightly increased the potency (EC50 = 0.19 μM), while significantly reducing cytotoxicity (CC50 > 300 μM, the highest tested concentration). As expected, the G6871A mutant virus was resistant to compound 1200 (data not shown). (ii) The pyrrolidine ring of lycorine was opened (compounds1193 and 1194). (iii) C7 of lycorine was oxidized to a carbonyl group (compound 1197). Compounds from (ii) and (iii) modifications had lower antiviral potencies. These results clearly indicate that modifications at the two hydroxyl groups of lycorine could be used to improve the antiviral profile.
Table 1.
Antiviral activities of lycorine analogues
aCC50 values were derived from Vero cells using an MTT assay.
bEC50 values were derived from viral titer reduction assays. Vero cells were infected with WNV (0.1 MOI) in the presence of various concentrations of each compound. Viral titers at 42 h p.i. were determined by plaque assays, as described in Materials and methods.
Discussion
A number of approaches have been reported for development of flavivirus antiviral agents (Shi, in press). The goals of this study were to establish the anti-flavivirus activity of lycorine and to analyze lycorine-resistant flavivirus in cell culture. Previous studies showed that lycorine inhibits poliovirus (Ieven et al., 1982), SARS-CoV (Li et al., 2005), herpes simplex virus (Renard-Nozaki et al., 1989), and vaccinia virus (Zhou et al., 2003). The current study has extended the antiviral spectrum to flaviviruses. In addition, we showed that lycorine does not inhibit an alphavirus, WEEV, and a rhabdovirus, VSV, (Fig. 3). The latter results, together with findings of an early study showing that lycorine does not inhibit HIV-1 at a noncytotoxic concentration (Szlavik et al., 2004), indicate the selectivity of the antiviral spectrum of the compound. As a proof-of-principle, we showed that modifications of the lycorine compound, especially at the two hydroxyl groups, could dramatically reduce its cytotoxicity, while improve its potency (Table 1). The results suggest that lycorine represents one class of inhibitor that could potentially be developed for flavivirus therapeutics.
Three WNV luciferase-reporting replicon-based assays were used to dissect the inhibitory step(s) of lycorine. First, a VLP-infection-based assay allowed identification of inhibitors of viral entry and replication (Fig. 1C). Second, use of Replicon-containing cell lines allowed screening for inhibitors of viral replication (Fig. 4A). Third, a transient replicon assay allowed differentiation between inhibitors of translation and inhibitors of RNA synthesis (Fig. 4B). Analysis of the compound in the above systems indicated that whereas lycorine only weakly reduces viral translation (< 30%, as indicated by luciferase activity), it significantly suppresses RNA synthesis (> 99%, as indicated by luciferase activity). The reduction of RNA synthesis could be caused by the compound-mediated suppression of viral translation. Alternatively, lycorine could suppress both translation and RNA synthesis, leading to the dramatic reduction of RNA synthesis. To differentiate between the above two possibilities, we performed a time-of-addition experiment. The results showed that lycorine gradually declined its anti-WNV activity, when the time of addition was varied from 0 to 10 h p.i.; the compound completely lost the inhibitory activity when added at > 10 p.i. (Fig. 4C). A single round of flavivirus infection is about 12 h in duration, with viral translation peaking around 2–6 h p.i. and RNA synthesis peaking around 7–12 h p.i. (Chambers et al., 1990, Puig-Basagoiti et al., 2005). If lycorine only inhibited the step of translation, it would have completely lost its antiviral activity when added at a time point earlier than 10 h p.i. (i.e., at 6 h p.i.). Therefore, the time-of-addition results clearly indicate that the compound inhibits a step beyond viral translation. These data, together with the identification of the resistance determinant in the 2K peptide (Fig. 6), led us to conclude that lycorine suppresses WNV mainly through suppression of viral RNA synthesis.
Our data are in contrast to poliovirus results, which indicated that ∼ 90% of viral translation was suppressed by 10 μg/ml (equivalent to 31 μM) lycorine (Ieven et al., 1982). Since the cytotoxicity of lycorine at 31 μM was not shown in the poliovirus study, experiments are needed to exclude the possibility that the strong inhibition of viral translation was due to compound-mediated cytotoxicity. We performed the mode-of-action analysis at < 1.5 μM of lycorine, a concentration that allowed > 90% cell viability. In our system, lycorine at 31 μM decreased the viability of Vero cells by ∼ 50% (Fig. 2A).
Two approaches were taken to identify potential the viral target(s) of lycorine. The first approach was to test the compound in biochemical enzyme assays using recombinant protease (with NS2B), NTPase, RdRp, and MTase proteins. None of the enzyme activities were suppressed by lycorine. The second approach was to select compound-resistant WNV. We selected resistant WNVs in cell culture. Engineering of the mutations (recovered from the resistant viruses) into an infectious clone (Fig. 6) and a replicon (Fig. 8A) of WNV allowed us to map the resistance determinant to a single amino-acid change (Val9Met) in the 2K peptide. It should be noted that the resistance results do not necessarily indicate that the 2K peptide is the direct target for lycorine. The 2K peptide mutation could exert its resistance phenotype through enhancement of viral replication so that, in the presence of inhibitor, viral replication could still be sustained at a level sufficient to generate virus. The latter scenario is supported by the observations that the 2K peptide mutation enhanced RNA synthesis of WNV replicon as well as the growth kinetics of WNV in Vero and C6/36 cells (Fig. 8). However, it should be pointed out that the 2K mutation-mediated enhancement of viral replication only partially contributes to the lycorine resistance. The latter conclusion was supported by the results that the 2K mutation increased replication by only 2 to 4-fold in the absence of lycorine (Fig. 8B), whereas the same mutation increased replication by > 100-fold in the presence of the inhibitor (Fig. 7A).
How does the 2K peptide mutation enhance WNV RNA replication? Flavivirus 2K peptide spans the ER membrane with its N- and C-terminal tails on the cytoplasmic and ER lumen sides, respectively (Miller et al., 2007). The cleavage at the 2K-NS4B junction by host signalase requires a prior cleavage at the NS4A-2K junction by viral NS2B/NS3 protease (Lin et al., 1993). The regulated cleavages at the NS4A-2K-NS4B sites play a role in rearranging cytoplasmic membranes (Miller et al., 2007, Roosendaal et al., 2006). The exact function of the 2K peptide in induction of membrane arrangement may differ among flaviviruses. In KUNV, individual expression of NS4A-2K resulted in membrane rearrangements that most resembling virus-induced structures, while removal of the 2K domain led to a less profound membrane rearrangement (Roosendaal et al., 2006). In contrast, expression of DENV-2 NS4A without 2K peptide resulted in altered membranes resembling virus-induced structures, whereas expression of NS4A-2K did not induce comparable membrane arrangement (Miller et al., 2007). Since the rearranged membranes form the scaffold for the viral replication complex, the Val9Met mutation may affect the 2K peptide-mediated membrane alterations, leading to an increase in RNA replication.
Alternatively, since the cleavages of NS4A-2K-4B intermediate are critical for the NS4A- and NS4B-mediated inhibition of interferon signaling (Munoz-Jordan et al., 2005), the Val9Met mutation in the 2K peptide may affect the processing of the NS4A-2K-4B polyprotein, resulting in a more efficient defense against the host immune response. The weakened interferon response allowed the virus to replicate to a higher level. This hypothesis is supported by our observations that the same 2K peptide mutation was recovered from the WNV that was able to perform superinfections (Zhang and Shi, unpublished data). In the later study, we found that the 2K peptide mutant virus showed improved capability to replicate in cells that contain persistently replicating replicon of WNV. One caveat to this hypothesis is that our lycorine resistance study was performed in Vero cells, in which the interferon gene has been deleted (Mosca and Pitha, 1986). More experiments are required to dissect the molecular mechanism of the 2K peptide that is operative in flavivirus replication or in evasion of the host immune response.
Materials and methods
Cells and viruses
Baby hamster kidney cells (BHK-21) and African green monkey kidney cells (Vero) were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum in 5% CO2 at 37°C. Aedes albopictus C6/36 cells were grown in Eagle's minimal essential medium (EMEM) with 10% FBS and 1% non-essential amino acid at 28°C. A reporting Vero cell line containing a persistently replicating WNV or DENV-1 replicon (Rluc-Neo-Rep; Fig. 4A) was cultured in DMEM with 10% FBS and 1 mg/ml of G418. WNV was derived from a full-length infectious cDNA clone of an epidemic strain 3356 (Shi et al., 2002a, Shi et al., 2002b). YFV (17D vaccine strain), DENV-2 (New Guinea C strain), WEEV (strain Cova 746), and VSV (New Jersey serotype) were used for antiviral assays, as described previously (Puig-Basagoiti et al., 2006).
Lycorine and analogues
Lycorine was purchased from BIOMOL Research Laboratories Inc. A panel of seven lycorine analogues (Table 1) was synthesized based on previous methodology: compounds 1180 (Lee et al., 2007), 1181 (Nakagawa, Uyeo, and Yajima, 1956), 1193 (Lee et al., 2007), 1194 (Lee et al., 2007), 1197 (Shu et al., 2002), and 1200 (Nakagawa et al., 1956). The method for synthesis of compound 1198 will be described elsewhere. The structure and purity (> 95%) of each compound were established by nuclear magnetic resonance. All compounds were dissolved in dimethyl sulfoxide (DMSO), and were tested at 1% DMSO final concentration. Mock-treated reactions were incubated with 1% DMSO without lycorine or any of its analogues.
VLP infection assay
VLPs of WNV and DENV-1 were prepared by trans-supply of viral structural proteins to replicon RNAs. Each replicon contains a Renilla luciferase (Rluc) and foot-and-mouth disease virus 2A sequence in the position where the viral structural genes (CprME) were deleted (Rluc2A-Rep; Fig. 1B). The construction and characterization of the WNV and DENV-1 Rluc2A-Rep were reported previously (Lo et al., 2003, Puig-Basagoiti et al., 2006). The structural genes of each virus were cloned into an SFV expression vector (Invitrogen) (Liljestrom and Garoff, 1991) at a unique Bam HI site, resulting in SFV-CprME (Fig. 1B). A Kozak sequence and a stop codon were engineered at the 5′ and 3′ ends of the CprME fragment, respectively. A double-transfection protocol was used to generate the VLPs (Khromykh et al., 1998, Puig-Basagoiti et al., 2005). Briefly, BHK-21 cells (8 × 106) were first electroporated with 10 μg of Rluc2A-Rep RNA in a 0.4-cm cuvette with the GenePulser apparatus (Bio-Rad) using settings of 0.85 kV and 25 μF, pulsing three times at 3-sec intervals. The transfected cells were resuspended in DMEM with 10% FBS and incubated at 37°C with 5% CO2 for 24 h. The replicon-transfected cells were then electroporated with 10 μg of SFV-CprME RNA of the corresponding virus, at the settings identical to those used for the first transfection. At 24 h after the second transfection, culture supernatants were collected and centrifuged to remove cellular debris. The VLPs in the supernatants were aliquoted and stored at-80°C. The titers of VLPs (in focus-forming units [FFU]/ml) were estimated by infection of Vero cells in a four-chamber Lab-Tek Chamber Slide (Nalge Nunc International) with serial dilutions of the culture fluid, followed by counting of immunofluorescence assay (IFA)-positive cell foci at 18 and 24 h p.i. for WNV and DENV-1, respectively. For IFA, immune mouse ascites fluid of WNV or DENV-1 (American Type Culture Collection) and goat anti-mouse immunoglobulin G conjugated with Texas Red were used as primary and secondary antibodies, respectively.
For VLP-based antiviral assays, a monolayer of naive Vero cells (2 × 104 cells per well in 96-well plate) was infected at 1 FFU/cell for WNV VLP, and at 0.01 FFU/cell for DENV-1 VLP. Lycorine was immediately added to the VLP-infected cells. At 24 h p.i., the cells were washed twice with cold PBS, lysed in 20 μl of 1× Renilla luciferase lysis buffer for 20 min, and assayed for luciferase activities using a Renilla luciferase assay kit (Promega). The luciferase signals were measured with a Veritas Microplate Luminometer (Promega). Average results of three or more independent experiments are presented here.
Reporting replicon cell line assay
Vero cells containing persistently replicating replicons of WNV (Lo et al., 2003) or DENV-1 (Puig-Basagoiti et al., 2006) were constructed previously. Each replicon contained two reporter genes: a Renilla luciferase (Rluc) and a neomycin phosphotransferase gene (Neo), resulting in Rluc-Neo-Rep (Fig. 4A). For antiviral assays, Rluc-Neo-Rep-containing Vero cells (2 × 104 in 100 μl) were seeded per well of 96-well plates in DMEM with 10% FBS without G418. Lycorine was added to the medium at 24 h post-seeding. After 24 or 48 h of lycorine-treatment, the cells were lysed and assayed for luciferase activities as described above.
Transient replicon assay
A transient replicon assay was used to quantify compound-mediated inhibition of viral translation and suppression of RNA synthesis (Deas et al., 2005). Briefly, 10 μg of Rluc2A-Rep RNA of WNV or DENV-1 (Fig. 4B) was electroporated into BHK-21 cells (8 × 106) as described above. The transfected cells were suspended in 25 ml of DMEM with 10% FBS. Cell suspension (1 ml) was seeded into 12-well plates, immediately treated with lycorine, and assayed for luciferase activities at 2, 4, and 6 h p.t. (representing viral translation), and at 24, 30, and 48 h p.t. (representing RNA synthesis). For quantification of compound-mediated inhibition, relative luciferase activities were presented, with the luciferase activity derived from the mock-treated cells set as 100%.
Viral titer reduction assay and cytotoxicity assay
Viral titer reduction assays were performed to examine the antiviral activities of lycorine and its analogues in WNV, DENV-2, YFV, WEEV and VSV. Approximately 6 × 105 Vero cells per well were seeded in a 12-well plate. After incubation for 24 h, the cells were infected with individual virus (0.1 MOI) and treated immediately with one of the compounds at the indicated concentration. For WNV, DENV-2, YFV, and WEEV, samples of culture medium were collected at 42 h post-infection. For VSV, culture medium was collected at 16 h post-infection. All collected samples were stored at − 80°C, and viral titers were determined by plaque assays on Vero cells. A double-overlayer protocol was followed for plaque assays (Puig-Basagoiti et al., 2006). All assays were performed in triplicate. Cytotoxicity of each compound was examined using a cell proliferation-based MTT assay (American Type Culture Collection) as described previously (Puig-Basagoiti et al., 2006).
Generation and sequencing of WNV resistant to lycorine
Three independent lineages of lycorine-resistant WNV were generated by passaging of the WT WNV (derived from an infectious cDNA clone) on Vero cells, with increasing concentrations of lycorine. For the first six passages, Vero cells in 12-well plates were infected with WNV (derived each time from the previous passage) at an MOI of 0.1 in the presence of 0.8 μM lycorine or 1% DMSO (as a negative control). Passages 7 to 12 were selected with 1.2 μM lycorine. For each passage, viral supernatants were harvested at 42 h p.i.; viral titers were quantified by plaque assays; viruses were analyzed in resistance assays to monitor the improvement of resistance, through comparison of the viral titers from the lycorine-treated infections with the viral titers from the mock-treated infections. The selections were terminated at passage 12, when no further improvement of the resistance was observed (see details in Results).
Lycorine-resistant WNVs from passage 12 were subjected to genome-length sequencing for identification of accumulated mutation(s). Virion RNAs were extracted from culture supernatants using RNeasy kits (QIAGEN). Viral RNAs were amplified by RT-PCR using SuperScript III one-step RT-PCR kits (Invitrogen). The PCR products were gel-purified and subjected to DNA sequencing. Sequences of the 5′- and 3′-terminal nucleotides of the viral genomes were determined by rapid amplification of cDNA ends (RACE). The 5′RACE was performed using the FirstChoice RLM-RACE kit (Ambion). The 3′RACE was performed as previously described (Tilgner and Shi, 2004).
Construction of cDNA plasmids of WNV containing various mutations
WNV genome-length cDNA clones with specific mutations were constructed by using a modified pFLWNV (Shi et al., 2002a, Shi et al., 2002b) and two shuttle vectors. Shuttle vector A was constructed by engineering the Bam HI-Sph I fragment from the pFLWNV (representing the upstream end of the T7 promoter [for RNA transcription of genome-length RNA] to nucleotide position 3627 of the WNV genome; GenBank no. AF404756) into the pACYC177 vector containing a modified cloning cassette (Zhou et al., 2007). A QuikChange II XL site-directed mutagenesis Kit (Stratagene) was used to engineer the mutations in the E gene into the shuttle vector A. The mutated DNA fragment was cut-and-pasted back into the pFLWNV clone at the Bam HI and Stu I sites (nucleotide position 2591). Shuttle vector B was constructed by engineering Kpn I-Xba I fragment (representing nucleotide 5341 through the 3′ end of the genome) into a pcDNA3.1(+) vector. The G6871A mutation was engineered into the shuttle vector B using the QuikChange II XL site-directed mutagenesis Kit. The mutated DNA fragment was pasted back into the pFLWNV clone and into the cDNA clone of WNV Rluc2A replicon at the Bsi WI and Spe I sites (nucleotide positions 5780 to 8022). All constructs were verified by DNA sequencing.
RNA transcription and transfection
Both genome-length RNA and replicon RNA were in vitro transcribed from corresponding cDNA plasmids that were linearized with XbaI. A T7 mMessage mMachine kit (Ambion) was used for RNA synthesis described before (Shi et al., 2002a). Both replicon and genome-length RNAs were electroporated into BHK-21 cells as described above. For transfection of genome-length RNA, culture fluids were collected every 24 h until apparent cytopathic effect was observed (day 4 to day 5 post-transfection). The viruses in the supernatants were aliquoted and stored at − 80°C.
Acknowledgments
We thank Wadsworth Center's Molecular Genetics Core for the DNA sequencing and Cell Culture Facility for the maintenance of BHK-21 and Vero cells. This work was supported partially by federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health, under contract NOI-AI-25490, and by NIH grants U01 AI061193 and U54-AI057158 (Northeast Biodefense Center).
References
- Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Best S.M., Morris K.L., Shannon J.G., Robertson S.J., Mitzel D.N., Park G.S., Boer E., Wolfinbarger J.B., Bloom M.E. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 2005;79(20):12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T.J., Hahn C.S., Galler R., Rice C.M. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay U., Chaudhuri L., Das S., Kumar Y., Ghosal S. Activation of lymphocytes by lycorine-1-O-beta-d-glucoside. Pharmazie. 1984;39(12):855–856. [PubMed] [Google Scholar]
- De Leo P., Dalessandro G., De Santis A., Arrigoni O. Metabolic responses to lycorine in plants. Plant Cell Physiol. 1973;14:487–496. [Google Scholar]
- Deas T.S., Binduga-Gajewska I., Tilgner M., Ren P., Stein D.A., Moulton H.M., Iversen P.L., Kauffman E.B., Kramer L.D., Shi P.Y. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 2005:4599–4609. doi: 10.1128/JVI.79.8.4599-4609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Miller R.H., Lai C.J. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 1993;67(4):2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler D., Kuno G., Markoff L. Flaviviruses. In: Knipe D.M., Howley P.M., editors. 5th. vol. 1. Lippincott William and Wilkins; Philadelphia, PA: 2007. pp. 1153–1253. (Fields virology). [Google Scholar]
- Guo J., Hayashi J., Seeger C. West nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 2005;79(3):1343–1350. doi: 10.1128/JVI.79.3.1343-1350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt K.J., Westaway E.G., Khromykh A.A. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods. 2001;92(1):37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- Ieven M., Vlietinck A.J., Vanden Berghe D.A., Totte J., Dommisse R., Esmans E., Alderweireldt F. Plant antiviral agents. III. Isolation of alkaloids from Clivia miniata Regel (Amaryllidaceae) J. Nat. Prod. 1982;45(5):564–573. doi: 10.1021/np50023a009. [DOI] [PubMed] [Google Scholar]
- Jones C., Patkar C., Kuhn R. Construction and applications of yellow fever virus replicons. Virology. 2005;331(2):247–259. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- Khromykh A.A., Varnavski A.N., Westaway E.G. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 1998;72:5967–5977. doi: 10.1128/jvi.72.7.5967-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer L., Li J., Shi P. West Nile virus. Lancet Neurol. 2007;6:171–182. doi: 10.1016/S1474-4422(07)70030-3. [DOI] [PubMed] [Google Scholar]
- Kummerer B.M., Rice C.M. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 2002;76(10):4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.S., Venkatesham U., Rao C.P., Lam S.H., Lin J.H. Preparation of secolycorines against acetylcholinesterase. Bioorg. Med. Chem. 2007;15:1034–1043. doi: 10.1016/j.bmc.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Leung D., Schroder K., White H., Fang N.X., Stoermer M., Abbenante G., Martin J., P.R., Y., Fairlie D. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Li H., Clum S., You S., Ebner K.E., Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 1999;73(4):3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., Zhang X., Hua S.N., Yu J., Xiao P.G., Li R.S., Tan X. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljestrom P., Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/technology. 1991;9(12):1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- Lin C., Amberg S.M., Chambers T.J., Rice C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 1993;67(4):2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Thiel H.J., Rice C.M. Flaviviridae: the virus and their replication. In: Knipe D.M., Howley P.M., editors. Fourth ed. Lippincott William and Wilkins; 2007. (Fields Virology). [Google Scholar]
- Liu W.J., Chen H.B., Khromykh A.A. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 2003;77(14):7804–7813. doi: 10.1128/JVI.77.14.7804-7813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Hu W.X., He L.F., Ye M., Li Y. Effects of lycorine on HL-60 cells via arresting cell cycle and inducing apoptosis. FEBS Lett. 2004;578(3):245–250. doi: 10.1016/j.febslet.2004.10.095. [DOI] [PubMed] [Google Scholar]
- Liu W., Wang X., Mokhonov V., Shi P., Randall R., Khromykh A. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 2005;79(3):1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Y., Tang L.J., Zhang G.P., Hu W.X. Treatment of lycorine on SCID mice model with human APL cells. Biomed. Pharmacother. 2007;61(4):229–234. doi: 10.1016/j.biopha.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L., Tilgner M., Bernard K., Shi P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus using a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77(18):10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L., Tilgner M., Shi P.Y. A potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 2003;77(23):12901–12906. doi: 10.1128/JVI.77.23.12901-12906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Kastner S., Krijnse-Locker J., Buhler S., Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007;282(12):8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- Mosca J.D., Pitha P.M. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 1986;6(6):2279–2283. doi: 10.1128/mcb.6.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan J.L., Sanchez-Burgos G.G., Laurent-Rolle M., Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 2003;100(24):14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan J.L., Laurent-Rolle M., Ashour J., Martinez-Sobrido L., Ashok M., Lipkin W.I., Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005;79(13):8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, Y., Uyeo, S., and Yajima, H., (1956). The double bond in lycorine. Chemistry and Industry (London, United Kingdom), 1238–9.
- Puig-Basagoiti F., Deas T.S., Ren P., Tilgner M., Ferguson D.M., Shi P.Y. High-throughput assays using luciferase-expressing replicon, virus-like particle, and full-length virus for West Nile virus drug discovery. Antimicrob. Agents Chemother. 2005;49(12):4980–4988. doi: 10.1128/AAC.49.12.4980-4988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Basagoiti F., Tilgner M., Forshey B., Philpott S., Espina N., Wentworth, Goebel S., Masters P.S., Falgout B., Ren P., Ferguson, Shi P.Y. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 2006;50(4):1320–1329. doi: 10.1128/AAC.50.4.1320-1329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T., Zhou Y., Li H., Shi P. West nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80(17):8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard-Nozaki J., Kim T., Imakura Y., Kihara M., Kobayashi S. Effect of alkaloids isolated from Amaryllidaceae on herpes simplex virus. Res. Virol. 1989;140(2):115–128. doi: 10.1016/s0923-2516(89)80089-5. [DOI] [PubMed] [Google Scholar]
- Roosendaal J., Westaway E.G., Khromykh A., Mackenzie J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 2006;80(9):4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, P.Y., (in press). Novel therapeutics against West Nile virus. West Nile encephalitis virus infection: viral pathogenesis and the host immune response in M. S. Diamond (ed), Springer Publisher.
- Shi P.Y., Tilgner M., Lo M.K. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology. 2002;296:219–233. doi: 10.1006/viro.2002.1453. [DOI] [PubMed] [Google Scholar]
- Shi P.Y., Tilgner M., Lo M.K., Kent K.A., Bernard K.A. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 2002;76(12):5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P.Y., Chen L.K., Chang S.F., Yueh Y.Y., Chow L., Chien L.J., Chin C., Yang H.H., Lin T.H., Huang J.H. Potential application of nonstructural protein NS1 serotype-specific immunoglobulin G enzyme-linked immunosorbent assay in the seroepidemiologic study of dengue virus infection: correlation of results with those of the plaque reduction neutralization test. J. Clin. Microbiol. 2002;40(5):1840–1844. doi: 10.1128/JCM.40.5.1840-1844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlavik L., Gyuris A., Minarovits J., Forgo P., Molnar J., Hohmann J. Alkaloids from Leucojum vernum and antiretroviral activity of Amaryllidaceae alkaloids. Planta Med. 2004;70(9):871–873. doi: 10.1055/s-2004-827239. [DOI] [PubMed] [Google Scholar]
- Tan B.H., Fu J., Sugrue R.J., Yap E.H., Chan Y.C., Tan Y.H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216(2):317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- Tilgner M., Shi P.Y. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 2004;78(15):8159–8171. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrener P., Tamura J.K., Collett M.S. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 1993;67(2):989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G., Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184(2):707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- Wong S.J., Boyle R.H., Demarest V.L., Woodmansee A.N., Kramer L.D., Li H., Drebot M., Koski R.A., Fikrig E., Martin D.A., Shi P.Y. An immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections, and form flavivirus vaccination. J. Clin. Microbiol. 2003;41(9):4217–4223. doi: 10.1128/JCM.41.9.4217-4223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Deng L., Kashanchi F., Brady J.N., Shatkin A.J., Kumar A. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. U. S. A. 2003;100(22):12666–12671. doi: 10.1073/pnas.1835726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K., Shi P., Li H. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]