Significance
Anthropogenic effects on the environment are ubiquitous and have enormous impacts on individual and ecosystem health. It is widely accepted that environmental change affects disease distribution, but how it may affect parasite-driven evolution remains elusive. Our results provide experimental evidence that parasites play a major role in ecosystem dynamics and, as a result, can affect selection in subsequent host generations. This role is further modified by the prevailing environmental conditions that affect disease dynamics in two ways: through altered ecological opportunities for disease and through altered evolutionary effects on the host.
Keywords: eco-evolutionary dynamics, three-spined stickleback, host–parasite interaction, Gyrodactylus, eutrophication
Abstract
Host resistance to parasites is a rapidly evolving trait that can influence how hosts modify ecosystems. Eco-evolutionary feedbacks may develop if the ecosystem effects of host resistance influence selection on subsequent host generations. In a mesocosm experiment, using a recently diverged (<100 generations) pair of lake and stream three-spined sticklebacks, we tested how experimental exposure to a common fish parasite (Gyrodactylus spp.) affects interactions between hosts and their ecosystems in two environmental conditions (low and high nutrients). In both environments, we found that stream sticklebacks were more resistant to Gyrodactylus and had different gene expression profiles than lake sticklebacks. This differential infection led to contrasting effects of sticklebacks on a broad range of ecosystem properties, including zooplankton community structure and nutrient cycling. These ecosystem modifications affected the survival, body condition, and gene expression profiles of a subsequent fish generation. In particular, lake juvenile fish suffered increased mortality in ecosystems previously modified by lake adults, whereas stream fish showed decreased body condition in stream fish-modified ecosystems. Parasites reinforced selection against lake juveniles in lake fish-modified ecosystems, but only under oligotrophic conditions. Overall, our results highlight the overlapping timescales and the interplay of host–parasite and host–ecosystem interactions. We provide experimental evidence that parasites influence host-mediated effects on ecosystems and, thereby, change the likelihood and strength of eco-evolutionary feedbacks.
Integrating ecosystem changes with rapid species adaptation is at the heart of modern evolutionary theory and an emerging eco-evolutionary synthesis (1–3). This integration crucially depends on understanding how phenotypic evolution can affect community structure and ecosystem functions (4). When the phenotypic effects of organisms on ecosystems are sufficiently large and persistent, an eco-evolutionary feedback may emerge if the organism-mediated environmental modifications become an important agent of selection that affects evolution of subsequent generations (1). Whereas this perspective has recently received much attention (e.g., refs. 5–8), little is known about how interactions between organismal traits and biotic as well as abiotic drivers of ecosystem change govern the occurrence and strength of these feedbacks (9).
Parasites play key roles in ecosystems (10, 11) and evolutionary dynamics (12) because they are ubiquitous and can have strong effects on host fitness. Host–parasite interactions can evolve rapidly (12–15) and depend strongly on prevailing environmental conditions (16–18). As a result, host–parasite and host–ecosystem interactions may evolve in tandem, functionally linking evolutionary and ecological processes (19–21). For instance, variation in the composition of prey communities can be strongly modified by hosts, but it can also influence the exposure of hosts to trophically transmitted parasites (22). Feedbacks between host evolution and ecosystem dynamics may emerge when resistance evolves rapidly and influences the effects of hosts on ecosystems. Current eco-evolutionary theory recognizes that the presence and strength of feedbacks depend on a balance between the effects of both organisms and external environmental drivers on ecosystems (18). In freshwater ecosystems, nutrient loading by humans not only alters patterns of nutrient cycling (23, 24), but can also threaten population persistence (25) and disrupt ongoing species divergence by changing selection regimes (26). Furthermore, nutrient loading can increase parasite prevalence and change evolutionary trajectories of host–parasite interactions (27–29). Although the ecological and evolutionary effects of nutrient loading are well studied, little is known about how it affects feedbacks among hosts, parasites, and ecosystems.
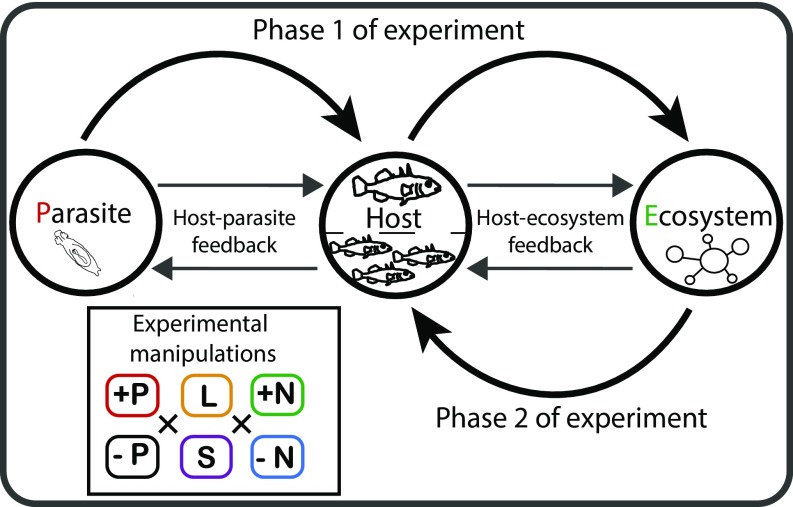
To test for the combined effects of nutrient inputs and parasites on host–ecosystem feedbacks, we performed a two-phase mesocosm experiment where we manipulated the presence of parasites, the host ecotype, and the level of nutrient loading (Fig. 1). In phase 1, we tested whether wild-caught lake and stream sticklebacks differed in parasite resistance, gene expression profiles, metabolic condition, diet, and ecosystem effects. Because we used wild-caught fish, we did not distinguish between ecosystem modifications originating from either genetic effects or plasticity (6, 30). In phase 2, we removed the adult fish, and tested whether the ecosystem modifications by adult fish in phase 1 altered selection pressures (measured as differences in relative survival) on the next host generation. This next generation consisted of a juvenile population with equal proportions of lake, stream, and hybrid juveniles (Fig. 1). Because these juveniles were reared in common-garden conditions, we could test for the effects of adult-mediated ecosystem modifications, while controlling for rearing history and prior exposure to parasites.
Fig. 1.
Conceptual background and experimental design. During the first experimental phase, we investigated how host–parasite interactions affect surrounding ecosystems with different nutrient loadings. We characterized interactive effects of three experimental contrasts: parasite presence vs. absence (P: +P/-P), lake vs. stream host ecotype (H: L/S) and high vs. low ecosystem nutrients (E: +N/-N) throughout different biological levels. In phase 2, we tested for host–ecosystem feedbacks focusing on the next host generation and assessed selection against different host genetic backgrounds and gene expression of survivors.
Forty outdoor aquatic mesocosm ecosystems were set up with a mixture of sediments and invertebrates from multiple lakes and streams in Switzerland. We added nutrients only once before the start of the experiment to manipulate the productivity of these ecosystems [environmental contrast (E), high vs. low nutrients]. We used recently diverged (<100 generations) ecotypes of lake and stream three-spined sticklebacks because these ecotypes [host contrast (H), lake vs. stream] are genetically differentiated (31, 32) and have different effects on mesocosm ecosystems (6). For phase 1 of the experiment, we manipulated parasite exposure of adults by disinfecting wild-caught fish and just before their introduction to the mesocosms, reinfecting half of the hosts with exactly four individuals of Gyrodactylus spp., a monogenean ectoparasite [parasite contrast (P), exposed vs. nonexposed]. Each parasite-exposed fish received two individual parasites each from lake and stream origin to control for potential local (co)adaptation (33, 34). Gyrodactylus reproduces on the fish, is transmitted directly between fish hosts, and can affect host condition and fitness (35). Each of the eight factorial combinations of parasite exposure, host ecotype, and nutrient level was replicated five times.
After 7 wk, we removed the adult fish and began phase 2 by adding juveniles to the same mesocosms that had been modified by the adults. These juvenile fish were bred by in vitro fertilization using wild-caught parents and were reared on a common food source in the laboratory. Because these common-garden juveniles were not the offspring of the adults used during phase 1, we avoided possible confounding transgenerational priming effects of parasite resistance (36). Measuring variation in survival, body condition, and gene expression of these juveniles allowed us to test for an eco-evolutionary feedback by evaluating whether ecosystem modifications during phase 1 altered selection pressures during phase 2.
To confirm that the effects of ecotype and parasite exposure on gene expression were not solely due to plasticity (particularly in phase 1), we performed an additional common-garden experiment in the following year by using laboratory-reared adult lake and stream fish from the same cohort as the second generation of the main experiment. To this end, we set up 12 identical outdoor tanks without sediment or zooplankton and exposed 17 laboratory-raised adult sticklebacks, in six groups of 2–3 individuals, to Gyrodactylus whereas another 17 served as control, unexposed fish (Materials and Methods and SI Appendix, Figs. S1 and S2 and Table S4).
Results and Discussion
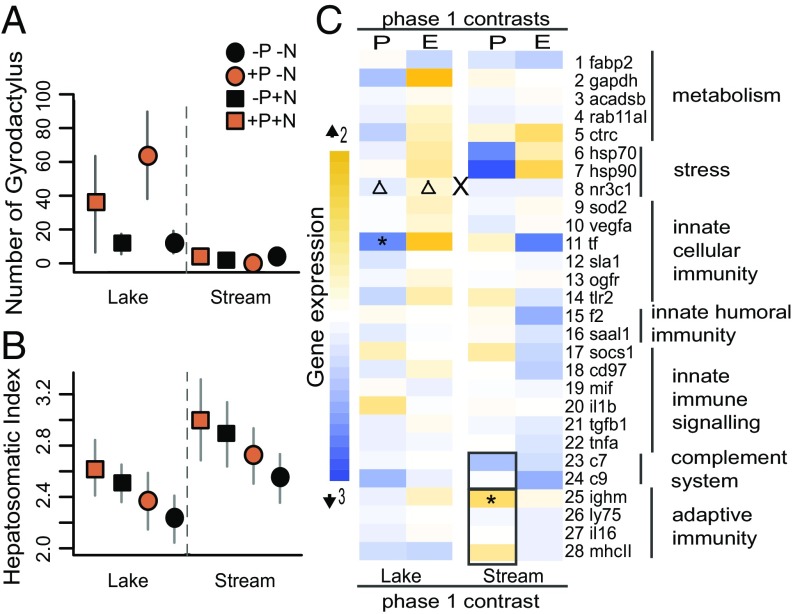
At the end of phase 1 (7 wk duration), stream fish carried fewer individual parasites than lake fish (infection intensities, defined as ΣGyrodactylus/Σexposed fish (infected + noninfected): i.i.L+P = 49.8 ± 19.1, i.i.S+P = 2.67 ± 0.85, Fig. 2A, PxH effect, SI Appendix, Table S1, infection prevalence: prevL+P = 63.0%, prevS+P = 49.5%). We observed similar infection intensity and prevalence patterns in the wild, lake fish being infected with higher numbers of Gyrodactylus than stream fish [i.i.Lwild = 30.4 ± 5.23, i.i.Swild = 4.68 ± 1.75, n = 40, H effect: χ2 = 30.22, P < 0.001, generalized linear mixed effects model (GLMM)], and showing comparable infection prevalence (prevLakewild = 57.1%, prevStreamwild = 63.2%). Although parasites were also present at low levels in control mesocosms, experimentally exposed fish showed significantly higher infection intensities (i.i.+P = 26.5 ± 9.88, i.i.-P = 7.2 ± 1.9; Fig. 2A). Gyrodactylus numbers were highest on lake fish in ecosystems with low nutrient loading (i.i.Lake+P+N = 35.4 ± 28.4, i.i.Lake+P-N = 64.2 ± 25.7; χ2 = 7.470, P = 0.006; Fig. 2A), suggesting that productive environments allow the less-resistant fish ecotype to compensate and reduce costs of parasitism.
Fig. 2.
Multilevel parasite and nutrient effects on sticklebacks in phase 1. Infection intensities with significant interaction of parasite exposure and host ecotype (PxH, n = 159; A). Fish condition assessed by hepatosomatic index with effects of ecosystem nutrients (E) and infection intensity (i.i., n = 159; B). Data are presented as means ± SEM. Data in A and B from ref. 43. Gene expression responses (C), from threefold down-regulation to twofold up-regulation in parasitized vs. control manipulations (P) and high vs. low nutrient levels (E). Significant expression changes for gene groups are highlighted by black outlines (lake: n = 18, stream: n = 20, test on tank averages), for single genes after Benjamini–Yekutieli correction for multiple testing (n = 146, lake: n = 66, stream: n = 80, test on individuals) indicated by asterisks (first level effect), triangles (two-way interaction) or X (three-way interaction). See SI Appendix, Tables S1 and S3.
To characterize the molecular phenotypes of differential parasite load between fish ecotypes, we quantified expression of 28 metabolic, immune, and stress response genes. We selected (i) genes from a previous transcriptomic study based on strong differential expression between fish ecotypes and between infection states (37) and (ii) genes associated with responses to Gyrodactylus in other fish species (see SI Appendix, Table S2 for gene specific references). In phase 1, Gyrodactylus exposure of adults differently affected gene expression profiles of the two stickleback ecotypes (Fig. 2C, PxH and PxHxE effects, SI Appendix, Table S3): Stream fish up-regulated genes of the adaptive immune system (P effect, P = 0.004) and down-regulated genes of the complement system [P effect, P = 0.024, permutational multivariate analyses of variance (perMANOVAs)]. By contrast, lake fish did not modify the expression of entire gene groups, but significantly down-regulated two genes: the antibacterial transferrin a and a glucocorticoid receptor involved in the general stress response [tf, P effect, P = 0.008; nr3c1, PxE effect, P = 0.002, linear mixed effect models (LMMs)]. The differential gene expression profiles and infection patterns indicate that stream fish have evolved stronger immune responses against this parasite, enabling them to limit infection better than lake fish. This differential resistance could potentially be achieved via mechanisms involving recognition of Gyrodactylus antigens by immune cell receptors (38). The observed contrasting immune gene expression responses and strong expression differences between the ecotypes (H effects throughout most genes; SI Appendix, Table S3) support the hypothesis that parasite-mediated selection between habitat types contributes to adaptive population divergence of lake and stream ecotypes (39) and corroborate the strong immune gene expression differences between wild lake and stream sticklebacks reported in a recent study (40).
Overall, we found no persistent effects of nutrient loading on gene expression profiles of sticklebacks in phase 1 (SI Appendix, Table S3). However, we found that a stress response gene (nr3c1) encoding a glucocorticoid receptor, which initiates stress responses upon cortisol binding, was indirectly affected through an interaction of parasite exposure, nutrient loading, and host ecotype (PxHxE effect, P = 0.006, LMMs). This effect was driven by up-regulation in response to parasite pressure and down-regulation in high nutrient environments in lake fish, further highlighting how the tight interaction of biotic and abiotic selection pressures can lead to population specific patterns of gene expression.
To test whether the ecotype effects of molecular phenotypes were due to genetic differences, rather than due to differences in history of infection in the wild, we performed an additional common-garden experiment where we quantified gene expression of laboratory-reared adults originating from the same laboratory populations of juveniles used for phase 2. Using the same 28 genes, we found that gene expression generally differed between ecotypes (perMANOVA, H effect: F1,8 = 3.859, P = 0.041, SI Appendix, Table S4). Furthermore, metabolism genes showed an ecotype-specific expression response to the parasite exposure (PxH effect: F1,8 = 11.20, P = 0.041; SI Appendix, Fig. S1). These expression differences between ecotypes and expression responses to Gyrodactylus in stream fish were conserved between experiments (SI Appendix, Fig. S2). This consistency demonstrates that genetic differences between the lake and stream stickleback ecotypes (32) predictably influence their molecular phenotype, and that the effects observed during phase 1 of the mesocosm experiment are likely due to both genetic differences and plasticity. The importance of metabolism genes for ecotype differences in the response to parasite exposure is also consistent with a previous study, despite the analysis of different immune organs (37).
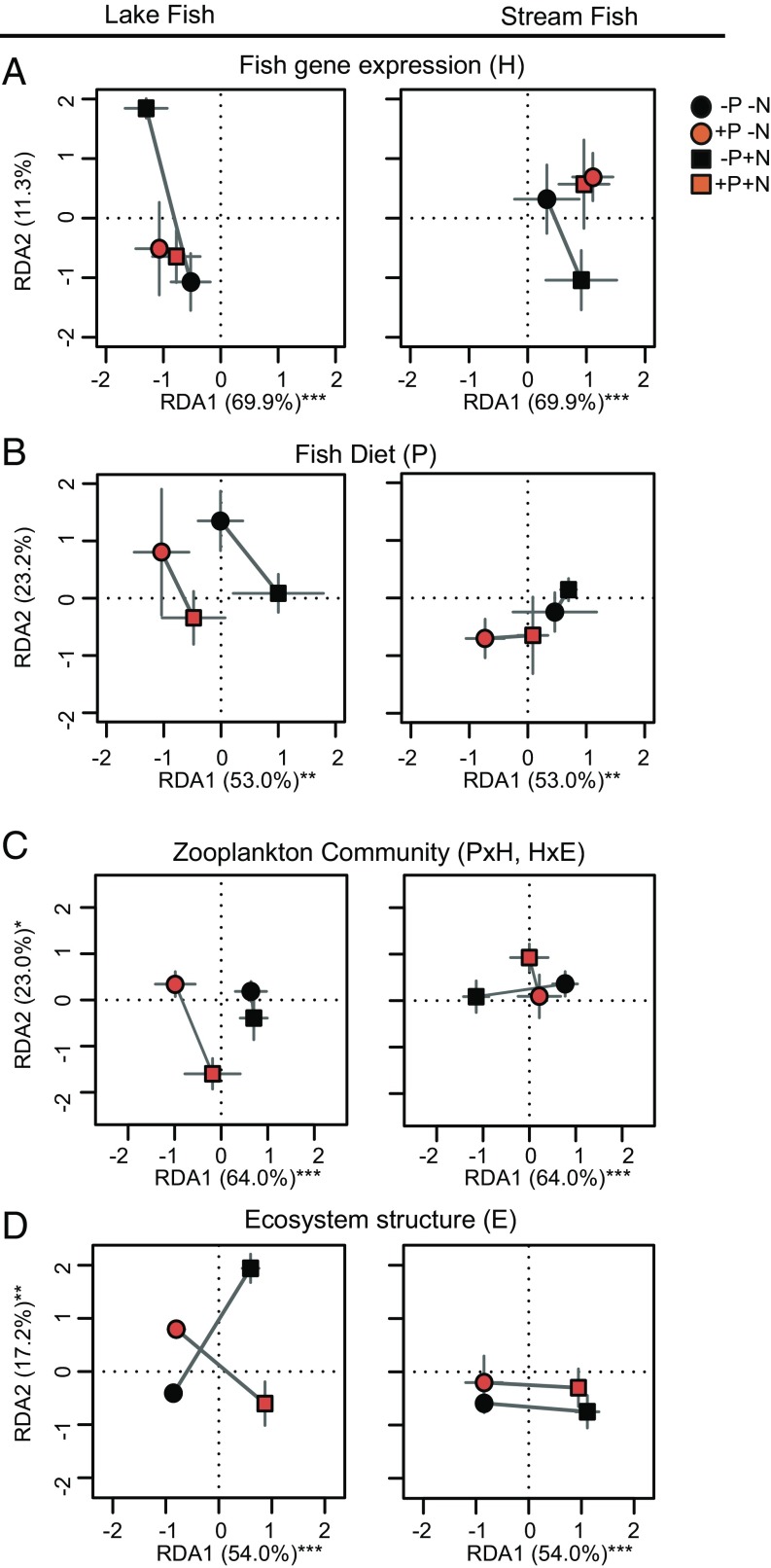
In phase 1 of the mesocosm experiment, we found a cost of parasitism such that neither host ecotype was completely tolerant to Gyrodactylus (41), indicated by a decrease of the hepatosomatic index (HSI) (42) with infection intensity in both ecotypes (infection intensity effect, P = 0.050; SI Appendix, Table S1 and figure 3A in ref. 43). In addition, parasite exposure caused fish to feed on different prey (P effect, R2 = 0.064, P = 0.020, diet composition RDA; Fig. 3B and SI Appendix, Table S5). Specifically, parasite-exposed individuals ate more cyclopoid copepods and fewer nymphs than control fish (SI Appendix, Table S5 and ref. 43). Such a diet shift could be caused either by direct parasite-mediated effects on feeding performance (44) or by changes in host feeding behavior to meet the nutritional requirements for coping with parasite infection (45).
Fig. 3.
Parasite effects from genes to ecosystem during phase 1 of the experiment. Gene expression (A), diet composition (B), zooplankton communities (C) and ecosystem parameters (D) are summarized by redundancy analyses (RDA; SI Appendix, Table S5) and shown as experimental group means ± SEM. Significant treatment effects for summarized data at each level are pointed out in figure headers. Percentages are explained variance by RDA axes, and asterisks indicate significance of RDA axes, *P < 0.05, **P < 0.01, ***P < 0.001. Data in B from ref. 43.
Given that parasites had effects on both the condition and diet of lake and stream sticklebacks, we hypothesized that parasite exposure might further influence how sticklebacks modify other aspects of their ecosystems. We found that the composition of the zooplankton community in the mesocosms was best predicted by the interaction between the fish ecotypes and the presence of Gyrodactylus (PxH effect, R2 = 0.067, P = 0.028, RDA; Fig. 3C and SI Appendix, Table S5). This effect might have been mediated by a differential top-down trophic effect of the stickleback ecotypes on the abundance of copepods in different nutrient and parasite environments (PxHxE effect, P = 0.042; SI Appendix, Table S5). Further down the food chain, the abundance of rotifers (Lepadellidae), which are a common prey of copepods, was also significantly affected by differences in how stickleback ecotypes responded to parasite exposure (i.e., a PxH effect, P = 0.017; SI Appendix, Table S5). Interestingly, interactive effects of hosts and parasites were also evident for abiotic ecosystem conditions. For example, despite the strong effects of our initial nutrient manipulation on the mesocosm ecosystems (i.e., E effects are common; SI Appendix, Table S5), the exposure of sticklebacks to parasites significantly altered the distribution of nutrients (e.g., dissolved nutrients, total nutrients, DOC) within the mesocosm ecosystems (PxE effect, R2 = 0.048, P = 0.019, nutrient concentration RDA; SI Appendix, Table S5). A previous mesocosm experiment using these same ecotypes of sticklebacks found that genetic background and plasticity interactively affected prey community structure and ecosystem conditions (6). Whereas both experiments found significant ecotype effects on a wide range of ecosystem metrics, the specific outcomes and dynamics differ between experiments. In both experiments, adult lake fish decreased copepod abundance more than stream fish in the short term (i.e., 3–7 wk). In the previous experiment, however, this effect was reversed after 12 wk (6). In general, mesocosms are only an approximation of natural ecosystems, and so the extent to which those effects are visible in nature remains unknown. In our experimental ecosystems, results suggest far-reaching consequences of parasitism (P effects) and host–parasite interactions (PxH effects) that extend well beyond the direct effects on host immunity, condition, and diet. In phase 2 of our experiment, we tested whether such ecosystem effects alter selection regimes in the next host generation.
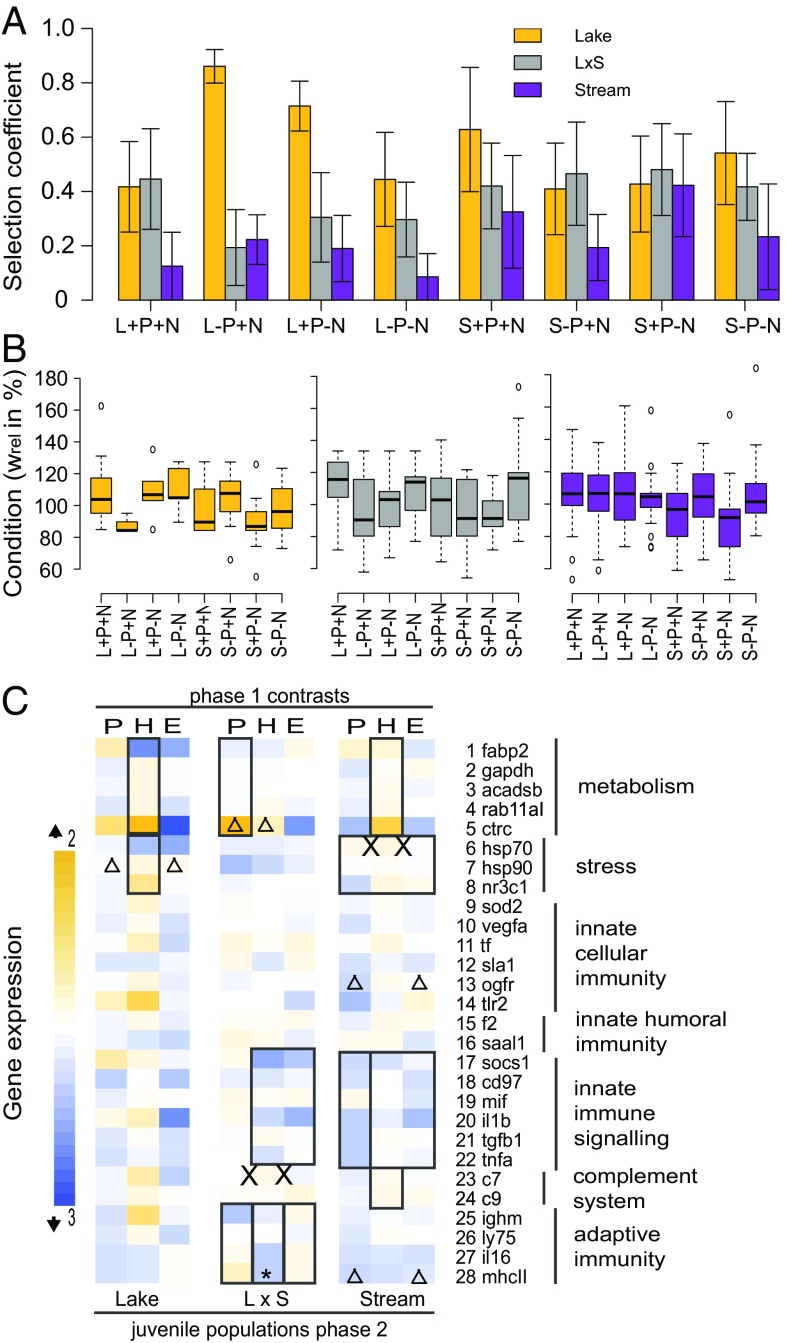
To initiate phase 2, we introduced lake, stream, and hybrid juvenile fish (laboratory-bred F1) into the tanks previously modified by the adult fish. At the end of phase 2 (13 wk duration), juvenile fish were collected and genotyped to quantify variation in survival depending on lake, hybrid, and stream fish origin (SI Appendix, Table S6). Overall, lake juveniles had a lower survival rate than either stream or hybrid juveniles (χ2 = 67.56, P < 0.001, Pearson’s χ2 test; SI Appendix, Fig. S3). Selection against lake juveniles was linked to a three-way interaction between treatment combinations in the first phase, namely parasite exposure of adults, host ecotype, and initial nutrient additions (PxHxE effect, P = 0.013; Fig. 4A and SI Appendix, Table S7). More specifically, selection against lake juveniles was higher in ecosystems previously manipulated by lake adults, particularly when these adults were either exposed to parasites in low nutrient mesocosms or unexposed to the parasite in high nutrient mesocosms. By comparison, the selection against stream and hybrid juveniles did not vary with the adult treatments in phase 1 (SI Appendix, Table S7). Among survivors however, stream juveniles had a lower body condition in ecosystems modified by parasite-exposed stream fish (PxH effect, P = 0.031; Fig. 4B and SI Appendix, Table S7). Together, the observed variation in survival rate and body condition show that both lake and stream ecotypes either have a survival disadvantage (lake juveniles) or a lower condition (stream juveniles) in ecosystems manipulated by adults of the same ecotype. Such effects could be due to differential depletion of preferred prey items, in particular by adults under parasite pressure. It is also possible that parasites persisted in the mesocosms in phase 2 and had differential effects on the juvenile genotypes (see SI Appendix, SI Discussion for further extrapolations from three-way interactions).
Fig. 4.
Effects of ecosystem modifications on second-phase fish. Selection coefficients (S = change in frequency relative to frequency of fittest genotype, subtracted from 1, within each tank; ref. 55) against different stickleback genetic backgrounds. Means ± SEM across five replicated tanks in ecosystems modified by phase 1 manipulations are shown (A). Within lake fish, selection is shaped by an interaction of all previous ecosystem manipulations (PxHxE, n = 39). The fittest genotype in each tank has a selection coefficient of 0 (Materials and Methods and SI Appendix, Table S7). Fish condition assessed by relative weight, showing the significant PxE effect on hybrid condition and PxH effect on stream fish condition (B, lake: n = 73, hybrid: n = 160, stream: n = 184, SI Appendix, Table S7). Gene expression profiles of survivors summarized by experimental manipulation in phase 1 and for different ecotype backgrounds of the juvenile fish in phase 2 (C). Expression responses in parasitized vs. control tanks (P), fish introduced to the previous lake vs. stream tanks (H), and high vs. low nutrient boost tanks (E) from threefold down-regulation (blue) to twofold up-regulation (yellow). Significant regulatory changes for gene groups are highlighted by black outlines (lake: n = 22, hybrid: n = 32, stream: n = 34, test on tank averages), for single genes after Benjamini–Yekutieli correction for multiple testing (lake: n = 32, hybrid: n = 79, stream: n = 109, test on individuals) indicated by asterisks (first level effect), triangles (two-way interaction), or X (three-way interaction). See SI Appendix, Table S8.
The body condition of hybrid juveniles was unaffected by the adult ecotype, but they had a lower condition in mesocosms where adult fish had been exposed to parasites at low nutrient loading and in parasite control tanks at high nutrient loading (PxE effect, P = 0.001; Fig. 4B and SI Appendix, Table S7). The dependence of hybrid juvenile condition on the interaction between parasite exposure and nutrient loading during phase 1 suggests that parasites might mediate selection against hybrids via changes in the ecosystems. Variation in the strength of selection against hybrids can influence the persistence of local adaptation, and influence the likelihood of biodiversity loss via reverse speciation (26, 46). For sticklebacks, parasite-mediated selection against hybrids has been both suggested (39) and experimentally demonstrated (38), however independently of the ecosystem effects of sticklebacks. Our experiment suggests a previously unexplored cross-generational effect of parasites, whereby parasites influence how hosts modify their ecosystems, altering selection on a subsequent generation (Figs. 3 and 4). Further experiments could test whether such an effect might be even stronger in a natural environment, where multiple generations of juvenile and adult stickleback co-occur (47). Our results also illustrate a potential mechanism underlying eco-evolutionary feedbacks, namely one where host-mediated ecosystem modifications affect selection and relative fitness of a subsequent host generation.
In addition to the effects of ecosystem modifications in phase 1 on relative juvenile survival in phase 2, we found effects on the expression of metabolism genes, general stress response, and innate immune signaling across juvenile ecotypes in phase 2 (Fig. 4C). In the modified ecosystems, the innate immune signaling of hybrid and stream juveniles showed an overall lower expression of genes in the high nutrient environments established in phase 1 (HxE effects; Fig. 4C and SI Appendix, Table S8). This lowered expression suggests that high nutrient environments shift either the cues or the trade-offs for investments in immune signaling by different host ecotypes. Additionally, stream juveniles exhibited differential regulation of the mhcII gene based on the parasite and nutrient treatments of phase 1 (PxE effect, P = 0.004; Fig. 4C and SI Appendix, Table S8). Major histocompatibility complex (MHC) class II genes are part of the adaptive immune system. They are involved in antigen recognition, and specific MHC alleles are correlated with Gyrodactylus resistance (38). If stream fish have previously evolved under high prevalence of this parasite or in the presence of virulent parasite strains, altering the baseline expression level of mhcII might be an adaptive response to reduce parasite spread and explain the selective advantage of this ecotype (Fig. 4A). The cross-generational effects of our parasite manipulation could also have been caused by the persistence of parasites in the mecocosms after the adults were removed. In this case, the regulatory response of juveniles may reflect the stronger parasite resistance of stream sticklebacks. In natural populations, the translation of parasite effects across generations, mediated by host-modified ecosystems, might be combined with transgenerational immune priming when hosts inherit epigenetic signals of their parents’ previous infections (36, 48). However, we can rule out this possibility in our experiment because juveniles were not the direct offspring of phase 1 adults. Instead, the cross-generational effects we observed were solely mediated by how the presence and infection status of hosts affected the subsequent rearing environment of juveniles.
Overall, our results show that the presence of parasites and the evolution of differential parasite resistance can influence host performance (e.g., diet and condition), and these parasite-induced performance differences can have cascading effects on community structure and ecosystem function. Variation in both parasite resistance and external environmental conditions can mediate the strength of eco-evolutionary feedbacks, and this feedback can be detected at the level of molecular phenotypes and ecosystem characteristics. That host-mediated modifications of the environment caused transgenerational effects on molecular phenotypes and differential selection among ecotypes warrants reconsidering the nature and importance of soft selection (9) and suggests that eco-evolutionary feedbacks might play an underappreciated role in adaptation. In light of our results, the effects of environmental change on infectious disease and on adaptive population divergence (26, 49) are more closely linked than previously considered.
Materials and Methods
Animal Collection and Treatment of Phase 1 Fish.
We collected three-spined sticklebacks (Gasterosteus aculeatus) with hand nets from two stream sites in the canton of St. Gallen, Switzerland (47.321131°N, 09.562395°E and 47.355822°N, 09.603133°E), and with minnow traps at one location on the shore of Lake Constance (47.484830°N, 09.542923°E). Fish collection and experiments were approved by local authorities (canton of St. Gallen fishing authorities and Veterinäramt of Kanton Luzern under permit LU03/12EE). Twenty sticklebacks each of stream and lake origin were euthanized directly to assess Gyrodactylus spp. prevalence in the natural populations. All experimental fish were disinfected by baths in 1:4,000 diluted Formalin on three consecutive days (modified from ref. 33). Experimental infection was achieved 7 d later by manual transfer of Gyrodactylus spp. individuals from nondisinfected sticklebacks collected from the same lake and stream populations. Two individual parasites from each of the lake and stream environments were transferred. Additional details are available in SI Appendix, SI Materials and Methods, Section SI.1.
Experimental Setup and First Phase Sampling.
The mesocosms were plastic tanks of 1 m3, filled with gravel, sand, and sediment collected from Lake Lucerne and a nearby stream, and lake water and a concentrated zooplankton inoculum from Lake Lucerne and Lake Constance. The full factorial cross design of Parasites × Host ecotype × Ecosystem Nutrients was replicated in five blocks for a total of 40 mesocosms. Within each block, we established contrasting nutrient environments by adding different amounts of nutrient solution containing NaNO3 and HNa2PO4 into high and low nutrient tanks, respectively (E contrast). For the first phase of the experiment, we introduced three-spined sticklebacks of either lake or stream origin to establish the host ecotype contrast (H).
We collected ecosystem data such as physico-chemical (e.g., turbidity, nutrient concentrations) and biological (e.g., chlorophyll levels in water and periphyton) properties of the ecosystems and sampled the zooplankton communities 6 wk after fish introduction to the mesocosms and removed the fish 1 wk later. Fifty-seven of 278 sticklebacks died during the first experimental phase and were collected from the mesocosms upon detection. Mortality differed between host ecotypes, being higher among lake fish, but did not vary with other treatments (χ2 test, H: χ2 = 4.164, P = 0.041, P: χ2 = 0.233, P = 0.629, E: χ2 = 0.002, P = 0.966; SI Appendix, Table S1). After euthanasia of the fish in 1 M MS-222, Gyrodactylus specimen were counted on each fish before morphological measurements and dissection. Additional details are available in SI Appendix, SI Materials and Methods, Section SI.2.
Introduction and Sampling of Phase 2 Fish.
After removal of phase 1 fish, groups of juvenile laboratory-bred F1 sticklebacks of lake, hybrid, and stream background were introduced into each tank modified throughout phase 1 of the experiment. These juvenile groups were standardized for family backgrounds within experimental blocks and ratio of stream, hybrid, and lake fish across all experimental tanks (n = 19–39 per tank; SI Appendix, Table S6). Hybrid crosses were done in either direction, seven with stream females, five with lake females. Ecosystems were all handled equally at this stage. All surviving fish were caught 3 mo after the juvenile phase 2 fish were introduced to the mesocosms. As with phase 1 fish, after euthanasia in a 1 M MS-222 solution, Gyrodactylus specimen were counted on each fish before length and weight measurements and removal of spleens and livers for gene expression assays. Only 10 of the 407 scanned individuals were infected with Gyrodactylus at the end of the experiment, with no significant effects of any previous treatment on infection levels in this second generation (binomial GLMMs, all χ2 < 2.03, all P > 0.15; SI Appendix, Table S7). Additional details are available in SI Appendix, SI Materials and Methods, Section SI.3.
Common Garden Experiment.
To validate that part of the ecotype effect during phase 1 that was based on genetic differences between lake and stream sticklebacks, we conducted a separate common garden experiment. This experiment ran for 5 wk and consisted of 34 laboratory-raised adult fish kept in 12 identical outdoor tanks. Half of these fish had a genetic lake background and the other half descended from stream fish. Again, half of the experimental groups were exposed to Gyrodactylus on an individual basis. Gene expression data were collected from their spleens as a comparison with the wild-caught fish from the first phase of the mesocosm experiment. Additional details are available in SI Appendix, SI Materials and Methods, Section SI.4.
Molecular Analyses.
We performed gene expression analyses with RNA extracted from spleens and combined spleens and livers for adults and juveniles, respectively. Because transcriptome analyses have been conducted with lake and stream three-spined sticklebacks (37), we used a target gene approach, measuring relative mRNA levels in microfluidic quantitative PCR assays of 28 target genes. Origin of surviving juveniles was determined by parentage analysis in Colony (50), using seven microsatellite markers (51) (Stich5196, Stich4170, Stich1125, Stich1097, Stich7033, STN18, STN75). Additional details are available in SI Appendix, SI Materials and Methods, Section SI.5.
Statistical Analyses.
All statistical analyses were performed in R version 3.1.0 (52). The following model structure was used to test for the effects of phase 1 experimental treatments: parasite exposure (P), host ecotype (H), ecosystem nutrient levels (E), and their interactions as fixed structure with block as a random factor. Univariate analyses on individual fish characteristics such as parasite burden, fish condition, gene expression, and survival also included tank identity nested within block as a random effect. Fish condition for phase 1 fish was calculated as the HSI = 1,000 × liver wet-mass (mg)/fish mass (mg) and for phase 2 fish as relative weight Wrel (53). HSI was tested with an LMM by using infection intensity as well as the experimental treatments as fixed structure.
Diet, zooplankton communities, ecosystem parameters and gene expression were tested for experimental treatment effects in RDAs and univariate (G)LMMs. Gene expression was analyzed as ΔCt values (54) and further assessed by perMANOVAs on functional gene groups. Juvenile stocking differences between tanks (19–39 per tank) were statistically accounted for by including tank as a random factor in individual based tests and by including stocking numbers in tank based tests for phase 2 analyses. Survival differences between lake, hybrid, and stream juveniles were tested with a Pearson’s χ2 test. Effects of phase 1 treatments on juvenile survival were tested in binomial GLMMs on survival rates from each tank. We calculated the selection coefficient S against each juvenile ecotype as the change in frequency of the ecotype relative to the frequency of the fittest genotype, subtracted from 1, within each tank (55). Effects of phase 1 ecosystem modifications on viability selection were tested in LMMs for each juvenile ecotype separately. Additional details are available in SI Appendix, SI Materials and Methods, Section SI.6.
Supplementary Material
Acknowledgments
We thank D. Steiner, D. Hohmann, N. Sommer, C. Federer, M. Heckwolf, T. Ballesteros, S. Urbanski, N. Kertesz, A. Taverna, and B. Kienholz and the Swiss Federal Institute of Aquatic Research and Technology Kastanienbaum community for assistance in the laboratory, mesocosm garden, and the field. We also thank J. Raeymaekers, L. Becks, O. Seehausen, two anonymous reviewers, and the editor for their helpful feedback on earlier versions of this manuscript. This project was funded by Lead Agency of the German Science Foundation (DFG) Grant EI841/4-1 (to C.E.) and Swiss National Science Foundation Grant 139326 (to B.M.). The project was enabled by the stickleback cluster of the DFG priority program 1399 “Host Parasite Coevolution” and supported by DFG Grant EI841/6-1 (to C.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.P.H. is a guest editor invited by the Editorial Board.
Data deposition: The results from this paper are available through Dryad (doi: 10.5061/dryad.5q783).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619147114/-/DCSupplemental.
References
- 1.Schoener TW. The newest synthesis: Understanding the interplay of evolutionary and ecological dynamics. Science. 2011;331(6016):426–429. doi: 10.1126/science.1193954. [DOI] [PubMed] [Google Scholar]
- 2.Pelletier F, Garant D, Hendry AP. Eco-evolutionary dynamics. Philos Trans R Soc Lond B Biol Sci. 2009;364(1523):1483–1489. doi: 10.1098/rstb.2009.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendry AP. Eco-Evolutionary Dynamics. Princeton Univ Press; Princeton: 2016. [Google Scholar]
- 4.Matthews B, et al. Toward an integration of evolutionary biology and ecosystem science. Ecol Lett. 2011;14(7):690–701. doi: 10.1111/j.1461-0248.2011.01627.x. [DOI] [PubMed] [Google Scholar]
- 5.Beckerman AP, Childs DZ, Bergland AO. Eco-evolutionary biology: Feeding and feedback loops. Curr Biol. 2016;26(4):R161–R164. doi: 10.1016/j.cub.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Matthews B, Aebischer T, Sullam KE, Lundsgaard-Hansen B, Seehausen O. Experimental evidence of an eco-evolutionary feedback during adaptive divergence. Curr Biol. 2016;26(4):483–489. doi: 10.1016/j.cub.2015.11.070. [DOI] [PubMed] [Google Scholar]
- 7.Rudman SM, Schluter D. Ecological impacts of reverse speciation in threespine stickleback. Curr Biol. 2016;26(4):490–495. doi: 10.1016/j.cub.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Kasada M, Yamamichi M, Yoshida T. Form of an evolutionary tradeoff affects eco-evolutionary dynamics in a predator-prey system. Proc Natl Acad Sci USA. 2014;111(45):16035–16040. doi: 10.1073/pnas.1406357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reznick D. Hard and soft selection revisited: How evolution by natural selection works in the real world. J Hered. 2016;107(1):3–14. doi: 10.1093/jhered/esv076. [DOI] [PubMed] [Google Scholar]
- 10.Kuris AM, et al. Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature. 2008;454(7203):515–518. doi: 10.1038/nature06970. [DOI] [PubMed] [Google Scholar]
- 11.Lafferty KD, et al. Parasites in food webs: The ultimate missing links. Ecol Lett. 2008;11(6):533–546. doi: 10.1111/j.1461-0248.2008.01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid-Hempel P. Evolutionary Parasitology: The Integrated Study of Infections, Immunology, Ecology and Genetics. Oxford Univ Press; Oxford: 2011. [Google Scholar]
- 13.Summers K, et al. Parasitic exploitation as an engine of diversity. Biol Rev Camb Philos Soc. 2003;78(4):639–675. doi: 10.1017/s146479310300616x. [DOI] [PubMed] [Google Scholar]
- 14.Eizaguirre C, Lenz TL, Kalbe M, Milinski M. Rapid and adaptive evolution of MHC genes under parasite selection in experimental vertebrate populations. Nat Commun. 2012;3:621–626. doi: 10.1038/ncomms1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dargent F, Scott ME, Hendry AP, Fussmann GF. Experimental elimination of parasites in nature leads to the evolution of increased resistance in hosts. Proc R Soc B Biol Sci. 2013;280(1773):20132371. doi: 10.1098/rspb.2013.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolinska J, King KC. Environment can alter selection in host-parasite interactions. Trends Parasitol. 2009;25(5):236–244. doi: 10.1016/j.pt.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Mostowy R, Engelstädter J. The impact of environmental change on host-parasite coevolutionary dynamics. Proc R Soc B Biol Sci. 2011;278(1716):2283–2292. doi: 10.1098/rspb.2010.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JN. The Geographic Mosaic of Coevolution. Univ of Chicago Press; Chicago: 2005. [Google Scholar]
- 19.Frickel J, Sieber M, Becks L. Eco-evolutionary dynamics in a coevolving host-virus system. Ecol Lett. 2016;19(4):450–459. doi: 10.1111/ele.12580. [DOI] [PubMed] [Google Scholar]
- 20.Penczykowski RM, Laine A-L, Koskella B. Understanding the ecology and evolution of host-parasite interactions across scales. Evol Appl. 2015;9(1):37–52. doi: 10.1111/eva.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohannan BJM, Lenski RE. Linking genetic change to community evolution: Insights from studies of bacteria and bacteriophage. Ecol Lett. 2000;3(4):362–377. [Google Scholar]
- 22.Marcogliese DJ, Cone DK. Food webs: A plea for parasites. Trends Ecol Evol. 1997;12(8):320–325. doi: 10.1016/S0169-5347(97)01080-X. [DOI] [PubMed] [Google Scholar]
- 23.Smith VH, Schindler DW. Eutrophication science: Where do we go from here? Trends Ecol Evol. 2009;24(4):201–207. doi: 10.1016/j.tree.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Elser JJ, et al. Shifts in lake N:P stoichiometry and nutrient limitation driven by atmospheric nitrogen deposition. Science. 2009;326(5954):835–837. doi: 10.1126/science.1176199. [DOI] [PubMed] [Google Scholar]
- 25.Donohue I, Jackson AL, Pusch MT, Irvine K. Nutrient enrichment homogenizes lake benthic assemblages at local and regional scales. Ecology. 2009;90(12):3470–3477. doi: 10.1890/09-0415.1. [DOI] [PubMed] [Google Scholar]
- 26.Vonlanthen P, et al. Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature. 2012;482(7385):357–362. doi: 10.1038/nature10824. [DOI] [PubMed] [Google Scholar]
- 27.Johnson PTJ, et al. Aquatic eutrophication promotes pathogenic infection in amphibians. Proc Natl Acad Sci USA. 2007;104(40):15781–15786. doi: 10.1073/pnas.0707763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budria A, Candolin U. How does human-induced environmental change influence host-parasite interactions? Parasitology. 2014;141(4):462–474. doi: 10.1017/S0031182013001881. [DOI] [PubMed] [Google Scholar]
- 29.McKenzie VJ, Townsend AR. Parasitic and infectious disease responses to changing global nutrient cycles. EcoHealth. 2007;4:384–396. [Google Scholar]
- 30.Lundsgaard-Hansen B, Matthews B, Vonlanthen P, Taverna A, Seehausen O. Adaptive plasticity and genetic divergence in feeding efficiency during parallel adaptive radiation of whitefish (Coregonus spp.) J Evol Biol. 2013;26(3):483–498. doi: 10.1111/jeb.12063. [DOI] [PubMed] [Google Scholar]
- 31.Lucek K, Roy D, Bezault E, Sivasundar A, Seehausen O. Hybridization between distant lineages increases adaptive variation during a biological invasion: Stickleback in Switzerland. Mol Ecol. 2010;19(18):3995–4011. doi: 10.1111/j.1365-294X.2010.04781.x. [DOI] [PubMed] [Google Scholar]
- 32.Marques DA, et al. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 2016;12(2):e1005887. doi: 10.1371/journal.pgen.1005887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raeymaekers JAM, Wegner KM, Huyset T, Volckaert FAM. Infection dynamics of the monogenean parasite Gyrodactylus gasterostei on sympatric and allopatric populations of the three-spined stickleback Gasterosteus aculeatus. Folia Parasitol (Praha) 2011;58(1):27–34. doi: 10.14411/fp.2011.003. [DOI] [PubMed] [Google Scholar]
- 34.De Roij J, Harris PD, MacColl ADC. Divergent resistance to a monogenean flatworm among three-spined stickleback populations. Funct Ecol. 2011;25:217–226. [Google Scholar]
- 35.Bakke TA, Cable J, Harris PD. The biology of gyrodactylid monogeneans: The “Russian-doll killers”. Adv Parasitol. 2007;64:161–376. doi: 10.1016/S0065-308X(06)64003-7. [DOI] [PubMed] [Google Scholar]
- 36.Kaufmann J, Lenz TL, Milinski M, Eizaguirre C. Experimental parasite infection reveals costs and benefits of paternal effects. Ecol Lett. 2014;17(11):1409–1417. doi: 10.1111/ele.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenz TL, Eizaguirre C, Rotter B, Kalbe M, Milinski M. Exploring local immunological adaptation of two stickleback ecotypes by experimental infection and transcriptome-wide digital gene expression analysis. Mol Ecol. 2013;22(3):774–786. doi: 10.1111/j.1365-294X.2012.05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eizaguirre C, Lenz TL, Kalbe M, Milinski M. Divergent selection on locally adapted major histocompatibility complex immune genes experimentally proven in the field. Ecol Lett. 2012;15(7):723–731. doi: 10.1111/j.1461-0248.2012.01791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eizaguirre C, Lenz TL, Traulsen A, Milinski M. Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol Lett. 2009;12(1):5–12. doi: 10.1111/j.1461-0248.2008.01247.x. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, et al. Transcriptome profiling of immune tissues reveals habitat-specific gene expression between lake and river sticklebacks. Mol Ecol. 2016;25(4):943–958. doi: 10.1111/mec.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Råberg L, Graham AL, Read AF. Decomposing health: Tolerance and resistance to parasites in animals. Philos Trans R Soc Lond B Biol Sci. 2009;364(1513):37–49. doi: 10.1098/rstb.2008.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chellappa S, Huntingford FA, Strang RHC, Thomson RY. Condition factor and hepatosomatic index as estimates of energy status in male three-spined stickleback. J Fish Biol. 1995;47(5):775–787. [Google Scholar]
- 43.Anaya-Rojas JM, et al. The association of feeding behaviour with the resistance and tolerance to parasites in recently diverged sticklebacks. J Evol Biol. 2016;29(11):2157–2167. doi: 10.1111/jeb.12934. [DOI] [PubMed] [Google Scholar]
- 44.Lefèvre T, et al. The ecological significance of manipulative parasites. Trends Ecol Evol. 2009;24(1):41–48. doi: 10.1016/j.tree.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Ponton F, et al. Hosts use altered macronutrient intake to circumvent parasite-induced reduction in fecundity. Int J Parasitol. 2011;41(1):43–50. doi: 10.1016/j.ijpara.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Seehausen O, Takimoto G, Roy D, Jokela J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol. 2008;17(1):30–44. doi: 10.1111/j.1365-294X.2007.03529.x. [DOI] [PubMed] [Google Scholar]
- 47.Bell MA, Foster SA. The Evolutionary Biology of the Threespine Stickleback. Oxford Univ Press; Oxford: 1994. [Google Scholar]
- 48.Roth O, Klein V, Beemelmanns A, Scharsack JP, Reusch TBH. Male pregnancy and biparental immune priming. Am Nat. 2012;180(6):802–814. doi: 10.1086/668081. [DOI] [PubMed] [Google Scholar]
- 49.Becker DJ, Streicker DG, Altizer S. Linking anthropogenic resources to wildlife-pathogen dynamics: a review and meta-analysis. Ecol Lett. 2015;18(5):483–495. doi: 10.1111/ele.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones OR, Wang J. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour. 2010;10(3):551–555. doi: 10.1111/j.1755-0998.2009.02787.x. [DOI] [PubMed] [Google Scholar]
- 51.Kalbe M, et al. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc Biol Sci. 2009;276(1658):925–934. doi: 10.1098/rspb.2008.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Core Team 2014. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
- 53.Froese R. Cube law, condition factor and weight-length relationships: History, meta-analysis and recommendations. J Appl Ichthyology. 2006;22(4):241–253. [Google Scholar]
- 54.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamilton MB. Population Genetics. 1st Ed Wiley-Blackwell; Chichester, UK: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.