Abstract
Previous studies in our laboratory have shown a differential activation of the mitogen-activated protein kinases (MAPKs) in primary bone marrow-derived macrophages following infection with pathogenic Mycobacterium avium compared to the activation following infection with nonpathogenic Mycobacterium smegmatis. Additionally, M. smegmatis-infected macrophages produced significantly elevated levels of tumor necrosis factor alpha (TNF-α) compared to the levels produced by M. avium-infected macrophages. The TNF-α production was dependent on both p38 and extracellular signal-regulated kinase 1/2 (ERK 1/2) activation. However, the macrophage transcription factors downstream of the MAPKs, which were required for TNF-α production, remained undefined. In this study we determined that the transcription factor cyclic AMP response element binding protein (CREB) is significantly more activated in M. smegmatis-infected macrophages than in M. avium-infected macrophages. We also found that CREB activation was dependent on p38 and protein kinase A but not on ERK 1/2 or calmodulin kinase II. Moreover, mutating the cAMP-responsive element on the TNF-α promoter resulted in significantly diminished promoter activity following M. smegmatis infection but not M. avium infection. The inability of macrophages infected with M. avium to sustain MAPK activation and to produce high levels of TNF-α was due, in part, to an increase in serine/threonine phosphatase PP2A activity. Our studies are the first to demonstrate an important role for the transcription factor CREB in TNF-α production by mycobacterium-infected macrophages, as well as a role for M. avium's induction of PP2A phosphatase activity as a mechanism to limit macrophage activation.
Mycobacterium avium is an opportunistic pathogen that affects people with suppressed immune systems, particularly people with late-stage human immunodeficiency virus or chronic lung diseases. In AIDS patients, M. avium is commonly disseminated and can involve almost any internal organ, especially the liver, spleen, and bone marrow. Moreover, M. avium is responsible for increased morbidity and mortality in human immunodeficiency virus-infected individuals (6).
M. avium is a facultative intracellular pathogen that resides within the phagosome of the host macrophage. The macrophage is the first line of defense against invading microorganisms, and it functions to phagocytose and subsequently destroy these invaders within a phagolysosome. However, following phagocytosis, M. avium, like other species in the genus Mycobacterium, including Mycobacterium tuberculosis and Mycobacterium leprae, has been shown to halt the maturation of the phagosome through coordinated blocking of lysosome fusion with the phagosome. Thus, the M. avium-containing phagosome fails to acquire lysosomal lytic enzymes, nor does it acidify, due to a scarcity of the proton ATPases (25). However, macrophages have another inherent defense mechanism against mycobacterial diseases. Macrophages are stimulated to secrete a large panel of inflammatory cytokines and immune modulators, including tumor necrosis factor alpha (TNF-α), following phagocytosis of bacteria. Studies with knockout mice have shown that TNF-α is critical for controlling an M. avium infection (1). TNF-α has also been shown to be important in control of M. avium infections in immunocompetent patients (31).
The importance of TNF-α in a mycobacterial infection is underscored by the fact that nonpathogenic mycobacteria induce significantly more TNF-α production by bone marrow-derived macrophages (BMMφ) than pathogenic mycobacteria (2, 14). Studies in our laboratory have shown that the differential regulation of the p38 and ERK 1/2 mitogen-activated protein kinase (MAPK) pathways in conjunction with the Ca2+-calmodulin-calmodulin kinase II (CaM-CaMKII) and cyclic AMP (cAMP)-protein kinase A pathways could account for the increased TNF-α production observed in BMMφ infected with the nonpathogenic bacillus M. smegmatis compared to the production in cells infected with the pathogenic M. avium 724. We showed that M. smegmatis induced significantly higher p38 and ERK 1/2 activation than M. avium 724 in infected BMMφ and that inhibiting either of these MAPKs significantly impaired the mycobacterium-induced TNF-α production (24). These results led us to question whether the overall suppression of these important signal transduction pathways during an M. avium 724 infection results in a limited TNF-α transcriptional response by the BMMφ.
Transcriptional activation of TNF-α is highly controlled, and the transcriptional apparatus that forms on the promoter has proven to be cell type specific, as well as stimulus specific (15). The promoter has numerous transcription factor binding sites that are shared by transcription factors; thus, the cell is able to discriminate between stimulating signals to form distinct complexes. Initiation of transcription and the extent of TNF-α production are thought to be dependent on the enhancesome complex that forms on the promoter (15). There are many transcription factors that are known to be involved in TNF-α production, including NFAT, ATF-2, Jun, Ets/Elk, SP-1, CBP/p300, NF-κB, and the cyclic AMP response element binding protein (CREB) (15, 29, 30). CREB is a known downstream target of both the cAMP-dependent protein kinase A (PKA) and MAPK pathways (8, 16). We therefore hypothesized that the different TNF-α responses by macrophages to pathogenic and nonpathogenic mycobacteria result, at least in part, from differential activation of the transcription factor CREB.
CREB's role in the inflammatory process has been well studied. As its name implies, this compound is closely associated with the classical intracellular second messenger cAMP. Following an increase in intracellular cAMP levels, PKA is activated. PKA can then passively diffuse into the nucleus and induce gene activation by phosphorylating CREB at serine 133 (16). Phosphorylation at this residue, in combination with the activation of other signaling pathways, promotes the recruitment of the transcriptional coactivator CREB binding protein (CBP) and its paralogue, p300. CREB has also been shown to be activated by many other kinases, including pp90rsk, protein kinase C, p38, Akt, mitogen- and stress-activated protein kinase 1, CaMKII, and MAPK-activated protein kinase 2 (11, 13, 22, 26, 27, 32). CREB binds as a dimer to a conserved cAMP-responsive element (CRE) palindrome, TGACGTCA, or to a half-site CRE motif (CGTCA) (19). Promoters containing either the palindromic or half-site motif of CRE regulate genes involved in glucose homeostasis, growth factor-dependent cell survival, learning, memory, and immune regulation, among other processes (18). The murine TNF-α promoter contains a CRE site at positions −100 to −107 (GenBank accession no. AB062426). However, a role for CREB in macrophage TNF-α promoter activity following a mycobacterial infection has not been defined. In the present study, we examined the level of CREB activation and the role of CREB in TNF-α expression in macrophages following pathogenic and nonpathogenic mycobacterial infections.
Our data demonstrate that M. smegmatis infection leads to significantly greater induction of CREB phosphorylation in macrophages than the induction observed in M. avium 724-infected cells and that this phosphorylation is dependent on p38 and PKA activation. TNF-α promoter activity following an M. smegmatis infection but not an M. avium infection was dependent on the CREB-specific binding nucleotides in the CRE site. Moreover, we found evidence for M. avium 724 down-regulation of TNF-α production through increased activation of the serine/threonine phosphatase PP2A.
MATERIALS AND METHODS
Bone marrow macrophage isolation and culture.
BMMφ, which were used in all experiments, were isolated from 6- to 8-week-old BALB/c mice as previously described (24). Briefly, bone marrow was isolated, and fibroblasts and mature macrophages were removed by selective adhesion. The monocytes were cultured in Dulbecco's modified Eagle's medium (GIBCO BRL, Grand Island, N.Y.) supplemented with 20 mM HEPES (Mediatech Cellgro, Herndon, Va.), 10% fetal bovine serum (GIBCO BRL), 100 U of penicillin (BioWhittaker, Walkersville, Md.) per ml, 100 μg of streptomycin (BioWhittaker) per ml, 1× l-glutamine (Mediatech Cellgro), and 20% L-cell supernatant as a source of macrophage colony-stimulating factor. After 4 days of incubation, fresh medium was supplied to the BMMφ, and mature macrophages were harvested on day 7 and frozen at −140°C. Thawed macrophages were cultured on non-tissue culture plates for 3 to 7 days, passaged, and allowed to recover for 3 to 6 days, and then they were replated at a concentration of approximately 3 × 105 cells/35-mm tissue culture plate for Western blot experiments and at a concentration of 1 × 105 cells/well in glass Lab-TekII chamber slides (Nalge Nunc International Corp., Naperville, Ill.) for immunofluorescence analysis. The cells were allowed to adhere for 24 h prior to infection.
For all experiments, mycobacteria were added to macrophages on ice and incubated for 10 min, which allowed the mycobacteria to settle onto the cells, and then the preparations were incubated at 37°C in 5% CO2 for the times indicated below. Culture medium without antibiotics or L-cell supernatant was used in place of complete medium during the infections. For the 9-h infections, the BMMφ were incubated for 4 h with the mycobacteria and dimethyl sulfoxide (DMSO) or inhibitors and washed with phosphate-buffered saline (PBS) three times, fresh medium with or without inhibitors was added, and the preparations were incubated for an additional 5 h. All tissue culture reagents were found to be negative for endotoxin contamination by using either the E-Toxate assay (Sigma) or the QCL-1000 endotoxin test (Cambrex Bio Science, Walkersville, Md.).
Inhibitor treatments.
The inhibitors were purchased from Calbiochem (La Jolla, Calif.), reconstituted in sterile, endotoxin-tested DMSO, and added 1 h before infection at the following concentrations: H89, 20 μM; SB203580, 20 μM; PD98059, 10 μM; and okadaic acid (OA), 10 nM. KN62 was used at a concentration of 10 μM and added 30 min before infection. The concentration of DMSO used was the same as the concentration in the vehicle control. For all inhibitors, a dose response was observed in relation to ERK 1/2 phosphorylation, and the concentrations used in subsequent studies were chosen based on the dose response and the results of previous studies with macrophages (10, 17, 24, 33). None of the inhibitors used had a significant effect on the uptake of the mycobacteria by the macrophages.
Bacterial culture.
To generate M. avium 724 stocks, the mycobacteria (generously provided by Andrea Cooper, Trudeau Institute, Saranac Lake, N.Y.) were passaged through a mouse to ensure virulence, and a single colony was used to inoculate Middlebrook 7H9 medium (Difco, Sparks, Md.) supplemented with 100 mM glucose (Sigma), 0.06% oleic acid (Fisher Scientific, Fair Lawn, N.J.), 5.0% (wt/vol) bovine albumin (Sigma), 0.5% Tween 20 (Fisher Scientific), and 150 mM NaCl (Fisher Scientific) (GOATS). The bacteria were grown for 10 days at 37°C with vigorous shaking, resuspended in Middlebrook medium containing GOATS with 15% glycerol, divided into aliquots, and stored at −80°C. Frozen stocks were quantitated by serial dilution onto Middlebrook 7H10 agar containing GOATS. M. smegmatis strain MC2155 (generously provided by Rich Groger, Washington University, St. Louis, Mo.) was grown in Middlebrook medium containing GOATS at 37°C for 2 to 4 days. Frozen stocks were prepared as described above for M. avium. All reagents used to grow mycobacteria were found to be negative for endotoxin contamination by using the E-Toxate assay (Sigma) and the QCL-1000 endotoxin test (Cambrex Bio Science).
Mycobacterial FITC labeling.
To generate fluorescein isothiocyanate (FITC)-labeled bacterial stocks, M. avium 724 and M. smegmatis were grown as described above. Before freezing, cultures were pelleted and washed with Hanks' buffered saline solution (GIBCO) supplemented with 1% bovine serum albumin (BSA) (ICN Biochemicals Inc., Aurora, Ohio). Cultures were resuspended in boric acid buffer (pH 9.2) containing 1.5 mg of FITC powder (Sigma) per ml dissolved in tissue-culture-grade DMSO (Sigma) and incubated at 37°C for 2 h. Cultures were washed four times with Hanks' buffered saline solution supplemented with 1% BSA to remove any residual buffer and pelleted. FITC-labeled cultures were resuspended and frozen as described above. Frozen stocks were quantified by serial dilution.
Mycobacterial infection.
To complement opsonize the mycobacteria, appropriate concentrations of mycobacteria were suspended in macrophage culture medium containing 10% purified protein derivative-negative human serum as a source of complement components and incubated for 2 h at 37°C (4). The human serum was obtained from the same donor for all experiments. The same concentration of human serum was added to uninfected controls for all experiments.
Infection assays evaluated by fluorescence microscopy were performed with each stock of mycobacteria to determine the infection ratio needed to infect approximately 80% of the macrophages. Briefly, BMMφ were plated on glass coverslips and infected with different doses of mycobacteria in triplicate. Infections were halted at either 1 or 4 h, and the preparations were fixed in methanol-acetone (1:1) and washed with PBS. Slides were visualized by fluorescence microscopy, and the level of infection was quantitated by counting the cells infected in at least four fields per replicate. No fewer than 100 cells per replicate were counted.
Immunofluorescence.
For immunofluorescence experiments, cells were infected as described above and then fixed in 4% paraformaldehyde (Sigma) in PBS at 4°C at designated times. Fixed cells were then permeabilized at −20°C with 100% methanol, blocked with 0.2% BSA and 0.02% gelatin (Sigma), and washed with PBS-0.1% Triton X-100. Immunofluorescence staining and confocal microscopy were performed as described previously (5). Briefly, the cells were incubated with the primary antibody against phospho-CREB (Ser133) (Cell Signaling, Beverly, Mass.) and then were incubated overnight at 4°C in PBS-23% goat serum (Gibco) at a 1:200 dilution. The antibody against lysosome-associated membrane protein 1 (1D4B) was obtained from The Developmental Hybridoma Bank (University of Iowa) and was used at a 1:100 dilution. All secondary antibodies were obtained from Chemicon International (Temecula, Calif.) and were used at a dilution of 1:400 in PBS-23% goat serum.
Each confocal experiment included uninfected BMMφ. Uninfected controls were imaged first, and the same confocal microscope settings were used for the subsequent samples. Z-series were obtained for each sample, and more than 100 cells were counted per experimental condition. Cells that had distinct CREB nuclear staining were scored positive. Scoring within experiments was performed blind with respect to experimental conditions.
Plasmids.
The luciferase reporter construct containing the mouse (BALB/c) TNF-α(−1200 bp) promoter was prepared by standard PCR-subcloning techniques. The nucleotide fragment from position −1200 to position 2 of the mouse TNF-α promoter was PCR amplified by using genomic DNA of BALB/c mice as the template and two flanking primers, 5′-TAGCTAGCCCATCTGTGAAACCCAATAAACCTCT-3′ (sense) and 5′-GCAAGCTTGGGAGCTTCTGCTGGCTGG-3′ (antisense). The primer sequences were derived from a Mus musculus Tnf gene for the 5′ regulatory region (GenBank accession no. AB062426). The PCR fragment obtained was inserted into the plasmid vector pGEM-T (pGEM-T Easy vector system; Promega, Madison, Wis.). The fragment containing the TNF-α(−1200 bp) promoter was subcloned as an NheI and HindIII fragment into the pGL3 basic luciferase vector (Promega). The mutant construct for the CRE site of the TNF-α(−1200 bp) promoter luciferase reporter was prepared by site-directed mutagenesis. The CRE site (positions −100 to −107) was mutated by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's protocol from 5′-TGAGATCA-3′ to 5′-TGAGAGTA-3′. The mutagenic primers utilized were 5′-CGCTTCCTCCACATGAGAGTATGGTTTTCTCCACCAAG-3′ and 5′-CTTGGTGGAGAAAACCATACTCTCATGTGGAGGAAGCG-3′. The mutation was confirmed by sequencing.
RAW 264.7 cell culture, transfection, and luciferase assay.
Murine macrophage cell line RAW 264.7 was grown in Dulbecco's modified Eagle's medium (GIBCO BRL) supplemented with 20 mM HEPES (Fisher Scientific), 10% fetal bovine serum (GIBCO BRL), 100 U of penicillin (Bio Whittaker) per ml, and 100 μg of streptomycin (Bio Whittaker) per ml. RAW 264.7 cells were plated onto 96-well plates at a concentration of 3 × 104 cells/well for 24 h prior to transfection. Transfection was performed by using FuGene6 (Boehringer-Mannheim) according to the manufacturer's protocol. Each well was cotransfected with the wild-type (TNF1200-pGL3-luc) or CRE mutant TNF-α promoter (CREmt TNF1200-pGL3-luc) firefly luciferase reporter or the basic firefly luciferase reporter without the TNF-α promoter (Basic-pGL3-luc). The Renilla luciferase reporter (phRL-SV40) was used as a transfection control. After 6 h of transfection, the cell medium was replaced with fresh medium, and 24 h later, the cells were infected with mycobacteria. The plates were kept on ice for 10 min after the bacteria were added and then incubated at 37°C in 5% CO2 for various times. For the times longer than 4 h, the cells were incubated with the mycobacteria for 4 h, washed with PBS, and then incubated with fresh medium for the additional time. Twenty-four hours after infection or treatment, the luciferase assay was performed by using the manufacturer's protocols (dual luciferase reporter assay system; Promega). The firefly luciferase activities were corrected for Renilla luciferase activity and normalized to the activities of cells transfected with Basic-pGL3-luc.
Western blot analysis.
At designated times, the treated BMMφ were removed from the incubator and placed on ice. The culture medium was collected and saved for subsequent enzyme-linked immunosorbent assays (ELISAs), and the cells were washed three times with ice-cold PBS containing 1 mM pervanadate. The cells were then treated for 5 to 10 min with ice-cold lysis buffer (150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 1 mM pervanadate, 1 mM EDTA, 1% Igepal, 0.25% deoxycholic acid, 1 mM NaF, 50 mM Tris-HCl; pH 7.4). The cell lysates were removed from the plates and stored at −20°C. Equal amounts of protein, as determined by using the Micro BCA protein assay (Pierce, Rockford, Ill.), were loaded onto sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels, electrophoresed, and transferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The membranes were blocked in Tris-buffered saline with 0.05% Tween 20 (TBST) supplemented with 5% powdered milk and then incubated with primary antibodies against phospho-p38, total p38, phospho-ERK 1/2, or total ERK 1/2 obtained from Cell Signaling. The blots were washed with TBST and incubated with a secondary antibody, either horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin (Pierce) in TBST containing 5% powdered milk. The bound antibodies were detected by using SuperSignal West Femto enhanced chemiluminescence reagents (Pierce). Densitometry was performed for some blots by using an LKB Bromma Ultroscan XL enhanced laser densitometer with the GelScan XL software (Pharmacia LKB Biotechnology, Uppsala, Sweden).
Nuclear extraction and electrophoretic mobility shift assays.
After stimulation, nuclear extracts were prepared by using the NE-PER nuclear and cytoplasmic extraction reagent protocol (Pierce). The protein concentration was determined by the BCA assay as described above for the Western blots.
Electrophoretic mobility shift assay (EMSA) probes were created by first biotinylating the 3′ end of the single-stranded oligonucleotides by using a biotin 3′ end DNA labeling kit (Pierce) according to the manufacturer's protocol. The biotinylated oligonucleotides were annealed by boiling them for 1 min and then slowly cooling them to room temperature for 1 h. The consensus nucleotide sequence for CREB was 5′-AGAGATTGCCTGACGTCAGACAGCTAG-3′.
The EMSA binding reactions were performed by utilizing a LightShift chemiluminescent EMSA kit (Pierce). Specifically, 3 μg of nuclear extract was incubated in 1× binding buffer containing 2.5% glycerol, 50 mM KCl, 5 mM MgCl2, 5 mM EDTA, and biotinylated probe with or without a 200-fold excess of unlabeled probe for 30 min at room temperature. The complexes were separated on a 6% polyacrylamide-0.5× Tris-borate-EDTA gel.
Protein phosphatase assay.
The activities of serine/threonine phosphatases were measured with a nonradioactive, malachite green-based phosphatase assay kit (Promega). The reaction buffer and the substrate used in this assay were designed to specifically detect the activity of PP2A phosphatase. The assay was performed according to the manufacturer's protocol. Briefly, 3.5 × 105 BMMφ were plated on 35-mm TC plates. The cells were treated with OA and infected as described above. The proteins were then extracted with modified radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml), the protein concentration was determined by the BCA assay, and then the preparation was transferred to phosphate-free PP2A buffer (250 mM imidazole [pH 7.2], 1 mM EGTA, 0.1% β-mercaptoethanol, 0.5 mg of BSA per ml) on Sephadex G-25 columns. Approximately 10 μg of total cellular protein was incubated in the presence of the substrate RRA(pT)VA for 20 min. The amount of phosphate released was calculated from a standard curve according to the manufacturer's instructions.
ELISA.
The levels of TNF-α secreted into the culture medium by infected macrophages were measured by using an OptEia mouse TNF-α ELISA kit (PharMingen, San Diego, Calif.). Culture medium collected from the macrophages was analyzed for cytokines according to manufacturer's instructions, and the cytokine concentrations were determined by using TNF-α standard curves.
Statistical analysis.
Statistical significance was determined by the paired two-tailed Student t test and a one-way analysis of variance at a level of significance (P value) of <0.05 by using the InStat/Prism software.
RESULTS
Significant accumulation of phosphorylated CREB in the nuclei of macrophages at 1 h postinfection with mycobacteria.
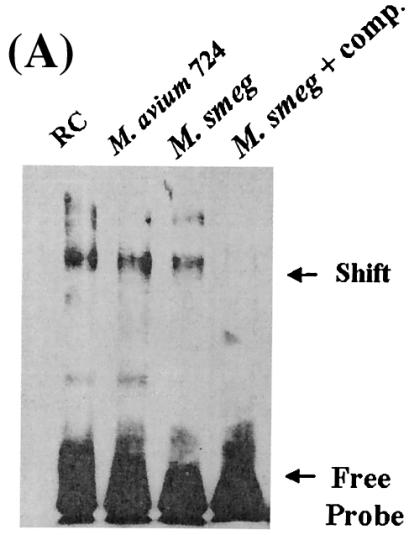
Previous studies showed that the p38, ERK 1/2, and PKA pathways play a role in the regulation of TNF-α production following M. smegmatis infection (33). We were interested in identifying the target(s) of these pathways at the level of transcription and therefore focused on CREB, a well-known substrate for PKA and MAPKs. To determine if CREB was capable of binding a CREB consensus sequence following a mycobacterial infection, we infected BMMφ with the pathogenic M. avium 724 strain and the nonpathogenic M. smegmatis for 1 h and evaluated its DNA binding activity by using an electrophoretic mobility shift assay. As shown in Fig. 1A, nuclear extracts from uninfected resting cells (RC), as well as mycobacterium-infected BMMφ, exhibited binding to the CRE oligonucleotide. To show specificity for the consensus sequence, the binding activity was competed out with unlabeled CRE oligonucleotide. These experiments were repeated with RAW 264.7 cells, and the results were comparable (data not shown).
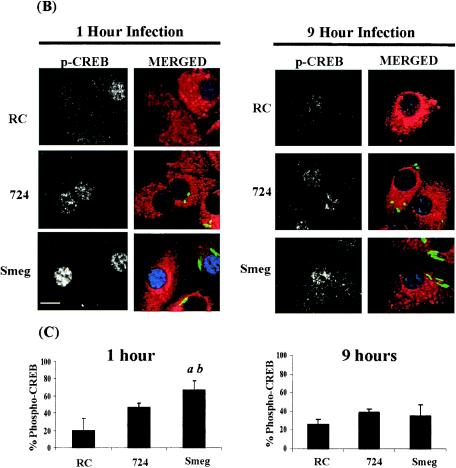
FIG. 1.
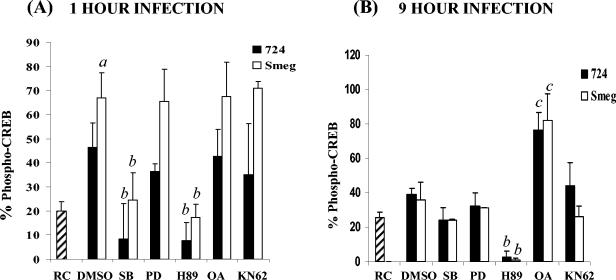
Increased CREB phosphorylation in M. smegmatis-infected BMMφ compared to the CREB phosphorylation in M. avium 724-infected BMMφ at 1 h postinfection. (A) EMSA following 1 h of infection with or without a 200-fold excess of cold competitor CRE oligonucleotide. (B) Macrophages infected for 1 or 9 h with either FITC-labeled M. avium 724 or FITC-labeled M. smegmatis as described in Materials and Methods and then fixed and stained for phosphorylated CREB. Cells were labeled with anti-phosphorylated CREB antibody (blue) and anti-lysosome-associated membrane protein 1 antibody (red) and then examined by confocal microscopy. The greyscale images for phosphorylated CREB labeling are shown on the left, and the merged images are shown on the right. Bar = 1 μm. (C) Cells with phosphorylated CREB in their nuclei were quantitated (a minimum of 100 cells were counted per condition) and compared to the RC control. The results are representative of at least four separate experiments. a, significant compared to RC (P < 0.001); b, significant compared to M. avium 724 (P < 0.05). p-CREB, phosphorylated CREB; Smeg, M. smegmatis.
To investigate whether there were differences in CREB activation, we infected BMMφ with M. avium 724 or M. smegmatis for 1 h and assayed for phosphorylated CREB. As shown in Fig. 1B, there was an increase in the level of phospho-CREB in the nuclei of Mycobacterium-infected BMMφ compared to the level in RC. We also noted that the relative level of phospho-CREB in the nuclei of FITC-labeled M. smegmatis-infected BMMφ was greater than the level observed for M. avium 724-infected cells, as shown by the increased intensity of phosphorylated CREB in the nucleus. We determined the percentage of cells containing nuclear phospho-CREB for RC and infected BMMφ (Fig. 1C). Approximately 20% of the RC had phospho-CREB. However, only M. smegmatis induced significantly more phosphorylated CREB than RC, and 70% of the cells contained nuclear phosphorylated CREB. This level was also significantly higher than the level observed in M. avium 724-infected cells (50%).
In previous studies, it was shown that M. smegmatis induced a dramatic increase in intracellular cAMP levels at 6 to 9 h postinfection. In contrast, in M. avium 724-infected macrophages the cAMP levels remained comparable to those in RC (33). Although CREB is generally known as an early response transcription factor (18), we hypothesized that the increased cAMP level might act on CREB at later times and thus increase TNF-α production. We therefore looked at CREB activation following 9-h M. smegmatis and M. avium 724 infections. As shown in Fig. 1B, there was significantly less activated CREB in the mycobacterium-infected cells at 9 h than at 1 h postinfection. We again determined the percentage of cells with nuclear phosphorylated CREB and found no significant difference between mycobacterium-infected cells and RC (Fig. 1C).
Mutation of the CRE site in the TNF-α promoter inhibits M. smegmatis-induced promoter activity in macrophages.
It was demonstrated previously that BMMφ infected with M. smegmatis produce significantly more TNF-α than M. avium 724-infected cells (24, 33). In the present study, we examined the effects of mycobacterial infections on the transcription of TNF-α. For these experiments, we transfected RAW 264.7 cells with a luciferase reporter containing the mouse (BALB/c) TNF-α(−1200 bp) promoter. Like bone marrow-derived macrophages, RAW 264.7 cells infected with M. smegmatis showed increased TNF-α production compared to M. avium 724-infected cells (Fig. 2A). Consistent with the production of TNF-α, RAW 264.7 cells infected with M. smegmatis showed higher firefly luciferase activity than cells infected with M. avium (Fig. 2B).
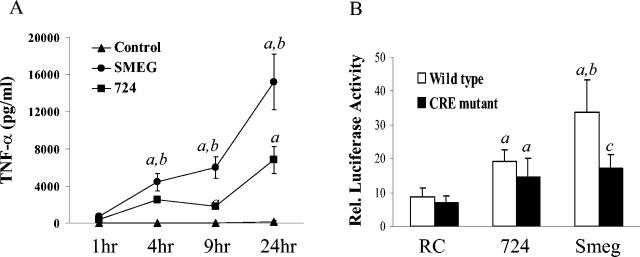
FIG. 2.
CRE site on the TNF-α promoter is required for the increased luciferase activity observed in M. smegmatis-infected RAW 264.7 cells. (A) RAW 264.7 cells were infected with mycobacteria, and supernatants collected after 1, 4, 9, and 24 h were used for TNF-α ELISA. (B) RAW 264.7 cells that were transfected with either Basic-pGL3-luc, TNF 1200-pGL3-luc, or CREmt TNF1200-pGL3-luc in combination with phRL-SV40-luc were infected with mycobacteria as described in Materials and Methods. Twenty-four hours after the mycobacterial infections, cell extracts were assayed by using a dual luciferase assay. The results are the ratio of firefly luciferase activity to Renilla luciferase activity and were normalized to the results for cells transfected with Basic-pGL3-luc. The data shown are representative of the results of at least two separate experiments. a, significantly higher than RC levels (P < 0.05); b, significant difference between M. smegmatis and M. avium 724 (P < 0.05); c, significant difference between wild type and CRE mutant (P < 0.05). Smeg and SMEG, M. smegmatis.
To confirm the importance of the CRE site for TNF-α promoter activity, we introduced a 2-bp mutation into this CRE site. The resulting CRE mutant was previously confirmed not to bind to transcription factors ATF-2, CREB, and AP-1 (23). As shown in Fig. 2B, the mutation of the CRE site significantly reduced M. smegmatis-induced luciferase expression but not M. avium-induced luciferase expression. Taken together, these results indicate that M. avium 724 and M. smegmatis differentially regulate the expression of TNF-α at the transcriptional level and that the CRE site promotes M. smegmatis-initiated expression of TNF-α.
Elevated phosphatase activity in M. avium 724-infected BMMφ compared to that in M. smegmatis-infected cells.
The lower levels of CREB activation seen in M. avium 724-infected BMMφ, combined with previous data showing all-around lower levels of MAPK, PKA, and CaM-CaMKII activities in these infected BMMφ, suggest either that M. smegmatis activates BMMφ to a greater extent or that M. avium 724 actively down-regulates the macrophage response or that there is a combination of both. A potential mechanism for down-regulation is through the activation of MAPK-specific phosphatases. The MAPKs are inactivated through dephosphorylation of either the Tyr or Thr residue, and many phosphatase families have been shown to function in this capacity (28).
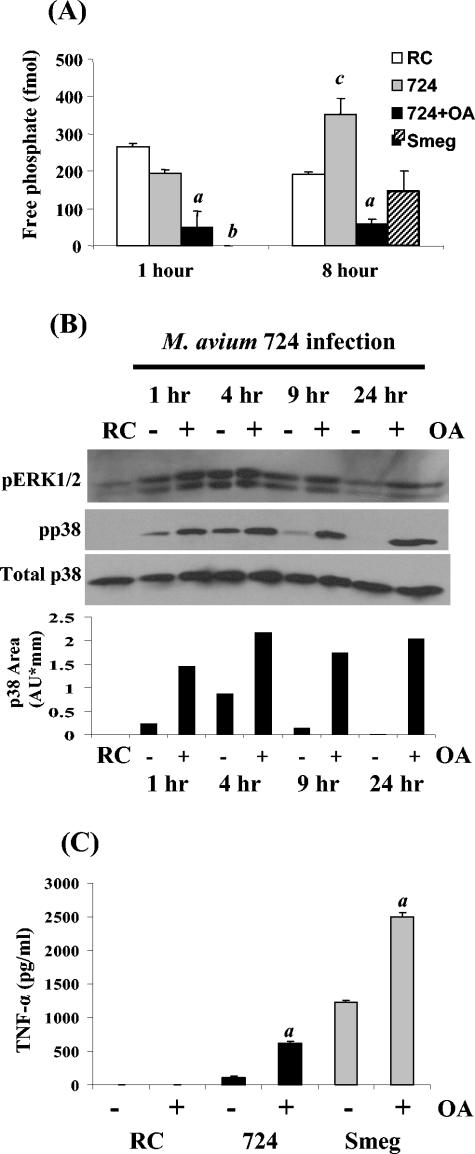
A phosphatase that is an important negative regulator of a macrophage inflammatory response and MAPK activation is the serine/threonine phosphatase PP2A (21). Therefore, we measured PP2A activity in infected BMMφ. As shown in Fig. 3A, M. avium 724 maintained levels of PP2A activity that were near the RC levels at 1 h postinfection. In contrast, no PP2A activity was detected in M. smegmatis-infected BMMφ at this time. After 8 h, M. avium 724-infected BMMφ had significantly more phosphatase activity than RC or M. smegmatis-infected cells, indicating an active process by which M. avium 724 suppresses macrophage activation. To show the specificity of the assay, treatment of M. avium 724-infected BMMφ with the inhibitor OA at concentrations that inhibit the serine/threonine-specific PP2A (3, 9) led to a significant decrease in phosphatase activity (Fig. 3A).
FIG. 3.
M. avium 724 infection promotes BMMφ PP2A activity, leading to decreased MAPK phosphorylation and TNF-α production. (A) Macrophages that were treated with 10 nM OA or the DMSO vehicle control 1 h prior to M. avium 724 or M. smegmatis infection were harvested at 1 and 8 h postinfection. Cell lysates were obtained and screened for phosphatase activity as described in Materials and Methods. (B) Macrophages were treated with OA (+) or the DMSO vehicle control (−) for 1 h prior to infection. Cell lysates, obtained at 1, 4, 9, and 24 h postinfection, were probed for activated ERK 1/2 and p38. Total p38 blots were electrophoresed to show equal protein loading, and the relative densities of the p38 bands were analyzed by densitometry. (C) Supernatants from BMMφ 9 h postinfection were analyzed for the presence of TNF-α by ELISA. The data are representative of the results of three separate experiments. a, significantly different from no OA treatment (P < 0.05); b, significantly lower than M. avium 724 and RC levels (P < 0.05); c, significantly higher than RC levels (P < 0.05). Smeg, M. smegmatis; AU, arbitrary units.
To determine if PP2A was regulating the MAPKs p38 and ERK 1/2, we treated M. avium 724-infected cells with OA. Figure 3B shows an increased p38 activation at all times and a dramatic increase at 9 and 24 h in OA-treated cells (as shown by densitometry). Likewise, in OA-treated macrophages, ERK 1/2 activation was increased significantly at 24 h postinfection. Moreover, TNF-α production was increased sixfold in M. avium 724-infected BMMφ following inhibitor treatment (Fig. 3C).
Treatment of M. smegmatis-infected BMMφ with OA also caused increases in p38 phosphorylation at 9 h (data not shown) and increased TNF-α levels twofold over untreated control levels (Fig. 3C). These data indicate that PP2A plays a major role in regulating TNF-α production in macrophages and that pathogenic M. avium 724 significantly up-regulates its activity.
CREB activation following mycobacterial infection is p38 and PKA dependent.
Our data indicate that M. avium 724 suppresses MAPK activation and TNF-α production through increased activation of PP2A. We were therefore interested in determining whether the diminished CREB activation observed in M. avium 724-infected BMMφ was a result of increased PP2A activity. We were also interested in defining the signaling pathways upstream of CREB activation. The inhibitors SB203580, PD98059, H89, and KN62 were used to inhibit the p38, MEK1/2, PKA, and CaMKII pathways, respectively. The BMMφ were also treated with OA to inhibit PP2A. Using the quantitative method described above, we found that at 1 h postinfection, CREB activation was p38 and PKA dependent in both M. smegmatis- and M. avium 724-infected BMMφ. However, inhibition of the ERK 1/2 (by blocking MEK1/2 activity) or CaMKII pathway had no effect on the level of CREB activation (Fig. 4A). This is in contrast to the TNF-α production seen after treatment with the inhibitors, as it has been shown that inhibition of p38, MEK1/2, PKA, and CaMKII leads to decreased TNF-α production (24, 33), providing further evidence that many signal transduction pathways converge to induce TNF-α transcription. This also suggests that although ERK 1/2 does not play a role in CREB activation, it likely activates other components of the transcription machinery that are important for full TNF-α production.
FIG. 4.
CREB phosphorylation is PKA and p38 dependent following mycobacterial infection. Macrophages were treated with SB203580 (SB) (p38 inhibitor), PD98059 (PD) (MEK1 inhibitor), H89 (PKA inhibitor), OA (PP2A inhibitor), KN62 (CaMKII inhibitor), or the DMSO vehicle control for 1 h prior to infection with M. avium 724 or M. smegmatis. Cells were infected for 1 h (A) or 9 h (B) as described in Materials and Methods. Cells were fixed, stained, and visualized by confocal microscopy as described in the legend to Fig. 1, and the number of cells containing phosphorylated CREB in the nucleus was determined. The data are representative of the results of three separate experiments. a, significantly greater than M. avium 724 and RC (P < 0.05); b, significantly less than mycobacteria and DMSO control (P < 0.05); c, significantly greater than mycobacteria and DMSO control (P < 0.05). Smeg, M. smegmatis.
OA treatment also had no effect on CREB phosphorylation and a very limited effect on p38 phosphorylation at 1 h postinfection, indicating that although PP2A is important in TNF-α production, it does not regulate CREB at this early time. However, by 9 h, the level of CREB phosphorylation was dramatically increased following OA treatment (Fig. 4B), suggesting that PP2A is important in regulating the RC level of CREB activation and in maintaining CREB mostly in its inactivated, dephosphorylated form.
DISCUSSION
Our previous studies indicated that BMMφ infected with nonpathogenic mycobacteria produce significantly elevated levels of proinflammatory mediators, such as TNF-α, compared to the levels observed in cells infected with the pathogenic M. avium. This was due to differential activation of various signaling pathways, including the MAPKs p38 and ERK 1/2, as well as PKA. However, the link between the activation of these pathways and the transcriptional response was not defined in these experiments. The present study began to bridge this information gap.
Clearly, TNF-α production is highly regulated. The TNF-α promoter has binding sites for a number of different transcription factors which, along with other regulator proteins, such as p300/CBP, form an enhancesome that dictates the transcriptional response. Of the various transcription factors that bind to the TNF-α promoter, we focused on CREB since its activation has been shown to be regulated by PKA and the MAPKs. Indeed, we found that phosphorylated CREB was present at increased levels in M. smegmatis-infected BMMφ compared to RC or M. avium-infected cells, which correlated with the results of our signaling studies. Not surprisingly, PKA was required for the phosphorylation of Ser133, a site previously shown to be necessary for CREB activation (16). The MAPK p38 was also found to be important in this response. Inhibiting either kinase resulted in a significant decrease in CREB phosphorylation. This also explains why the level of CREB phosphorylation was only minimally above RC levels in M. avium-infected BMMφ since previous studies indicated only limited production of cAMP in M. avium-infected BMMφ (33). Therefore, the robust early p38 activation observed in BMMφ infected with M. avium (24) in itself is not sufficient to induce CREB activation.
Our data show that the M. smegmatis-induced CREB phosphorylation occurs within 1 h after infection but that the levels return to the RC levels by 9 h postinfection. This also is consistent with the hypothesis that CREB activation is an early response transcription factor. However, the CREB binding site (i.e., CRE) on the TNF-α promoter is clearly important in the transcriptional response to M. smegmatis infection since mutation of this binding site resulted in a significant decrease in promoter activity. The minimal CREB activation observed at 9 h was unexpected since previous studies indicated that M. smegmatis induces high levels of cAMP at 6 to 9 h postinfection, and we hypothesized that this cAMP, via PKA, acts on CREB. This suggests that the elevated levels of cAMP in M. smegmatis-infected BMMφ do not activate PKA (and therefore only the basal level of PKA activates CREB at 9 h) or that the PKA activated specifically during an M. smegmatis infection is not available to phosphorylate CREB. We favor the former hypothesis. Although PKA is traditionally thought of as the target of cAMP, recently other cAMP-binding proteins, particularly a family of guanine nucleotide exchange factors, have been identified (7). These guanine nucleotide exchange factors have been designated Epac 1 and Epac 2, and they selectively activate Rap1 in a cAMP-dependent but PKA-independent manner (12). Future experiments should determine if M. smegmatis infection induces increased Rap1 activity at later times postinfection. Macrophages infected with M. avium, however, do not show elevated levels of cAMP at later times and likely have only minimal Rap1 activity.
Based on our data, we hypothesize that the early CREB activation observed in M. smegmatis-infected BMMφ is important not only in the initial transcription from the TNF-α promoter but also for the subsequent promoter activity. We suggest that the initial CREB binding is important in recruiting other components, such as p300/CBP, which are known to bind CREB and also to stabilize transcriptional complexes (20). In contrast, M. avium infection leads to nominal TNF-α promoter activity, in part due to minimal CREB activation but mostly due to limited MAPK activation observed at later times postinfection. A diagram encompassing our data and predictions is shown in Fig. 5.
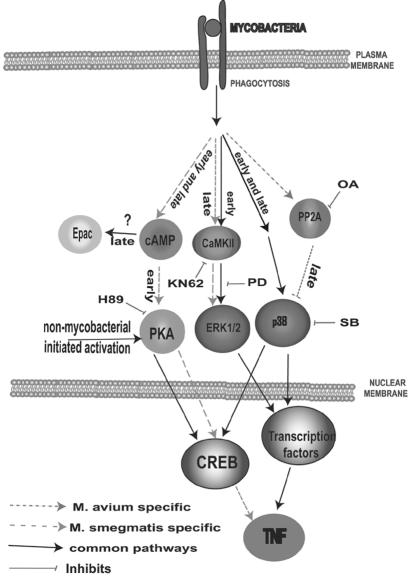
FIG. 5.
Diagram of the signaling pathways that lead to CREB activation and TNF-α production. Early cAMP production and PKA activation following an M. smegmatis infection leads to CREB phosphorylation. This also requires the early activation of p38. Later production of cAMP associated with an M. smegmatis infection does not lead to CREB phosphorylation, and we hypothesize that this cAMP does not work through PKA but works through other cAMP targets (e.g., the GTPase exchange factor Epac). The limited activation of CREB, which is not mycobacterium specific, functions only through PKA (observed 9 h postinfection). CREB activation promotes M. smegmatis-induced TNF-α production but not M. avium-induced TNF-α production. Both M. smegmatis and M. avium infections lead to early calmodulin kinase (CaMKII)-dependent activation of ERK 1/2, as well as activation of p38. However, at later times postinfection, calmodulin kinase-dependent ERK 1/2 activation is specific to M. smegmatis-infected BMMφ. In contrast, the limited p38 activation associated with M. avium-infected macrophages results from increased activation of the Ser/Thr phosphatase PP2A. The prolonged activation of the MAPKs associated with M. smegmatis-infected BMMφ leads to increased TNF-α production through as-yet-undefined transcription factors. SB203580 (SB) is a p38 inhibitor; PD98059 (PD) is an MEK1 inhibitor; H89 is a PKA inhibitor; OA is a PP2A inhibitor; and KN62 is a CaMKII inhibitor.
Our results which indicate that there is increased PP2A phosphatase activity in M. avium-infected macrophages provide, at least in part, a mechanism to explain the differences in MAPK activities observed between M. smegmatis-infected BMMφ and M. avium-infected BMMφ. The observation that p38 activity in M. avium-infected BMMφ was elevated by OA treatment to levels that approximate those induced by M. smegmatis suggests that PP2A is a major factor in down-regulating p38 activity following mycobacterial infection. In turn, the elevated p38 activity in OA-treated cells results in increased TNF-α production. This is consistent with previous studies which showed a 50% reduction in TNF-α production in mycobacterium-infected BMMφ following treatment with a p38-specific inhibitor (24). Based on the present studies and previous work (24, 33), we suggest that the differential activation of the MAPKs in M. smegmatis-infected BMMφ and M. avium-infected BMMφ stems from two different effects. First, as mentioned above, increased PP2A activation in M. avium-infected macrophages results in down-regulation of p38 activation. Second, M. avium infection does not induce calmodulin kinase activity and cAMP production, which is required for the prolonged ERK 1/2 activation observed in M. smegmatis-infected macrophages (33). The early cAMP activation and subsequent PKA activation following an M. smegmatis infection are also responsible for the increased CREB phosphorylation observed in the infected BMMφ. The result of these signaling processes is activation of CREB and other, as-yet-undefined transcription factors, which leads to increased TNF-α production. Future studies will focus on defining the additional transcription factors which regulate TNF-α production and identifying other immune modulators which are affected by the differential activation of the macrophage signaling responses upon mycobacterial infection.
Acknowledgments
This work was funded by grants AI52439 and AI56979 to J.S.S. from the National Institutes of Health. S.K.R. was funded through a predoctoral fellowship from the Walther Cancer Institute, Indianapolis, Ind.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Bekker, L. G., S. Freeman, P. J. Murray, B. Ryffel, and G. Kaplan. 2001. TNF-alpha controls intracellular mycobacterial growth by both inducible nitric oxide synthase-dependent and inducible nitric oxide synthase-independent pathways. J. Immunol. 166:6728-6734. [DOI] [PubMed] [Google Scholar]
- 2.Beltan, E., L. Horgen, and N. Rastogi. 2000. Secretion of cytokines by human macrophages upon infection by pathogenic and non-pathogenic mycobacteria. Microb. Pathog. 28:313-318. [DOI] [PubMed] [Google Scholar]
- 3.Bialojan, C., and A. Takai. 1988. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 256:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohlson, S. S., J. A. Strasser, J. J. Bower, and J. S. Schorey. 2001. Role of complement in Mycobacterium avium pathogenesis: in vivo and in vitro analyses of the host response to infection in the absence of complement component C3. Infect. Immun. 69:7729-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshans, R. L., S. Szanto, L. van Aelst, and C. D'Souza-Schorey. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20:3685-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. May1999, posting date. You can prevent MAC (disseminated Mycobacterium avium complex disease): a guide for people with HIV infection. [Online.] Centers for Disease Control and Prevention, Atlanta, Ga. http://www.cdc.gov/hiv/pubs/brochure/oi_mac.htm.
- 7.Chin, K. V., W. L. Yang, R. Ravatn, T. Kita, E. Reitman, D. Vettori, M. E. Cvijic, M. Shin, and L. Iacono. 2002. Reinventing the wheel of cyclic AMP: novel mechanisms of cAMP signaling. Ann. N. Y. Acad. Sci. 968:49-64. [DOI] [PubMed] [Google Scholar]
- 8.Cho, M. K., Y. H. Cho, G. H. Lee, and S. G. Kim. 2004. Induction of cyclooxygenase-2 by bovine type I collagen in macrophages via C/EBP and CREB activation by multiple cell signaling pathways. Biochem. Pharmacol. 67:2239-2250. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, P., S. Klumpp, and D. L. Schelling. 1989. An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. FEBS Lett. 250:596-600. [DOI] [PubMed] [Google Scholar]
- 10.Cuschieri, J., D. Gourlay, I. Garcia, S. Jelacic, and R. V. Maier. 2003. Modulation of endotoxin-induced endothelial function by calcium/calmodulin-dependent protein kinase. Shock 20:176-182 [DOI] [PubMed] [Google Scholar]
- 11.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Rooij, J., F. J. Zwartkruis, M. H. Verheijen, R. H. Cool, S. M. Nijman, A. Wittinghofer, and J. L. Bos. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396:474-477. [DOI] [PubMed] [Google Scholar]
- 13.Du, K., and M. Montminy. 1998. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273:32377-32379. [DOI] [PubMed] [Google Scholar]
- 14.Falcone, V., E. B. Bassey, A. Toniolo, P. G. Conaldi, and F. M. Collins. 1994. Differential release of tumor necrosis factor-alpha from murine peritoneal macrophages stimulated with virulent and avirulent species of mycobacteria. FEMS Immunol. Med. Microbiol. 8:225-232. [DOI] [PubMed] [Google Scholar]
- 15.Falvo, J. V., A. M. Uglialoro, B. M. Brinkman, M. Merika, B. S. Parekh, E. Y. Tsai, H. C. King, A. D. Morielli, E. G. Peralta, T. Maniatis, D. Thanos, and A. E. Goldfeld. 2000. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell. Biol. 20:2239-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, G. A., and M. R. Montminy. 1989. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675-680. [DOI] [PubMed] [Google Scholar]
- 17.Iwahashi, H., A. Takeshita, and S. Hanazawa. 2000. Prostaglandin E2 stimulates AP-1-mediated CD14 expression in mouse macrophages via cyclic AMP-dependent protein kinase A. J. Immunol. 164:5403-5408. [DOI] [PubMed] [Google Scholar]
- 18.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 19.Mayr, B. M., G. Canettieri, and M. R. Montminy. 2001. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc. Natl. Acad. Sci. USA 98:10936-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McManus, K. J., and M. J. Hendzel. 2001. CBP, a transcriptional coactivator and acetyltransferase. Biochem. Cell Biol. 79:253-266. [PubMed] [Google Scholar]
- 21.Miskolci, V., S. Castro-Alcaraz, P. Nguyen, A. Vancura, D. Davidson, and I. Vancurova. 2003. Okadaic acid induces sustained activation of NFkappaB and degradation of the nuclear IkappaBalpha in human neutrophils. Arch. Biochem. Biophys. 417:44-52. [DOI] [PubMed] [Google Scholar]
- 22.Nemeth, Z. H., S. J. Leibovich, E. A. Deitch, B. Sperlagh, L. Virag, E. S. Vizi, C. Szabo, and G. Hasko. 2003. Adenosine stimulates CREB activation in macrophages via a p38 MAPK-mediated mechanism. Biochem. Biophys. Res. Commun. 312:883-888. [DOI] [PubMed] [Google Scholar]
- 23.Newell, C. L., A. B. Deisseroth, and G. Lopez-Berestein. 1994. Interaction of nuclear proteins with an AP-1/CRE-like promoter sequence in the human TNF-alpha gene. J. Leukoc. Biol. 56:27-35. [DOI] [PubMed] [Google Scholar]
- 24.Roach, S. K., and J. S. Schorey. 2002. Differential regulation of the mitogen-activated protein kinases by pathogenic and nonpathogenic mycobacteria. Infect. Immun. 70:3040-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]
- 26.Sun, P., H. Enslen, P. S. Myung, and R. A. Maurer. 1994. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 8:2527-2539. [DOI] [PubMed] [Google Scholar]
- 27.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 28.Theodosiou, A., and A. Ashworth. 2002. MAP kinase phosphatases. Genome Biol. 3(7):REVIEWS 3009.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai, E. Y., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai, E. Y., J. Jain, P. A. Pesavento, A. Rao, and A. E. Goldfeld. 1996. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 16:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vankayalapati, R., B. Wizel, B. Samten, D. E. Griffith, H. Shams, M. R. Galland, C. F. Von Reyn, W. M. Girard, R. J. Wallace, Jr., and P. F. Barnes. 2001. Cytokine profiles in immunocompetent persons infected with Mycobacterium avium complex. J. Infect. Dis. 183:478-484. [DOI] [PubMed] [Google Scholar]
- 32.Xing, J., D. D. Ginty, and M. E. Greenberg. 1996. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273:959-963. [DOI] [PubMed] [Google Scholar]
- 33.Yadav, M., S. K. Roach, and J. S. Schorey. 2004. Increased mitogen-activated protein kinase activity and TNF-alpha production associated with Mycobacterium smegmatis- but not Mycobacterium avium-infected macrophages requires prolonged stimulation of the calmodulin/calmodulin kinase and cyclic AMP/protein kinase A pathways. J. Immunol. 172:5588-5597. [DOI] [PubMed] [Google Scholar]