Abstract
Post-transcriptional and post-translational modifications of factors involved in translation are very important for the control and accuracy of protein biosynthesis. Among these factors, tRNAs harbor the largest variety of grafted chemical structures, which participate in tRNA stability or mRNA decoding. Here, we focused on Trm112 protein, which associates with four different eukaryotic methyltransferases modifying tRNAs (Trm9 and Trm11) but also 18S-rRNA (Bud23) and translation termination factor eRF1 (Mtq2). In particular, we have investigated the role of Trm112 in the Trm11–Trm112 complex, which forms 2-methylguanosine at position 10 on several tRNAs and thereby is assumed to stabilize tRNA structure. We show that Trm112 is important for Trm11 enzymatic activity by influencing S-adenosyl-L-methionine binding and by contributing to tRNA binding. Using hydrogen-deuterium eXchange coupled to mass spectrometry, we obtained experimental evidences that the Trm11–Trm112 interaction relies on the same molecular bases as those described for other Trm112–methyltransferases complexes. Hence, all Trm112-dependent methyltransferases compete to interact with this partner.
INTRODUCTION
Protein synthesis relies on the decoding of messenger RNAs (mRNAs) by the ribosome assisted by many protein factors and transfer RNAs (tRNAs) to produce corresponding polypeptides. The tRNAs are central adaptor molecules in this process. First, they recognize the mRNA codon triplet present in the ribosomal A-site through their anticodon loop. Second, they bring the corresponding amino acid attached to their 3΄-CCA end into the ribosomal peptidyl transferase center (PTC) during elongation. The tRNAs are transcribed by RNA polymerase III into precursor molecules, which undergo many maturation steps including removal of leader sequences, addition of CCA at the 3΄ end, splicing and modification of several nucleotides (1). The frequency and the diversity of post-transcriptional modifications in tRNAs is the largest of all RNAs and in some bacteria, it is estimated that 1% of the total genome is involved in tRNA modifications (2). Depending on their location on the tRNA molecules, these modifications are assumed to play different roles. Modifications located in the tRNA anticodon loop are important for efficient and accurate codon recognition (3). In particular, nucleotides at position 34 are very often heavily modified leading to complex chemical structures, which allow a single tRNA molecule to recognize several codons through a process known as ‘wobble pairing’. Some modifications are important for proper recognition of tRNAs by amino-acyl tRNA synthetases (4). Finally, many modifications are located in the tRNA structural core and are considered to confer correct folding and to stabilize the tRNA tertiary structure (5,6). Among tRNA modifications, methylations catalyzed by tRNA methyltransferases (MTase) are by far the most abundant. The majority of tRNA MTases uses S-adenosyl-L-methionine (SAM) as methyl donor group and these enzymes are classified according to their catalytic domain into two groups (7). Class I MTases are characterized by the presence of a Rossmann fold domain to accommodate the SAM, while class IV enzymes adopt a topological-knot structure and are members of the SPOUT (SpoU-TrmD) superfamily (8).
Most of the MTases modifying tRNAs have been identified in bacteria and yeast but much less experimental characterization is available for archaeal and human counterparts. However, growing evidences link tRNA MTases to human diseases such as cancers and metabolic syndromes (9,10). Recently, an increasing number of MTases has been found to exist as multiprotein complexes in eukaryotes. For instance, the m2G10, Cm32, Nm34, mcm5U34, m7G46 and m1A58 modifications are respectively formed by the Trm11-Trm112, Trm7-Trm732, Trm7-Trm734, Trm9-Trm112, Trm8-Trm82 and Trm61-Trm6 holoenzymes in Saccharomyces cerevisiae (the catalytic MTase subunit is underlined; (11–17)). Four of these modifications are found in bacteria where their formation requires only one protein. The role of the partner protein varies between these different enzymes as it can participate in tRNA binding (Trm6, Trm732, Trm734), MTase activation (Trm82) or MTase stability and solubility (Trm82, Trm112). Interestingly, the two modifications (m2G10 and mcm5U34), which are absent in bacteria require the Trm112 protein. The formation of the N2-methylguanosine (m2G), found at position 10 from 18 tRNAs in yeast S. cerevisiae, is catalyzed by the Trm11–Trm112 complex (13) while the Trm9–Trm112 complex catalyzes the methylation of 5-carboxymethyluridine (cm5U) to form 5-methoxycarbonylmethyl-uridine (mcm5U) at position 34 of some tRNAs (15,18). Interestingly, the Trm112 zinc-binding protein interacts with and activates two additional class I MTases, whose substrates are also involved in translation. The Bud23–Trm112 complex catalyzes the N7-methylation of guanosine 1575 of 18S rRNA and is important for efficient ribosomal small subunit synthesis (19–21). The Mtq2-Trm112 methylates translation termination factor eRF1 on the amide group of the glutamine side chain from the universally conserved GGQ motif, which enters into the PTC and triggers the release of newly synthesized proteins (22–24). Trm112 is essential for enzymatic activity of these MTases in eukaryotes by mechanisms that remain largely unknown (15,18,20,21,23,25–27). It has been shown that S. cerevisiae Trm112 solubilizes Mtq2, Trm9 and Bud23 upon-co-expression in Escherichia coli by masking hydrophobic regions from these MTases (15,18,20,21,23,24,28) and stabilizes at least Mtq2 and Bud23 in vivo (20,29). In addition, human Trm112 ortholog (hereafter named TRMT112) was shown to be necessary for HEMK2α (hereafter annotated MTQ2), the human Mtq2 ortholog, to bind SAM (24).
Here, we focused on Trm11 from S. cerevisiae. This protein was shown to require its Trm112 partner to be active in vivo in yeast (13) while two archaeal orthologous proteins from Pyrococcus abyssii (PAB1283 or PabTrm-G10) and Thermococcus kodakarensis (TkoTrm11) catalyzing the formation of m2G prior to m2,2G at position 10, are active on their own (30–32). These proteins adopt a similar domain organization with a predicted N-terminal RNA-binding THUMP domain and a C-terminal SAM-dependent class I MTase domain (13,30,33). THUMP domains are fused or interact with various catalytic domains or subunits involved in the modification of bases present in the tRNA aminoacyl stem or D- and T-arms (34–39). The Trm11 MTase domain harbors the (D/N/S)PP(F/Y/W/H) consensus motif found in many MTases modifying exocyclic amines from bases or glutamine side chains (22,40,41). Using various biochemical and biophysical methods, we investigated the role of Trm112 on the function of Trm11. We show that the tight interaction between Trm112 and Trm11 is mediated by hydrophobic interactions and, using known structures of Trm112-MTase complexes together with hydrogen/deuterium eXchange coupled to mass spectrometry (HDX-MS), we propose a model of the Trm11–Trm112 complex. We also show that Trm112 activates Trm11 by strongly stimulating SAM binding to this protein and provide evidence that Trm112 contributes to tRNA binding by the Trm11–Trm112 enzyme.
MATERIALS AND METHODS
Cloning, mutagenesis, expression and purification of proteins
The YOL124c ORF was amplified by polymerase chain reaction (PCR) from S. cerevisiae (strain S288C) genomic DNA as a template using y-trm11-eag1-poly and y-trm11-not1-poly oligonucleotides (Supplementary Table S1). The PCR product was then cloned into the pKHS vector (42) to yield plasmid pMG526 encoding for Trm11 fused to a C-terminal His6-tag (Supplementary Table S2). Plasmids pMG745, pMG476 and pMG747 encoding C-terminally His6-tagged Trm11 D238A, D286R and D291A mutants, respectively, were obtained by one-step site-directed mutagenesis of pMG526 using oligonucleotides oMG301/oMG302, oMG303/oMG304 and oMG305/oMG306 (Supplementary Tables S1 and 2), respectively, according to Zheng et al. (43). All the oligonucleotides and plasmids used in this study are listed in Supplementary Tables S1 and 2. The plasmid encoding Trm112 R53E mutant fused to a C-terminal His6-tag is a kind gift from Dr V. Heurgué-Hamard.
All proteins or protein complexes were (co-)expressed in E. coli BL21-Gold (DE3) strain in 1 L of auto-inducible terrific broth media (ForMedium AIMTB0260) supplemented with adequate antibiotics, first at 37°C for 3 h then at 15°C overnight. The cells were harvested by centrifugation and resuspended in 30 ml of buffer A (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol). Cell lysis was completed by sonication and clarification was performed by centrifugation. Proteins or protein complexes were first purified on Ni-NTA resin (Qiagen) followed by HiTrap Heparin (GE Healthcare) and finally size-exclusion chromatography using a Superdex75 16/60 column (GE Healthcare) pre-equilibrated in buffer A. For purification of His-tagged Trm112 from S. cerevisiae, the Heparin column was replaced by an anion exchange chromatography (HiTrap Q, GE Healthcare).
In vitro transcription of tRNA
The pILE plasmid used as template for tRNAIle(AAU) (hereafter named tRNAIle) transcription was a generous gift from Dr B.Collinet (I2BC; Univ. Paris-Saclay; Orsay; France). Plasmid pMG755 coding the tRNAIle G10C:C25G mutant, respectively, was obtained from pILE plasmid using oMG318 and oMG319 oligonucleotides. Wild type and G10C:C25G mutant tRNAIle were transcribed from 2 μg of the NdeI linearized pILE and pMG755 plasmids, respectively. PCR products were directly used as templates for transcripts A and B using corresponding oligonucleotides (Supplementary Table S1). Transcription reactions were performed overnight at 37°C with 50U T7 RNA polymerase in 120 mM Hepes-KOH pH 7.6, 20 mM MgCl2, 20 mM DTT, 2 mM spermidine, 5 mM of each nucleotide triphosphate. Transcripts were purified using denaturing urea 10% polyacrylamide gel. tRNAs were visualized by UV shadowing and were extracted with elution buffer (20 mM Tris–HCl pH 7.5, 250 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.1% sodium dodecyl sulphate). Transcripts were precipitated by adding 2.5 volumes of 100% ethanol and resuspended in 8 mM MgCl2. Prior to use, the tRNA substrates were heat-denatured at 65°C for 8 min and annealed at room temperature for 15 min.
In vitro RNA methylation assay
Methylation was performed in 50 μl by mixing 1 pmol of wild-type enzyme with 10 pmol of tRNA in phosphate buffer (50 mM K-Phosphate pH 7.5, 0.1 mM EDTA, 10 mM NH4Cl, 10 mM MgCl2, 2 μM SAM) containing 0.02 μM of [methyl-3H] SAM (Perkin Elmer) at 30°C. Reaction was stopped by precipitation with cold trichloroacetic acid (TCA) 5% and filtered on glass fiber filter (GF/C filter, GE Healthcare). Tritium incorporation was measured by scintillation counter. The initial velocities (Vi) were calculated using the equation CtRNA = Vi*(1-exp(-nt))/n where CtRNA is the concentration of methylated tRNA, t the time in minutes, Vi the initial enzyme cycling velocity and n the relaxation rate constant of Vi by fitting the experimental spots with the software ORIGIN® according to Cao et al. (44).
Steady-state kinetics analyses were performed by using various tRNA concentrations (from 0.2 to 2 μM). The initial rate calculated for each tRNA concentration was plotted as a function of the tRNA concentration. The data were fit to the Michaelis–Menten equation using the software ORIGIN® and the Km, kcat and kcat/Km values were determined from the curve fitting. All these experiments were performed at least in triplicates.
ITC
Isothermal titration calorimetry (ITC) experiments were performed at 10°C or 20°C using an ITC-200 microcalorimeter (MicroCal). All proteins were prepared in 20 mM Tris–HCl pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol. For all titration experiments, 20 injections of 2 μl of titrant (either Trm112 or SAM) were added to the target (either Trm11 or Trm11–Trm112) at intervals of 180 s. The heat of dilution of the titrant was determined from the peaks measured after full saturation of the target by the titrant. A theoretical curve assuming a one-binding site model calculated with the ORIGIN® software gave the best fit to the experimental data. This software uses the relationship between the heat generated by each injection and ΔH (enthalpy change), Ka (association binding constant), n (the number of binding site per monomer), the total target concentration, and the free and total titrant concentrations.
Denaturing and non-denaturing mass spectrometry
Prior to MS analysis, samples were desalted using 0.5 ml Zeba™ Spin Columns (Life Technologies, Pierce Biotechnology, Rockford, IL, USA) in 200 mM ammonium acetate pH 7.5.
For non-denaturing MS analysis, samples were directly injected after desalting, whereas denaturing conditions were obtained by an additional 2-fold dilution with 50% acetonitrile (ACN), 0.1% formic acid. In both cases, analyses were performed on a LCT (Waters, Manchester, UK) coupled to an automated chip-based nano-electrospray source (Triversa Nanomate, Advion Biosciences, Ithaca, NY, USA) and running in positive ion mode. The instrument parameters in denaturing conditions were as follow: cone voltage was set to 30 V, with an interface pressure of 1.5 mbar. In non-denaturing MS conditions, the cone voltage was increased to 120 V and the interface pressure to 6 mbar. The instrument was calibrated with a 2 μM solution of horse heart myoglobin (Sigma-Aldrich) for denaturing MS and with a 2 mg/ml cesium iodide solution in 50% isopropanol for non-denaturing MS. Data analysis was performed with MassLynx 4.1 (Waters, Manchester, UK).
Automated Hydrogen-Deuterium eXchange coupled to mass spectrometry (HDX-MS)
HDX-MS experiments were performed on a Synapt-G2 HDMS (Waters, Manchester, UK) coupled to a Twin HTS PAL dispensing and labeling robot (LEAP Technologies, Carborro, NC, USA) via a NanoAcquity system with HDX technology (Waters, Manchester, UK). Briefly, 3 μl of protein at 60 μM were diluted in 57 μl of protonated (for peptide mapping) or deuterated buffer (20 mM Tris pH 7.5, 200 mM NaCl, 10 μM Zn acetate) and incubated at 20°C for 0, 0.5, 1, 5, 10 and 30 min. A total of 55 μl were then transferred to vials containing 55 μl of pre-cooled quenching solution (100 mM glycine at pH 2.6). After a 30 s quench, 105 μl were injected to a 100 μl loop. Proteins were digested on-line with a 2.1 × 30 mm Poros Immobilized Pepsin column (Life Technologies/Applied Biosystems, Carlsbad, CA, USA). Peptides were desalted for 2 min on a C18 pre-column (Acquity UPLC BEH 1.7 μM, VANGUARD) and separated on a C18 column (Acquity UPLC BEH 1.7 μM, 1.0 × 100 mm) by a linear gradient (2–40% ACN in 7 min). Experiments were run in triplicates and protonated buffer was injected between each sample to wash the column and avoid cross-over contamination.
Peptide identification was performed with ProteinLynx Global Server (PLGS, Waters, Manchester, UK) based on the MSE data acquired on the non-deuterated samples. Peptides were filtered in DynamX 2.0 with the following parameters: peptides identified in at least 3 acquisitions, 0.2 fragments per amino-acid, intensity threshold 1000. Structural representation of the differential uptake was performed using Pymol (DeLano Scientific).
Generation of S. cerevisiae Trm11–Trm112 model
A 3D-structure model for S. cerevisiae Trm11 region encompassing residues 1 to 406 was generated with the I-TASSER server using the coordinates file from archaeal Trm11 from T. kodakarensis, whose crystal structure was recently published (32), together with an alignment between the Trm11 sequences from both organisms (Supplementary Figure S1;(32)). The model accuracy has been assessed using the MetaMQAPII server (45), yielding GDT_TS (for global distance test, total score) value of 47.5 and predicting an overall rmsd of 3.5 Å. The structure of the Trm11–Trm112 model was obtained by superimposing the coordinates of our Trm11 model onto Bud23 in the crystal structure of Bud23–Trm112 complex (21). The coordinates of the S. cerevisiae Trm11 model and of the Trm11–Trm112 model are available as supplementary files.
RESULTS AND DISCUSSION
The S. cerevisiae Trm11–Trm112 complex expressed in E. coli is active
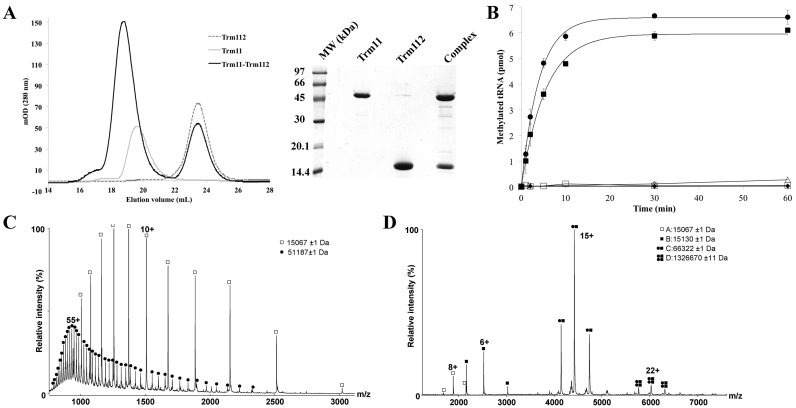
Previous studies have reported that Trm11 or the Trm11–Trm112 complex produced in E. coli were enzymatically inactive while the Trm11–Trm112 obtained from cell-free synthesis using wheat-germ extracts was active (13,46). Furthermore, yeast extracts obtained from either trm11Δ or trm112Δ strains were unable to modify tRNAs in vitro (13). Altogether, this suggested that either other protein components or post-translational modifications were needed for the complex to be active. To characterize the MTase activity of Trm11 more deeply, we first over-expressed a C-terminally His-tagged version of this protein alone in E. coli. Unlike the three other yeast MTases (Mtq2, Trm9 and Bud23) known to interact with Trm112 that are mostly found insoluble when expressed alone (15,20,23), Trm11 is produced as a soluble protein and can be purified to homogeneity and in large quantities following three chromatography steps (Figure 1A). Its tRNA MTase activity was then tested by measuring the transfer of [methyl-3H] from SAM onto an in vitro transcribed yeast tRNAIle(AAU) (hereafter named tRNAIle) but Trm11 proved to be completely inactive alone (Figure 1B). This lack of activity cannot be attributed to protein unfolding as the circular dichroism (CD) spectrum recorded on this protein is typical of well-folded proteins containing α-helices and β-strands (Supplementary Figure S2A). As Trm112 was shown to be necessary for Trm11 activity in vivo (13), we expressed and purified Trm112 in E. coli (Figure 1A). The Trm11 and Trm112 proteins were mixed together (in the presence of a small molar excess of Trm112) and subjected to analytical size-exclusion chromatography. This yielded to a major peak containing both proteins and eluting with a smaller volume than Trm11 or Trm112 alone (Figure 1A). This indicates that Trm11 and Trm112 form a complex in vitro and that Trm11 alone does not form aggregates in solution. Interestingly, the resulting complex proved to be enzymatically active while as expected Trm112 alone was not (Figure 1B). Finally, as the co-expression of human TRMT112 with MTQ2 led to a significantly more active complex than the one reconstituted from isolated proteins (25), we co-expressed C-terminally His-tagged Trm11 together with an untagged version of Trm112 in E. coli. Trm112 was retained in stoichiometric amounts with Trm11 on Ni-NTA resin, indicating that both proteins interact together. This complex was further purified on Heparin HiTrap column and finally on size-exclusion chromatography column without any dissociation, indicating a stable complex. Time-course measurements of [methyl-3H] incorporation revealed that the co-expressed complex is only slightly more active that the reconstituted one (Figure 1B). We further confirmed that the observed enzymatic activity was specific for Trm11 by co-expressing Trm112 together with Trm11 D291A mutant, which was previously shown to abrogate m2G10 formation in vivo (13). The resulting mutant complex was purified similarly to the wild-type and was completely inactive (Table 1). This loss of activity does not appear to result from misfolding as this mutant binds SAM with a similar affinity as the wild-type complex (Table 2 and Supplementary Figure S3). Finally, to confirm that the purified complex modifies tRNAIle at position G10, we synthesized a tRNAIle variant by inverting the G10:C25 base pair into C10:G25 to maintain the tRNA cloverleaf structure. This C10:G25 tRNA variant was not a substrate of the Trm11-Trm112 complex, in agreement with the fact that this holoenzyme specifically modifies G10 into m2G10 (Figure 1B).
Figure 1.
The yeast Trm11–rm112 holoenzyme produced in Escherichia coli is active. (A) Reconstitution of Trm11–Trm112 complex from isolated proteins. Left: Chromatograms obtained following injection of Trm11 (light gray line), Trm112 (dashed line) or a mixture of Trm11 and Trm112 (black line) on a Superdex 75 10/300 column (GE Healthcare). Right: SDS-PAGE (15%) analysis of the protein content of the fraction corresponding to the center of the main peak observed on each chromatogram. (B) Time-course enzymatic activity measurements for Trm112 (filled diamonds), Trm11 (open triangles), reconstituted Trm11–Trm112 complex (filled squares) and co-expressed Trm11–Trm112 complex (filled spheres) on tRNAIle. For all these experiments, 20 nM of enzyme and 200 nM of tRNA substrates were used. The measurements performed with wild-type complex on tRNAIle C10:G25 variant is also shown (open squares). The curves obtained after fitting of the experimental data with equation given in the ‘Materials and Methods’ section are shown by lines. (C) MS analysis of Trm11–Trm112 complex in denaturing conditions (Pi = 1.5 mbar; Vc = 30 V). (D) MS analysis of Trm11–Trm112 complex in non-denaturing conditions (Pi = 6 mbar; Vc = 120 V).
Table 1. Enzymatic analysis of Trm11–Trm112 mutants.
| Mutants | Km for tRNA (μM) a | kcat for tRNA (s−1) a | kcat/Km (μM−1.s−1) |
|---|---|---|---|
| Trm11–Trm112 WT b | 0.159 ± 0.015 | 0.019 ± 0.003 | 0,120 |
| Trm11 D238A–Trm112 b | ND | ND | ND |
| Trm11 D291A–Trm112 b | ND | ND | ND |
| Trm11–Trm112 A106E b | 0.353 ± 0.034 | 0.029 ± 0.006 | 0.082 |
| Trm11–Trm112 E107K c | 0.494 ± 0.053 | 0.0074 ± 0.0005 | 0.015 |
| Trm11–Trm112 I118E d | 1.71 ± 0.16 | 0.0173 ± 0.004 | 0.010 |
| Trm11–Trm112 Y120D d | 1.46 ± 0.19 | 0.0082 ± 0.003 | 0.006 |
aThese values were determined by fitting the data using the Michaelis–Menten equation.
bThe kinetics were performed with 20 nM of enzyme.
cThe kinetics were performed with 30 nM of enzyme.
dThe kinetics were performed with 60 nM of enzyme.
ND: Not Detectable.
Table 2. Summary of thermodynamics parameters of interactions between several Trm11–Trm112 complexes and SAM studied by ITC at 20°C.
| Target a | Kd (μM) | Stoichiometry (n) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|
| Trm11–Trm112 b | 1.9 ± 0.3 | 0.91 | −11.6 | −3.9 |
| Trm11 D238A–Trm112 c | ND | ND | ND | ND |
| Trm11 D291A–Trm112 c | 3.4 ± 0.6 | 1.2 | −8.1 | −0.75 |
| Trm11–Trm112 A106E d | 3.6 ± 0.5 | 1.1 | −9.6 | −2.3 |
| Trm11–Trm112 E107K b | 2.6 ± 0.1 | 1.1 | −11.8 | −4.3 |
| Trm11–Trm112 I118E b | 3.5 ± 0.5 | 1.06 | −13.2 | −5.9 |
| Trm11–Trm112 Y120D b | 2.8 ± 0.3 | 1.11 | −10.8 | −3.4 |
aExperiments performed with Trm11–Trm112 complexes concentrated at 28–36 μM.
bExperiment performed with SAM at a concentration of 400 μM.
cExperiment performed with SAM at a concentration of 257.7 μM.
dExperiment performed with SAM at a concentration of 385.5 μM.
ND: Not Detectable.
Next, we used non-denaturing MS to characterize the oligomeric state of the Trm11-Trm112 complex in solution. First, we performed MS analysis under denaturing conditions (Figure 1C). The molecular weight measured for Trm112 (15067.6 ± 1.2 Da) corresponded precisely to the theoretical one calculated from the gene sequence (15066.2 Da). Two species are observed for Trm11, a minor form (51111.6 ± 2.3 Da) corresponding to the His-tagged Trm11 theoretical molecular weight (51108.8 Da), and a major form (51187.0 ± 1.7 Da) corresponding to the minor form with a β-mercaptoethanol (present in the purification buffer) adduct (+76 Da) on one of the three Trm11 cysteine residues. Second, we analyzed the same complex under non-denaturing conditions and were able to detect four different species (Figure 1D). Species A has a molecular weight of 15067 ± 1 Da and corresponds to Trm112. Species B (15130 ± 1Da, i.e. A+63 Da) is compatible with Trm112 with a zinc atom attached to its Zn-binding domain as observed in the crystal structures of this protein (21,23). Species C (66322 ± 1Da) has a molecular weight close to that expected for a heterodimeric Trm11-Trm112 complex in the presence of zinc (15130 + 51187 i.e. 66317 Da). Finally, the fourth very minor species (132670 ± 11Da) could correspond to a dimer of species C, likely due to experimental artifact (e.g. buffer exchange) as further analyses by SEC-MALLS indicated only the presence of the heterodimer in solution (Supplementary Figure S2B). This analysis also indicates that although Trm112 can exist in two forms when not associated to Trm11 in our non-denaturing MS conditions, i.e. alone or associated with one zinc atom, only the zinc-bound form is found associated with Trm11. This suggests that zinc might be important for Trm11 binding to Trm112, most probably by maintaining the three-dimensional structure of Trm112 zinc-binding domain, which in the other Trm112-MTase complexes is directly interacting with the MTase domains (see below, (21,24,28)).
Altogether, these results indicate that the heterodimeric Trm11-Trm112 complex produced in E. coli is active and that Trm112 is essential for Trm11 MTase activity. This also indicates that both proteins are necessary and sufficient for m2G10 formation on tRNAs and that no post-translational modifications on Trm11-Trm112 complex are required to perform this tRNA modification. In addition, this complex is also active on tRNAs lacking any post-transcriptional modification but we cannot exclude that some tRNA modifications will enhance Trm11-Trm112 catalytic activity.
The Trm112–Trm11 interaction mode is reminiscent of the other Trm112–MTase complexes
The ability to purify Trm11 alone and in large quantities offers for the first time the opportunity to study the interaction mode of Trm112 with one of its MTase partners using biophysical approaches. Indeed, as the three other MTases interacting with Trm112 are largely insoluble in E. coli when expressed alone, this was not possible before.
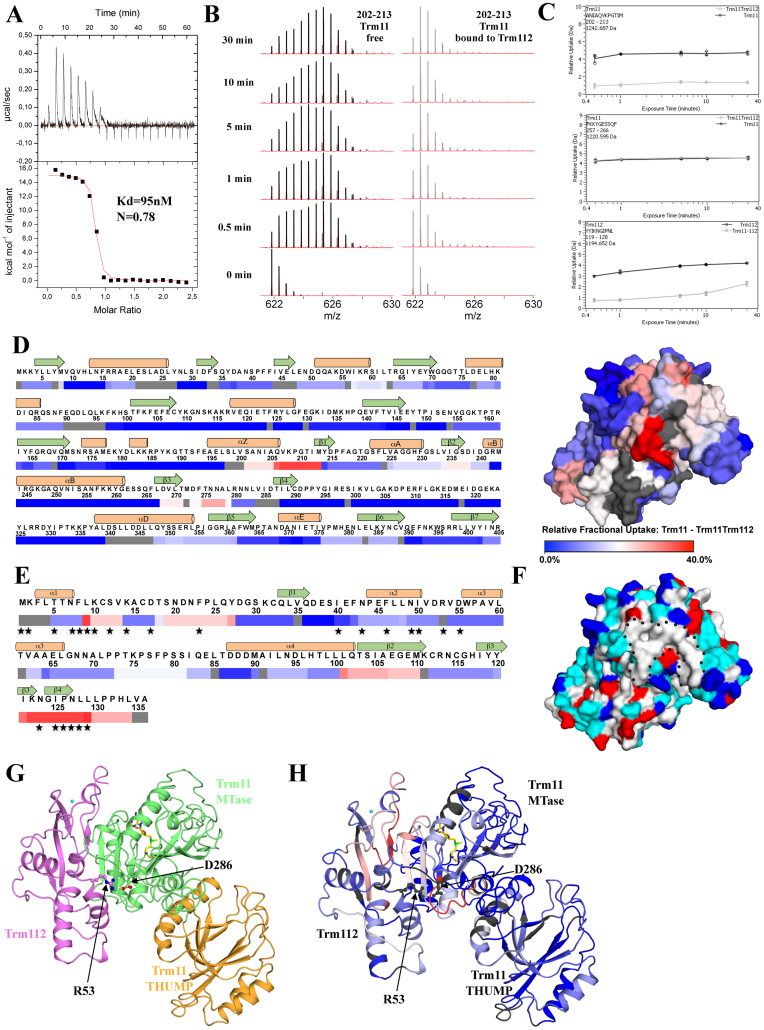
First, using ITC, we characterized the interaction between both proteins. This was performed at 10°C as Trm11 precipitated at 20°C after few minutes. Titration of freshly purified Trm11 by successive addition of Trm112 indicates that the two proteins interact tightly with a measured Kd of 95 nM (Table 3; Figure 2A and Supplementary Figure S4A). Furthermore, the binding is largely endothermic (ΔH = 15.1 kcal/mol) and is entropically driven (TΔS = 24.1 kcal/mol), indicating that the association between both proteins is mainly ruled by hydrophobic interactions. This observation corroborates structural analyses of other Trm112-MTase complexes (21,24,28), showing that Trm112 contacts a large hydrophobic patch at the surface of these MTases.
Table 3. Summary of thermodynamics parameters of interactions studied by ITC at 10°C.
| Target a | Titrant | Kd (μM) | Stoichiometry (n) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|---|
| Trm11 | Trm112 b | 0.095 ± 0.030 | 0.78 | 15.1 | 24.1 |
| Trm11 D286R | Trm112 b | ND | ND | ND | ND |
| Trm11 | Trm112 R53E b | ND | ND | ND | ND |
| Trm11 | SAM c | ND | ND | ND | ND |
| Trm11–Trm112 | SAM c | 1 ± 0.1 | 0.98 | -6.7 | 1.1 |
aExperiments performed with Trm11 or Trm11–Trm112 complexes concentrated at 28–50 μM.
bExperiment performed with Trm112 at a concentration of 391.5–443 μM.
cExperiment performed with SAM at a concentration of 257.7 μM.
ND: Not Detectable.
Figure 2.
Characterization of the interaction between Trm11 and Trm112. (A) ITC of Trm112 binding to Trm11. Upper panel: ITC data obtained by injecting Trm112 into Trm11. Lower panel shows the fit to the binding curve with resulting affinity (Kd) and stoichiometry (N). (B) Hydrogen-Deuterium exchange monitored on the doubly charged peptide 202–213 from Trm11, alone (left, black) or in the presence of Trm112 (right, gray) at 0, 0.5, 1, 5, 10 and 30 min (bottom to the top). (C) HDX kinetics of three representative peptides from Trm11 (top and middle) and Trm112 (bottom). Black circles and gray boxes correspond to data extracted from isolated and complexed forms, respectively. The error bars represent standard deviations from the mean of three replicates. (D) Differential HDX heat map for free Trm11 compared to Trm11 bound to Trm112. Colored from 0% (blue) to 40% (red) difference of deuterium uptake after 5 min (Left). Secondary structure elements as observed in our model of the Trm11 protein structure are shown above the sequence. (Right) Mapping of differential heat map at the surface of the Trm11 model structure using the same color code. Residues in gray were not covered by peptide mapping. (E) Differential HDX heat map for free Trm112 compared to Trm112 bound to Trm11. Colored from 0% (blue) to 40% (red) difference of deuterium uptake after 5 min. Trm112 secondary structure elements as observed in Saccharomyces cerevisiae Bud23-Trm112 crystal structure are shown above the sequence. Trm112 residues that are involved in the interaction with Bud23 MTase are identified by black stars below the sequence. Residues not covered by peptide mapping are shown in gray. (F) Mapping of the distribution of residues at the surface of the model of Trm11 structure. Residues color code: hydrophobic aromatic and aliphatic (G, A, V, L, I, F, Y, W, M and P) in white, positively charged (K and R) in blue, negatively charged (D and E) in red, polar (N, Q, H, S and T) in cyan. Same orientation as panel D. The hydrophobic patch is delineated by black dotted lines. (G) Ribbon representation of the model of the Trm11–Trm112 complex, considering that Trm11 interacts with Trm112 in a similar way as Bud23 (21). Trm112 is colored purple while Trm11 THUMP and MTase domains are colored light brown and green, respectively. The zinc atom bound to Trm112 is shown as a cyan sphere. A SAM molecule bound to the Trm11 MTase domain is shown as yellow sticks. Side chains from Trm11 Asp286 and Trm112 Arg53 are shown as ball and sticks. (H) Mapping of differential HDX heat map (after 5 min) on the ribbon representation of the Trm11–Trm112 complex. Colored from 0% (blue) to 40% (red) difference of deuterium uptake. Same orientation as panel G. Residues not covered by peptide mapping are shown in gray.
Next, we used HDX-MS to identify regions from Trm11 and Trm112 proteins affected upon heterodimer formation. This method allows identifying regions, which either are protected from the solvent upon complex formation or undergo conformational changes resulting in differences in solvent accessibility. We thus compared the deuterium incorporation rates of the Trm11–Trm112 complex to those of the isolated partners. Pepsin digestion generated numerous peptides, of which 96 (Trm11) and 37 (Trm112) could be monitored upon deuteration, resulting in ∼95% sequence coverages for both proteins, with redundancies of 2.89 and 4.29 for Trm11 and Trm112, respectively (Supplementary Figure S5). This redundant set of data highlighted that upon dimer formation, regions encompassing residues 201–213, 225–241 and 268–279 from Trm11 exhibit significantly decreased exchange rates compared to Trm11 alone (Figure 2B–D and Supplementary Figure S6). This indicates that Trm112 interacts mainly with the MTase domain (predicted to start around residue 190) from Trm11. However, we cannot exclude that Trm11 THUMP domain (predicted to encompass approximately residues 1–185) also contacts Trm112. The same analysis highlights residues 8–13, 19–27, 101–110 and 121–134 from Trm112 as exhibiting the most decreased exchange rates in the context of the heterodimer (Figure 2C and E, Supplementary Figure S7). Interestingly, most of these Trm112 regions encompass residues involved in the interaction with Bud23 (21).
In order to map these peptides on Trm11 structure and to propose a model of the Trm11–Trm112 complex, we generated a model of S. cerevisiae Trm11 3D-structure using the recently published crystal structure of archaeal Trm11 from T. kodakarensis (sharing 20% sequence identity and 46.5% sequence similarity with Trm11 (32)), as a template. In this Trm11 model, the peptides from the MTase domain that have been identified by HDX-MS are localized (i) from the end of helix αZ to the beginning of strand β1 (region 201–213), (ii) in the region encompassing the C-terminal half of helix αA, strand β2 and the loop connecting strand β2 to helix αB (region 225–241) and (iii) in strand β3 and the first half of the loop connecting strands β3 and β4 (region 268–279) (Figure 2D). Interestingly, these regions form a hydrophobic patch at the surface of our Trm11 model (Figure 2D and F) and structurally match with those from Mtq2, Bud23 or Trm9 MTases that are involved in the interaction with Trm112 in the other complexes solved to date (21,24,28). This supports a competitive binding mode of all these MTases to the same region of Trm112 and is in agreement with previous observations that Trm11 over-expression in yeast reduces the amount of Trm112 co-immunoprecipitated with Trm9 (47). The superimposition of the Trm11 model onto Bud23 MTase domain in the crystal structure of S. cerevisiae Bud23–Trm112 complex shows that the Trm11 and Trm112 peptides identified by HDX-MS are near each other (Figure 2G and H). Our experimental approach does not allow a precise identification of residues directly involved in the Trm11–Trm112 interface but rather highlights an area surrounding the residues engaged in complex formation. To further validate our model and obtain more precise information on residues involved at the interface, we mutated Asp286 from Trm11 into Arg (D286R) and Arg53 from Trm112 into Glu (R53E). This later mutant was previously shown to inactivate Mtq2 enzymatic activity (24). Although Trm11 D286 residue could not be monitored in our MS experiments (Figure 2D and E), the corresponding residue from Bud23 (D112) forms a salt-bridge with Arg53 from Trm112 in the crystal structure of the Bud23–Trm112 complex (21). Furthermore, its mutation into Gly (D112G) results in the loss of Bud23 interaction with Trm112 according to yeast two-hybrid experiments (48) while the D112R mutant results in almost undetectable levels of Bud23 in vivo and loss of 18S rRNA methylation, two phenotypes associated with disruption of Bud23–Trm112 complex (21). This Trm11 D286R mutant was purified and did not interact anymore with Trm112 according to ITC measurements, further supporting our model (Table 3 and Supplementary Figure S4B). Similarly, the Trm112 R53E mutant did not interact with wild-type Trm11 as tested by ITC (Table 3 and Supplementary Figure S4C). These data support that Trm11 interacts with Trm112 in the same way as the three other MTases known to interact with Trm112 (21,24,28).
Trm112 stimulates SAM binding to Trm11 and contributes to tRNA binding.
The availability of the purified Trm11 protein also renders possible the detailed study of the molecular mechanisms responsible for the activation of Trm11 by Trm112. As it was previously observed that TRMT112 confers SAM binding capacity to MTQ2 (24), we first analyzed whether or not Trm112 had the same effect on Trm11 using ITC. While neither Trm11 nor Trm112 alone interact with SAM in our conditions (Supplementary Figure S4D), the wild-type Trm11–Trm112 complex displays a high affinity (Kd values of 1–2 μM depending on the temperature used to perform ITC experiments) for SAM and a 1:1 stoichiometry (Tables 2 and 3; Supplementary Figure S3A and 4D). To confirm that Trm11 is responsible for SAM binding, we mutated Asp238 from Trm11 into Ala as the corresponding Asp in class I MTase is known to form bidentate hydrogen bonds with the 2΄ and 3΄ hydroxyl groups of the SAM ribose moiety. As expected, this Trm11 D238A–Trm112 mutant complex does not bind SAM (Table 2 and Supplementary Figure S3) and is enzymatically inactive (Table 1), confirming that Trm11 is responsible for SAM binding. Hence, these experiments show that Trm112 is required for SAM binding to Trm11. Crystals structures of Mtq2–Trm112 and Bud23–Trm112 complexes bound to SAM have shown that the loop connecting strands β3 and β4 from these class I MTases is sandwiched between Trm112 on one side and SAM on the other. Hence, based on the proposed similar binding modes between Trm112 and its four MTase partners ((21,24,28), this study), we suggest that Trm112 allows SAM to bind to Trm11 by stabilizing the loop connecting Trm11 strands β3 and β4 (encompassing residues 235–265). Interestingly, the deuterium incorporation rates of the corresponding region in HDX-MS experiments are indeed affected upon binding of Trm11 to Trm112 (Figure 2D and H).
Next, we investigated in more details the role of Trm112 on tRNA methylation by Trm11. In addition to its role in SAM binding to Trm11, we cannot exclude that Trm112 also contributes to substrate binding. Indeed, the Trm112–MTase complexes are structurally similar to the bacterial RmlA(I) rRNA MTase, whose class I MTase domain is fused to an N-terminal Zn-binding domain, which has been suggested to interact with the rRNA substrate (49). When the MTase domains from Trm112–MTase complexes and RmlA(I) are superimposed, the Zn-binding domains from Trm112 and RmlA(I) match together (24). Interestingly, the A106E, E107K, I118E and Y120D Trm112 mutants located at the surface of the Zn-binding domain were previously shown to affect eRF1 methylation by Mtq2, further supporting a role of Trm112 in substrate binding (24). We tested whether these Trm112 mutants complexed to wild-type Trm11, affected tRNA MTase activity in vitro. All these complexes could be purified using the same protocol as for the wild-type complex and proved to be stable. They all bind SAM with affinities comparable to that of the wild-type complex (Kd of 2–3.6 μM; Table 2 and Supplementary Figure S3), strongly supporting the fact that they are correctly folded. These four mutants are all affected in their enzymatic activity and we performed kinetic analyses to determine whether these mutations affected tRNA binding or catalysis (Table 1). Compared to wild-type complex, the kcat values for these mutants were marginally affected (Table 1), indicating that these residues contribute modestly or not at all to the catalysis. On the opposite, all these mutants were affected in Km supporting a role of these residues in substrate binding (Table 1). However, whether these Trm112 residues contact directly tRNA substrates or have an allosteric effect on Trm11 active site remains to be clarified. We thus conclude that beyond its role in conferring SAM binding capacity to Trm11, Trm112 also contributes directly or indirectly to tRNA binding. Hence, Trm112 belongs to a long list of proteins containing a Zn-binding domain that participates in RNA binding (50–52).
The tRNA CCA tail is important for methyltransferase activity
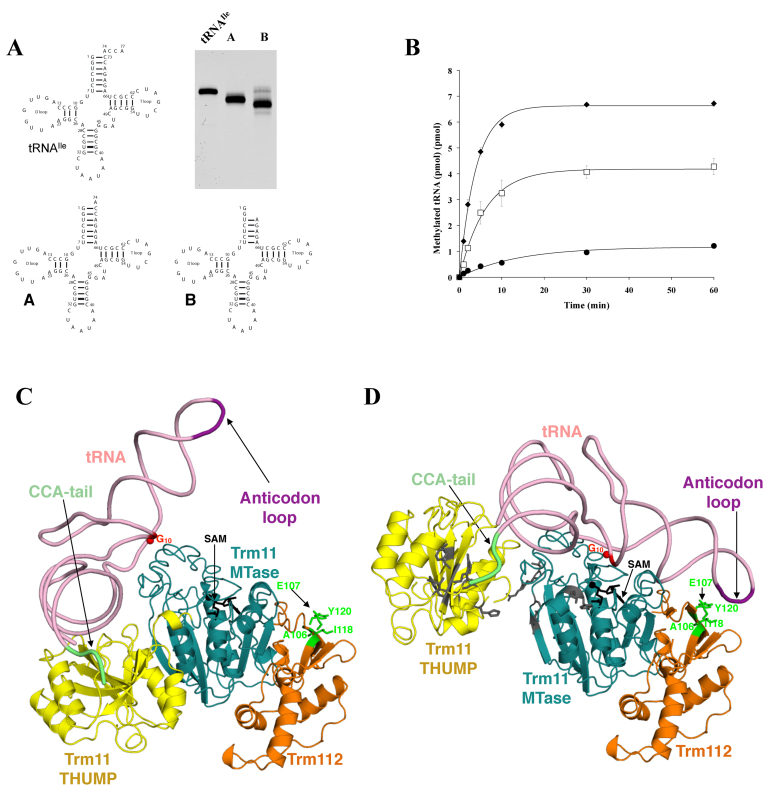
We next investigated the importance of the 3΄-CCA end from the aminoacyl stem loop for tRNA modification. Indeed, THUMP domains have been identified by bioinformatics analyses or structural approaches in several tRNA-modifying enzymes, which are primarily involved in modification of nucleotides located in the aminoacyl acceptor stem-loop. Among these proteins, Trm14/TrmN are MTases forming m2G at position 6 of tRNAPhe (37,53), ThiI is responsible for the formation of 4-thiouridine at position 8 of some bacterial tRNAs (38), the archaeal CDAT8 catalyzes the C-to-U editing at position 8 (36), Trm11 and archaeal PabTrm-G10 act on G10 (13,30,31). Furthermore, S. cerevisiae Tan1, which is only made of a THUMP domain, has recently been shown to associate with Kre33 acetyltransferase to form N4-acetylcytidine at position 12 of tRNASer and tRNALeu (34,39). Altogether, this led to the suggestion that the THUMP domain might recognize a specific tRNA region and orient the central three-dimensional core of the tRNA molecule toward the catalytic domain (30). This was recently confirmed by the crystal structure of Thermotoga maritima ThiI bound to a truncated tRNA, which revealed that the THUMP domain binds both the single stranded 3΄-CCA end and the double-stranded acceptor stem (38). We thus synthesized two tRNAIle transcripts lacking either the 3΄-CCA end (transcript A) or the 3΄-CCACCA sequence (transcript B; Figure 3A). Interestingly, the removal of the 3΄-CCA end (transcript A) significantly decreased the activity of Trm11–Trm112 (Figure 3B). This result highlights the importance of the tRNA 3΄-CCA end. Transcript B, whose acceptor stem-loop should be partly destabilized due to the removal of base pairs G1:C73 and G2:C72 upon deletion of the 3΄-CCACCA sequence, is an even poorer substrate compared to the intact tRNA (Figure 3B). This indicates that any destabilization of the acceptor stem-loop seriously affects Trm11 enzymatic activity. Altogether, our analysis supports that Trm11-Trm112 complex recognizes the 3΄ CCA tail and the extremity of the aminoacyl stem from the tRNA.
Figure 3.
Trm11–Trm112 recognizes the intact tRNA molecule. (A) Schematic representation of full-length yeast tRNAIle and two truncated tRNA molecules (A and B). Analysis of the various transcripts by denaturing 10% polyacrylamide gel electrophoresis. The gel was stained by ethidium bromide. (B) Enzymatic activity of Trm11–Trm112 complex on intact tRNAIle (black diamond), transcripts A (open square) and B (filled circle). The curves obtained after fitting of the experimental data with equation given in the ‘Materials and Methods’ section are shown by solid lines. (C) Preliminary model of the Trm11–Trm112–tRNA complex. The SAM molecule modeled into Trm11 MTase domain is shown as black sticks. The tRNA G10 position is shown by a red sphere. Trm112 residues mutated in this study are shown as sticks in green. (D) Model of the Trm11–Trm112–tRNA complex obtained after rotation of the THUMP domain relative to the MTase domain. Same color code as panel C. Residues from Saccharomyces cerevisiae Trm11 corresponding to TkoTrm11 residues mutated by Hirata and coworkers are shown as gray sticks (32).
Based on our current results and on the crystal structure of ThiI complexed to a tRNA fragment, we generated a model of the Trm11–Trm112–tRNA complex. We first superimposed the structure of the ThiI THUMP domain complexed to a tRNA fragment onto the structure of the THUMP domain in our model of the Trm11–Trm112 complex. We then superimposed the crystal structure of intact yeast tRNAPhe (54) onto the ThiI-bound tRNA aminoacyl acceptor stem loop. In this preliminary Trm11–Trm112–tRNA model, the G10 nucleotide from the tRNA is more than 20Å away from the SAM methyl group to be transferred (Figure 3C). However, conformational changes of the various partners, as suggested for the Pus10–tRNA complex (38), would be sufficient to bring G10 nucleotide in the close vicinity of the SAM molecule (Figure 3D). Such changes could consist in tRNA bending (accompanied by base flipping), Trm11 THUMP–tRNA subcomplex rearrangement relative to Trm11 MTase domain or both. This supports a role of the Trm11 THUMP domain in sensing the length between the tRNA 3΄-CCA end and the G10 nucleotide to be modified in order to project this later in the enzyme active site. Furthermore, in this model, Ile118 and Tyr120 residues from Trm112, whose substitution by Glu and Asp, respectively, results in a 10-fold increase in Km value (Table 1), come in the vicinity of the tRNA anticodon loop. However, it remains unclear whether these residues directly contact tRNA as suggested by our model or enhance indirectly the affinity of Trm11 for tRNA substrate. This will be the subject of future research.
CONCLUSION
In this study, we have characterized more deeply the enzymatic activity of the Trm11–Trm112 tRNA MTase complex from S. cerevisiae yeast and have enlightened multiple roles for Trm112 in this complex. Indeed, we show that Trm112 is required for the Trm11 catalytic subunit to be active, for SAM binding to Trm11 but is also important for tRNA binding. We provide strong experimental evidences supporting that Trm112 interacts with the Trm11 MTase via the same binding mode as the one previously described from Mtq2–Trm112, Bud23–Trm112 and Trm9–Trm112 crystal structures. Hence, these four enzymes directly compete to interact with Trm112. Based on these similarities between these four Trm112–MTase complexes, our study further suggests that Trm112 could also contribute to the binding of Trm9–Trm112 and Bud23–Trm112 enzymes to their respective RNA substrates.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Dr V. Heurgué-Hamard (IBPC, CNRS, Paris, France) for sharing plasmids with us and for critical reading of the manuscript. We thank Dr M. Nicaise (I2BC, CNRS, University of Paris-Saclay, Orsay, France) and Dr M. Blaud (LCRB, CNRS, University of Paris V-Descartes, Paris, France) for their help with ITC and CD measurements, respectively. We thank Shounak Bhogale for his help with protein purification.
Footnotes
Present addresses:
Julien Marcoux, IPBS, CNRS, UMR 5089, 205 Route de Narbonne, 31077 Toulouse, France.
Jean-Michel Saliou, Center for Infection and Immunity of Lille (CIIL), Institut Pasteur de Lille, CNRS UMR 8204, INSERM U1019, Université Lille Nord de France, 1 rue du Pr. Calmette, 59000 Lille, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National pour la Recherche Scientifique (CNRS) (to M.G.); ATIP-AVENIR program (to M.G.); Agence Nationale pour la Recherche (ANR) [ANR14-CE09-0016-02] (to M.G.); Ecole Polytechnique (to M.G.); CNRS (to S.C.); University of Strasbourg (to S.C.). S.C. thanks GIS IBiSa, the Région Alsace and the Communauté Urbaine de Strasbourg for the financial support in purchasing the Synapt G2 HDMS and the HDX equipments, respectively. Funding for open access charge: ANR [ANR14-CE09-0016-02].
Conflict of interest statement. None declared.
REFERENCES
- 1.Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Genes Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F.. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Yacoubi B., Bailly M., de Crecy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 4.Putz J., Florentz C., Benseler F., Giege R.. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1994; 1:580–582. [DOI] [PubMed] [Google Scholar]
- 5.Hopper A.K., Phizicky E.M.. tRNA transfers to the limelight. Genes Dev. 2003; 17:162–180. [DOI] [PubMed] [Google Scholar]
- 6.Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 7.Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014; 5:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anantharaman V., Koonin E.V., Aravind L.. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002; 4:71–75. [PubMed] [Google Scholar]
- 9.Shimada K., Nakamura M., Anai S., De Velasco M., Tanaka M., Tsujikawa K., Ouji Y., Konishi N.. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009; 69:3157–3164. [DOI] [PubMed] [Google Scholar]
- 10.Swinehart W.E., Jackman J.E.. Diversity in mechanism and function of tRNA methyltransferases. RNA Biol. 2015; 12:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson J., Phan L., Hinnebusch A.G.. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov A., Martzen M.R., Phizicky E.M.. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002; 8:1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purushothaman S.K., Bujnicki J.M., Grosjean H., Lapeyre B.. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005; 25:4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leulliot N., Chaillet M., Durand D., Ulryck N., Blondeau K., van Tilbeurgh H.. Structure of the yeast tRNA m7G methylation complex. Structure. 2008; 16:52–61. [DOI] [PubMed] [Google Scholar]
- 15.Mazauric M.H., Dirick L., Purushothaman S.K., Bjork G.R., Lapeyre B.. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 2010; 285:18505–18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guy M.P., Podyma B.M., Preston M.A., Shaheen H.H., Krivos K.L., Limbach P.A., Hopper A.K., Phizicky E.M.. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA. 2012; 18:1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy M.P., Phizicky E.M.. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014; 11:1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C., Huang B., Anderson J.T., Bystrom A.S.. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PLoS One. 2011; 6:e20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White J., Li Z., Sardana R., Bujnicki J.M., Marcotte E.M., Johnson A.W.. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell. Biol. 2008; 28:3151–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figaro S., Wacheul L., Schillewaert S., Graille M., Huvelle E., Mongeard R., Zorbas C., Lafontaine D.L., Heurgue-Hamard V.. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol. Cell. Biol. 2012; 32:2254–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letoquart J., Huvelle E., Wacheul L., Bourgeois G., Zorbas C., Graille M., Heurgue-Hamard V., Lafontaine D.L.. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5518–E5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heurgue-Hamard V., Champ S., Mora L., Merkulova-Rainon T., Kisselev L.L., Buckingham R.H.. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005; 280:2439–2445. [DOI] [PubMed] [Google Scholar]
- 23.Heurgue-Hamard V., Graille M., Scrima N., Ulryck N., Champ S., van Tilbeurgh H., Buckingham R.H.. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 2006; 281:36140–36148. [DOI] [PubMed] [Google Scholar]
- 24.Liger D., Mora L., Lazar N., Figaro S., Henri J., Scrima N., Buckingham R.H., van Tilbeurgh H., Heurgue-Hamard V., Graille M.. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011; 39:6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figaro S., Scrima N., Buckingham R.H., Heurgue-Hamard V.. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008; 582:2352–2356. [DOI] [PubMed] [Google Scholar]
- 26.Fu D., Brophy J.A., Chan C.T., Atmore K.A., Begley U., Paules R.S., Dedon P.C., Begley T.J., Samson L.D.. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010; 30:2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Songe-Moller L., van den Born E., Leihne V., Vagbo C.B., Kristoffersen T., Krokan H.E., Kirpekar F., Falnes P.O., Klungland A.. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010; 30:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letoquart J., Tran N.V., Caroline V., Aleksandrov A., Lazar N., van Tilbeurgh H., Liger D., Graille M.. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 2015; 43:10989–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sardana R., Johnson A.W.. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol. Biol. Cell. 2012; 23:4313–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armengaud J., Urbonavicius J., Fernandez B., Chaussinand G., Bujnicki J.M., Grosjean H.. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004; 279:37142–37152. [DOI] [PubMed] [Google Scholar]
- 31.Gabant G., Auxilien S., Tuszynska I., Locard M., Gajda M.J., Chaussinand G., Fernandez B., Dedieu A., Grosjean H., Golinelli-Pimpaneau B. et al. . THUMP from archaeal tRNA:m22G10 methyltransferase, a genuine autonomously folding domain. Nucleic Acids Res. 2006; 34:2483–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirata A., Nishiyama S., Tamura T., Yamauchi A., Hori H.. Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 2016; 44:6377–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravind L., Koonin E.V.. THUMP–a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001; 26:215–217. [DOI] [PubMed] [Google Scholar]
- 34.Johansson M.J., Bystrom A.S.. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA. 2004; 10:712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCleverty C.J., Hornsby M., Spraggon G., Kreusch A.. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007; 373:1243–1254. [DOI] [PubMed] [Google Scholar]
- 36.Randau L., Stanley B.J., Kohlway A., Mechta S., Xiong Y., Soll D.. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009; 324:657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fislage M., Roovers M., Tuszynska I., Bujnicki J.M., Droogmans L., Versees W.. Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res. 2012; 40:5149–5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann P., Lakomek K., Naumann P.T., Erwin W.M., Lauhon C.T., Ficner R.. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014; 42:6673–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S., Langhendries J.L., Watzinger P., Kotter P., Entian K.D., Lafontaine D.L.. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015; 43:2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bujnicki J.M. Phylogenomic analysis of 16S rRNA:(guanine-N2) methyltransferases suggests new family members and reveals highly conserved motifs and a domain structure similar to other nucleic acid amino-methyltransferases. FASEB J. 2000; 14:2365–2368. [DOI] [PubMed] [Google Scholar]
- 41.Graille M., Heurgue-Hamard V., Champ S., Mora L., Scrima N., Ulryck N., van Tilbeurgh H., Buckingham R.H.. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005; 20:917–927. [DOI] [PubMed] [Google Scholar]
- 42.Eckert K., Saliou J.M., Monlezun L., Vigouroux A., Atmane N., Caillat C., Quevillon-Cheruel S., Madiona K., Nicaise M., Lazereg S. et al. . The Pih1-Tah1 cochaperone complex inhibits Hsp90 molecular chaperone ATPase activity. J. Biol. Chem. 2010; 285:31304–31312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng L., Baumann U., Reymond J.L.. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao W., De La Cruz E.M.. Quantitative full time course analysis of nonlinear enzyme cycling kinetics. Sci. Rep. 2013; 3:2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pawlowski M., Gajda M.J., Matlak R., Bujnicki J.M.. MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics. 2008; 9:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okada K., Muneyoshi Y., Endo Y., Hori H.. Production of yeast (m2G10) methyltransferase (Trm11 and Trm112 complex) in a wheat germ cell-free translation system. Nucleic Acids Symp. Ser. (Oxf). 2009; 53:303–304. [DOI] [PubMed] [Google Scholar]
- 47.Studte P., Zink S., Jablonowski D., Bar C., von der Haar T., Tuite M.F., Schaffrath R.. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008; 69:1266–1277. [DOI] [PubMed] [Google Scholar]
- 48.Sardana R., Zhu J., Gill M., Johnson A.W.. Physical and functional interaction between the methyltransferase Bud23 and the essential DEAH-box RNA helicase Ecm16. Mol. Cell Biol. 2014; 34:2208–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das K., Acton T., Chiang Y., Shih L., Arnold E., Montelione G.T.. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:4041–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall T.M. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005; 15:367–373. [DOI] [PubMed] [Google Scholar]
- 51.Hamill S., Wolin S.L., Reinisch K.M.. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15045–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loughlin F.E., Gebert L.F., Towbin H., Brunschweiger A., Hall J., Allain F.H.. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2012; 19:84–89. [DOI] [PubMed] [Google Scholar]
- 53.Menezes S., Gaston K.W., Krivos K.L., Apolinario E.E., Reich N.O., Sowers K.R., Limbach P.A., Perona J.J.. Formation of m2G6 in Methanocaldococcus jannaschii tRNA catalyzed by the novel methyltransferase Trm14. Nucleic Acids Res. 2011; 39:7641–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi H., Moore P.B.. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000; 6:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.