Abstract
Human cells contain five topoisomerases in the nucleus and cytoplasm, but which one is the major topoisomerase for mRNAs is unclear. To date, Top3β is the only known topoisomerase that possesses RNA topoisomerase activity, binds mRNA translation machinery and interacts with an RNA-binding protein, FMRP, to promote synapse formation; and Top3β gene deletion has been linked to schizophrenia. Here, we show that Top3β is also the most abundant mRNA-binding topoisomerase in cells. Top3β, but not other topoisomerases, contains a distinctive RNA-binding domain; and deletion of this domain diminishes the amount of Top3β that associates with mRNAs, indicating that Top3β is specifically targeted to mRNAs by its RNA binding domain. Moreover, Top3β mutants lacking either its RNA-binding domain or catalytic residue fail to promote synapse formation, suggesting that Top3β requires both its mRNA-binding and catalytic activity to facilitate neurodevelopment. Notably, Top3β proteins bearing point mutations from schizophrenia and autism individuals are defective in association with FMRP; whereas one of the mutants is also deficient in binding mRNAs, catalyzing RNA topoisomerase reaction, and promoting synapse formation. Our data suggest that Top3β is the major topoisomerase for mRNAs, and requires both RNA binding and catalytic activity to promote neurodevelopment and prevent mental dysfunction.
INTRODUCTION
Topoisomerases are ‘magicians of the DNA world’, solving critical topological problems generated in many essential biological processes on DNA, including replication, transcription and segregation (1,2). These enzymes have a unique activity to catalyze DNA strand passage reactions, which enables them to relax supercoils generated during unwinding of duplex DNA by replication and transcription machinery, and to segregate chromosomes during cell division. Topoisomerases are universally present in all species (3), and that their dysfunction can cause genomic instability, defective cell proliferation, abnormal development, shortened life-span, lethality and human diseases (1,4–6). For example, depletion of topoisomerase I (Top1) from human cells can induce DNA breaks, chromosome aberrations and replication defects (7). Inhibition of topoisomerase 2α (Top2α) activity, or disruption of its recruitment to chromatin, can lead to defective chromosome segregation during mitosis (8,9).
Unlike the well-characterized DNA topoisomerases, RNA topoisomerases have received little attention for many years. In fact, the first RNA topoisomerase in eukaryotes was discovered only a few years ago, and it was shown to be capable of catalyzing RNA cleavage (4) as well as RNA strand passage reactions (10). To date, RNA topoisomerase activity has been observed in Type IA topoisomerases from all three domains of life (bacteria, archaea and eukarya) (10–13). This broad prevalence is similar to that of DNA topoisomerases, suggesting that RNA topoisomerase activity could provide growth advantage for its host so that it is retained through millions of years of evolution similarly as DNA topoisomerases. Interestingly, some of the most well-known Type IA topoisomerases, Top1 and Top3 of Escherichia coli and Top3 of yeast, all have dual activities for RNA and DNA (11–13), so that they may solve topological problems for both nucleic acids. In human, only one of the two Type IA enzymes, Top3β, possesses dual activity for both RNA and DNA, whereas its paralog, Top3α, has activity only for DNA (10). This difference in activity for RNA is largely due to a conserved RNA-binding motif, RGG-box, which is present only in Top3β but not Top3α; and deletion of this motif disrupts the RNA topoisomerase activity of Top3β (10).
Increasing evidence has revealed that Top3β regulates translation of mRNAs important for neurodevelopment and mental health together with other RNA binding proteins (RBPs). First, Top3β has been isolated in a complex with TDRD3 (Tudor domain-containing 3) (4,10,14), and this complex biochemically and genetically interacts with FMRP (4,10), an RBP that is inappropriately silenced in Fragile X syndrome and regulates translation of mRNAs important for neuronal function and autism (15). Second, the interaction between Top3β-TDRD3 and FMRP is impaired by a patient-derived FMRP mutation (10,16); and Top3β gene deletion has also been linked to schizophrenia and cognitive defects (4). Moreover, de novo single nucleotide variants (SNVs) of Top3β gene have been identified in schizophrenia and autism patients (17,18). Third, Top3β resembles TDRD3 and FMRP in its association with active-translating polyribosomes (4,10,16), and this association is conserved from human, chicken to fruit flies (12). Fourth, Top3β binds many mRNAs in vivo, and some of them are encoded by genes with neuronal functions related to schizophrenia and autism (10). Expression of one such gene, ptk2/FAK (protein kinase 2/focal adhesion kinase) is reduced in synapses at neuromuscular junctions (NMJs) of Top3β mutant flies. Synapse formation is defective in both flies and mice deleted for Top3β, as observed in FMRP mutant animals. These data suggest that Top3β works with FMRP and TDRD3 to regulate expression of mRNAs important for neurodevelopment and mental health.
Here, we investigate three basic questions regarding RNA topoisomerases in animals. Question one, human cells contain five topoisomerases in nucleus and/or cytoplasm, Top1, Top2α, Top2β, Top3α and Top3β. Which one is the major mRNA-binding topoisomerase in cells? We demonstrate that Top3β is the major mRNA-binding topoisomerase in human cells; and it is targeted to mRNAs mainly by its distinctive RNA-binding domain, which is absent in other topoisomerases. This finding leads to us further investigate the mechanism of how Top3β functions. Question two, is the RNA binding and catalytic activity of Top3β needed for its function in vivo? Question three, do Top3β variants from mental disorder patients disrupt its activities and functions? We show that both RNA-binding and catalytic activity is needed for Top3β to promote synapse formation in Drosophila. Moreover, when the de novo SNV from an autism patient is introduced into Top3β mutant flies, its function in synaptic formation is disrupted, providing evidence that Top3β function is linked to mental health.
MATERIALS AND METHODS
mRNA-binding protein capture assay
HEK293 cells were cultured as described previously (10). The mRNA-binding protein capture assay was performed using HEK 293 cells as described (19). Briefly, HEK 293 cells were transfected with plasmids expressing GFP-Top3β or its variants. After 48 h, the cells were exposed to 300 MJ/cm2 UV254 and harvested. The cells were lysed in lysis buffer (100 mM Tris, pH7.5, 150 mM LiCl, 0.1% lithium dodecyl sulfate (LiDS), 1% NP-40, 10 mM ethylenediaminetetraacetic acid (EDTA), 5 mM dithiothreitol (DTT)) and spun at 17 000 g. The supernatant was added with equal volume binding buffer (100 mM Tris, pH 7.5, 850 mM LiCl, 2% LiDS, 10 mM EDTA, 5 mM DTT) and incubated with Magnetic Oligo(dT) Beads (TAKARA) for 30–60 min at RT. The RNA–protein-captured beads were washed with WB1 buffer (100 mM Tris, pH 7.5, 500 mM LiCl, 1% LiDS, 10 mM EDTA, 5 mM DTT) 3 times and WB2 (20 mM Tris, pH 7.5, 200 mM LiCl, 1 mM EDTA, 1 mM DTT) twice. The RNA–protein complex was eluted with 20 μl EB buffer (20 mM Tris, pH 7.5) at 80°C for 2 min, and RNA was removed by RNase A. Each eluted mixture was then analyzed by immunoblotting together with their corresponding cell lysate.
RNA topoisomerase assay
The RNA topoisomerase assay for human Top3β has been previously described (10,12). We performed the experiment exactly as the previous method (12).
Cloning and expression of human Top3β variants
Human Top3β-R472Q and C666R mutants were made following the protocol of QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). The following oligos were used:
C666R_f: CTACAAGGAGCTCCGCCGCCCTCTGGATGAC;
C666R_r: GTCATCCAGAGGGCGGCGGAGCTCCTTGTAG;
R472Q_f: CTGCCCACTTGCCAGCAGGGTGATGCCTTCCCTG;
R472Q_r: CAGGGAAGGCATCACCCTGCTGGCAAGTGGGCAG.
Flag-tagged human Top3β and its mutants (R472Q and C666R) were expressed and purified as described previously (10). Briefly, HEK 293 cells were transfected with pcDNA constructs of human Top3β and its mutants using polyethylenimine. Cells were incubated in CO2 incubator shaker at 130 rpm, 5% CO2 for 72 h. Cells were then harvested, washed 2 times with cold phosphate buffered saline (PBS) and lysed in 3.5 volume of lysis buffer containing 20 mM Tris pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP40, 10 mM NaF and protease cocktail (Roche) on ice for 30 min. Two volumes of cold 20 mM Tris pH 7.5 were added to the cell lysate, and the diluted lysate was centrifuged at 18 000 rpm at 4°C for 30 min. The supernatant was incubated with the anti-Flag M2-agarose beads (Sigma) at 4°C for 3 h. Beads were washed three times with cold washing buffer (50 mM Tris pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP40, 1% EDTA and protease inhibitor cocktail), and once with cold elution buffer (25 mM Tris pH 7.5, 100 mM NaCl and 10% glycerol) for 5–8 min per wash. Flag-tagged proteins were eluted from anti-Flag M2 agarose beads in elution buffer with 200 μg/ml 3X Flag peptide (Sigma).
Immunoprecipitation (IP) and immunoblotting
Subcellular fractionation of HEK293 cell extracts into cytoplasmic and nuclear extracts have been described previously (10). IP using Flag antibody was performed as described earlier (10). Immunoblotting and various antibodies used in this study have also been described (10). Anti-humTop1 antibodies were from Abcam (ab3825); anti-humTop2α and anti-humTop2β antibodies were from Bethyl (A300-054A and A300-950A, respectively); anti-humTop3α antibodies were generated as described early (20). ImageJ software was used to quantify immunoblotting images.
Generation of Drosophila mutants
For generation of mutant flies carrying C660 R mutation in Top3β, pMT/V5-Flag-Top3β construct served as template to introduce the required mutation. The template was mutated using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and a set of primers gTgTATCgCgAgTTCAAgCgCCCgCTggACgACTTTg and gATCAAAgTCgTCCAgCgggCGCTTgAACTCgCg, respectively, following the manufacture's protocol. The mutation was confirmed by sequencing. A Top3β fragment carrying the C660R mutation was excised out from PMT/V5 Vector using NotI and XbaI restriction enzymes (New England Biolab) and sub-cloned into NotI and XbaI digested pBID-UASC vector (Addgene).
Morphological analyses of Drosophila neuromuscular junction
The NMJ analysis was performed as described previously (10). Briefly, the third instar wandering Drosophila larvae were dissected in cold Schneider's Drosophila Medium (Invitrogen). Then samples were fixed in 1× PBS with 4% paraformaldehyde (pH 7.4) for 25 min at room temperature. They were then rinsed three times with 1× PBT (phosphate-buffered saline in Triton X-100 (0.2% Triton X-100 in 1× PBS)) and blocked with 1% normal goat serum for 1 h at room temperature. Primary and secondary antibodies were diluted with 1× PBT with 1% normal goat serum and incubated with the samples overnight at 4°C and for 2 h at room temperature, respectively. The primary antibodies used in this study were: anti-DLG (4F3, 1:500 dilution, http://dshb.biology.uiowa.edu/4F3-anti-discs-large) from the Developmental Studies Hybridoma Bank (University of Iowa, IA, USA) and DyLight 549 anti–horseradish peroxidase (1:500 dilution) from Jackson Immuno Research (http://www.jacksonimmuno.com/pdf/lots/98340.pdf). Stained preparations were imaged with the Zeiss confocal microscopy system LSM 710 (Zeiss). The number of synaptic boutons and branches from NMJ4 segments 3–5 (n ≥ 20) of the different genotypes were quantified. The means and the standard errors of means (s.e.m.) of the boutons and branches were calculated. The P-values were obtained using Student's t-test (pairwise). Three independent experiments were performed, and the results are reproducible.
RESULTS
Top3β is the major mRNA-binding topoisomerase in human cells
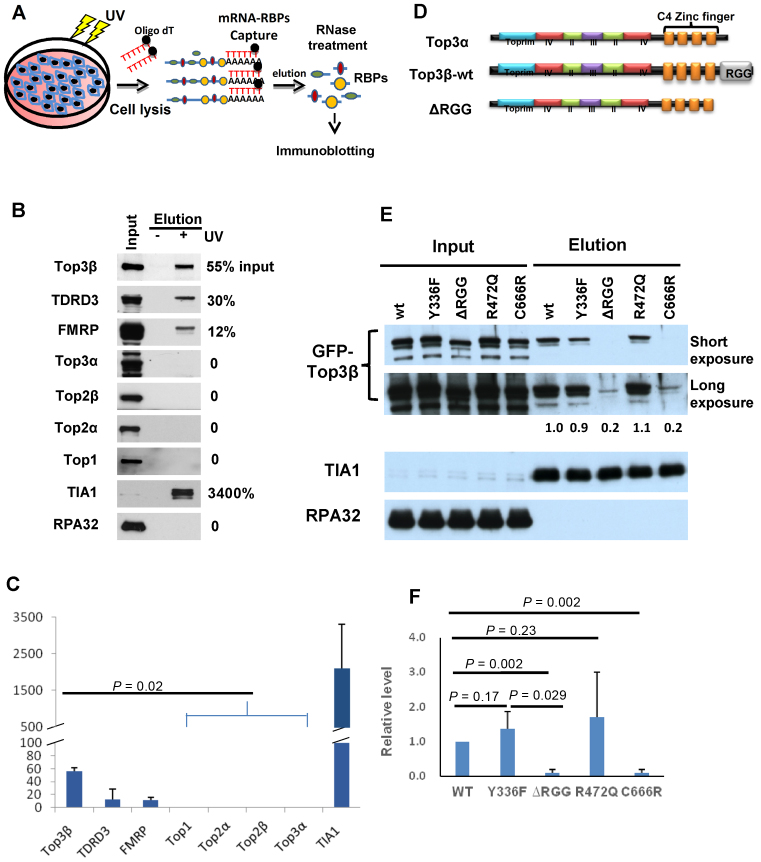
Three of the five known human topoisomerases (Top1, Top2α and Top3β; but not Top2β and Top3α) have been captured as RBPs directly binding to mRNAs in cells by two independent studies using UV-based crosslinking protocols coupled with mass spectrometry (21,22). We investigated which one is the major mRNA-binding topoisomerase in human HEK293 cells by modifying one of the protocols (21): we similarly performed UV-crosslinking of RBPs to mRNAs, captured the mRNA-bound RBPs using oligo-dT beads, but substituted the mass spectrometry with immunoblotting for protein identification (Figure 1A). One advantage of the immunoblotting is that it can detect the levels of an RBP in both the captured mRNA–protein complexes and the input extract on the same gel. The relative ratio between the two levels should correlate with the percentage of the RBP that binds mRNAs in total extract. As positive controls, three known RBPs that have been previously captured by mRNA-binding studies (21,22)—TIA1(23), FMRP (15) and TDRD3 (13)—were all detected by our approach, at relative ratios of greater than 10% (Figure 1B and C). As a negative control, RPA32, a subunit of a single-strand DNA binding complex RPA, was absent, with a relative ratio of 0%.
Figure 1.
Top3β is a major mRNA-binding topoisomerase and requires its RGG RNA-binding domain to bind mRNAs in vivo. (A) Schematic representation of an mRNA-binding protein capture assay modified from previous studies (21,22). mRNA and its RNA binding proteins (RBPs) were crosslinked by UV treatment in cells. After cell lysis, the mRNA-RBP complexes were captured by oligo-dT beads. After washing, the complexes were eluted and treated with RNase. The topoisomerases in RBPs were identified by immunoblotting. (B) Immunoblotting images and (C) quantification of mRNA-binding proteins captured on oligo-dT beads from lysates of HEK293 cells untreated or treated with UV. In (B), the relative ratios between the immunoblotting signals in the captured complex (lane 3) and the input (lane 1) were shown on the right. Only 0.1% of the input extract was loaded on the gel, whereas 50% of the eluted mRNA-bound RBPs were loaded. In (C), the graph shows the means of the relative ratios between the signals of the eluted proteins and the input from three independent experiments. The error bars indicate standard deviation. P-values between Top3β and other topoisomerases are calculated using Student's t-test. (D) Schematic representation of human Top3α, Top3β-wildtype and its RGG-deletion mutant (ΔRGG) proteins. The conserved core domains and the non-conserved CTDs, including Zn-fingers (orange boxes) and RGG-box, are indicated. Notably, the domain structure of Top3α and Top3β are highly similar, except the RGG-box which is only present in the former but not latter. (E) Immunoblotting images and (F) quantification from the mRNA-binding protein capture assay showing that GFP-tagged Top3β-ΔRGG and C666R mutants have strongly reduced mRNA binding activity after transfection in HEK293 cells, whereas Y336F and R472Q mutants retained the activity. In (B), for each Top3β mutant, the relative ratio of its immunoblotting signal in the captured mRNA complex versus the signal of the wild-type protein was shown between the images. A positive (TIA1) and negative (RPA) control was shown. In (F), the graph shows the means of the relative ratios between the immunoblotting signal of each mutant and that of the wild-type protein from three independent experiments. The error bars indicate standard deviation. P-values between Top3β and other topoisomerases are calculated using Student's t-test.
We performed our analyses on all five human topoisomerases. Top3β was detected in mRNA–protein complexes at a relative ratio of about 50% when compared to its level in the input extract (Figure 1B and C). In contrast, none of the other topoisomerases was detected in the same complexes (relative ratio is 0), even though their signals in the input were comparable to that of Top3β. These data suggest that Top3β is the major RNA topoisomerase working on mRNAs in human cells. We noted that the relative ratio of Top3β is similar to those of TDRD3 and FMRP, but substantially less than that of TIA1, which are in agreement with the notion that Top3β, TDRD3 and FMRP interact with each other to bind mRNAs (4,10).
Top3β requires its distinctive RNA-binding domain to bind mRNAs in vivo
We investigated the question why Top3β, but not other topoisomerases, is the most abundant mRNA-binding topoisomerase. A distinguishing feature of Top3β is the presence of an RGG RNA-binding domain, which is not found in Top3α and other human topoisomerases (Figure 1D; data not shown). We therefore investigated whether this domain is required by Top3β to bind mRNAs using the same method described above (Figure 1A). We transfected expression vectors encoding GFP-tagged Top3β wild-type or RGG-box deletion mutant (ΔRGG) protein into HEK293 cells, and found that the mutant protein was present at a substantially reduced level (about 5-fold less) in the mRNA-bound RBPs than that of the wild-type protein (Figure 1E and F). As an internal control, the level of TIA1 was comparable in mRNA-bound RBPs isolated from cells transfected with the two vectors. These data indicate that the RGG–RNA binding domain is critical for targeting Top3β mRNAs in vivo.
It should be noted that a putative nuclear localization sequence (NLS), PNPRRPKDK, is present at the C-terminus of RGG-box of Top3β based on a prediction program, PSORT II. This sequence was removed in our RGG-box-deletion construct, raising a possibility that the observed effect may be due to impairment of Top3β nuclear localization. We consider this possibility unlikely because majority of mRNAs are present in the cytoplasm, so that excluding Top3β from the nucleus should not limit its access to mRNAs. In addition, this NLS is absent in Drosophila Top3β, so that the observed phenotype for the Drosophila RGG-deletion mutant should not be due to perturbation of nuclear localization. Moreover, the same prediction program also detected the presence of at least another putative NLS in the middle of Top3β protein (RPRK at amino residue 377). Future experiments are needed to specifically delete these putative NLSs and RGG-box to elucidate their roles in nuclear localization, mRNA binding and other functions.
We also transfected the catalytic-inactive point mutant of Top3β (Y336F), which is completely deficient in both DNA and RNA topoisomerase activity, into the same cell line. We found that this mutant was present at a level similar to that of the wild type, but substantially higher than that of the ΔRGG mutant, in eluted mRNA–protein complexes (Figure 1E and F), indicating that the catalytic mutant largely retained the mRNA binding activity.
Top3β requires both RNA binding and topoisomerase activity to promote synapse formation
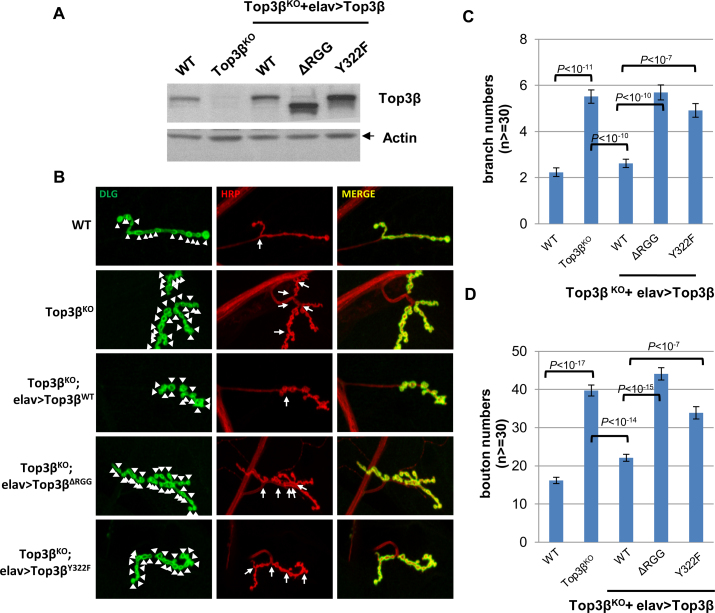
Top3β has at least two RNA-related biochemical activities: an mRNA binding activity that depends on the RGG-box (as described above), and RNA topoisomerase activity that requires the catalytic Tyr residue (10). We investigated which activity is needed to rescue the abnormal synapse formation at NMJs of Drosophila top3β mutant (10). We generated transgenic flies ectopically expressing Flag-tagged Top3β wildtype, RGG-box deletion and catalytic Tyr mutant proteins in the neurons of top3β deletion background (Figure 2A). Whereas ectopically-expressed Top3β-wildtype protein largely rescued the abnormally higher numbers of synaptic branches and boutons in the top3β mutant, the RGG-box deletion mutant (ΔRGG) and the catalytic mutant Y322F did not (Figure 2B, C and D). These data indicate that Top3β requires both its RNA binding activity and topoisomerase activity to promote synapse formation.
Figure 2.
Top3β requires its distinctive RNA-binding domain and catalytic activity to promote synapse formation in Drosophila. (A) Immunoblotting shows the expression levels of Top3β wild type and various mutant proteins in brain extracts of Top3β-knockout and transgenic flies. (B) Representative immunofluorescence images of neuromuscular junctions at muscle 4 (NMJ4) of wandering third instar Drosophila larvae of different genotypes as indicated. The NMJ4 was co-labeled with a presynaptic marker (anti-HRP, red) and a postsynaptic marker (anti-DLG, green). The arrowheads mark synaptic boutons, and the arrows mark branches. Quantification of (C) synaptic branches and (D) boutons at NMJ4 from segments 3, 4 and 5 of both sides of wandering third instar larvae (n ≥ 30). The graphs show the means of the bouton or branch numbers, and error bars represent standard errors of mean from three independent experiments. The P-values shown above each bar were calculated using Student's t-test.
Two Top3β variants found in schizophrenia and autism patients disrupt protein functions
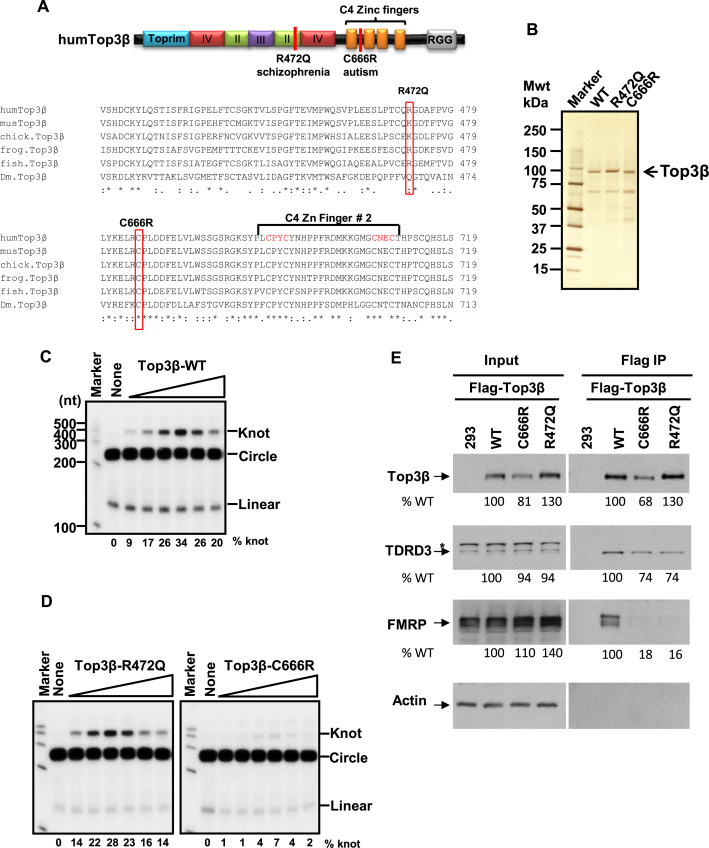
Genomic deletion of Top3β has been linked to schizophrenia (4). In addition, Top3β has been shown to interact with FMRP, a protein silenced in Fragile X syndrome, which is a leading cause of autism. Because autism and schizophrenia share common etiology factors (24,25), we investigated whether Top3β point mutations are similarly associated with mental disorders. Indeed, independent genome-wide sequencing studies have identified two de novo SNVs of Top3β, C666R and R472Q, in autism and schizophrenia patients, respectively (17,18), but absent in their unaffected siblings or parents. The C666R variant substituted a strictly conserved Cys residue in the C-terminal Zn-finger domain with a charged residue, whereas R472Q replaced a positively-charged residue (either R or K) conserved in the core domains of vertebrate Top3β with a non-charged residue (Q) (Figure 3A). Because these variants significantly altered the property of highly conserved amino acid residues, we examined whether they disrupted functions of Top3β using both in vitro and in vivo assays.
Figure 3.
Two de novo single nucleotide variants (SNVs) of Top3β from schizophrenia and autism patients are defective in association with FMRP; and one SNV is also defective in RNA topoisomerase activity. (A) Schematic representation (top), and sequence alignment (bottom), of protein sequences of two de novo SNVs from schizophrenia and autism patients (17,18). Red colored vertical lines indicate locations of the mutations. Alignment of Top3β from several higher eukaryotic organisms was generated by Clustal W2.1. The identical residues are indicated by asterisks, whereas conserved residues are marked with columns. The species aligned are: human (gene identification (gi): 47678723), mouse (gi: 6755851), chicken (gi: 57525152), frog (gi: 148228452), fish (gi: 326667668), Drosophila (gi: 7290697). (B) A silver-stained gel shows purified recombinant human Top3β proteins of wild type and two de novo single nucleotide variants R472Q and C666R from a schizophrenia and autism patient, respectively. (C) Autoradiographs of the RNA topoisomerase assay of the wild type and the (D) two de novo SNVs of Top3β from schizophrenia (R472Q) and autism (C666R) patients. The closed circular substrate (circle), the knot product (knot) and linear RNA were indicated. The percentage of the knot converted from the circle was listed below each lane. These assays have been done three times, and the data are reproducible. (E) IP-western analyses show that Flag-Top3β-C666R and Flag-Top3β-R472Q co-immunoprecipitated with about 5-fold less amount of FMRP, and about 30% less amount of TDRD3 compared to the wild-type protein after transfection into HEK293 cells. Quantification of the immunoblotting images was performed using ImageJ software. The relative percentages of the input and IP signals for the two mutants were calculated using the signals of the wild-type protein as the standard (artificially set as 100%), and were listed below each image. The level of Flag-Top3β-C666R mutant is lower than that of the wild-type protein (about 20% less) in the input extract, suggesting that this mutation modestly reduces the stability of the protein. An asterisk marks a cross-reactive polypeptide. The experiments have been repeated twice, and the data are reproducible.
First, we expressed and purified flag-tagged recombinant proteins for the wild type and two variants of Top3β (R472Q and C666R) using HEK293 cells (Figure 3B), and then tested their activity using the RNA topoisomerase assay as described previously (10,12). In this assay, a synthetic closed circular RNA substrate is converted to a trefoil knot by the strand-passage activity of an RNA topoisomerase, such as Top3β. The knot produced can be distinguished from the circle substrate by denaturing urea-polyacrylamide gel electrophoresis. Consistent with previous findings, wild-type Top3β converted up to about 30% of the RNA circle to knot (Figure 3C). Similarly, the R472Q mutant exhibited conversion efficiency comparable to that of the wild-type protein (Figure 3D). Notably, the C666R variant converted about 5- to 10-fold less circle to knot at various protein concentrations tested (Figure 3D), indicating that this variant is strongly defective in its RNA topoisomerase activity.
Second, we examined whether C666R and R472Q mutants have reduced mRNA binding activity in vivo using the RBP capture assay described above. We found that the C666R variant showed reduced mRNA binding activity (about 5-fold reduction), whereas the R472Q variant displayed activity similar to that of the wild-type protein (Figure 1E and F). Previous studies have shown that Top3β localizes in both nuclear and cytoplasmic fractions (4,10). One possibility for the observed reduction of C666R in binding to mRNAs is that the mutant protein may be misfolded and thus mis-localized in the nucleus, which precludes it from access to the mRNAs in the cytoplasm. To exclude this possibility, we examined the subcellular localization of these mutants by immunoblotting. The two mutant proteins were detected in both nuclear and cytosolic fractions, and their distribution in these fractions was comparable to that of the wild-type protein (Supplementary Figure S1), which argue against this possibility. However, the level of the C666R mutant protein was somewhat lower than that of the wild type (Supplementary Figure S1, and also Figure 3E below). We therefore cannot rule out a possibility that a small fraction of C666R mutant protein may be misfolded and degraded.
Third, we tested whether the two variants have normal association with FMRP and TDRD3 by transfecting both variants into HEK293 cells and performing co-immunoprecipitation assay as described previously (10). Both variants co-immunoprecipitated with about 5-fold less amounts of FMRP, as well as about 30% less amounts of TDRD3, when compared to the wild-type protein (Figure 3E). These data suggest that the two variants are strongly deficient in association with FMRP, but are only slightly defective in association with TDRD3.
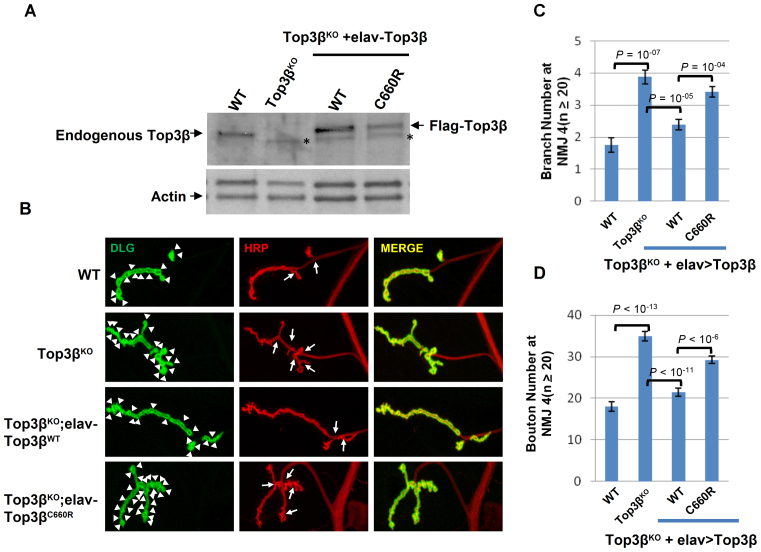
Finally, we generated transgenic flies expressing Drosophila Top3β-C660R variant, which corresponds to the human Top3β-C666R mutant, in the top3β deletion background (Figure 4A). We found that this variant displayed reduced ability to rescue the higher number of synaptic branches and boutons observed at the NMJs of top3β mutant (Figure 4B–D). For example, whereas the ectopically-expressed Drosophila Top3β-wild type protein suppressed the mean branch number from 3.9 to 2.4 (lane 2–3), the C660R variant suppressed the number from 3.9 to 3.4; suppression of synaptic boutons was 35 to 21 for the wild type, and 35 to 29 for the C660R mutant. The data provide in vivo evidence that C666R is a partial loss-of-function mutant. We were unable to test the R472Q variant, because this residue is variable in invertebrates, and its corresponding residue in Drosophila Top3β happens to be Q (Figure 3A).
Figure 4.
A de novo SNV of Top3β from an autism patient is defective in promoting synapse formation in Drosophila. (A) Western blot images show expression levels of Top3β wild type and mutant proteins in extracts of adult brains from the indicated genotypes. (B) Representative immunofluorescence images of NMJs at muscle 4 (NMJ4) of wandering third instar Drosophila larvae of different genotypes as indicated. The NMJ4 was co-labeled with a presynaptic marker (anti-HRP, red) and a postsynaptic marker (anti-DLG, green). The arrowheads mark synaptic boutons, and the arrows mark branches. (C and D) Quantification of synaptic branches and boutons at NMJ4 from segments 3, 4 and 5 of both sides of wandering third instar larvae (n ≥ 20). The graphs show the means of the boutons and branches, and error bars represent standard errors of mean from three independent experiments. The P-values shown above each bar were calculated using Student's t-test.
DISCUSSION
Top3β is the major RNA topoisomerase for mRNAs in human cells
Human cells have five topoisomerases that exist in nucleus and/or cytoplasm. One basic question is how many of them can bind mRNAs and function as RNA topoisomerases. We and others have previously shown that Top3β possesses RNA topoisomerase activity in vitro and binds many mRNAs in vivo (4,10). Here, we demonstrate that only Top3β and its two interacting partners (TDRD3 and FMRP) were detected in cellular RBPs that directly bind mRNAs, whereas the other four topoisomerases (Top1, Top2α, Top2β and Top3α) were undetectable. We conclude that Top3β is the major RNA topoisomerase working on mRNAs in human cells.
Our data have similarity and difference when compared to two previous screens using UV-crosslinking coupled with mass spectrometry to identify mRNA-binding RBPs in human cells (21,22). The similarity is that all three studies have identified Top3β and its two partners, TDRD3 and FMRP, but not Top3α and Top2β, in mRNA-binding RBPs. These data together support the proposal that Top3β, but not Top3α, is an RNA topoisomerase that functions in mRNA translation (10). The difference is that the two previous screens, but not the current study, have identified Top1 and Top2α as mRNA-binding RBPs. This raised a possibility that these two enzymes may have RNA topoisomerase activity and work on mRNA, which needs to be addressed in future studies. Nevertheless, our inability to detect Top1 and Top2α implies that the percentage of these two topoisomerases stably associating with mRNAs might be quite low, so that they could not be detected by our immunoblotting-based approach.
The RGG–RNA binding domain is critical for targeting Top3β to mRNAs
Why Top3β differs from the other topoisomerase in mRNA association? We found that the amount of Top3β that binds mRNAs in cells was strongly decreased (about 5-fold) by deletion of its RGG RNA-binding domain. This finding is consistent with our earlier data that the RNA topoisomerase activity of Top3β is severely disrupted by deletion of the same domain (10). Together, these data indicate that the RGG domain plays a crucial role in targeting Top3β to RNA both in vitro and in vivo.
We have examined the protein sequences of all human topoisomerases for the presence of RGG domain and other RNA-binding motifs, and found that Top3β is the only one with a recognizable RNA-binding domain (data not shown) (10). We therefore propose that the main reason for Top3β being the major mRNA-binding topoisomerase is because of its unique RNA-binding domain, which targets the topoisomerase to mRNAs. Other topoisomerases lack such an RNA-targeting domain, so that their main substrate is most likely DNA, but not RNA. We should point out that both interacting partners of Top3β, TDRD3 and FMRP, also possess RNA-binding activity (13,15) and could thus contribute to targeting of Top3β to mRNAs. Moreover, TDRD3 has been shown to directly interact with the exon-junction complex through its EBM motif (exon-junction complex binding motif) (4), and several other mRNA translation factors through its TUDOR domain (4,10,26). Depletion of TDRD3 or mutation of either its EBM or TUDOR domains reduce the amount of TDRD3–Top3β complex that associates with translating polyribosomes (4,12). Thus, Top3β may be targeted to mRNAs by at least three mechanisms: either through its own RGG-box, or its partner's RNA binding domains, or the protein binding motifs of TDRD3 (TUDOR and EBM). Existence of several targeting mechanisms may explain why the Top3β mutant deleted of the RGG-box still has residual mRNA binding activity in cells.
It should be noted that most of Top3β in cells co-fractionates with free mRNAs or mono-ribosomal fractions, whereas only a small amount of Top3β associates with the translating polyribosome fractions (4,10,12). The amount of Top3β associating with polyribosomes is strongly decreased when TDRD3 is depleted, or when the EBM or TUDOR domains of TDRD3 are mutated (4,10,12). The data imply that binding of Top3β to mRNAs through its RGG domain is insufficient for its recruitment to the mRNA translation machinery; and this recruitment requires additional interactions mediated by TUDOR and EBM domains of TDRD3.
Top3β requires both RNA binding and catalytic activity to function in neurodevelopment
We have previously shown that the RGG-domain deletion mutant of Top3β is defective in catalyzing RNA topoisomerase reaction. Our current data that the RGG-domain is required for targeting of Top3β to mRNAs in cells and for promoting neurodevelopment in Drosophila provide in vivo evidence for importance of this RNA-binding domain in Top3β function. Moreover, we demonstrated that the Top3β catalytic Tyr mutant, which had deficient RNA topoisomerase activity but largely normal mRNA binding activity, failed to promote neurodevelopment in flies. Thus, the topoisomerase activity of Top3β is also required for its function in neurodevelopment, whereas the RNA-binding activity alone is insufficient.
It should be pointed out that although Top3β possesses RNA topoisomerase activity in vitro (10), binds mRNAs in cells and requires its RGG RNA-binding domain and catalytic activity to promote neurodevelopment, there is so far no direct evidence to prove that Top3β actually catalyzes topoisomerase reactions on mRNAs in cells. In fact, deletion of the Top3β-RGG domain or mutation of its catalytic residue can impair both RNA and DNA topoisomerase activity of the enzyme (4,10,13,27). Thus, it remains to be determined whether it is the DNA or RNA topoisomerase activity of Top3β that is important for neurodevelopment.
Finally, we demonstrated that two de novo SNVs of human Top3β from schizophrenia and autism patients are both impaired in FMRP association, which are reminiscent of the previous findings that a Fragile X patient-derived mutation disrupts the association between FMRP and Top3β-TDRD3 complex (10,16). Because Fragile X syndrome is a known cause of autism (28,29) and Top3β gene deletion is linked to schizophrenia (4), our data suggest that the association between Top3β-TDRD3 and FMRP is important to prevent both of these mental disorders. Notably, whereas the C666R variant from the autism patient has not been detected in the general population, the R472Q from the schizophrenia patient has been detected, with a frequency of 0.0004 (NCBI SNP database). It will be interesting to study if the individuals carrying this minor allele may have increased risk of developing schizophrenia. Moreover, the C666R variant has additional defects in mRNA binding, in RNA topoisomerase activity, and in promoting synapse formation, suggesting that variants that impair RNA binding and topoisomerase activities of Top3β can lead to abnormal neurodevelopment and increased risk to mental dysfunction. Consistent with this notion, copy number variants of both Top3β and TDRD3 have been reported in individuals with autism spectrum disorders (listed in http://autismkb.cbi.pku.edu.cn). Micro-duplication of the genomic region containing Top3β gene has also been reported for a patient with mental retardation (30). Future studies are needed to identify genes regulated by the Top3β–TDRD3 complex, which could provide targets for therapeutic intervention.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr T.S. Hsieh for providing Drosophila Top3β flies and antibody, and Dr D. Schlessinger for critical reading of the manuscript. The authors also thank an anonymous reviewer for indicating presence of a putative nuclear localization sequence at the C-terminus of Top3β.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institute on Aging, National Institutes of Health [Z01 AG000657-08 in part]; National Basic Research Program of China [2013CB911002]; National Natural Science Foundation of China [31271435]. Funding for open access charge: National Institute on Aging, NIH [Z01 AG000657-08].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002; 3:430–440. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y., Sun Y., Huang S.N., Nitiss J.L.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forterre P., Gribaldo S., Gadelle D., Serre M.C.. Origin and evolution of DNA topoisomerases. Biochimie. 2007; 89:427–446. [DOI] [PubMed] [Google Scholar]
- 4.Stoll G., Pietilainen O.P., Linder B., Suvisaari J., Brosi C., Hennah W., Leppa V., Torniainen M., Ripatti S., Ala-Mello S. et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013; 16:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan K.Y., Greenwald R.J., Mohanty S., Sharpe A.H., Shaw A.C., Wang J.C.. Development of autoimmunity in mice lacking DNA topoisomerase 3beta. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:9242–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan K.Y., Wang J.C.. Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5717–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao Z.H., Player A., Shankavaram U., Wang Y.H., Zimonjic D.B., Lorenzi P.L., Liao Z.Y., Liu H., Shimura T., Zhang H.L. et al. Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 2007; 67:8752–8761. [DOI] [PubMed] [Google Scholar]
- 8.Sumner A.T. Inhibitors of topoisomerase II delay progress through mitosis and induce a doubling of the DNA content in CHO cells. Exp. Cell Res. 1995; 217:440–447. [DOI] [PubMed] [Google Scholar]
- 9.Dykhuizen E.C., Hargreaves D.C., Miller E.L., Cui K., Korshunov A., Kool M., Pfister S., Cho Y.J., Zhao K., Crabtree G.R.. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013; 497:624–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Fox D. 3rd et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013; 16:1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Di Gate R.J., Seeman N.C.. An RNA topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:9477–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad M., Xue Y., Lee S.K., Martindale J.L., Shen W., Li W., Zou S., Ciaramella M., Debat H., Nadal M. et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016; 44:6335–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siaw G.E., Liu I.F., Lin P.Y., Been M.D., Hsieh T.S.. DNA and RNA topoisomerase activities of Top3beta are promoted by mediator protein Tudor domain-containing protein 3. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E5544–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., McBride K.M., Hensley S., Lu Y., Chedin F., Bedford M.T.. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell. 2014; 53:484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011; 146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linder B., Plottner O., Kroiss M., Hartmann E., Laggerbauer B., Meister G., Keidel E., Fischer U.. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 2008; 17:3236–3246. [DOI] [PubMed] [Google Scholar]
- 17.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A. et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012; 74:285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu B., Ionita-Laza I., Roos J.L., Boone B., Woodrick S., Sun Y., Levy S., Gogos J.A., Karayiorgou M.. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012; 44:1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castello A., Horos R., Strein C., Fischer B., Eichelbaum K., Steinmetz L.M., Krijgsveld J., Hentze M.W.. System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc. 2013; 8:491–500. [DOI] [PubMed] [Google Scholar]
- 20.Meetei A.R., Sechi S., Wallisch M., Yang D., Young M.K., Joenje H., Hoatlin M.E., Wang W.. A multiprotein nuclear complex connects Fanconi anemia and bloom syndrome. Mol. Cell. Biol. 2003; 23:3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 22.Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 23.Bauer W.J., Heath J., Jenkins J.L., Kielkopf C.L.. Three RNA recognition motifs participate in RNA recognition and structural organization by the pro-apoptotic factor TIA-1. J. Mol. Biol. 2012; 415:727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilman S.R., Chang J., Xu B., Bawa T.S., Gogos J.A., Karayiorgou M., Vitkup D.. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat. Neurosci. 2012; 15:1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan P.F., Magnusson C., Reichenberg A., Boman M., Dalman C., Davidson M., Fruchter E., Hultman C.M., Lundberg M., Langstrom N. et al. Family history of schizophrenia and bipolar disorder as risk factors for autism. Arch. Gen. Psychiatry. 2012; 69:1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goulet I., Boisvenue S., Mokas S., Mazroui R., Cote J.. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 2008; 17:3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson-Sali T., Hsieh T.S.. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7974–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhakar A.L., Dolen G., Bear M.F.. The pathophysiology of fragile X (and what it teaches us about synapses). Annu. Rev. Neurosci. 2012; 35:417–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santoro M.R., Bray S.M., Warren S.T.. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 2012; 7:219–245. [DOI] [PubMed] [Google Scholar]
- 30.Tarsitano M., Ceglia C., Novelli A., Capalbo A., Lombardo B., Pastore L., Fioretti G., Vicari L., Pisanti M.A., Friso P. et al. Microduplications in 22q11.2 and 8q22.1 associated with mild mental retardation and generalized overgrowth. Gene. 2014; 536:213–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.