Abstract
Isoforms of human miRNAs (isomiRs) are constitutively expressed with tissue- and disease-subtype-dependencies. We studied 10 271 tumor datasets from The Cancer Genome Atlas (TCGA) to evaluate whether isomiRs can distinguish amongst 32 TCGA cancers. Unlike previous approaches, we built a classifier that relied solely on ‘binarized’ isomiR profiles: each isomiR is simply labeled as ‘present’ or ‘absent’. The resulting classifier successfully labeled tumor datasets with an average sensitivity of 90% and a false discovery rate (FDR) of 3%, surpassing the performance of expression-based classification. The classifier maintained its power even after a 15× reduction in the number of isomiRs that were used for training. Notably, the classifier could correctly predict the cancer type in non-TCGA datasets from diverse platforms. Our analysis revealed that the most discriminatory isomiRs happen to also be differentially expressed between normal tissue and cancer. Even so, we find that these highly discriminating isomiRs have not been attracting the most research attention in the literature. Given their ability to successfully classify datasets from 32 cancers, isomiRs and our resulting ‘Pan-cancer Atlas’ of isomiR expression could serve as a suitable framework to explore novel cancer biomarkers.
INTRODUCTION
RNA-sequencing technologies have enabled the discovery of novel categories of non-coding RNA (ncRNAs) (1). Among ncRNAs, microRNAs (miRNAs) are the best studied to date (2–9), having been linked to a wide range of processes (10–17) as well as conditions and diseases (18–20), including cancer (21,22). Their important roles and relatively easy quantification have made miRNAs ideal biomarker candidates (23–26) for tumor classification (27,28).
Recently, we made three contributions to the miRNA field. First, we discovered 3 707 novel human miRNAs most of which are primate-specific and exhibit tissue-specific expression (29). Second, we demonstrated that miRNA isoforms (isomiRs) are produced constitutively in human tissues and their expression depends on tissue type, tissue state, disease subtype and a person's sex, population origin, and race (30,31). Third, we showed that the level of transcription is not the main determinant of isomiR relative abundance but the isomiR levels depend on secondary features such as their lengths and their 5΄ or 3΄ termini (31). We also showed computationally and experimentally that different isomiRs of the same miRNA can target different genes and pathways, a finding that greatly extends the gamut of the regulatory events that are mediated by miRNA loci (31).
These findings suggest that a complex process drives the expression of isomiRs. Thus, we hypothesized that information about the isomiRs that are present in a tissue may suffice to permit accurate sample classification in a pan-cancer setting. Specifically, we evaluated whether ‘binarized isomiR profiles’ can distinguish among multiple cancer types. On a related note, an earlier application of binarized signatures to protein-coding transcripts reported promising results (32–34). For this project, we focused on The Cancer Genome Atlas (TCGA) repository. TCGA represents an ideal framework for testing our hypothesis, because it makes available small RNA-sequencing profiles for more than 11 000 samples from 32 cancer types (35–55).
MATERIALS AND METHODS
Data acquisition and correction
We quantified the TCGA isomiR expression data of 10 271 TCGA datasets representing 32 cancer types. From the whole TCGA cohort, 1 134 datasets were skipped because they are annotated as ‘potentially problematic’ (TCGA data portal, file_annotations.txt files of 28 October 2015). In order to do this, we downloaded the public loci-based isoform.quantification.txt files from the TCGA datasets (downloaded from the TCGA data portal https://tcga-data.nci.nih.gov on 6 August 2015) and converted them to be molecule/sequence based. Importantly, our pool of candidate biomarker miRNA loci includes miRBase as well as those hairpin arms of miRBase for which we reported recently that they are expressed in various tissues (29). Prior to the analysis, we applied corrections to account for mature sequences that could originate from any of several known miRNA paralogs (56). We also corrected for the fact that the isoform.quantification.txt files made available by TCGA often list only a subset of possible loci of miRNA paralogs (56). Importantly, even though we counted the expression of miRNA paralogs once (thereby avoiding multiple counting), we maintained the labels of all possible paralogs throughout the analysis.
We only included samples corresponding to primary solid tumors (sample infix ‘01’ in the TCGA sample barcode), except for acute myeloid leukemia (LAML) where blood-derived samples were used (sample infix ‘03’).
‘Binarized isomiR’ and ‘binarized miRNA-arm’ profiles
We generate binarized isomiR profiles for a given sample (dataset) by labeling its top 20% most expressed isomiRs ‘present.’ All other isomiRs are labeled ‘absent’ from the dataset. Drawing the line at the top 20% represents a threshold of ∼10 reads per million, which is stringent (Supplementary Figure S1). We generate binarized miRNA-arm profiles for a given dataset by labeling the arm ‘present’ if and only if at least one isomiR originating from the arm is ‘present;’ otherwise, we label the arm ‘absent.’ IsomiRs mapping to the arms of (multiple) miRNA paralogs are merged into meta-arms, i.e. collections of arms all of which share the union of produced isomiRs.
Expression profiles of miRNA arms
For a given miRNA arm, we generated the arm's expression profile by summing the expression of all the isomiRs that are produced by the arm.
Statistical and machine learning analyses
Statistical analyses were done in R version 3.3.0 (57) and Python version 2.7. X2 tests were performed and P-values were corrected to false discovery rate (FDR) values. We called an isomiR or miRNA-arm feature ‘discriminatory,’ when the absolute difference between the percentage (%) of datasets containing the feature in one cancer type but not the other was ≥80%. Hamming distance was calculated with the hamming.distance function of the e1071 package (58), while all other distance metrics of hierarchical clustering (HCL) were performed with the hcluster function of the amap package (59). Visualization of dendrograms was performed with the dendextend package of R (60). Networks were visualized using the igraph package in R (61).
Support vector machines (SVMs) (62,63) were run with the svm function of the e1071 package in R (58) with linear kernel function and with allowed probability predictions (see Supplementary Methods). The variable importance (VI) scores were computed separately for each isomiR, or for each miRNA arm, as the average of the squared values of the weights across all pairwise SVM comparisons (64) and then were scaled to one by dividing by the maximum score. RandomForest was run with the H2O package in R (65).
Significance analysis of microarrays (SAM) (66) was used to identify differentially expressed (DE) isomiRs between the tumor and normal tissue samples in eight cancer types that contained enough (>35) normal samples. Independently for each cancer type, the numeric expression profiles were filtered so that only isomiRs with significant expression, i.e. the isomiR to be marked as ‘present,’ in more than 75% of the samples were used. SAM was run at 5 000 permutations and an FDR cutoff filter of 0.00%. We note that on average 593 isomiRs were included in the analysis of each cancer (905 unique isomiRs across all eight cancers) and an average of 54% of those were found to be DE in each cancer type (727 unique DE isomiRs across all eight cancers), either up- or downregulated.
Retrieval of PubMed entries
For this step, we specifically used those miRNA loci that have entries in the Gene database of National Center for Biotechnology Information and retrieved the number of PubMed entries associated with each miRNA gene (current as of 7 October 2016) using Biopython (67). For each PubMed entry, we also retrieved the title, converted all characters to lowercase and searched for the strings ‘biomarker’ or ‘signature’.
Additional validation using non-TCGA datasets
We downloaded several, publicly available (29,68–70), non-TCGA tumor datasets and generated the miRNA expression profile for each (See Supplementary Methods for more details). We subsequently binarized the expression profiles and used an SVM classifier trained on the TCGA tumor samples of six cancer types to classify each of the datasets.
RESULTS
A ‘Pan-cancer Atlas of IsomiR Expression’ and statistics of binarized isomiRs
We processed 10 271 normal and tumor TCGA datasets and identified 7 466 isomiRs that arise from 807 arms and 767 miRNA loci (Supplementary Tables S1 and 2). The latter include miRBase loci and novel human miRNA genes that we reported previously (29). We intentionally focused on binary isomiR profiles, i.e. profiles that simply label an isomiR or miRNA arm as ‘present’ or ‘absent’ (Supplementary Tables S1 and 2). We make available this complete Pan-Cancer Atlas of isomiR expression in the Supplementary Tables.
We found that the majority (90.2%) of the 7 466 isomiRs are present in ≤50% of the analyzed datasets. A mere 48 of the 7 466 isomiRs are in all datasets (Supplementary Figure S2A). Interestingly, 11 of the 48 isomiRs are from the let-7 family. Other ubiquitous isomiRs arise from widely-studied loci including mir-21, mir-29, mir-30, the mir-17/92 cluster. For individual miRNA loci, the distribution of their isomiRs varied greatly across samples. For example, let-7 isomiRs were ‘dichotomized:’ one subset is present in nearly all datasets whereas a second subset is in fewer than 25% of the datasets (Supplementary Figure S2B). Moreover, 77.5% of the 7 466 isomiRs are in at least two of the 32 TCGA cancer types (Supplementary Figure S2C).
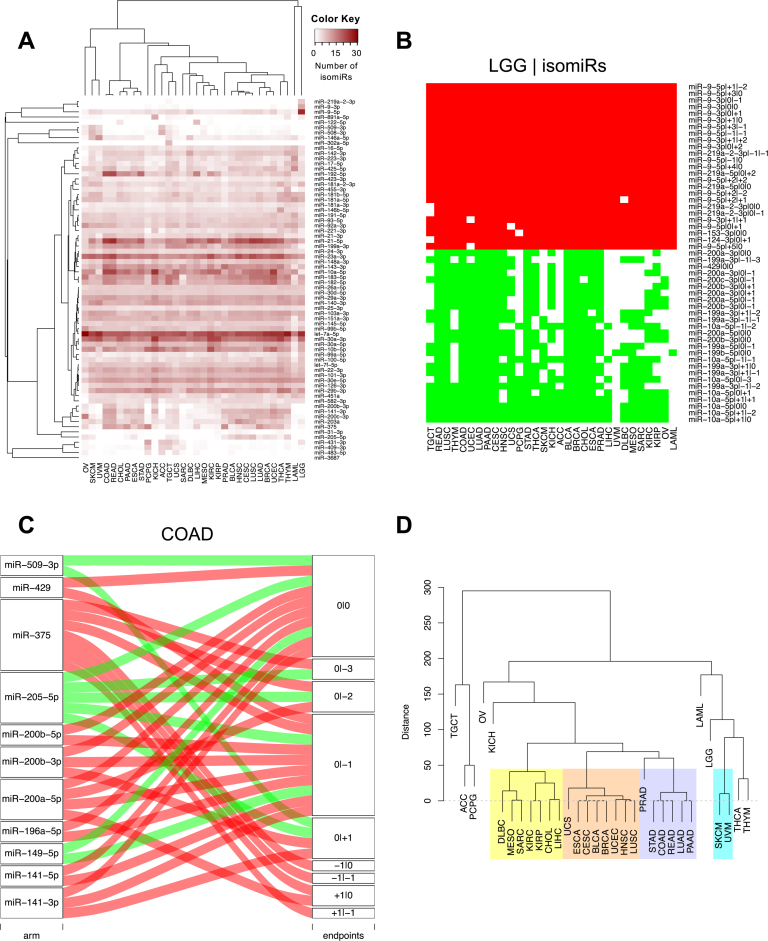
Figure 1A shows a heatmap of the number of isomiRs arising from the 70 highest-yielding miRNA arms. Let-7a-5p is consistently present in numerous isomiRs across all 32 cancer types. MiR-21-5p and miR-30a-3p produce many isomiRs in most of the analyzed cancers. Ovarian cancer (OV) in the case of miR-21-5p and LAML in the case of miR-30a-3p are striking exceptions to this observation. Also of note is the fact that several arms produce numerous isomiRs in select cancers only: e.g. characteristically, the 5p and 3p arms of mir-9 produce numerous isomiRs in lower glade glioma (LGG). Lastly, we found the mean expression of a miRNA arm correlates very well with the mean number of isomiRs that arise from this locus across all the tumor samples in a given cancer (average Spearman correlation coefficient across cancer types: 0.976)—see also (31) for a similar finding in our previous isomiR study that analyzed only a small subset of the TCGA breast cancer (BRCA) datasets. In Supplementary Figure S2D we show a specific example of this correlation for bladder cancer (BLCA).
Figure 1.
Differentially present isomiRs among different cancer types. (A) Heatmap of the number of isomiRs per miRNA arm. Shown are data from the 70 highest-yielding miRNA arms. The darker the color of each cell, the higher the number of isomiRs that the respective arm (rows of the heatmap) produces in the respective cancer type (columns of the heatmap). (B) Differential presence of isomiRs in lower grade glioma (LGG) tumor samples as compared to the rest of cancer types. Red indicates that the isomiR (row) was found as ‘present’ in LGG as compared to the respective cancer type (column), while green indicates ‘absence’ in LGG. The data from all possible pairwise comparisons is included in Supplementary Table S3. (C) Overlap between miRNA arms and isomiRs in the comparison of colon adenocarcinoma (COAD) with the rest of the cancer types. Red indicates that both the isomiR and arm were ‘present’ in COAD as compared to at least one other cancer type, green indicates they were ‘absent’. For example, miR-205-5p is ‘absent’ in COAD as well as its five isomiRs, miR-205-5p|0|0, miR-205-5p|0|−3, miR-205-5p|0|−2, miR-205-5p|0|−1 and miR-205-5p|0|+1. (D) Hierarchical clustering (HCL) (complete method) considering the number of differentially present isomiRs as the distance between cancer types. Colored clusters are described in the main text.
IsomiR production and miRNA-arm production are cancer dependent
We studied the binary differences of presence/absence of abundant isomiRs by conducting all pairwise comparisons among 32 cancers.
We discovered several isomiRs that are significantly present in one cancer and absent from many of the remaining cancers (Supplementary Table S3). LGG offers a characteristic such example. As mentioned above (Figure 1A), isomiRs from the miR-9-3p arm are present in LGG samples and absent from nearly all other cancers. On the contrary, isomiRs of miR-10a-5p and the mir-200 family are largely absent from LGG and present in 71–93% of the other cancers (Figure 1B). Another example can be seen in Supplementary Table S3: the mir-302 family and the mir-371/372/373 cluster express several isomiRs that are nearly exclusive to testicular germ cell tumors (TGCT) (71).
Use of ‘binarized miRNA-arm profiles’ largely replicated the results we obtained with ‘binarized isomiRs’ (Supplementary Table S4). In some cases, miRNA arms inherit the specificity of the isomiRs they produce: e.g. the miR-215-5p arm is specific to colon adenocarcinoma (COAD) (Supplementary Table S3) as are the isomiRs it produces (Supplementary Tables S3 and 4). This is visually summarized in Figure 1C. The left hand-side column of this figure includes the miRNA arms, as boxes, that are differentially present in the comparisons of COAD with at least one of the other cancer types. The right hand-side column of the figure shows isomiRs as boxes labeled with isomiR coordinate-pairs (see Loher et al. (30) for more details on this notation). If a miRNA arm box is connected to an isomiR box, then this is taken to mean that the specific isomiR from this arm is differentially present in COAD as compared to at least one other cancer type. The color of the line that connects the two boxes indicates whether the miRNA arm and the specific isomiR are ‘present’ in COAD (red color) or ‘absent’ (green color).
Next we examined how well HCL can classify the 32 cancer types when we use the cardinality of isomiRs that are differentially present between two cancers as a distance metric (Supplementary Table S3). The resulting dendrogram is shown in Figure 1D. In it, several interesting clusters can be seen. One cluster (light purple background) comprises almost all the adenocarcinomas including pancreatic ductal (PAAD) and prostate adenocarcinoma (PRAD). A second cluster (light orange background) comprises BRCA and BLCA (72) along with the squamous cell carcinoma of lung (LUSC) and head-and-neck (HNSC) (73). A third cluster (light yellow background) includes renal clear cell carcinoma (KIRC), renal papillary cell carcinoma (KIRP), hepatocellular carcinoma (LIHC) and cholangiocarcinoma (CHOL). Note also the clustering of the uveal (UVM) and skin (SKCM) melanomas (cyan background).
However, this univariate analysis is not suitable for tackling the multidimensional question of cancer classification, as it does not reveal the widespread and significant differences among cancers that are observed at the isomiR level (Supplementary Table S3).
Conventional multivariate clustering cannot separate all cancer types
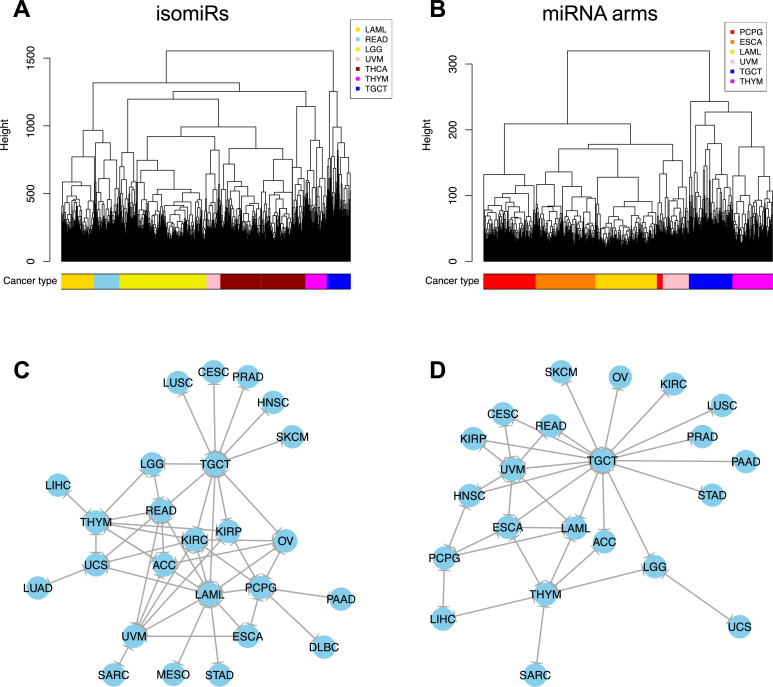
We used multivariate statistics to test whether the binarized isomiR and binarized miRNA-arm profiles can be used for tumor discrimination and classification at the sample level. Doing so allowed us to discriminate the samples from up to seven cancers using HCL with binarized isomiR profiles as features and Hamming distance as a metric (Figure 2A). Using binarized miRNA-arm profiles as features did not improve discrimination (Figure 2B).
Figure 2.
Multivariate HCL on the binary expression vectors. (A and B) HCL (Hamming distance as metric) on the isomiR (A) or miRNA arm (B) profile of samples from different cancer types. The leaves of the dendrogram are tumor datasets. The colored bar indicates the cancer type of the respective sample. (C and D) Networks of all potential pairwise discriminations using HCL (hamming distance as metric) on the isomiR (C) or miRNA arm profile (D). Two nodes (cancers) are connected if and only if the corresponding datasets are found to form two separate clusters in the respective comparison, with the datasets of one cancer clustered distinctly from the other.
To investigate the upper limit of discrimination power that can be achieved by using HCL + Hamming distance, we performed all pairwise comparisons and examined whether each cancer's samples clustered together. Figure 2C (binarized isomiRs) and D (binarized miRNA arms) show the results. Not surprisingly, the isomiR profiles (Figure 2C) help distinguish among more cancers by comparison to miRNA-arm profiles (Figure 2D). In the shown networks, LAML, TGCT, thymoma (THYM) and UVM act as ‘central hubs.’ In other words, these three cancers are easily distinguishable from several other cancers. The absence of nodes such as, e.g. COAD and thyroid cancer (THCA) from these networks highlights the limited ability of this clustering model.
Binarized isomiR profiles can discriminate among cancers
At the interface of biology and machine learning (63,74,75), SVMs have been the tool of choice for many multi-class classification problems (62,76,77). For our multi-cancer classification, we followed an SVM-based approach analogous to PhyloPythia (78,79), our classifier of metagenomes.
We ran a Monte-Carlo cross-validation approach for 1 000 iterations. In each iteration, we trained the SVM classifier using a random sample comprising 60% of the datasets. We used the remaining 40% of the datasets for testing (See ‘Materials and Methods’ section and Supplementary Data). Supplementary Table S5 contains an example of the probability vectors for the test datasets as well as the confusion matrix from one iteration. It is evident that correctly-classified datasets receive high probabilities (∼0.9 or higher). We note that the SVM models produced at each iteration are fairly similar to one another (Supplementary Figure S3A and B), which indicates that the training process is highly stable.
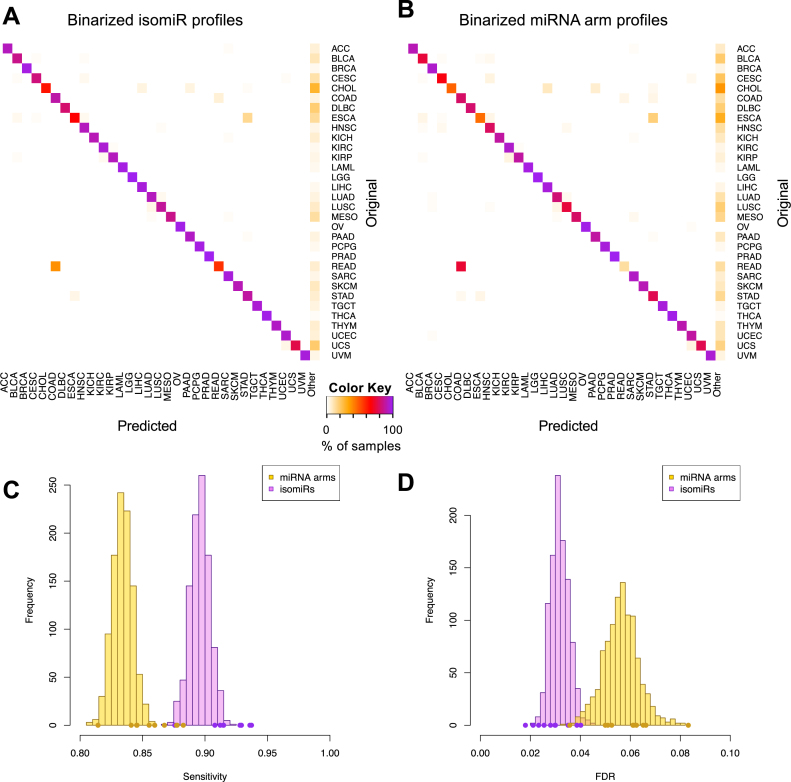
Figure 3A is a heatmap of the average prediction performance of the SVMs that used binarized isomiR profiles as features. Each row designates the test sample's cancer type. Each column designates the cancer type predicted by the classifier. The perfect classifier should not generate any non-diagonal entries (specific) or any entries in the ‘Other’ category (sensitive). As Figure 3A shows, the binarized isomiR features can discriminate among cancer types and classify samples correctly. One instance of seemingly decreased performance involves several rectum adenocarcinoma (READ) tumors that are ‘misclassified’ as COAD. This is not a mistake but rather an expected result, because READ and COAD tumors are molecularly similar and their distinction is largely driven by anatomy (35). We also note that the classifier shows an ability to very effectively distinguish cancer types that originate from the same organ, such as lung adenocarcinoma (LUAD) and LUSC, and the three kidney tumors, KIRP, KIRC and chromophobe renal cell carcinoma (KICH). Figure 3B shows the counterpart heatmap for SVMs that used binarized miRNA-arm profiles as features. Despite the sharp decrease in the amount of used information that results from using the miRNA-arm profiles, classification performance remains high. This suggests that simply examining whether a miRNA-arm produces one or more isomiRs above threshold can achieve satisfactory classification performance.
Figure 3.
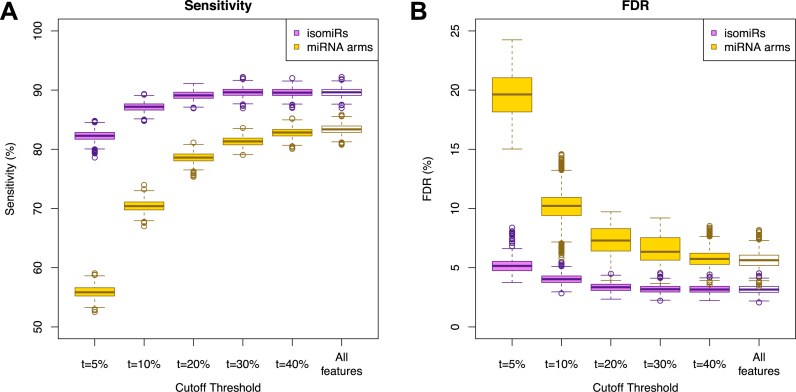
Support vector machines (SVMs) correctly classify 32 cancer types. (A and B) SVM classification using the binarized isomiR (A) or the miRNA arm (B) expression profile. Each row of the heatmap represents the original and each column the predicted cancer class. The color of each cell in the heatmap is proportional to the percentage (%) of samples originally as the cancer type in the respective row to be predicted as the cancer type of the respective column. The % is calculated as the average across 1 000 iterations. (C and D) Sensitivity (C) and FDR (D) scores for the SVM models built using the binarized isomiR (magenta) or miRNA arm (yellow) expression profiles. The points at the bottom of the distribution represent the sensitivity (C) and FDR (D) scores from the 10-fold cross-validation analysis.
Figure 3C and D depict the distribution of the sensitivity and FDR values respectively that were achieved by each of the 1000 SVMs that were built. Evidently, binarized isomiR profiles are very effective features, achieving an average pan-cancer sensitivity of 90% versus 83% when binarized miRNA-arm profiles are used. Even at 83%, the pan-cancer sensitivity of SVMs that leverage miRNA-arm information is high in absolute terms. The binarized miRNA-arm performance is particularly notable considering the difficulty of the task and how little information is actually available to these SVM models. The concomitant FDR scores further corroborate the effectiveness of the classification. SVM models that use the binarized isomiRs exhibit a mean FDR of 3% versus 6% for the models that use the binarized miRNA arms.
Lastly, we validated the SVM-based classification by shuffling dataset labels and by standard 10-fold cross-validation (Supplementary Figure S3 and Supplementary Methods). We also note that the differences in the number of datasets across cancer types did not affect the SVMs’ performance (Supplementary Figure S4 and Supplementary Methods).
Expression-based profiles perform less well than binarized isomiR profiles
In light of the ability of the binarized profiles to correctly classify samples, we hypothesized that taking into account the actual expression levels of the various isomiRs and miRNA arms would improve the classification. In order to generate a more comprehensive picture, we also considered the scenario where only the expression levels of the archetype miRNA (0|0 isomiRs; for details on isomiR nomenclature see (30,31)) were used as features. To enable a direct performance comparison with the classifiers we described already, we used the exact same iterative SVM approach as above, only this time we used as features the actual expression profiles of the (i) isomiRs, (ii) miRNA arms and (iii) archetype miRNA, respectively. Classification results, and the sensitivity and FDR scores were calculated as above.
Supplementary Figure S5 shows the results of this analysis. In all cases, use of the expression profiles resulted in a diminished ability to classify samples: the sensitivity was considerably lower and the FDR considerably higher, compared to the classifiers with binarized features. The classifier that used isomiR expression profiles achieved an average sensitivity of 67% with an FDR of 6%. When miRNA expression profiles were used instead, the classifier's sensitivity decreased to 64%, whereas the FDR increased to 8%. Lastly, we note that when the used features comprised the expression profile of only the archetype miRNAs, the classifier's sensitivity decreased further to 60% (t-test P-value < 10−4). Note added to the proofs: when we applied isomiR expression standardization across the training samples, the classifier's sensitivity improved to 83% at an FDR of 4% (data not shown) but continued to trail the performance of the classifier that used binarized features.
The most discriminatory isomiRs and miRNA arms are not the most studied
As the SVM attempts to identify the best-separating hyperplane in the multi-dimensional space, some of the features (isomiRs or miRNA arms) are given more weight than others. To elucidate these special features, we trained the SVM yet again, this time using the binarized profiles from all 9 291 primary tumor TCGA datasets and extracted the VI score for each feature (see ‘Materials and Methods’ section).
Our analysis shows that two isomiRs of miR-205-5p are deemed most important by the isomiR-based SVM classifiers (Supplementary Table S6), followed by several isomiRs from both arms of mir-141. Notably, SVM models, built on binarized isomiR features and on binarized miRNA-arm features respectively, tend to agree with regard to the genomic loci that each model deems important, e.g. mir-205, mir-141, mir-200c (Supplementary Tables S6 and 7).
To validate these findings, we used the RandomForest algorithm because of its ability to identify significant variables for classification (80). The VI scores from RandomForest are strongly and positively correlated with the VI scores from the SVM models: the Spearman rho correlation coefficient is 0.886 (P-value < 0.01). For the models using miRNA-arm features, the correlation of the VI scores improves to 0.932 (P-value < 0.01). The validation of the SVM conclusions by an independent algorithm adds further support to the relevance of using binarized profiles.
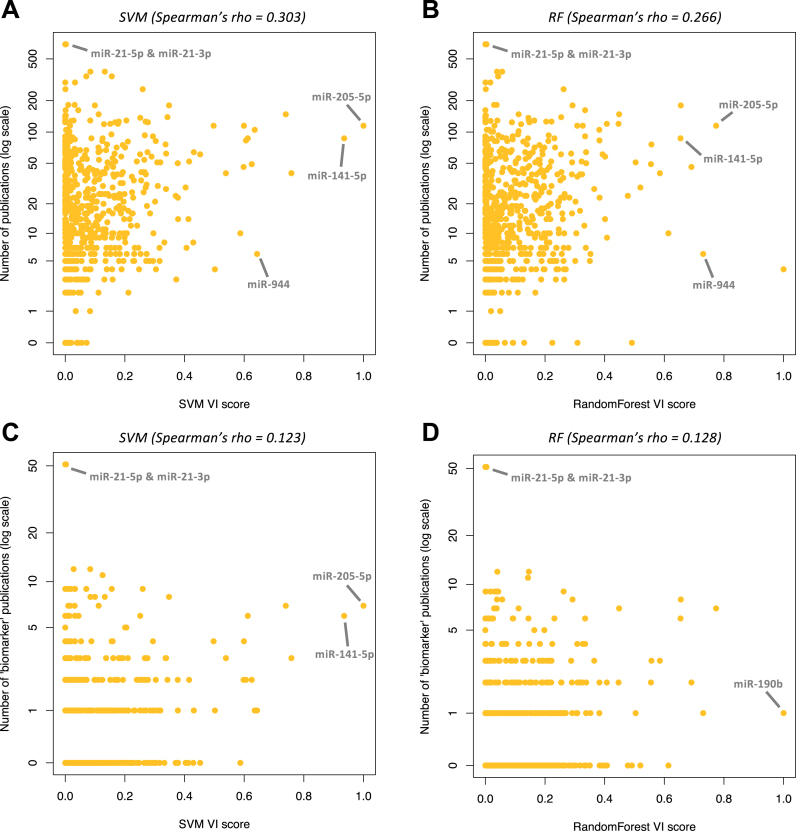
Having confirmed the VI scores, we associated the corresponding molecules with the number of PubMed entries (see ‘Materials and Methods’ section). We calculated a mean of 30 publications per mature miRNA arm. Figure 4A and B show the results for SVMs and RandomForest respectively. Strikingly, mir-21 is associated with the highest number (689) of publications. However, both SVM and RandomForest assign a considerably low VI score to mir-21's two arms, with regard to their discriminatory power. Conversely, both SVM and RandomForest deem miR-944, with only six PubMed entries, as one of the most important for cancer classification (Supplementary Table S7). Other highly discriminatory miRNAs with few PubMed entries include miR-194-3p, miR-192-3p and miR-135a-5p with 10, 46 and 29 publications respectively.
Figure 4.
The number of existing publications does not correlate with a miRNA's importance for classification. (A and B) Number of publications against the variable importance (VI) score as calculated in the SVM (A) or RandomForest (B) classification model based on the binary miRNA arm profiles. Spearman correlation coefficients: for SVM: 0.303, for RandomForest: 0.266. (C and D) Number of publications with the string ‘biomarker’ or ‘signature’ in the title against the VI score as extracted from the SVM (C) or RandomForest (D) models that were trained with the binarized miRNA-arm profiles. Spearman correlation coefficients: 0.123 for SVM and 0.128 for RandomForest.
A similarly weak correlation characterizes the number of PubMed entries and the number of times a miRNA arm is found differentially present between two cancer types (Supplementary Figure S6A). We note that the miRNA arms (or isomiRs) with the most impact on cancer classification are those found to be differentially present in many cancer-type comparisons (Supplementary Figure S6B and C). This means that, for the purpose of multivariate SVM-based classification, isomiRs and miRNA arms that are present in (or absent from, respectively) only one cancer type are not the most valuable. Nonetheless, such isomiRs and miRNA arms do remain important for the biology of the cancer in which they are exclusively present or from which they are exclusively absent. RandomForest models produced similar results for both isomiRs and miRNA arms (Supplementary Figure S6D and E).
One could argue that the rationale for studying a miRNA locus in the first place lies in its role as a producer of oncogenic or tumor-suppressing miRNAs and not its utilization as a biomarker. We thus filtered the publication lists and kept only articles that contained the string ‘biomarker’ or the string ‘signature’ in the title and correlated with the VI scores as extracted from the SVM (Figure 4C) and RandomForest (Figure 4D) algorithms. The results show a very weak correlation of the public literature with the importance of specific miRNA loci as multi-cancer classification features. Even this biomarker-specific list of publications remains biased in favor of mir-21, whereas the most discriminatory miRNAs have fewer than 10 publications.
Use of a reduced set of features preserves the ability to classify with binarized profiles
Since only a relatively small number of isomiRs and miRNA arms have high VI scores, we examined whether we could obtain reasonable classification using a reduced set comprising the most important isomiR or miRNA-arm features.
We selected among the most important isomiRs from the SVM and RandomForest models by thresholding at five different values: top 5%, top 10%, top 20%, top 30% and top 40%. For each selection, we used the resulting isomiRs to train 2 000 multi-cancer SVM-based classifiers: 1 000 made use of binarized isomiR profiles and another 1 000 of binarized miRNA-arm profiles. Figure 5 shows the resulting sensitivity and FDR for each choice of cutoff threshold. Binarized isomiRs maintain their ability to correctly classify datasets, even after a >15× reduction in the number of used features. Indeed, when using the top 5% (456) most important isomiRs, and following 1 000 training/testing iterations, we observed an average sensitivity of 82% (from the original 90%) at an average FDR of 5% (from the original 3%) (Figure 5).
Figure 5.
Boxplots of the sensitivity (A) and FDR (B) for the SVM classification using reduced lists of features for different cutoff thresholds t for the most important isomiRs or miRNA arms. The distributions with the full isomiR and miRNA profiles are also included for comparison.
We also repeated the analysis and built our SVM models using the most important binarized miRNA-arm features. Even the considerably reduced signature of 47 miRNA arms (top 10%) maintained a reasonable ability to classify samples exhibiting a sensitivity of 70% (from the original 83% when all features are used), but a rather increased FDR of 10% (from the original 6% when using all features) (Figure 5).
The most discriminatory isomiRs are differentially expressed in normal versus cancer comparisons
During the training of the SVM classifiers, we had excluded normal or metastatic tissue samples. We investigated the effectiveness of our pan-cancer classifier and studied its predictions when it was presented with these samples. To this end, we used the SVM model that was trained with all of the TCGA tumor datasets. As can be seen from Supplementary Figure S7, the resulting classification exhibited a tissue-specific component. In other words, many of the normal and metastatic samples were classified to the cancer type of the organ of origin. For organs that give rise to distinct types of cancers, the normal samples clustered with one of the organ's cancers: e.g. normal lung samples were labeled as ‘LUAD,’ and normal kidney samples as ‘KIRP.’ In some instances, as was the case with COAD and READ, the samples were labeled ‘Other,’ indicating uncertainty by the algorithm as to how to best classify them.
The findings of this first foray suggest that the classifier may also capture a component that relates to the state of the tissue/organ of origin. We hypothesized that the most discriminatory isomiR features across cancers are also deregulated between the normal and the cancerous state of the corresponding tissue. To test this hypothesis, we focused on the eight cancer types for which an adequate number of normal samples are available as part of the TCGA repository. We performed SAM analyses and identified those isomiRs that were DE between the normal and the tumor samples (Supplementary Table S8). We observed that isomiRs with high VI scores happen to also be DE in these comparisons.
Binarized miRNA-arm profiles from TCGA can correctly classify non-TCGA datasets
In spite of our extensive tests, there remains the formal possibility that our models have been over-fitted to TCGA and are not extendible to non-TCGA datasets. To investigate this possibility, we sought public non-TCGA datasets that were generated by independent sequencing platforms or microarrays.
Before continuing, it is important to stress that, from a statistical point of view, the ideal classifier should be trained using datasets that are a balanced representation not only of the cancer types, but also of the data generating platforms (e.g. deep sequencing as well as microarrays). This is not feasible with the currently available miRNA data, because data for many cancer types are either under-represented or absent from public repositories. In addition, internal control standards, like spike-in standards (81), among and within sequencing and microarray platforms, which would ensure a consistent binarization of the features, are lacking. Moreover, the set of features (in this case, miRNA arms or isomiRs) should be significantly large to adequately account for technical (e.g. library preparation) or biological (e.g. race, population, disease subtype) variations and biases among datasets.
Taking the above considerations into account, we reduced the number of cancer types in our analysis while keeping the same number of features (miRNA arms), in order to increase statistical power. We selected six commonly detected and deadly cancer types with large sample sizes in the TCGA cohort: BRCA, LAML, LIHC, PRAD, COAD and LUAD. We then trained an SVM classifier using the binarized miRNA-arm profiles and all tumor samples that are available in the TCGA repository for these six cancer types. The resulting classifier was used to label publicly available datasets (29,68–70) (see Supplementary Methods for details) that were acquired independently from TCGA and from a diverse set of platforms, including microarrays and RNA-seq with a different deep-sequencing chemistry (ABI SOLiD). Table 1 shows the results of this analysis. As can be seen, the TCGA-trained SVM classifier performed well and correctly classified usually more than 90% of the samples in each of the project. Supplementary Table S9 contains the prediction probabilities for each sample. These validation steps demonstrate the utility of binarized miRNA arm profiles of TCGA datasets for training an SVM classifier that does not exhibit overfitting to the TCGA project and can be used to analyze data from other projects and platforms.
Table 1. Predictions of non-TCGA datasets using the SVM model trained with the binarized miRNA arm profiles of the TCGA tumor samples.
| Accession number | Platform | Cancer type | Samples correctly classified (%) |
|---|---|---|---|
| GSE35834 | Affymetrix miRNA Array | COAD | 26 (84%) |
| GSE36802 | Affymetrix miRNA Array | PRAD | 16 (76%) |
| GSE53159 | Affymetrix miRNA Array | COAD | 29 (91%) |
| GSE67138 | Affymetrix miRNA Array | LIHC | 52 (91%) |
| GSE67139 | Affymetrix miRNA Array | LIHC | 110 (96%) |
| SRP034550 | ABI SOLiD sequencing | PRAD | 5 (100%) |
| SRP034557 | ABI SOLiD sequencing | BRCA | 2 (100%) |
DISCUSSION
We sought to determine whether binarized isomiR and binarized miRNA-arm profiles can be used to classify datasets in a pan-cancer setting. If validated independently, these features could become potential biomarkers. Our work was spurred by previous observations that miRNA profiles can be tissue-specific (27–29,82,83) and cell type differences can be described adequately by the presence (or absence) of RNA transcripts (33,34). Our analysis led to several observations.
For instance, we discovered isomiRs with cancer-specific expression. For example, the isomiRs of mir-9, a miRNA that is highly expressed in the nervous system and has important roles in neuronal development and diseases (84,85), are present exclusively in LGG datasets. Similar comments can be made for LGG and the isomiRs of mir-219, a miRNA that is implicated in neural differentiation processes (86). Also, isomiRs from the mir-302 family, which has important roles in stem cell pluripotency and cell reprogramming (87,88) are present in TGCT exclusively.
We also identified miRNA loci with cancer-specific expression. Examples include the novel miRNA ID00737-3p (29) for THCA, mir-671 for OV and other. OV is particularly interesting in that a large number of isomiRs and miRNA arms, which are expressed in other cancer types, are absent from OV. On the contrary, tumors of the male reproductive system, TGCT, are characterized by a gain of isomiRs and miRNAs as compared to all other tumor types.
Molecules with tissue-specific expression are invaluable biomarker candidates. Consequently, ubiquitous isomiRs and miRNA arms are suboptimal choices in this regard (89). Two characteristic examples include mir-21 and let-7a: both loci produce many abundant isomiRs in most of the studied TCGA datasets and all 32 cancer types. The miRNA molecules studied to date exhibit mixed expression patterns across tissues, which previously led to the proposed use of miRNA ‘panels’ as biomarkers (90–92).
In a pan-cancer setting, we found that an SVM-based classifier using binarized isomiR profiles as features can label datasets accurately (≥90% sensitivity, FDR<5%). SVMs using binarized isomiR features outperformed SVMs using binarized miRNA-arm features (Figure 3), even when the number of features was reduced by 15× (Figure 5). Use of binarized miRNA-arm features lowered the prediction rates. Conceivably, this is because modeling at the miRNA arm level inherently discards information that may be reflecting underlying biological events (93–95). This area remains largely unexplored and the number of reports on the discriminatory power of isomiRs is currently limited (31,96).
We stress that causative links should not be inferred based on our analysis. Additionally, the identified tissue-specific isomiRs and miRNA arms are not guaranteed to be expressed only when the tissue enters its cancer state. However, we found that the most important isomiRs for the discrimination of various cancer types were also present in normal tissue but, importantly, were DE in the cancer state. These results suggest a tissue-specific trajectory of deregulation from the normal to the cancer state (97). Similar to previous studies (28), further resolving this question hinges upon the availability of ‘normal’ samples. Such samples do not currently exist in adequate numbers and for multiple tissues. As the TCGA initiative focused on tumor classifications, only a limited number of normal samples were included. Thus, alternative sources will be needed to embark to addressing these questions.
Supplementary Material
ACKNOWLEDGEMENTS
Author Contributions: I.R. conceived the study. A.G.T. and I.R. designed and supervized the study with contributions from I.C. and E.L.. A.G.T., R.M. and P.L. downloaded and corrected the expression and clinical data. A.G.T. performed the analyses with contributions from R.M. and P.L.. A.G.T. and I.R. wrote the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
W. M. Keck Foundation (to I.R., in part); Institutional Funds. Funding for open access charge: Institutional Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Veneziano D., Di Bella S., Nigita G., Lagana A., Ferro A., Croce C.M.. Noncoding RNA: current deep sequencing data analysis approaches and challenges. Hum. Mutat. 2016; 37:1283–1298. [DOI] [PubMed] [Google Scholar]
- 2. Aalto A.P., Pasquinelli A.E.. Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 2012; 24:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 4. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clark M.B., Choudhary A., Smith M.A., Taft R.J., Mattick J.S.. The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem. 2013; 54:1–16. [DOI] [PubMed] [Google Scholar]
- 6. Clerget G., Abel Y., Rederstorff M.. Small non-coding RNAs: a quick look in the rearview mirror. Methods Mol. Biol. 2015; 1296:3–9. [DOI] [PubMed] [Google Scholar]
- 7. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 8. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winter J., Jung S., Keller S., Gregory R.I., Diederichs S.. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009; 11:228–234. [DOI] [PubMed] [Google Scholar]
- 10. Dumortier O., Fabris G., Van Obberghen E.. Shaping and preserving beta-cell identity with microRNAs. Diabetes Obes. Metab. 2016; 18(Suppl. 1):51–57. [DOI] [PubMed] [Google Scholar]
- 11. Edelstein L.C., McKenzie S.E., Shaw C., Holinstat M.A., Kunapuli S.P., Bray P.F.. MicroRNAs in platelet production and activation. J. Thromb. Haemost. 2013; 11(Suppl. 1):340–350. [DOI] [PubMed] [Google Scholar]
- 12. Flowers E., Won G.Y., Fukuoka Y.. MicroRNAs associated with exercise and diet: a systematic review. Physiol. Genomics. 2015; 47:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hilz S., Modzelewski A.J., Cohen P.E., Grimson A.. The roles of microRNAs and siRNAs in mammalian spermatogenesis. Development. 2016; 143:3061–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeon T.I., Osborne T.F.. miRNA and cholesterol homeostasis. Biochim. Biophys. Acta. 2016; 1861:2041–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee R.C., Feinbaum R.L., Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 16. Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G.. The 21-nucleotide let-7 RNA regulates developmental timing in caenorhabditis elegans. Nature. 2000; 403:901–906. [DOI] [PubMed] [Google Scholar]
- 17. Sparks E., Wachsman G., Benfey P.N.. Spatiotemporal signalling in plant development. Nat. Rev. Genet. 2013; 14:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehta A., Baltimore D.. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016; 16:279–294. [DOI] [PubMed] [Google Scholar]
- 19. Mogilyansky E., Rigoutsos I.. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013; 20:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su Y., Wu H., Pavlosky A., Zou L.L., Deng X., Zhang Z.X., Jevnikar A.M.. Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death Dis. 2016; 7:e2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calin G.A., Croce C.M.. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006; 6:857–866. [DOI] [PubMed] [Google Scholar]
- 22. Di Leva G., Garofalo M., Croce C.M.. MicroRNAs in cancer. Annu. Rev. Pathol. 2014; 9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Backes C., Sedaghat-Hamedani F., Frese K., Hart M., Ludwig N., Meder B., Meese E., Keller A.. Bias in high-throughput analysis of miRNAs and Implications for biomarker studies. Anal. Chem. 2016; 88:2088–2095. [DOI] [PubMed] [Google Scholar]
- 24. Cortez M.A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A.K., Calin G.A.. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011; 8:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. et al. . MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838. [DOI] [PubMed] [Google Scholar]
- 26. Pimentel F., Bonilla P., Ravishankar Y.G., Contag A., Gopal N., LaCour S., Lee T., Niemz A.. Technology in MicroRNA Profiling: circulating MicroRNAs as noninvasive cancer biomarkers in breast cancer. J. Lab. Autom. 2015; 20:574–588. [DOI] [PubMed] [Google Scholar]
- 27. Rosenfeld N., Aharonov R., Meiri E., Rosenwald S., Spector Y., Zepeniuk M., Benjamin H., Shabes N., Tabak S., Levy A. et al. . MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008; 26:462–469. [DOI] [PubMed] [Google Scholar]
- 28. Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M. et al. . A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Londin E., Loher P., Telonis A.G., Quann K., Clark P., Jing Y., Hatzimichael E., Kirino Y., Honda S., Lally M. et al. . Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E1106–E1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loher P., Londin E.R., Rigoutsos I.. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014; 5:8790–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Telonis A.G., Loher P., Jing Y., Londin E., Rigoutsos I.. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015; 43:9158–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shmulevich I., Zhang W.. Binary analysis and optimization-based normalization of gene expression data. Bioinformatics. 2002; 18:555–565. [DOI] [PubMed] [Google Scholar]
- 33. Tuna S., Niranjan M.. Classification with binary gene expressions. J. Biomed. Sci. Eng. 2009; 02:390–399. [Google Scholar]
- 34. Zilliox M.J., Irizarry R.A.. A gene expression bar code for microarray data. Nat. Methods. 2007; 4:911–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015; 517:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015; 161:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013; 368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013; 499:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014; 507:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014; 513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014; 159:676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015; 163:1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cancer Genome Atlas Research Network. Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R., Cooper L.A., Rheinbay E., Miller C.R., Vitucci M. et al. . Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015; 372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cancer Genome Atlas Research Network. Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R. et al. . Integrated genomic characterization of endometrial carcinoma. Nature. 2013; 497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cancer Genome Atlas Research Network. Linehan W.M., Spellman P.T., Ricketts C.J., Creighton C.J., Fei S.S., Davis C., Wheeler D.A., Murray B.A., Schmidt L. et al. . Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016; 374:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M. et al. . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016; 164:550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., McLellan M., Yau C., Kandoth C. et al. . Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015; 163:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H., Buhay C., Kang H., Kim S.C., Fahey C.C. et al. . The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014; 26:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zheng S., Cherniack A.D., Dewal N., Moffitt R.A., Danilova L., Murray B.A., Lerario A.M., Else T., Knijnenburg T.A., Ciriello G.. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016; 29:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chu A., Robertson G., Brooks D., Mungall A.J., Birol I., Coope R., Ma Y., Jones S., Marra M.A.. Large-scale profiling of microRNAs for The Cancer Genome Atlas. Nucleic Acids Res. 2016; 44:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. R Core Team R: a Language and Environment for Statistical Computing. 2014; Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 58. Meyer D., Dimitriadou E., Hornik K., Weingessel A., Leisch F.. e1071: Misc functions of the department of statistics, probability theory group (formerly: E1071), TU Wien. 2015; R package version 1.6-7.
- 59. Lucas A. amap: another multidimensional analysis package. 2014; R package version 0.8-14.
- 60. Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015; 31:3718–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Csardi G., Nepusz T.. The igraph software package for complex network research. InterJournal, Complex Systems. 2006; 1695:1–9. [Google Scholar]
- 62. Noble W.S. What is a support vector machine. Nat. Biotechnol. 2006; 24:1565–1567. [DOI] [PubMed] [Google Scholar]
- 63. Kourou K., Exarchos T.P., Exarchos K.P., Karamouzis M.V., Fotiadis D.I.. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015; 13:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Guyon I., Weston J., Barnhill S., Vapnik V.. Gene selection for cancer classification using support vector machine. Mach. Learn. 2002; 46:389–422. [Google Scholar]
- 65. Aiello S., Kraljevic T., Maj P. contributions from the H2O.ai team . h2o: R Interface for H2O. 2016; R package version 3.8.2.6.
- 66. Tusher V.G., Tibshirani R., Chu G.. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. . Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen D.L., Wang Z.Q., Zeng Z.L., Wu W.J., Zhang D.S., Luo H.Y., Wang F., Qiu M.Z., Wang D.S., Ren C. et al. . Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014; 60:598–609. [DOI] [PubMed] [Google Scholar]
- 69. Lin P.C., Chiu Y.L., Banerjee S., Park K., Mosquera J.M., Giannopoulou E., Alves P., Tewari A.K., Gerstein M.B., Beltran H. et al. . Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013; 73:1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pizzini S., Bisognin A., Mandruzzato S., Biasiolo M., Facciolli A., Perilli L., Rossi E., Esposito G., Rugge M., Pilati P. et al. . Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013; 14:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A. et al. . A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv. Exp. Med. Biol. 2007; 604:17–46. [DOI] [PubMed] [Google Scholar]
- 72. Damrauer J.S., Hoadley K.A., Chism D.D., Fan C., Tiganelli C.J., Wobker S.E., Yeh J.J., Milowsky M.I., Iyer G., Parker J.S. et al. . Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3110–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hoadley K.A., Yau C., Wolf D.M., Cherniack A.D., Tamborero D., Ng S., Leiserson M.D., Niu B., McLellan M.D., Uzunangelov V. et al. . Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014; 158:929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Angermueller C., Parnamaa T., Parts L., Stegle O.. Deep learning for computational biology. Mol. Syst. Biol. 2016; 12:878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. LeCun Y., Bengio Y., Hinton G.. Deep learning. Nature. 2015; 521:436–444. [DOI] [PubMed] [Google Scholar]
- 76. Tsirigos A., Rigoutsos I.. A sensitive, support-vector-machine method for the detection of horizontal gene transfers in viral, archaeal and bacterial genomes. Nucleic Acids Res. 2005; 33:3699–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yang Z.R. Biological applications of support vector machines. Brief Bioinform. 2004; 5:328–338. [DOI] [PubMed] [Google Scholar]
- 78. McHardy A.C., Martin H.G., Tsirigos A., Hugenholtz P., Rigoutsos I.. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods. 2007; 4:63–72. [DOI] [PubMed] [Google Scholar]
- 79. Warnecke F., Luginbuhl P., Ivanova N., Ghassemian M., Richardson T.H., Stege J.T., Cayouette M., McHardy A.C., Djordjevic G., Aboushadi N. et al. . Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007; 450:560–565. [DOI] [PubMed] [Google Scholar]
- 80. Statnikov A., Wang L., Aliferis C.F.. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics. 2008; 9:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M., Gingeras T.R., Oliver B.. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011; 21:1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M. et al. . A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007; 129:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McCall M.N., Jaffee H.A., Zelisko S.J., Sinha N., Hooiveld G., Irizarry R.A., Zilliox M.J.. The Gene expression barcode 3.0: improved data processing and mining tools. Nucleic Acids Res. 2014; 42:D938–D943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Coolen M., Katz S., Bally-Cuif L.. miR-9: a versatile regulator of neurogenesis. Front. Cell Neurosci. 2013; 7:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yuva-Aydemir Y., Simkin A., Gascon E., Gao F.B.. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011; 8:557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hudish L.I., Galati D.F., Ravanelli A.M., Pearson C.G., Huang P., Appel B.. miR-219 regulates neural progenitors by dampening apical Par protein-dependent Hedgehog signaling. Development. 2016; 143:2292–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Barroso-del Jesus A., Lucena-Aguilar G., Menendez P.. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009; 8:394–398. [DOI] [PubMed] [Google Scholar]
- 88. Gao Z., Zhu X., Dou Y.. The miR-302/367 cluster: a comprehensive update on its evolution and functions. Open Biol. 2015; 5:150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shi J. Considering exosomal miR-21 as a biomarker for cancer. J. Clin. Med. 2016; 5, doi:10.3390/jcm5040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hornick N.I., Huan J., Doron B., Goloviznina N.A., Lapidus J., Chang B.H., Kurre P.. Serum exosome MicroRNA as a minimally-invasive early biomarker of AML. Sci. Rep. 2015; 5:11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang Y., Liang J., Di C., Zhao G., Zhao Y.. Identification of miRNAs as potential new biomarkers for nervous system cancer. Tumour Biol. 2014; 35:11631–11638. [DOI] [PubMed] [Google Scholar]
- 92. Xin H., Li X., Yang B., Zhang L., Han Z., Han C.. Blood-based multiple-microRNA assay displays a better diagnostic performance than single-microRNA assay in the diagnosis of breast tumor. Tumour Biol. 2014; 35:12635–12643. [DOI] [PubMed] [Google Scholar]
- 93. Boele J., Persson H., Shin J.W., Ishizu Y., Newie I.S., Sokilde R., Hawkins S.M., Coarfa C., Ikeda K., Takayama K. et al. . PAPD5-mediated 3΄ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11467–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Koppers-Lalic D., Hackenberg M., Bijnsdorp I.V., van Eijndhoven M.A., Sadek P., Sie D., Zini N., Middeldorp J.M., Ylstra B., de Menezes R.X. et al. . Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014; 8:1649–1658. [DOI] [PubMed] [Google Scholar]
- 95. Starega-Roslan J., Witkos T.M., Galka-Marciniak P., Krzyzosiak W.J.. Sequence features of drosha and dicer cleavage sites affect the complexity of isomiRs. Int. J. Mol. Sci. 2015; 16:8110–8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Koppers-Lalic D., Hackenberg M., de Menezes R., Misovic B., Wachalska M., Geldof A., Zini N., de Reijke T., Wurdinger T., Vis A. et al. . Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016; 7:22566–22578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schaefer M.H., Serrano L.. Cell type-specific properties and environment shape tissue specificity of cancer genes. Sci. Rep. 2016; 6:20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.