Abstract
Although all Type II restriction endonucleases catalyze phosphodiester bond hydrolysis within or close to their DNA target sites, they form different oligomeric assemblies ranging from monomers, dimers, tetramers to higher order oligomers to generate a double strand break in DNA. Type IIP restriction endonuclease AgeI recognizes a palindromic sequence 5΄-A/CCGGT-3΄ and cuts it (‘/’ denotes the cleavage site) producing staggered DNA ends. Here, we present crystal structures of AgeI in apo and DNA-bound forms. The structure of AgeI is similar to the restriction enzymes that share in their target sites a conserved CCGG tetranucleotide and a cleavage pattern. Structure analysis and biochemical data indicate, that AgeI is a monomer in the apo-form both in the crystal and in solution, however, it binds and cleaves the palindromic target site as a dimer. DNA cleavage mechanism of AgeI is novel among Type IIP restriction endonucleases.
INTRODUCTION
Type II restriction endonucleases (REases) belong to four different nuclease families: PD-(D/E)XK, PLD, HNH and GYI-IYG (1,2). PD-(D/E)XK family REases which recognize palindromic DNA sequences assemble into different oligomeric structures to generate double strand breaks in DNA (2,3). Orthodox Type IIP REases are arranged as dimers and each monomer contains an active site that acts on one DNA strand within a symmetrical target site. Tetrameric restriction enzymes are composed of two primary dimers that are similar to those of orthodox REases (4). Tetrameric REases require binding to two target sites simultaneously and cleave four phosphodiester bonds in a concerted manner. Intermediate variants, exemplified by Ecl18kI, BsaWI and SgrAI, exist as dimers in the apo form, but cleave DNA as tetramers (Ecl18kI and BsaWI) or ‘run-on’ oligomers (SgrAI) (3,5,6). Monomeric Type II restriction enzymes interact with their palindromic (MspI and HinP1I) or pseudo-palindromic (BcnI and MvaI) sites as monomers: a single protein subunit makes contacts with both parts of the palindromic target site (7–10). Monomeric REases contain a single active site and cleave both target strands sequentially (8,11). On the other hand, the Type IIS restriction enzyme FokI is a monomer, composed of two domains: the N-terminal DNA recognition domain, which recognizes asymmetric sequence 5΄-GGATG-3΄ as a monomer, and the C-terminal PD-(D/E)XK nuclease domain that contains a single active site and lacks sequence specificity (12). To achieve a double strand break in DNA the catalytic domains from two separate monomers associate to form a dimer with two active sites (13). The second catalytic domain can come from the FokI monomer in solution or bound to another target site. The dimerization interface between FokI catalytic domains is small and the dimer formed between DNA-bound and unbound FokI monomers is presumably unstable; therefore a single site DNA is cleaved by FokI at a low rate (14,15). On DNA with two recognition sites, the FokI dimer is presumably formed by two DNA-bound FokI monomers. Consequently, FokI cleaves DNA substrates with two copies of its recognition sequence more rapidly than DNA containing one target site (14).
Restriction endonuclease AgeI from Agrobacterium gelatinovorum recognizes a palindromic DNA sequence 5΄-A/CCGGT-3΄ and cleaves it as indicated by ‘/’. AgeI is a part of a typical Type II restriction-modification system composed of a restriction endonuclease and a DNA methyltransferase (http://rebase.neb.com/rebase/rebase.html). It belongs to a well-characterized group of REases termed here the CCGG-family which contain a conserved CCGG tetranucleotide within their target sites and share a cleavage pattern of a 4-base 5΄-overhang. The CCGG-family REases exhibit a variety of DNA cleavage mechanisms: they exist as dimers, tetramers or oligomers and require one, two or three DNA target site copies for optimal DNA cleavage (6). AgeI shares a significant sequence similarity (24% identical, 41% similar aa) with the BsaWI REase which recognizes a related DNA sequence 5΄-W/CCGGW-3΄ (W stands for A or T) (6). To establish the molecular mechanism of DNA cleavage we performed structural and biochemical characterization of AgeI. First, we solved crystal structures of AgeI in apo- and DNA-bound forms. We show that in DNA-free form AgeI is a monomer both in the crystal and in solution. Next, we demonstrate that in the DNA-bound form in the crystal AgeI is a dimer and shows a conserved interaction pattern with the CCGG tetranucleotide characteristic for other CCGG-family enzymes, although in AgeI only a part of the R-(D/E)R motif conserved between the CCGG-family proteins is employed for the target recognition. We further show that AgeI also dimerizes in solution upon DNA binding and cleaves DNA as a dimer supporting the structural model. Taken together, structural and biochemical data suggest that AgeI uses a DNA cleavage mechanism unique for Type IIP REases but similar to that of IIS restriction enzyme FokI.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides used in this study were synthesized by Metabion. DNA oligonucleotides used in crystallization, mutagenesis, DNA binding and cleavage studies are presented in Supplementary Table S1. Oligoduplexes for crystallization were assembled by slow annealing from 95°C to room temperature in a buffer (10 mM Tris–HCl (pH 8.0 at 25°C), 50 mM NaCl).
Cloning and mutagenesis
A plasmid pRRSAgeIRM was constructed by inserting a 2.1 kb BamHI PCR fragment bearing ageIRM genes into pRRS expression vector. AgeI mutants were obtained by the modified QuickChange Mutagenesis Protocol (16). The ageIR gene was cloned into pET-Duet expression vector with N-terminal His6-tag (His-AgeI) using standard procedures. Sequencing of the entire genes of the mutants confirmed that only the designed mutations had been introduced.
Protein expression and purification
The wild type (wt) and mutant AgeI proteins were expressed in Escherichia coli ER2566 cells carrying plasmids pACYC-HpaII.M(CmR), pVH1 (KnR) and pRRSAgeIRM(ApR) (or a corresponding plasmid, containing the mutant ageIR gene), were grown in the LB medium supplemented with 30 μg/ml chloramphenicol (Cm), 25 μg/ml kanamycin (Kn) and 50 μg/ml carbenicillin (Cb). pACYC-HpaII.M plasmid carrying HpaII m5C methylase specific for the CCGG sequence (modified cytosine underlined) was used to protect host DNA from AgeI cleavage. Plasmid pVH1 (KnR, lacIq (17)) ensured transcription repression of the ageIR gene in the absence of an inductor. Protein expression was performed by cultivation for 4 h at 30°C in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). To isolate AgeI the cells were re-suspended in the Purification Buffer 1 (10 mM K-phosphate (pH 7.4), 0.1 M NaCl, 1 mM EDTA, 7 mM 2-mercaptoethanol) supplemented with 2 mM phenylmethanesulfonylfluoride (PMSF) and sonicated. The supernatant was subjected to a subsequent chromatography on Heparin-sepharose, Blue-sepharose and Q-Sepharose (GE Healthcare). Fractions containing the target protein were dialyzed against the Storage buffer (30 mM Tris–HCl (pH 7.4), 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 50% glycerol) and stored at −20°C.
The His-AgeI protein was expressed in E. coli ER2566 cells carrying plasmids pACYC-HpaII.M (CmR), pVH1 (KnR) and pET-Duet-His-AgeIR (ApR). Cells were grown as described above for the wt AgeI expressing cells. For His-AgeI isolation the cells were initially re-suspended in the Purification Buffer 2 (20 mM Tris–HCl pH 8.0, 500 mM NaCl) supplemented with 2 mM PMSF and sonicated. The supernatant was subjected for a subsequent chromatography on HiTrap chelating HP and HiTrap Heparin columns (GE Healthcare). Fractions containing His-AgeI were dialyzed against the Storage buffer and stored at −20°C. His-AgeI DNA binding and cleavage properties were similar to those of wt AgeI (data not shown).
Concentrations of the wt and mutant proteins were determined by measuring UV absorbance at 280 nm using extinction coefficient 24410 M−1 cm−1 calculated by the ProtParam tool (http://web.expasy.org/protparam/). All protein concentrations are given in terms of monomer if not stated otherwise.
Crystallization and structure determination
For crystallization AgeI was dialyzed against the Crystallization buffer (10 mM Tris–HCl (pH 7.5), 200 mM NaCl) and concentrated to 6–10 mg/ml. AgeI–DNA complexes for crystallization were prepared by mixing concentrated AgeI protein with DNA oligoduplex SP11 or SP13 (Supplementary Table S1) in 1:1.2 ratio to a final protein concentration of 8.3 mg/ml. Crystallization was performed by sitting drop technique at 20°C. We obtained crystals of apo-AgeI diffracting to 2.5 Å resolution and three different crystal forms of AgeI–DNA complexes: form I and form II with 11 bp oligonucleotide SP11 (Supplementary Table S1), and form III with 13 nt oligonucleotide SP13. The crystals of the apo-protein belong to P212121 space group and diffract X-rays at 2.5 Å resolution. Form I crystals belong to the space group P6122 and diffract X-rays to 2.7 Å resolution, form II—space group P212121 and diffract to 1.5 Å resolution; form III crystals belong to space group P21 and diffract to 2.4 Å resolution. Crystallization and cryo-protection conditions are given in Supplementary Table S2. Diffraction data were collected at EMBL DESY (Germany) beamlines BW7B (form I, native and Hg derivative), X13 (form III) and X12 (form II), in-house Rigaku MICROMAX-007 HF (form II Hg derivative), MAX Lab I911-3 (Sweden) beamline (apo). MOSFLM (18), SCALA and TRUNCATE (19) were used for data processing.
AgeI–DNA structure was solved by AutoRikshaw in P6122 space group (form I) by SIRAS protocol of AutoRikshaw (20) using Hg derivative (soak in 1 mM HgCl2). Four heavy atoms were found using the programs SHELXCD (21,22). The occupancy of all substructure atoms was refined using the program BP3 (23). The initial phases were improved using density modification and phase extension by the program DM (24). A partial alpha-helical model (212 residues) was produced using the program HELICAP (25). However, only a partial model containing N-terminal domain and DNA could be built using Coot (26). The partial model was used in molecular replacement (MR) by MOLREP (27) in P21 and P212121 space groups; however phases were not sufficient for model improvement. We collected additional dataset of Hg derivative (0.2 mM (C2H5HgO)2HPO2 (EMP)) of crystal form II. Heavy atom sites were found using program HARA (S.G., unpublished), SIRAS phases were calculated using MLPHARE (28). However, SIRAS phases also were weak and did not allow improving the model. Therefore we performed map averaging by DM of MR map and SIRAS map of the crystal form II which lead to a better interpretable map. The model was improved manually and subjected to automated molecular replacement by AutoRikshaw: molecular replacement with MOLREP, refinement with REFMAC and model rebuilding by ARP/wARP (25), which built 490 residues of 556. Manual model rebuilding and DNA placement in all models was performed in COOT and the structures were refined with REFMAC5 (29) and phenix.refine.1.8.3 (30). The final model (one protein subunit) was used in the MR (MOLREP) in the crystal form III. The model of the crystal form III was used in MR to solve apo-AgeI structure. Data collection and refinement statistics is presented in Table 1.
Table 1. Data collection and refinement statistics.
| Crystal form | I-native | I-Hg | II-native | II-Hg | III native | apo native |
|---|---|---|---|---|---|---|
| DNA | SP11 | SP11 | SP11 | SP11 | SP13 | — |
| preSP-complex | SP-complex | apo | ||||
| Data collection statistics | ||||||
| Space group | P6122 | P6122 | P212121 | P212121 | P21 | P212121 |
| A (Å) | 111.4107 | 110.7967 | 66.0026 | 65.7569 | 50.5872 | 37.2130 |
| B (Å) | 111.4107 | 110.7967 | 79.8060 | 79.9872 | 87.0933 | 59.2690 |
| C (Å) | 216.6102 | 216.5802 | 101.6550 | 102.4873 | 66.5213 | 120.9154 |
| α/β/γ | 90/90/120 | 90/90/120 | 90/90/90 | 90/90/90 | 90/99.825/90 | 90/90/90 |
| Wavelength | 0.84230 | 0.84230 | 0.99985 | 1.5418 | 0.8150 | 0.9100 |
| X-ray source | DESY/DORIS BW7B | DESY/DORIS BW7B | DESY/DORIS X12 | MICROMAX-007 HF | DESY/DORIS X13 | MAX-lab I911-3 |
| Total reflections | 203263 | 296802 | 694292 | 245909 | 79694 | 56770 |
| Unique reflections | 21497 | 10545 | 84560 | 17243 | 21165 | 9795 |
| Resolution range (Å) | 49.51–2.71 | 11.07–3.50 | 65.00–1.50 | 43.15–2.60 | 31.48–2.35 | 34.23–2.50 |
| Completeness* (%) | 96.2 (74.6) | 100 (100) | 97.8 (100.0) | 100 (99.9) | 95.2 (86.3) | 100.0 (98.8) |
| Multiplicity* | 9.5 (9.1) | 29.3 (28.1) | 8.2 (5.0) | 14.3 (14.1) | 3.8 (3.6) | 5.8 (4.8) |
| Anomalous multiplicity* | 5.1 (4.8) | 15.9 (15.9) | 4.3 (2.5) | 7.7 (7.4) | 1.9 (1.8) | 2.9 (2.4) |
| I/σ* | 4.5 (1.9) | 3.2 (1.8) | 6.9 (3.8) | 9.0 (3.2) | 4.8 (1.3) | 5.8 (1.1) |
| R(merge)a (%) | 13.2 (40.6) | 19.0 (39.1) | 6.2 (19.7) | 7.1 (23.5) | 5.6(19.8) | 11.8 (64.7) |
| B(iso) from Wilson (Å2) | 54.7 | 9.7 | 17.1 | 48.5 | 36.5 | 41.5 |
| Refinement statistics | ||||||
| Resolution range (Å) | 1.50–40.27 | 2.40–25.46 | 2.50–33.33 | |||
| Reflections work (non-anomalous)/ test | 159783 (84472)/ 16072 | 40461 (21116)/4145 | 17824 (9756)/1737 | |||
| Protein/DNA/solvent atoms | 4496/444/524 | 4253/526/175 | 2126/–/19 | |||
| R-factor/R-free (%) | 15.17/18.52 | 17.88/21.73 | 21.42/25.60 | |||
| R.m.s.d. bond lengths (Å)/ angles (°) | 0.06/1.022 | 0.005/0.916 | 0.002/0.676 | |||
| Ramachandran core/allowed/disallowed region (%) | 96.72/3.28/0 | 97.28/2.72/0 | 95.56/4.44/0 | |||
| PDB ID | 5DWA | 5DWB | 5DWC | |||
a Values for the outer shell are given in parenthesis.
The contact surfaces buried between the two molecules were calculated using PISA server (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) (31). Protein–DNA contacts were analyzed by NUCPLOT (32). All molecular scale representations were prepared using MOLSCRIPT (33) and RASTER3D (34) software.
Gel filtration
DNA duplex SP (Supplementary Table S1) and His-AgeI were used for gel filtration. Gel filtration was carried out at room temperature on an ÄKTA Avant 25 system (GE Healthcare) using a Superdex 75 HR 10/30 column (GE Healthcare) pre-equilibrated with 20 mM Tris–HCl (pH 8.0), 0.2 M NaCl and 5 mM CaCl2. Protein (5–35 μM concentration) and protein–DNA (protein concentration 7 μM, SP DNA (Supplementary Table S1) concentration 3.5–140 μM) samples were prepared in 100 μl of the above indicated buffer. Protein elution from the column was monitored by measuring absorbance at 280 nm. The apparent molecular mass was evaluated from the elution volume using a series of standards (Gel filtration Calibration Kit from GE Healthcare).
Small angle X-ray scattering (SAXS) experiments
The SAXS data were collected at the P12 EMBL beam line on PETRAIII storage ring. Before the SAXS experiment AgeI preparation was run through the Superdex200 16/600 column (GE Healthcare) to remove possible aggregates and to exchange a buffer for SAXS measurements (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl2). The protein was concentrated using centrifugal concentrators (Pierce) with MWCO 10 kDa. The AgeI sample (3.1 mg/ml) was divided in two aliquots, the first one was used directly to collect data on apo-AgeI, while in the second AgeI was mixed with an equimolar amount of SP13 oligoduplex before data collection. The main SAXS data collection parameters are presented in the Supplementary Table S3. Data automatically averaged and reduced at the beam line (35) were further processed using PRIMUS and GNOM packages (36). Two scattering curves of apo-AgeI were merged with PRIMUS. Ab initio modelling was performed by DAMMIN (37). Ten pseudoatomic models were averaged by DAMAVER package (38). Experimental SAXS data were compared with crystal structures using CRYSOL (39) and SUPCOMB (40).
Electrophoretic mobility shift assay (EMSA)
DNA binding by AgeI was analyzed by the electrophoretic mobility shift assay (EMSA) using the 33P-labeled specific (SP), non-canonical (NC) or non-specific (NSP) oligoduplex (Supplementary Table S1). DNA (final concentration 1 nM) was incubated with proteins (final concentrations varied from 0.5 to 50 nM monomer) for 15 min in 20 μl of the Binding buffer containing 40 mM Tris-acetate (pH 8.3 at 25°C), 5 mM Ca-acetate, 0.1 mg/ml BSA and 10% (v/v) glycerol at room temperature (22°C). To determine a stoichiometry of the specific AgeI–DNA complex the 33P-labeled specific oligoduplex (10 nM) was incubated with wt AgeI and His-AgeI or their mixture (final concentration 10 nM monomer). Free DNA and protein–DNA complexes were separated by electrophoresis using 8% (w/v) acrylamide gels (29:1 acrylamide/bisacrylamide in 40 mM Tris-acetate, pH 8.3 at 25°C, and 5 mM Ca-acetate). Electrophoresis was run at room temperature for 3 h at ∼6 V/cm or 8 h at ∼8 V/cm in the stoichiometry experiments. Radiolabeled DNA was detected and quantified using the Cyclone phosphorimager and the OptiQuant software (Packard Instrument). Apparent Kd values for DNA binding were determined as described (41).
DNA cleavage assay
The specific catalytic activity of AgeI and mutant proteins was evaluated using phage λ DNA that contains 13 AgeI targets. Varied protein amounts (from 1 × 10−5 mg/ml up to 1 × 10−1 mg/ml) were incubated with 1 μg λ DNA in 50 μl of 33 mM Tris–HCl (pH 7.5 at 37°C), 10 mM MgCl2 and 0.1 mg/ml BSA for 1 h at 37°C. Reactions were terminated by the addition of 20 μl ‘STOP’ solution (75 mM EDTA, pH 9.0, 0.3 % SDS, 0.01 % bromophenol blue and 50 % (v/v) glycerol) and heating at 70°C for 20 min. Cleavage of the pUC18 plasmid DNA (10 nM) lacking AgeI targets was performed at 37°C in the Reaction buffer (33 mM Tris-acetate (pH 7.9 at 37°C), 66 mM K-acetate, 10 mM Mg-acetate, 0.1 mg/ml BSA) containing wt AgeI or the D177A mutant (150 nM dimer). The samples were collected at timed intervals and the reaction was quenched by the addition of ‘STOP’ solution. The DNA was analyzed by agarose gel electrophoresis (0.8% and 1% (w/v) for phage λ and pUC18 DNA, respectively).
Cleavage of the 33P-labeled specific oligoduplex SP (200 nM) (see Supplementary Table S1) was performed at 25°C in the Reaction buffer incubating with various concentrations (from 1 nM up to 50 nM in terms of dimer) of AgeI. DNA cleavage of wt AgeI (10 nM) was stimulated by addition of the D142A mutant (40 nM). The samples were collected at timed intervals and the cleavage reaction was quenched with a loading dye solution (95% v/v formamide, 25 mM EDTA, 0.01% bromphenol blue). Separation of the DNA hydrolysis products was performed by denaturing PAGE: the 20% polyacrylamide gel (acrylamide/N,N’-methylenebisacrylamide 29:1 (w/w)) in Tris-borate containing 8.5 M urea was run at 30 V/cm. Radiolabeled DNA was detected and quantified as described above. In multiple turnover conditions the DNA cleavage rates were determined from the linear parts of the reaction progress curves by a linear regression.
Data analysis
The KYPLOT 2.0 software (42) was used for cleavage rate calculations.
RESULTS
Apo–AgeI structure
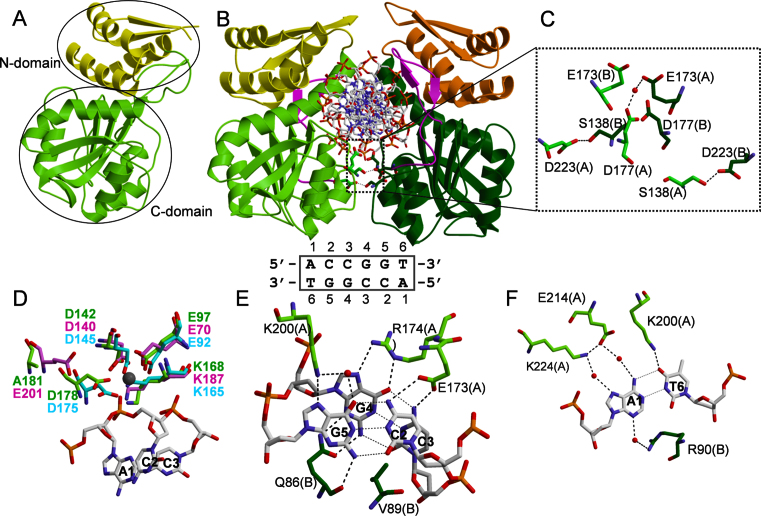
To elucidate the structural organization of AgeI, we performed crystallographic analysis of the enzyme in apo- and DNA-bound forms (see ‘Materials and Methods’, Table 1). Surprisingly, in the crystal in the absence of DNA AgeI is a monomer (Figure 1A). Indeed, in the crystal each pair of protein molecules interact with each other using different interfaces meanwhile in the homodimer protein–protein interaction interfaces are supposed to be identical. The examined crystal contacts by PISA server also exclude the presence of the dimer. The AgeI monomer is composed of two domains: an N-terminal domain (residues 1–82, termed N-domain) and a C-terminal domain (residues 83–278, termed C-domain). The N-domain comprises an N-terminal β hairpin and three α helices H1–H3 (Supplementary Figure S1A). One loop (residues 8–10) in the N-domain is disordered. The C-domain possesses typical for PD-(D/E)XK REases six-stranded β sheet flanked by five α helices (Figure 1A). The AgeI monomer structure is similar to the individual subunits of dimeric/tetrameric restriction enzymes SgrAI, Bse634I (1KNV), Cfr10I (1CFR), NgoMIV (1FIU) (DALI Z-scores 11, 10.5, 10.3, 10.2, respectively) which share the conserved CCGG tetranucleotide in their target sites. AgeI shows the closest structural similarity to BsaWI (4ZSF): DALI Z-score for the PD-(D/E)XK domain is 14.9. BsaWI recognizes a degenerate sequence 5΄-WCCGGW-3΄ where one of the variants matches the AgeI recognition sequence. Moreover, AgeI and BsaWI protein sequences show high similarity: 24% identical and 41% similar amino acids, respectively (Supplementary Figure S1A). BsaWI is also composed of two domains; however in BsaWI the N-domains are swapped and contribute to the protein dimerization, while in AgeI the N-domain makes only intra-subunit contacts (Supplementary Figure S1B) (6).
Figure 1.
AgeI apo- and DNA-bound structures. (A) Apo–AgeI monomer structure. N-domain is coloured yellow, C-domain is green. (B) Specific AgeI–DNA complex. N-domains are colorued yellow and orange, C-domains are coloured light green and dark green. AgeI structural element (residues 197–224) involved in the recognition of the outer A:T base pair is coloured magenta. The dimerization interface is boxed. AgeI recognition sequence is shown below (numbering is the same as in the structures). (C) Close-up view of the dimerization contacts. (D) AgeI active site residues (green) overlaid with NgoMIV (4ABT, pink) and BsaWI (4ZSF, cyan) active sites. Ca2+ ion from NgoMIV-substrate structure is shown as grey sphere. (E) CC:GG recognition by AgeI. (F) Recognition of the outer A:T base pair by AgeI. Note in (E) and (F) that both protein subunits contribute for the recognition of one half-site of the target 5΄-ACCGGT-3΄ target through hydrogen bonds in the major and minor grooves.
Structures of Agel–DNA complexes
To determine the AgeI structure in the DNA-bound form we solved crystal structures of two AgeI–DNA complexes with 13 bp (SP13) and 11 bp (SP11) oligoduplexes containing the 5΄-ACCGGT-3΄ target (Supplementary Table S1). AgeI forms a dimer bound to a DNA duplex in both crystal forms, however these complexes are not identical. Based on the contacts made to DNA we term these complexes specific (SP-complex, with SP13) and pre-specific (preSP-complex, with SP11) and they will be discussed separately.
Specific AgeI–DNA complex
Overall structure
The asymmetric unit of the SP-complex crystal contains two protein and two DNA chains which correspond to the AgeI dimer bound to a DNA duplex (Figure 1B). Protein subunits of the dimer are very similar (r.m.s.d. over Cα atoms is 0.36 Å). Main structural differences between AgeI in the apo-form and SP-complex are at the N-terminus of helix H4 (residues 82–87), fragment 118–139 and loop 7–11, which become ordered in the DNA-bound protein (Supplementary Figure S2A).
Dimer structure
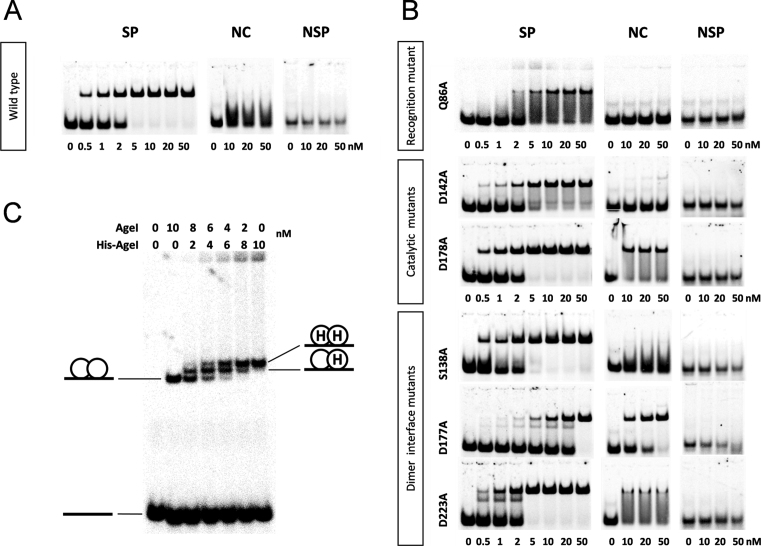
In the SP-complex AgeI dimer almost completely encircles DNA (Figure 1B). However, the dimerization interface is rather small (buried surface area 619 Å2) compared to the average dimer interface of 1600 (±400) Å2 (43). Dimerization is achieved through the two pairs of helices H6 and H7 of the C-domains, similar to Cfr10I, Bse634I, NgoMIV primary dimers (44–46). Although the N-domains are located close to each other, they make only water-mediated contacts. Analysis of the dimer interface suggests that residues S138, D177 and D223 could be involved in the intersubunit contacts (Figure 1C). Surprisingly, alanine replacement of S138 and D223 residues results in the wt or 5-fold activity decrease, respectively (Table 2), while the D177A variant exhibits 50-fold decrease in cleavage activity of λ DNA. Importantly, the cleavage specificity of the D177A mutant is relaxed (Supplementary Figure S3A) and it even linearizes plasmid DNA lacking the AgeI recognition sequence (Supplementary Figure S3B).
Table 2. DNA binding and cleavage properties of AgeI mutants.
| Protein | Mutation | DNA binding (Kd, nM)a | Specific activity, %b | ||
|---|---|---|---|---|---|
| SP | NC | NSP | |||
| wt | 0.9 ± 0.2 | >50 | >50 | 100 | |
| Q86A | Recognition | 4.6 ± 1.4 | >50 | >50 | 8 |
| D142A | Catalytic | 2.4 ± 0.8 | >50 | >50 | n.d. |
| D178A | Catalytic | 1.0 ± 0.3 | ∼5 | >50 | 50 |
| S138A | Dimer interface | 1.2 ± 0.2 | >50 | >50 | 100 |
| D177A | Dimer interface | 19.5 ± 5.6 | 14.7±3.1 | >50 | 2c |
| D223A | Dimer interface | 1.2 ± 0.4 | ∼5 | >50 | 20 |
aThe Kd values were determined by EMSA (see Figure 3 and ‘Materials and Methods’ for the details) using the specific (SP), the non-canonical (NC) or the non-specific (NSP) oligoduplex (Supplementary Table S1).
bThe specific activity of AgeI variants was estimated using λ DNA, n.d.—DNA cleavage activity was not detected under our experimental conditions (see ‘Materials and Methods’).
cThe D177A mutant shows a strong star activity (Supplementary Figure S3).
Active site
The active site of AgeI is composed of residues E97, D142, K168 and D178 (Figure 1D) which overlay with the active site residues of NgoMIV, BsaWI (Figure 1D) and other CCGG family enzymes (not shown) in the superimposed structures. The signature feature of the CCGG family enzymes is the permutated PD-KX12-13(E/D) active site motif that differs from the canonical PD-(D/E)XK sequence (41,45). D178 of AgeI matches with D175 of BsaWI, but does not overlap with the catalytic E201 residue of NgoMIV and the other CCGG enzymes (Figure 1D, Supplementary Figure S5). The D178A mutant of AgeI retains 50% of wt activity (Table 2). On the other hand, D142A mutant is inactive but binds to the specific DNA with the similar affinity as wt AgeI (Table 2).
CCGG recognition
Each subunit of AgeI recognizes one half-site of the target sequence 5΄-ACCGGT-3΄ from the major groove side and makes contacts in the minor groove to another half-site. E173 side chain makes hydrogen bonds to N4 atoms of neighboring C2 and C3 cytosines in the CC:GG half-site, while R174 makes bidentate hydrogen bond to the N6 and N7 atoms of inner guanine (G4) from the same half-site (Figure 1E). To complete the CC:GG recognition network in the major groove, K200 makes direct and water mediated hydrogen bonds to the N7 and O6 atoms of the guanine G5, respectively. In the minor groove Q86 from the other protein subunit makes hydrogen bonds to both guanines G4 and G5 using a main chain O and side chain atoms (Figure 1E). This minor grove interaction seems to be important: Q86A replacement significantly compromises DNA binding and cleavage activity compared to wt enzyme (Table 2). Interestingly, the side chain of V89 is inserted between the cytosine C2 and cytosine C3 bases in the minor groove and presumably ensures proper angle to position N4 atoms for the hydrogen-bonding by the side chain of E173 (Figure 1E).
A:T recognition
AgeI residues interacting with the outer A:T base pair reside on a structural element (residues 197–224) that protrudes from the conserved catalytic core and is unique for AgeI (Figure 1B, Supplementary Figure S1A). The outer A1 adenine base is recognized by K224 and E214 residues through the water mediated hydrogen bonds to the N7 and N6 atoms, respectively (Figure 1F). The side chain of K200 makes hydrogen bond to the O4 atom of the thymine T6 base. From the minor groove side, R90 of the other subunit makes water mediated hydrogen bond to the N3 atom of adenine A1 (Figure 1F).
Pre-specific AgeI–DNA complex
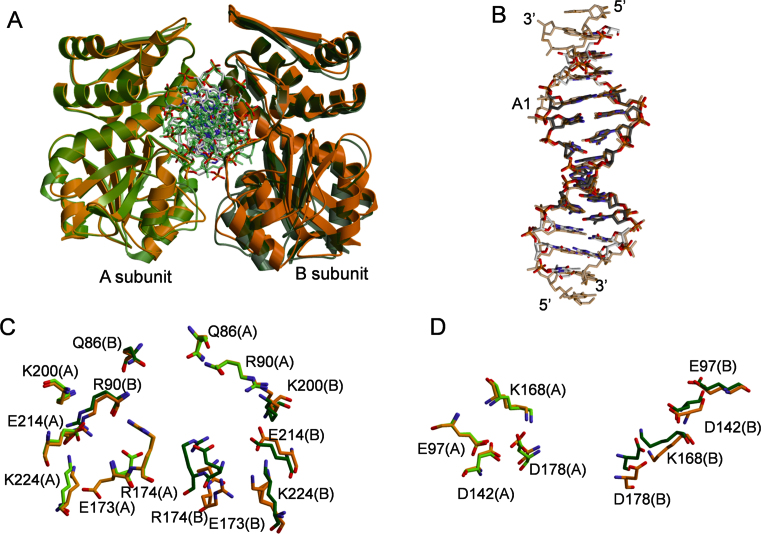
AgeI forms a dimer in a complex with SP11 oligoduplex (Figure 2A). Overall structure of AgeI in complex with SP11 oligoduplex is similar to that of the SP-complex. However, after superposition of SP- and preSP complexes over A-subunits, the positions of B-subunits of the SP- and preSP-dimers are slightly different (Figure 2A). Moreover, the buried surface area in the preSP-complex is smaller than in the SP-complex (∼400 Å2 versus ∼600 Å2). The loop containing S138 adopts different conformation resulting in the loss of S138–D223 contact at the dimer interface (Supplementary Figure S2B and C). Noteworthy, the putative AgeI dimer interface is smaller than other protein-protein contacts in the crystal (data not shown). In the case of preSP complex, the largest protein-protein interface in the crystal is asymmetric (involves different regions in two protein molecules), while the second one is symmetric and similar to that of the SP complex.
Figure 2.
Comparison of AgeI SP- and preSP-complexes. (A) Overlay of the SP (green) and preSP (orange) complexes. (B) Overlay of the DNA molecules from SP- and preSP-complexes. (C) Overlay of the DNA recognition residues. (D) Overlay of the active site residues.
Protein subunits in the preSP-complex are not identical (r.m.s.d. Cα 0.97 Å) mainly due to conformational differences located in the DNA binding cleft and the dimer interface. The A-subunit in the preSP-complex is more similar to the SP-complex subunits (r.m.s.d. 0.83 Å); the main differences are in the conformation of the N-terminal hairpin (residues 7–11), 119–141 helix–loop fragment and 203–225 fragment (DNA recognition element) (Supplementary Figure S2B). The differences in the conformation of the B-subunit in the preSP complex are more pronounced (r.m.s.d. 1.08 Å): additional conformational differences are located at the recognition-dimerization helix H6 (residues 172–191) (Supplementary Figure S2C). DNA conformation in both AgeI complexes is similar across the target site region, except the conformation of A1 in the vicinity of the B-subunit (Figure 2B). While the differences between the protein conformations in the SP and the preSP complexes are small, we expect them to be reproducible since we have refined all structures using the same protocol, employing Rfree values to guard against over-refinement.
Contacts with DNA
In the SP-complex DNA binding by AgeI dimer buries ∼5800 Å2 of a surface area, while in the preSP-complex buried surface area is a bit smaller - ∼5200 Å2. Differences of DNA binding in SP- and preSP-complexes occur both in the contacts with heterocyclic bases and backbone (Supplementary Figure S2D). In the preSP-complex not all specific contacts to the target site bases are present comparing to the SP-complex (Figure 2C, Supplementary Table S4). In the major groove only K200 contacts to G5 and T6 bases are conserved in the SP- and preSP-complexes. Other DNA recognizing residues possess different conformations in the SP-complex and in at least one subunit of the preSP-complex. The side chain of E173(A) points away from DNA and is located close to D223(A) residue, while E173(B) side chain is disordered. Conformations of R174, E214 and K224 in the A-subunit of the preSP-complex are the same as in the SP-complex, but in the B-subunit R174 and K224 moved away from DNA, while the side chain of E214 residue acquires a different conformation. On the other hand, DNA minor groove contacts in the preSP- and SP-complexes are identical. Contacts with phosphates at the 3΄ region of each strand involving mostly residues from the N-domain are conserved in both complexes, but differ at the 5΄ region including the scissile phosphate (Supplementary Figure S2D, Supplementary Table S5).
Active site
Active site residues of the preSP- and SP-complexes overlap well in the superimposed A-subunits, however only E97 and D142 overlay in the subunits B (in the dimer superimposed over A-subunits) (Figure 2D).
Oligomeric state of AgeI in solution
The oligomeric assembly of AgeI in solution was analyzed by gel filtration (Supplementary Table S6, Supplementary Figure S4A). Apo–AgeI at 5–35 μM loading concentrations interval elutes from the column as a single peak corresponding to Mw 25–29 kDa, which is close to the Mw of AgeI monomer (32.5 kDa). AgeI–DNA complex (AgeI:DNA ratio varied from 1:0.5 to 1:20) elutes as a peak corresponding to Mw 64.3–67.2 kDa (Supplementary Table S6). This value is in between of the calculated Mws of AgeI monomer and dimer bound to a DNA duplex (32.5 + 18.4 kDa = 50.9 kDa and 2 × 32.5 + 18.4 kDa = 83.4 kDa, respectively) indicating that AgeI does not form a stable dimer complex with DNA under the gel filtration conditions.
Next we used small angle X-ray scattering (SAXS) to estimate the oligomeric assembly of AgeI (Supplementary Figure S4B–D, Supplementary Table S3). Theoretical scattering curves were calculated from the apo and SP-complex structures and compared with experimental SAXS data (Supplementary Figure S4B and D). Monomeric apo-AgeI and dimeric SP-complex structures fit well the SAXS data of apo–AgeI and AgeI–SP13 complex in solution, indicating that the protein adopts similar conformations both in the crystals and in solution (Supplementary Figure S4B and D).
DNA binding studies
DNA binding by AgeI was analyzed by EMSA using the cognate SP oligoduplex containing the 5΄-ACCGGT-3΄ target, and a non-specific NSP oligoduplex (Supplementary Table S1, Figure 3). To evaluate the specificity of AgeI variants we also performed DNA binding experiments with the non-canonical NC DNA duplex containing the 5΄-ACCGGA-3΄ sequence that differs from the target site by 1 bp (underlined). Wt AgeI and mutants form complexes with the specific SP oligoduplex (at 2–20 nM concentrations), but show only weak or no binding to a non-specific NSP oligoduplex (Figure 3A and B, Table 2). The specific AgeI–DNA complex is supposed to be an AgeI dimer bound to a single oligoduplex similar to that observed in the crystal (Figure 1B). Interestingly, dimerization interface mutants D177A and D223A show faint bands with increased mobility in the EMSA compared to that of the proposed specific AgeI dimer–DNA complex (Figure 3B). It cannot be excluded that these faint bands correspond to the specific AgeI–DNA complex, where a protein monomer is bound to the specific oligoduplex. Wt AgeI, Q86A, S138A and D142A mutants did not form stable complexes with a non-canonical NC DNA, while D177A, D178A and D223A mutants bind to it with similar affinity as to SP DNA (Figure 3B, Table 2). This indicates that D177, D223 (dimerization interface) and D178 (active site) residues, which are not directly involved in the DNA recognition, are important for the REase specificity.
Figure 3.
DNA binding analysis of AgeI. (A, B) EMSA of specific (SP), non-canonical (NC) or non-specific (NSP) oligoduplex binding (Supplementary Table S1). The reactions contained 1 nM of the 33P-labeled oligoduplex and the protein at the concentrations indicated below. The samples were separated by native PAGE and analyzed as described in ‘Materials and Methods’. (C) EMSA of DNA binding by AgeI and His-AgeI mixture. The reactions contained 10 nM of the 33P-labeled specific oligoduplex (Supplementary Table S1) and wt AgeI and His-AgeI (possesses a His6-tag at the N-terminus) protein mixtures at concentrations indicated above each lane. After 15 min incubation at room temperature, the samples were subjected to the native PAGE and analyzed as described in ‘Materials and Methods’. The cartoons illustrate different AgeI–DNA complexes.
In order to determine the stoichiometry of the specific AgeI–DNA complex DNA binding experiments were performed, where the specific oligoduplex was incubated with wt AgeI, His-AgeI (contains a His6-tag at the N-terminus) or their mixture (Figure 3C). In the gel, electrophoretic mobility of the specific His-AgeI–DNA complex was reduced compared to that of the wt AgeI–DNA complex due to the presence of the His6-tag (18 amino acids, ∼2 kDa). In the case of wt AgeI and His-AgeI mixtures, three different protein–DNA complexes were observed: two of them showed electrophoretic mobilities corresponding to those of specific complexes of wt AgeI and His-AgeI homodimers, respectively, while the third one exhibited an intermediate electrophoretic mobility that presumably corresponds to the wt AgeI–His-AgeI heterodimer (Figure 3C). This experiment provides a direct evidence that AgeI in solution binds to the DNA target as a dimer.
DNA cleavage
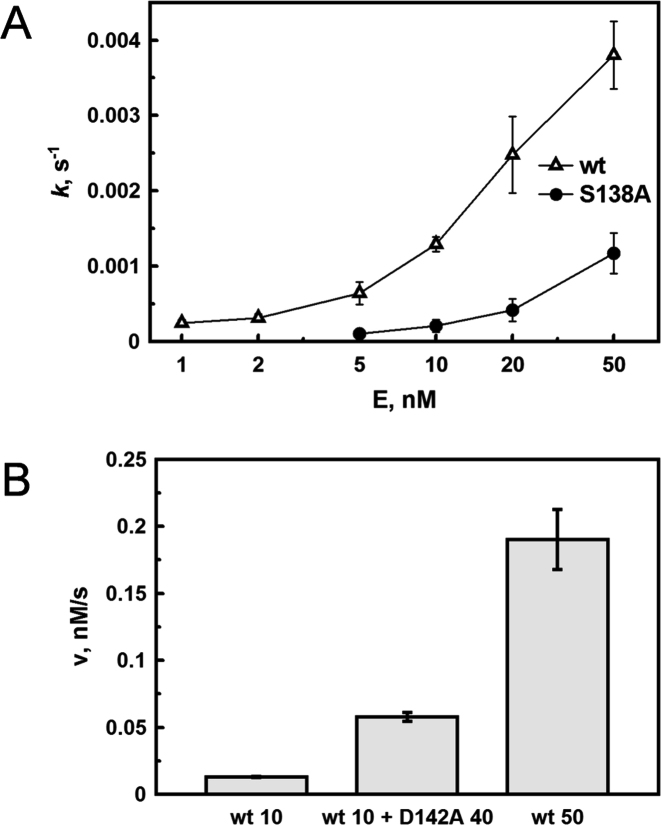
According to the solution and crystallographic data apo-AgeI is a monomer, but binds the target sequence as a dimer indicating that monomeric AgeI should dimerize for DNA cleavage. Therefore concentration dependence of DNA cleavage by wt AgeI and the dimerization interface mutants might be different. To test this hypothesis we analyzed concentration dependence of DNA cleavage by wt AgeI and the S138A dimerization mutant under the steady-state conditions using the specific oligoduplex SP (Supplementary Table 1) as a substrate. Dependence of the cleavage rate constant on the wt AgeI protein concentration is shown in Figure 4A. A similar dependence is observed in the case of the S138A mutant; however, the curve is shifted to higher protein concentrations. This suggests that the S138A mutation at the dimerization interface shifts the monomer-dimer equilibrium towards monomer. Therefore, increased concentrations of the S138A mutant are required to achieve the same cleavage rate as in the case of wt AgeI. Indeed, using a high enzyme concentration (1000 nM) the cleavage rates of wt AgeI and the S138A mutant are similar indicating that the S138A mutation specifically affects the dimerization of AgeI (data not shown). This finding is in accordance with the λ DNA cleavage data, where a specific activity of the S138A mutant is the same as wt enzyme (Table 2). Moreover, DNA cleavage by wt AgeI is stimulated by addition of the inactive D142A mutant (Figure 4B). In this experiment, the concentration of the cleavage competent active sites (wt AgeI) remains the same, however, due to a higher final protein concentration (because of the addition of inactive D142A) the monomer–dimer equilibrium is shifted towards the catalytically competent heterodimer resulting in a faster DNA cleavage (Figure 4B).
Figure 4.
DNA cleavage by AgeI. (A) Concentration dependence of DNA cleavage by wt AgeI and the S138A mutant. (B) Stimulation of the wt AgeI cleavage activity by the D142A mutant. Cleavage of the specific oligoduplex SP (200 nM) (see Supplementary Table S1) was performed under multiple turnover conditions as described in ‘Materials and Methods’. wt (10) and wt (50) corresponds to reaction rates obtained at 10 and 50 nM concentrations of wt AgeI, respectively; wt (10) + D142A (40) corresponds to reaction rates obtained at 10 nM wt and 40 nM D142A mutant.
DISCUSSION
Comparison to CCGG-family restriction endonucleases
Active site
The active site of AgeI is composed of E97, D142, K168, D178 residues and is similar to the other CCGG-family enzymes, which contain a permutated canonical PD-(D/E)XK motif (Figure 1D). In AgeI, the spatial position of the second acidic residue D178 is similar to D175 of BsaWI but differs from other structurally characterized CCGG-family enzymes (6). Surprisingly, D178A mutant retains ∼50% activity compared to wt AgeI, while the structurally equivalent BsaWI D175A mutant is inactive (Table 2) (6). Alanine replacement of the second acidic residue also produce inactive variants in Cfr10I (E204Q, (47)), Bse634I (E212A, M.Z., unpublished data), EcoRII (E337A, (48)) while Ecl18kI E195A mutant retains only 4% of the DNA cleavage activity (41). On the other hand, PspGI E173A mutant retains 12% (49), NgoMIV E201A 25% of the catalytic activity (V.S., unpublished data).
CCGG recognition
All CCGG-family restriction enzymes characterized so far use the residues from the conserved R-(D/E)R motif to recognize the central CCGG tetranucleotide (Supplementary Figure S5). Surprisingly, CCGG recognition by AgeI differs from the enzymes belonging to the same family. Only E173 and R174 residues corresponding to the two last residues (underlined) from the R-(D/E)R motif are conserved in the AgeI sequence (Supplementary Figure S5). The first arginine of the R-(D/E)R motif (underlined) which is used for the recognition of G in the first C:G bp in the other CCGG-family enzymes is replaced by K200 in AgeI. K200 is located on the distinct structural element and also makes hydrogen bond with the outer T6 base (Figure 1E and F). In the crystal, AgeI also makes contacts with the CCGG tetranucleotide in the minor groove. Q86 from both AgeI subunits recognize the central CCGG tetranucleotide making hydrogen bonds to the GG dinucleotide of one half-site (Figure 1E). Structural comparison of AgeI with other CCGG-family REases revealed that this minor groove contact is conserved in BsaWI, Ecl18kI, EcoRII-C, PspGI and SgrAI. Apart of the conserved recognition R-(D/E)R motif, all these enzymes possess conserved N or Q residues (Q86 in AgeI, N81 in BsaWI, Q114 in Ecl18kI, N260 in EcoRII, Q94 in PspGI, N92 in SgrAI), which make contacts to GG bases in the minor groove (Supplementary Figures S5 and S6) (17,50–52). Corresponding contacts are absent in the REases NgoMIV and Bse634I which approach DNA mostly from the major groove side (46,53). Therefore we assume that GG recognition in the minor groove by the conserved N/Q residues might be a universal mechanism for the CCGG-family REases.
Recognition of the outer base pairs
To recognize outer A:T bp AgeI uses residues K200, E214 and K224 from the unique structural element (residues 197–224) (Figure 1F). Only contact of K200 to O4 of T6 base is direct, other contacts are water-mediated and may be regarded as less important for outer base pair discrimination. To investigate an impact of these contacts on the AgeI specificity we performed in silico mutagenesis of the outer bp of the AgeI recognition sequence (Supplementary Figure S7A). Replacement of the A1:T6 bp by C1:G6 or G1:C6 bp is incompatible with the DNA structure observed in the crystal: there is a steric clash between O2 atom of C base and N2 atom of G base in the minor groove (Supplementary Figure S7A). Moreover, there is a steric clash between the R90 side chain and both N2 atom of G base and O2 atom of C base in C1:G6 variant and R90 side chain and O2 atom of the C base in the case of G1:C6 base pair (Supplementary Figure S7A). In addition, the N4 atom of C (G1:C6 bp) cannot form a hydrogen bond with NZ of K200 (both are hydrogen bond donors). In the case of T1:A6 bp there are no clashes between DNA bases or with R90 side chain, however direct hydrogen bond could not be formed between NZ atom of K200 and N6 of A base (Supplementary Figure S7A). Therefore we assume that one direct and three water-mediated hydrogen bonds together with indirect readout could discriminate the outer A1:T6 bp.
Interestingly, in the superimposed AgeI and NgoMIV structures the side chains of K200 and E214 of AgeI overlay with the side chains of D34 and R227 of NgoMIV (5΄-GCCGGC-3΄), respectively (Supplementary Figure S7B). NgoMIV residues R227 and D34 are involved in the contacts with the outer G:C bp (46). This suggests that the spatial position of the residues/atoms interacting with the outer bp is conserved, despite they come from different structural elements. In the minor groove, R90 makes a water-mediated hydrogen bond to the first base A1 of the recognition sequence (Figure 1F). Similar conserved minor groove contacts are made by BsaWI, NgoMIV and Bse634I (6).
AgeI is similar to BsaWI
AgeI recognition sequence 5΄-ACCGGT-3΄ is related to the BsaWI target 5΄-WCCGGW-3΄. AgeI and BsaWI proteins share 24% identical and 41% similar amino acids (Supplementary Figure S1A). Thus it is not surprising that their structures are also similar. Both proteins are composed of two domains: the N-terminal helical domain and the C-terminal catalytic domain. The catalytic C-domains of AgeI and BsaWI are similar (Supplementary Figure S1). The active sites of the proteins also superimpose very well (Figure 1D). The main difference between the C-domains is the additional structural element of AgeI, which is involved in DNA recognition (Supplementary Figure S1). The N-domains of AgeI and BsaWI also adopt very similar structures, whereas the AgeI N-domain contains the additional hairpin, which is absent in BsaWI (Supplementary Figure S1B). In BsaWI the N-domains are involved in the dimerization contacts, they are swapped between the protein subunits and ensure a very large dimerization interface (∼2000 Å2) (6). Differently, AgeI is the monomer in the absence of DNA and the N-domains are in contact with the C-domain of the same protein subunit (Figure 1A). Interestingly, in the overlaid dimers the N-domain of BsaWI overlaps with the N-domain of AgeI from the different subunit indicating that the relative N-C domain position in the proteins is also conserved (Supplementary Figure S1B). The domain swap in BsaWI might be related to the thermostability of the protein. BsaWI comes from a thermophilic bacterium Bacillus stearothermophillus with an optimal growth temperature of 55°C and at higher temperatures it is important to ensure the stability of the dimer for the enzyme function (http://rebase.neb.com/rebase/rebase.html). In contrast, AgeI, which exists as a monomer and forms dimer only in DNA-bound form, was identified in bacterium Agrobacterium gelatinovorum, which grows at 26°C (http://rebase.neb.com/rebase/rebase.html). Indeed, a thermal stability of BsaWI (melting temperature (Tm) 70.6°C) is significantly higher compared to that of AgeI (Tm = 41.6°C) (M.Z., data not shown).
DNA cleavage mechanism of AgeI
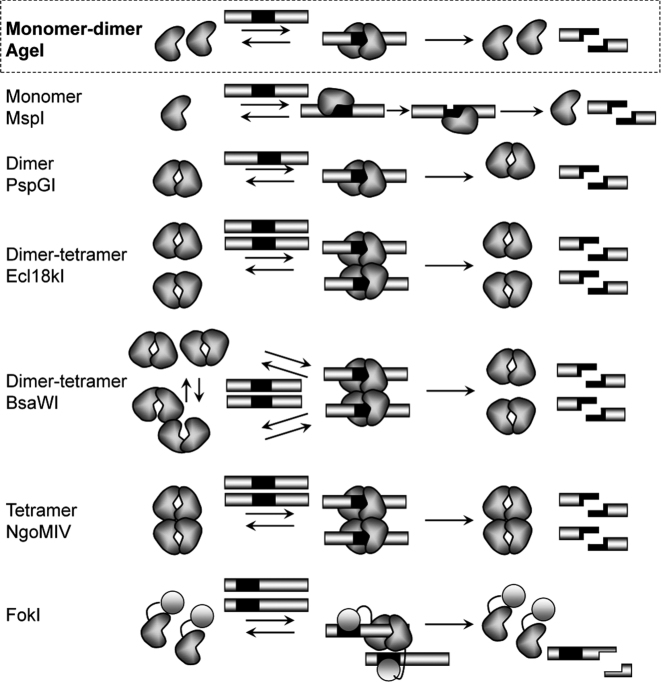
Structural and biochemical data allow us to propose DNA cleavage mechanism of AgeI (Figure 5). Crystal structure, gel filtration and SAXS data show, that AgeI in the apo-form is a monomer (Figure 1A, Supplementary Figure S4). The apo–AgeI protein purified from the A.gelatinovorum also eluted from gel-filtration column at a size consistent with it being a monomer (54). On the other hand, DNA-bound AgeI in the crystal shows a dimer; however, the intersubunit interface is rather small and contains only few hydrogen bonds (Figure 1B and C). In the DNA complex, each AgeI subunit interacts with both halves of the palindromic target site: it makes contacts to one-half-site from the major groove side and it contacts the other half-site in the minor groove (Figure 1E and F). On the other hand, kinetic studies indicate that AgeI dimerization is required for DNA cleavage (Figure 4). To reconcile structural and biochemical data, we propose that AgeI monomers dimerize upon DNA binding and each subunit than cleaves a phosphodiester bond on the opposite strands of the target sequence (Figure 5). Such mechanism seems to be unique among the restriction enzymes recognizing palindromic target sites (Figure 5). Monomeric restriction endonucleases like MspI typically bind symmetric target sites asymmetrically and cleave two DNA strands sequentially (7). Most of REases, like PspGI, are stable dimers in the apo form and each subunit within a dimer cuts phosphodiester bonds on the opposite strands of DNA target (49). Some REases, as exemplified by Cfr10I, Bse634I and NgoMIV, are homotetramers, which bind and cleave two target sites simultaneously (45,46,55). Other restriction enzymes like Ecl18kI, SgrAI and BsaWI are dimers in the apo form and make tetramers or higher order oligomers when bound to DNA (3,5,6). DNA cleavage mechanism of AgeI is most similar to that of the Type IIS enzyme FokI. However, FokI, differently from AgeI, is composed of two separate domains for DNA binding and cleavage. FokI is a monomer in solution but after binding to the target site through the binding domain, FokI nuclease domains form a dimer and cleave both DNA strands at one of the target sites (Figure 5).
Figure 5.
Oligomeric forms and DNA cleavage mechanisms of Type II restriction endonucleases interacting with (pseudo)palindromic sequences and Type IIS enzyme FokI. Black rectangles represent recognition sequence.
Coupling of dimerization, recognition and catalysis
Structural and biochemical studies of AgeI revealed a tight interconnected network of amino acid residues involved in dimerization, recognition and catalysis (Supplementary Figure S1A). The S138 residue, which makes hydrogen bond to D223 of the other subunit (Figure 1C) is positioned in the vicinity to the catalytic D142 residue. On the other hand, D223 is located next to the K224 residue, which is involved in the recognition of the A1 base, and makes water-mediated contact to the oxygen atom of DNA backbone (Figure 1F, Supplementary Table S5). The D177 residue in the dimerization helix H9 is located close both to the recognition residues E173, R174 and the catalytic D178 residue. Importantly, mutations of the dimer interface residues D177 and D223 and active site residue D178 affect not only on the DNA cleavage activity, but also on the DNA binding specificity of AgeI (Table 2, Figure 3). D177A, D178A and D223A mutants bind non-canonical DNA with the similar affinity as cognate DNA. Moreover, D177A mutant exhibits relaxed cleavage specificity (Supplementary Figure S3). D177 is involved in the water-mediated hydrogen bond with E173 from the other subunit (Figure 1C), and these interactions presumably ensure the proper orientation of E173 side chain for the CC recognition (Figure 1E). In the US patent it was claimed that two AgeI mutants, S201A and R139A show reduced star activity (56). R139 to A substitution may affect the interaction of S138 with D223 of the second subunit and activation of AgeI dimer on non-cognate sites (star sites). S201 is located next to the A/T bp recognition residue K200 and makes water mediated hydrogen bond to the backbone phosphate (Supplementary Figure S2); the loss of the –OH side chain group by S to A substitution altered (decreased) the star activity on non-cognate sites. The study of the wt AgeI enzyme structure and mechanism of sequence recognition will help us understand AgeI mutants with reduced star activity and improved fidelity. Altered dimer interface interactions of REases and/or loss of DNA backbone interactions may turn out to be a general mechanism to engineer these enzymes to high fidelity.
Since AgeI cuts DNA as a dimer, it is important that both subunits of the dimer make all required contacts to the specific target site prior to DNA cleavage. In the SP- and preSP-complex structures contacts within the minor groove and 3΄- end phosphates of the target are conserved in both complexes. On the contrary, specific contacts with the target site in the major groove and 5΄-end phosphates at the of the recognition sequence are made only in the SP-complex (Supplementary Figure S2D). Therefore, we assume that the preSP-complex resembles an intermediate complex of the enzyme searching for the target site. It is likely, that AgeI forms a weak dimer on the non-specific DNA and scans DNA from the minor groove searching for the target site using Q86 and R90 residues (Figure 1E and F). Finally, the catalytically competent dimer is formed on the target by making base-specific contacts in the major groove and positioning the active site residues near the scissile phosphate. Such target search mechanism can be shared by BsaWI, Ecl18kI and EcoRII REases, which also employ the conserved N/Q residue in the minor groove for the CC:GG recognition (6).
ACCESSION NUMBERS
Coordinates and structure factors are deposited under PDB ID 5DWA (preSP-complex), 5DWB (SP-complex) and 5DWC (apo).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr M. Groves and Dr G. Bourenkov at EMBL beamlines BW7B, X12 and X13 at the DORIS storage ring, P12 at PetraIII storage ring, Hamburg, Germany, and Dr R. Appio at MAX II I-911-3 beamline Lund, Sweden for the invaluable help with the beamline operation. We thank S. Valinskyte for purification of the AgeI mutants.
Footnotes
Present address: Virginija Jovaisaite, Laboratory for Integrative and Systems Physiology, École Polytechnique Fédérale de Lausanne, EPFL, CH-1015 Lausanne, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Council of Lithuania [MIP-41/2013]; SAXS measurements, performed on P12 Beamline of EMBL Hamburg Outstation on PETRA III storage ring at DESY and data collection at MAX II beamline have received funding from the European Community's Seventh Framework Programme [FP7/2007–2013] under BioStruct-X [283570]. Funding for open access charge: Research Council of Lithuania.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bujnicki J.M. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the “midnight zone" of homology. Curr. Protein Pept. Sci. 2003; 4:327–337. [DOI] [PubMed] [Google Scholar]
- 2. Pingoud A., et al. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005; 62:685–707. [DOI] [PubMed] [Google Scholar]
- 3. Lyumkis D., et al. Allosteric regulation of DNA cleavage and sequence-specificity through run-on oligomerization. Structure. 2013; 21:1848–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siksnys V., Grazulis S., Huber R.. Pingoud A. Structure and Function of the Tetrameric Restriction Enzymes, in Restriction endonucleases. 2004; Springer; 237–260. [Google Scholar]
- 5. Zaremba M., et al. DNA synapsis through transient tetramerization triggers cleavage by Ecl18kI restriction enzyme. Nucleic Acids Res. 2010; 38:7142–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tamulaitis G., et al. Functional significance of protein assemblies predicted by the crystal structure of the restriction endonuclease BsaWI. Nucleic Acids Res. 2015; 43:8100–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu Q.S., Roberts R.J., Guo H.C.. Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure. Protein Sci. 2005; 14:2590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horton J.R., et al. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006; 34:939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sokolowska M., et al. Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007; 369:722–734. [DOI] [PubMed] [Google Scholar]
- 10. Kaus-Drobek M., et al. Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically. Nucleic Acids Res. 2007; 35:2035–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sasnauskas G., et al. Target site cleavage by the monomeric restriction enzyme BcnI requires translocation to a random DNA sequence and a switch in enzyme orientation. Nucleic Acids Res. 2011; 39:8844–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wah D.A., et al. Structure of FokI has implications for DNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bitinaite J., et al. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10570–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bath A.J., et al. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002; 277:4024–4033. [DOI] [PubMed] [Google Scholar]
- 15. Catto L.E., et al. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006; 34:1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng L., Baumann U., Reymond J.L.. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bochtler M., et al. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006; 25:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leslie A.G.W. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:48–57. [DOI] [PubMed] [Google Scholar]
- 19. CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- 20. Panjikar S., et al. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 2005; 61:449–457. [DOI] [PubMed] [Google Scholar]
- 21. Sheldrick G.M., Hauptman H.A., Weeks C.M., Miller R., Uson I.. Arnold MGRAE. Ab initio phasing, in International Tables for Macromolecular Crystallography. 2001; Kluwer Academic Publishers; 333–345. . [Google Scholar]
- 22. Schneider T.R., Sheldrick G.M.. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002; 58:1772–1779. [DOI] [PubMed] [Google Scholar]
- 23. Pannu N.S., McCoy A.J., Read R.J.. Application of the complex multivariate normal distribution to crystallographic methods with insights into multiple isomorphous replacement phasing. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:1801–1808. [DOI] [PubMed] [Google Scholar]
- 24. Cowtan K. Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr. D Biol. Crystallogr. 1999; 55:1555–1567. [DOI] [PubMed] [Google Scholar]
- 25. Morris R.J., Zwart P.H., Cohen S., Fernandez F.J., Kakaris M., Kirillova O., Vonrhein C., Perrakis A., Lamzin V.S. et al. . Breaking good resolutions with ARP/wARP. J. Synchrotron Rad. 2004; 11:56–59. [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 27. Vagin A., Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:22–25. [DOI] [PubMed] [Google Scholar]
- 28. Otwinowski Z. Maximum likelihood refinement of heavy atom parameters. Proceedings of the CCP4 Study Weekends. 1991; 80–86. [Google Scholar]
- 29. Murshudov G., Vagin A., Dodson E.. Application of maximum likelihood refinement in the refinement of protein structures. Proceedings of Daresbury Study Weekend. 1996. [Google Scholar]
- 30. Afonine P.V., et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 32. Luscombe N.M., Laskowski R.A., Thornton J.M.. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997; 25:4940–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kraulis P. Molscript-a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991; 24:946–950. [Google Scholar]
- 34. Merritt E.A., Murphy M.E.. Raster3D Version 2.0 A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994; 50:869–873. [DOI] [PubMed] [Google Scholar]
- 35. Franke D., Kikhney A.G., Svergun D.I.. Automated acquisition and analysis of small angle X-ray scattering data. Nuclear Instrum. Methods Phys. Res. A. 2012; 689:52–59. [Google Scholar]
- 36. Konarev P.V. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003; 36:1277–1282. [Google Scholar]
- 37. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volkov V.V., Svergun D.I.. Uniqueness of it ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003; 36:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Svergun D., Barberato C., Koch M.H.J.. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995; 28:768–773. [Google Scholar]
- 40. Kozin M., Svergun D.. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001; 34:33–41. [Google Scholar]
- 41. Tamulaitis G., Solonin A.S., Siksnys V.. Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett. 2002; 518:17–22. [DOI] [PubMed] [Google Scholar]
- 42. Yoshioka K. KyPlot - a user-oriented tool for statistical data analysis and visualization. Comp. Stat. 2002; 17:425–437. [Google Scholar]
- 43. Lo Conte L., Chothia C., Janin J.. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999; 285:2177–2198. [DOI] [PubMed] [Google Scholar]
- 44. Bozic D., et al. Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 A resolution. J. Mol. Biol. 1996; 255:176–186. [DOI] [PubMed] [Google Scholar]
- 45. Grazulis S., et al. Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res. 2002; 30:876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deibert M., et al. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000; 7:792–799. [DOI] [PubMed] [Google Scholar]
- 47. Skirgaila R., et al. Structure-based redesign of the catalytic/metal binding site of Cfr10I restriction endonuclease reveals importance of spatial rather than sequence conservation of active centre residues. J. Mol. Biol. 1998; 279:473–481. [DOI] [PubMed] [Google Scholar]
- 48. Tamulaitis G., Mucke M., Siksnys V.. Biochemical and mutational analysis of EcoRII functional domains reveals evolutionary links between restriction enzymes. FEBS Lett. 2006; 580:1665–1671. [DOI] [PubMed] [Google Scholar]
- 49. Pingoud V., et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003; 329:913–929. [DOI] [PubMed] [Google Scholar]
- 50. Golovenko D., et al. Structural mechanisms for the 5΄-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 2009; 37:6613–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szczepanowski R.H., et al. Central base pair flipping and discrimination by PspGI. Nucleic Acids Res. 2008; 36:6109–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dunten P.W., et al. The structure of SgrAI bound to DNA; recognition of an 8 base pair target. Nucleic Acids Res. 2008; 36:5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manakova E., et al. Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease. Nucleic Acids Res. 2012; 40:6741–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mizuno H., et al. Purification, properties and determinations of recognition sequence and cleavage site of restriction endonuclease from "Agrobacterium gelatinovorum" IAM 12617, a marine bacterium (AgeI). Agric. Biol. Chem. 1990; 54:1797–1802. [PubMed] [Google Scholar]
- 55. Siksnys V., et al. The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol. 1999; 291:1105–1118. [DOI] [PubMed] [Google Scholar]
- 56. Zhu Z. Patent U. High fidelity restriction endonucleases. 2013; Ipswich, MA: New England Biolabs, Inc; 1–78. Patent US 8,628,945, pp.. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.