Abstract
Complete and accurate genome assembly and annotation is a crucial foundation for comparative and functional genomics. Despite this, few complete eukaryotic genomes are available, and genome annotation remains a major challenge. Here, we present a complete genome assembly of the skin commensal yeast Malassezia sympodialis and demonstrate how proteogenomics can substantially improve gene annotation. Through long-read DNA sequencing, we obtained a gap-free genome assembly for M. sympodialis (ATCC 42132), comprising eight nuclear and one mitochondrial chromosome. We also sequenced and assembled four M. sympodialis clinical isolates, and showed their value for understanding Malassezia reproduction by confirming four alternative allele combinations at the two mating-type loci. Importantly, we demonstrated how proteomics data could be readily integrated with transcriptomics data in standard annotation tools. This increased the number of annotated protein-coding genes by 14% (from 3612 to 4113), compared to using transcriptomics evidence alone. Manual curation further increased the number of protein-coding genes by 9% (to 4493). All of these genes have RNA-seq evidence and 87% were confirmed by proteomics. The M. sympodialis genome assembly and annotation presented here is at a quality yet achieved only for a few eukaryotic organisms, and constitutes an important reference for future host-microbe interaction studies.
INTRODUCTION
Malassezia species are commensal yeasts and the predominant fungi colonizing the human skin (1–3). They have been associated with several common inflammatory skin conditions and can also cause systemic infections (4). To better understand the molecular basis of host-microbe interactions in these diseases, it is important to establish a high-quality catalog of genes and proteins encoded by Malassezia species. We have previously reported a draft genome sequence and a preliminary gene set for Malassezia sympodialis (5), which is implicated in atopic dermatitis (4). However, this genome assembly was primarily based on short-read sequencing and therefore highly fragmented, comprising 156 contigs (in 66 scaffolds), although the nuclear genome only consists of eight chromosomes (6). In addition, genes were chiefly inferred by computational prediction based on the assembled genome sequence and comparison with protein sequences from other organisms. A set of 1536 expressed sequence tags from Malassezia globosa was used for training gene predictors and assessing predictions, but no other Malassezia-specific transcript or protein data were incorporated (5,7).
The ultimate proof of a gene being protein coding is experimental validation of the encoded protein products. Development of mass spectrometry (MS) based proteomics has made it possible to perform such experiments in a comprehensive manner. In MS-based proteomics, proteins are digested into peptides using proteolytic enzymes, such as trypsin, and analyzed by MS. The resulting mass spectra are interpreted by comparison to a theoretical peptide spectra library, generated by in silico sequence digestion of known and predicted proteins of the studied organism (8).
Proteogenomics is an emerging field in which proteomics and genomics data are combined to improve genome annotation and study impact of genome variations at the protein level. Unbiased discovery of protein-coding regions can be performed by interpreting mass spectra through comparison to a database of the hypothetical peptide sequences obtained by translating a genome sequence in all six reading frames (9). If candidate splice junctions are available from RNA sequencing (RNA-seq), they can be included in the database for discovery of novel splice junction peptides (10). Unlike conventional MS data analysis, this approach does not rely on a reference protein database and can therefore detect previously unannotated coding regions. Improvements in throughput and proteome coverage of MS-based proteomics has potentiated the use of protein evidence to improve gene annotation in many organisms such as Campylobacter concisus (11), Saccharomyces cerevisiae (12,13), Arabidopsis thaliana (14), mouse (9,15) and human (9,16). In contrast to these previous proteogenomics studies, our present study combines proteomics and RNA-seq for genome-wide annotation as part of an integrative workflow. The earlier studies primarily used proteomic data to confirm gene models and discover missing genes after annotation by RNA-seq or homology based means.
When annotating large genomes, proteogenomics is challenging because protein-coding regions constitute a minor part of these genomes and inclusion of hypothetical peptides from non-coding regions may increase the search space several hundred times. In this scenario, it is necessary to restrict database size to maintain an acceptable false discovery rate (FDR) (17), e.g. using isoelectric points of peptides to reduce the database sizes (9). Proteogenomics is particularly applicable to fungal genomes without the need for database reduction because they are small and gene-dense (18,19).
Several aspects of the M. sympodialis genome architecture could not be resolved through short-read sequencing (5), e.g. telomeric and centromeric regions, mating-type loci and mitochondrial genome (mtDNA) structure. Assembly of such regions can reveal new features and biological insights. A distinguishing feature of the M. sympodialis mtDNA is the presence of a 5.9 kb inverted repeat containing the ATP9 gene and tRNAs for methionine, leucine and arginine (5). Large inverted repeats (LIRs) are uncommon in basidiomycete mtDNAs, although a 4 kb LIR encoding Nad4 has been identified in the white button mushroom Agaricus bisporus (20) and a 2.4 kb LIR, harboring plasmid-related sequences and encoding tRNAs, has been found in the poplar mushroom Agrocybe aegerita (21). Species of the ascomycete genus Candida have LIRs that facilitate inter-conversion between circular and linear mtDNA architectures and may produce multiple mtDNA isomers through flip-flop recombination (22). It is not currently known whether the mitochondrial LIR in M. sympodialis has a similar function.
The majority of basidiomycetous species have tetrapolar mating systems in which the P/R locus (encoding the pheromone and pheromone receptors) and HD locus (encoding transcription factors that govern sexual development) are located on different chromosomes and segregate independently during sexual reproduction (23–25). In contrast, in some basidiomycetes such as Cryptococcus neoformans, the mating system is bipolar and the P/R and HD loci have fused to form a large mating-type (MAT) locus that segregates as a single continuous unit during sexual reproduction (26). While recombination within the MAT locus is generally repressed during sexual reproduction, likely due to both the extensive sequence divergence as well as chromosomal rearrangements that are typically present between MAT alleles of compatible mating types, non-crossover recombination (such as gene conversion) has been observed to occur within the MAT locus in C. neoformans (27). Interestingly, in Malassezia species the P/R and HD MAT loci organization differs from both tetrapolar and bipolar mating systems. Specifically, studies have shown that while the two MAT loci are located on the same chromosome, they are not tightly linked, but instead are separated by large syntenic conserved chromosome regions that do not appear to be involved in mating (5,28). This novel MAT organization has been termed a ‘pseudo-bipolar’ mating system to reflect that, while the two MAT loci are linked, recombination can still occur between the P/R and HD regions to generate novel mating type configurations (29). It is not known how linkage between the P/R and HD loci was initially established, or to what extent recombination occurs in the region encompassing the P/R and HD loci during sexual reproduction. Because extant sexual reproduction has yet to be observed for any Malassezia species in a laboratory setting, evidence of recombination involving the MAT loci has only been provided based on population genetics studies of natural isolates.
Here, we used single molecule real-time (SMRT) DNA sequencing on the PacBio RS II system to obtain complete chromosome sequences for the M. sympodialis reference strain (ATCC 42132) and four selected M. sympodialis clinical isolates. In the sequenced M. sympodialis genomes, we identified the presence of all four possible allele combinations of two linked but recombining mating-type loci, detected telomeres and predicted centromere regions on all chromosomes, and found evidence for multiple mtDNA arrangements. Additionally, we present a high-quality reference genome annotation for M. sympodialis in terms of both completeness and accuracy, produced by a novel genome annotation workflow followed by manual curation. The workflow integrated several computational gene predictors, transcriptome sequencing and mass spectrometry based proteomics data. The annotation obtained contains 4493 protein-coding genes, 957 more than in our previous M. sympodialis annotation (5) and it is exceptionally well supported by transcriptome and proteome data. The M. sympodialis gene catalog resulting from this work constitutes a high-quality reference for future studies of host-microbe interactions with Malassezia species.
MATERIALS AND METHODS
M. sympodialis isolates
M. sympodialis ATCC strain 42132 were used in addition to four clinical isolates obtained from the skin of two healthy individuals and two patients with atopic eczema at the Dermatology Unit, Karolinska University Hospital, Stockholm, Sweden. See detailed protocol in (5).
DNA extraction
M. sympodialis ATCC 42132 and the four clinical isolates were cultured on Dixon agar (30) plates modified to contain 1% (vol/vol) Tween 60, 1% (wt/vol) agar and no oleic acid (mDixon) at 32°C and contamination was excluded using blood and Sab-oxide agar plates. After 4 days, cells were harvested using a loophole and suspended in 20 ml phosphate buffered saline and counted by the trypan blue exclusion method (31). DNA was extracted using the QIAGEN Genomic-tip 500/G kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions with some modifications. Briefly, ∼4 × 1010 cells were used for each extraction and two extractions were pooled onto one QIAGEN Genomic-tip500/G. The lysing incubation was carried out on a shaker at 30°C for ∼22 h in lysing buffer Y1 containing 10 mM Tris. The protease treatment was incubated for 3 h at 50°C. The DNA was analyzed on a 1% agarose gel and the concentration was measured with NanoDrop (NanoDrop Technologies, Wilmington, DE, USA).
Genome sequencing and assembly
DNA was sheared into 10 kb fragments using a Genemachines HydroShear Instrument (Digilab, Marlborough, MA, USA). SMRTbells were constructed and sequenced according to the manufacturer's instructions (Pacific Biosciences, Menlo Park, CA, USA). Sequencing was performed on a PacBio RS II sequencer with 3 h movie-time, using 3 SMRT cells for strain ATCC 42132 and 2 SMRT cells for each isolate.
Reads were assembled using the SMRT Analysis HGAP3 assembly pipeline. For strain ATCC 42132, 679 Mb of subreads longer than 3 kb were used for preassembly and 395 Mb corrected reads (average read length 5 kb) were used to assemble the genome with the Celera assembler, followed by polishing with Quiver. The isolate genomes were assembled using the same parameters and similar amounts of data. To assess the completeness of the assemblies, read coverage profiles were inspected and contig ends analyzed for repetitive sequence motifs.
To assess read coverage, SMRT reads were mapped to the genome assemblies using the MEM algorithm in BWA version 0.7.12 (Li 2013 arXiv, http://arxiv.org/abs/1303.3997), with parameter ‘–x pacbio’. Illumina and 454 reads from our previous study (5) were similarly mapped using BWA-MEM with default options. For reads with multiple equally good matches, one was picked at random to avoid overestimating coverage of repeat regions. Coverage profiles were computed with IGVtools (32). The new ATCC 42132 assembly was compared to our previously published assembly (5) using the tool r2cat (33). For centromere prediction, GC content was computed at 25 bp intervals in windows of 250 bp. GC3 content was computed in windows of 10 genes, using the final gene annotation from this study.
RNA extraction
M. sympodialis (ATCC 42132) was cultured on mDixon agar plates as described for DNA extraction above. After 2 or 4 days the cells were suspended in diethylpyrocarbonate water, harvested by centrifugation 1000 g for 5 min, resuspended in diethylpyrocarbonate water and counted. Between 1 × 108 and 4 × 109 cells were harvested by centrifugation. The pellets were resuspended in 600 μl Buffer RLT from the RNeasy kit (QIAGEN) and added to ∼600 μl of acid-washed 0.4–0.6 mm silica beads. The cells were disrupted in a Precellys 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France), using 3 cycles (6000 rpm, 3 × 30 s). The tubes were cooled on ice after each cycle. RNA was extracted using the RNeasy kit following the instructions from the manufacturer (Qiagen), including on-column DNase digestion.
RNA sequencing
Seven RNA-seq libraries were used (Supplementary Table S1). Two of these were prepared from poly(A)-selected RNA using the Illumina TruSeq sample preparation kit (Catalog ID RS-122-2001, Illumina, San Diego, CA, USA) in an automated procedure as previously described (34). The remaining five libraries were prepared from RNA treated with RiboMinus (Thermo Fisher Scientific, Waltham, MA, USA) using a modification of the Illumina TruSeq sample preparation kit to achieve strand-specificity as previously described (35). Clustering was performed on an Illumina cBot cluster generation system using a HiSeq paired-end read cluster generation kit according to the manufacturer's instructions. All libraries were sequenced on an Illumina HiSeq 2000 as paired-end reads to 100 bp. Base conversion was done using Illumina OLB version 1.9. Adapter sequences were removed with cutadapt version 1.2.1 (36). For de novo transcriptome assembly, we additionally trimmed low-quality sequence (using cutadapt option –q 15) and excluded reads shorter than 36 nt after trimming.
RNA-seq read mapping and splice junction discovery
Reads were mapped to the M. sympodialis genome assembly with STAR version 2.3.0e (37) using a two-pass workflow that increases accuracy of alignment across introns (38). In the first pass, reads were mapped to discover an initial set of splice junctions. All reads were then realigned in a second pass, using the splice junction set from the first pass to guide alignment. Reads mapped to the highly expressed ribosomal repeat on chromosome 5 and the mitochondrial large ribosomal RNA gene were excluded from all analyses.
The putative splice junctions reported by STAR were pooled across all seven RNA-seq libraries and filtered to retain high-confidence junctions for proteogenomic mapping. Specifically, we required junctions to have canonical splice site dinucleotides (GT–AG, GC–AG or AT–AC), support from at least 10 RNA-seq reads and spliced alignments extending at least 20 bp into the putative exons on each side (Supplementary Figure S1). These characteristics are reported by STAR, and thresholds were chosen by comparison to M. sympodialis gene models predicted without using RNA-seq data (5).
De novo transcript assembly from RNA-seq data
Four RNA-seq libraries with high strand-specificity were used for transcript assembly (Supplementary Table S1). Strand-specific sequencing typically produces a small proportion of misoriented reads (39). We therefore filtered the reads by analyzing the STAR alignments to the genome and excluding spliced reads for which mapping orientation disagreed with splice site dinucleotide sequences. The data from all four libraries were combined and assembly conducted with Trinity version 2013-11-10 (40). The option –jaccard_clip was enabled to minimize fusion artifacts, using bowtie version 1.0.0 (41) for alignment. Transcript sequences were mapped to the genome using BLAT version 34 (36), requiring 95% identity and allowing introns up to 2000 bp. BLAT results were filtered by running the associated program pslReps with default parameters.
The PASA pipeline version 2.0.1 (42) was applied to identify likely protein-coding regions in the assembled transcripts. PASA clusters transcripts by genomic location and invokes the program TransDecoder to find coding regions. PASA was executed according to the guidelines for strand-specific RNA-seq, requiring stringent overlap (30 bp) for transcript clustering. Untranslated regions (UTRs) were stripped from PASA-inferred gene models. Only models with open reading frames (ORFs) that began with a start codon and ended with a stop codon were used for further analysis.
Genome-guided transcript assembly from RNA-seq data
Transcripts were also assembled with a genome-guided approach. The resulting models were used alongside the Trinity models to support manual annotation, but not included in the automated annotation pipeline. For each of the strand-specific RNA-seq libraries (Supplementary Table S1), reads were mapped to the genome with TopHat version 2.0.8b (43) (using Bowtie version 2.1.0 (44) as the alignment engine), followed by transcript assembly with Cufflinks version 2.1.1 (45). The intron size range was set to 10–2000 bp for both programs and TopHat micro-exon search was enabled. We used TopHat instead of the STAR alignments described above, because the latter contain soft-clipping operations, which are not understood by Cufflinks. Inspection of initial Cufflinks results using the WebApollo genome browser, in comparison to the other data used in this study, revealed an abundance of fusion artifacts, i.e. Cufflinks transcript models comprising multiple adjacent genes. One explanation is that Cufflinks was developed for less compact vertebrate genomes. The occurrence of such artifacts was substantially reduced by setting the parameter overlap-radius to 1 and limiting the amount of input data by processing each of the four samples separately.
Proteogenomics analysis
The MS data have been described previously (5) and deposited in PRIDE under accession PXD003773. Peptide spectra were searched against a customized database using the SEQUEST algorithm (46) in Proteome Discoverer 1.4 (maximum two missed cleavage sites allowed). Peptide spectra matches were filtered at 1% FDR, estimated with the Percolator algorithm (47). The customized database was constructed by combining peptide sequences from (i) the complete genome sequence translated in all six reading frames, (ii) splice junctions extracted from the previously published M. sympodialis gene set (5), (iii) candidate splice junctions discovered by RNA-seq as detailed above and (iv) known Bos taurus proteins downloaded from UniProt. Spectra matching B. taurus peptides were regarded as contaminants from bovine serum used in the culture medium and therefore excluded. To generate splice junction peptide sequences, 2 × 75 bp flanking nucleotide sequences were taken from splice junction sites (previously annotated or identified from RNA-seq data). If a previously annotated exon was shorter than 75 bp, the whole exon sequence was extracted. Three-frame translation was done on the extracted nucleotide sequences. In silico trypsin digestion of splice junction sequences was performed with no miscleavage allowed between consecutive arginine or lysine and no trypsin cut before proline. Peptides with six or more amino acids and spanning the junction sites were kept.
Computational genome annotation
Protein-coding gene structures were first inferred computationally using the pipeline MAKER version 2.31 (48,49). Transcripts assembled from RNA-seq data with Trinity, MS peptides (excluding those mapped to multiple loci) and the Swiss-Prot database (release 2014_02) of manually reviewed protein sequences were used as evidence. Swiss-Prot comprised 542 503 sequences, including 40 from Malassezia species (37 from M. globosa and 3 from M. furfur), but none from M. sympodialis.
Three different MAKER workflows were tested (see Supplementary Table S2). The results of workflow 3 formed the basis for the final curated gene models, whereas workflows 1 and 2 were used for comparison. Workflow 1 represented a basic pipeline run without RNA-seq or peptide evidence (Supplementary Table S2, run 1). Workflows 2 and 3 were more complex, each comprising three MAKER runs (2a–c and 3a–c), in order to improve performance by retraining gene predictors between runs. These two workflows were identical, with the exception that only workflow 3 made use of peptide data.
The gene predictor GeneMark-ES (50) was used in all MAKER runs, trained on the genome sequence only according to an established protocol (51). The gene predictors SNAP (52) and Augustus (53,54) were used in both second and third iterations, trained on gene sets from the preceding iteration. In the first iterations in workflows 2 and 3, candidate coding regions identified by RNA-seq (see PASA analysis above) were provided to MAKER (via the option pred_gff), such that the initial SNAP and Augustus training sets were based on RNA evidence. The following MAKER options were common to all runs: est2genome = 0, protein2genome = 0, keep_preds = 0, min_protein = 10, single_exon = 1, correct_est_fusion = 1. Note that setting keep_preds = 0 ensures that only gene predictions with supporting evidence are retained. Setting single_exon = 1 enables MAKER to consider evidence from single-exon transcripts in the absence of protein evidence at the same loci. As described in results, however, RNA-seq evidence was not sufficient for detection of single-exon genes at some loci, where genes were revealed only after addition of peptide evidence.
Manual genome annotation
Gene models from MAKER run 3c (Supplementary Table S2) were manually curated using the JBrowse (55) plugin WebApollo (56). To assist manual annotation, multiple evidence tracks were configured in JBrowse, including all evidence provided to MAKER; GeneMark-ES, SNAP and Augustus gene predictions from MAKER; RNA-seq read alignments, candidate introns and strand-specific read coverage; transcript sequences assembled by Cufflinks; known and predicted proteins from other fungi; and the previously published M. sympodialis annotation (5). The primary aim was to annotate protein-coding regions. UTR boundaries were annotated when there was sufficient supporting data. Minor isoforms were generally not considered. In the absence of peptide evidence, genes were required to have an ORF longer than 300 bp and mean RNA-seq read count above 10. Singleton peptides with a SEQUEST Xcorr score below 2 were not considered sufficient evidence to annotate genes.
Gene naming
A conservative procedure was used to assign descriptions and gene names, such that automated name assignment only was carried out for genes with high-confidence orthologs in the S. cerevisiae protein sequence database from SGD (57). A gene name was transferred to M. sympodialis only when there was a reciprocal best S. cerevisiae BLASTP hit with E-value < 10−5, >80% coverage of both query and target and >50% identity. Other genes with BLASTP hits (E-value < 10−5) were given a description of the form ‘Similar to S. cerevisiae protein …’. Other genes with peptide evidence from mass spectrometry were given the description ‘uncharacterized protein’, and remaining genes annotated as coding but having with only RNA-seq support were described as ‘hypothetical protein’.
Direct comparison of annotations
The previously published annotation (5) was transferred to our new genome assembly using the liftOver program (58) and then compared to our current annotation using ParsEval (59). A chain file for liftOver was constructed by aligning the old and new genome assemblies with BLAT and processing the alignments according to the instructions on the UCSC Genome Browser wiki (http://genomewiki.ucsc.edu/index.php/Minimal_Steps_For_LiftOver). We used the gt stat command from genometools (60) to validate the GFF3 format and check phase numbers for CDS features. After that, ParsEval was run in HTML output mode using previously published annotation as reference and current annotation as prediction.
Pfam analysis
Pfam analysis was conducted using interproscan-5.11-51.0 with default parameters, retaining matches with E-value < 10−10. Matches to reverse transcriptase, integrase, virus-related, unknown and uncharacterized domains were ignored. The best scored (lowest E-value) Pfam domain was counted for each gene. Protein sequences of S. cerevisiae were downloaded from the SGD website (57). Protein sequences of other fungi were downloaded from the NCBI website: Candida albicans strain WO-1 (61) (bioproject 16371), C. neoformans H99 (62) (bioproject 411) and Ustilago maydis strain 521 (63) (bioproject 1446).
MAT loci
The MAT loci (HD and PR) of the four clinical M. sympodialis isolates (Table 3), as well as for strain ATCC 42132, were identified by searching the genome assemblies for matches to the M. sympodialis HD and PR sequences in GenBank (accessions JX964802.1 and JX964848.1). The identified sequences were extracted from the genome assemblies and aligned using the program ClustalX (64). Phylogeny was constructed using the maximum likelihood algorithm implemented in MEGA version 6.06 (65).
Table 3. HD and PR allele combinations for M. sympodialis strain ATCC 42132 and four clinical isolates.
| Strain | Diagnosis | Mating type | HD locus | PR locus |
|---|---|---|---|---|
| ATCC 42132** | a1b1 | HD1 | PR1 | |
| KS004** | HC | a2b2 | HD2 | PR2 |
| KS024 | HC | a2b3 | HD3 | PR2 |
| KS292** | AE | a1b2 | HD2 | PR1 |
| KS327** | AE | a2b1 | HD1 | PR2 |
Isolates highlighted with ** represent all four possible allele combinations between the HD (HD1 and HD2) and PR (PR1 and PR2) loci. HC: healthy controls; AE: atopic eczema patients.
RESULTS
A complete and gapless reference genome assembly for M. sympodialis
Through long-read sequencing (100x coverage, 3 SMRT cells) of the M. sympodialis (ATCC strain 42132) genome, we obtained an assembly comprising nine contigs, which correspond to eight nuclear chromosomes and one mitochondrial genome. The nuclear contig sizes sum to 7.75 Mb and closely match the chromosome sizes previously observed by pulsed-field gel electrophoresis (PFGE) (6,66) (Table 1). These contigs are fully collinear with our previous assembly (Supplementary Figure S2) that was based on short-read data using other sequencing technologies (Illumina and 454) and comprised 156 contigs (5). A repeated 7 bp sequence was identified on both ends of the eight nuclear chromosomes. This telomere sequence is TTAACAC at the 5΄-end, and its reverse complement GTGTTAA at the 3΄-end. No internal telomere repeats were identified (Supplementary Figure S3). The new assembly contains no sequence gaps and it only has one unresolved region in the nuclear genome: the ribosomal repeat on chromosome 5, which was assembled in a short six-copy version with about 5 to 6 times higher read coverage than the rest of the chromosome, indicating a ribosomal copy number of 30 to 36. Read coverage was otherwise even across the nuclear contigs (Supplementary Figure S4). Thorough genome annotation, as detailed below, identified only one error (a mononucleotide stretch missing one base), which caused a frame shift and was corrected. We screened for additional base-level errors by comparison to the independent Illumina and 454 reads (5), but no credible errors were identified (data not shown; note that the lack of evidence for allelic variation is consistent with the hypothesis that M. sympodialis is haploid (6,66)). The genome assembly was also in excellent agreement with transcript sequences independently assembled from RNA-seq data (described below). More than 97% of RNA contigs longer than 300 bp were mapped to the genome and these displayed very high similarity to the genome sequence (mean identity 99.96% within aligned regions). Taken together, these analyses demonstrate that our M. sympodialis reference assembly is highly accurate.
Table 1. M. sympodialis nuclear chromosome sizes and predicted centromeres.
| Chr | Size in current assembly (bp) | Size estimate from PFGE (Mb) | Putative centromeric region (CEN) | GC content of 250 bp trough | Size of CEN (bp) |
|---|---|---|---|---|---|
| 1 | 1 508 930 | 1.55 | 786 541–787 061 | 16.4% | 520 |
| 2 | 1 381 175 | 1.37 | 355 760–355 841 | 20.0% | 81 |
| 3 | 1 353 702 | 1.37 | 237 534–238 686 | 15.6% | 1152 |
| 4 | 1 203 350 | 1.17 | 418 202–418 728 | 15.2% | 526 |
| 5 | 709 412 | 0.75 | 125 056–125 220 | 18.0% | 164 |
| 6 | 634 681 | 0.62 | 101 950–102 502 | 14.4% | 552 |
| 7 | 517 958 | 0.53 | 431 542–431 987 | 13.2% | 445 |
| 8 | 438 251 | 0.47 | 24 694–25 564 | 18.4% | 870 |
| Total | 7 747 459 | 7.83 | n.a. | n.a. | 4310 |
Centromeres of many fungal species including S. cerevisiae are AT-rich as compared to the rest of the genome (67,68). GC3-troughs (regions with low GC content at third positions in codons) correlate with centromere loci in several yeast species, specifically in Yarrowia lipolytica (69). Kapoor et al. (70) provided corroboration of this and further observed that global GC-troughs (regions of the chromosome that have the lowest GC content) correspond precisely with centromere loci in Candida lusitaniae. Considering that M. sympodialis possesses a genome of comparable size to those species in which GC3/GC troughs are found to be associated with centromeres, we performed a similar in silico analysis to predict centromere regions in M. sympodialis. We found that each chromosome had precisely one locus with a sharply lower GC content (≤ 20% in all cases) (Supplementary Figure S5). The next lowest trough has GC content above 30% in all but one case. Each of these loci corresponds with a local trough in GC3 content. In addition, these GC troughs bear very low nucleotide-composition similarity to any other region on the chromosome (Supplementary Figure S5). Based on this analysis, we predict that these unique regions with global GC troughs are the centromere regions in M. sympodialis (Table 1). However, further experimental validation is required.
Genome annotation combining RNA-seq and proteogenomics
To achieve an accurate and complete genome annotation, we developed a novel genome annotation workflow integrating RNA-seq and proteomics data (Figure 1). For RNA-seq data generation, we applied two different enrichment methods (Supplementary Table S1) to sequence both mRNA and non-coding RNA. In total, we obtained 71 million RNA-seq read pairs mapping to genomic regions other than the highly expressed ribosomal loci. RNA-seq is well suited for discovery of splice junctions, which are difficult to identify from genomic sequence alone. In total, we obtained 6786 putative splice junctions (excluding low-confidence junctions; see Materials and Methods), of which 5169 (76%) were novel, i.e. absent from the previous annotation that was produced without using RNA-seq (5). Candidate transcript sequences were assembled from RNA-seq reads, mapped to the genome and scanned for ORFs. This identified a conservative set of 2683 likely protein-coding genes, which served as an initial set for training gene prediction programs to recognize M. sympodialis gene structures.
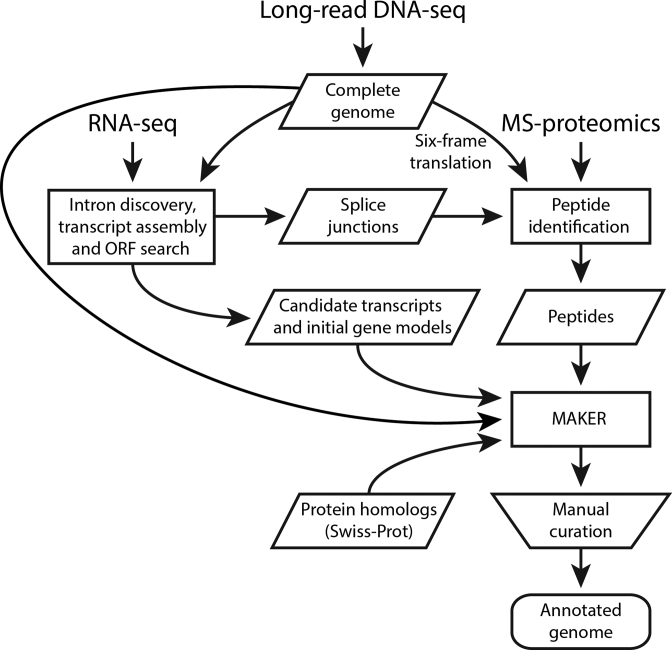
Figure 1.
Integrative genome annotation workflow. Data from four different sources (long-read DNA sequencing, RNA-seq, MS-based proteomics and Swiss-Prot reviewed proteins) were integrated using an evidence-based genome annotation framework (MAKER). Transcripts were assembled from RNA-seq reads using Trinity and PASA was used to identify likely protein-coding regions to provide gene models for initial gene predictions. Three ab initio gene predictors (GeneMark-ES, Augustus and SNAP) were included in MAKER. Augustus and SNAP were iteratively trained based on MAKER-generated gene models (see Materials and Methods and Supplementary Table S2). The computationally inferred gene structures were manually curated. Shapes are used according to workflow figure standards (rectangles show processes, data are in parallelograms, the trapezoid indicates a manual step and the rounded rectangle represents output).
To obtain peptide data for genome annotation, we performed proteogenomics analysis using a previously generated comprehensive proteomics data set for M. sympodialis (5). Here, we re-analyzed this data set by interpreting the mass spectra against an expanded and more accurate peptide database, including (i) all peptides from a six-frame translation of the new genome assembly, (ii) the putative splice junction spanning peptides from our earlier annotation (5) and (iii) the 5169 novel candidate splice junctions discovered by RNA-seq as described above. At an estimated 1% FDR, 35 998 unique M. sympodialis peptides were identified, and 829 of these mapped to splice junctions. To assess the extent to which these peptide data cover the proteome, independently of any annotation, we divided the nuclear genome into 2 kb windows (M. sympodialis is thought to harbor approximately 1 gene per 2 kb (5)) and counted the number of unique peptides per window (Supplementary Figure S6). Only 5.5% of windows lacked peptides entirely and 90% of windows had at least two mapped peptides, indicating that the proteomics data can be expected to provide direct evidence of translation for the great majority of protein-coding genes. The only larger region lacking peptides is a 0.5 Mb region on chromosome 5, corresponding to the incompletely resolved ribosomal RNA repeat (Supplementary Figure S6). In a complementary analysis, we calculated how many ORFs in the nuclear genome were supported by peptides when randomly subsampling different proportions of the complete peptide data set. This indicated that the number of supported ORFs is nearing saturation (Supplementary Figure S7). Note that we do not expect peptide coverage of all protein-coding genes. The detection of a given protein depends on multiple factors in a MS-experiment; mainly on protein abundance, but also on protein sequence, since for successful MS-detection tryptic peptides (cleaving on arginine or lysine) in a certain length interval need to be obtained (here, we used search parameters including peptides between 6 and 50 amino acids). Moreover, ionization properties of the generated peptides render some peptides difficult to detect from a complex peptide mixture by MS. Finally, not all protein-coding genes are expected to generate proteins in the culture conditions used.
We then customized the integrative genome annotation framework MAKER (48) to infer gene structures based on evidence from three different sources: transcripts assembled from strand-specific RNA-seq data, peptides from the proteogenomic search and the full Swiss-Prot database of manually reviewed protein sequences from all domains of life (Figure 1). Note that, although we refer to these genes as predicted, we configured MAKER to output only gene predictions supported by RNA-seq, peptide and/or homology evidence. The resulting gene structures were manually curated (Figure 2) according to the guidelines described in Materials and Methods.
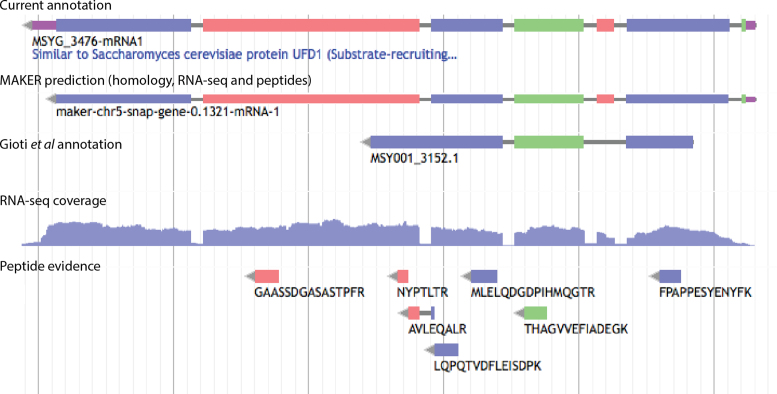
Figure 2.
Gene annotation facilitated by RNA-seq and peptide evidence. Screenshot from the WebApollo genome annotation editor showing a locus where RNA-seq and peptide evidence improved gene annotation compared to the previous annotation described by Gioti et al. (5). The 5΄-UTR and protein-coding segments were identified by the MAKER-based pipeline integrating RNA-seq and peptide data. Manual curation added a 3΄-UTR (uppermost track). The colors of exons and peptides indicate reading frame, such that exons and peptides with the same color are in the same reading frame. UTRs are indicated in purple and introns in gray. RNA-seq coverage is shown for the genomic minus strand (i.e. the strand of the annotated gene) and indicates the number of read pairs at each base.
Integration of RNA-seq and proteogenomics facilitates highly accurate annotation
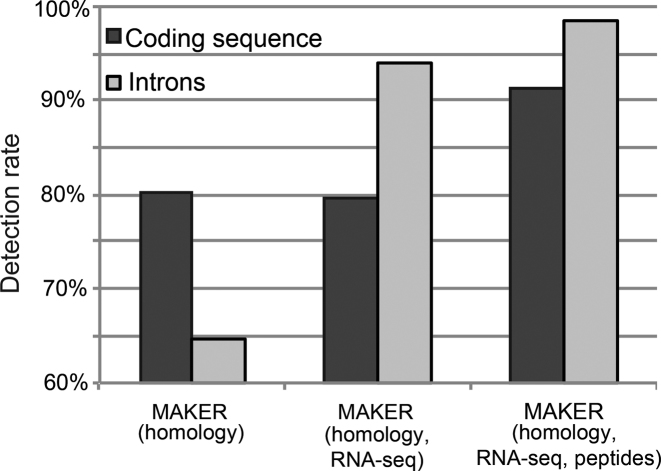
To assess the benefits of including peptide data in gene prediction, we ran MAKER both with and without peptide evidence. We additionally compared these results to our previously published annotation (5), which primarily consists of MAKER gene predictions based on homology evidence, but no RNA-seq or proteomics data. We found that addition of RNA-seq evidence only slightly increased the total number of protein-coding genes predicted, but revealed 2585 (156%) more introns supported by RNA-seq reads (Figure 3 and Table 2, columns 1 and 2). This comparison illustrates the value of including transcriptome sequencing for accurate annotation of intron-containing genomes. Rather than identifying more introns, the integration of additional peptide data in MAKER facilitated the identification of substantially more protein-coding regions (Figure 3 and Table 2, columns 2 and 3). In total, 4113 genes were predicted, 14% more than the number of genes predicted using RNA-seq and homology data only. There was a corresponding 15% increase in the total amount of nucleotide sequence predicted as protein-coding (5.35 to 6.14 Mb, see Table 2). In accordance with more introns annotated, mean exon and intron sizes were decreased, and fewer extremely short or long introns were included (Table 2 and Supplementary Figure S8).
Figure 3.
Increases in coding sequence and intron detection through addition of RNA-seq and proteomics data. Percentages were calculated using the values (length of coding sequences and total number of introns) from the manually curated annotation as denominator.
Table 2. Characteristics of M. sympodialis gene sets.
| Published (MAKER with homology evidence) (5) | MAKER with homology and RNA-seq evidence | MAKER with homology, RNA-seq and peptide evidence | Manually curated annotation | |
|---|---|---|---|---|
| Protein-coding genes | 3536 | 3612 | 4113 | 4493 |
| Gene density (genes/kb)1 | 0.46 | 0.46 | 0.53 | 0.58 |
| Coding sequence (Mb) | 5.40 | 5.35 | 6.14 | 6.72 |
| Coding exons | 6995 | 8453 | 9212 | 9793 |
| Introns | 3462 | 5030 | 5267 | 5350 |
| Mean exon size (bp)2 | 772 | 635 | 669 | 687 |
| Mean intron size (bp) | 65 | 52 | 50 | 30 |
| Genes supported by peptides | 3176 | 3176 | 3674 | 3891 |
| Introns supported by RNA-seq | 1661 (48%) | 4246 (84%) | 4275 (81%) | 5271 (99%) |
| Out-frame peptides | 4658 (13%) | 5453 (15%) | 1796 (5%) | 338 (1%) |
1Gene density was computed relative to the size of the corresponding genome assembly (7.71 Mb for the draft assembly of Gioti et al. (5) and 7.79 Mb for the current assembly).
2Excluding untranslated regions.
Compared to the gene set acquired with RNA-seq and homology data, 497 genes were annotated at novel loci by MAKER when including peptide data. To investigate why these genes were missed without peptide evidence, we examined multiple features: protein length, RNA-seq read coverage, intron and exon numbers and UTRs. First, these genes are not particularly short as one may suspect (mean length 538 aa, compared to 498 aa for the entire final gene set). However, we found that 249 of the 497 genes were merged into neighboring genes with long UTRs when peptide evidence was excluded. Among the other 248 missed genes, 188 are single-exon genes (based on manual annotation of the corresponding loci). It is well recognized that single-exon genes are hard to distinguish based on RNA-seq data alone, because a certain background level of intronless read coverage commonly exists, for biological and technical reasons (e.g. run-through transcription from neighboring genes or imperfect strand-specificity). Of the remaining 60 missed genes, many had either very low RNA-seq coverage (<10 reads per gene) or no underlying gene prediction. To exemplify these issues, Supplementary Figure S9 shows four genes that were predicted only when peptide evidence was used. Overall, provision of peptide data helps MAKER overcome these problems and improves it to be a more robust and sensitive platform for discovery of protein-coding genes.
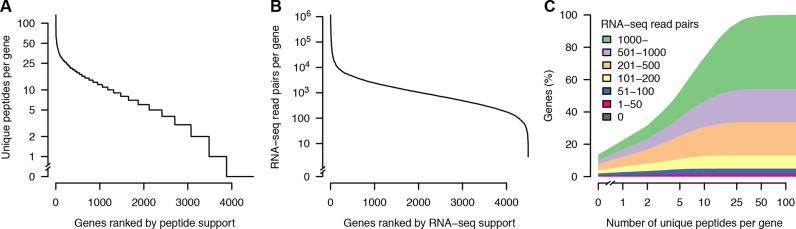
Manual curation resulted in a further 9% increase in the number of protein-coding genes to 4493 and a corresponding 9% increase in total coding sequence (Table 2). All 4493 genes were supported by RNA-seq reads and only 611 (14%) lacked peptide support (Figure 4, panel A and B). The inter-connection between the number of unique peptides and RNA-seq reads is shown in Figure 4C. Of the 611 genes without peptide support, 344 were similar to S. cerevisiae proteins (BLASTP E-value < 10−5) or domains characterized in other proteins (Pfam E-value < 10−10). We carried out a systematic comparison between previously published (5) and current annotation. In total, we identified 957 more protein-coding genes, including 862 genes in novel loci, i.e. regions without genes in the previous annotation (5). These new genes include homologs to catalytic enzymes, transporter proteins and transcription factors from S. cerevisiae (see classification of these genes in Supplementary Figure S10). There were only 1264 genes with identical amino acid sequences between the two annotations, and only 649 of these have perfect matches in gene structure including UTRs. Thus, our new gene catalog includes changes to 64% of previously annotated protein sequences. These statistics show that our current annotation constitutes a major improvement over the previous annotation, not only in identifying novel genes, but also in accuracy of gene structures.
Figure 4.
Experimental support for the final set of protein-coding genes. (A) Number of unique peptides per gene. (B) Number of RNA-seq read pairs per gene. (C) Relation between peptide and RNA-seq support. The uppermost curve shows the cumulative distribution of the number of unique peptides per gene, for all protein-coding genes. Genes were additionally stratified by the number of supporting strand-specific RNA-seq read pairs, and the area under the curve colored accordingly (inset legend). To be conservative, read pairs were only counted if uniquely mapped within annotated coding sequences, i.e. reads containing UTRs or other non-coding sequences were excluded. Note the use of logarithmic scale for y-axis (A and B) and x- axis (C).
In- and out-frame peptide analysis indicate that virtually all coding genes have been annotated
Peptides identified by genome-wide six reading frame (6RF) search are direct evidence of ORF translation, independent of any annotation. The peptides falling outside annotated protein-coding regions indicate potentially incorrect exon boundaries, missed genes or coding exons, and can thus be used to assess indirectly the completeness of a genome annotation. It was found that 4246 (14%) peptides from 6RF search mapped outside annotated protein-coding regions in our previously published annotation (5), indicating that a substantial number of genes had been missed. The number of such out-frame peptides dramatically decreased (14% to 5%) when using peptide data in MAKER annotation, while such improvement was not observed in MAKER annotation using RNA-seq and homology data only (see Table 2). After manual curation, only 338 (0.94%) peptides mapped outside protein-coding regions or in a different reading frame. The confidence score of these out-frame peptides were significantly lower than those of in-frame peptides (P < 10−15, two-tailed t-test) and their score distribution resembles the decoy peptide hit distribution (Supplementary Figure S11), indicating that these 338 cases are likely false peptide matches. The remaining 35 450 peptides confirmed the reading frame and strand of annotated genes. Thus, our curated annotation captures all genes that have robust evidence in the proteomics data set.
Protein domain analysis confirms accuracy of M. sympodialis gene annotation
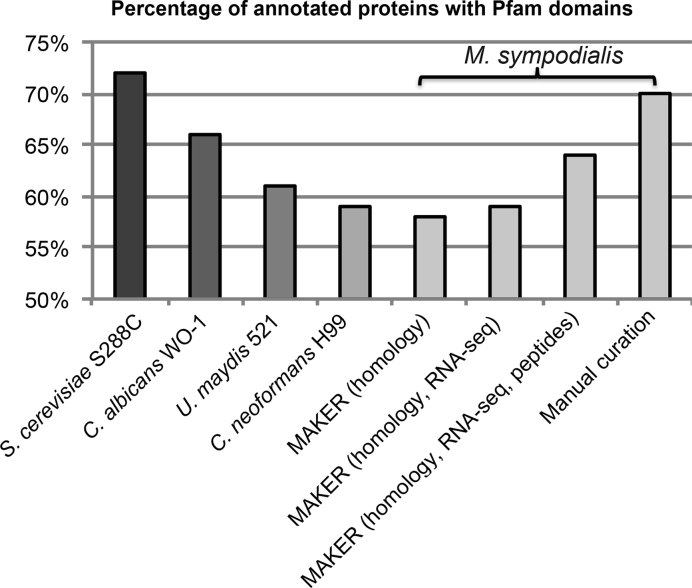
We further assessed the quality of annotation by searching for conserved protein domains (Pfam domains) in the protein sequences from different annotation approaches (71), under the assumption that domains will be relatively more detectable in a well-annotated genome. The number of proteins with Pfam domain matches detected in M. sympodialis was increased by integration of RNA-seq and peptide data in genome annotation, and was highest in the final manually curated gene set (Figure 5). For reference purposes, we carried out the same analysis for our previously published M. sympodialis annotation (5) and four other well-annotated fungi: S. cerevisiae (57), Candida albicans (61), C. neoformans (62) and U. maydis (63). Apart from S. cerevisiae, M. sympodialis contained the highest percentage of proteins with Pfam domains among the selected fungal species (Figure 5). Besides annotation quality, the high proportion of M. sympodialis proteins with Pfam domains (70%) compared to other species likely reflects the evolution in Malassezia species of compact genomes with a high proportion of conserved essential genes (28).
Figure 5.
Pfam domain content in different annotation sets compared to reference species. The percentage of proteins with Pfam domains in M. sympodialis annotation was calculated using the total number of genes after manual curation as denominator. The numbers of M. sympodialis proteins with Pfam domains identified from different annotations sets were 2595 in the Gioti annotation (MAKER with homology evidence) (5), 2647 in MAKER annotation with homology and RNA-seq evidence, 2903 in MAKER annotation with homology, RNA-seq and peptide evidence, and 3173 after manual annotation.
Evidence for multiple mitochondrial genome arrangements in M. sympodialis
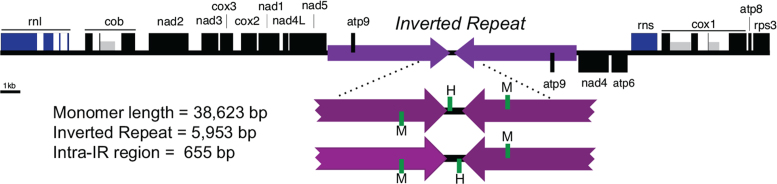
We previously assembled and annotated a 38.6 kb sequence representing the M. sympodialis mtDNA (5). The SMRT reads confirmed this sequence, with the exception of a single base insertion in an intergenic region (an A at position 9822). As previously described, the M. sympodialis mtDNA contains a large inverted repeat of 5.9 kb separated by an intra-repeat region of 655 bp (5). Interestingly, SMRT read assembly produced a greater-than-unit length alternative mtDNA contig of 65.8 kb that contains two copies of the inverted repeat region having different flanking regions (Supplementary Figure S12). The SMRT reads were not sufficiently long to verify the existence of these different configurations; however, the shorter length of the intra-repeat region allowed for confirmation of two orientations relative to the flanking IR (Figure 6). This indicates that the repeated regions undergo homologous recombination that inverts the intra-IR region. By inference, recombination may occur between distal repeats in multimeric molecules, which could produce multiple genomic configurations as predicted by the longer 65.8 kb assembly (Supplementary Figure S12). Similar evidence for mitochondrial genome variability was also observed in the sequencing data from the four clinical isolates (Supplementary Figure S13). The inverted repeats are present in the majority of Malassezia species that have been analyzed (Kennell J.C. et al., manuscript in preparation) as well as in Candida species (72). Inverted repeats in mtDNAs have been demonstrated to mediate inter-conversion between linear and circular forms of the mitochondrial genome in Candida species (72), but similar analyses have not yet been carried out in Malassezia species.
Figure 6.
Evidence of multiple mitochondrial genome configurations. The physical map of the mitochondrial DNA (mtDNA) is displayed in a linear form, beginning with the rnl gene. Rectangles indicate genes or exons of highly conserved protein-coding regions (black), ribosomal RNAs (blue) and intron-encoded homing endonuclease genes (grey). The unit-length, monomeric mtDNA contains a large inverted repeat (purple), separated by an intra-repeat region. The intra-repeat and flanking region is shown below, with the position of tRNAs met (M) and his (H) indicated in green. SMRT reads demonstrated that the intra-repeat region exists in two orientations relative to the inverted repeats.
Evidence for sexual reproduction in M. sympodialis from comparative analysis of mating-type loci
Our analysis of the M. sympodialis draft genome sequence (5) provided evidence for an unusual mating type locus configuration termed pseudo-bipolar. Specifically, the two MAT loci (HD and PR) are physically linked, but sufficiently far apart that recombination can occur between the two and thus drive meiosis. Here, we further examined this unusual genomic configuration using the new complete M. sympodialis ATCC 42132 genome sequence. In addition, we SMRT sequenced the genomes of four M. sympodialis clinical isolates selected based on previous PCR and sequence analysis (5) to test if the two MAT loci were linked in all four possible allele combinations. These four genomes were independently assembled, resulting in the same number of chromosomes with no major structural differences or gaps (Supplementary Figure S14).
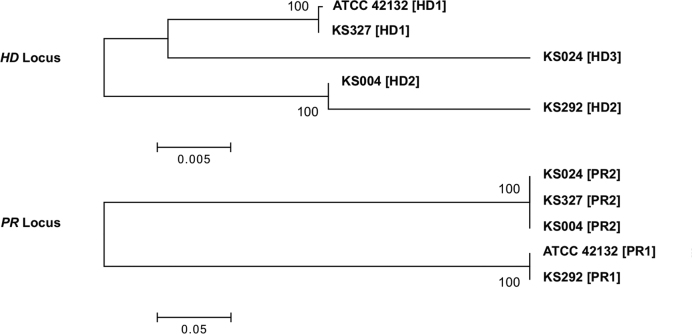
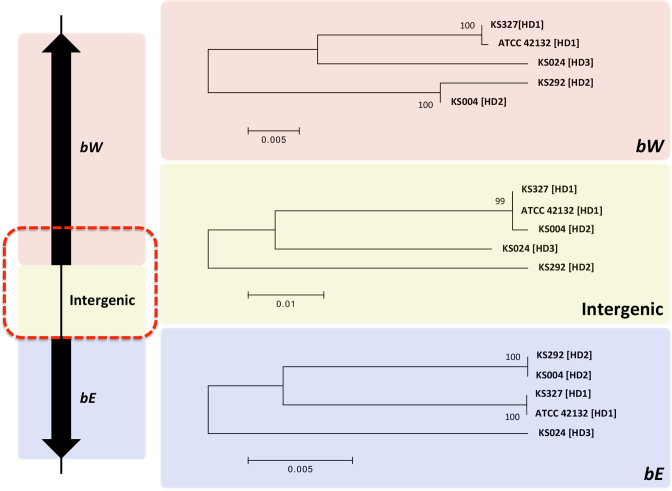
We found the MAT loci to be similarly organized in the four selected clinical M. sympodialis isolates and strain ATCC 42132. That is, the two MAT loci are located on the same chromosome (chr1) and are ∼145 kb apart from each other. For the PR locus, only two sequence clusters were identified among strain ATCC 42132 and the four clinical isolates, corresponding to the PR1 and PR2 alleles (Figure 7). For the HD locus, while polymorphisms are present between alleles of any pair of isolates, phylogenetic analysis showed that the five alleles form two well supported clusters (HD1 and HD2) that each contain two alleles, with the allele from isolate KS024 (HD3) being significantly different from either cluster (Figure 7). It should be pointed out that while the sequences at the HD locus for isolates KS004 and KS292 cluster together, significant polymorphism is present between the two alleles. Interestingly, when the three components of the HD locus (the bW and bE genes, as well as the intergenic region between the two genes) were analyzed separately, it was clear that the majority of the polymorphisms between the HD alleles of isolates KS004 and KS292 are contributed by the divergence present in the intergenic region, where the KS004 allele (HD2) clustered together with the HD1 alleles (Figure 8). Additionally, closer inspection of the bW alleles showed that the polymorphisms between the isolates KS004 and KS292 are restricted to a small region at the 5΄ end of the gene, where the allele in KS004 is similar to the HD1 sequences, consistent with the observation for the intergenic region. Thus, it appears that the HD2 allele of isolate KS004 has a mosaic structure, where although the majority of the allele is composed of HD2 sequence, the intergenic region and the 5΄ end of the gene bW are more similar to HD1 sequence (Figure 8). This could be the result of a homogenization process, such as gene conversion, which may have occurred during sexual reproduction. Additionally, our analysis showed that the five genomes represent all four possible allele combinations of the HD (HD1 and HD2) and PR (PR1 and PR2) loci (see Table 3), which is consistent with the scenario where sexual reproduction is extant in the natural population and reshuffles the allele combinations.
Figure 7.
Phylogeny of the MAT loci and mating type designations of the M. sympodialis isolates. Phylogenetic relationships among the five sequenced M. sympodialis genomes (Table 3) at the HD and PR loci. The allele designation for each genome is shown in brackets. Scale bars indicate the number of substitutions per site. Bootstrap values are based on 1000 replications.
Figure 8.
Phylogeny of the three components of the HD locus. Shown on the left is a schematic diagram of the HD locus. The red dashed rectangle indicates the region where the HD2 allele of isolate KS004 is identical to the HD1 allele (see text). Shown on the right are the phylogenies of the five M. sympodialis genomes (see Table 3) for each of the three components of the HD locus. Scale bars indicate the number of substitutions per site. Bootstrap values are based on 1000 replications.
DISCUSSION
In this study, we described the gap-free genome sequences of M. sympodialis ATCC 42132 and four clinical M. sympodialis isolates based on high-coverage, long-read sequencing. The long sequence reads were critical for assembly of complete chromosome sequences. These data further confirmed that M. sympodialis mating type loci undergo recombination and revealed the existence of multiple mitochondrial genome arrangements. Although only a handful of gapless eukaryotic genomes have been reported so far, more complete genomes will be anticipated as long-read sequencing technologies are increasingly applied and improved (73). Besides the complete genome assemblies, we also attained a comprehensive and highly accurate genome annotation for M. sympodialis, using a novel annotation workflow integrating RNA sequencing and proteogenomics followed by manual curation (Figure 1). As demonstrated, RNA-seq data were particularly useful in detecting introns and provide initial gene sets for accurate model training in gene prediction. Proteogenomics data made the annotation pipeline even more robust and accurate, by distinguishing genes with overlapping UTRs and enabling discovery of single-exon genes that are hard to distinguish from transcriptional noise, as well as genes that ab initio predictors missed and genes with little RNA-seq evidence. Furthermore, the RNA-seq and peptide data also facilitated accurate manual curation. As a result, 4493 protein-coding genes were annotated, representing a 27% increase over the previously published gene set (5) and revealing 862 novel protein-coding loci. Compared to the previously published M. sympodialis annotation (5), our new integrative strategy resulted in changes to 64% of protein sequences and explained >4000 peptides (14% of all identified peptides) mapping outside previously annotated protein-coding regions. All genes and 99% of introns in our current annotation were supported by RNA-seq reads and 87% of protein-coding genes were confirmed by peptide level evidence.
RNA-seq data have been widely used in evidence based genome annotation to improve accuracy and current annotation tools are specifically designed to utilize RNA data. Although some programs, such as MAKER, can make use of peptide data, this information is not fully exploited. Large-scale MS-based proteomics is becoming a widely accessible method, with a cost comparable to that for RNA-seq, and the amount of proteomics data in public databases is rapidly increasing. We therefore advocate further development of current gene predictors to make best use of readily available proteomics data, to improve genome annotation in various organisms. Gene prediction algorithms should be extended to integrate information provided by MS-based proteomics, such as reading frame and identification scores of peptides.
We have demonstrated the utility of peptide data for annotating a small eukaryotic genome. The same strategy should be applicable to large genomes, if the proteogenomic analysis is adapted to limit the amount of false peptide matches, using, e.g. rational database reduction and class-specific FDR estimation (17). We recently used those techniques to discover 98 and 52 novel coding loci in human and mouse genome through proteogenomics (9). In our opinion, the integrative genome annotation approach presented here should be broadly applied to newly sequenced genomes and to refine previous genome annotations.
The M. sympodialis gene catalog resulting from this work can in the future be used as a high quality reference to study a range of biological questions, e.g. regarding host-microbe interactions, and assist genome annotation of closely related fungal species.
DATA AVAILABILITY
The M. sympodialis ATCC 42132 genome annotation, as well as genome assemblies and underlying DNA sequence reads for strain ATCC 43132 and the four clinical isolates, have been deposited in the European Nucleotide Archive under accession number PRJEB13283. The RNA-seq data have been deposited in ArrayExpress under accession E-MTAB-4589. The proteomics data have been deposited in PRIDE under accession PXD003773.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Gustav Wikberg, Karolinska University Hospital, Stockholm, for providing clinical samples of Malassezia. The authors thank Marc P. Hoeppner, Henrik Lantz and Jacques Dainat from the NBIS assembly and annotation service for advice and setting up a WebApollo server for manual curation. The authors are grateful to the Malassezia Research Consortium and Sanela Kjellkvist (Science for Life Laboratory, Stockholm) for helpful discussions. The authors acknowledge support from Science for Life Laboratory and the National Genomics Infrastructure (NGI) for assistance with massively parallel sequencing. Computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project b2010045.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council [to J.Le. and A.S.]; Swedish Foundation for Strategic Research [to J.Le.]; Karolinska Institutet (KID) [to Y.Z. and J.Le.]; Cancer and Allergy Association [to A.S.]; the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet [to A.S. and J.Le.]; National Institutes of Health [R01 grant AI50113-12, R37 grant AI39115-19 to S.S., B.B. and J.H.]; Procter & Gamble Co. and A*STAR/IMB [to T.D.]; Knut and Alice Wallenberg Foundation to the Wallenberg Advanced Bioinformatics Infrastructure [to P.E.]; PRISM 12th plan project at IMSc Chennai [to R.S.]; JNCASR [to S.R.S.]; DBT and SERB, Govt. of India [to K.S.]. Funding for open access charge: Swedish Research Council [2015–04622].
Conflict of interest statement. During initial relevant work T.D. was, but is no longer supported by the Procter & Gamble Company. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1. Oh J., Byrd A.L., Deming C., Conlan S., Program N.C.S., Kong H.H., Segre J.A.. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014; 514:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J.A., Schoenfeld D., Nomicos E., Park M., Kong H.H. et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013; 498:367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gemmer C.M., DeAngelis Y.M., Theelen B., Boekhout T., Dawson T.L. Jr. Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J. Clin. Microbiol. 2002; 40:3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saunders C.W., Scheynius A., Heitman J.. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog. 2012; 8:e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gioti A., Nystedt B., Li W.J., Xu J., Andersson A., Averette A.F., Munch K., Wang X.Y., Kappauf C., Kingsbury J.M. et al. Genomic insights into the atopic eczema-associated skin commensal yeast malassezia sympodialis. MBio. 2013; 4, doi:10.1128/mBio.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boekhout T., Kamp M., Gueho E.. Molecular typing of Malassezia species with PFGE and RAPD. Med. Mycol. 1998; 36:365–372. [DOI] [PubMed] [Google Scholar]
- 7. Xu J., Saunders C.W., Hu P., Grant R.A., Boekhout T., Kuramae E.E., Kronstad J.W., DeAngelis Y.M., Reeder N.L., Johnstone K.R. et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:18730–18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aebersold R., Mann M.. Mass spectrometry-based proteomics. Nature. 2003; 422:198–207. [DOI] [PubMed] [Google Scholar]
- 9. Branca R.M., Orre L.M., Johansson H.J., Granholm V., Huss M., Perez-Bercoff A., Forshed J., Käll L., Lehtiö J.. HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat. Methods. 2014; 11:59–62. [DOI] [PubMed] [Google Scholar]
- 10. Sheynkman G.M., Shortreed M.R., Frey B.L., Smith L.M.. Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq. Mol. Cell Proteomics. 2013; 12:2341–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deshpande N.P., Kaakoush N.O., Mitchell H., Janitz K., Raftery M.J., Li S.S., Wilkins M.R.. Sequencing and validation of the genome of a Campylobacter concisus reveals intra-species diversity. PLoS One. 2011; 6:e22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oshiro G., Wodicka L.M., Washburn M.P., Yates J.R. 3rd, Lockhart D.J., Winzeler E.A.. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002; 12:1210–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yagoub D., Tay A.P., Chen Z., Hamey J.J., Cai C., Chia S.Z., Hart-Smith G., Wilkins M.R.. Proteogenomic discovery of a small, novel protein in yeast reveals a strategy for the detection of unannotated short open reading frames. J. Proteome Res. 2015; 14:5038–5047. [DOI] [PubMed] [Google Scholar]
- 14. Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S.. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008; 320:938–941. [DOI] [PubMed] [Google Scholar]
- 15. Xing X.-B., Li Q.-R., Sun H., Fu X., Zhan F., Huang X., Li J., Chen C.-L., Shyr Y., Zeng R. et al. The discovery of novel protein-coding features in mouse genome based on mass spectrometry data. Genomics. 2011; 98:343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khatun J., Yu Y., Wrobel J.A., Risk B.A., Gunawardena H.P., Secrest A., Spitzer W.J., Xie L., Wang L., Chen X. et al. Whole human genome proteogenomic mapping for ENCODE cell line data: identifying protein-coding regions. BMC Genomics. 2013; 14:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nesvizhskii A.I. Proteogenomics: concepts, applications and computational strategies. Nat. Methods. 2014; 11:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohanta T.K., Bae H.. The diversity of fungal genome. Biol. Proced. Online. 2015; 17:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galagan J.E., Henn M.R., Ma L.J., Cuomo C.A., Birren B.. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 2005; 15:1620–1631. [DOI] [PubMed] [Google Scholar]
- 20. Ferandon C., Xu J., Barroso G.. The 135 kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal Genet. Biol. 2013; 55:85–91. [DOI] [PubMed] [Google Scholar]
- 21. Ferandon C., Chatel Sel K., Castandet B., Castroviejo M., Barroso G.. The Agrocybe aegerita mitochondrial genome contains two inverted repeats of the nad4 gene arisen by duplication on both sides of a linear plasmid integration site. Fungal Genet. Biol. 2008; 45:292–301. [DOI] [PubMed] [Google Scholar]
- 22. Valach M., Farkas Z., Fricova D., Kovac J., Brejova B., Vinar T., Pfeiffer I., Kucsera J., Tomaska L., Lang B.F. et al. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 2011; 39:4202–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raper J.R. Genetics of sexuality in higher fungi. 1966; NY: Ronald Press Co. [Google Scholar]
- 24. Ni M., Feretzaki M., Sun S., Wang X., Heitman J.. Sex in fungi. Annu. Rev. Genet. 2011; 45:405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heitman J., Sun S., James T.Y.. Evolution of fungal sexual reproduction. Mycologia. 2013; 105:1–27. [DOI] [PubMed] [Google Scholar]
- 26. Lengeler K.B., Fox D.S., Fraser J.A., Allen A., Forrester K., Dietrich F.S., Heitman J.. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell. 2002; 1:704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun S., Hsueh Y.P., Heitman J.. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012; 8:e1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu G., Zhao H., Li C., Rajapakse M.P., Wong W.C., Xu J., Saunders C.W., Reeder N.L., Reilman R.A., Scheynius A. et al. Genus-Wide Comparative Genomics of Malassezia Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin. PLoS Genet. 2015; 11:e1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coelho M.A., Sampaio J.P., Goncalves P.. A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete. PLoS Genet. 2010; 6:e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gueho E., Midgley G., Guillot J.. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996; 69:337–355. [DOI] [PubMed] [Google Scholar]
- 31. Gehrmann U., Qazi K.R., Johansson C., Hultenby K., Karlsson M., Lundeberg L., Gabrielsson S., Scheynius A.. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses–novel mechanisms for host-microbe interactions in atopic eczema. PLoS One. 2011; 6:e21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Husemann P., Stoye J.. r2cat: synteny plots and comparative assembly. Bioinformatics. 2010; 26:570–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stranneheim H., Werne B., Sherwood E., Lundeberg J.. Scalable transcriptome preparation for massive parallel sequencing. PLoS One. 2011; 6:e21910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sigurgeirsson B., Emanuelsson O., Lundeberg J.. Analysis of stranded information using an automated procedure for strand specific RNA sequencing. BMC Genomics. 2014; 15:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17.1:10–12. [Google Scholar]
- 37. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Engström P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Rätsch G., Goldman N., Hubbard T.J., Harrow J., Guigo R. et al. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013; 10:1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levin J.Z., Yassour M., Adiconis X., Nusbaum C., Thompson D.A., Friedman N., Gnirke A., Regev A.. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010; 7:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011; 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K. Jr, Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003; 31:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eng J.K., McCormack A.L., Yates Iii J.R.. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994; 5:976–989. [DOI] [PubMed] [Google Scholar]
- 47. Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J.. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007; 4:923–925. [DOI] [PubMed] [Google Scholar]
- 48. Holt C., Yandell M.. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011; 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sanchez Alvarado A., Yandell M.. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008; 18:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ter-Hovhannisyan V., Lomsadze A., Chernoff Y.O., Borodovsky M.. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008; 18:1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Borodovsky M., Lomsadze A.. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinformatics. 2011; 35, doi:10.1002/0471250953.bi0406s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004; 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stanke M., Steinkamp R., Waack S., Morgenstern B.. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004; 32:W309–W312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stanke M., Schoffmann O., Morgenstern B., Waack S.. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006; 7:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee E., Helt G.A., Reese J.T., Munoz-Torres M.C., Childers C.P., Buels R.M., Stein L., Holmes I.H., Elsik C.G., Lewis S.E.. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013; 14:R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M. et al. Life with 6000 genes. Science. 1996; 274:546–567. [DOI] [PubMed] [Google Scholar]
- 58. Kuhn R.M., Haussler D., Kent W.J.. The UCSC genome browser and associated tools. Brief. Bioinformatics. 2013; 14:144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Standage D.S., Brendel V.P.. ParsEval: parallel comparison and analysis of gene structure annotations. BMC Bioinformatics. 2012; 13:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gremme G., Steinbiss S., Kurtz S.. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013; 10:645–656. [DOI] [PubMed] [Google Scholar]
- 61. Butler G., Rasmussen M.D., Lin M.F., Santos M.A., Sakthikumar S., Munro C.A., Rheinbay E., Grabherr M., Forche A., Reedy J.L. et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009; 459:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Janbon G., Ormerod K.L., Paulet D., Byrnes E.J. 3rd, Yadav V., Chatterjee G., Mullapudi N., Hon C.C., Billmyre R.B., Brunel F. et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014; 10:e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kamper J., Kahmann R., Bolker M., Ma L.J., Brefort T., Saville B.J., Banuett F., Kronstad J.W., Gold S.E., Muller O. et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006; 444:97–101. [DOI] [PubMed] [Google Scholar]
- 64. Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 65. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013; 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boekhout T., Bosboom R.W.. Karyotyping of Malassezia yeasts: Taxonomic and epidemiological implications. Syst. Appl. Micorbiol. 1994; 17:146–153. [Google Scholar]
- 67. Roy B., Sanyal K.. Diversity in requirement of genetic and epigenetic factors for centromere function in fungi. Eukaryot. cell. 2011; 10:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mehta G.D., Agarwal M.P., Ghosh S.K.. Centromere identity: a challenge to be faced. Mol. Genet. Genomics. 2010; 284:75–94. [DOI] [PubMed] [Google Scholar]
- 69. Lynch D.B., Logue M.E., Butler G., Wolfe K.H.. Chromosomal G + C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol. Evol. 2010; 2:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kapoor S., Zhu L., Froyd C., Liu T., Rusche L.N.. Regional centromeres in the yeast Candida lusitaniae lack pericentromeric heterochromatin. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:12139–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J. et al. The Pfam protein families database. Nucleic Acids Res. 2012; 40:D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Valach M., Pryszcz L.P., Tomaska L., Gacser A., Gabaldon T., Nosek J.. Mitochondrial genome variability within the Candida parapsilosis species complex. Mitochondrion. 2012; 12:514–519. [DOI] [PubMed] [Google Scholar]
- 73. Faino L., Seidl M.F., Datema E., van den Berg G.C., Janssen A., Wittenberg A.H., Thomma B.P.. Single-molecule real-time sequencing combined with optical mapping yields completely finished fungal genome. MBio. 2015; 6, doi:10.1128/mBio.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The M. sympodialis ATCC 42132 genome annotation, as well as genome assemblies and underlying DNA sequence reads for strain ATCC 43132 and the four clinical isolates, have been deposited in the European Nucleotide Archive under accession number PRJEB13283. The RNA-seq data have been deposited in ArrayExpress under accession E-MTAB-4589. The proteomics data have been deposited in PRIDE under accession PXD003773.