Abstract
Eubacterial translation initiation involves assembly of tRNAfMet, mRNA, initiation factors (IFs) and 30S ribosome in a 30S pre-initiation complex (30S pre-IC), which rearranges and joins 50S ribosome to form 70S IC. Upon releasing IFs, 70S IC becomes elongation-competent 70S. The direct recruitment of initiator tRNA (tRNAfMet) into the ribosomal P-site, crucial in accurate initiation of translation, is attributed to two conserved features of tRNAfMet: (i) formylation of amino acid attached to it and, (ii) the presence of three consecutive G-C base pairs (3GC base pairs) in the anticodon stem. However, the precise roles of these two conserved features of tRNAfMet during the various steps of initiation remain unclear. Using natural and engineered tRNAs, we show that the 3GC pairs license tRNAfMet transitions from 30S to 70S IC and then to elongation-competent 70S by release of IF3. Of the 3GC pairs, the middle GC pair (G30-C40), or merely G30 (in a specific context) suffices in this role and is essential for the sustenance of Escherichia coli. Furthermore, rescue of formylase deficient E. coli by overproduced tRNAfMet reveals that the feature of formylation licenses initial targeting of tRNAfMet to 30S ribosome.
INTRODUCTION
Initiator tRNAs are special in their direct binding to the ribosomal P-site, to decode the start codon and to determine the reading frame in an mRNA during the regulatory step of translation initiation. On the other hand, elongator tRNAs enter the ribosome at the A-site to decode the subsequent codons (1). In all domains of life, initiator tRNAs are characterized by a highly conserved feature of the three consecutive G-C base pairs in the anticodon stem (at 29:41, 30:40 and 31:39 positions; referred to as GC/GC/GC or 3GC pairs), which facilitate its P-site targeting. Eubacterial initiator tRNAs (tRNAfMet) are also branded by the presence of a mismatch at the top of the acceptor stem (C1:A72 in Escherichia coli). This feature together with the 2:71 and 3:70 base pairs forms a major determinant in recognition of tRNAfMet by Fmt, the formylating enzyme (2,3).
Initiation of translation occurs in various stages such as the binding of initiation factors (IFs), tRNAfMet and mRNA to 30S ribosome to form 30S pre-initiation complex (30S pre-IC), its conversion into 30S IC, docking of 50S subunit to the 30S IC to form 70S IC, followed by the release of IFs to finally convert the 70S IC to an elongation competent 70S complex (4,5). In the 30S IC, tRNAfMet is bound in P/I state (between the P/E and the P/P states). Docking of the 50S subunit at this stage creates 70S IC where the tRNAfMet is still in the P/I state. It was proposed that upon GTP hydrolysis, a rotational movement accommodates tRNAfMet in the classical P/P state. The rotation is unique as it occurs in a direction opposite to the commonly encountered movement of the tRNA from P/P state to P/E state. In the cryo- EM structure of the 30S IC, the anticodon stem comprising the 3GC pairs was tilted toward the E-site raising the possibility of their role in achieving this conformation (6).
Recently, we observed significant binding of tRNAfMet lacking the 3GC pairs to 30S ribosomes. However, its presence in the elongation competent 70S complex was highly compromised (7). Although this observation suggested a primary role of 3GC pairs in the later stages of initiation (in transition of tRNAfMet from 30S IC to 70S), the contribution of individual GC pairs in this role was not investigated. While both the features of formylation, and the 3GC pairs are crucial in targeting tRNAfMet to the P-site (3,8–10), it is unknown if these two features contribute concurrently or sequentially during the various stages of initiation.
Moreover, even though the GC/GC/GC pairs are extraordinarily conserved in the initiator tRNAs, there are naturally occurring exceptions to this rule. In some of the mycoplasmal and α-proteobacterial species, the first and/or the third GC pair may be found as AU and GU pairs, respectively (11). Intriguingly, these variants (au/GC/GC; GC/GC/gu; or au/GC/gu; variant/mutant pairs are shown in small letters hereafter) still retain the feature of the three consecutive R:Y (purine:pyrimidine) pairs. Thus, could the ‘rule’ be a degenerate one, consisting of three consecutive R:Y pairs? Furthermore, a more recent observation in yeast showed that when the middle GC pair was changed to CG, unlike in E. coli, it still allowed its survival (12). These observations demand a systematic mutational analysis of the 3GC pairs to further understand their role in translation initiation.
Both the genetic and structural studies have suggested that the 3GC pairs are scrutinized by the evolutionarily conserved residues (G1338 and A1339) of 16S rRNA (13,14). A1339 and G1338 make type I and type II A-minor interactions with the 1st and the 2nd GC pairs, respectively (13,14). However, the details of these interactions with the natural variants of the initiator tRNAs lacking a full complement of the 3GC pairs have not been investigated. Also, while initiation factors 3 (IF3) has been shown to be crucial in discriminating against the 3GC pairs and proposed to act via G1338 and A1339 (14) its mechanism has remained unclear.
In this study, we have carried out extensive mutational analysis of the 3GC pairs and shown that the middle G (G30) is the most crucial feature of the 3GC pairs in tRNAfMet in E. coli. Further, we show that initial binding of tRNAfMet to the 30S IC is mainly facilitated by formylation whereas the crucial role of the 3GC pairs begins at the subsequent stages.
MATERIALS AND METHODS
Bacterial strains, plasmids, DNA oligomers and growth conditions
The bacterial strains, plasmids and the DNA oligomers are listed in Supplementary Tables S1, 2 and 3 respectively. E. coli KL16 and its derivatives were grown in Luria-Bertani (LB) liquid or LB-agar plates containing 1.5% bacto-agar (Difco). Unless mentioned otherwise, media were supplemented with ampicillin (Amp, 100 μg/ml), chloramphenicol (Cm, 30 μg/ml), kanamycin (Kan, 25 μg/ml) or tetracycline (Tet, 7.5 μg/ml) when required. Sinorhizobium medicae WSM419 was grown in LB medium.
Site directed mutagenesis
Using DNA oligomers (Supplementary Table S3) containing the desired mutations and appropriate plasmid background, inverse polymerase chain reaction (PCR) was carried out, the amplicon was digested with DpnI and transformed into E. coli TG1. Desired mutants were confirmed by plasmid DNA sequencing. The templates used differed according to the need. The pTrcmetYau/GC/gu was used for generating mutations in the anticodon stem of the initiator tRNA with CAU anticodon (wild-type anticodon). To generate another set of mutants with CUA anticodons (to check for in vivo initiation efficiencies), pCATam1metYCUA/3GC was used as template.
Bacterial growth analysis
Bacterial growth was monitored either on plates or by growth curve assays. For plate assays cultures grown overnight were streaked on desired agar plates and incubated at desired temperatures for various time intervals and imaged using a gel doc (Alpha Imager, Alpha Innotech). For growth curve analysis, bacterial cultures (four replicates/colonies of each strain) were grown in LB with the desired antibiotic(s) until they reached saturation at 37°C. Then a 200 μl volume of a hundred to a thousand fold dilution (10−2–10−3) in the required medium was taken in honeycomb plates and placed in automated Bioscreen C growth reader. O.D.600 was measured every 1 h. Mean O.D. values and standard deviations were calculated and the data were plotted with time on X-axis against O.D.600 on Y-axis using GraphPad Prism software.
Generation of strains sustained with mutant initiator tRNAs
The strains lacking metY gene (KL16ΔmetY::cm) were transformed with plasmids harboring mutant initiator tRNA genes. In this background, deletion of metZWV locus using KanR marker was attempted by P1 mediated transductions (11). Briefly, P1 phage lysate raised on KL16ΔmetZWV::kan was used to infect KL16ΔmetY::cm/pmetY* (*indicates various mutants of metY used in this study, Supplementary Table S2) and selected for KanR transductants. The transductants obtained were confirmed for deletion of metZWV locus by PCR using flanking primers metZWV Fp and metZWV Rp. The wild-type locus gives ∼300 bp amplicon while the knockouts give ∼1.5 kb amplicon. Further verification was done by northern blotting by checking for the absence of metZWV encoded tRNAfMet1.
Isolation of tRNA
Total tRNA preparations from various strains were isolated under cold and acidic conditions to preserve the ester linkage between the amino acid and the tRNA (15). For preparations under neutral conditions, the cells were resuspended in 1× TE (10 mM Tris-HCl pH 8.0, 1 mM Na2 ethylenediaminetetraacetic acid (EDTA)) and extracted with water saturated phenol. Rest of the procedure was the same. The tRNA prepared was completely deacylated.
Acid urea PAGE
The tRNA samples (2–4 μl) were mixed with equal volumes of acid-urea dye (0.1 M sodium acetate (pH 5.0), 10 mM Na2 EDTA, 8 M urea, 0.05% bromophenol blue and 0.05% xylene cyanol) and separated on 6.5% polyacrylamide gel containing 8 M urea and 0.1 M sodium acetate (pH 5.0 buffer). The gel was run at 4°C till the bromophenol blue reached the bottom of the gel, and electroblotted onto a nytran membrane at 15V for 20 min (15). In some cases, the tRNA preparations were treated with 10 mM CuSO4 in 100 mM Tris–HCl (pH 8.0) to deacylate aminoacyl-tRNA (formylated tRNA is resistant) or with 100 mM Tris–HCl (pH 9.0) to deacylate both the formylaminoacyl and the aminoacyl forms of tRNA (16,17).
Native PAGE
The total tRNA was prepared under neutral pH conditions and separated on 15% polyacrylamide gel (pH 8.0 using 1× Tris-Borate-Ethylenediaminetetraacetic acid (TBE)). The gel was run at room temperature till the xylene cyanol reached the bottom of the gel, and electroblotted onto a nytran membrane (15).
Polysome profiling
The total ribosomal preparations from E. coli were carried out essentially as described (18). Translation was inhibited by 0.3 μM tetracycline prior to harvesting the cells, and chilled on ice-salt mix. The buffer 1 contained 10% sucrose. In case of Sinorhizobial study, the cell extract was prepared by sonication. Approximately 10 to 20 O.D.260 were used for analysis on 20–40% (w/v) (15–35% in some cases) sucrose gradients (buffered in 20 mM HEPES-KOH, pH 7.5, 50 mM NH4Cl, 10 mM MgCl2, 4 mM β-mercaptoethanol) prepared using a BioComp gradient master (BioComp Instruments) in polyclear tubes (SETON, catalog no. 7022). Samples were loaded on gradients and spun in an ultracentrifuge using SW55 rotor at 45 000 rpm for 2–3 h at 4°C as described in the figure legends. Gradients were fractionated using a BioComp gradient fractionator fitted with BIO-RAD Econo UV monitor at a flow rate of 0.3 mm per second. Fractions of 200–300 μl volume were collected using fraction collector (BIO RAD Model 2110). In some cases, pooled fractions corresponding to various ribosomal subunits were manually collected by monitoring the absorption trace at 254 nm.
Analysis of tRNA
To analyze tRNA from various fractions of the polysome profiles, the total RNAs were extracted from the fractions using hot phenol saturated with sodium acetate, pH 4.5 and precipitated with ethanol. The RNA were fractionated on native polyacrylamide gel electrophoresis, and transferred on to nytran membrane.
Northern blotting
The tRNAs were fixed onto the nytran membrane by UV cross linking at 120 mJ/cm2 (CL1000-UV products). The membranes were blocked using pre-hybridization buffer containing yeast total RNA and Denhardt's solution (1% bovine serum albumin, 1% ficoll, 1% polyvinylpyrrolidone 40). The northern blot analysis was performed using a 5΄-32P end-labeled DNA oligomers. For analyzing tRNAs, various probes such as D loop ini tRNA (complementary to tRNAfMet D loop positions 2–27), Met33 (complementary to positions 25–39), UV5G (complementary to tRNAfMet2 positions 40–56), met elongator D loop (complementary to tRNAMet D loop positions 2–27) were used. The blots were exposed to phosphor-imager screen and analyzed on BioImage Analyzer (FLA5100, Fuji Film).
Western blotting
Equal amounts of total proteins were separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis with 5% stacking gel and 12% resolving gel. The proteins were transferred onto polyvinylidenedifluoride membrane activated by dipping in methanol and keeping in the transfer buffer (1× sodium dodecyl sulphate (SDS) running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.3) with 15% methanol). The transfer was set up in a BioRad semi-dry trans-blot apparatus at 15 V for 2 h. After transfer, the membrane was blocked with 5% skimmed milk in 1× TBST (20 mM Tris–HCl, pH 7.5, 0.9% NaCl and Tween 20 (0.2% v/v)) and kept at 4°C overnight or 3–4 h rocking at room temperature. It was then washed with TBS-Tween for 10 min. Primary antibody was added (1:10 000, polyclonal antiIF3) and incubated for rocking for 3–5 h at room temperature. After incubation, three 10 min washes were performed with TBS-Tween (20 mM Tris–HCl pH 7.5, 0.9% NaCl and Tween 20 (0.2% v/v)) followed by addition of secondary antibody (1:3000 α-rabbit IgG-HRP, Genei) and incubated for 2–3 h. Three successive washes were performed with TBS-Tween for 10 min each. Blot was developed using ECL reagent (Millipore) and scanned in a Chem Doc (GE).
RESULTS
Generation of mutations in the anticodon stem of tRNAfMet
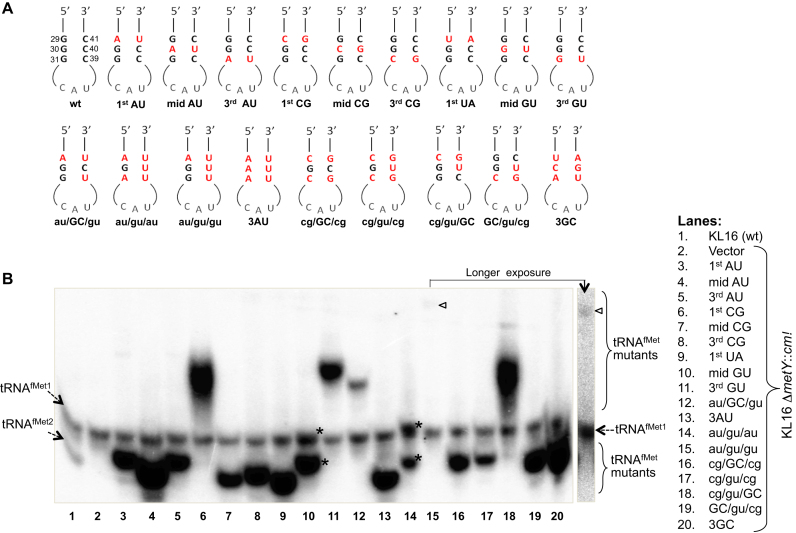
E. coli contains four initiator tRNA (tRNAfMet) genes. Three of these, metZ, metW and metV, encoding tRNAfMet1 (∼75% of total tRNAfMet) are present as single operon metZWV at 63.5΄ while the fourth gene metY, encoding tRNAfMet2 (∼25% of total tRNAfMet), is present at 71.5΄ in the genome. In E. coli K, the two tRNAs differ only at position 46, where mG is found in tRNAfMet1 and A in tRNAfMet2 allowing their separation on a native gel (lane 1; Figure 1B). The naturally occurring tRNAfMet variants possessing au/GC/GC, GC/GC/gu or au/GC/gu in the 3GC region indicated degeneracy of the ‘3GC rule’ in the form of purine:pyrimidine (R:Y) pairs. To test this, we introduced mutations (Figure 1A) in the plasmid borne metY gene (3,19). One set of the mutants comprised single base pair changes retaining R:Y pairing (au/GC/GC, GC/au/GC, GC/GC/au, GC/gu/GC and GC/GC/gu; also referred to as 1st AU, mid AU, 3rd AU, mid GU and 3rd GU, respectively), or resulting in Y:R pairs (cg/GC/GC, GC/cg/GC, GC/GC/cg, and ua/GC/GC; also referred to as 1st CG, mid CG, 3rd CG and 1st UA, respectively). Another set of mutants contained multiple changes (au/GC/gu, au/gu/gu, au/gu/au, au/au/au (3AU mutant), cg/GC/cg, cg/gu/cg, cg/gu/GC, GC/gu/cg and ua/cg/au (3GC mutant)).
Figure 1.
Generation and analysis of 3GC mutants of tRNAfMet. (A) Sequences of the anticodon stem loops of the mutants used in this study. (B) Expression analysis of the mutant tRNAfMet. Approximately 10 μg of total tRNA preparations from E. coli KL16, or KL16ΔmetY::cm (retains tRNAfMet1, as indicated) harboring plasmids containing the mutant metY genes were separated on native polyacrylamide gel electrophoresis (PAGE), transferred onto nytran membrane and hybridized with D-loop ini tRNA probe (complementary to 2–27 nt position) common to all tRNAfMet. tRNAMet (detected by met elongator D loop probe) serves as internal loading control. The two conformer of mid GU and au/gu/au tRNAs are marked by asterisks (*), and the band corresponding to au/gu/gu has been indicated by an arrowhead. Longer exposure of lane 15 has also been shown.
Northern blot analysis of the metY derived tRNAfMet mutants from E. coli KL16ΔmetY host showed their accumulation in the cell (Figure 1B). Except for the au/gu/gu mutant, all others were produced in amounts equal to or greater than tRNAfMet1 (representing ∼75% of cellular tRNAfMet) (Figure 1B). Introduction of two GU wobble pairs (au/gu/gu mutant) led to a drastic decrease in its accumulation (Figure 1B, lane 15, indicated by arrow head). Also, the mutants differed in their mobility. The mid GU and the au/gu/au mutant tRNAs migrated as two distinct conformers (lanes 10 and 14, indicated by ‘*’) one of which migrated very close to tRNAfMet1.
Checking for the essentiality of 3GC pairs in E. coli
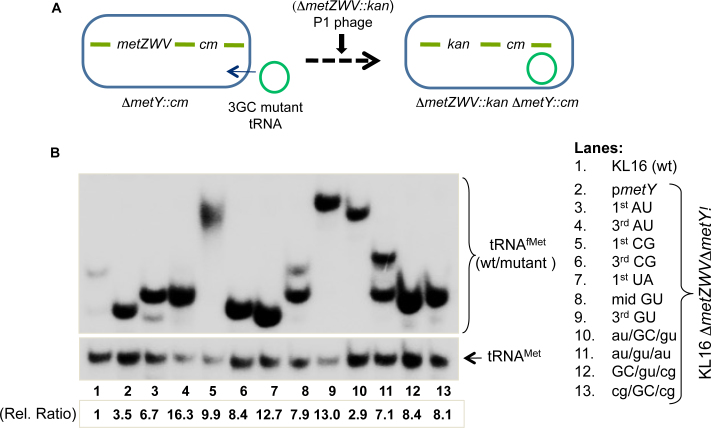
To check for the functionality of the mutants, we used the ‘four-gene knockout’ assay, a strategy where we knockout all four of the chromosomal tRNAfMet genes (11). E. coli KL16 ΔmetY::cm strains harboring plasmids borne metY mutants were transduced with the P1 phage raised on E. coli KL16 ΔmetZWV::kan (Figure 2A). The wild-type (pmetY) was used as a positive control in these experiments. As the presence of functional tRNAfMet is essential for E. coli survival, the occurrence of transductants with the 1st AU, 3rd AU, 1st CG, 3rd CG, 1st UA, mid GU, 3rd GU, au/GC/gu, au/gu/au, GC/gu/cg and cg/GC/cg on kanamycin (Kan) plates (not shown) indicated that these mutant tRNAfMet functioned in E. coli to sustain its growth. Deletion of metZWV was verified by a diagnostic PCR (Supplementary Figure S1). To further validate the absence of wild-type tRNAfMet, northern blotting of total tRNAs from the transductants was carried out using a radiolabeled DNA oligomer probe (Figure 2B). As expected, the band corresponding to tRNAfMet1 was missing (compare Figures 2B with 1B). The blot was also probed for the elongator tRNAMet as loading control. The mutants which could not sustain the cell (as there were no transductants obtained for deletion of metZWV) could not be included in this analysis (compare Figures 2B with 1B). Taken together, we observed that the mutants excepting mid AU, mid CG, 3AU, au/gu/gu, cg/gu/cg, cg/gu/GC and 3GC; sustained E. coli for its growth implying that the middle GC pair is crucial for tRNAfMet function.
Figure 2.
Survival of E. coli on 3GC mutant tRNAfMet. (A) The ‘four-gene knockout’ strategy to check for the ability of mutant tRNAfMet to sustain E. coli. Among the two loci coding for tRNAfMet, the strain deleted for metY (ΔmetY::cm) was transformed with plasmid encoding mutant tRNAfMet; then the deletion of metZWV locus was attempted by P1 mediated transduction of ΔmetZWV::kan locus using Kan selection. (B) Northern blot analysis to confirm sustenance of the cell on the mutant tRNAfMet. Total tRNAs from cells sustained on wild-type or mutant metY genes were prepared and separated on native PAGE, transferred onto nytran membrane and probed with D-loop ini tRNA probe against tRNAfMet. For loading control, elongator tRNAMet was also probed with met elongator D loop probe, complementary to 2–27 nt position). Cellular abundances (relative ratios) of the initiator tRNAs in the strains were calculated relative to total tRNAfMet in wild-type strain (lane 1), after normalization with the corresponding tRNAMet.
Even though the tRNAfMet mutants having substitutions in the 3GC base pairs are not expected to be defective in their aminoacylation/formylation, the mutants that did not sustain metZWV locus deletion in the ‘four-gene knockout’ assay (midAU, midCG, 3AU, cg/gu/cg, cg/gu/GC, 3GC) were, nonetheless, checked for their aminoacylation/formylation status (Supplementary Figure S2). Expectedly, the mid AU, mid CG, 3AU, cg/gu/cg, cg/gu/GC and 3GC tRNA mutants were aminoacylated/formylated as well as the mutants that sustained E. coli for its growth. Thus, the failure of the mid AU, mid CG, 3AU, cg/gu/cg, cg/gu/GC and 3GC mutants in sustaining E. coli growth related to their deficiency in initiation. We may add that, because of its very low level accumulation in cell, the status of the au/gu/gu mutant could not be discerned. However, as an additional validation, we tested the initiation activities of the tRNA mutants using our CATam1 reporter based in vivo initiation assays (20,21). As shown in Supplementary Figure S3, by and large, the mutants that failed to sustain E. coli for its growth also showed lower initiation activity. The au/gu/gu mutant showed no detectable activity.
In yet another approach, we checked for the functionality of the mutant tRNAs, by the rescue of cold sensitivity of KL16 ΔmetZWV, a strain highly compromised for its growth at 22°C due to the deficiency of functional tRNAfMet (22). The plasmids encoding tRNAfMet mutants were introduced into ΔmetZWV strain and the transformants were checked for growth at 22°C by plate assay (Supplementary Figure S4A) or in broth (Supplementary Figure S4B). Most of the mutants that functioned in the four-gene knockout assay (Figure 2) functioned in this assay. However, the cg/GC/cg mutant (functional in the four-gene knockout assay) did not rescue the cold sensitive phenotype of the host, suggesting that it functioned poorly. The mutants lacking G30 were found to be toxic even at 37°C (Supplementary Figure S4A, sectors 4, 7, 13, 20, 21). The reasons for toxicity by these mutants in the ΔmetZWV strain have not been investigated any further in this study.
Inability of 3AU tRNAfMet to sustain E. coli growth suggests that mere presence of the three consecutive R:Y pairs is not sufficient for its function in initiation. Interestingly, the observations also showed that the middle GC pair could be changed to GU wobble pair. That the mid GU but not the mid AU is functional indicated that G30 is the most crucial base in the 3GC pairs for tRNAfMet function in E. coli. Interestingly, incorporation of G30 into the 3AU mutant (as in au/gu/au) sustained the cell growth suggesting that, at least in this context, the presence of single G, the G30 is sufficient. However, the mutants such as au/gu/gu, cg/gu/GC and cg/gu/cg (retaining G30) were found to be nonfunctional. The native gel analysis of au/gu/gu mutant showed not only its poor accumulation in the cell but also a significant decrease in its mobility compared to other mutants, implying severe conformational instability/change in this mutant (Figure 1B, lane 15). And, in the cases of cg/gu/GC or cg/gu/cg mutants, lack of purine/purine stacking between positions 29 and 30 might have affected accessibility of the wobble paired G30 for its recognition. Such a notion is reinforced by the observation that the cg/GC/cg mutant where the position of G30 would be fixed due to its Watson-Crick pairing with C40 supported E. coli growth.
Growth analysis of strains sustained on mutant initiator tRNAs
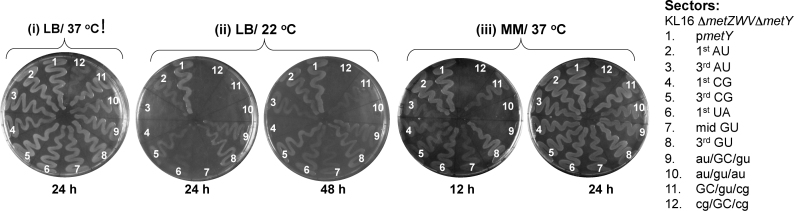
The four-gene knockout, and the rescue of the cold sensitivity assays revealed that among the 3GC pairs, only the middle GC; and in fact, primarily the G30 (in an appropriate context) is the most crucial element for the tRNAfMet function. Why should then the organisms retain the 3GC pairs? At 37°C, except for the cg/GC/cg mutant harboring strain (Figure 3i, sector 12), all others (sectors 1–11) showed similar growth on LB agar or broth (Supplementary Figure S5A). However, at 22°C not only the cg/GC/cg but also the 3rd AU, 1st CG, 1st UA, mid GU, au/gu/au, GC/gu/cg mutants showed poor to very poor growth (Figure 3ii, sectors 3, 4, 6, 7, 10-12). The mid GU, au/GC/gu, au/gu/au and cg/GC/cg showed growth deficiencies in minimal medium even at 37°C (Figure 3iii, sectors 7, 9, 10 and 12). Phenotypes of the various mutant tRNAfMet supported strains under different growth conditions are summarized in Supplementary Table S4. These observations suggest that while the middle GC or the G30 (in appropriate contexts) are adequate to sustain growth of E. coli, the presence of the full complement of 3GC confers fitness advantage to sustain growth under varied conditions.
Figure 3.
Growth of the strains sustained on the tRNAfMet mutants. Growth analysis of strains sustained on the wild-type or mutant tRNAfMet on LB agar at 37°C (i), at 22°C (ii) and on minimal media agar (MM) at 37°C (iii). The saturated cultures were streaked on plates containing ampicillin (Amp) and incubated at the indicated temperatures for the indicated periods.
Impact of increased cellular levels of the tRNAfMet mutants on growth
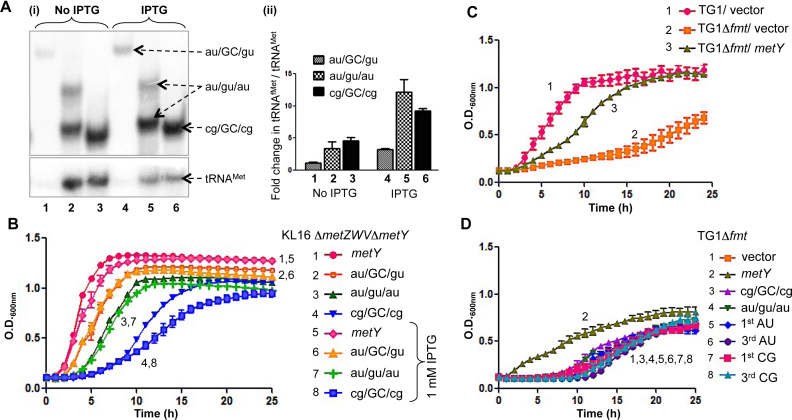
The two important features of tRNAfMet, the formylation of the amino acid attached to it and the 3GC pairs in its anticodon stem, have been generally ascribed as crucial in recruiting tRNAfMet to the P-site of 30S IC. To further investigate the role of the 3GC pairs, cellular levels of the three different mutants (au/GC/gu, au/gu/au and cg/GC/cg) which sustained E. coli for growth with increasing degrees of defect, were overexpressed by IPTG induction (Figure 4Ai and ii). However, there were no improvements in the growth phenotypes of the strains (Figure 4B, compare curves 2 with 6, 3 with 7 and 4 with 8). These observations imply that the rate limiting step in poor activity of the 3GC mutants is unlikely to be their initial binding to 30S ribosomes, and are consistent with our earlier report that tRNAfMet lacking the 3GC pairs is primarily defective in transiting to the 70S stage (7). In contrast, when we used Fmt deficient strain (Δfmt), which fails to formylate tRNAfMet, overexpression of wild-type initiator tRNA compensated for the lack of its formylation (Figure 4C, compare curve 2 with 3) suggesting that the formylation of tRNAfMet indeed helps in its initial binding to 30S ribosome (23,24). Formylation is known to increase the affinity of tRNAfMet to IF2, which in turn assists its initial binding to 30S. Not surprisingly, the 3GC mutants that are defective at a downstream step fail to rescue the growth of even the Fmt deficient strain (Figure 4D, compare curves 1 with 3–8).
Figure 4.
Effect of over-expression of wild-type or mutant tRNAfMet deficient in the conserved features of 3GC pairs or formylation. (A) Analysis of expression of tRNAfMet mutants. Total tRNAs from the strains supported by au/GC/gu, au/gu/au or cg/GC/cg tRNAfMet mutants, grown in LB or LB with 1 mM IPTG were separated on native PAGE, transferred onto nytran membrane and probed with tRNAfMet and tRNAMet specific probes (i). The blots were quantified using BioImage Analyzer (FLA5000; Fuji). The pixel values of tRNAfMet were normalized to that of corresponding tRNAMet and the fold differences with respect to au/GC/gu in LB (lane 1) were plotted (ii). (B) Effect of induction of expression of tRNAfMet mutants (au/GC/gu, au/gu/au or cg/GC/cg) by 1 mM IPTG, on the growth of Escherichia coli strains. (C) Overexpression of wild-type tRNAfMet in E. coli TG1Δfmt strain. TG1Δfmt strain was either transformed with vector control or a plasmid borne metY gene (metY) and monitored for growth. (D) Overexpression of 3GC mutant tRNAfMet in TG1Δfmt strain. TG1Δfmt strain was transformed with the plasmid (vector) or the plasmids containing metY or various mutants of metY and monitored for growth. For growth analysis, the strains were grown to saturation; diluted 1000-fold with LB containing Amp and the growth was monitored using Bioscreen C growth reader.
In vivo binding of mutant initiator tRNAs to the ribosomes
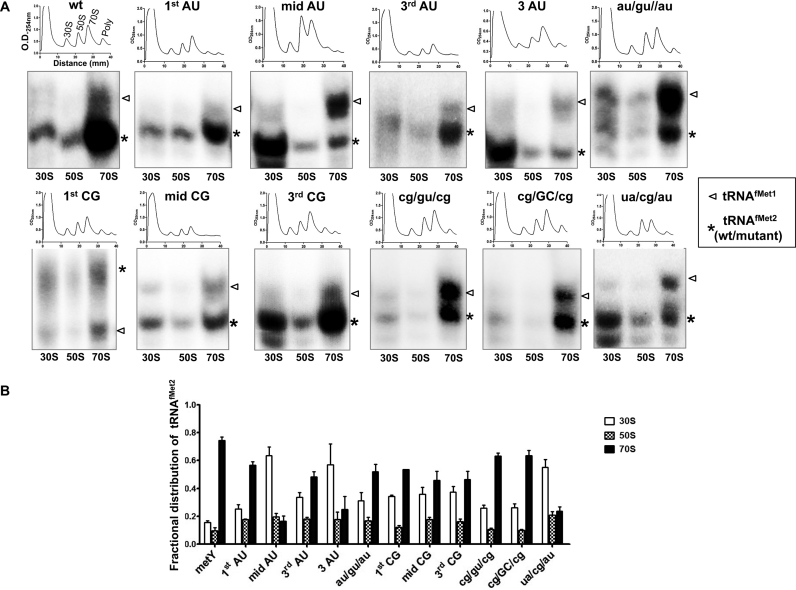
We have earlier shown that changing the 3GC pairs to those found in the elongator tRNAMet (ua/cg/au) led to its severely decreased efficiency to enter the 70S complex (7). Whether tRNAfMet mutants with middle GC alone would be sufficient in this function or if the flanking GC pairs also make a contribution was not investigated. Hence, we prepared polysomes from the ΔmetY strains harboring various plasmid borne tRNAfMet mutants and checked for their abundance in the 30S, 50S and 70S fractions (Figure 5A). The wild-type tRNAfMet2 as well as the mutants that functioned in E. coli, transited into 70S ribosomes. The mutants such as mid AU, 3AU and ua/cg/au, which failed to function in E. coli were highly defective in entering 70S (Figure 5A and B). As these experiments were done in the strains with intact metZWV locus, analysis of the transition of tRNAfMet1 served as an internal positive control (Figure 5A, shown by arrow head). Surprisingly, the mutants which could not sustain E. coli such as cg/gu/cg and mid CG, also accumulated in the 70S population. In cg/gu/cg, the most crucial G30; and in mid CG (GC/cg/GC), the minor groove determinants are similar to mid GC and the two flanking GC pairs are still present suggesting that either of these elements facilitated transition of tRNAfMet into the 70S. However, there must be some additional stage(s) in the initiation pathway that eventually convert this complex into an elongation competent 70S, for which the presence of G30 in the correct context or the mid GC is essential, as these mutants are not able to sustain E. coli. Importantly, this observation highlights that the role of the 3GC pairs is not limited to only transiting tRNAfMet into the 70S but also at a step that converts the 70S complex into an elongation competent complex.
Figure 5.
In vivo binding of tRNAfMet mutants to the ribosomes. (A) Total ribosomal cell-free extracts from KL16ΔmetY containing plasmid borne metY genes (wild-type, wt; or mutants) were separated on 15–35% sucrose gradient and their profiles were analyzed at 254 nm. The tRNAs from 30S, 50S and 70S fractions were separated on native PAGE and analyzed by northern blotting using a probe specific to tRNAfMet. The metZWV encoded tRNAfMet1 was used as internal control (shown by arrowhead). All tRNAfMet mutants were derived from tRNAfMet2 and indicated by asterisk. (B) Quantification of fractional distribution of tRNAfMet2 (wt or mutants) within ribosomal subunits and 70S. Individual tRNA intensities were divided with the total tRNAfMet intensity in the corresponding profile to obtain the fractional distribution.
Cross talk between the 3GC pairs and IF3
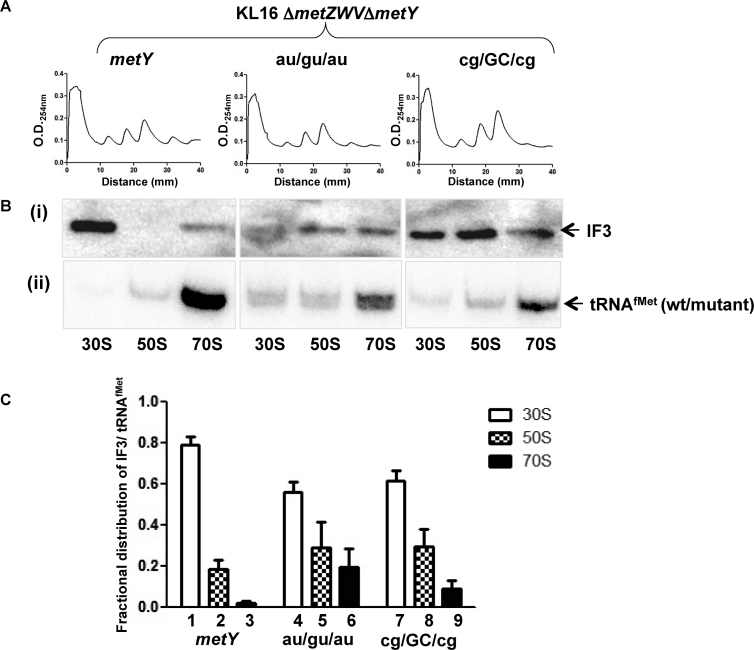
To further study the importance of the 3GC pairs, we analyzed polysome profiles of the strains sustained on the wild-type (metY) or the tRNAfMet mutants such as au/gu/au and cg/GC/cg in a ΔmetZWVΔmetY strain (deleted for all four of its chromosomal tRNAfMet genes). In repeated experiments, compared to the polysomes in the strain sustained on plasmid borne metY, we observed less abundance of polysomes in the strain sustained on au/gu/au and their even lesser abundance in the strain sustained on the cg/GC/cg mutants (Figure 6A and Supplementary Figure S6). This observation suggests that although these mutants form the 70S initiation complexes, they are slow in entering the elongation phase highlighting the importance of the 3GC pairs for efficient initiation.
Figure 6.
Polysome profiles and abundance of IF3/tRNAfMet in the ribosomes. (A) Polysome profiles from E. coli sustained on wild-type, au/gu/au or cg/GC/cg tRNAfMet. Ribosomal cell-free extracts were layered over 15–35% sucrose gradient and the profiles were monitored by absorbance at 254 nm. (B) Immunoblot (i) and northern (ii) analyses to determine abundance of IF3 (i), and tRNAfMet (ii) in 30S, 50S and 70S ribosomal fractions. (C) Quantification of fractional distribution of IF3 with respect to tRNAfMet levels in free subunits and 70S fractions.
Further, even though the mechanism remains unclear, IF3 is involved in the selection of tRNAfMet via the 3GC pairs. To investigate the connection between the two, we analyzed the abundance of IF3 as well as the tRNAfMet (wild-type, or the mutants (au/gu/au and cg/GC/cg)) in the free subunits and 70S complexes. There was a slightly increased abundance of IF3 in the 70S peaks for the two mutants (Figure 6Bi, ii and 6C). In contrast, the abundance of tRNAfMet mutants (au/gu/au and cg/GC/cg) themselves showed a significant reduction in the 70S peak (Figure 6Bii). Considering that the 70S population would also contain 70S IC, the level of IF3 relative to the initiating tRNAfMet in the 70S fraction is more for the tRNAfMet mutants lacking the 3GC pairs. It may also be said that the significant presence of IF3 as well as the tRNAfMet mutants themselves in the 50S subunit is most likely a consequence of their release from 70S complexes during the course of sucrose density gradient centrifugation.
We then analyzed the status of IF3 in the presence of other 3GC mutants having varied defects in their transition into 70S IC. Although both the mid AU and mid CG mutants did not sustain the cell (hence the use of strain with intact metZWV), between the two, the later entered 70S IC more efficiently. Interestingly, the strain with mid CG had more IF3 bound in 70S than the one with mid AU (Supplementary Figure S7). Similar observation was made with the cg/GC/cg and cg/gu/cg mutants. While both of these mutants transited to 70S IC, only the cg/GC/cg sustained the cell. The cg/gu/cg was defective in the efficient release of IF3 from the 70S IC (Supplementary Figure S7). These observations suggest that the 3GC pairs may be needed for the formation of functional elongation competent 70S, for example by IF3 release. Thus, while IF3 is important in preferential selection of tRNAfMet onto the 30S, ironically the 3GC base pairs may drive, at least in the final stages of initiation, eviction of IF3 from the 70S IC to allow it to convert into an elongation competent complex.
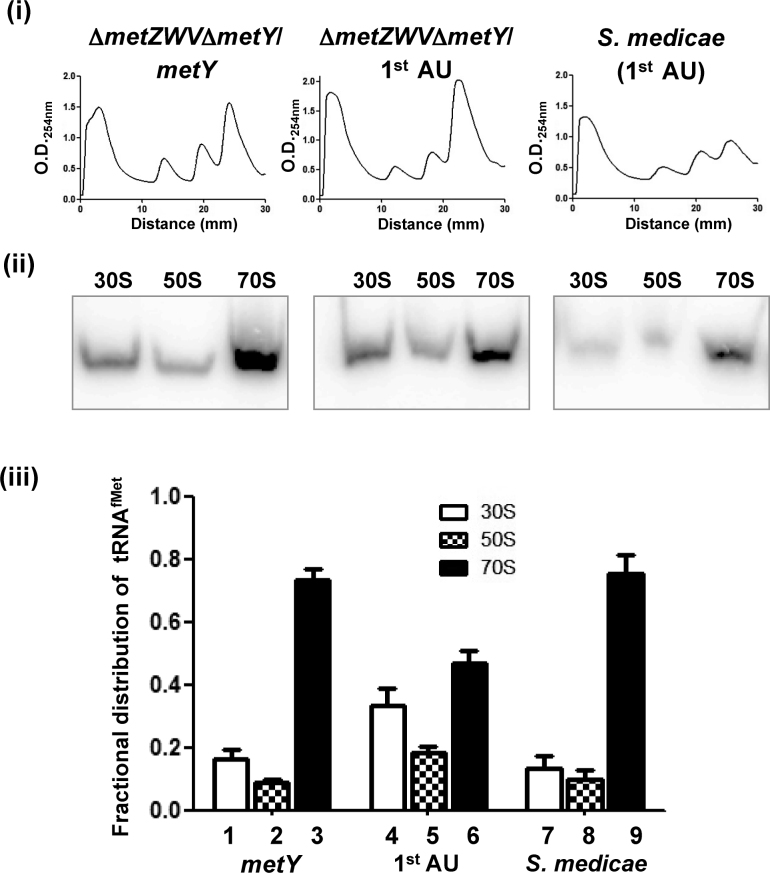
Mutant initiator tRNA abundance in rhizobial ribosomes
The 1st AU is found naturally in all α-proteobacteria and few species of mycoplasmas. In vivo experiments using E. coli show that this variant does have a defect in initiation (Figure 3 and Supplementary Figure S3) as well as a slight defect in transiting to the 70S complex (Figure 5). This poses a question of how α-proteobacteria could do well with a tRNAfMet variant less efficient in translation? To investigate this, we isolated ribosomes from S. medicae WSM419 and checked for the presence of the 1st AU variant tRNAfMet in the ribosomal populations (Figure 7i and ii). E. coli strains sustained on either the 1st AU or the wild-type (metY) tRNAfMet were used as controls. The occurrence of the 1st AU tRNAfMet was higher in the 70S ribosomes from S. medicae compared to those in E. coli (Figure 7iii). In fact, the occurrence of the 1st AU tRNAfMet in rhizobial 70S ribosomes was as good as that of the wild-type tRNAfMet in E. coli 70S suggesting that the rhizobial ribosomes co-evolved to efficiently utilize the 1st AU variant.
Figure 7.
Co-evolution of ribosomes to utilize natural variants of initiator tRNA. Binding of 1st AU mutant tRNAfMet in E. coli and S. medicae WSM419 ribosomes. (i) Total ribosomal cell-free extracts of E. coli KL16ΔmetZWVΔmetY sustained on wild-type metY or the 1st AU metY or from S. medicae were separated on 15–35% sucrose gradient and analyzed by O.D.254. (ii) The tRNAs from the individual peaks were analyzed by northern blotting using tRNAfMet probe (the same probe specific to E. coli was used as it differed only by single nucleotide with S. medicae initiator tRNA). (iii) The fractional distribution of tRNAfMet across the profile.
DISCUSSION
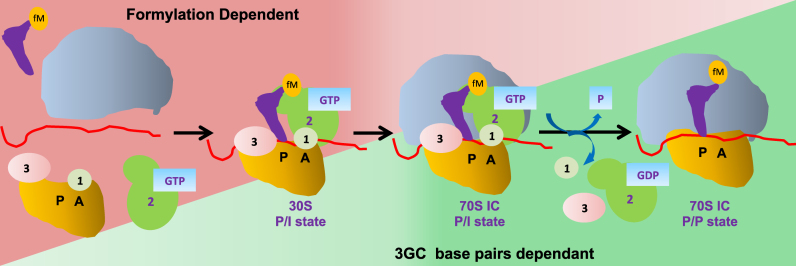
One of the highly conserved features of the initiator tRNAs, across the three domains of life, is the presence of 3GC pairs in their anticodon stems. The second feature of formylation is also highly conserved but specific to eubacterial tRNAfMet. Our systematic mutational analysis of the 3GC pairs in tRNAfMet in E. coli suggests that within these pairs, the G30 of the middle GC is extremely important for the tRNAfMet function in initiation. However, the flanking GC pairs contribute to the function of G30 and their presence confers growth advantage to the host (Figure 3 and Supplementary Table S4). Further, our study unravels the major roles of the two highly conserved features of the eubacterial tRNAfMet. Formylation is most crucially required for the initial recruitment of tRNAfMet to the 30S ribosome, whereas the 3GC pairs play their essential role at the later stages in initiation in transiting tRNAfMet from the 30S pre-IC to the 70S complex (Figure 8). Our results do not imply that the 3GC pairs make no contributions in the early stages or that formylation in the later stages of initiation; they only highlight their most crucial roles.
Figure 8.
Steps involved in the translation initiation in eubacteria. The tRNAfMet selection in P-site is scrutinized at two checkpoints (initial binding to 30S and its transition into 70S). The initial recruitment of tRNAfMet to the ribosomal P-site is facilitated by formylation. The tRNAfMet transition from the 30S initiation complex (IC) to 70S IC is facilitated by the 3GC pairs in the anticodon stem. In the absence of 3GC pairs, the transition into 70S complex not favored. Thus, contribution of formylation (shown as a gradient in Red) is crucial mainly for the formation of 30S IC and lesser at the later stages, while the role of 3GC base pairs (shown as gradient in green) is more critical for transition of the tRNA from 30S IC to the 70S and not so much at the initial stages of 30S IC assembly.
The observation that a defect in formylation of tRNAfMet can be overcome by the increased amount of tRNAfMet supports that the primary role of formylation is to enhance tRNAfMet targeting to the ribosomal P-site at an early stage of tRNAfMet binding to the ribosome, a stage where such an approach (of mass action) would be most efficient in bypassing a defect. In a similar experiment, overexpression of the tRNAfMet lacking the 3GC pairs does not rescue the growth defect of the strains sustained on the 3GC mutants. Taken together with the observation that such mutants accumulate in the 30S ribosome population, we suggest that initial binding of the 3GC mutant tRNAfMet to the 30S subunit is unlikely to be rate limiting in compromising their activity in initiation. Instead, a major role of the 3GC base pairs is at a stage subsequent to their binding to the 30S ribosome. Also, overexpression of tRNAfMet mutated in the 3GC pairs (even those containing two GC pairs) did not rescue the growth defect of a strain deficient for Fmt. This observation further supports the importance of the 3GC pairs in tRNAfMet function at a step subsequent to its initial binding. Such a division of labor of the two distinct features of the tRNAfMet appears well suited to allow the initiator but not the elongator tRNAs to pass through the two critical checkpoints during the process of initiation, i. e. facilitation of initial binding by formylation and transition of the bound initiator tRNA to the elongation competent ribosomes by the 3GC pairs. This finding of ours also provides a rationale for why the elongator tRNAs, even those with the two GC pairs, fail to function as initiator tRNAs.
In previous studies it was reported that the highly conserved residues G1338/A1339 of 16S rRNA interact with the minor groove of initiator tRNA in the 3GC region (13,14). In addition, our studies reveal that among the 3GC pairs, the middle GC and particularly the G30 of this pair, is essential for tRNAfMet function as changing it to GU but not to AU (mid AU) allowed it to function. The observation that the au/gu/au tRNAfMet but not the 3AU (au/au/au) tRNAfMet sustained the cell also shows that the presence of G30 is not only necessary for tRNAfMet to function but it may also contribute to direct interactions with 16S rRNA. Interestingly, growth of yeast strains could be sustained with the mutant initiator tRNAs such as mid AU, mid CG lacking the middle GC pair indicating that it is not crucial in this organism (12). However, whether the flanking GC base pairs compensate for the deficiency of the middle GC pair has not been explored in yeast. As noted in our study, the flanking GC pairs do contribute to tRNAfMet function to enhance E. coli growth. Although it may also be noted that initiation in eukaryotes is assisted by about a dozen initiation factors (as opposed to 3 in eubacteria), making the requirement of 3GC pairs somewhat different in yeast (25–27).
Changing the middle GC pair to AU, rendered tRNAfMet mutant highly inefficient in entering the 70S complex. However, changing the same pair to CG did not severely affect its ability to enter the 70S complex. This may be due to the fact that the minor groove determinants are indistinguishable in case of CG and GC pairs (28), explaining their efficiency in transiting to 70S complex (also see below). Further, the flanking GC pairs (the 1st and the 3rd) do contribute at this step as the abundance of 3GC mutant tRNA (ua/cg/au) in the 70S reduced drastically. It should also be noted that although the mid CG mutant is able to enter the 70S stage efficiently, this tRNA does not sustain E. coli for its growth indicating that there might be additional roles the 3GC pairs play subsequent to their role in transiting the tRNA to the 70S IC. Likewise, while the cg/GC/cg and cg/gu/cg mutants were present to a similar abundance in 70S, only the former of the mutants sustained E. coli for its growth. This observation also supports for an additional role of the 3GC pairs in converting the initial 70S complex to an elongation competent complex (Figure 8). As shown in Figure 6, one such role could be to evict the IF3 from the 70S IC to allow it to convert into an elongation competent complex.
Earlier in vitro studies have shown an IF3 dependent rejection of 3GC mutant tRNAfMet in 30S subunit but these studies were carried out with either anticodon stem loop alone (29) or elongator tRNA which lack formylation (30). In one of the studies, IF3 was shown to reject both the initiator as well as the elongator tRNAs from 30S to a similar extent (31). The presences of IF3 in 70S ribosomes suggest that the release of IF3 may occur after the formation of the 70S IC, in agreement with the single molecule studies (4,32,33). Recently, helix 69 of 50S subunit has been shown to be important for the eviction of IF3 from 70S IC, thus supporting the release of IF3 from the 70S stage (34). Interestingly, in the strains sustained on the au/gu/au or cg/GC/cg mutants, retention of the IF3 was more in the 70S complex compared to the strain sustained on the wild-type tRNAfMet. Thus, it may well be that release of IF3 requires conformational changes in 70S IC, which depend on the presence of the 3GC pairs. Plausibility that the 3GC pairs change the conformation of the G1338/A1339 loop in 16S rRNA, which in turn facilitates the release of IF3 from the 70S cannot be ruled out.
The decreases in polysome population (as assessed from the disome peaks) in the cg/GC/cg mutant sustained cells suggest that there may be a defect in the entry of the initiation complex into the elongation phase even though the mutant was not significantly defective in entering the 70S stage. This observation further highlights the importance of the flanking GC pairs in tRNAfMet for its role at a step in initiation that makes elongation competent 70S (from 70S IC). Whether such a role of the 3GC pairs is in moving tRNAfMet from its P/I state to P/P state (4) or in disassembling the Shine-Dalgarno (S.D.) and an S.D. contact (35) necessary for the step of translocation subsequent to the first peptide bond formation or otherwise, is not known.
From our in vivo studies, it is clear that the 3GC pairs play an important role in transiting tRNAfMet from 30S pre-IC to the elongation competent 70S stage. Evolutionarily, it seems that the 3GC pairs were acquired to enhance discrimination between the initiator and elongator tRNAs for an appropriate recognition of start codon to avoid random events of translation. In case of mycoplasma and rhizobia, there may be other mechanisms that compensate for the lack of the 3GC pairs, allowing them to retain the variant initiator tRNAs. Efficient transition of variant tRNAfMet (1st AU) into 70S complex by rhizobial ribosomes suggests that they might have co-evolved to utilize variant tRNAfMet efficiently. As already noted, initiation in yeasts can dispense with even the crucial requirement of the middle GC pair. Exploration of these mechanisms would be of interest to understand the diversity in biological systems.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our laboratory colleagues for their suggestions on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology (DBT), Ministry of Science and Technology; Department of Science and Technology (DST), Ministry of Science and Technology; DST J.C. Bose Fellowship (to U.V.); Shyama Prasad Mukherjee SRF of the Council of Scientific and Industrial Research, New Delhi (to S. Sh.); University Grants Commission (UGC) of India, New Delhi DS Kothari Fellowship (to S. Sa.). DST Fund for Improvement of Science and Technology Infrastructure (FIST) level II infrastructure and UGC Centre of Excellence supports are acknowledged. Funding for open access charge: Government of India through DBT and DST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gualerzi C.O., Pon C.L.. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990; 29:5881–5889. [DOI] [PubMed] [Google Scholar]
- 2.Seong B.L., RajBhandary U.L.. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:8859–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RajBhandary U.L. Initiator transfer RNAs. J. Bacteriol. 1994; 176:547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall R.A., Aitken C.E., Puglisi J.D.. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol. Cell. 2009; 35:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomsic J., Vitali L.A., Daviter T., Savelsbergh A., Spurio R., Striebeck P., Wintermeyer W., Rodnina M.V., Gualerzi C.O.. Late events of translation initiation in bacteria: a kinetic analysis. EMBO J. 2000; 19:2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julian P., Milon P., Agirrezabala X., Lasso G., Gil D., Rodnina M.V., Valle M.. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011; 9:e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shetty S., Nadimpalli H., Shah R.A., Arora S., Das G., Varshney U.. An extended Shine-Dalgarno sequence in mRNA functionally bypasses a vital defect in initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E4224–E4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varshney U., Lee C.P., RajBhandary U.L.. From elongator tRNA to initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R.. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989; 17(Suppl.): r1–r172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seong B.L., RajBhandary U.L.. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samhita L., Shetty S., Varshney U.. Unconventional initiator tRNAs sustain Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:13058–13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J., Munoz A., Kolitz S.E., Saini A.K., Chiu W.L., Rahman H., Lorsch J.R., Hinnebusch A.G.. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev. 2014; 28:502–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selmer M., Dunham C.M., Murphy F.V.T., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 14.Lancaster L., Noller H.F.. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005; 20:623–632. [DOI] [PubMed] [Google Scholar]
- 15.Varshney U., Lee C.P., RajBhandary U.L.. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991; 266:24712–24718. [PubMed] [Google Scholar]
- 16.Sarin P.S., Zamecnik P.C.. On the stability of aminoacyl-S-Rna to nucleophilic catalysis. Biochim. Biophys. Acta. 1964; 91:653–655. [DOI] [PubMed] [Google Scholar]
- 17.Schofield P., Zamecnik P.C.. Cupric ion catalysis in hydrolysis of aminoacyl-tRNA. Biochim. Biophys. Acta. 1968; 155:410–416. [DOI] [PubMed] [Google Scholar]
- 18.Mawn M.V., Fournier M.J., Tirrell D.A., Mason T.L.. Depletion of free 30S ribosomal subunits in Escherichia coli by expression of RNA containing Shine-Dalgarno-like sequences. J. Bacteriol. 2002; 184:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandal N., RajBhandary U.L.. Escherichia coli B lacks one of the two initiator tRNA species present in E. coli K-12. J. Bacteriol. 1992; 174:7827–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varshney U., Lee C.P., Seong B.L., RajBhandary U.L.. Mutants of initiator tRNA that function both as initiators and elongators. J. Biol. Chem. 1991; 266:18018–18024. [PubMed] [Google Scholar]
- 21.Varshney U., RajBhandary U.L.. Initiation of protein synthesis from a termination codon. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapoor S., Das G., Varshney U.. Crucial contribution of the multiple copies of the initiator tRNA genes in the fidelity of tRNA(fMet) selection on the ribosomal P-site in Escherichia coli. Nucleic Acids Res. 2011; 39:202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson A.I., Zorzet A., Kanth A., Dahlstrom S., Berg O.G., Andersson D.I.. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6976–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillon J.M., Heiss S., Soutourina J., Mechulam Y., Laalami S., Grunberg-Manago M., Blanquet S.. Interplay of methionine tRNAs with translation elongation factor Tu and translation initiation factor 2 in Escherichia coli. J. Biol. Chem. 1996; 271:22321–22325. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch A.G., Lorsch J.R.. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 2012; 4:a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drabkin H.J., Estrella M., Rajbhandary U.L.. Initiator-elongator discrimination in vertebrate tRNAs for protein synthesis. Mol. Cell. Biol. 1998; 18:1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopka M.L., Goodsell D.S., Han G.W., Chiu T.K., Lown J.W., Dickerson R.E.. Defining GC-specificity in the minor groove: side-by-side binding of the di-imidazole lexitropsin to C-A-T-G-G-C-C-A-T-G. Structure. 1997; 5:1033–1046. [DOI] [PubMed] [Google Scholar]
- 29.Hartz D., McPheeters D.S., Gold L.. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989; 3:1899–1912. [DOI] [PubMed] [Google Scholar]
- 30.Antoun A., Pavlov M.Y., Lovmar M., Ehrenberg M.. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell. 2006; 23:183–193. [DOI] [PubMed] [Google Scholar]
- 31.Antoun A., Pavlov M.Y., Lovmar M., Ehrenberg M.. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006; 25:2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milon P., Konevega A.L., Gualerzi C.O., Rodnina M.V.. Kinetic checkpoint at a late step in translation initiation. Mol. Cell. 2008; 30:712–720. [DOI] [PubMed] [Google Scholar]
- 33.Mangroo D., RajBhandary U.L.. Mutants of Escherichia coli initiator tRNA defective in initiation. Effects of overproduction of methionyl-tRNA transformylase and the initiation factors IF2 and IF3. J. Biol. Chem. 1995; 270:12203–12209. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q., Fredrick K.. Roles of helix H69 of 23S rRNA in translation initiation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:11559–11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uemura S., Dorywalska M., Lee T.H., Kim H.D., Puglisi J.D., Chu S.. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature. 2007; 446:454–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.