Abstract
Over recent decades, substantial efforts have been made to understand the interactions between host genomes and transposable elements (TEs). The impact of TEs on the regulation of host genes is well known, with TEs acting as platforms of regulatory sequences. Nevertheless, due to their repetitive nature it is considerably hard to integrate TE analysis into genome-wide studies. Here, we developed a specific tool for the analysis of TE expression: TEtools. This tool takes into account the TE sequence diversity of the genome, it can be applied to unannotated or unassembled genomes and is freely available under the GPL3 (https://github.com/l-modolo/TEtools). TEtools performs the mapping of RNA-seq data obtained from classical mRNAs or small RNAs onto a list of TE sequences and performs differential expression analyses with statistical relevance. Using this tool, we analyzed TE expression from five Drosophila wild-type strains. Our data show for the first time that the activity of TEs is strictly linked to the activity of the genes implicated in the piwi-interacting RNA biogenesis and therefore fits an arms race scenario between TE sequences and host control genes.
INTRODUCTION
Transposable elements (TEs) are mobile sequences that can be highly abundant in genomes (1). First described by B. McClintock in the 1950s (2), TEs have a high impact on genome dynamics, and are undoubtedly major players in genome evolution (1,3). Despite the increasing amount of transcriptomic data being produced for many species, very few studies have performed genome-wide analyses of the transcription levels of TEs (4–7). Such knowledge gap is partly due to the low levels of transcription of TEs in normal conditions, but also to the fact that one given TE family may be represented by several sequences, making more difficult to have an accurate idea of TE transcription levels.
In Drosophila, a category of small RNAs called piwi-interacting RNAs (piRNAs) are involved in the control of TEs in germline and somatic cells (8–11) and participate in transcriptional and post-transcriptional control of TEs (12). The disruption of the piRNA biogenesis pathway leads to TE mobilization (transcription and transposition), DNA breaks and sterility (13). Understanding the way TE activity is regulated thus requires to have an accurate knowledge of piRNA abundances which could then be associated with TE mRNA levels. Currently, no available method is dedicated to both the analysis of TE expression and piRNA production, associated with differential expression analysis with statistical relevance, for both model and non-model species with non-annotated genomes.
Presently, one tool is available to analyze piRNAs that is based on the approach proposed by Brennecke (10,14). This tool is suited for the analysis of well annotated genomes. However, the methodology that is applied may lead to a loss of information. The first step consisting in a strict mapping at a unique position on the reference genome makes two strong assumptions. Firstly, retaining only reads mapping with no mismatch implies that the corresponding small RNA displays a perfect match with the regulated TE sequences. Secondly, retaining only reads mapping at unique positions when they are supposed to target repeated sequences assumes that only particular small RNA can be generated by only one given position. Other major problems are that this step completely relies on the quality of the genome sequence and assembly, and that it cannot be directly applied when a TE family is absent from the reference genome but exists in the genomes of other strains. Moreover, the association between piRNAs and the TE family is made by comparing the reads to TE consensus sequences and allowing up to three mismatches, which corresponds to a divergence of approximately 10%. The consensus sequence in itself represents an average sequence of a given family and may result in a sequence that is not present in the genome. A consensus will be representative of the family only if the copies used to build it are very similar, which is the case for the majority of the Drosophila melanogaster families, but it is not the case in other Drosophila genomes, such as the sister species Drosophila simulans (15). The same is true when determining TE expression from mRNA reads.
In this article we propose a different approach implemented in the pipeline TETOOLS which is dedicated to the analysis of the TE transcriptome, and takes into account the sequence diversity at the TE copy level, using a complete list of all available TE copies from an organism. This pipeline provides quantitative information for both small and messenger RNAs, performing differential expression analyses among different samples using the DESeq2 program (16). It can be used for non-model organisms with no annotated reference genome but for which a list of TE copies is available. When this list is not available, TETOOLS can be jointly used with a dedicated tool for TE identification from raw reads, such as DnaPipeTE (17), RepeatExplorer (18) or other TE identification tools if the genome is assembled (see as a review (19)). The pipeline is user friendly and is available for use in Galaxy (20).
We applied TEtools to explore TE regulation in D. simulans wild-type strains. In this species, TE sequences belonging to the same family are very diverse and the activity of TEs depends on the strain studied (21–24). Several hypotheses have been proposed to understand the origin and evolution of the intra-specific variability of TEs (25–28), but none has integrated in a satisfying way the high variability uncovered in genes involved in the piRNA pathway (GIPPs) (both at the DNA sequence (29,30) and transcription levels (31)). Indeed, we propose that the natural variation of TEs is due to variability in the piRNA pathway, which evolves very rapidly and constitutes a genomic immune pathway (29–31). We sequenced mRNAs and small RNAs in several wild-type strains of D. simulans and used TEtools to analyze TE expression levels and the production of corresponding piRNAs. Our results show, for the first time, a negative relationship between TE and GIPP activities and provide insights into the dynamics of TEs in their natural context.
MATERIALS AND METHODS
Biological material
Four wild-type strains of D. simulans were used; these strains originated from various regions around the world: Chicharo (Portugal), Makindu (Kenya), Mayotte (Indian Ocean island) and Zimbabwe. We also included the main source of the reference genome sequence (w501). This last strain originated from the USA and was obtained from the UC San Diego Drosophila Stock Center. Flies were kept in the lab at 24°C in regular fruit fly medium.
Thirty pairs of ovaries were dissected in phosphate buffered saline. Total RNA was extracted using the RNeasy kit (Qiagen) followed by RNase treatment (DNA free kit, Ambion). Two replicates were performed for each strain and the overall qualities were assessed using the Bioanalyzer 2100 (Agilent).
Illumina library production and mRNA sequencing
The TruSeq RNA sample Preparation v2 kit (Illumina Inc., California, USA) was used according to the manufacturer's protocol with the following modifications. Poly-A-containing mRNA molecules were purified from 1 μg of total RNA using poly-T oligo-attached magnetic beads. The purified mRNA was fragmented by the addition of the fragmentation buffer and heated to 94°C in a thermocycler for 4 min. A fragmentation time of 4 min was used to yield library fragments of 250–500 bp. First-strand cDNA was synthesized using random primers to eliminate the general bias towards the 3΄ end of the transcript. Second-strand cDNA synthesis, end repair, A-tailing and adapter ligation were performed in accordance with the manufacturer's supplied protocols. Purified cDNA templates were enriched by 15 cycles of polymerase chain reaction (PCR) for 10 s at 98°C, 30 s at 65°C and 30 s at 72°C using the PE1.0 and PE2.0 primers and the Phusion DNA polymerase (NEB, USA). Each indexed cDNA library was verified and quantified using a DNA 100 Chip on a Bioanalyzer 2100 and then mixed equally with six different samples. The final library was quantified by real-time PCR with the KAPA Library Quantification Kit for Illumina Sequencing Platforms (Kapa Biosystems Ltd, South Africa), adjusted to 10 nM in water and provided to the Get-PlaGe core facility (GenoToul platform, INRA Toulouse, France http://www.genotoul.fr) for sequencing. The final mixed cDNA library was sequenced using the Illumina mRNA-Seq paired-end protocol on a HiSeq2000 sequencer for 2 × 100 cycles. Each sample provided between 30 and 55 million reads (SRX1287831, SRX1287832, SRX1287833, SRX1287834 and SRX1287843).
Small RNA extraction and sequencing
Small RNAs from D. simulans ovaries were manually isolated in HiTrap Q HP anion exchange columns (GE Healthcare) as described in Grentzinger and Chambeyron (32). Library construction and 50 nt read sequencing were performed by Fasteris SA (Switzerland) on an Illumina HiSeq 2500 instrument. Libraries from the Makindu and Chicharo strains were previously published (33). The small RNA library of the Mayotte strain is available under the accession number SRX1287860. The poly-A tails attached to the sequence before sequencing to obtain 50nt RNA were removed using UrQt (–N A) before other analysis (34).
Gene transcript analysis
D. simulans gene sequences were obtained from FlyBase (ftp.flybase.net/genomes/Drosophila_simulans/dsim_r1.4_F B2014_03/fasta/dsim-all-gene-r1.4.fasta.gz). RNA-seq reads were trimmed to remove poor quality nucleotides using UrQt (–t 25) (34) and then aligned against D. simulans genes using Tophat2 (35). Alignment counts were performed on sorted bam files using eXpress (36), and differential expression was assessed using DESeq2 (16). We used a 0.05 FDR threshold value for significance. All subsequent calculations were performed on the DESeq2 normalized read counts. Genetic Euclidian distance matrices were computed on the 10 samples using the R dist() function with default parameters on normalized read counts. We retrieved D. melanogaster orthologs using the gene_orthologs_fb_2014_06.tsv.gz file from FlyBase and used the corresponding gene IDs to obtain gene ontology data from FlyBase.
To test whether genes of the piRNA pathway (GIPPs) are more frequently differentially expressed than other genes, we randomly sampled 10 000 sets of 19 genes in the complete list of genes (because our list of GIPPs is made of 19 genes) and determined the proportion of differentially expressed genes for each set. We then compared this empirical distribution of the proportion of differentially expressed genes to the value observed for GIPPs.
TE transcript analyses
Fasta sequences of TE copies and rosette file construction
To be as exhaustive as possible concerning the identification of TE copies in the D. simulans genome, we retrieved the copies from the two D. simulans sequenced genomes. The first genome was produced in 2007 (37) and corresponded to a hybrid assembly of sequences from five different strains. The second genome was produced in 2013 (38) and corresponded to the sequencing of the majority strain (w501) present in the 2007 version. We used the RepeatMasker program (39) using a custom library of TE references to identify the hits in the genome. The sequences of each copy were obtained using the tool ‘One code to find them all’ (40) (sequences available upon request). The rosette file (available as Supplementary Data) was generated using the sequence names of each copy by adding a column corresponding to the TE (sub)family and a column corresponding to the TE class, which represented 36 046 copies associated with 793 (sub)families.
The TEtools pipeline
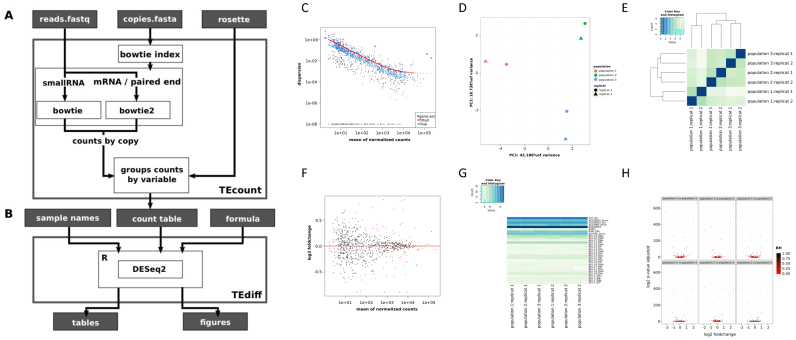
To determine the read count corresponding to each TE family, we used the first module of TEtools (TEcount) with the TE (sub)family column in the rosette file as the variable (Figure 1A). The output table from this module was used in the second module (TEdiff) to perform the differential expression analyses (Figure 1B). The module TEdiff outputs a table of TE families (or any other variables specified in the rosette file) that are differentially expressed among the various conditions/strains, as well as various graphics on the quality of the analysis and the results corresponding to DESeq2 analyses. As an example, we put on Figure 1 (C to H) the graphics corresponding to an mRNA analysis of three of our strains. Figure 1C corresponds to the model goodness of fit of the data that takes into account the within-group variability and that corresponds to the dispersion plot of the data estimates (black), the fit to a trend curve to the maximum likelihood estimates to capture the dependence of these estimates on average expression strength (red) and the maximum a posteriori estimates used in testing (blue). Figure 1D and E show the principal component analysis (PCA) of the different samples and the heatmap of the sample-to-sample distances, respectively. These two figures allow to verify that the replicates of a given sample are congruent and may also provide information concerning the grouping of the samples based on the divergence of the variable (TE family expression for example). The heatmap gives additional information over similarities and dissimilarities between samples concerning the variation of TE expression, which do not appear on the PCA. Figure 1F shows a MA plot of all samples, which displays the log2 fold changes of all TEs between all samples according to the mean normalized read counts. The TEs with an adjusted P-value < 0.1 are shown in red and correspond to the differentially expressed TEs. A heatmap corresponding to the expression levels of each variable (TE families for example) for the various samples and replicates is provided (Figure 1G). This allows to visualize the differences between samples and which variables are implicated. The volcano plots of all pairwise sample comparisons are provided with red dots corresponding to differentially expressed variables (TE families for example) between the two considered samples (Figure 1H).
Figure 1.
Workflow of the TEtools pipeline and the different outputs that can be obtained. (A) Details of the TEcount module, which uses reads in fastq format, TE sequences in fasta format and a rosette file (see text) as input. (B) Details of the TEdiff module, which uses DESeq2 to perform the differential analysis of expression and produces result files in tables and figures. Examples of the various figures produced by the TEdiff module are presented from C to H. (C) Model goodness of fit of the data. (D) Principal component analysis of the different samples with their replicates. (E) Heatmap of the various samples. (F) MA plot of all samples. The red dots correspond to significant differences. (G) Heatmap corresponding to the expression levels of each variable for the various samples and replicates. (H) Volcano plots of all pairwise sample comparisons. The figures were obtained with three strains from our mRNA data.
Identification of ping-pong signatures
The identification of ping-pong signatures was performed using the tool Small RNA Signatures (41) after mapping the piRNA reads from each strain onto all TE reference sequences using bowtie (42).
RESULTS
A new approach for the automatic transcriptomic analysis of TEs
We developed TEtools, which is a new pipeline to perform analyses of the differential amounts of mRNAs and piRNAs from TE copies across different samples. This tool can be used to analyze factors such as different strains, conditions and tissues. This pipeline is implemented in two different modules.
The first module (TEcount, Figure 1A) is a python script that performs the mapping of all reads from the RNA-seq dataset to a large list of TE sequences representing different copies, and produces a list of read counts. The use of a list of TE copies provided by the user rather than a sequenced genome or TE consensus sequences has two advantages. First, we can work with TE families not present in the sequenced genome and with non-annotated genomes. Second, the reads are more likely to map with fewer mismatches onto the TE copy than onto the TE consensus sequence (43). This second point can be critical for piRNA analysis for which the read size is small, and a few mismatches can make a difference between mapped and unmapped reads. In contrast to other analytical pipelines, we set the mapper bowtie (42) to its most sensitive option (–best) to position the maximum number of reads along the TE copies. The parameters of the mapper are set to randomly choose a position for a read mapping at multiple positions with the same score. With these settings and a list of TE copies, we can include more reads than other approaches as they discard reads mapping at multiple positions and reads with non-perfect mapping along the genome. The higher number of reads obtained gives more power for subsequent differential expression analyses. The third input of the TEcount module is a rosette file that contains the names of each TE copy. This simple tabular text file can be easily built to group the TE copies by family or any other criteria (i.e. super-family, or even according to other features, such as germline or somatic cell specificity). TEcount produces a list of read counts corresponding to the chosen criteria in the rosette file. We stress the fact that TEtools uses raw counts in contrast to other piRNA analysis pipelines, which allows the system to avoid biased normalization and to lower the number of false positives for the subsequent differential analyses (44). An option is also available to filter by size and place read counts that could correspond to siRNAs (21 nt-long reads) into a separate file. The novelty of TEtools is that it intends to integrate the TE intra-family sequence diversity that was observed in some genomes. Thus, the expected outcome is a higher number of aligned reads compared to the use of only consensus sequences, as already existing software do. However, in genomes that show low intra-family sequence diversity for TEs—such as D. melanogaster—we expect the outcomes of both tools not to be significantly different. We used TEtools on our dataset using a list of consensus sequences instead of the full set of TE insertions. The total number of TE aligned reads was then 20% lower to what we got using the full set of TE insertions (2 175 381 versus 1 780 985), reinforcing the relevance of our procedure.
The second module of the TEtools pipeline (TEdiff) is an R script (45) that performs a differential analysis of the read counts using DESeq2 (46) (Figure 1B). TEdiff requires only the list of counts computed by TEcount, a description of each sample (i.e. names and replicates) and a formula specifying the conditions under which to perform the differential analyses. Then, TEdiff outputs a table of TE families (or any other variables specified in the rosette file) that are differentially expressed among the various conditions/strains. Our tool also uses a logarithmic transformation of read counts (using the Rlog function of DESeq2) to output various graphics on the quality of the analysis and the results (i.e. volcano plots and expression heatmaps) that are ready for interpretation (Figure 1C–H).
TEtools was first intended to study small RNA data. However, this tool can also be used to study any type of RNA-seq data, with the possibility of using bowtie2 (47) instead of bowtie for better mapping of mid-length or long reads and paired-end reads (Figure 1A). To use bowtie2 on paired-end reads, the user must specify the size of the insert and the mapper is set to its most sensitive option (–very-sensitive).
To facilitate the use of TEtools and its adoption, the pipeline has been implemented as a Galaxy package (20). All the modules of TEtools, which are distributed under the GNU General Public License version 3 (https://github.com/l-modolo/TEtools), can also be used with a command line interface.
Gene transcription reflects the geographical distribution of strains
Our dataset was generated from five wild-type strains of D. simulans. Four strains of natural origin (Chicharo, Makindu, Mayotte and Zimbabwe) were chosen because they were known to present variable proportions of some TEs, different levels of TE transcripts and different amounts of piRNAs (22,24,33,48,49). We also included w501, which is the most represented strain in the 2007 D. simulans sequenced genome (37).
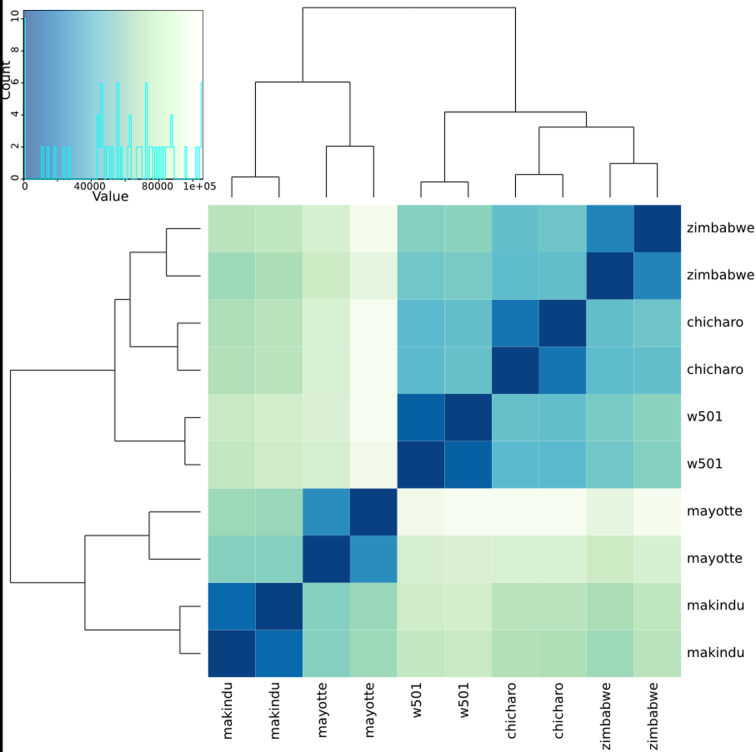
Hierarchical clustering on the sample-to-sample distances from normalized gene counts (Figure 2) first clusters samples per replicate of the same strain and then groups them together with two strains from the ancestral area (Mayotte and Makindu) and strains from the derived area (w501 and Chicharo) (50). This geographical pattern is reinforced by the significant correlation between the geographical distance (in km) and genetic distance calculated from the read counts (see ‘Materials and Methods’ section, Mantel test, r = 0.434, P-value = 0.016).
Figure 2.
Heatmap of sample-to-sample distances. This heatmap was built using DESeq2 on normalized gene read counts. Strains are clustered by replicate and the analysis separates strains from derived (w501 (USA) and Chicharo (Portugal)) and ancestral (Mayotte and Makindu (Kenya)) areas.
Globally, we found that 7416 genes out of a total of 16 169 genes were differentially expressed between the five strains. When we considered the geographical structure (derived versus ancestral areas), we found 3188 differentially expressed genes between the two groups. The top 20 differentially expressed genes belonged to biological categories such as antennal morphogenesis, DNA repair, epigenetic modifications and eye morphogenesis (Supplementary Data).
TE expression is variable across D. simulans wild-type strains
As previously mentioned, most of the analyses performed to date on TE and gene expression were performed on D. melanogaster strains. In this species, copies of TEs are mostly identical (15,51,52), which is not the case for most genomes and especially for other Drosophila genomes (15). For instance, D. simulans harbors a majority of degraded and deleted copies (15,48). Thus, the use of the latter organism as a model requires access to all the TE sequence diversity data and hence to use TEtools. All figures and the complete tables produced by the TEtools pipeline are available as supplemental data (Supplementary Data; Supplementary Data).
The PCA discriminates the different strains and the positions of the replicates are consistent in this system (Supplementary Data), indicating that we can globally discriminate between the five different strains based on TE variability. This finding supports previous observations using other experimental approaches concerning the variability in TE expression between natural strains on a global scale (22,25,53,54). According to the normalized read counts, we observe that the most highly expressed TE (sub)families are the same in all strains (Supplementary Data). These (sub)families correspond to the Long Terminal Repeat (LTR) retrotransposons Gypsy-28_DAn, and Gypsy-12_DVir and to the non-LTR retrotransposon Jockey3_DSim, which together represent more than 20% of the total TE reads for the different strains (20.48% in w501, 23.49% in Chicharo, 24.04% in Makindu, 25.03% in Mayotte and 23.88% in Zimbabwe).
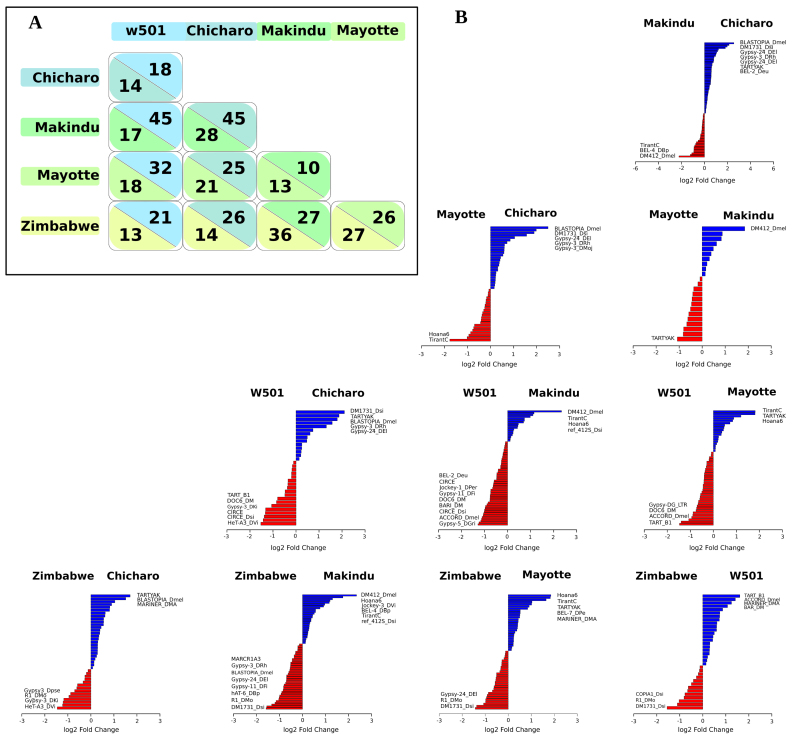
Pairwise differential analyses allowed us to identify several significant TE (sub)families as differentially expressed (Figure 3). The numbers of these TE (sub)families are indicated in Figure 3A. For example, we can observe that many TE (sub)families are differentially expressed between Makindu and three other strains w501, Chicharo and Zimbabwe (62, 73 and 63 TE (sub)families, respectively). Conversely, only 23 TE (sub)families are differentially expressed between Makindu and Mayotte. In Figure 3B, the log2-fold changes for each differentially expressed TE family for these pairwise comparisons is represented. Clearly, the expression of some TE (sub)families is specific for a given strain compared to the other strains. For example, DM412_Dmel is always more highly expressed in Makindu than in the other strains. The same is true for BLASTOPIA_Dmel in Chicharo and R1_DMo in Zimbabwe.
Figure 3.
Differentially expressed TEs between strain pairs. (A) Numbers of differentially expressed TE (sub)families between strains. The comparisons were performed between pairs of strains. Numbers above the diagonal indicate the numbers of more highly expressed TEs for the strains in columns, numbers above the diagonal indicate the numbers of more highly expressed TEs for the strains in rows. Each color corresponds to a different wild-type strain. (B) Pairwise log2-fold change for each differentially expressed TE family. The names of the most differentially expressed TEs are indicated. Blue and red indicate the sense of the comparison.
These data show that the TE transcript levels are significantly different between strains. However, the correlation between genetic distances calculated on TE read counts and geographic distances is weaker than when considering genes (Mantel test, r = 0.385, P-value = 0.036) (see Results previous section).
piRNA amounts are positively correlated to TE transcript amounts
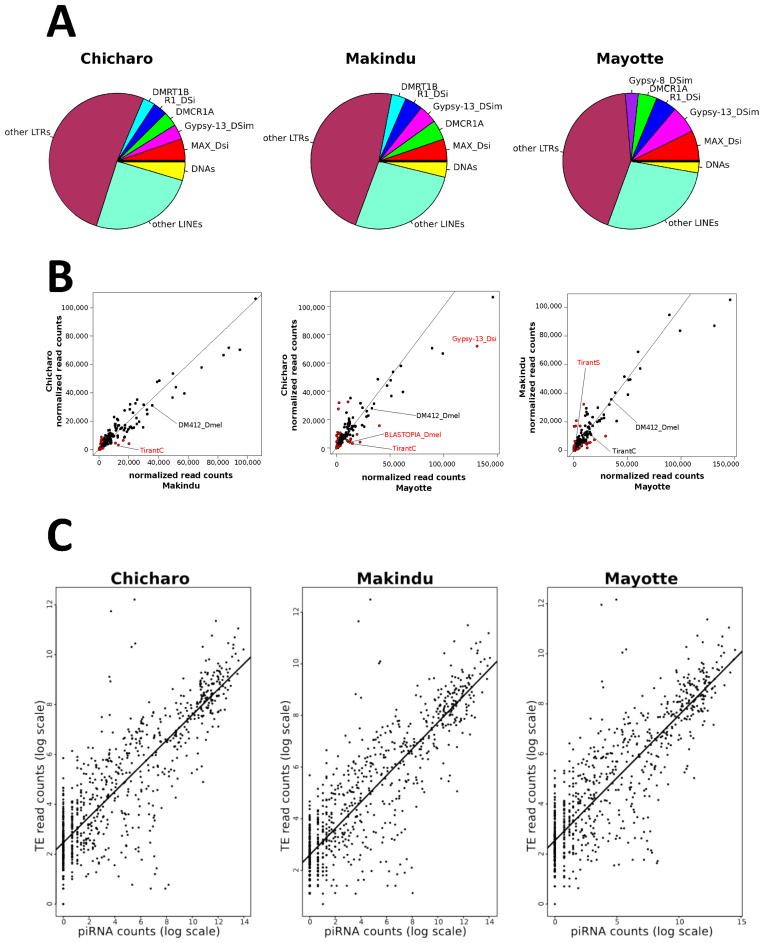
To deepen our study of TE dynamics, we used piRNA sequencing data previously obtained for three of our wild-type strains (see (33) for Chicharo and Makindu) and we performed small RNA sequencing in one additional strain, Mayotte. These data were analyzed using TEtools and all figures and complete tables produced are available as supplemental data (Supplementary Data). Because the piRNA data were not produced with replicates, DESeq2 could not provide a statistical result on the differential expression analysis. We compared the expression of the piRNAs based on their normalized read counts and observed that the most targeted TEs by piRNAs were the same for all strains (Supplementary Data). These TEs correspond to the LTR retrotransposons MAX_Dsi and Gypsy-13_DSim and to the non-LTR retrotransposons R1_Dsi and DMCR1A. The piRNAs of these four elements correspond to 18.54, 15.77 and 23.26% of all piRNA reads in Makindu, Chicharo and Mayotte, respectively (Figure 4A).
Figure 4.
Normalized piRNA read count analysis. (A) piRNA production in the different strains. The more abundant piRNAs are identified in the picture and are the same in all the strains. (B) Comparison of the normalized piRNA read counts for each pair of strains. Red dots indicate piRNAs with a log2-fold change >1. The black line corresponds to the 1:1 ratio line. As an example we indicate some TEs that display differential mRNA expression levels (see Figure 3). (C) Positive correlation between TE read counts and piRNA read counts for the different three strains. Pearson correlation tests on log transformed read counts: Chicharo: r = 0.857, P-value < 2.10−16, Makindu, r = 0.866, P-value < 2.10−16 and Mayotte: r = 0.860, P-value < 2.10−16
The pairwise comparison of the piRNA normalized read counts for each TE family is depicted on Figure 4B. This approach allows us to analyze the piRNA production of specific TEs that display differential mRNA expression levels across the three strains (i.e. the LTR retrotransposons DM412_Dmel, TirantC and BLASTOPIA_Dmel as highlighted in Figure 4B). In these cases, the log2-fold changes in the piRNAs corresponding to these elements are higher than 1.5 (output from TEdiff). For example, in the comparison between Chicharo and Mayotte, the piRNAs targeting the TirantC element exhibit a log2-fold change of 1.84, with more piRNAs targeting TirantC in the Mayotte strain than in the Chicharo strain. The same is true for this element in the comparison between Chicharo and Makindu, which is in agreement with our experimental knowledge of this TE (33).
The silencing of TEs depends on two distinct piRNA pathways that specifically trigger either somatic or germline-expressed TEs. Primary piRNAs are produced from genomic clusters and are implicated in the somatic regulation of TEs. Secondary piRNAs are either produced from TE transcripts that participate in the ping-pong amplification loop or are maternally transmitted from the mother to the embryo. One way to distinguish primary from secondary piRNAs is to identify the ping-pong signature. We estimated the proportion of piRNAs implicated in the ping-pong loop for 10 representative TEs with high piRNA production log fold changes (>|1|) (Supplementary Data). We observe that a ping-pong signal is detectable for most of the considered TEs. Additionally, the ping-pong signature is dependent not only on the TEs but also on the strain. For example, no ping-pong signal is detectable in the Chicharo strain when considering the LTR retrotransposon TirantC as is expected from previous experimental work (33). Moreover, a ping-pong signature for this element is detected for the Mayotte strain, which we previously described as having only somatic transcripts (49). The TirantS, which is a structural variant specific to D. simulans that was previously described as non-transcribed (22,55), has a very weak ping-pong signature, which is expected for non-active TEs. DOC and Gypsy-13_Dsim present the highest proportion of piRNAs with ping-pong signatures, suggesting that these TEs are probably highly transcribed in the germ line.
One hypothesis to explain the variability in copy numbers between different natural strains links the expression of TEs to the amount of piRNAs (27). Kelleher and Barbash tested this model in two strains of D. melanogaster. In the present study, using three strains of D. simulans, we found a significant positive correlation between TE read counts and piRNA read counts for each strain (Pearson correlation tests on log transformed read counts: Chicharo: r = 0.857, P-value < 2.10−16, Makindu, r = 0.866, P-value < 2.10−16 and Mayotte: r = 0.860, P-value < 2.10−16, Figure 4C). This finding illustrates a general trend for which an increase in TE transcripts is associated with an increase in piRNA production. This result is expected because secondary piRNAs are implicated in the regulation due to the ping-pong amplification loop. Thus, we searched for ping-pong signatures in the most highly expressed elements. In Supplementary Data, we show that the signature is strong for most of the TEs that have the highest amount of total piRNAs. Moreover, this analysis also reveals TE families that have no associated piRNAs but have reads in the RNA-seq data (197 (sub)families in Chicharo, 186 in Makindu and 222 in Mayotte). This result could indicate that these TEs are absent from piRNA clusters in these specific strains.
TE expression is negatively correlated with piRNA pathway gene activity
The analysis of our dataset provides a demonstration of the huge natural variability in TE expression. Indeed, we find significant variation in the levels of TE transcripts between strains and this is correlated with the corresponding piRNA production levels. In a previous study, we showed that GIPPs also displayed high transcription and sequence variability (31). Therefore, we sought to confirm the GIPP variability in the present dataset and explore its relationship with TE expression variability.
We focused on subsets of genes involved in the piRNA pathway and used other genes involved in the siRNA and immune pathways for comparison (see Supplementary Data for the complete lists of genes). We find that the piRNA pathway genes are more frequently differentially expressed than other random sets of genes (piRNA pathway 19/19 versus total dataset 7416/16 169, P-value = 0, see ‘Materials and Methods’ section). Therefore, the analysis of the present dataset confirms the existence of high intra-specific variability for GIPPs.
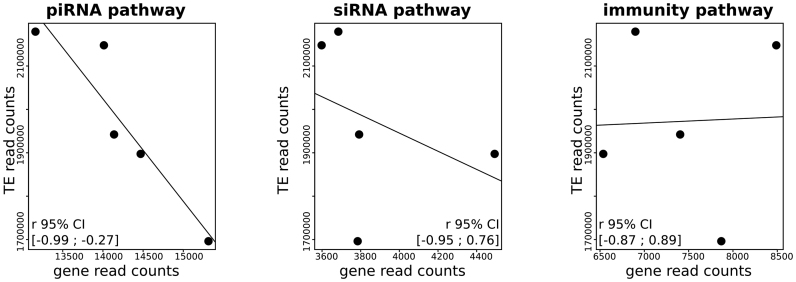
Subsequently, we tested whether the variability in TE expression was related to GIPP activity estimated by the amount of transcripts. Based on the sum of the read counts for each category of sequences, we find a strong negative correlation between the activity of GIPPs and the global TE expression (Pearson correlation test, r = −0.93, P-value = 0.022, Figure 5). No significant correlations are found between TE expression and the activity of the siRNA pathway genes (Pearson correlation test, r = −0.38, P-value = 0.530) or between TE expression and the activity of immune genes (Pearson correlation test, r = 0.04, P-value = 0.953).
Figure 5.
Negative correlation between the sum of TE read counts and the sum of GIPP read counts. No significant correlations are observed when considering genes of the siRNA pathway or genes of the immunity. Confidence intervals (95%) for Pearson correlation coefficients are mentioned at the bottom of each graph.
DISCUSSION
Advantages of TEtools
In this manuscript, we present a new analysis pipeline dedicated to the analysis of TE expression for both messenger and small RNAs. Contrary to previous approaches, this method places emphasis on the TE copies rather than on consensus sequences. This approach allows us to consider more reads and thus to reduce the loss of information because we take into account reads mapping at several positions on the genome and the individual copy variability. Moreover, this pipeline uses raw counts as proposed by Anders and Huber (16), which is a less biased approach than other normalization methods used for RNA-seq data. The pipeline also allows the use of various types of mapper and expression analysis software. In the current version we use bowtie/bowtie2 and DESeq2, but the use of alternative programs is also possible.
TEtools relies on DESeq2 for the differential expression analysis, which works well when the differentially expressed sequences account for a small amount of the total number of reads. All other differential expression programs available to date behave the same way. DESeq2 first adjusts the geometric means of the read counts across samples. This approach is valid if the potential differences reflect differences in the sample sizes that are not biologically relevant. Therefore, our procedure is valuable for the majority of transcriptome studies in which a few TE families are differentially expressed. However, in very specific cases in which one sample could be expected to display higher expression levels of all TE families (and thus increased total numbers of TE reads), the DESeq2 approach will not be relevant because differences in the geometric means of the read counts will be expected to be biologically different. In such cases, we advise pooling the count files obtained for genes and TEs separately (we recommend using TEcount to obtain the read counts) and performing the differential expression analysis on the pooled count file. When we applied the latter procedure to the present data, the results were comparable to those obtained using TEdiff on the TE reads alone (data not shown).
TE and gene expression exhibit strain differentiation but with specific dynamics
Gene transcription variation among species and populations has been previously described in D. melanogaster and D. simulans (56–58). Our study on D. simulans wild-type strains shows that variation in gene transcription is important and is sufficient to separate strains from the ancestral area (50) from strains from the derived areas.
Our data also suggest that genes that are differentially expressed between the ancestral and derived areas belong to functional categories linked to antennal morphogenesis, DNA repair, epigenetic modifications and eye morphogenesis. Some of these genes could be associated with specific different environments and could be linked to local adaptations, but further experiments are necessary to link expression levels to phenotypic features.
Previous works on TE dynamics showed that D. simulans strains harbored different numbers of TEs and different TE activities, suggesting that strains could be well distinguished based on TE dynamics (22,24,53,59). However, these previous studies were performed on a small scale. The present analysis allowed a genome-wide confirmation of these results. We find that the variability uncovered for TEs does not follow geographical patterns as strongly as genes. We propose that the regulation of TE expression evolves faster than the regulation of expression of the rest of the genome, thereby starting to erase more rapidly the geographical structures inherited from the worldwide colonization process. This faster evolution of TE expression regulation is consistent with the work by Song et al. (28), which showed that piRNA cluster expression was more variable than protein-coding gene expression in 16 inbred lines of D. melanogaster.
These data also raise the question of the interaction between TEs and gene expression. Several decades ago, McClintock (2) and Britten (60) proposed that TEs participated in gene regulatory networks and provided regulatory regions; this finding was recently confirmed (61–63). More recently, TE insertions were shown to affect the chromatin structure of nearby genes via the spread of chromatin silencing marks (i.e. H3K9me3) that may affect gene expression (6,33,64). Considering that TE expression evolves faster than protein-coding gene expression and that TEs can contribute to the modulation of gene expression through epigenetic processes, then TEs appear to be potential fundamental actors of genome expression diversification and thus adaptation (65). Further studies are necessary to elucidate the interactions between TEs and gene expression in different genetic backgrounds in a genome-wide manner.
piRNA production is positively correlated with TE expression
Previous works on TE dynamics attempted to relate piRNA production to TE copy numbers (26,28,66) but found no significant correlation. A previous analysis of wild-type strains of D. simulans showed that TE copy numbers were not correlated with GIPP expression (31). Song et al. (28) found the same result for D. melanogaster inbred lines. Taking advantage of the present dataset, we tested whether piRNA production was related to TE expression instead of TE copy numbers. Indeed, only active (expressed) TE copies are the targets of piRNA inhibition. We find a significant positive correlation between piRNA production and TE expression. The most highly expressed TE families display the highest quantity of piRNAs and vice versa. This result is consistent with the work of Kelleher and Barbash (27), which was performed on two strains of D. melanogaster. However, this result concerns only TE families controlled in the germline by secondary piRNAs.
GIPP activity can explain TE activity
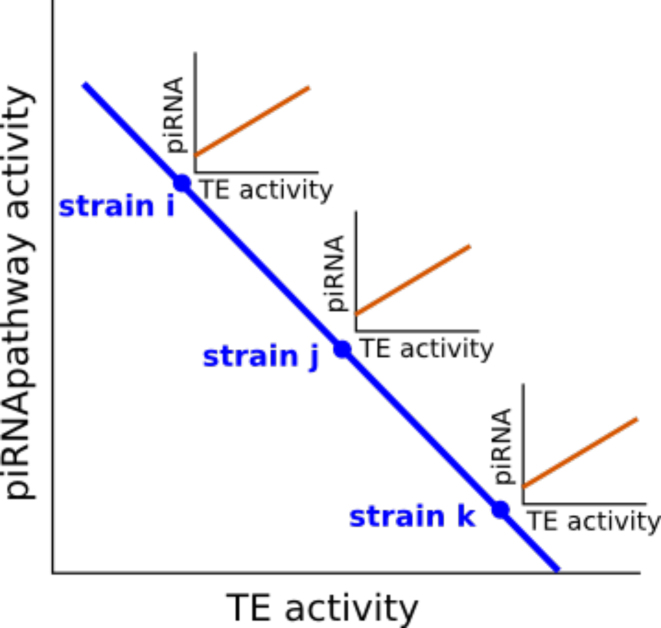
We found a strong negative correlation between GIPP activity and TE expression. This result indicates that TE expression is higher in strains in which effectors of the piRNA pathway are weakly transcribed and vice versa. This is a characteristics of the genome of each given strain. We have also shown in this work a positive correlation between TE transcription and piRNA production. This result reflects a property of TE families. Thus, the two above mentioned correlations are not incompatible but deal with different levels of variability. TE global activity varies between strains, inversely to the activity of the piRNA pathway. In addition, within the genome of each strain, at the TE family level, the production of piRNAs is positively correlated to the transcription level of TEs (Figure 6). This model can conciliate differences in copy numbers between strains that are not associated with piRNA pathway activity or piRNA production, since it considers the same evolutionary scale.
Figure 6.
Proposed model to integrate the inside genome regulation of TEs and the strain differences in the TE transcript amounts. Each strain has a specific activity of TEs that is negatively associated with the piRNA pathway efficiency. At a different level, inside each genome strain the activity of TEs is positively associated with the production of piRNAs.
The negative correlation that we find between GIPP activity and TE expression fits perfectly with the Red Queen hypothesis (67): the pathogen/host relationship is embodied by the ‘pathogenic’ TEs and the piRNA pathway which acts as a genomic defense against them. We previously explored this issue, using TE copy number data and this did not allow us to find any correlation between TEs and GIPP activity (31). At that time, we proposed that the evolutionary time scales were not compatible because TE copy number includes recent as well as very ancient TE insertion events, whereas GIPP activity is highly dynamic on a short time scale. The transcriptomes that we analyzed here provide us with data from compatible evolutionary time scales and reveal a relationship between TEs and GIPPs. Therefore, TEs and GIPPs do appear to follow the same evolutionary dynamics and are involved in an antagonistic, rapidly evolving relationship. Natural variability in the GIPPs (31) may be envisioned as tightly linked to natural variability in TEs and their dynamics in natural strains (25,49). We believe this is a very strong result, which has to be considered in future evolutionary studies of TEs. We propose that this arms race may drive strain divergence and be implicated in the beginning of speciation.
ACCESSION NUMBERS
SRX1287831, SRX1287832, SRX1287833, SRX1287834, SRX1287843 and SRX1287860.
Supplementary Material
ACKNOWLEDGEMENTS
The work was performed using the computing facilities of the CC LBBE/PRABI and the galaxy.prabi.fr web service. We thank P. Veber, S. Chambeyron and R. Rebollo for useful discussions, and A. Gibert, C. Goubert, N. Burlet and S. Martinez for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche [Exhyb ANR-14-CE19-0016-01 to C.V.]; Fondation pour la Recherche Médicale [DEP20131128536 to C.V.]; CNRS; Institut Universitaire de France (to C.V.). Funding for open access charge: Agence National de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1. Biémont C., Vieira C.. Genetics: junk DNA as an evolutionary force. Nature. 2006; 443:521–524. [DOI] [PubMed] [Google Scholar]
- 2. McClintock B. Induction of instability at selected loci in maize. Genetics. 1953; 38:579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kidwell M.G., Lisch D.R.. Perspective: transposable elements, parasitic DNA, and genome evolution. Evol. Int. J. Org. Evol. 2001; 55:1–24. [DOI] [PubMed] [Google Scholar]
- 4. Lipatov M., Lenkov K., Petrov D.A., Bergman C.M.. Paucity of chimeric gene-transposable element transcripts in the Drosophila melanogaster genome. BMC Biol. 2005; 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deloger M., Cavalli F.M.G., Lerat E., Biémont C., Sagot M.-F., Vieira C.. Identification of expressed transposable element insertions in the sequenced genome of Drosophila melanogaster. Gene. 2009; 439:55–62. [DOI] [PubMed] [Google Scholar]
- 6. Sienski G., Dönertas D., Brennecke J.. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012; 151:964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graveley B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L., Artieri C.G., van Baren M.J., Boley N., Booth B.W. et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011; 471:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aravin A.A., Hannon G.J.. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008; 73:283–290. [DOI] [PubMed] [Google Scholar]
- 9. Vagin V.V., Klenov M.S., Kalmykova A.I., Stolyarenko A.D., Kotelnikov R.N., Gvozdev V.A.. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004; 1:54–58. [PubMed] [Google Scholar]
- 10. Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J.. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008; 322:1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saito K., Siomi M.C.. Small RNA-mediated quiescence of transposable elements in animals. Dev. Cell. 2010; 19:687–697. [DOI] [PubMed] [Google Scholar]
- 12. Sienski G., Batki J., Senti K.-A., Dönertas D., Tirian L., Meixner K., Brennecke J.. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015; 29:2258–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le Thomas A., Rogers A.K., Webster A., Marinov G.K., Liao S.E., Perkins E.M., Hur J.K., Aravin A.A., Tóth K.F.. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013; 27:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han B.W., Wang W., Zamore P.D., Weng Z.. piPipes: a set of pipelines for piRNA and transposon analysis via small RNA-seq, RNA-seq, degradome- and CAGE-seq, ChIP-seq and genomic DNA sequencing. Bioinformatics. 2015; 31:593–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lerat E., Burlet N., Biémont C., Vieira C.. Comparative analysis of transposable elements in the melanogaster subgroup sequenced genomes. Gene. 2011; 473:100–109. [DOI] [PubMed] [Google Scholar]
- 16. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goubert C., Modolo L., Vieira C., ValienteMoro C., Mavingui P., Boulesteix M.. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti). Genome Biol. Evol. 2015; 7:1192–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Novák P., Neumann P., Pech J., Steinhaisl J., Macas J.. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics. 2013; 29:792–793. [DOI] [PubMed] [Google Scholar]
- 19. Lerat E. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity. 2010; 104:520–533. [DOI] [PubMed] [Google Scholar]
- 20. Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J. et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005; 15:1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vieira C., Fablet M., Lerat E., Boulesteix M., Rebollo R., Burlet N., Akkouche A., Hubert B., Mortada H., Biémont C.. A comparative analysis of the amounts and dynamics of transposable elements in natural populations of Drosophila melanogaster and Drosophila simulans. J. Environ. Radioact. 2012; 113:83–86. [DOI] [PubMed] [Google Scholar]
- 22. Fablet M., McDonald J.F., Biémont C., Vieira C.. Ongoing loss of the tirant transposable element in natural populations of Drosophila simulans. Gene. 2006; 375:54–62. [DOI] [PubMed] [Google Scholar]
- 23. Mugnier N., Biémont C., Vieira C.. New regulatory regions of Drosophila 412 retrotransposable element generated by recombination. Mol. Biol. Evol. 2005; 22:747–757. [DOI] [PubMed] [Google Scholar]
- 24. Rebollo R., Horard B., Begeot F., Delattre M., Gilson E., Vieira C.. A snapshot of histone modifications within transposable elements in Drosophila wild type strains. PLoS ONE. 2012; 7:e44253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vieira C., Lepetit D., Dumont S., Biémont C.. Wake up of transposable elements following Drosophila simulans worldwide colonization. Mol. Biol. Evol. 1999; 16:1251–1255. [DOI] [PubMed] [Google Scholar]
- 26. Lu J., Clark A.G.. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010; 20:212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelleher E.S., Barbash D.A.. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol. Biol. Evol. 2013; 30:1816–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song J., Liu J., Schnakenberg S.L., Ha H., Xing J., Chen K.C.. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol. Evol. 2014; 6:2786–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kolaczkowski B., Hupalo D.N., Kern A.D.. Recurrent adaptation in RNA interference genes across the Drosophila phylogeny. Mol. Biol. Evol. 2011; 28:1033–1042. [DOI] [PubMed] [Google Scholar]
- 30. Obbard D.J., Gordon K.H.J., Buck A.H., Jiggins F.M.. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009; 364:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fablet M., Akkouche A., Braman V., Vieira C.. Variable expression levels detected in the Drosophila effectors of piRNA biogenesis. Gene. 2014; 537:149–153. [DOI] [PubMed] [Google Scholar]
- 32. Grentzinger T., Chambeyron S.. Fast and accurate method to purify small noncoding RNAs from Drosophila ovaries. Methods Mol. Biol. 2014; 1093:171–182. [DOI] [PubMed] [Google Scholar]
- 33. Akkouche A., Grentzinger T., Fablet M., Armenise C., Burlet N., Braman V., Chambeyron S., Vieira C.. Maternally deposited germline piRNAs silence the tirant retrotransposon in somatic cells. EMBO Rep. 2013; 14:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Modolo L., Lerat E.. UrQt: an efficient software for the Unsupervised Quality trimming of NGS data. BMC Bioinformatics. 2015; 16:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L.. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011; 12:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drosophila 12 Genomes Consortium. Clark A.G., Eisen M.B., Smith D.R., Bergman C.M., Oliver B., Markow T.A., Kaufman T.C., Kellis M., Gelbart W. et al. Evolution of genes and genomes on the drosophila phylogeny. Nature. 2007; 450:203–218. [DOI] [PubMed] [Google Scholar]
- 38. Hu T.T., Eisen M.B., Thornton K.R., Andolfatto P.. A second-generation assembly of the drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013; 23:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smit A., Hubley R., Green P.. RepeatMasker Open-4.0. 2013. [Google Scholar]
- 40. Bailly-Bechet M., Haudry A., Lerat E.. ‘One code to find them all’: a perl tool to conveniently parse RepeatMasker output files. Mob. DNA. 2014; 5:13. [Google Scholar]
- 41. Antoniewski C. Computing siRNA and piRNA overlap signatures. Methods Mol. Biol. 2014; 1173:135–146. [DOI] [PubMed] [Google Scholar]
- 42. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Caboche S., Audebert C., Lemoine Y., Hot D.. Comparison of mapping algorithms used in high-throughput sequencing: application to Ion Torrent data. BMC Genomics. 2014; 15:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dillies M.-A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N., Keime C., Marot G., Castel D., Estelle J. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013; 14:671–683. [DOI] [PubMed] [Google Scholar]
- 45. R Core Team. R: A Language and Environment for Statistical Computing. 2013; Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 46. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rebollo R., Lerat E., Kleine L.L., Biémont C., Vieira C.. Losing helena: the extinction of a drosophila line-like element. BMC Genomics. 2008; 9:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Akkouche A., Rebollo R., Burlet N., Esnault C., Martinez S., Viginier B., Terzian C., Vieira C., Fablet M.. tirant, a newly discovered active endogenous retrovirus in Drosophila simulans. J. Virol. 2012; 86:3675–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lachaise D., Silvain J.-F.. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica. 2004; 120:17–39. [DOI] [PubMed] [Google Scholar]
- 51. Kaminker J.S., Bergman C.M., Kronmiller B., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M. et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002; 3, RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lerat E., Rizzon C., Biémont C.. Sequence divergence within transposable element families in the Drosophila melanogaster genome. Genome Res. 2003; 13:1889–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vieira C., Biémont C.. Geographical variation in insertion site number of retrotransposon 412 in Drosophila simulans. J. Mol. Evol. 1996; 42:443–451. [DOI] [PubMed] [Google Scholar]
- 54. Vieira C., Piganeau G., Biémont C.. High copy numbers of multiple transposable element families in an Australian population of Drosophila simulans. Genet. Res. 2000; 76:117–119. [DOI] [PubMed] [Google Scholar]
- 55. Fablet M., Lerat E., Rebollo R., Horard B., Burlet N., Martinez S., Brasset E., Gilson E., Vaury C., Vieira C.. Genomic environment influences the dynamics of the tirant LTR retrotransposon in Drosophila. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009; 23:1482–1489. [DOI] [PubMed] [Google Scholar]
- 56. Zhao L., Wit J., Svetec N., Begun D.J.. Parallel Gene Expression Differences between Low and High Latitude Populations of Drosophila melanogaster and D. simulans. PLoS Genet. 2015; 11:e1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Müller L., Hutter S., Stamboliyska R., Saminadin-Peter S.S., Stephan W., Parsch J.. Population transcriptomics of Drosophila melanogaster females. BMC Genomics. 2011; 12:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee Y.C.G. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet. 2015; 11:e1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Biémont C., Nardon C., Deceliere G., Lepetit D., Loevenbruck C., Vieira C.. Worldwide distribution of transposable element copy number in natural populations of Drosophila simulans. Evol. Int. J. Org. Evol. 2003; 57:159–167. [DOI] [PubMed] [Google Scholar]
- 60. Britten R.J. DNA sequence insertion and evolutionary variation in gene regulation. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:9374–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Casacuberta E., González J.. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013; 22:1503–1517. [DOI] [PubMed] [Google Scholar]
- 62. Rebollo R., Romanish M.T., Mager D.L.. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu. Rev. Genet. 2012; 46:21–42. [DOI] [PubMed] [Google Scholar]
- 63. Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008; 9:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shpiz S., Ryazansky S., Olovnikov I., Abramov Y., Kalmykova A.. Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the drosophila germline. PLoS Genet. 2014; 10:e1004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fablet M., Vieira C.. Evolvability, epigenetics and transposable elements. Biomol. Concepts. 2011; 2:333–341. [DOI] [PubMed] [Google Scholar]
- 66. Castillo D.M., Mell J.C., Box K.S., Blumenstiel J.P.. Molecular evolution under increasing transposable element burden in Drosophila: a speed limit on the evolutionary arms race. BMC Evol. Biol. 2011; 11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liow L.H., Van Valen L., Stenseth NC.. Red Queen: from populations to taxa and communities. Trends Ecol. Evol. 2011; 26:349–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.