Abstract
In bacteria, the start site and the reading frame of the messenger RNA are selected by the small ribosomal subunit (30S) when the start codon, typically an AUG, is decoded in the P-site by the initiator tRNA in a process guided and controlled by three initiation factors. This process can be efficiently inhibited by GE81112, a natural tetrapeptide antibiotic that is highly specific toward bacteria. Here GE81112 was used to stabilize the 30S pre-initiation complex and obtain its structure by cryo-electron microscopy. The results obtained reveal the occurrence of changes in both the ribosome conformation and initiator tRNA position that may play a critical role in controlling translational fidelity. Furthermore, the structure highlights similarities with the early steps of initiation in eukaryotes suggesting that shared structural features guide initiation in all kingdoms of life.
INTRODUCTION
In bacteria, during the initiation phase of protein synthesis three initiation factors (IFs: IF1, IF2 and IF3), the messenger RNA (mRNA), and initiator tRNA (fMet–tRNA) assemble in a 30S pre-initiation complex (preIC) in which proper codon–anticodon base pairing has not yet taken place (1–3). The actual mRNA start codon recognition progresses through several checkpoints the first of which entails a first order conformational change of the 30S preIC that allows codon–anticodon base pairing in the P-site that produces a ‘locked’ 30S initiation complex (30SIC) provided that both composition and structure of the 30S preIC are canonical (2). The resulting 30SIC is structurally competent for joining the large ribosomal subunit (50S) to form the 70S ribosomal initiation complex (70SIC), a process in which IF1 and IF3 dissociate from the ribosome whereas IF2 remains ribosome-bound until it has delivered the acceptor end of fMet-tRNA in a position amenable for the formation of the first peptide bond (4,5). The transition from the ‘unlocked’ 30S preIC to the ‘locked’ 30SIC is antagonized by IF3 and IF1 if the former complex contains a non-canonical element (e.g. an initiation triplet other than AUG, GUG or UUG). In this case, IF3 remains more tightly bound to the 30S subunit and association with the 50S subunit is impaired (2,6).
Recent data have shown that GE81112, a specific inhibitor of the initiation phase of protein synthesis (7), binds near the anticodon stem loop of fMet–tRNA and while allowing the formation of the 30S preIC it blocks the locking step that marks the transition to 30SIC; therefore, GE81112 represents a powerful tool to trap the 30S subunit stalled in a preIC state characterized by only partial P-site codon–anticodon base-pairing and tighter IF3 binding (8). These properties of GE81112 were exploited in this study to produce a 30S preIC in which IF1, IF2, IF3 and the fMet–tRNA can be visualized on the 30S subunit as to reveal for the first time the structural details of this early translation initiation intermediate. In fact, whereas bacterial complexes representative of the late steps of the initiation process, namely the 30SIC and 70SIC have been studied by cryo-EM (9–15), no structural information is so far available for the 30S preIC.
MATERIALS AND METHODS
Assembly of the 30S IC
Escherichia coli ribosomes, ribosomal subunits and translational factors were prepared as previously described (16,17). Using purified components (Supplementary Figure S1), the 30SIC was assembled by incubating the 30S ribosomal subunits with GE81112 for 10 min at 37°C in Buffer IC (10 mM Tris–HCl (pH 7.7), 7 mM MgCl2, and 60 mM NH4CH3CO2) prior to addition of the mRNA construct containing a model Shine-Dalgarno sequence (AAG UUA ACA GGU AUA CAU ACU AUG UUU ACG AUU ACU ACG AUC Sigma Genosys), fMet–tRNA, GTP and IF1, IF2, IF3, and continuing the incubation at 37°C for an additional 10 min. Throughout this incubation buffer conditions are kept constant and the final factor concentrations are 1 μM 30S, 20 μM GE81112, 4 μM mRNA, 2.8 μM fMet–tRNA, 50 μM GTP, 3.2 μM IF1, 1.6 μM IF2 and 3.2 μM IF3.
Electron microscopy
The 30SIC sample was vitrified using a Vitrobot (FEI) by diluting the reconstituted 30S IC in Buffer IC containing 20 μM GE81112, 4 μM mRNA, 2.8 μM fMet–tRNA and 50 μM GTP onto Quantifiol R2/2 grids. The samples were then imaged under low dose conditions at 74 183× magnification using a JEOL JEM 2200FS + Ultrascan 4K CCD camera (CIC bioGUNE; Bilbao) yielding a pixel size of 2.02 Å/pixel.
Single particle analysis and structure determination
Contrast transfer function parameters were estimated using CTFFIND4 (18). Semi-automated particle selection was performed in RELION 1.4 yielding a total of 174 412 particles from which poorly aligning particles were removed using the 2D reference-free classification and 3D classification procedures also in the RELION software package (19,20). The remaining 74 848 particles were used to initiate a 3D refinement using a low-pass filtered (40 Å) empty 30S subunit as the starting model. The ensuing volume was subjected to 3D classification using local searches of 5° and the resulting three classes were then refined and post-processed within RELION 1.4 (Supplementary Figure S2) (19). The 30S IC-1 and 30S IC-2 volumes were interpreted using a combination of manual rigid body docking and automated multibody docking with the UCSF Chimera and SITUS software packages (21,22). Specifically, for IF1 the cryo-EM density did not have enough features for automated docking with SITUS to yield a consistent position so that for this factor we used the model observed in the 30S-IF1 crystal structure (23). The position of IF3 relative to the 30S subunit was kept constant in both models. Segmentation of the 30S IC-1 and 30S IC-2 volumes used the Segger plug-in for UCSF Chimera (24). The following structures deposited in the PDB were used as rigid body models: 4YBB, E. coli 30S head and body; 3JCN, E. coli IF2 and fMet–tRNA; 1HR0, T. thermophilus IF1; 2IFE, E. coli IF3CTD; 1TIF, Geobacillus stearothermophilus IF3NTD. When ligands were fitted as multiple independent bodies (i.e. domains G2-C1 and C2 of IF2, residues 1–72 and 73–76 of the tRNA) the conformation of the linking residues were minimized. All figures were prepared with UCSF Chimera (21). The 30S IC-1 and 30S IC-2 volumes and fitted models are deposited in the EMDB (EMD-3494 and EMD-3495) and PDB (5ME0 and 5ME1).
RESULTS
Overview of the structures
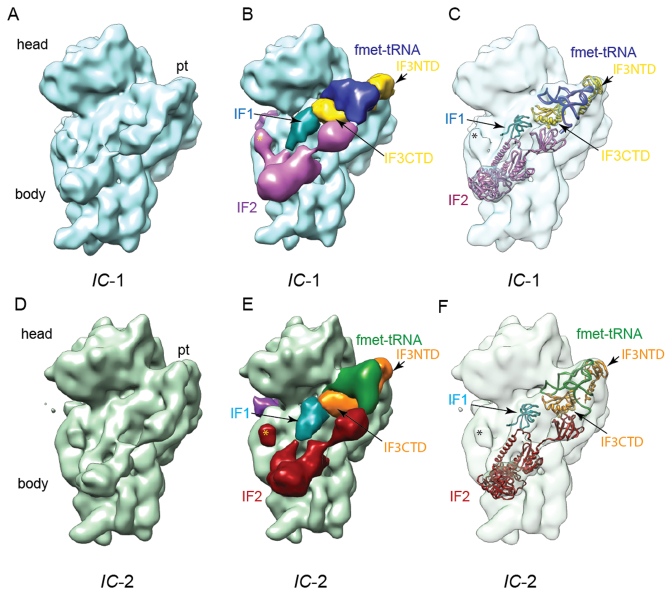
The stalled E. coli 30S preIC was assembled in vitro by incubating, in the presence of GE81112, purified 30S subunits, IFs, fMet–tRNA, GTP and an mRNA construct containing a model Shine-Dalgarno sequence (Supplementary Figure S1A; Materials and Methods) (6). The binding conditions (i.e. buffers and relative concentration of 30S ligands) were adapted from those established to follow the kinetics of initiation complex formation using variations of the FRET signal as a measurable (8). The resulting sample showed good dispersion with no aggregation as seen in the representative micrograph (Supplementary Figure S1B). An initial reconstruction of this complex showed strong densities accounting for all added ligands; namely IF1, IF2, IF3 and fMet–tRNA (Supplementary Figure S2). Because it was evident in the cryo-EM density map that the individual IFs and the tRNA displayed variable occupancies and/or conformations, a 3D classification/refinement protocol was employed (Supplementary Figure S2). Through this approach two distinct classes of complex were distinguished (Supplementary Figure S2) with an overall resolution of 13.5 Å, each characterized by well-defined density for fMet–tRNA and IF1 as well as individual domains of IF2 and IF3 (Figure 1 and Supplementary Figure S3). These classes, referred to as 30S IC-1 (31% of total data) and 30S IC-2 (36% of total data; Supplementary Figure S2) both reveal a binding site of IF3 not previously seen on the 30S subunit and show two distinct fMet–tRNA positions.
Figure 1.
Overview of the two distinct 30S preIC states observed in the presence of GE81112 by cryo-EM. The (A–C) 30S IC-1 and (D–F) 30S IC-2 maps are shown as viewed from the 50S ribosomal subunit interface. (A and D) The volumes are rendered as a solid surface with the 30S landmarks, head, body and platform (pt) indicated. (B and E) The volumes have been segmented using Segger (24) and the segmented densities are uniquely colored and labeled to reflect their correspondence with the 30S ligands: fMet–tRNA, IF1, IF2, IF3CTD and IF3NTD. (C and F) The volumes are rendered as a transparent surface and the models corresponding to the 30S ligands (fMet–tRNA, IF1, IF2, IF3CTD and IF3NTD) are shown as ribbons (see Materials and Methods). In panels B–C and E–F the star indicates density that we attribute to the G1 domain of IF2 while the dark purple density located on the back of the subunit near the mRNA entrance channel is attributed to the N1/N2 domain of IF2.
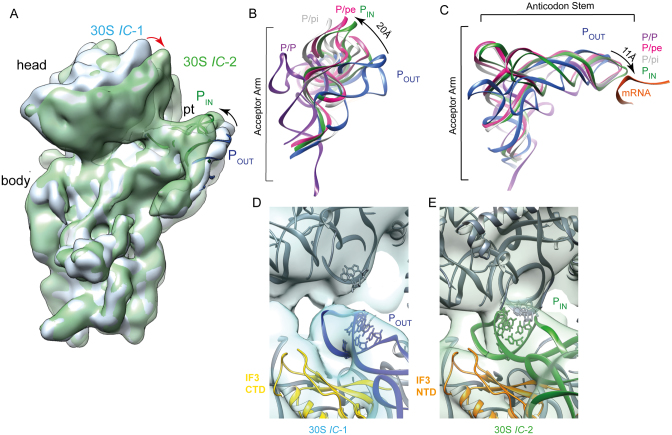
Position of fMet–tRNA in IC-1 and IC-2
The most notable difference between the IC-1 and IC-2 maps is the position of the fMet–tRNA. In the 30S IC-2 volume (green transparent surface, Figure 2A), the fMet–tRNA is positioned within the 30S P-site in an orientation similar to that of the P/pe-tRNA seen in the 70S initiation complex I (70SIC I; Figure 2B and C) (9), whereas in the 30S IC-1 (blue solid surface, Figure 2A) the elbow region of initiator tRNA is tilted toward the E-site with an ∼20 Å displacement of the tRNA elbow (compare blue and green ribbon in Figure 2B). Furthermore, in the 30S IC-1, the anticodon stem–loop is not fully accommodated into the 30S P-site (Figure 2B and C), being shifted out by ∼11 Å compared to the anticodon loop of the fMet–tRNA in the 30S IC-2, or in the 70SIC I and II structures (9) (Figure 2C). Strikingly, similar tRNA binding positions, with respect to the anticodon loop displacement, have been observed in eukaryotic initiation complexes where the two sites, termed PIN and POUT, differ by about 7 Å with respect to the extent to which they enter into the P-site (25,26). Following the same nomenclature, we term the tRNA position observed in IC-2 and IC-1 PIN and POUT, respectively. In eukaryotes, the shift between the PIN and POUT state is hypothesized to represent the conformational change that occurs during 40S scanning to facilitate start site selection (25,27–30). This step is functionally similar to the process of initiation triplet recognition occurring during the 30S preIC to 30SIC transition (1–3), although the actual pathway of initiation and many of the factors involved differ greatly between eubacteria and eukaryotes. The cryo-EM density maps also indicate that fMet–tRNA accommodation into the PIN site is accompanied by a closure of the 30S head around the fMet–tRNA (Figure 2A; red arrow) so that G1338 and A1339 of 16S rRNA could form the characteristic A-minor interactions with the G-C base pairs of initiator tRNA anticodon stem only with the PIN tRNA (Figure 2D and E). This interaction was shown to increase the stability of the complexes containing P-site bound fMet–tRNA (31,32). In yeast equivalent interactions are made between conserved G-C pairs in the anticodon stem–loop of yeast initiator tRNA and 18S rRNA residues (G1575 and A1576) and presumably stabilize the PIN state (25,33,34). This observation furthers the structural parallels seen in the initiation complexes when eubacteria and eukaryotes are compared.
Figure 2.
The binding sites of the fMet–tRNA in 30S IC-1 and 30S IC-2 structures. (A) The 30S IC-1 (blue solid surface) and 30S IC-2 (green transparent surface) have been aligned on the 30S body to highlight the movement of the fMet–tRNA (black arrow) and 30S head (red arrow). The fMet–tRNA positions in the 30S IC-2 and 30S IC-1 volumes are designated PIN (green ribbon) and POUT (blue ribbon), respectively. (B and C) Orthogonal views of the tRNAs that have been aligned relative to the 30S body and represent the PIN-tRNA (30S IC-2, this study), POUT-tRNA (30S IC-1, this study), P/ei-tRNA (70SIC-I, PDBID 3JCN), P/pi-tRNA (70SIC-II, PDBID 3JCJ) and P/P-tRNA (PDBID 4V9D). The mRNA as seen in the 70SIC-I structure is indicated as an orange ribbon. The displacement of the elbow (B) and anticodon loop (C) as the fMet–tRNA shifts from the POUT to the PIN site is indicated with an arrow. The POUT-tRNA (D) and PIN-tRNA (E) are shown inside the 30S IC-1 and 30S IC-2 volumes along with the surrounding elements of the 30S subunit and IF3CTD (yellow ribbon). 16S rRNA (G1338-A1339) and fMet–tRNA (positions 29–31 and 39–41) residues that have been established to form stabilizing interactions are drawn in an all atom representation to illustrate that the two are in proximity only in the 30S IC-2 model.
Conformation of the initiation factors in IC-1 and IC-2
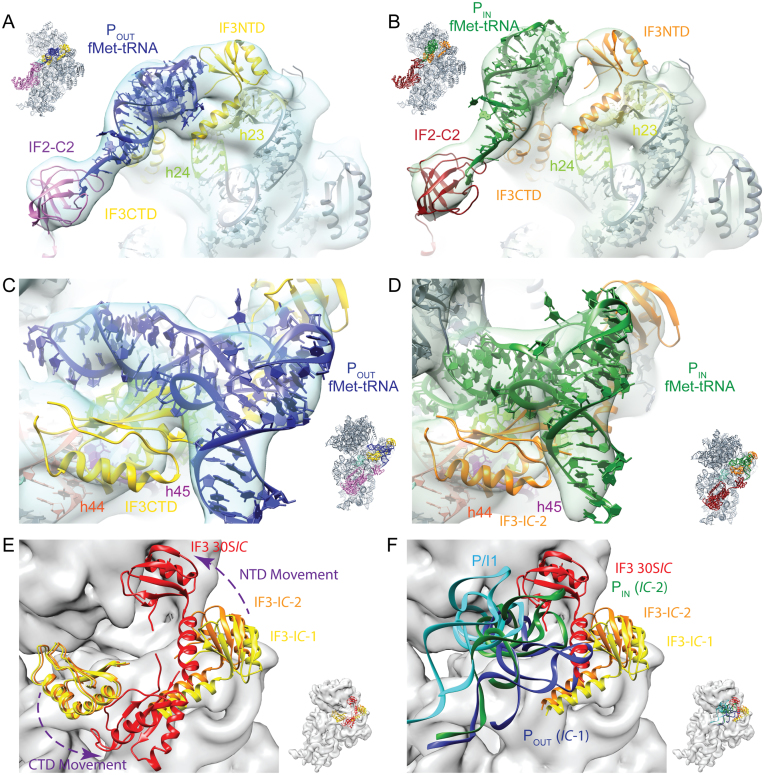
In the IC-1 and IC-2 maps IF1 is positioned on the top of helix 44 adjacent to helix 18 in agreement with previous structural investigations of the 30S initiation complexes (11,14,15,23). IF2 is positioned on the interface of the 30S subunit (Figure 1) with the C2 domain contacting the formylmethionyl moiety attached to the CCA end of the fMet–tRNA (Figure 3A and B). As seen in Supplementary Figure S4 the position of IF2 on the 30S subunit and the arrangement of its individual G2, G3, C1 and C2 domains is similar to that seen in 30S and 70S initiation complexes (9,11–15,23). In addition to these domains, E. coli IF2 has a long N-terminal extension (residues 1–381) composed of three domains, N1, N2 and G1 (35,36). Density suitable to accommodate the G1 domain (marked with a star in Figure 1B and E) emerges from the G3 domain and extends up toward helix 16 (bases 414–417 and 428–430) and the C-terminal tail of S12. The G1 domain (N-domain in T. thermophilus) is dynamic as in two 70S initiation complexes (12,13) the G1 domain was seen interacting similarly with helix 16 while in a T. thermophilus 30S initiation complex (14) the G1 domain was seen extending away from the 30S subunit. In both 30S IC-1 and IC-2 maps, a second larger density (purple density in Figure 1B and E) is located at the entrance to the mRNA channel suitable to account for parts of the N1 and N2 domains which were shown by NMR to consist of a small ordered fold and a larger unstructured region (37).
Figure 3.
fMet–tRNA in 30S IC-1 and 30S IC-2 structures is bounded by IF3NTD and IF3CTD. The IF3NTD (A and B) and IF3CTD (C and D)were placed by rigid body docking in to 30S IC-1 (blue transparent surface) and 30S IC-2 (green transparent surface). In panels A–B, 16S rRNA helices 23 (h23, residues 684–705) and 24 (h24, residues 783–800) that form an interface with IF3NTD are colored pale yellow and pale green, respectively. In panels C–D, 16S rRNA helices 24 (h24, residues 783–800), 44 (h44, residues 1401–1408 and 1494–1501) and 45 (h45, residues 1506–1529) that form an interface with IF3CTD are colored pale green, red-orange, and purple, respectively. The orientation of the model in each panel is indicated by the inset. (E and F) The IC-1, IC-2 and 30SIC (11) models have been aligned on the 30S subunit body to highlight the different conformations of IF3 (E) and illustrate that IF3NTD follows the elbow region of the tRNA as it moves from the POUT (blue), to PIN (green), to the P/I1 (cyan; (11)) site (F). In panel F it is notable that the P/I1 site seen by Julián et al. is distinct from both the PIN, POUT sites seen here and this difference may result from the IF3 movement seen in panel E. IF3 as seen in the IC-1 and IC-2 states is colored yellow and orange, respectively, while IF3 as observed in the 30SIC by Julián et al. is colored red. Panels E and F are from the same perspective but in E the fMet–tRNAs are not shown for clarity, while in F the IF3CTD is omitted and the tRNAs are illustrated as colored ribbons. The model corresponding to the 30S ribosomal subunit has been rendered as a gray surface. The purple arrows indicate the relative movement of the IF3CTD and IF3NTD. PDB files corresponding to the 30SIC (EMD-1771) were kindly provided by Mikel Valle.
In the 30S IC-1 and 30S IC-2 maps, the fMet–tRNA is bounded by IF3 (Figure 3). IF3 consists of two domains (IF3NTD and IF3CTD) joined by a flexible linker (38). Among other activities, IF3CTD promotes translational fidelity by discriminating canonical vs. non-canonical start sites while the role of the NTD seems to be restricted to modulating the affinity of the factor for the 30S subunit (39). In the 30S IC-1 and 30S IC-2 maps IF3NTD approaches the fMet–tRNA near the elbow region and the 30S platform near 16S rRNA helices 23 and 24 (Figure 3A and B), while IF3CTD is positioned between IF1, the anticodon stem of the fMet–tRNA, and helices 24, 44 and 45 (Figure 3C–D). This IF3 contact pattern is consistent with hydroxyl radical probing experiments (40), the protection of rRNA residues in h23 and h24 from chemical modification (41), and the observation that IF3 disrupts a crosslink between U793 (h24a) and G1517 (h45) (42). Furthermore the arrangement of IF3CTD and IF3NTD in the 30S preIC (Figure 3E) is unique compared to the binding positions observed in previous cryo-EM investigations of initiation complexes (10–12). Specifically, when the position of IF3NTD is compared among the IC-1, IC-2 and 30SIC (11) models it can be seen that IF3NTD follows the fMet–tRNA rotation by maintaining an interaction with the elbow region of the tRNA but largely loses contact with the 30S platform (Figure 3E and F). This could represent the molecular basis for the reduction of the IF3 affinity for the ribosome upon formation of a canonical 30SIC in preparation for the dissociation of the factor during 70SIC formation. This finding is also in full agreement with the proposed mechanism of IF3 dissociation (39,43). This entails the initial loss of contact between IF3NTD and the 30S subunit leaving the factor 30S-bound via its CTD with a corresponding increase by about two orders of magnitude Kdiss of the 30S-IF3 complex (39,43). Similarly, when compared to IC-1 and IC-2, IF3CTD in the 30SIC (11) is displaced by ∼30 Å and occupies a position lower on the 30S body (Figure 3F). These varied IF3 conformations are in agreement with prior chemical probing and structural studies that have shown that the two domains of IF3 can occupy several sites on the ribosome during translational initiation (10–12,40,43–46) and recent single molecule FRET experiments indicate that there are at least 3 major conformations of the two domains of IF3 on the ribosome (5). As seen in Figure 3C–F, IF3CTD occupies a similar position on the 30S subunit in IC-1 and IC-2 while the fMet–tRNA rotates to accommodate into the 30S P-site.
The IF3CTD binding site in IC-1 and IC-2
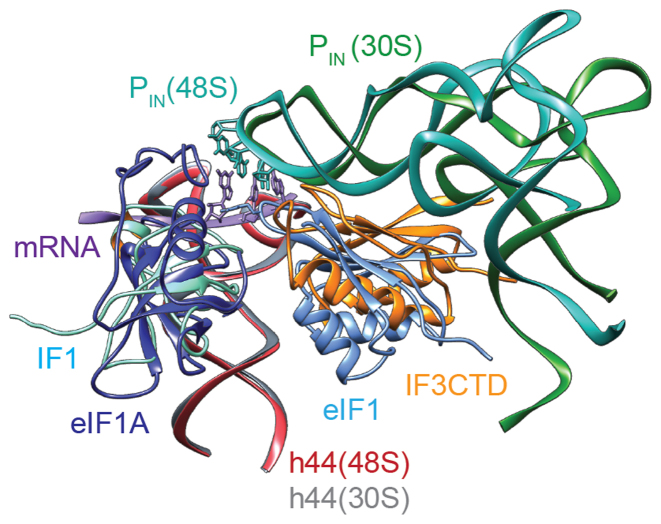
The positioning of IF3 in the 30S IC-1 and 30S IC-2 maps reveals that the 30S-IF3CTD interface involves α-helix 3 and the preceding loop 6 (Figure 3C and D; Supplementary Figure S5) which is in agreement with mutagenesis experiments that demonstrated the importance of residues in these secondary structure elements in ribosomal interactions (47). Similarly, in the 30S IC-1 and 30S IC-2 models we observe that the two conserved loops connecting ß-strand 6 and α-helix 4 (loop 7) and ß-strands 7 and 8 (loop 9) of IF3CTD are oriented toward the anticodon loop of the fMet–tRNA and IF1 (Figure 3C and D; Supplementary Figure S5) in agreement with the observation that residues of loop 7 are involved in mRNA interactions (47). The positioning of IF3CTD and the proximity of its loops toward the anticodon triplet is reminiscent of eIF1 in a yeast 48S initiation complex (Figure 4) where an arginine at the tip of loop 1 of eIF1, Arg36, interacts with the codon–anticodon base-pair (25). This similarity is noteworthy as although eIF1, like IF3, plays a role in maintaining translational fidelity by distinguishing against non-AUG initiation triplets, these two factors have a completely different topology (Supplementary Figure S6) and do not share any sequence homology (48). Our investigation reveals that despite their difference, in agreement with their functional role, they occupy similar binding sites on the small ribosomal subunit (Figure 4) (46,48).
Figure 4.
Comparison between the 30S IC-2 and the partial yeast 48S preinitiation complex (py48S). Models representing the 30S IC-2 and py48S (PDB ID: 3J81) have been aligned using residues of the 16S/18S rRNA (body) to highlight the fact that IF3CTD and eIF1 occupy overlapping sites relative to the small ribosomal subunit in their respective structures.
DISCUSSION
The translational inhibitor GE81112 was shown to target and block the transition that leads to the formation of a stable, ‘locked’ 30S initiation complex from a labile ‘unlocked’ complex (8). In the 30S-GE81112 X-ray structure, GE81112 was seen to be bound close to the anticodon stem loop and found to induce local conformational changes in both the h44/45 interface and anti-codon stem loop precluding canonical codon–anticodon interactions (8). This property of GE81112 was exploited in this study to stall the initiation process, allowing us to observe the formation of two complexes, IC-1 and IC-2, in which, unlike that seen in previous studies (11,14,15), the tRNA is either in a PIN or POUT position and the three initiation factors are present. In particular, IF3 is positioned on the 30S in a manner not yet observed in previous cryo-EM investigations (10–12) (Figure 3E and F) of bacterial initiation complexes but consistent with data from hydroxyl radical probing experiments (40). The proximity of IF3CTD and the fMet–tRNA, in particular in the region of the codon–anticodon interaction, is consistent with the role of IF3CTD in maintaining translational fidelity by rejecting non-canonical complexes.
In IC-1, the fMet–tRNA is in a POUT site with the anticodon displaced from the 30S P-site and therefore unlikely to canonically base-pair with the mRNA while the open conformation of the 30S head precludes formation of stabilizing interactions between G1338-A1339 and the anticodon stem (Figure 2); this is consistent with the labile tRNA binding seen in the 30S preIC state stabilized by GE81112 (8). It is important to note, that although we are not able to resolve GE81112 or its associated local conformational changes in the anti-codon stem loop and the h44/45 interface, the significant differences seen in the arrangement of both the fMet–tRNA and IF3 with respect to the structure of the 30SIC presented by Julián et al. (11) (Figure 3F) indicate that GE81112 is functional in our system and the observed states are a consequence of its action. During the revision of this manuscript, initiation complexes prepared in the absence of GE81112 were reported by Hussain et al. (49). A comparison of the structures presented in the two studies, show that the IC-1 and IC-2 complexes reported here are similar to the PIC-II and PIC-2 series of complexes described by Hussain et al., namely they carry an fMet–tRNA and the IF3CTD is positioned at the top of h44 (position 1 as defined by Hussain and coworkers). In the study of Hussain et al., IF3CTD moves to a second position lower on h44 (position 2) in the later stages of the initiation pathway (Supplementary Figure S7A). This agrees with the results presented here that suggest the IC-1 and IC-2 states are stalled early in the initiation pathway by the action of GE81112.
Although these states, particularly at a local structural level, might be specific to the action of GE81112 and distinct from those occurring during canonical initiation, several considerations indicate that the POUT structure corresponds to the physiological structure of the 30S preIC formed as a kinetic intermediate in the formation of the bona fide 30SIC. Indeed, the first kinetic analyses carried out to elucidate controversial mechanistic aspects of the translational pathway demonstrated that translational initiation begins with the obligatory formation of a 30S initiation complex (50). Moreover, initial rate kinetic analyses indicated that binding of mRNA and initiator tRNA to the small ribosomal subunit does not follow a defined pathway but occurs in a stochastic order (1) regardless of the presence of a Shine-Dalgarno sequence in the mRNA (51). A corollary of these analyses was that the formation of a 30SIC was preceded by the formation of a labile 30S preIC in which the two ligands mRNA and fMet–tRNA are present on the ribosomal subunit but are not interacting (i.e. base pairing) with each other. The first order transition from a 30S preIC to 30SIC was identified as the rate-limiting step in the formation of a bona fide initiation complex (1). These premises were further confirmed by more recent stopped-flow kinetic analyses (3). A 30S preIC with the postulated characteristics was actually detected when a complex containing non-interacting initiator tRNA and rpsO mRNA on the 30S subunit was identified as an intermediate in the translational regulation of ribosomal protein S15 (52). However, because the latter complex was obtained in the absence of initiation factors, the first physical evidence for the existence of the 30S preIC was obtained analyzing the complete complex stalled by GE81112 in an ‘unlocked’ conformation (8). The defective P-site codon–anticodon base-pairing between 30S-bound fMet–tRNA and mRNA was demonstrated by the altered accessibility to hydroxyl radical cleavage of the mRNA codon and fMet–tRNA anticodon (8). Nevertheless, no crystallographic data were obtained to support further the biochemical results because the complex analysed by X-ray crystallography did not contain a mRNA and the initiator tRNA anticodon stem loop was represented by the spur of a second 30S subunit occupying the P-site. Finally, recent data demonstrate that at a cold-shock temperature (i.e. 15°C) an ‘unlocked’ 30S preIC having the same properties as the POUT complex stalled by GE81112 is prevented by IF3 from undergoing the transition to ‘locked’ 30SIC when the initiator tRNA is bound in response to a non-cold shock mRNA (Giuliodori, A.M., Spedalieri, G., Fabbretti, A. and Gualerzi, C.O., manuscript in preparation). Finally, in the recent work of Hussain et al.(49), the anticodon loop in the PIC-2B complex is observed in an unusual position being partially displaced into the E site, which although not exactly the same as the POUT tRNA observed here is similar in the range of movement (Supplementary Figure S7B). For these reasons we regard the POUT complex seen in the presence of GE81112 as a genuine complex corresponding to that formed along the translation initiation pathway and consider the present evidence an important step forward in the physical characterization of the so far elusive structure of the 30S preIC.
In IC-2, the anticodon loop of the fMet–tRNA in the PIN position enters deeper into the 30S P-site with the anticodon stem resting on top of IF3CTD (Figure 3C and D; Supplementary Figure S5). This binding mode is similar to that of the eukaryotic initiator tRNA and eIF1, a functional homologue of IF3, in the yeast 48S initiation complex (Figure 4) where Hussain et al. propose that eIF1 promotes translational fidelity by destabilizing the PIN tRNA in a manner that is only overcome by base-pairing with the AUG codon (25). Because of this common binding mode it is likely that the mechanism of inhibition of non-canonical complexes formation at the start site is similar in bacteria and eukaryotes. In this regard, part of the conformational change that drives the unlocked preIC to locked 30SIC transition involves the movement of the fMet–tRNA from the POUT to PIN site where the tRNA is destabilized and rejected by IF3 if a non-canonical codon–anticodon duplex is present in the 30S P-site. On the contrary, when a canonical codon–anticodon is present there are concurrent conformational changes that transform the 30S preIC to a 30SIC which could include, for example, the POUT to PIN fMet–tRNA movement, a change in the h44/45 interface (8), and a conformational change that weakens the 30S-IF3 interaction (2,8).
The structures of the bacterial pre-initiation complexes presented here (IC-1 and IC-2) draw structural parallels between bacterial and eukaryotic initiation complexes, indicating that despite the striking differences between the initiation processes, there are shared structural features that guide start site selection during initiation of protein synthesis in all kingdoms of life although different protein factors have been developed to facilitate this process.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Initial funding for this work was provided by the European Commission Contract [QLRT-2001-00892 ‘Ribosome inhibitors’ to C.O.G., L.B.]; Subsequent work in Camerino was made possible by generous gifts of chemicals and fine biochemical precursors and other support from colleagues and friends around the world. Additional support was provided by Bizkaia:Talent and the European Union's Seventh Framework Program [Marie Curie Actions; COFUND; to S.R.C., T.K.]; Marie Curie Action Career Integration Grant [PCIG14-GA-2013-632072 to P.F.]; Ministerio de Economía Y Competitividad Grant [CTQ2014-55907-R to P.F., S.R.C.]. Funding for open access charge: Ministerio de Economía y Competitividad Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gualerzi C., Risuleo G., Pon C.L.. Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry. 1977; 16:1684–1689. [DOI] [PubMed] [Google Scholar]
- 2.Milon P., Maracci C., Filonava L., Gualerzi C.O., Rodnina M.V.. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat. Struct. Mol. Biol. 2012; 19:609–615. [DOI] [PubMed] [Google Scholar]
- 3.Milon P., Carotti M., Konevega A.L., Wintermeyer W., Rodnina M.V., Gualerzi C.O.. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010; 11:312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomsic J., Vitali L.A., Daviter T., Savelsbergh A., Spurio R., Striebeck P., Wintermeyer W., Rodnina M.V., Gualerzi C.O.. Late events of translation initiation in bacteria: a kinetic analysis. EMBO J. 2000; 19:2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyal A., Belardinelli R., Maracci C., Milon P., Rodnina M.V.. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res. 2015; 43:10700–10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Teana A., Pon C.L., Gualerzi C.O.. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandi L., Fabbretti A., La Teana A., Abbondi M., Losi D., Donadio S., Gualerzi C.O.. Specific, efficient, and selective inhibition of prokaryotic translation initiation by a novel peptide antibiotic. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabbretti A., Schedlbauer A., Brandi L., Kaminishi T., Giuliodori A.M., Garofalo R., Ochoa-Lizarralde B., Takemoto C., Yokoyama S., Connell S.R. et al. Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding. Proc. Natl. Acad. Sci. U.S.A. 2016; doi:10.1073/pnas.1521156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprink T., Ramrath D.J.F., Yamamoto H., Yamamoto K., Loerke J., Ismer J., Hildebrand P.W., Scheerer P., Bürger J., Mielke T. et al. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci. Adv. 2016; 2:e1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCutcheon J.P., Agrawal R.K., Philips S.M., Grassucci R.A., Gerchman S.E., Clemons W.M., Ramakrishnan V., Frank J.. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:4301–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julián P., Milon P., Agirrezabala X., Lasso G., Gil D., Rodnina M.V., Valle M.. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011; 9:e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen G.S., Zavialov A., Gursky R., Ehrenberg M., Frank J.. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005; 121:703–712. [DOI] [PubMed] [Google Scholar]
- 13.Myasnikov A.G., Marzi S., Simonetti A., Giuliodori A.M., Gualerzi C.O., Yusupova G., Yusupov M., Klaholz B.P.. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 2005; 12:1145–1149. [DOI] [PubMed] [Google Scholar]
- 14.Simonetti A., Marzi S., Billas I.M.L., Tsai A., Fabbretti A., Myasnikov A.G., Roblin P., Vaiana A.C., Hazemann I., Eiler D. et al. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15656–15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonetti A., Marzi S., Myasnikov A.G., Fabbretti A., Yusupov M., Gualerzi C.O., Klaholz B.P.. Structure of the 30S translation initiation complex. Nature. 2008; 455:416–420. [DOI] [PubMed] [Google Scholar]
- 16.Brandi L., Dresios J., Gualerzi C.O.. Assays for the identification of inhibitors targeting specific translational steps. New Antibiotic Targets, Methods In Molecular Medicine™. 2008; 142, Totowa: Humana Press; 87–106. [DOI] [PubMed] [Google Scholar]
- 17.Pawlik R.T., Littlechild J., Pon C., Gualerzi C.. Purification and properties of Escherichia coli translational initiation factors. Biochem. Int. 1981; 2:421–428. [Google Scholar]
- 18.Rohou A., Grigorieff N.. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheres S.H.W. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 2014; doi:10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 22.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2010; 2:21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Hartsch T., Wimberly B.T., Ramakrishnan V.. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001; 291:498–501. [DOI] [PubMed] [Google Scholar]
- 24.Pintilie G., Chen D.-H., Haase-Pettingell C.A., King J.A., Chiu W.. Resolution and probabilistic models of components in CryoEM maps of mature P22 bacteriophage. Biophys. J. 2016; 110:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain T., Llácer J.L., Fernandez I.S., Munoz A., Martin-Marcos P., Savva C.G., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014; 159:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Hellen C.U.T., Pestova T.V., Frank J.. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013; 153:1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lind C., Åqvist J.. Principles of start codon recognition in eukaryotic translation initiation. Nucleic Acids Res. 2016; doi:10.1093/nar/gkw534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini A.K., Nanda J.S., Lorsch J.R., Hinnebusch A.G.. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev. 2010; 24:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maag D., Algire M.A., Lorsch J.R.. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J. Mol. Biol. 2006; 356:724–737. [DOI] [PubMed] [Google Scholar]
- 30.Hinnebusch A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011; 75:434–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancaster L., Noller H.F.. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005; 20:623–632. [DOI] [PubMed] [Google Scholar]
- 32.Shetty S., Bhattacharyya S., Varshney U.. Is the cellular initiation of translation an exclusive property of the initiator tRNAs?. RNA Biol. 2015; 12:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong J., Munoz A., Kolitz S.E., Saini A.K., Chiu W.-L., Rahman H., Lorsch J.R., Hinnebusch A.G.. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev. 2014; 28:502–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J., Nanda J.S., Rahman H., Pruitt M.R., Shin B.-S., Wong C.-M., Lorsch J.R., Hinnebusch A.G.. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes Dev. 2008; 22:2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eiler D., Lin J., Simonetti A., Klaholz B.P., Steitz T.A.. Initiation factor 2 crystal structure reveals a different domain organization from eukaryotic initiation factor 5B and mechanism among translational GTPases. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15662–15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gualerzi C.O., Pon C.L.. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell. Mol. Life Sci. 2015; 72:4341–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laursen B.S., Mortensen K.K., Sperling-Petersen H.U., Hoffman D.W.. A conserved structural motif at the N terminus of bacterial translation initiation factor IF2. Exp. Appl. Acarol. 2003; 278:16320–16328. [DOI] [PubMed] [Google Scholar]
- 38.Biou V., Shu F., Ramakrishnan V.. X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix. EMBO J. 1995; 14:4056–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrelli D., La Teana A., Galofaro R., Spurio R., Pon C.L., Gualerzi C.O.. Translation initiation factor IF3: two domains, five functions, one mechanism?. EMBO J. 2001; 20:4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dallas A., Noller H.F.. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001; 8:855–864. [DOI] [PubMed] [Google Scholar]
- 41.Moazed D., Samaha R.R., Gualerzi C.O., Noller H.F.. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 1995; 248:207–210. [DOI] [PubMed] [Google Scholar]
- 42.Shapkina T.G., Dolan M.A., Babin P., Wollenzien P.. Initiation factor 3-induced structural changes in the 30 S ribosomal subunit and in complexes containing tRNA(f)(Met) and mRNA. J. Mol. Biol. 2000; 299:615–628. [DOI] [PubMed] [Google Scholar]
- 43.Fabbretti A., Pon C.L., Hennelly S.P., Hill W.E., Lodmell J.S., Gualerzi C.O.. The real-time path of translation factor IF3 onto and off the ribosome. Mol. Cell. 2007; 25:285–296. [DOI] [PubMed] [Google Scholar]
- 44.Elvekrog M.M., Gonzalez R.L.. Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat. Struct. Mol. Biol. 2013; 20:628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pioletti M., Schlünzen F., Harms J., Zarivach R., Glühmann M., Avila H., Bashan A., Bartels H., Auerbach T., Jacobi C. et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001; 20:1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomakin I.B., Kolupaeva V.G., Marintchev A., Wagner G., Pestova T.V.. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003; 17:2786–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrelli D., Garofalo C., Lammi M., Spurio R., Pon C.L., Gualerzi C.O., Teana A.L.. Mapping the active sites of bacterial translation initiation factor IF3. J. Mol. Biol. 2003; 331:541–556. [DOI] [PubMed] [Google Scholar]
- 48.Lomakin I.B., Shirokikh N.E., Yusupov M.M., Hellen C.U.T., Pestova T.V.. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006; 25:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussain T., Llácer J.L., Wimberly B.T., Kieft J.S., Ramakrishnan V.. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell. 2016; 167:133–137.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blumberg B.M., Nakamoto T., Goldberg I.S.. Kinetic evidence for the obligatory formation of a 30S initiation complex in polyphenylalanine synthesis initiated with N-acetylphenylalanyl-5RNA. Biochemistry. 1975; 14:2889–2894. [DOI] [PubMed] [Google Scholar]
- 51.Calogero R.A., Pon C.L., Canonaco M.A., Gualerzi C.O.. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philippe C., Eyermann F., Bénard L., Portier C., Ehresmann B., Ehresmann C.. Ribosomal protein S15 from Escherichia coli modulates its own translation by trapping the ribosome on the mRNA initiation loading site. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:4394–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.