Abstract
RAD52 is a homologous recombination (HR) protein that is conserved from bacteriophage to humans. Simultaneously attenuating expression of both the RAD52 gene, and the HR and tumor suppressor gene, BRCA2, in human cells synergistically reduces HR – indicating that RAD52 and BRCA2 control independent mechanisms of HR. We have expressed the human RAD52 gene (HsRAD52) in budding yeast strains lacking the endogenous RAD52 gene and found that HsRAD52 supports repair of DNA double-strand breaks (DSB) by a mechanism of HR that conserves genome structure. Importantly, this mechanism of HR is independent of RAD51, which encodes the central strand exchange protein in yeast required for conservative HR. In contrast, BRCA2 exerts its effect on HR in human cells together with HsRAD51, potentially explaining the synergistic effect of attenuating the expression of both HsRAD52 and BRCA2. This suggests that multiple mechanisms of conservative DSB repair may contribute to tumor suppression in human cells.
INTRODUCTION
Throughout phylogeny, the homologous recombination (HR) apparatus plays a critical role in rescuing deleterious DNA lesions (1). The near simultaneous appearance of the HR and DNA replication apparatus early in the evolution of life on Earth indicates the importance of this DNA repair function (2). Inactivation of both copies of the HR genes, BRCA1, BRCA2 or RAD51 leads to early embryonic lethality in mice – suggesting that much of the HR apparatus is essential for survival in mammals (3–8).
In keeping with the ancient origin and critical importance of the HR apparatus, many of its components are conserved throughout phylogeny (Supplementary Table S1). Key among these components is the DNA strand exchange protein, which is critical for HR mechanisms that conserve genome structure (9–12). RAD51 is the central strand exchange protein from yeast to humans (13,14). In human cells, the function of HsRAD51 in conservative HR requires an array of additional proteins, including the tumor suppressor and recombination mediator, BRCA2 (15–19). Loss of BRCA2 function correlates with decreases in genome stability and viability (20).
RAD52 is another HR protein that is conserved from bacteriophage to humans (21–23). Like BRCA2, HsRAD52 has the ability to interact with DNA and HsRAD51 (15,17,24–27). However, mutations in the BRCA2 gene have substantial effects on HR, genome stability and cell viability in human cells, while mutations in the HsRAD52 gene have relatively minor effects (20). In order to address this paradox, Powell et al. simultaneously attenuated the expression of both BRCA2 and HsRAD52 in human cells, and observed synergistic decreases in HR and cell viability consistent with BRCA2 and HsRAD52 controlling distinct mechanisms of HR. Further, these data suggest that HsRAD52 could play a substantial role in suppressing tumorigenesis when the function of BRCA2 is compromised.
HsRAD52 is structurally and functionally similar to the Rad52 protein of budding yeast, suggesting that yeast might be a suitable model organism with which to further investigate the function of HsRAD52 (25,26,28–30). Here, we report that the expression of a chromosomally integrated cDNA of HsRAD52 in rad52-null mutant yeast strains supported substantial levels of resistance to ionizing radiation and HR. Intriguingly, HsRAD52 was found to support conservative HR in the absence of RAD51, which is integral to the function of the yeast RAD52 gene (31,32). Molecular analysis indicated that HsRAD52 could not interact with Rad51 in yeast cells, but could associate with the end of a broken chromosome capable of repair by conservative HR.
MATERIALS AND METHODS
Yeast strains
All strains used in this study were isogenic, and are listed in Supplementary Table S2. Standard techniques for yeast strain construction, and growth were used (33).
Plasmids
Plasmids (Supplementary Table S3) were constructed for supplementary expression of the RAD52-FLAG gene, and the study of interactions between proteins using the yeast two-hybrid system, and co-immunoprecipitation (co-IP). Established techniques for plasmid construction and amplification were used (34). The plasmid for supplementary expression of RAD52-FLAG, pLAY864, was constructed by inserting the RAD52-FLAG coding sequence between the ADH1 promoter and terminator sequences in the URA3 marked centromere plasmid, pLAY606. The yeast two-hybrid plasmids were constructed in the multi-copy vectors pGBT9, which contains sequences encoding the DNA binding domain of the yeast Gal4 protein and the TRP1 selectable marker, and pGAD424, which contains sequences encoding the transcription activation domain of Gal4 and the LEU2 selectable marker (Clontech, Mountain View, CA, USA). The plasmids pGBT9-RAD52 and pGAD424-RAD52 were the gifts of Michael Lisby, Ph.D. The plasmids pGBT9-HsRAD52, and pGAD424-HsRAD52 were the gifts of Aaron Adamson, Ph.D. and Susan Neuhausen, Ph.D. The G8-MYC9 sequence that encodes a series of eight glycine residues linked to nine MYC epitopes was obtained from the plasmid pBS-G8-MYC9, which was the gift of Mark Boldin, Ph.D.
Analysis of cellular protein levels
Levels of Rad52-FLAG, and HsRAD52-FLAG proteins were determined by Western blot analysis, using a modification of the protocol of Sambrook and Russell (34). Whole cell extracts were prepared by glass bead disruption of yeast cells of the appropriate genotypes. Equivalent levels of levels of total protein were separated on acrylamide gels and transferred to nylon. Protein signals were revealed by probing with anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO, USA) and anti-GAPDH/Clone GA1R (Aviva Systems Biology, San Diego, CA, USA) primary antibodies and goat anti-mouse HRP conjugated secondary antibody (Thermo Scientific, Rockford, IL, USA), followed by chemiluminescent signal propagation and detection on X-ray film.
Assaying ionizing radiation resistance
Single colonies of yeast strains of the appropriate genotype were used to inoculate a minimum of 10 five milliliter YPD (1% yeast extract, 2% bacto peptone, 2% dextrose) liquid cultures that were grown to mid-log at 30°C before washing and resuspension in 5 ml of distilled water. Cell numbers were assessed by hemacytometer and the suspensions submitted to various doses of ionizing radiation in a 60Co irradiator before appropriate dilutions were plated onto YPD, incubated at 30°C for three days and the resulting colonies counted. Percent viability was calculated by dividing the number of colonies arising on the plates by the number of cell bodies plated and multiplying by 100. The 95% confidence intervals, and t-test values were determined using Prism (GraphPad, San Diego, CA, USA).
Assaying DSB-stimulated ectopic gene conversion (EGC)
Single colonies of strains of the appropriate genotype that contained the his3-Δ3΄-HOcs allele at the HIS3 locus on chromosome XV, and the his3-ΔMscI allele at the LEU2 locus on chromosome III were used to inoculate a minimum of 10 one, or five milliliter cultures of YPGL (1% yeast extract, 2% bacto peptone, 3% glycerol, 3% lactic acid) medium and grown overnight at 30°C. Strains containing the plasmids pLAY606 or pLAY864 were, instead used to inoculate 10 one milliliter cultures of synthetic complete medium lacking uracil and containing glycerol and lactate. Double-strand break (DSB) formation at the HIS3 locus was induced by addition of 20% galactose (2% final) to each culture and 4 h of incubation at 30°C. Appropriate dilutions of each culture were plated onto YPD to determine viability, and onto synthetic complete medium lacking histidine to select for recombinants. Recombination frequencies were determined by dividing the number of His+ prototrophic colonies by the number of viable cells plated after three days incubation at 30°C. Due to the opposite orientations of the his3-Δ3΄-HOcs and his3-ΔMscI alleles relative to their respective centromeres on chromosomes XV and III, all His+ prototrophs are the result of the repair of the DSB by either gene conversion, or a double crossover. Mean ectopic gene conversion (EGC) frequencies, 95% confidence intervals and t-test values were determined using Prism. Genomic DNA was prepared from select His+ recombinants from separate platings and submitted to Southern blot analysis (35).
Assaying interaction between proteins by yeast two-hybrid analysis
The yeast strain, Y187 (Clontech, Mountain View, CA, USA) was transformed with pGBT9 and pGAD424, or their derivatives. Single transformant colonies were used to inoculate a minimum of 10 five milliliter cultures of synthetic complete medium lacking leucine and tryptophan, grown overnight at 30°C and final cell densities determined by absorbance at 600 nm. Levels of expression of the gal1::lacZ gene in the transformants were determined by measuring the β-galactosidase activities in the cells using a standard protocol. Levels of o-nitrophenol released determined by absorbance at 420 nm. Absorbance at 550 nm was determined to correct for light scattering by cell debris. Specific activities were rendered in Miller Units, which were determined using the following formula: 1000 × (OD420 – 1.75 × OD550)/reaction time (minutes) × reaction volume (milliliters) × OD600. Mean specific activities, 95% confidence intervals and t-test values were determined using Prism.
Assaying interaction between proteins by co-immunoprecipitation (co-IP)
Single colonies of yeast strains expressing Rad52-FLAG or HsRAD52-FLAG from genomically integrated fusion genes, and Rad51-MYC, Rad52-MYC or HsRAD52-MYC from fusion genes on plasmids containing the appropriate combination of tagged alleles were used to inoculate cultures of synthetic complete medium lacking uracil and grown overnight. The overnight cultures were used to inoculate YPD cultures that were grown to mid-log at 30°C. Whole cell extracts were prepared by silica bead disruption and pre-cleared by the addition of pre-blocked protein A/G agarose beads (Pierce Thermo-Fisher Scientific, Waltham, MA, USA). Pre-cleared extracts were mixed with either anti-FLAG M2 antibody (Sigma-Aldrich, St. Louis, MO, USA) or anti-MYC clone 9E10 (Santa Cruz Biotechnology, Dallas, TX, USA), followed by the addition of pre-blocked protein A/G beads. Immunoprecipitates were eluted from the beads and loaded onto acrylamide gels, the proteins separated by electrophoresis and then electrophoretically transferred to nylon. Protein signals were revealed using either a HRP-conjugated anti-FLAG antibody, or the mouse anti-MYC clone 9E10 primary antibody and HRP-conjugated goat anti-mouse secondary antibody as described previously.
Assaying interaction between proteins and genomic DNA by chromatin immmunoprecipitation (ChIP)
ChIP analysis was performed as previously described (36). YPGL cultures of yeast strains of the appropriate genotype were grown to the appropriate density and an aliquot taken for the initial, 0 time point. HO endonuclease was induced in the remaining culture by addition of galactose (2% final). Cells were collected at additional time points with cell numbers adjusted to equal the number collected at time 0. Cells were fixed with formaldehyde prior to lysis with a Beadbug Homogenizer (Benchmark Scientific, Edison, NJ, USA), and sonication with a Misonix Sonicator 3000 (Cole-Palmer, Vernon Hills, IL, USA). Sonicated extracts were pre-cleared with pre-blocked Protein A/G agarose beads, at which point aliquots of chromatin were immediately collected and frozen at −80°C. Chromatin bound by Rad52-FLAG or HsRAD52-FLAG was immunoprecipitated from chromatin aliquots with anti-FLAG M2 antibody, and loaded onto pre-blocked Protein A/G beads. Protein–DNA complexes were eluted from the beads with 3XFLAG peptide (ApexBio Technology, Houston, TX, USA). Protein–DNA crosslinks were thermally reversed with simultaneous RNAse treatment prior to digestion of the protein with proteinase K, DNA extraction with 1:1 phenol–chloroform and precipitation with ethanol. Input and ChIP DNA samples were resuspended in 1X TE and stored at 4°C.
Levels of specific genomic DNA sequences retained were determined by qPCR. Sequences from the target gene (his3-Δ3΄-HOcs) were detected using the primers HIS3MscI-f (5΄ – TGC TCT GGC CAA GCA TTC – 3΄) and HIS3MscI-r (5΄ – CAG TAG GGC CTC TTT AAA AG – 3΄) that amplify a 127 bp region approximately 300 bp upstream of the HO cut site. This primer set does not amplify the equivalent region of the his3-ΔMscI allele as the HIS3MscI-f primer does not anneal there (A. Clear – unpublished results). As a reference gene, the SAM1 locus was detected using the primers SAMF-(-192) (5΄ – CAC TCT GGT ATC GAT GAA A – 3΄) and SAMR-(-106) (5΄ – CGA TGA ATA ACA GAC AAC AC – 3΄) that amplify a 105 bp region upstream of the SAM1 coding sequence, which was found to be similarly enriched under all experimental conditions (A. Clear – unpublished results).
A complete reaction consisted of 2.5 μl undiluted ChIP DNA or 1:50 Input DNA, 1 μl each of appropriate primers (5 μM stock), 8 μl ddH2O and 12.5 μl 2x qPCR Master Mix (BioPioneer, San Diego, CA, USA; with HotStart Taq DNA Polymerase, SYBR Green I, 3 mM MgCl2) for a total reaction volume of 25 μl. Each qPCR was performed in triplicate on a BioRad CFX96 Touch Real-Time PCR Detection System with the following cycling conditions: Step 1, 95°C, 10 min; Step 2, 95°C, 30 s; Step 3, 56°C, 30 s; Step 4, 72°C, 30 s; Step 5, Detection; return to step 2 and repeat 39 times; Step 6, melt curve analysis from 65°C to 95°C. Data were analyzed using the Bio-Rad CFX Manager 3.1. Cq values were reported in triplicate and outliers were defined as those outside a range of 0.5 cycles and were excluded from calculations. No-template control samples were confirmed to yield Cq values outside the acceptable range (Cq > 35).
Fold enrichment values represent the degree of Rad52-FLAG or HsRAD52-FLAG occupancy at the target gene (his3Δ’3-HOcs) relative to occupancy at the reference gene (SAM1), as calculated by the Livak method, and reported relative to time point 0 (37). Results from experimental strains were then normalized to results obtained with a control strain without FLAG-tagged proteins. The mean values for fold change in degree of occupancy for each genotype that resulted from a minimum of eight technical trials (at the qPCR stage) and a minimum of three independent experiments were plotted with standard deviations. Statistical significance was determined with the t-test using Prism.
RESULTS
Expression of a single, genomic copy of the HsRAD52 cDNA in budding yeast produces a stable HsRAD52 protein
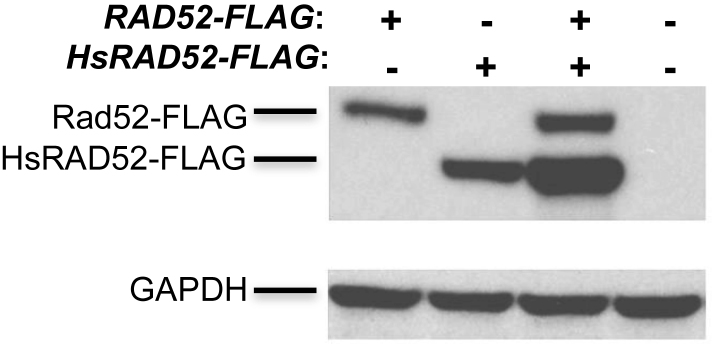
HsRAD52 and Rad52 are extensively similar at the primary sequence level, sharing several stretches of identical amino acids (Supplementary Figure S1). This is consistent with HsRAD52 and Rad52 sharing similar functional properties, and suggests that HsRAD52 and Rad52 might exert similar effects in yeast cells. In order to study the effects of HsRAD52 in budding yeast cells, a cDNA encoding full-length HsRAD52 or a C-terminally FLAG-tagged recombinant was inserted into the ADH1 locus such that expression was controlled by the ADH1 promoter and terminator sequences. Western blots of whole cell extracts from strains containing the adh1::HsRAD52-FLAG allele displayed a 49 kDa peptide consistent with that expected for HsRAD52-FLAG while extracts from strains containing the RAD52-FLAG allele yielded a 55 kDa peptide consistent with Rad52-FLAG. Levels of HsRAD52 were equal to, or above that of Rad52-FLAG (Figure 1). This indicates that strains containing the adh1::HsRAD52 allele express a stable HsRAD52 protein at a level that is at least equal to that of the endogenous Rad52 protein.
Figure 1.
HsRAD52 is stable in yeast cells Proteins from whole cell extracts of yeast cultures derived from the spores of a tetratype tetrad obtained through sporulation of an adh1::HsRAD52-FLAG/ADH1 RAD52-FLAG/RAD52 diploid (ABX3600) were separated on gels, electroblotted and probed with anti-FLAG or anti-GAPDH antibodies. Genotypes of the spores are depicted at the top of the figure. Signals corresponding to the 55 kDa Rad52-FLAG, 49 kDa HsRAD52-FLAG and 37 kDa GAPDH proteins are denoted on the left side of the figure.
HsRAD52 increases resistance to ionizing radiation of rad52 mutant cells
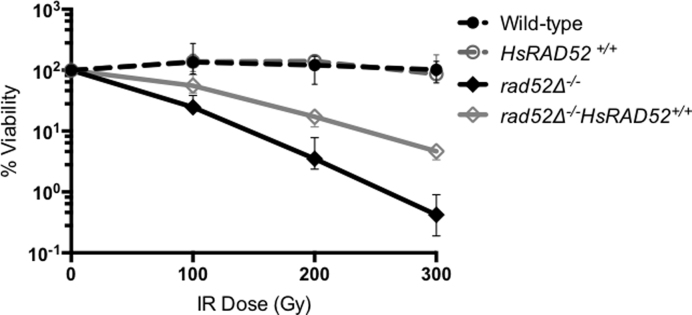
Over-expression of HsRAD52 in monkey cells increases their resistance to ionizing radiation (IR), indicating that HsRAD52 can support the repair of DNA damage induced by exposure to IR (38). Budding yeast RAD52 has also been implicated in IR resistance, as rad52 mutants are acutely sensitive to IR due to a defect in the repair of IR-induced DSBs (39,40). These observations suggest that both HsRAD52 and Rad52 function in the repair of IR-induced DNA lesions. We tested if expression of HsRAD52 can support IR resistance in yeast and found that expression in rad52-null mutant diploid yeast cells led to a statistically significant (P < 0.0001) 12-fold increase in resistance to 300 Gy of IR, which results in ∼30 DSBs per diploid yeast genome (Figure 2; Supplementary Table S4) (40). These results indicate that HsRAD52 has a limited but significant capacity to repair IR-induced DNA lesions in yeast.
Figure 2.
HsRAD52 confers ionizing radiation resistance to rad52 mutant cells Suspensions of wild-type and mutant diploid cells grown to mid-log were counted by hemacytometer before exposure to 100, 200 or 300 Gy of ionizing radiation. Appropriate dilutions of unirradiated and irradiated cells were plated onto YPD. Percent survivorship was calculated by dividing the number of colonies that arose by the number of cell bodies plated and multiplying by 100. The mean percent survivorship from a minimum of 10 independent determinations for each genotype and 95% confidence interval was plotted against levels of radiation exposure. Strains used in this analysis: WT – ABX3566; rad52−/− - ABX3568; HsRAD52+/+ - ABX3569; rad52−/− HsRAD52+/+- ABX3570.
HsRAD52 supports repair of a DSB by conservative HR that is independent from the central strand exchange gene, RAD51
Resistance to ionizing radiation in budding yeast correlates closely with the ability to propagate a HR mechanism that conserves genome structure (41). Accordingly, RAD52 plays a critical role in the repair by conservative HR of genomic DSBs catalyzed by the HO-endonuclease that simulate but have distinct repair requirements from IR-induced DSBs (42,43). The ability of HsRAD52 to confer IR resistance to rad52 mutant yeast suggests that HsRAD52 could support the repair of a HO-catalyzed DSB by conservative HR (Figure 2).
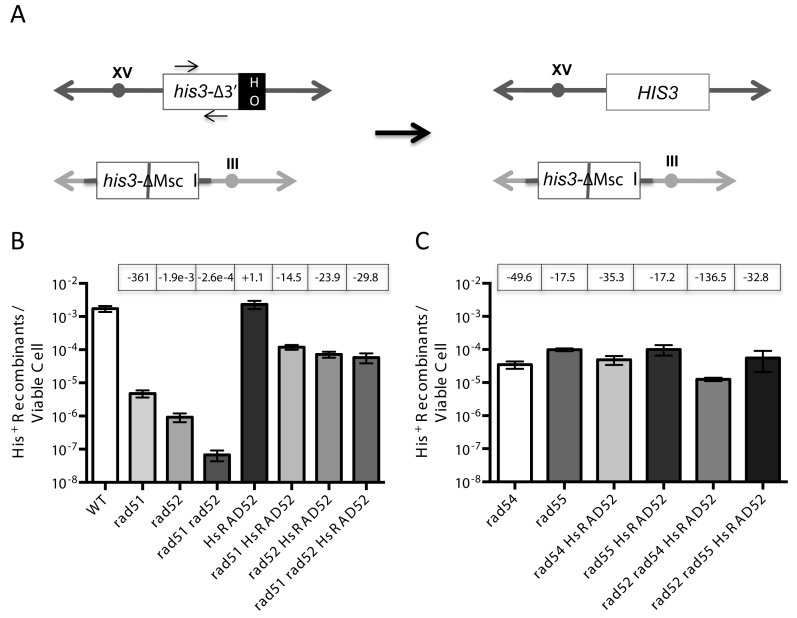
To study this we employed an assay of EGC stimulated by an HO-catalyzed DSB that involves the selection of recombinants, permitting low frequencies of recombination to be readily quantitated. Histidine prototrophs were selected following DSB formation at the HIS3 locus on chromosome XV, and repair by the unidirectional transfer of information from a second, inactivated copy of HIS3 at the LEU2 locus on chromosome III, creating an intact HIS3 locus (Figure 3A).
Figure 3.
HsRAD52 suppresses the double-strand break (DSB)-stimulated ectopic gene conversion (EGC) defect in rad51 and rad52 mutant cells (A) Scheme depicting the his3 EGC assay. The his3-Δ3΄-HOcs substrate (white ‘his3Δ-3΄ box) at the HIS3 locus on chromosome XV (dark gray double ended arrow) substitutes a 127 bp DNA fragment containing an HO cut site (black ‘HO’ box) for 238 bp of the 3΄ end of the HIS3 coding sequence and flanking DNA. The his3-ΔMsc I substrate (white ‘his3-ΔMsc I’ box) at the LEU2 locus on chromosome III (light gray double ended arrow) is comprised of a 1.8 kb genomic clone containing the HIS3 gene that has been disrupted by the insertion of a 10 bp Not I linker into the Msc I site in the coding sequence (gray bar). These substrates share 413 bp of uninterrupted homology between the Msc I site and 5΄ end of the HOcs, and 443 bp between the 3΄ end of the HOcs and downstream Bam HI site. Repair of an HO-catalyzed DSB at the his3-Δ3΄-HOcs substrate by unidirectional transfer of information from the his3-ΔMsc I substrate (black arrow) creates an intact HIS3 gene. The positions of the primers used to detect the association of proteins with the HIS3 locus in chromatin immmunoprecipitation (ChIP) experiments are depicted as arrows above and below the white ‘his3Δ-3΄’ box. (B) The effect of HsRAD52 on frequencies of EGC in wild-type, and rad51, rad52 and rad51 rad52 mutant strainsSingle colonies of wild-type and mutant his3 DSB-stimulated ectopic gene conversion haploid strains grown on YPD plates were used to inoculate at least 10 one milliliter YPGL cultures per genotype, and grown overnight. After a period of induction of HO endonuclease appropriate dilutions of cells were plated onto YPD to assess viability, and medium lacking histidine to select for recombinants. EGC frequencies were determined by dividing the number of His+ recombinants by the number of viable cells plated. The mean recombination frequency from a minimum of 10 independent determinations for each genotype and 95% confidence intervals was plotted. Fold differences above (+) and below (-) the wild-type frequency are indicated in boxes above the graphed value for each genotype. Strains used in this analysis: WT – ABX3666-37B; rad51Δ - ABX3678-49B; rad52Δ - ABX3697-82D; rad51Δ rad52Δ - ABX3728-14A; HsRAD52 – ABX3703-36B; rad51Δ HsRAD52 – ABX3728-28A; rad52Δ HsRAD52 – ABM537; rad51Δ rad52Δ HsRAD52 – ABX3728-11C. (C) The effect of HsRAD52 on frequencies of EGC in rad54, rad55, rad52 rad54 and rad52 rad55 mutant strains Same as legend for panel B. Strains used in this analysis: rad54Δ - ABM562; rad55Δ - ABM571; rad54Δ HsRAD52 – ABM564; rad55Δ HsRAD52 – ABM568; rad52Δ rad54Δ HsRAD52 – ABM563; rad52Δ rad55Δ HsRAD52 – ABM570.
In budding yeast, Rad52 exerts its effect on DSB repair by conservative HR by directly interacting with the primary strand exchange protein, Rad51 and mediating its strand exchange activity (28,44). Several additional proteins are also critical mediators of strand exchange by Rad51, including the heterodimer comprised of the Rad51 paralogs, Rad55 and Rad57, and the translocase and chromatin remodeling factor, Rad54 (45–48). Accordingly, we examined the roles of RAD51, RAD54 and RAD55 in our investigation of DSB repair by conservative HR.
In wild-type cells, the frequency of repair of a DSB at the HIS3 locus by EGC was 1.73 × 10−3, which is very similar to frequencies of DSB-stimulated EGC that we obtained previously with other assay systems (Figure 3B; Supplementary Table S4) (49). The frequency of EGC was reduced 1900-fold in rad52 mutant cells, consistent with Rad52 being a critical factor in the repair of DSBs by conservative HR (43). The 360-fold reduced frequency of EGC in rad51 mutant cells indicated a similarly critical role for Rad51. Interestingly, the frequency of EGC was reduced 26,000-fold in rad51 rad52 double mutant cells, which is significantly lower (P < 0.0001) than the frequencies in either the rad51 or rad52 single mutant cells. This is consistent with Rad51 and Rad52 possessing a weak capacity to independently support DSB repair by conservative HR, as demonstrated previously for DSB repair by inter-chromatid HR (50). Importantly, over-expression of RAD52 did not result in significantly increased frequencies of EGC in rad51 mutant cells (p = 0.295), confirming the limited capacity of Rad52 to support conservative HR independently of Rad51 (Supplementary Figure S2; Supplementary Table S4). The 50-fold reduced frequency of EGC in rad54 mutant cells and the 18-fold reduced frequency in rad55 mutant cells indicate that the Rad51 mediators, Rad54 and Rad55, play important roles in DSB repair by conservative HR, as demonstrated previously (Figure 3C; Supplementary Table S4) (51,52).
We examined the function of HsRAD52 in EGC by determining HR frequencies in rad52 mutant cells expressing the HsRAD52 gene and found that it increased the frequency of HR 78-fold (Figure 3B; Supplementary Table S4). Southern blots of representative His+ recombinants from all strains confirmed repair of the DSB in the his3 allele on chromosome XV by the unidirectional transfer of information from the his3 allele on chromosome III (Supplementary Figure S3). These data indicate that HsRAD52 supports a mechanism of conservative DSB repair that is similar to that controlled by the endogenous yeast HR machinery.
While mechanistically similar, the EGC supported by HsRAD52 displayed a level of independence from the RAD51 gene that was distinct from that of the endogenous yeast HR machinery. Expression of HsRAD52 in rad51 single, and rad51 rad52 double mutant cells increased frequencies of EGC to levels that were similar to the frequency observed when HsRAD52 is expressed in rad52 single mutant cells (Figure 3B; Supplementary Table S4). Therefore, HsRAD52 substantially suppressed the EGC defects conferred by rad51 and rad52, indicating that HsRAD52 can work independently of Rad51 and Rad52 in the repair of DSBs by conservative HR.
In contrast to its ability to suppress the conservative HR defects of the rad51 and rad52 mutants, HsRAD52 had no significant effect on EGC in rad54 (p = 0.080), or rad55 (p = 0.918) single mutant cells (Figure 3C; Supplementary Table S4). However, Rad51 and Rad52 retain partial function in DSB repair by conservative HR in rad54 and rad55 mutant cells, which could obscure the function of HsRAD52 and prevent evaluation of the genetic interaction of HsRAD52 with RAD54 and RAD55 (51,52). In order to address this issue we examined the effect of expressing HsRAD52 in strains where RAD52, as well as RAD54 or RAD55 had been inactivated. Expression of HsRAD52 in rad52 rad55 double mutant cells resulted in frequencies of EGC that were not significantly different (p = 0.270) from those observed when HsRAD52 was expressed in rad52 single mutant cells. This indicates that Rad55 plays no significant role in the propagation of EGC by HsRAD52, further distinguishing HsRAD52-dependent conservative HR from that controlled by the endogenous HR machinery (Figure 3B and C; Supplementary Table S4). In contrast, expression of HsRAD52 in rad52 rad54 double mutant cells led to a frequency of EGC that was 6-fold lower than, and significantly different (P < 0.0001) from the frequency observed when it was expressed in rad52 single mutant cells. This indicates that Rad54 plays a limited role in HsRAD52-dependent conservative HR in budding yeast cells and that HsRAD52 works with at least one of the mediators of yeast strand exchange.
HsRAD52 can self-associate but cannot associate with Rad51
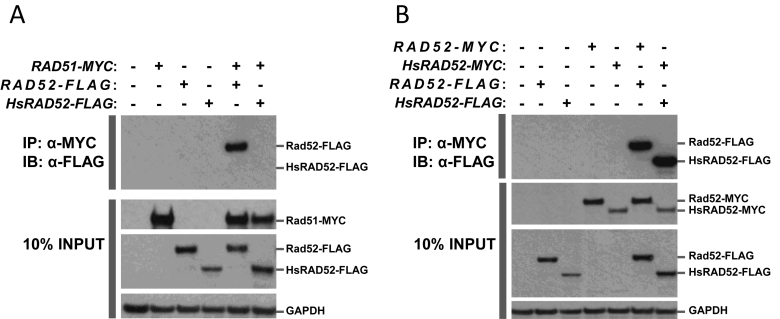
The results of previous experiments indicate that DSB-stimulated EGC in budding yeast requires that Rad52 interact physically with Rad51 (28,31). Conversely, our data indicated that HsRAD52 supported DSB-stimulated EGC independently from Rad51 (Figure 3B; Supplementary Table S4). This suggests that the function of HsRAD52 in EGC would not require an interaction with Rad51. In order to explore the interaction between HsRAD52 and Rad51 we employed the yeast two-hybrid and co-IP assays. Consistent with previously published results, our yeast two-hybrid experiments indicated robust interaction between Rad51 and Rad52, with β-galactosidase levels that were 100- to 1000-fold higher than background when appropriate fusions of both Rad51 and Rad52 to the Gal4 DNA binding and transactivation domains were present in the cell (Supplementary Figure S4) (28). In contrast, the presence of both Rad51 and HsRAD52 fusions led to expression levels that were not above background levels. This indicates that Rad51 interacts with Rad52, but not HsRAD52.
In order to more directly study the physical interaction between Rad51, and Rad52 or HsRAD52, we examined the ability of the proteins, tagged with the MYC or FLAG epitopes to interact by co-IP. Previous co-IP experiments demonstrated that Rad51 interacts with Rad52, and our experiments confirmed this result when extracts of strains containing both Rad51-MYC and Rad52-FLAG were immunoprecipitated with anti-MYC (Figure 4A) (44). Reciprocal co-IP experiments failed to reveal an interaction suggesting that the anti-FLAG antibody could not interact with and precipitate Rad52-FLAG from cell extracts while associated with Rad51-MYC (Supplementary Figure S4A). In contrast, co-IP with strains containing both Rad51-MYC and HsRAD52-FLAG failed to display an interaction between the proteins regardless of the antibody used for immunoprecipitation. These results indicate that Rad51 can interact physically with Rad52, but not HsRAD52, which is consistent with the yeast two-hybrid results (Supplementary Figure S3) (28).
Figure 4.
Intermolecular interactions between Rad51, Rad52 and HsRAD52Whole cell extracts were prepared from haploid yeast strains with or without chromosomal copies of RAD52-FLAG, or HsRAD52-FLAG, and with or without plasmid-borne copies of RAD51-MYC, RAD52-MYC or HsRAD52-MYC. Aliquots of extracts were incubated with anti-MYC antibody, and the immunoprecipitated proteins separated on SDS-PAGE gels, electroblotted and probed with anti-FLAG antibody. Aliquots of extract (10% of the volume submitted to immunoprecipitation) were also run directly on SDS-PAGE gels, electroblotted and probed with anti-MYC, anti-FLAG or anti-GAPDH antibodies. Proteins were visualized by treating blots with HRP-conjugated secondary antibodies and chemiluminescent detection reagents, followed by exposure to X-ray film. (A) Yeast Rad52 protein can interact with yeast Rad51 protein but human RAD52 protein cannotPresence of genomic copies of the RAD52-FLAG or HsRAD52-FLAG fusion genes, and/or a plasmid copy of RAD51-MYC in the yeast strains used to make the whole cell extracts are denoted with a (+) at the top of the figure. The top panel depicts proteins immunoprecipitated (IP) by anti-MYC antibody and immunoblotted (IB) with anti-FLAG antibody. The bottom three panels depict the proteins in 10% INPUT. Signals corresponding to the 55 kDa Rad52-FLAG, 49 kDa HsRAD52-FLAG, 44 kDa Rad51-MYC and 37 kDa GAPDH proteins are denoted on the right side of the figure. Strains used in this analysis: WT – W961-5A; RAD51-MYC – ABT821; RAD52-FLAG – ABM559; HsRAD52-FLAG – ABX3684-12B; RAD51-MYC RAD52-FLAG – ABT822; RAD51-MYC HsRAD52-FLAG – ABT836. (B) Both the yeast Rad52 and human RAD52 proteins self-interactPresence of genomic copies of the RAD52-FLAG or HsRAD52-FLAG fusion genes, and/or plasmid copies of RAD52-MYC or HsRAD52-MYC in the yeast strains used to make the whole cell extracts are denoted with a (+) at the top of the figure. The top panel depicts proteins IP by anti-MYC antibody and IB with anti-FLAG antibody. The bottom three panels depict the proteins in 10% INPUT. Signals corresponding to the 55 kDa Rad52-FLAG, 49 kDa HsRAD52-FLAG, 53 kDa Rad52-MYC, 47 kDa HsRAD52-MYC and 37 kDa GAPDH proteins are denoted on the right side of the figure. Strains used in this analysis: WT – W961-5A; RAD52-FLAG – ABM559; HsRAD52-FLAG – ABX3684-12B; RAD52-MYC – ABT823; HsRAD52-MYC – ABT839; RAD52-FLAG RAD52-MYC – ABT824; HsRAD52-FLAG HsRAD52-MYC – ABT838.
Monomers of Rad52 and HsRAD52 are capable of self-association and assembly into multimeric complexes that are proposed to have functional significance (29,53–57). As both RAD52 and HsRAD52 support HR in budding yeast, we used co-IP to address the capacity of Rad52 and HsRAD52 to undergo self-interaction in our strains. Extracts of strains containing both Rad52-FLAG and Rad52-MYC displayed co-IP when either anti-FLAG or anti-MYC was used for immunoprecipitation, and strains containing HsRAD52-FLAG and HsRAD52-MYC performed similarly (Figure 4B; Supplementary Figure S5B). This indicates that both Rad52 and HsRAD52 can self-associate in our strains, and suggests that multimerization could be a factor in the function of both Rad52 and HsRAD52 in HR in budding yeast.
HsRAD52 associates with DSBs that undergo repair by conservative HR
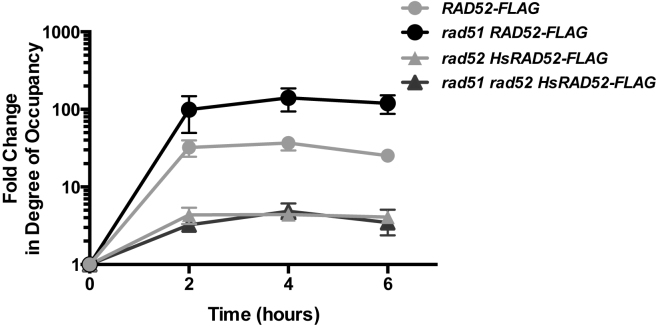
Previous investigations have shown that Rad52 associates with a DSB undergoing repair by conservative HR (51,52). As HsRAD52 also supports repair of a DSB by conservative HR, we used ChIP to examine the association of HsRAD52-FLAG and Rad52-FLAG with a DSB at the HIS3 locus that can be repaired by EGC (Figure 3A). Importantly, Rad52-FLAG and HsRAD52-FLAG supported frequencies of EGC that were less than 2-fold different from those supported by the untagged proteins, indicating that the ChIP experiments were reporting the behavior of the proteins under normal recombination conditions (Supplementary Table S4). Rad52-FLAG accumulated at the HIS3 locus following HO endonuclease cleavage of the his3-Δ3΄-HOcs substrate, peaking at a 37-fold level of enrichment after 4 h, which is consistent with the previously published results supporting a direct role for Rad52 in conservative DSB repair (Figure 5). In rad52 mutant cells expressing HsRAD52-FLAG, HsRAD52-FLAG accumulated at the HIS3 locus following DSB formation, with a peak level of enrichment of 4.4-fold after 4 h. This indicates that like Rad52, HsRAD52 associates with DSBs while propagating their repair in budding yeast cells. This peak level of enrichment of HsRAD52-FLAG, while significant may also indicate that HsRAD52 associates with DSBs more weakly than Rad52.
Figure 5.
Rad52 and HsRAD52 associate with a DSB undergoing repair by EGCWhole cell extracts prepared from cells containing the EGC assay components and expressing Rad52-FLAG or HsRAD52-FLAG were collected before and various times after induction of HO-endonuclease and subjected to ChIP using anti-FLAG antibody. Immunoprecipitated and input DNA was probed for sequences adjacent to the DSB at the HIS3 locus or uncut sequences at the SAM1 locus by qPCR. Fold changes in degree of occupancy by Rad52-FLAG or HsRAD52-FLAG at the HIS3 locus relative to the SAM1 locus were normalized to those of a control strain lacking FLAG-tagged proteins and plotted against time after the initiation of the expression of HO endonuclease. Relevant strain genotypes are indicated in the legend. Strains used in this analysis: Untagged – ABX3834-2D; RAD52-FLAG – ABX3844-23D; rad51Δ RAD52-FLAG – ABX3879-17B; rad52Δ HsRAD52-FLAG – ABX3885-17B; rad51Δ rad52Δ HsRAD52-FLAG – ABX3885-32C.
RAD52 and RAD51 together supported DSB repair by EGC, suggesting that the association of Rad52-FLAG with the DSB might be affected in rad51 mutant cells (Figure 3B). Interestingly, Rad52-FLAG displayed a statistically significant (P < 0.0001), 6-fold greater peak level of association with the DSB in rad51 mutant cells than in cells with a wild-type RAD51 gene, indicating that Rad51 can either partially inhibit association of Rad52 with DSBs, or displace Rad52 from them (Figure 5). In contrast, HsRAD52-FLAG displayed nearly identical kinetics of association, and peak levels of enrichment at the DSB when HsRAD52-FLAG was expressed in rad52 single, or rad51 rad52 double mutant cells (p = 0.923), consistent with HsRAD52 supporting EGC independently of RAD51.
DISCUSSION
By expressing a cDNA copy of the HsRAD52 gene in strains of budding yeast lacking an endogenous RAD52 gene we have observed partial suppression of their ionizing radiation sensitivity and HR defects. This HR was independent of most of the endogenous HR machinery, including the central yeast recombinase, Rad51, indicating that HsRAD52 drove mitotic HR. Consequently, these strains provide an excellent platform for studying the action of HsRAD52 in a living system. This is in contrast to studying HsRAD52 in mammalian systems, which is hampered by its minor role in intact cells and its requirement for the growth and viability of cells with mutations in canonical HR pathway genes (20,58–60).
While the N-termini of HsRAD52 and Rad52 are structurally conserved, the C-termini bear little similarity (Supplementary Figure S1). Importantly, the C-terminus of Rad52 is required for resistance of budding yeast to the radio-mimetic compound, methyl methanesulfonate and conservative DSB repair by the HR machinery (31,32,61). The C-terminus of Rad52 is also required for the physical interaction between Rad52 and Rad51, implicating this interaction in methyl methanesulfonate resistance and conservative DSB repair in budding yeast (28,62). HsRAD52 also interacts with HsRAD51 through its C-terminus, although the region of HsRAD52 that governs its interaction with HsRAD51 bears no significant amino acid sequence homology with the region of Rad52 that interacts with Rad51 (27). Given the lack of sequence homology between the C-termini of Rad52 and HsRAD52 we were surprised to observe that the expression of HsRAD52 substantially suppressed the defects in ionizing radiation sensitivity and HR conferred by the rad52-null mutation in budding yeast (Figures 2 and 3; Supplementary Table S4).
Genetic analysis demonstrated that the conservative HR supported by HsRAD52 was independent of both RAD51 and its paralog, RAD55, indicating that HsRAD52 worked independently from much of the strand exchange machinery in budding yeast, while the conservative HR supported by the endogenous RAD52 gene required both RAD51 and RAD55 (Figure 3). The relationship between RAD52 and RAD51 in conservative HR was reflected in the strong interaction between Rad52 and Rad51 observed in our yeast two-hybrid and co-IP assays, whereas the failure to detect an interaction between HsRAD52 and Rad51 supports our findings that HsRAD52 functions separately from RAD51 in support of conservative HR (Figure 4; Supplementary Figures S4 and S5). In keeping with the results of our HR and protein–protein interaction assays, the association of HsRAD52 with the his3-Δ3΄-HOcs substrate after DSB formation observed by ChIP was also independent of RAD51, supporting a direct role for HsRAD52 in conservative HR that occurs independently of the central strand exchange protein (Figure 5). Curiously, Rad52 was observed to associate at a 6-fold higher level with the his3-Δ3΄-HOcs substrate after DSB formation in rad51 mutant cells than in cells with a wild-type RAD51 gene, suggesting that Rad51 either inhibits some interaction of Rad52 with DSBs, or to some degree displaces Rad52 from DSBs in our strains. As this increased level of association of Rad52-FLAG is observed in a strain that displays a frequency of EGC 385-fold below that in a strain with a wild-type RAD51 gene (Supplementary Table S4), we conclude that unlike HsRAD52, the association of Rad52 with a DSB cannot support substantial levels of EGC without Rad51. This is further supported by the observation that over-expression of RAD52 does not increase the frequency of EGC in rad51 mutant cells (Supplementary Figure S2; Supplementary Table S4). These results suggest that HsRAD52 may have evolved to support conservative HR independently of HsRAD51 as the result of having been liberated from the role as mediator of HsRAD51 by BRCA2 (15,17,63). Alternatively, Rad52 may have lost the ability to work independently of Rad51 in conservative HR in the process of evolving the capacity to act as a Rad51 mediator.
In contrast to RAD51 and RAD55, RAD54 played a significant though limited role in the conservative HR supported by HsRAD52 (Figure 3C; Supplementary Table S4). RAD54 mediates multiple aspects of Rad51 function in vitro, and RAD51-dependent DSB repair by conservative HR in budding yeast, suggesting that RAD54 may support one or more stages of HsRAD52-dependent HR (51,52,64). In vitro studies of Rad54 have demonstrated that it can stimulate Rad51-mediated D-loop formation, which is thought to be required for heteroduplex formation during conservative DSB repair (12,65). Since in vitro studies have documented that HsRAD52 can propagate a variety of similar activities, this presents the fascinating possibility that the molecular basis for the HsRAD52-dependent conservative HR we have observed in budding yeast may be a strand exchange-like mechanism (66–71). Alternatively, the annealing activity of HsRAD52 may stimulate the formation of heteroduplex in tandem with the heteroduplex pump activity of Rad54 (65).
The capacity of HsRAD52 to support conservative HR in human cells that carry hypomorphic mutations in the canonical HR and tumor suppressor genes BRCA1, BRCA2 and PALB2 suggests a potential role for HsRAD52 in tumor suppression (20,58,72). While HsRAD52 may exert little if any effect when the canonical HR pathway is fully functional, genetic or epigenetic disruption of the canonical pathway would uncover the otherwise occult function of HsRAD52 in genome stabilization. If genome stabilization is the basis of the tumor suppressive effect of the canonical HR machinery, HsRAD52-dependent genome stabilization may also suppress tumorigenesis when the canonical HR machinery is attenuated. Genetic or epigenetic disruption of HsRAD52-dependent HR in cells with an attenuated canonical HR pathway might then enhance tumorigenesis, suggesting a potential role for HsRAD52 in altering the tumorigenicity of pathogenic variants of canonical HR pathway genes.
The synthetic lethality observed upon simultaneous attenuation of expression of HsRAD52 and canonical HR pathway genes in human cells suggests that inhibiting the activity of HsRAD52 in tumors with reduced canonical HR pathway function would be a worthwhile therapeutic goal (20,58,72). Several studies using small molecule or peptide aptamer inhibitors of HsRAD52 describe an inhibition of the growth of human tumor cells possessing attenuated canonical HR pathway function, demonstrating the utility of this strategy (73–75). High throughput screening with the ‘humanized’ yeast strains described here could be used to identify chemical compounds that inhibit the function of HsRAD52, while detailed studies of HsRAD52 function at the molecular level could reveal the specific effects of known inhibitors. The recent linkage of pathogenic germline mutations in canonical HR pathway genes to a broad range of pediatric cancers suggests that HsRAD52 inhibitors could ultimately play a prominent role in cancer treatment (76,77).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Boldin, A. Adamson, S. Neuhausen and M. Lisby for plasmids used in this study. The authors also thank P. Singh, M. Boldin, R.-J. Lin, L. Malkus, S. Neuhausen, J. Weitzel, J. Stark, R. Rothstein, W.-D. Heyer and W. Wright for helpful discussions. The authors further thank E. Wolf, P. Fischhaber, X. Zhao, W.-D. Heyer, M. Lieber, N. Pannunzio, D. Meyer and several anonymous reviewers for helpful comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
H. N. and Frances C. Berger Foundation Fellowship [to A. D. C.]; Morgan and Helen Chu Graduate Student Fellowship [to L. C. L.]; Molecular Biology Program of Pomona College; Beckman Research Institute of the City of Hope. Funding for open access charge: Beckman Research Institute of the City of Hope.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cox M.M. Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8173–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity. 2002; 88:125–141. [DOI] [PubMed] [Google Scholar]
- 3.Gowen L.C., Johnson B.L., Latour A.M., Sulik K.K., Koller B.H.. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 1996; 12:191–194. [DOI] [PubMed] [Google Scholar]
- 4.Lim D.S., Hasty P.. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996; 16:7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M.. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999; 4:511–518. [DOI] [PubMed] [Google Scholar]
- 6.Moynahan M.E., Pierce A.J., Jasin M.. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001; 7:263–272. [DOI] [PubMed] [Google Scholar]
- 7.Sharan S.K., Morimatsu M., Albrecht U., Lim D.S., Regel E., Dinh C., Sands A., Eichele G., Hasty P., Bradley A.. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997; 386:804–810. [DOI] [PubMed] [Google Scholar]
- 8.Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T.. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita T., Yoshimura Y., Yamamoto A., Murata K., Mori M., Yamamoto H., Matsushiro A.. A mouse homolog of the Escherichia coli recA and Saccharomyces cerevisiae RAD51 genes. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6577–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakash R., Zhang Y., Feng W., Jasin M.. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015; 7:a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Story R.M., Bishop D.K., Kleckner N., Steitz T.A.. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993; 259:1892–1896. [DOI] [PubMed] [Google Scholar]
- 12.Symington L.S., Rothstein R., Lisby M.. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014; 198:795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumann P., Benson F.E., West S.C.. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996; 87:757–766. [DOI] [PubMed] [Google Scholar]
- 14.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994; 265:1241–1243. [DOI] [PubMed] [Google Scholar]
- 15.Jensen R.B., Carreira A., Kowalczykowski S.C.. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010; 467:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Heyer W.D.. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008; 18:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Doty T., Gibson B., Heyer W.D.. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010; 17:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schutte M., da Costa L.T., Hahn S.A., Moskaluk C., Hoque A.T., Rozenblum E., Weinstein C.L., Bittner M., Meltzer P.S., Trent J.M. et al. . Identification by representational difference analysis of a homozygous deletion in pancreatic carcinoma that lies within the BRCA2 region. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5950–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N., Nguyen K., Seal S., Tran T., Averill D. et al. . Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994; 265:2088–2090. [DOI] [PubMed] [Google Scholar]
- 20.Feng Z., Scott S.P., Bussen W., Sharma G.G., Guo G., Pandita T.K., Powell S.N.. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer L.M., Koonin E.V., Aravind L.. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortensen U.H., Erdeniz N., Feng Q., Rothstein R.. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics. 2002; 161:549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploquin M., Bransi A., Paquet E.R., Stasiak A.Z., Stasiak A., Yu X., Cieslinska A.M., Egelman E.H., Moineau S., Masson J.Y.. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 2008; 18:1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuta R., LaSalle J.M., Cheng H.L., Shinohara A., Ogawa H., Copeland N., Jenkins N.A., Lalande M., Alt F.W.. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park M.S., Ludwig D. L., Stigger E., Lee S.-H.. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 1996; 271:18996–19000. [DOI] [PubMed] [Google Scholar]
- 26.Reddy G., Golub E.I., Radding C.M.. Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat. Res. 1997; 377:53–59. [DOI] [PubMed] [Google Scholar]
- 27.Shen Z., Cloud K.G., Chen D.J., Park M.S.. Specific interactions between the human RAD51 and RAD52 proteins. J. Biol. Chem. 1996; 271:148–152. [DOI] [PubMed] [Google Scholar]
- 28.Milne G.T., Weaver D.T.. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993; 7:1755–1765. [DOI] [PubMed] [Google Scholar]
- 29.Shen Z., Peterson S.R., Comeaux J.C., Zastrow D., Moyzis R.K., Bradbury E.M., Chen D.J.. Self-association of human RAD52 protein. Mutat. Res. 1996; 364:81–89. [DOI] [PubMed] [Google Scholar]
- 30.Sugiyama T., New J.H., Kowalczykowski S.C.. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manthey G.M., Bailis A.M.. Rad51 inhibits translocation formation by non-conservative homologous recombination in Saccharomyces cerevisiae. PLoS One. 2010; 5:e11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukamoto M., Yamashita K., Miyazaki T., Shinohara M., Shinohara A.. The N-terminal DNA-binding domain of Rad52 promotes RAD51-independent recombination in Saccharomyces cerevisiae. Genetics. 2003; 165:1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F., Fink G.R., Hicks J.B.. Methods in Yeast Genetics. 1986; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 34.Sambrook J., Russell D.W.. Molecular Cloning: A Laboratory Manual. 2001; 3rd edn NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 35.Hoffman C.S., Winston F.. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987; 57:267–272. [DOI] [PubMed] [Google Scholar]
- 36.Pannunzio N.R., Manthey G.M., Liddell L.C., Fu B.-X., Roberts C.M., Bailis A.M.. Rad59 regulates association of Rad52 with DNA double-strand breaks. Microbiologyopen. 2012; 1:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 38.Park M.S. Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem. 1995; 270:15467–15470. [DOI] [PubMed] [Google Scholar]
- 39.Resnick M.A. Genetic control of radiation sensitivity in Saccharomyces cerevisiae. Genetics. 1969; 62:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resnick M.A., Martin P.. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. 1976; 143:119–129. [DOI] [PubMed] [Google Scholar]
- 41.Resnick M.A. The repair of double-strand breaks in chromosomal DNA of yeast. Basic Life Sci. 1975; 5B:549–556. [DOI] [PubMed] [Google Scholar]
- 42.Moreau S., Morgan E.A., Symington L.S.. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001; 159:1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White C.I., Haber J.E.. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990; 9:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997; 272:28194–28197. [DOI] [PubMed] [Google Scholar]
- 45.Amitani I., Baskin R.J., Kowalczykowski S.C.. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006; 23:143–148. [DOI] [PubMed] [Google Scholar]
- 46.Hays S.L., Firmenich A.A., Berg P.. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugawara N., Ivanov E.L., Fishman-Lobell J., Ray B.L., Wu X., Haber J.E.. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995; 373:84–86. [DOI] [PubMed] [Google Scholar]
- 48.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997; 11:1111–1121. [DOI] [PubMed] [Google Scholar]
- 49.Bailis A.M., Maines S., Negritto M.T.. The essential helicase gene RAD3 suppresses short-sequence recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995; 15:3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pohl T.J., Nickoloff J.A.. Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol. Cell. Biol. 2008; 28:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugawara N., Wang X., Haber J.E.. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003; 12:209–219. [DOI] [PubMed] [Google Scholar]
- 52.Wolner B., van Komen S., Sung P., Peterson C.L.. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003; 12:221–232. [DOI] [PubMed] [Google Scholar]
- 53.Donovan J.W., Milne G.T., Weaver D.T.. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 1994; 8:2552–2562. [DOI] [PubMed] [Google Scholar]
- 54.Hays S.L., Firmenich A.A., Massey P., Banerjee R., Berg P.. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol. Cell. Biol. 1998; 18:4400–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shinohara A., Shinohara M., Ohta T., Matsuda S., Ogawa T.. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998; 3:145–156. [DOI] [PubMed] [Google Scholar]
- 56.Van Dyck E., Hajibagheri N.M., Stasiak A., West S.C.. Visualisation of human Rad52 protein and its complexes with hRad51 and DNA. J. Mol Biol. 1998; 284:1027–1038. [DOI] [PubMed] [Google Scholar]
- 57.Van Dyck E., Stasiak A.Z., Stasiak A., West S.C.. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999; 398:728–731. [DOI] [PubMed] [Google Scholar]
- 58.Lok B.H., Carley A.C., Tchang B., Powell S.N.. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013; 32:3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rijkers T., Van Den Ouweland J., Morolli B., Rolink A.G., Baarends W.M., Van Sloun P.P., Lohman P.H., Pastink A.. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998; 18:6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M.. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004; 24:9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boundy-Mills K.L., Livingston D.M.. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics. 1993; 133:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krejci L., Song B., Bussen W., Rothstein R., Mortensen U.H., Sung P.. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 2002; 277:40132–40141. [DOI] [PubMed] [Google Scholar]
- 63.van Veelen L.R., Essers J., van de Rakt M.W., Odijk H., Pastink A., Zdzienicka M.Z., Paulusma C.C., Kanaar R.. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat. Res. 2005; 574:34–49. [DOI] [PubMed] [Google Scholar]
- 64.Heyer W.D., Li X., Rolfsmeier M., Zhang X.P.. Rad54: the Swiss Army knife of homologous recombination?. Nucleic Acids Res. 2006; 34:4115–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wright W.D., Heyer W.D.. Rad54 Functions as a Heteroduplex DNA Pump Modulated by Its DNA Substrates and Rad51 during D Loop Formation. Mol. Cell. 2014; 53:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bi B., Rybalchenko N., Golub E.I., Radding C.M.. Human and yeast Rad52 proteins promote DNA strand exchange. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9568–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kagawa W., Kagawa A., Saito K., Ikawa S., Shibata T., Kurumizaka H., Yokoyama S.. Identification of a second DNA binding site in the human Rad52 protein. J. Biol. Chem. 2008; 283:24264–24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kagawa W., Kurumizaka H., Ikawa S., Yokoyama S., Shibata T.. Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem. 2001; 276:35201–35208. [DOI] [PubMed] [Google Scholar]
- 69.Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S.. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell. 2002; 10:359–371. [DOI] [PubMed] [Google Scholar]
- 70.Kumar J.K., Gupta R.C.. Strand exchange activity of human recombination protein Rad52. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9562–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Navadgi V.M., Dutta A., Rao B.J.. Human Rad52 facilitates a three-stranded pairing that follows no strand exchange: a novel pairing function of the protein. Biochemistry. 2003; 42:15237–15251. [DOI] [PubMed] [Google Scholar]
- 72.Lok B.H., Powell S.N.. Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin. Cancer Res. 2012; 18:6400–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chandramouly G., McDevitt S., Sullivan K., Kent T., Luz A., Glickman J.F., Andrake M., Skorski T., Pomerantz R.T.. Small-Molecule Disruption of RAD52 Rings as a Mechanism for Precision Medicine in BRCA-Deficient Cancers. Chem. Biol. 2015; 22:1491–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cramer-Morales K., Nieborowska-Skorska M., Scheibner K., Padget M., Irvine D.A., Sliwinski T., Haas K., Lee J., Geng H., Roy D. et al. . Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013; 122:1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan K., Cramer-Morales K., McElroy D.L., Ostrov D.A., Haas K., Childers W., Hromas R., Skorski T.. Identification of a Small Molecule Inhibitor of RAD52 by Structure-Based Selection. PLoS One. 2016; 11:e0147230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Couzin-Frankel J. A Cancer Legacy. Science. 2016; 351:440–443. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J., Walsh M.F., Wu G., Edmonson M.N., Gruber T.A., Easton J., Hedges D., Ma X., Zhou X., Yergeau D.A. et al. . Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. of Med. 2015; 373:2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.