Abstract
The biogenesis of ribosomes in vivo is an essential process for cellular functions. Transcription of ribosomal RNA (rRNA) genes is the rate-limiting step in ribosome biogenesis controlled by environmental conditions. Here, we investigated the role of folate antagonist on changes of DNA double-strand breaks (DSBs) landscape in mouse embryonic stem cells. A significant DSB enhancement was detected in the genome of these cells and a large majority of these DSBs were found in rRNA genes. Furthermore, spontaneous DSBs in cells under folate deficiency conditions were located exclusively within the rRNA gene units, representing a H3K4me1 hallmark. Enrichment H3K4me1 at the hot spots of DSB regions enhanced the recruitment of upstream binding factor (UBF) to rRNA genes, resulting in the increment of rRNA genes transcription. Supplement of folate resulted in a restored UBF binding across DNA breakage sites of rRNA genes, and normal rRNA gene transcription. In samples from neural tube defects (NTDs) with low folate level, up-regulation of rRNA gene transcription was observed, along with aberrant UBF level. Our results present a new view by which alterations in folate levels affects DNA breakage through epigenetic control leading to the regulation of rRNA gene transcription during the early stage of development.
INTRODUCTION
In eukaryotes, ribosomal RNA (rRNA) genes are the most actively transcribed genes. rRNA genes account for up to 80% of all cellular RNA transcription (1,2). Regulation of cell growth ultimately depends on the control of new ribosome synthesis while rRNA gene transcription is the major rate limiting step of ribosome biogenesis (3–5) and dysregulation of ribosome biogenesis has been implicated in many diseases (6–9).
In typical human cells, there are ∼400 copies of a 43-kb rRNA gene unit arranged in tandem repeats with a head-to-tail orientation on the five acrocentric chromosomes (10,11). Each rRNA gene unit can be divided into two major parts: coding region and non-coding intergenic spacer (IGS) region. The former contains the sequences that encoding 18S, 5.8S and 28S rRNAs, the latter contains a large number of repeats (LINEs, SINEs, Alu elements) and also the space promoter and core promoter of rRNA gene. 18S, 5.8S and 28S rRNAs result from the processing of one precursor transcript, pre-ribosomal RNA (pre-rRNA). The IGS contains a spacer promoter, the core promoter and a large number of simple repeats (10).
A lot of work have been done in the past focusing the relationship between chromatin structures and transcription. Several modifications of chromatin components including lysine-specific demethylase 2A (KDM2A), lysine-specific demethylase 1 (LSD1) have been identified (12–15). However, although the development of CHIP-seq technique has revealed relevant protein occupancy throughout the genome, the rRNA gene units with high repetitive regions are not included in reference genome assemblies. Only recently Zentner et al. (16) has established a high-resolution map of chromatin structure of the rRNA genes combining both the epigenetic marks and RNA-seq data, providing a useful tool for further studies on chromatin-mediated regulation of rRNA genes.
Recent works have highlighted the importance of epigenetic control of rRNA gene transcription. rRNA gene units are indicated to be the most fragile regions in the human genome. Tchurikov et al. (17) observed a strong link between hot spots of DNA double-strand breaks (DSBs) and epigenetic regulation of rRNA gene transcription. Hot spots of DSBs are located exclusively in IGS regions of rRNA genes and they coincide with regions possessing active chromatin marks-H3K4me3 (a promoter-specific histone modification associated with active transcription). DNA breaks serve as signals to recruit regulators required for transcription specificity and efficiency. Among them, poly(ADP-ribose) polymerase 1 (PARP1) and heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) are found to bind at hot spots of spontaneous DSBs in rRNA genes (18,19). A recently published crystal structure of a DSB underlies the DNA-dependent activation mechanism of human PARP1 (20).
Folate is a micronutrient that plays a key role in ensuring normal cellular proliferation, especially throughout early embryonic development (21). A worldwide Recommendation on Daily Allowance level of folate intake has been proposed (22). Enormous efforts have been made to understand the epidemiology and experimental evidence that links folate status to many diseases, including cancers, neurological conditions, and congenital malformations (23–27). Folate can serve as the source of epigenetic modifications, affecting the methylation of DNA, proteins and lipids through S-adenosylmethionine (SAM) mediated one-carbon transfer reaction. SAM is the unique methyl donor for many methylation modifications, including histone methylation (28–31). Although folate levels contribute to the accumulation of DSBs in the genome (22,32–35), the effects of folate metabolism disorder on levels of DSBs in rRNA gene units and its transcriptional regulation are unknown.
Here, we report the identification of a DSBs landscape in mouse embryonic stem cells (mESCs) using a folate antagonist (methotrexate, MTX) that has been implicated as a risk factor for neural tube defects (NTDs). We detected strong and significant DSB enrichment in rRNA genes in cells treated with MTX. Similar to the reported hot spots of DSBs in human rRNA genes in H1-hESCs, the hot spots of DSBs that we detected in mESCs were non-randomly distributed inside rRNA gene repeats and were located exclusively in several specific regions of IGS regions. We showed that the DNA breakage sites of rRNA gene units corresponding to histone H3K4me1 mark sites were exclusively elevated after MTX treatment. In addition, MTX treatment resulted in an increase of recruitment of UBF to rRNA genes leading to its elevated transcription. Our results indicated the importance of folinic acid in genomic stability and showed that folate deficiency leads to increases in rRNA gene transcription and over-expression of UBF in fetal NTDs. Together, our findings highlight how alterations in folate level affect DNA breakage through epigenetic control leading to the regulation of rRNA genes transcription during the early stage of development, and offer new insights into the etiology of NTDs.
MATERIALS AND METHODS
Cell cultures and methotrexate treatment
Mouse embryonic stem cells Sv/129 were kindly provided by Xuanwu Hospital (Beijing, China). The cells were maintained on mitotically inactivated primary mouse embryonic fibroblasts prior to cultivation under feeder-free conditions. Flasks were coated with 0.2% gelatin (Sigma-Aldrich) and the mESCs were maintained in complete growth medium: Dulbecco's modified Eagle's medium (Invitrogen), 0.1 mM β-mercaptoethanol (Life Technology), 0.1 mM non-essential amino acids (Invitrogen), 0.1 mM Glutamax (Invitrogen), 1000 U/ml mouse leukemia inhibitory factor (Millipore), 15% ES-cell qualified fetal bovine serum (Biochrom). The cells were maintained at 37°C in a humidified atmosphere with 5% CO2, and passaged every 2–3 days. A split ratio of 1:4 to 1:7 was performed.
After three passages under normal conditions, the colorimetric tetrazolium dye procedure, commonly referred to as the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, was performed to evaluate the cytotoxicity of methotrexate (MTX) concentrations from 0.001 to 100 μM at 24 h against mESCs. The cells were plated into 96-well plates at densities of 1 × 104 cells per well.
A total volume of 200 μl of sample treatment was added to each well and incubated for 24 h prior to addition of 20 μl of 5 mg/ml MTT (Sigma-Aldrich) to each well. The cells were then left to incubate for 4 h, after which DMSO solution was added to each well to dissolve the formazan crystals. The plates were read using a microplate reader at 570 nm (BioTek Instruments). The results were expressed as a percentage of the viability of control cells ± standard error of the mean (SEM) in three independent experiments and the half inhibition concentration (IC50) was calculated using the SPSS 16.0 package.
Cell cycle and apoptosis analyses by flow cytometry
For the flow cytometry, mESC cells were seeded overnight and treated with a serial gradient concentration of MTX around the IC50 value for 24 h. Untreated control cells were prepared simultaneously. After the treatment period, cells were harvested, washed, and stained using a BD Cycletest™ Plus DNA reagent kit (Becton Dickinson) according to the manufacturer's protocol. The cell cycle phase was analyzed using BD CellQuest Pro software by FACSCalibur™ (Becton Dickinson).
For apoptosis analyses, mESCs were trypsinized and washed with PBS, then resuspended in PBS, and ice-cold 100% ethanol was added dropwise to obtain a final ethanol concentration of 75% for fixing cells overnight at −20°C. Cells were centrifuged at 1000 × g at 4°C for 5 min, then washed with PBS and resuspended in propidium iodide (PI) working solution (PBS containing 100 μg/ml of RNase A, 50 μg/ml of PI and 0.1% Trition X-100) for 30 min in the dark at 4°C. Cells were filtered through a 35-m strainer cap before being subjected to fluorescence-activated cell sorter (FACS) analysis.
Detection of gamma-phosphorylated H2AX histone by flow cytometry
To analyze the relationship between DSBs and cell cycle, we performed gH2AX-flow cytometry combined with cell cycle analysis. In brief, MTX-treated mESCs and normal cells were collected, fixed with 100% methanol for 5 min, and then permeabilized with 0.1% PBS-Tween for 20 min. The cells were then incubated in 1× PBS containing 10% normal goat serum and 0.3 M glycine to block non-specific protein–protein interactions followed by primary antibody for 30 min. Secondary antibody coupled with DyLight 488 was added for 30 min in the dark. After washing with PBS, PI working solution was used for the cell cycle analysis. Isotype control antibody (1 μg/1 × 106 cells) was also used in the experiment. Acquisition of >10, 000 events was performed. BD CellQuest Pro software by FACSCalibur™ (BD, Franklin Lakes, USA) was used for the analysis.
Immunofluorescence
mESCs were seeded onto a glass bottom cell culture dish (NEST). On the next day, cells were incubated with 0.12 μM MTX for 24 h. Cells were washed with PBS and treated as follows: incubation with 100% methanol chilled to −20°C for 20 min. Then permeabilized in 0.5% Triton X-100 for 15 min at room temperature and blocked with 1× PBS containing 10% normal goat serum and 0.3 M glycine for 60 min.
Cells were incubated with primary antibody diluted in PBS containing 1% BSA and 0.1% Tween overnight at 4°C. After washing, secondary antibodies with Alexa Fluor 568 were used for 1 h incubation at room temperature in the dark. Nuclei were counterstained with DAPI. Images were captured on a Zeiss LSM710 confocal microscope.
Single-cell electrophoresis assay (comet assay)
The comet assay was performed as described (36). mESCs were seeded on six-well plates at a density of 4 × 105 cells/well. After 24 h incubation with MTX, cells from each well were trypsinized, pelleted, redissolved in PBS mixed with 0.8% LMP agarose (Invitrogen). A 100 μl aliquot of each cell suspension was spread onto a precoated glass slide and covered with a cover glass, and incubated for 30 min at 4°C. The cells were then lysed in pre-cold lysis buffer (2.5 mol/l NaCl, 0.1 mol/l Na2EDTA, 10 mmol/l Tris–HCl, 1% Triton X-100, 10% DMSO, pH 10) at 4°C for at least 2 h. Electrophoresis was performed on ice for 30 min at 25 V. The gels were neutralized twice with 0.4 M Tris–HCl (pH 7.5) for 5 min and stained with 2 μg/ml ethidium bromide in the dark. One hundred cells were sorted for measuring by image analysis (Nikon TE-2000S, Japan) equipped with a UV filter. DNA migration was assessed using the CASP software package. For a direct comparison of the influence of MTX on DNA damage, we used length of DNA tail and tail% DNA.
DSB enrichment workflow
mESCs were collected after MTX treatment. The DSB workflow is summarized in Figure 2A. The assay for preparing intact nuclei embedded in low melting point (LMP) agarose was performed as described previously (37). In brief, about 5 million mESCs as single-cell suspensions were centrifuged and washed by cold 1× PBS. Then, the cells were lysed in lysis buffer containing 10 mM Tris–Cl (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630. The suspension was centrifuge immediately at 500 × g for 10 min at 4°C. We carefully embedded the nuclei in 0.8% LMP agarose melted in sterile 50 mM EDTA, pH 8.0), and washed them with fresh LIDS buffer containing 10 mM Tris–Cl (pH 8.0), 100 mM EDTA, and 1% lauryl sulfate lithium salt overnight at 37°C. After washing overnight, the LMP agarose was washed twice by 10 mM Tris–Cl and 0.25 mM EDTA (pH 8.0), with 60 rpm shaking at room temperature. Blunt-ending the DSB ends was performed in 200 μl final volume using 10× T4 DNA polymerase buffer, 2.5 mM dNTPs, T4 DNA polymerase (5 U/μl), T4PNK (10U/μl), ATP (10 mM) and BSA (100×) for 4 h at room temperature.
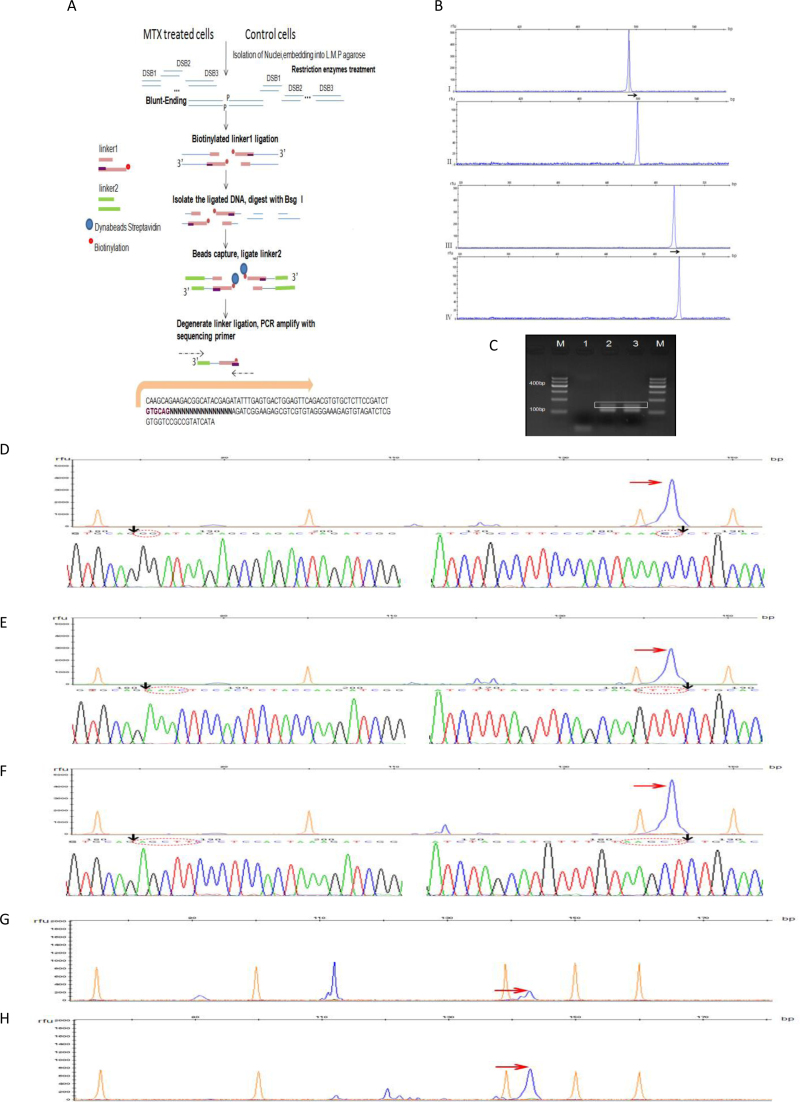
Figure 2.
DSBs enrichment workflow and specificity of the DNA DSBs (A) DSBs enrichment workflow by MTX treatment or Restriction endonuclease digestion for quality control. Fragments released from the streptavidin beads were amplified by PCR using sequencing primers and sequenced. (B) Quality control of in situ digestion and blunt-ending by capillary electrophoresis. The top two traces are for the endonuclease digestion and blunt-ending performed in liquid; the bottom two traces are in low melting point agarose gel. I represents the size of digestion product of a 567-bp fluorescence-labeled DNA fragment by restriction digestion while II shows the size of digestion product after blunt-ending; III and IV represent the above reactions respectively in low melting point agarose gel. The arrow in black represents the complete blunt-ending. X-axis represents the size of fragments(bp), Y-axis represents the detector signal of peak(rfu). (C) DSBs enrichment products separated by agarose gel electrophoresis indicated by white box. (D–F) Capillary electrophoresis to detect DSB enrichment products after SbfI (D), PmeI (E) and HindIII (F) digestion. TA clone sequencing confirmed the results. The arrow in red indicates the DSB enrichments on Capillary electrophoresis; the circle marked with red-dotted lines shows the restriction sites; the arrow in black shows the ligation point. X-axis represents the size of fragments(bp), while Y-axis represents the detector signal of peak(rfu). (G, H) Capillary electrophoresis to detect DSB enrichment products of normal mESCs cultured in complete medium (G) and cultured in complete medium with 0.12 μM MTX (H). The arrow in red indicates the DSB enrichments. X-axis represents the size of fragments(bp), while Y-axis represents the detector signal of peak(rfu).
The DNA was extracted carefully by phenol:chloroform and precipitated from the LMP agarose by alcohol. The DNA was ligated to 20 pmol biotinylated linker 1 in a 100 ul reaction volume containing 400 U of T4 DNA ligase by incubation at 16°C overnight. We used 0.8% UltraPure LMP Agarose gel containing 10 mM EDTA and 1× TBE to leave out the unligated linkers. After gel purification, the ligated DNA with linker 1 was treated with MmeI (NEB) according to the manufacturer's instructions. The biotinylated ends were captured with 50 μl of streptavidin-coated paramagnetic beads (Dynabeads® M-280 Streptavidin, Invitrogen), and the DNA was then ligated to 60 pmol linker 2 in a 100 μl reaction volume containing 400 U of T4 DNA ligase by incubation at 16°C for 4 h. The precipitate was gently resuspended in 100 μl of 0.15 M NaOH at room temperature for 10 min and eluted with 30 μl 10 mM Tris–Cl (pH 8.0).
For the PCR amplification, the suspension was prepared using a Dynal bead suspension, Phusion polymerase (NEB), sequencing primers, and dNTP. The PCR cycles were as follows: initial denaturation at 98°C for 2 min, and 24 cycles of (98°C for 10 s, 65°C for 30 s, 72°C for 30 s), followed by extension at 72°C for 5 min. The DSB enrichment products were obtained by electrophoresis and gel-purified.
The restricted enzyme SbfI, HindIII or PmeI digestion on nucleis of normal cells in 0.8% LMP agarose at 37°C incubated overnight are used as the control for capturing the expected cut sites of DSB.
The details of linker 1, linker 2 and primers sequences are listed in Supplementary Table S1. The linkers were amplified as follows: 90°C for 1 min, and 32 cycles of (90°C for 30 s; −0.5°C/cycle; 89.5°C for 30 s; −0.5°C/cycle; 89°C for 30 s; −0.5°C/cycle), followed by 40°C for 1 h and 4°C hold.
Next-generation sequencing
We assessed quality and quantity of DSB enrichment on a 2100 Bioanalyzer (Agilent) using a High Sensitivity DNA Kit (Agilent) and by qPCR using a Kapa Library Quantification Kit (Kapa Biosystems). We generated clusters on the Illumina flow cell using the automatic cBot station and the TruSeq PE Cluster Kit v3-cBot-HS. Sequencing was carried out by synthesis on an Illumina HiSeq 2000 platform using TruSeq SBS Kit v3-HS chemistry. Samples were sequenced and an average of 12 M reads was obtained.
Alignment and analysis of sequencing data
Data were first converted to FASTQ format using Illumina Casava 1.8 software. Quality evaluation was performed using FastQC 0.10.1 software. Joint sequences (FASTX) were removed from the sequencing data, then filter out length. Sequences that contained the correct enzyme digestion sites were retained based on them meeting the conditions encoded in our in-house Perl script. We mapped the sequence onto the mouse genome sequence and alignment (mm10), to obtain valid sequences that aligned to the mouse genome. The statistics for each mapped sample are presented in Supplementary Information, Excel S1.
To analyze the data in an unbiased fashion, the data were median-smoothed in 10K to 1000K (step of 10K) as the bin for getting the valid read counts of each sample. Statistics on fold-changes or P values among them was calculated by HYPGEOMDIST in R (38).
For hypergeometric test, the relative parameters were the following: total number of mapped reads in MTX treatment; total number of mapped reads in MTX treatment and the control; total number of mapped reads in sliding window in MTX treatment only; total number of mapped reads in the sliding window in MTX treatment and the control.
Determining the number of bins with P < 0.05 and location of the DSB peaks. Based on width of 43K, the distribution and data were presented. Also, the fold change and P values of each sample in each 43K unit were provided (Supplementary Excel S5). Individual sample replicates using the hypergeometric test for overlap between significant regions (3 versus 1; 3 versus 2; 4 versus 1; 4 versus 2) were presented in Supplementary Information, Supplementary Table S6 and Supplementary Figure S7. We computed the abundance of various DNA repeat families inside the regions susceptible to MTX as compared to the rest of the genome using the hypergeometric test.
We used ANNOVAR and DAVID tools for gene annotation in the DSB enrichment regions (Supplementary Information, Excel S2). The reproducibility of the regions identified as susceptible to MTX for biological replicates were analyzed by GenometriCorr R package (17,39) (Supplementary Information, Supplementary Figure S7).
The H3K4me1, H3K4me3, H3K9me3 and H3K27me3 sequences and the sequences from the four samples described above were aligned to the mouse rRNA genes sequences. ‘MM10_plus_rRNA genes’ to evaluate the distribution of DSBs and peak calling and to compare the statistical differences between samples and the methylation data as described (16). Use F-seq to detect the peak for DSBs, H3K27me3, H3K9me3, H3K4me3 and H3K4me1 in the entire rRNA genes unit calculated the fold-change in the number of reads, then use GenometriCorr to calculate the correlation and draw heatmap in R package between the peaks (16,17).
Data sets
The mouse rRNA genes repeat sequence was obtained from GenBank (accession no. BK000964 of the MM10 genome assembly). ChIP-seq data for mapping histone modifications in mouse rRNA genes were obtained from the ENCODE Broad Histone track in the UCSC Genome Browser (H3K4me1, SRR002255; H3K4me3, SRR006831; H3K9me3, SRR007435; H3K27me3, SRR006818).
Detection of ongoing rRNA genes transcription
To reveal active polymerase (pol) I transcription foci, ongoing rRNA synthesis was measured by in situ run-on assays (40). Briefly, mESCs were incubated in complete medium containing 2 mM 5-fluorouridine (FUrd, Sigma-Aldrich) for 20 min at 37°C in 5% CO2 and fixed with methanol for 20 min at −20°C. Then, the cells were subjected to acid hydrolysis using 4 M HCl in 0.1% PBS-Tween for 2 h at 37°C in a water bath to denature the DNA. The cells were then incubated in 0.1% PBS–Tween containing 1% BSA, 10% normal goat serum, and 0.3 M glycine for 1 h. Finally, the cells were incubated with BrdU antibody (ab8039) overnight at 4°C followed by the secondary antibody with DyLight 488 used at a 1/250 dilution for 1 h. Fluorescence images were obtained using a confocal microscope (Zeiss LSM meta) with a 63× oil objective NA 1.3. Images were collected and analyzed using the software of LAS X 2D Analysis (adjust threshold, measurement create report). The numbers of cells with FUrd signals remaining in the nuclei were counted.
ChIP-qPCR
A SimpleChIP™ Enzymatic Chromatin IP Kit (CST) was used for the ChIP assays according to the manufacturer's protocol. Formaldehyde cross-linked chromatin was obtained from about 4 × 107 cells. Cross-linked chromatin was immunoprecipitated with antibodies to H3K4me1, H3K27me3, H3K4me3, H3K9me3 and UBF. Non-specific mouse IgG and H3 were used for negative and positive controls, respectively.
DNA-protein complexes were analyzed by qPCR with specific primers to amplify multiple regions of the rRNA genes. Several pairs of primers flanking the coding sequence and intergenic spacer were designed. (The primers used in this experiment are listed in Supplementary Table S2.)
qPCR was performed using SYBR SuperMix (TransGen Biotech) according to the manufacturer's instructions. Amplification, data acquisition, and analysis were carried out using a 7500 Fast Real-Time PCR System (Applied Biosystems). The percentage of DNA brought down by ChIP (percent input) was calculated as follows:
Percent input = 2% × 2(C[T] 2% input sample − C[T] IP sample) where C[T] = Ct = threshold cycle of the PCR reaction. Three independent ChIP experiments were performed for each analysis.
Subjects
All clinical samples were from the Lvliang area of Shanxi Province in northern China with informed consent from the patients or their families. The enrolled pregnant women were diagnosed by trained local clinicians using ultrasonography and then registered. The surgical procedures were performed as previously described (41). The epidemiological method was described in detail in our previous publication (42). The Ethics Board of Capital Institute of Pediatrics approved the study protocol.
Detection of folate level
Brain tissue from fetuses was collected, and folate levels were determined using Chemiluminescent Immunoenzyme Assay Access II (Beckman Coulter, Fullerton, Germany) as previously described (43). Briefly, tissue samples were homogenized with extraction buffer (TRIS buffered saline, A16792, Beckman) and ultra-sonicated. The samples were then centrifuged and the supernatant were added to the sample vials for folic acid detection.
Transcription analysis by RT-qPCR
Total RNA was extracted from cultured cells or frozen tissue samples at −80°C using Trizol reagent (Invitrogen). 1 μg of RNA was reversed transcribed using random primers and 1 μl (total 20 μl) of the resultant single-strand cDNA was used as the template. The qPCRs were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems) using SYBR SuperMix (TransGen Biotech). The data were analyzed using the 2−ΔΔCt method. The primers used in this experiment are listed in Supplementary Table S3.
Western blotting
Total protein extracted from cells or frozen tissue samples at -80°C was prepared using Total Protein Extraction (TPE™) according to the manufacturer's instructions (Sangon Biotech).
Histone extraction was performed as described previously (44). Briefly, nuclei were incubated with four volumes of 0.2 N sulfuric acid (H2SO4) overnight at 4°C. The supernatant was precipitated with 33% trichloroacetic acid (final concentration) and centrifuged at 12 000 × g for 20 min. The obtained pellet was washed with cold acetone and then dissolved in distilled water. Samples were run on 12% SDS-PAGE gels and then electro-transferred onto difluoride membranes (Hybond ECL, Amersham Biosciences), which had been blocked in 10% non-fat skimmed milk. The membranes were incubated with primary antibody overnight at 4°C the treated with secondary antibodies. The bands were detected using an enhanced chemiluminescence technique (Amersham Biosciences).
siRNA transfection
mESCs were transfected with UBF siRNA (sc-29515, Santa Cruz) targeting mouse UBF1/2. Alternatively, cells were transfected with control siRNA (sc-37007, Santa Cruz) as an experimental control. Transfections were performed according to the manufacturer's instructions (Santa Cruz). After transfection, the medium was replaced and the cells were incubated for 48–96 h depending on the following experiments.
Nano string nCounter assay
The NanoString nCounter was used to detect the number of transcripts in human brain tissues. Total RNA was extracted following the manufacturer's instructions (Cat. no. 74104, Qiagen) and gene specific probes were designed by the manufacturer (NanoString Technologies).
Hybridizations were carried out according to the nCounter Element 24-plex Assay Manual (NanoString Technologies). Gene expression data were filtered using quality control (QC) criteria according to the manufacturer's recommendations. Raw counts of QC-passed samples were normalized using three reference genes as internal controls (GAPDH, CLTC and GUSB). All QC and normalization procedures were performed using nSolver Analysis Software v2.0; all data were log2-transformed before further analysis. The Student's t-test was used to compare normalized expression values between normal and NTDs.
Antibodies
The following antibodies were used: rabbit polyclonal antibody against phospho S139 gamma H2A.X (ab2893, Abcam), rabbit monoclonal antibody against alpha-tubulin (ab52866, Abcam), mouse monoclonal antibody against UBF (sc13125, Santa Cruz), RPA194 (sc48385, Santa Cruz), phospho S139 gamma H2A.X antibody (ab26350, Abcam), Anti-BrdU antibody (B8434, Sigma), sheep polyclonal antibody against BrdU (ab1893, Abcam), Alexa 488-conjugated goat anti-mouse IgG (A11029, Invitrogen), Alexa 568-conjugated goat anti-rabbit IgG (A11011, Invitrogen), DyLight 488 goat anti-mouse IgG (ab96879, Abcam), isotype mouse IgG antibody (ab91353, Abcam), alkaline phosphate-conjugated rabbit IgG (ZB5305, ZSGB) and HRP goat anti-mouse IgG (A0208, Beyotime).
Statistical analysis
All the experiments were repeated independently at least twice, and the data are presented as mean ± SD. Statistical significance was determined using the Student's t-test. A P-value of <0.05 was considered to be statistically significant and is presented as *P < 0.05 or **P < 0.01.
RESULTS
MTX-induced double-strand breaks in mESCs
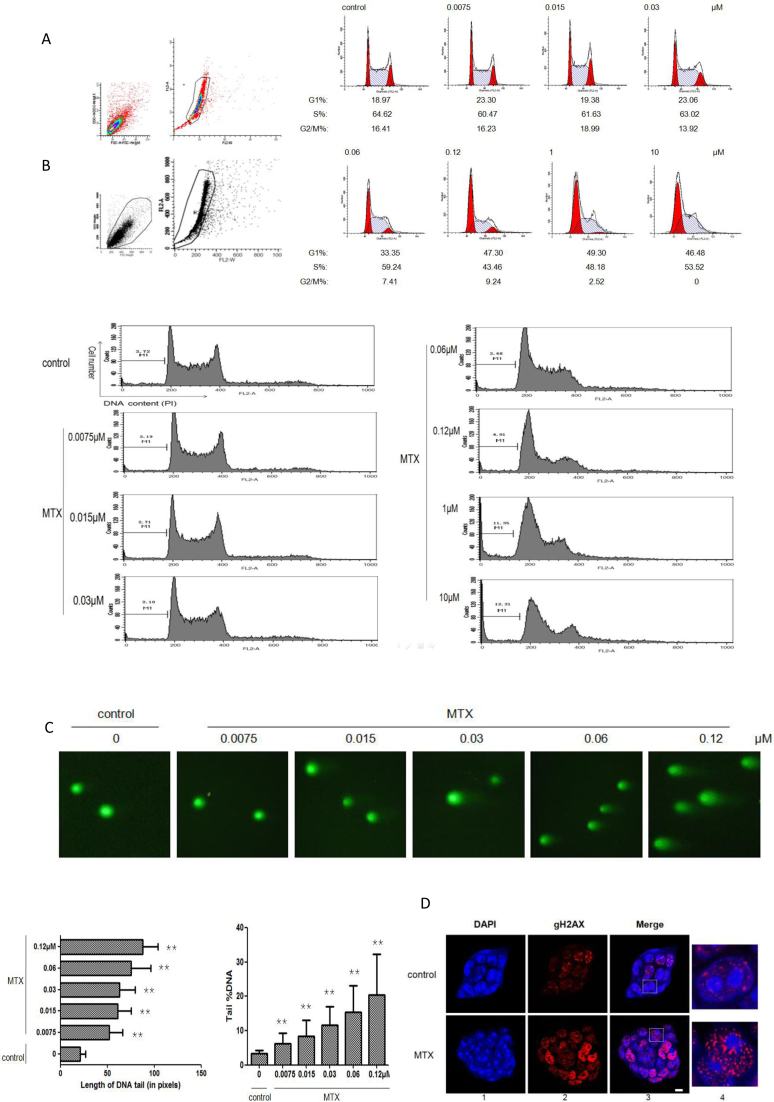
First, we established the appropriate MTX doses for the induction of DNA breaks in mESCs. The half inhibition concentration (IC50) of MTX was determined after 24 h incubation. In the MTX-treated cells, a remarkable decrease (about 30%) of cells in the S phase was visible at 0.12 μM MTX (Figure 1A), indicating transition to the G1/S checkpoint was delayed. Upon MTX treatment, significant number of cells was arrested in the G1 phase. To exclude the likelihood that the extensive fragmentation of nuclear DNA was the results of apoptosis (45), we performed quantitative analysis of the fractions of apoptotic cells after MTX treatment at several concentrations. No obvious apoptosis was detected at 0.12 μM MTX, while more than 10% of the cells were found to be apoptotic at 1 μM MTX (Figure 1B).
Figure 1.
Establishing MTX concentrations that induce DNA breaks and the effect of MTX on DNA. (A) Analysis of the cell cycle in mESCs with different doses of MTX after 24 h of treatment, by flow cytometry. The results were graded according to the cell cycle phase. (B) Rate of apoptosis in mESCs at different doses of MTX after 24 h of treatment, by flow cytometry. (C) Comet assays in mESCs at different doses of MTX after 24 h of treatment. The length of the DNA tail and tail %DNA for each dose of MTX are shown. Values are means ± SD (standard deviation) derived from three independent experiments. ** Indicates a statistically significant difference compared with the control (P < 0.01) based on Student's two-tailed t-test. (D) Representative images of nuclei with gH2AX foci in mESCs following 0.12 μM MTX for 24 h, by immunofluorescent staining gH2AX (panel 2). Nuclear DNA was stained by DAPI (panel 1). Merged images (panel 3) are shown. One representative nucleus with associated merged images (panel 4) are shown. Red: gH2AX; blue: DAPI staining. Bar, 5 μM.
Next, we performed a comet assay to detect DSBs with a MTX concentration gradient of 0–0.12 μM. The genotoxicity effect of MTX, measured as the length of DNA tail and percentage of DNA in the comet tail (tail %DNA) of mESCs, is shown in Figure 1C. The quantitative results clearly demonstrated that cells treated with the higher doses of MTX had increased mean levels of basal DNA damage, compared with the control cells. The highest percentage of DNA in the comet tail was observed after 24 h of incubation with 0.12 μM MTX. The differences between the treatment group and the control, in the lengths of the DNA tail and the tail %DNA, were statistically significant (Figure 1C). Furthermore, immunofluorescence of histone gamma-H2AX (gH2AX), a novel marker for DNA breakage, also revealed a substantial number of foci in mESCs after incubation with 0.12 μM MTX for 24 h (5.7 ± 10.6 and 35.6 ± 18.9 foci per nucleus for the control and MTX-treated cells respectively, n = 100; Figure 1D). Meanwhile, the total cell numbers between the control and 0.12 μM MTX treated group were comparable after 24 h (data not shown).
These results show that 0.12 μM MTX affects the cell cycle and can induce genome-wide DSBs in mESCs after 24 h without random fragmentation.
Hot spots of DSBs in rRNA genes units in MTX-treated mESCs
To pinpoint MTX-induced DNA breakage sites in the mESCs genome, we utilized a DSB enrichment protocol (Figure 2A). First, we performed restriction endonuclease digestion of nuclei from control cells which generated DSBs mimicking the effects of MTX, and used it as an enrichment quality control for capturing the expected cut sites of DSB. To minimize the risk of false positives, intact nuclei was embedded in low melting point (LMP) agarose. Direct in situ digestion and blunt-ending endonuclease-digested ends were used in the pilot experiment. To test the cleavage efficiency of HindIII in the LMP agarose gel, we prepared a 567-bp fluorescence-labeled DNA fragment that contained one HindIII restriction site. The fragment was digested by HindIII and blunt-ended in either liquid or agarose gel. The blunt-ended products were diluted in ultra pure water in multiples of 10, and then analyzed by capillary electrophoresis on a 3130xl Genetic Analyzer (Applied Biosystems). No differences between the in situ digestion and blunt-ending efficiency were detected. In both systems the genome was completely digested (Figure 2B). Excessive amounts of biotinylated linker 1 guaranteed effective ligation of the DSBs. After isolating the ligated DNA, linker 2 was attached to the captured genomic fragments. PCR amplification with sequencing primers was performed and the DSBs enrichment products were isolated as shown in Figure 2C.
The size of DSBs enrichment products obtained by SbfI, PmeI and HindIII digestion that simulated the DSBs after MTX treatment were analyzed by capillary electrophoresis. Data obtained using GeneMapper 4.1 revealed that the size of the digestion products peaked at about 144 bp, which was the predicted size of the PCR amplification products. Results from TA-clone sequencing showed that it corresponded to the correct (expected) double-strand break sites, but not single-strand breaks (Figure 2D–F, Supplementary material S1), demonstrating that the workflow specifically was capable of identifying DSBs at various genomic loci. We then applied the same method for the detection of DSBs induced by MTX. The DSBs enrichment products for the control and MTX-treated mESCs are shown in Figure 2G and H. The peak at 144 bp, which represents the spontaneous background DSBs in normal cells, was trivial compared with the peak at 112 bp, which represents linker self-ligation.
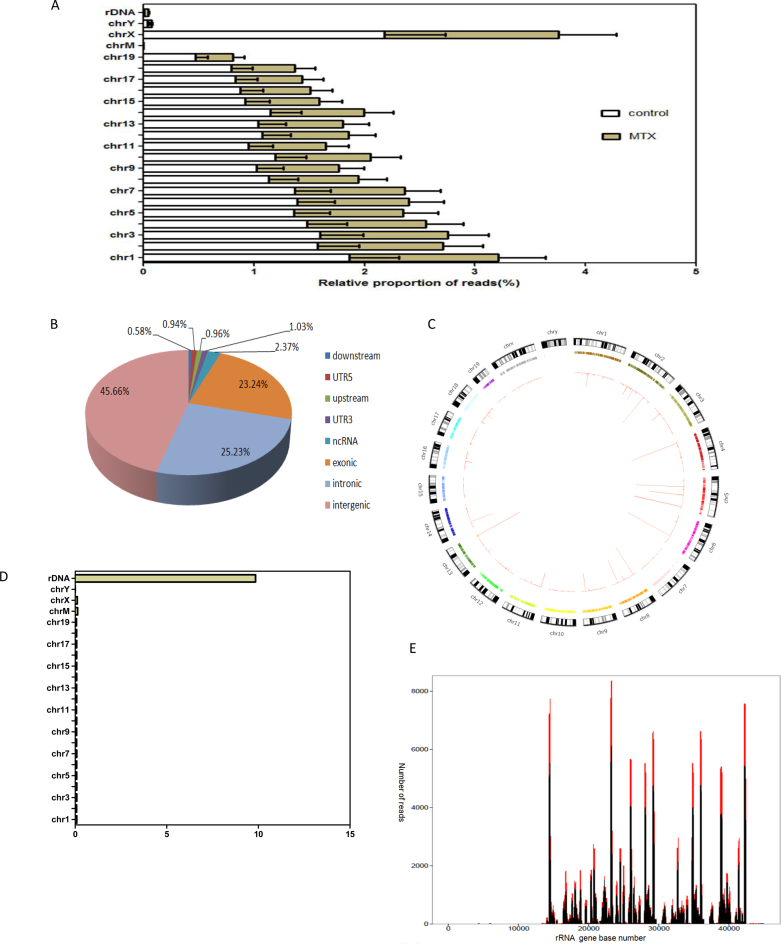
Chromosome analysis detected no significant difference in read density among the four samples (two controls and two MTX-treated mESCs), but the numbers of breakpoints increased with the length of the chromosome, with the exception of chromosome X (Figure 3A). After MTX treatment, the DSBs were most prominently enriched in intergenic regions of the genome (45.7%), followed by the intronic and exonic regions (25.2% and 23.24%, respectively) (Figure 3B). Gene information among above mentioned regions is presented in Supplementary Data, Excel S3). To display the large volumes of genomic DSBs data, we used Circos, a visualization tool that was developed by Krzywinski et al. (46) building on the established use of circular maps (Figure 3C, Supplementary Figure S1).
Figure 3.
Genome-wide hot spots of DSBs in rRNA genes units in MTX-treated mESCs (A) The relative proportion of reads of DSBs in chromosomes of mESCs from the MTX and control obtained by Illumina sequencing. (B) Genome-wide distribution of DSB sites induced by MTX. (C) Genome-wide MTX sensitivity landscape of DSBs in each chromosome in mESCs by hypergeometric test and visualized using Circos. (D) Significantly enriched DSBs in rRNA genes in mESCs compared to the rest of the genome calculated the fold-change in the number of reads. (E) Distribution of DSBs peak in the IGS regions of rRNA genes in mESCs detected with F-seq in complete medium and in complete medium with 0.12 μM MTX for 24 h. The control is in black and the treated is in red. X-axis represents the rRNA gene base number, Y-axis represents the read counts.
Repetitive DNA sequences, which tend to form hairpin-like secondary structures, were highly sensitive to breakage under replication stress (38). To investigate the association between DNA repeats and its susceptibility to MTX treatment, we applied RepeatMasker to compute the enrichment of mouse DNA repeat families compared with the rest of the genome. A strong and significant enrichment in rRNA gene units was detected (P = 2.8 × 10−89) using the hypergeometric test (Figure 3D, Supplementary Tables S4 and S5). Further analysis revealed that MTX treatment exclusively increased the spontaneous hot spots of DSBs in rRNA gene repeat units, compared with the control. Clearly, the profiles of the hot spots of DSBs exhibited a striking similarity in both the positions and the spans of the peaks between the MTX-treated and control mESCs (R2 = 0.995, P = 0, Figures 3E and 4F, Supplementary Excel S4).
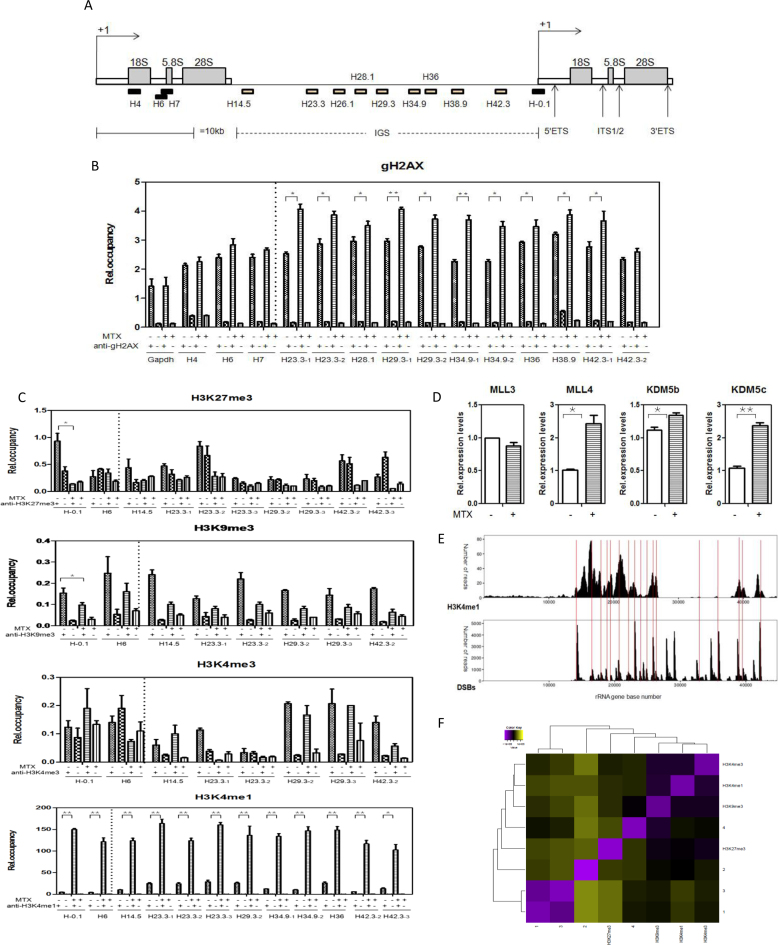
Figure 4.
Verification of hot spots of DSBs in rRNA genes units and the H3K4me1 modifications correlated with them (A) Schematic representation of a mouse rRNA genes repeat unit. Black squares represent regions that do not break and white squares represent hot spots of DSBs. Primer pairs (solid bars) and their approximate positions relative to the transcription start site are indicated. (B) Enrichment of gH2AX in rRNA genes breakage sites obtained with anti-gH2AX. The occupancy was determined by chromatin cross-linking and immunoprecipitation (ChIP) analysis using chromatin prepared from mESCs cultured in complete medium and complete medium with 0.12 μM MTX for 24 h. Chromatin DNA was quantitated by real-time PCR with primer sets, as indicated in (A). Values are means ± SD (standard deviation) derived from three independent experiments. *P < 0.05; **P < 0.01. (C) Binding of four histone modifications (H3K27me3, H3K9me3, H3K4me3, and H3K4me1) to the DSBs in rRNA genes after 0.12 μM MTX treatment compared with the control, determined by ChIP-qPCR analysis. *P < 0.05; **P < 0.01. (D) Expression changes of histone methyltransferases (MLL3, MLL4) and demethylases (KDM5b, KDM5c) in mESCs cultured in complete medium and complete medium with 0.12 μM MTX for 24 h, by RT-qPCR. Assays were performed in triplicate and the mean ± SD was calculated. *P < 0.05; **P < 0.01. (E) Comparison of H3K4me1 binding and DSBs peak in rRNA genes units detected with F-seq. The raw data of H3K4me1 in mESCs were from SRR002255 ES_H3K4me1_ChIP-Seq (UCSC Genome Browser). Thin red lines show the position of overlapped DSBs and H3K4me1 inside the IGS regions. X-axis represents the rRNA gene base number, Y-axis represents the read counts. (F) Correlation heatmap of pairwise comparisons using GenometriCorr R package between median signals for DSBs, H3K27me3, H3K9me3, H3K4me3 and H3K4me1 in the entire rRNA genes unit detected with F-seq.
Our data also showed that hot spots of DSBs in rRNA gene repeat units were not random; rather, they were located in several specific regions of the intergenic spacer (IGS) regions. The top four such regions were 14.5, 23.3, 29.3 and 42.3 kb in length (Figure 3E).
DNA breakage sites in rRNA gene units correspond to H3K4me1 sites
The nature of spontaneous DSBs in human rRNA gene units is unclear, but hot spot regions in rRNA gene repeat units show consistency with the histone H3K4me3 mark (17). Currently, it is not know whether this holds true in mouse rRNA gene units. However, MTX, an inhibitor of folate metabolism via dihydrofolate reductase, may influence methylation reactions related to histone modifications. To explore this, we first verified them using gH2AX ChIP assays to assess the level and position of DNA breakage sites in MTX treated cells. We found that DNA breakage sites in rRNA gene units expressed increased amount of gH2AX along the IGS regions compared with the non-IGS regions (Figure 4A, B).
Next, we examined the distribution of histone modifications along the hot spot regions. Within the IGS region, H3K4me1 was enriched and distributed at H23.3, H34.9 and H36 in mESCs treated with MTX, while H3K27me3, H3K9me3 and H3K4me3 showed no significant changes or were virtually absent in hot spot regions (Figure 4C), consistent with the previously published report (47,48). To further investigate the effect of MTX on H3K4me1 modification, we analyzed the histone methyltransferases and demethylases specific for H3K4. The RT-qPCR analysis showed an apparent increase in MLL4, KDM5b and KDM5c mRNA levels induced by MTX, indicating all three enzymes are required for the methylation status of H3K4me1 after MTX treatment (Figure 4D). The lysine-specific methyltransferases MLL3 and MLL4 are particularly important for maintaining H3K4me1 levels and H3K4 methylation can be reversed by members of the KDM5/Jarid family (H3K4me3/2 to H3K4me1) (49–52). However, the total proteins of H3K4me1 did not change in the mESCs, neither did the expressions of H3K27me3, H3K4me3, and H3K9me3 (Supplementary Figure S3).
Our data indicated that the distributions of hot spot of DSBs and H3K4me1 in the IGS regions were correlated in the mESCs (R2 = 0.2, P = 10−23, Figure 4E and F, Supplementary Excel S4). We also calculated the relationship between DSBs hot spots and the distributions of other histone modifications in rRNA gene units by aligning the high-throughput sequencing data to the rRNA genes. Pairwise linear regression analyses for the four samples were performed. The repressive H3K27me3 mark showed the lowest degree of correlation between samples based on the average R2 values using GenometriCorr R package analysis (Figure 4F, Supplementary Figure S2, Supplementary Excel S4). All the above results suggest that DNA breakage sites in rRNA gene units in mESCs may differ from those in human rRNA genes and that MTX influences the active chromatin state at rRNA gene loci by modulating histone posttranslational modification that correlates with DSB hot spots.
UBF recruitment to DNA breakage sites of rRNA gene units and rRNA genes transcription increased by MTX treatment
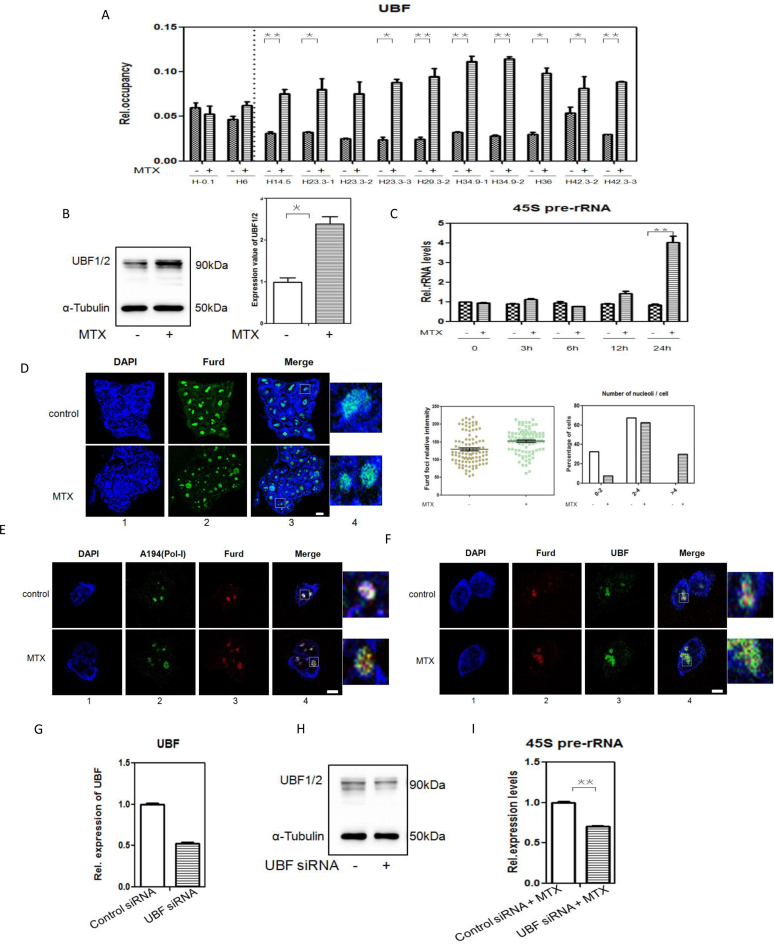
There is a general consensus that H3K4me1 may play a role in transcriptional activation. Additionally, MTX belongs to the first-category of chemotherapeutic drugs that impact rRNA gene transcription and subsequently ribosome biogenesis (53). rRNA gene transcription refers to the regulation of silenced and active rRNA genes, which depends on a number of key chromatin remodeling complexes. The cytoarchitectural upstream binding transcription factor (UBF) has been linked to decondensation and formation of an active chromatin environment at rRNA genes in mammals (54). Therefore, we tested whether UBF localized to DSBs of rRNA gene repeat units after MTX treatment. Using ChIP assays, we found that immune-precipitated UBF was almost exclusively associated with rRNA gene repeat units across DSBs induced by MTX, rather than those in the promoter (H-0.1) or transcribed regions (H-6). MTX treatment led to a more than twofold increase in occupancy of UBF throughout several hot spots of DSBs in IGS regions, especially the top four regions (Figure 5A). Consistently, results from western bolt analysis showed an elevated level of UBF protein after MTX treatment (Figure 5B).
Figure 5.
MTX promotes rRNA transcription and UBF specifically binds to DSB regions of rRNA genes (A) Binding of UBF to the DSBs in rRNA genes unit after 0.12 μM MTX treatment compared with the control, determined by ChIP-qPCR analysis. *P <0.05; **P <0.01. (B) Detection of UBF expression in mESCs cultured in complete medium and complete medium with 0.12 μM MTX treatment, by western blotting. α-Tubulin was used as a loading control. Positions of molecular weight markers are indicated. *P <0.05. (C) Expression levels of 45S pre-rRNA in mESCs cultured in complete medium with 0.12 μM MTX for 3, 6, 12 or 24 h, determined by RT-qPCR. The assays were performed in triplicate and the mean ± SD was calculated. **P <0.01. (D) Detection of ongoing rRNA synthesis by FUrd incorporation assays on mESCs subjected to 0.12 μM MTX for 24 h. Transcription was monitored by a FUrd pulse to observe incorporation into nascent nucleolar transcripts. FUrd detection with BrdU antibody was by confocal microscopy (panel 2). Nuclear DNA was stained by DAPI (panel 1). Merged images (panel 3) are shown. One representative nucleus with associated merged images (panel 4) are shown. Green: FUrd; blue: DAPI staining. Bar, 5 μM. Relative intensity of FUrd foci and the number of nucleoli per cell with FUrd foci were analyzed. The histogram shows the average number of nucleoli per cell based on an analysis of 93 cells from two independent experiments. (E) Co-localization between nucleolar proteins and rRNA transcripts by double-stained preparations of nucleolar markers (panel 2, Pol-I largest subunit A194) and FUrd pulse (panel 3, BrdU antibody).Nuclear DNA was stained by DAPI(panel 1).Merged images (panel 4) are shown. Green: A194(Pol-I); red: FUrd; blue: DAPI staining. Bar, 5 μM. (F) Co-localization between rRNA transcripts and UBF by double-stained preparations of FUrd pulse (panel 2, BrdU antibody) and UBF(panel3, upstream binding factor antibody).Nuclear DNA was stained by DAPI (panel 1). Merged images (panel 4) are shown. Green: UBF; red: FUrd; blue: DAPI staining. Bar, 5 μM. (G) Efficiency of siRNA mediated UBF depletion on mRNA level. The levels of the mRNAs UBF were analysed in mESCs with either a non-targeting siRNA (control siRNA) or siRNA directed against UBF(UBF siRNA). The assays were performed in triplicate and the mean ± SD was calculated. (H) Efficiency of siRNA mediated UBF depletion on protein level. mESCs were transfected with either control siRNA or UBF siRNA and UBF1/2 were determined 72 h post-transfection by Western-blotting. α-Tubulin was used as a loading control. Positions of molecular weight markers are indicated. (I) Expression levels of 45S pre-rRNA in mESCs with control siRNA or UBF siRNA cultured in complete medium with 0.12 μM MTX for 24 h, determined by RT-qPCR. The assays were performed in triplicate and the mean ± SD was calculated. **P <0.01.
UBF increases the rate of rRNA synthesis (55). In line with this, we found that 45S pre-rRNA genes transcription was increased at 24 h after MTX treatment (Figure 5C). Ongoing rRNA synthesis was assessed by fluorouridine (FUrd) incorporation in an in situ run-on assay and an increase in Furd positive foci number was observed after MTX treatment (Figure 5D and E), suggesting that active rRNA synthesis was up-regulated in these cells, compared with control cell. The correlation between the presence of UBF and rRNA genes transcriptional regulation was further evaluated. In the nucleoli of the cells, the presence of UBF and FUrd signals were found to be almost completely overlapped. An elevation of UBF staining intensity was observed in MTX-treated cells (Figure 5F). Additionally, siRNA technique was used to down-regulate UBF expression (Figure 5G and H). The results indicated that the rRNA genes transcription was profoundly reduced compared to the group with control siRNA transfection upon MTX treatment (Figure 5I). Therefore, we concluded that MTX treatment led to a predominant transcription activation of rRNA genes and that the insufficient presence of UBF binding along can attenuate rRNA genes transcription upon MTX treatment.
These data strongly indicated that increased distribution of UBF to DSBs of rRNA genes repeats with H3K4me1 foci promoted an open chromatin structure of the rRNA genes induced by MTX, thereby influencing rRNA genes transcription.
Supplementary folinic acid alleviates DSBs at the G1 phase of cell cycle induced by MTX and affects UBF binding at DNA breakage sites of rRNA gene loci
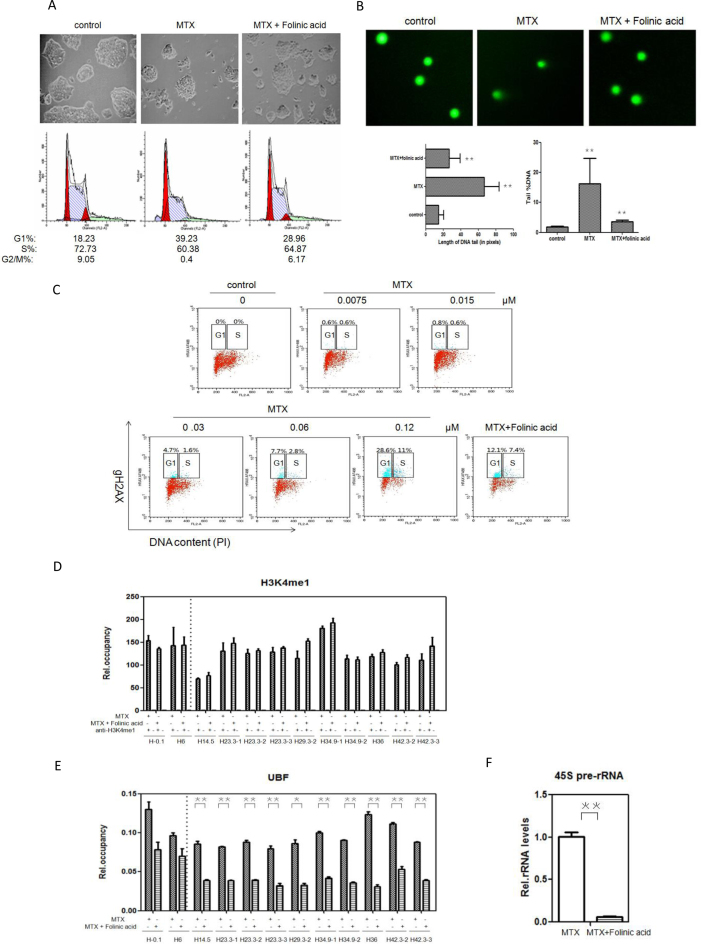
Previous studies have shown that supplementary folinic acid can be used as an antidote after MTX treatment to decrease toxicity in various cells and tissues (56–58). However, information about folinic acid antagonizing MTX-treated mESC is limited. In the current study, supplementary folinic acid (50 mg/l) was added to mESCs pre-treated with MTX. Our results showed that supplementary folinic acid led to incremental improvements in cell cycle and DNA damage in cells. Cells arrested in G1 phase by MTX treatment were rescued to enter the S and G2/M phases after supplementary folinic acid treatment. In addition, mESC colony formation was partially reversed (Figure 6A), and DNA damage was significantly reduced in the MTX-treated cells with the addition of folinic acid (Figure 6B). These findings further confirmed that G1/S checkpoint delay and the induction of DSBs by MTX in mESCs were closely associated with folate-deficiency.
Figure 6.
Effects of supplementary folinic acid on cell cycle and DSBs induced by MTX (A) mESC colony formation was partially reversed by folinic acid. mESCs were cultured in complete medium, complete medium with 0.12 μM MTX, and supplementary folinic acid (50 mg/L) for 24 h. The colonies were photographed at 100× resolution. Representative fields are shown. (B) Comet assay in mESCs by 0.12 μM MTX treatment and after supplementary folinic acid treatment. The length of DNA tail and tail %DNA are shown for each treatment. Values are means ± (standard deviation) derived from three independent experiments. ** Indicates statistically significant difference from control (P < 0.01) based on Student's two-tailed t-test. (C) Fraction of gH2AX positive cells in the indicated phase of the cell cycle following 0.12 μM MTX for 24 h or supplementary folinic acid. Data are from three independent experiments. (D) H3K4 methylation states at representative sites of DSBs in rRNA genes unit after supplementary folinic acid compared with MTX treatment alone, as determined by ChIP-qPCR. Values are means ± SD derived from three independent experiments. *P < 0.05; **P < 0.01. (E) Binding of UBF binding to the DSBs in rRNA genes unit after supplementary folinic acid compared with the MTX treatment alone, by ChIP-qPCR analysis. *P < 0.05; **P < 0.01. (F) Expression levels of 45S pre-rRNA determined by RT-qPCR in mESCs cultured in complete medium with 0.12 μM MTX for 24 h and supplementary folinic acid. The assays were performed in triplicate and the mean ± SD was calculated. **P < 0.01.
After MTX treatment, cells bearing DNA damage marker, gH2AX, were predominantly in the G1 phase, rather than the S phase (Figure 6C). However, such an effect was reversed by the addition of folinic acid, especially in the G1 phase. Together, these results indicate that supplementary folinic acid affected DSBs and targeted mainly the G1 phase.
The ChIP data revealed that in contrast with MTX treatment alone, H3K4me1 modification was distributed in hot spots of IGS regions, and did not appear to vary after supplementary folinic acid at 24 h (Figure 6D); however, the levels of UBF binding uniquely to the IGS hotspots were reduced (Figure 6E). Meanwhile, there was a sharp decline in 45S pre-rRNA genes transcription at 24 h (Figure 6F). The mRNA levels after supplementary folinic acid of MLL3, MLL4, KDM5b and KDM5c were also detected (Supplementary Figure S4).
Decreased UBF occupancy in rRNA gene units after supplementary folinic acid suggested that the association of UBF with rRNA genes in mESCs under low folate status may be necessary for the open chromatin structure within rRNA gene units, optimal for transcription activation. This result is consistent with previously published reports that UBF is responsible for maintaining active nucleolus organizer regions (NORs) in cells arrested in G0-G1 phase, but not S phase (54).
rRNA genes transcription is altered with increased UBF expression in low-folate NTD fetuses
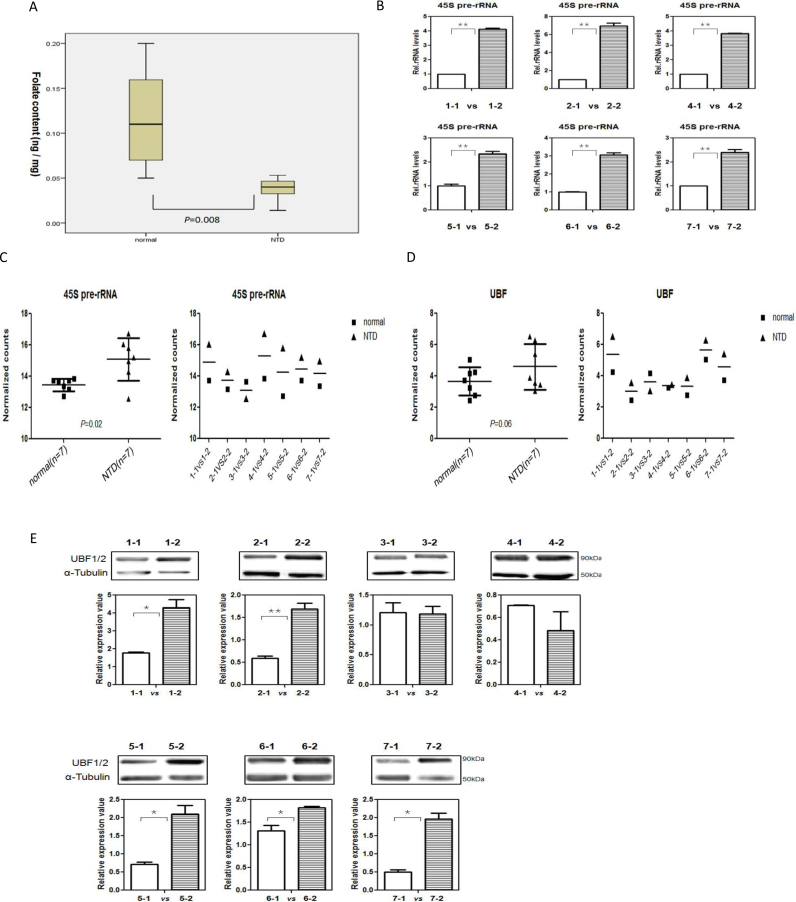
Normal nerve system development involves numerous cellular and molecular processes, and ribosome biogenesis is fundamental to all these processes. Abnormal synthesis of ribosomes has recently been reported to cause several developmental defects (9,59–61). To explore the potential effect of folate on ribosome biogenesis in NTDs, we evaluated rRNA genes transcription in low-folate NTD fetuses. Seven pairs of normal fetal brains and fetal brains from spina bifida were selected, based on best match in age and gender, as summarized in Table 1. Folate levels in tissues from individuals with spina bifida were lower than that in the controls (Figure 7A).
Table 1. Clinical manifestations of normal fetus and NTD fetus.
| Sample ID | Sample type | tissue | Folate of tissue (ng/mg) | Gender | Gestational weeks |
|---|---|---|---|---|---|
| 1-1 | Normal | Brain | 0.050 | Female | 39 |
| 2-1 | Normal | Brain | 0.090 | Female | 36 |
| 3-1 | Normal | Brain | 0.199 | Male | 30 |
| 4-1 | Normal | Brain | 0.110 | Female | 25 |
| 5-1 | Normal | Brain | 0.050 | Female | 39 |
| 6-1 | Normal | Brain | 0.120 | Male | 38 |
| 7-1 | Normal | Brain | 0.200 | Female | 30 |
| 1-2 | Spina bifida | Brain | 0.014 | Female | 40 |
| 2-2 | Spina bifida | Brain | 0.036 | Female | 33 |
| 3-2 | Spina bifida | Brain | 0.040 | Female | 32 |
| 4-2 | Spina bifida | Brain | 0.042 | Female | 27 |
| 5-2 | Spina bifida | Brain | 0.029 | Female | 38 |
| 6-2 | Spina bifida | Brain | 0.053 | Male | 38 |
| 7-2 | Spina bifida | Brain | 0.051 | Female | 32 |
Figure 7.
rRNA transcription increased with increased UBF expression in low-folate NTDs (A) Folate content in brain tissue of normal fetus and fetus with NTDs. (B) Expression levels of 45S pre-rRNA in the brain of NTD fetuses, determined by RT–TPCR. The values were normalized using the expression of beta-actin mRNA are a reference. Assays were performed in triplicate and the mean ± SD was calculated. *P < 0.05; **P < 0.01. (C) Expression levels of 45S pre-rRNA in the brain of NTD fetuses, determined by NanoString. P value was determined using the Student's t test. (D) The mRNA expression of UBF in the brain of NTD fetuses, determined by NanoString. P value was determined using the Student's t test. (E) Detection of UBF in the brain of normal fetus and low-folate NTDs for matched samples. Tubulin was used as a loading control. *P < 0.05; **P < 0.01.
Among all pairs of fetal brain tissue, the pre-45s rRNA was significantly overexpressed in spina bifida brain in six pairs (except for the 3-1/3-2 pair because of poor RNA quality) (Figure 7B). The difference in number of transcripts between two groups was further quantified using Nanostring and the results unambiguous testified the pre-45s rRNA overexpression in spina bifida brain samples (P = 0.02, Figure 7C). In the same experimental setting, although statistically insignificant, the number of transcripts of UBF in spina bifida brain showed a upward tendency (P = 0.06, Figure 7D). We further determined the UBF expression at protein level in samples with matched gestational weeks and gender. Interestingly, in five of the seven brains with NTDs, UBF was overexpressed (Figure 7E). In conclusion, we identified the up-regulation of rRNA genes transcription with increased UBF expression in low-folate NTD fetuses. This work is the first to demonstrate that, in NTDs, folate deficiency increased ribosome biogenesis and was accompanied by aberrant UBF expression. Whether this increased biogenesis has an impact on the rate and time sequence of protein synthesis in NTDs will be examined in future studies. These results confirm the importance of increased folate during early pregnancy when NTDs are likely to form.
DISCUSSION
It is generally accepted that DSBs represent the most deleterious form of DNA lesions (62). In this report, we identified intensive DSBs in rRNA genes of mESCs with folate deficiency and demonstrated that effect of folate deficiency on transcription activation of rRNA genes. In addition, we showed that in NTDs, folate deficiency with closely correlated with an increased rRNA gene transcription which was accompanied by aberrant UBF expression.
It has been previously reported that hot spots of DSBs exist at specific IGS regions in human (17), and similar findings in mouse rRNA gene repeat units are reported in this study. During mapping and analysis of epigenetic marks, the hot spot of DSBs in human rRNA genes coincide with regions that possess active chromatin marks (H3K4me3) along the rRNA gene repeat. In our study, the correlation between H3K4me3 and hot spots of DSBs in rRNA genes was quite poor (Supplementary Figure S2A and B). This difference can be explained by the large divergence in histone modification patterns in rRNA genes between human and mouse cells, especially for ESCs (48). Indeed in mESCs, the highest correlation occurred between the distribution of H3K9me3 and DSBs in rRNA gene repeat unit (R2 = 0.43, P = 10−57). The distribution of H3K9me3 and H3K4me1 also were highly correlated to DSBs (R2 = 0.65, P = 10−105). Both of these histone methylation modifications are involved in the transcription regulation of rRNA genes. There are two forms of rRNA gene repeats exist within the nucleolus: an open, highly-transcribed conformation and a state of transcriptional silence. The former is euchromatic, hypomethylated at CpG sites, and marked with active histone modifications (i.e., H3K4me1, H3K4me3, and H3K9ac), while the latter is heterochromatic, hypermethylated and marked with repressive histone markers (i.e. H3K27me3 and H3K9me3) (10,63). The key finding from our study is that inhibition of folate metabolism specifically targeted the active transcriptional histone modification H3K4me1, suggesting that epigenetic regulation of rRNA genes is unequivocal between different histone modifications. Furthermore, genomic studies have linked this modification to distal regulatory regions and it is widely distributed across broader regions (64). Fine mapping of the interaction between the regulatory regions and H3K4me1 on transcription of 45S rRNA genes, the most actively transcribed genes in eukaryotes, is warranted.
Both IGS regions and non-DSBs regions of rRNA genes are occupied with H3K4me1-active histone marker for up-regulated rRNA genes transcription, indicating a more open and loose chromatin structure formed by rRNA gene repeats within NORs after chromosomal breakage induced by MTX. It is reasonable to speculate that some breakages originate from the active rRNA gene units, while others originate from the silenced units. One limitation of this study is that the DSBs that we mapped in rRNA gene repeat units reflect aggregate signals at all the rRNA genes copies; that is, we cannot distinguish breaks that exist in active or inactive units or even in both units. Likewise, the ChIP-qPCR data that we have analyzed cannot discriminate the signals from active or inactive rRNA gene units. Therefore, the data presented in this study are not able to distinguish patterns of active and repressive histone marks occurred on independent repeats or coexisted on the same repeats.
In mammals, the regulation of silenced and active rRNA genes depends on a number of key chromatin remodeling complexes. UBF, a nucleolar-specific HMG-box-containing protein, has been shown to regulate the epigenetic silencing switch of rRNA gene clusters and transcription activation (10,50,65). We have shown that MTX induced large amounts of UBF in mESCs cells (Figure 5B). UBF is involved in the formation of the pre-initiation complex (PIC) at the rRNA gene promoter region, and is enriched across the transcribed region and non-transcribed spacer sequences but rarely found in the IGS regions of genomes (66–67). Strikingly, we found that UBF binds specially across the DSBs region of IGS without a promoter or transcribed region, supporting the idea that UBF plays a role in structurally organizing the rRNA gene repeat units as a specialized cytoarchitectural transcription factor that maintains undercondensed r-chromatin in active NORs (68). Additionally, it has been suggested that when UBF binds to the transcribed regions of rRNA genes, it represses rather than activates the transcription elongation of rRNA genes (55). After supplementary folinic acid, UBF showed a significant callback of binding across repeat regions while H3K4me1 showed no response. We speculate that the cellular UBF level is sensitive to folate deficiency during the G1 phase.
NTDs are severe congenital malformations in the central nervous system, which can be effectively prevented by the periconceptional consumption of folate (69–71). However, the mechanism about its various phenotype is still poorly understood. Recent reports indicate that abnormal ribosome biogenesis includes craniofacial anomalies. Craniofacial malformations are associated with over 700 human congenital syndromes. Abnormalities in the nucleolar protein NOL11 essential for optimal pre-rRNA genes transcription and processing were found to trigger cell apoptosis and contribute to craniofacial defects (9). In our study, during the early stage of development, mESCs in which folate metabolism was inhibited demonstrated robust rRNA gene transcription. It is unclear whether this compensation of ribosome biogenesis was related to folate deficiency and the development of NTDs. Our result defined NTDs in brain tissue from fetuses during gestational weeks 25–40, indicating the unusual rRNA genes transcription was sustained over the whole gestation period. Notably, up-regulated rRNA genes transcription occurred only in spina bifida tissue, but not in anencephalus where pre-rRNA genes transcription decreased substantially (data not show). These findings suggest the interesting possibility that ribosome biogenesis in NTD phenotypes may be different in different tissue phenotypes and raise the question of how different cell types can be affected by abnormality in the ubiquitous process of making ribosomes.
Taken together, our analyses provide a novel insight into chromosome breakages in rRNA gene units and the regulatory potential of these breakages for ribosome biogenesis under folate dysmetabolism. We identified the dysregulation of UBF in NTDs, which was involved in the up-regulation of rRNA genes transcription. This is the first report of abnormal ribosome biogenesis in NTDs, although the consequences of this abnormality are yet to be discovered. Our findings may offer a new approach to uncovering the complexity of NTDs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all of the woman who participated for their cooperation. We are grateful to all participating hosptials for their assistance in sample collection and recording of clinical information. Special thanks should be given to Nicola Crosetto, Gregory E. Crawford, Lingyun Song, Nickolai A. Tchurikov and Shan Wang for helpful discussion during the course of this project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of the P.R. China, National ‘973’ project [2013CB945404]; National Natural Science Foundation of China, Beijing, China [81270699]. Funding for open access charge: Ministry of Science and Technology of the P.R. China, National ‘973’ project [2013CB945404] and the National Natural Science Foundation of China, Beijing, China [No. 81270699].
Conflict of interest statement. None declared.
REFERENCES
- 1.Moss T., Langlois F., Gagnon-Kugler T., Stefanovsky V.. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life. Sci. 2007; 64:29–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Little R.D., Braaten D.C.. Genomic organization of human 5 S rRNA genes and sequence of one tandem repeat. Genomics. 1989; 4:376–383. [DOI] [PubMed] [Google Scholar]
- 3.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003; 17:1691–1702. [DOI] [PubMed] [Google Scholar]
- 4.Moss T., Stefanovsky V.Y.. At the center of eukaryotic life. Cell. 2002; 109:545–548. [DOI] [PubMed] [Google Scholar]
- 5.Bateman E., Paule M.R.. Regulation of eukaryotic ribosomal RNA transcription by RNA polymerase modification. Cell. 1986; 47:445–450. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield R.J., Stevenson T.J., Xing L., Newcomb T.M., Nelson B., Zeng W., Li X., Lu H.M., Lu H., Gonzalez K.D. et al. Congenital lethal motor neuron disease with a novel defect in ribosome biogenesis. Neurology. 2014; 82:1322–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCann K.L., Baserga S.J.. Mysterious ribosomopathies. Science. 2013; 341:849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon J., Jones N.C., Sandell L.L., Jayasinghe S.M., Crane J., Rey J.P., Dixon M.J., Trainor P.A.. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13403–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin J.N., Sondalle S.B., Del Viso F., Baserga S.J., Khokha M.K.. The ribosome biogenesis factor Nol11 is required for optimal rRNA genes transcription and craniofacial development in Xenopus. PLoS. Genet. 2015; 11:e1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McStay B., Grummt I.. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell. Dev. Biol. 2008; 24:131–157. [DOI] [PubMed] [Google Scholar]
- 11.Ransohoff R.M., Denker J.A., Takacs A.M., Hannon G.J., Nilsen T.W.. Organization and expression of 5S rRNA genes in the parasitic nematode, Brugia malayi. Nucleic Acids Res. 1989; 17:3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007; 447:407–412. [DOI] [PubMed] [Google Scholar]
- 13.Kustatscher G., Ladurner A.G.. Modular paths to ‘decoding’ and ‘wiping’ histone lysine methylation. Curr. Opin. Chem. Biol. 2007; 11:628–635. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka Y., Okamoto K., Teye K., Umata T., Yamagiwa N., Suto Y., Zhang Y., Tsuneoka M.. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010; 29:1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stavropoulos P., Hoelz A.. Lysine-specific demethylase 1 as a potential therapeutic target. Expert. Opin. Ther. Targets. 2007; 11:809–820. [DOI] [PubMed] [Google Scholar]
- 16.Zentner G.E., Saiakhova A., Manaenkov P., Adams M.D., Scacheri P.C.. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011; 39:4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tchurikov N.A., Fedoseeva D.M., Sosin D.V., Snezhkina A.V., Melnikova N.V., Kudryavtseva A.V., Kravatsky Y.V., Kretova O.V.. Hot spots of DNA double-strand breaks and genomic contacts of human rRNA genes units are involved in epigenetic regulation. J. Mol. Cell. Biol. 2015; 7:366–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchurikov N.A., Kretova O.V., Fedoseeva D.M., Sosin D.V., Grachev S.A., Serebraykova M.V., Romanenko S.A., Vorobieva N.V., Kravatsky Y.V.. DNA double-strand breaks coupled with PARP1 and HNRNPA2B1 binding sites flank coordinately expressed domains in human chromosomes. PLoS Genet. 2013; 9:e1003429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doege C.A., Inoue K., Yamashita T., Rhee D.B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W.B., Shih A. et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012; 488:652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langelier M.F., Planck J.L., Roy S., Pascal J.M.. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012; 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey L.B., Stover P.J., McNulty H., Fenech M.F., Gregory J.F., Mills J.L., Pfeiffer C.M., Fazili Z., Zhang M., Ueland P.M. et al. Biomarkers of nutrition for development-folate review. J. Nutr. 2015; 145:1636S–1680S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blount B.C., Mack M.M., Wehr C.M., MacGregor J.T., Hiatt R.A., Wang G., Wickramasinghe S.N., Everson R.B., Ames B.N.. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kibar Z., Bosoi C.M., Kooistra M., Salem S., Finnell R.H., De Marco P., Merello E., Bassuk A G., Capra V., Gros P.. Novel mutations in VANGL1 in neural tube defects. Hum. Mutat. 2009; 30:E706–E715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Chen J.. One-carbon metabolism and breast cancer: an epidemiological perspective. J. Genet. Genomics. 2009; 36:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duthie S.J. Epigenetic modifications and human pathologies: cancer and CVD. Proc. Nutr. Soc. 2011; 70:47–56. [DOI] [PubMed] [Google Scholar]
- 26.Blom H.J., Smulders Y.. Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J. Inherit. Metab. Dis. 2011; 34:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith AD., Smith SM., de Jager C.A., Whitbread P., Johnston C., Agacinski G., Oulhaj A., Bradley K.M., Jacoby R., Refsum H.. Homocysteine-lowering by B vitamins slows the rate of accelerated brain atrophy in mild cognitive impairment: a randomized controlled trial. PLoS One. 2010; doi:10.1371/pone.0012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tibbetts A.S., Appling D.R.. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010; 30:57–81. [DOI] [PubMed] [Google Scholar]
- 29.Martin C., Zhang Y.. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell. Biol. 2005; 6:838–849. [DOI] [PubMed] [Google Scholar]
- 30.Park LK., Friso S., Choi SW.. Nutritional influences on epigenetics and age-related disease. Proc. Nutr. Soc. 2012; 71:75–83. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q., Xue P., Li H., Bao Y., Wu L., Chang S., Niu B., Yang F., Zhang T.. Histone modification mapping in human brain reveals aberrant expression of histone H3 lysine 79 dimethylation in neural tube defects. Neurobiol. Dis. 2013; 54:404–413. [DOI] [PubMed] [Google Scholar]
- 32.Blount B.C., Ames B.N.. DNA damage in folate deficiency. Baillieres. Clin. Haematol. 1995; 8:461–478. [DOI] [PubMed] [Google Scholar]
- 33.Duthie SJ., Narayanan S., Blum S., Pirie L., Brand GM.. Folate deficiency in vitro induces uracil misincorporation and DNA hypomethylation and inhibits DNA excision repair in immortalized normal human colon epithelial cells. Nutr. Cancer. 2000; 37:245–251. [DOI] [PubMed] [Google Scholar]
- 34.Duthie S.J., Hawdon A.. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic aciddepletion in human lymphocytes in vitro. FASEB J. 1998; 12:1491–1497. [PubMed] [Google Scholar]
- 35.Duthie S.J., Grant G., Narayanan S.. Increased uracil misincorporation in lymphocytes from folate-deficient rats. Br. J. Cancer. 2000; 83:1532–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins A.R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004; 26:249–261. [DOI] [PubMed] [Google Scholar]
- 37.Song L., Crawford G.E.. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010; doi:10.1101/prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crosetto N., Mitra A., Silva M.J., Bienko M., Dojer N., Wang Q., Karaca E., Chiarle R., Skrzypczak M., Ginalski K. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods. 2013; 10:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Favorov A., Mularoni L., Cope L.M., Medvedeva Y., Mironov A.A., Makeev V.J., Wheelan S.J.. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput. Biol. 2012; 8:e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruhlak M., Crouch E.E., Orlov M., Montaño C., Gorski S.A., Nussenzweig A., Misteli T., Phair R.D., Casellas R.. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007; 447:730–734. [DOI] [PubMed] [Google Scholar]
- 41.Chen X., Shen Y., Gao Y., Zhao H., Sheng X., Zou J., Lip V., Xie H., Guo J. et al. Detection of copy number variants reveals association of cilia genes with neural tube defects. PLoS One. 2013; 8:e54492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X., Guo J., Lei Y., Zou J., Lu X. et al. Global DNA hypomethylation is associated with NTD-affected pregnancy: a case-control study. Birth Defects Res. A Clin. Mol. Teratol. 2010; 88:575–581. [DOI] [PubMed] [Google Scholar]
- 43.Chen S., Zhang Q., Bai B., Ouyang S., Bao Y., Li H., Zhang T.. MARK2/Par1b insufficiency attenuates DVL gene transcription via histone deacetylation in lumbosacral spina bifida. Mol. Neurobiol. 2016; doi:10.1007/s12035-016-0164-0. [DOI] [PubMed] [Google Scholar]
- 44.Shechter D., Dormann H.L., Allis C.D., Hake S.B.. Extraction, purification and analysis of histones. Nat. Protoc. 2007; 2:1445–1457. [DOI] [PubMed] [Google Scholar]
- 45.Nagata S. Apoptotic DNA fragmentation. Exp. Cell. Res. 2000; 256:12–18. [DOI] [PubMed] [Google Scholar]
- 46.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A.. Circos: an information aesthetic for comparative genomics. Genome Res. 2009; 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zentner G.E., Henikoff S.. High-resolution digital profiling of the epigenome. Nat. Rev. Genet. 2014; 15:814–827. [DOI] [PubMed] [Google Scholar]
- 48.Zentner G.E., Balow S.A., Scacheri P.C.. Genomic characterization of the mouse ribosomal DNA locus. G3 (Bethesda). 2014; 4:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng J., Blum R., Bowman C., Hu D., Shilatifard A., Shen S., Dynlacht B.D.. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell. 2014; 53:979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herz H.M., Mohan M., Garruss A.S., Liang K., Takahashi Y.H., Mickey K., Voets O., Verrijzer C.P., Shilatifard A.. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012; 26:2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blair L.P., Cao J., Zou M.R., Sayegh J., Yan Q.. Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers (Basel). 2011; 3:1383–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen H., Xu W., Guo R., Rong B., Gu L., Wang Z., He C., Zheng L., Hu X., Hu Z. et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016; 165:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burger K., Mühl B., Harasim T., Rohrmoser M., Malamoussi A., Orban M., Kellner M., Gruber-Eber A., Kremmer E., Hölzel M. et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J. Biol. Chem. 2010; 285:12416–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T.P., Quin J., Robb E., Wong L.H., Thomas W.G., Stefanovsky V. et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell. Biol. 2008; 183:1259–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panov K.I., Friedrich J.K., Russell J., Zomerdijk J.C.. UBF activates RNA polymerase I transcription by stimulatingv promoter escape. EMBO J. 2006; 25:3310–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen I.J. Prevention of high-dose-methotrexate neurotoxicity by adequate folinic acid rescue is possible even after central nervous system irradiation. Med. Hypotheses. 2007; 68:147–153. [DOI] [PubMed] [Google Scholar]
- 57.Shahin A.A., Ismail M.M., Saleh A.M., Moustafa H.A., Aboul-Ella A.A., Gabr H.M.. Protective effect of folinic acid on low-dose methotrexate genotoxicity. Z. Rheumatol. 2001; 60:63–68. [DOI] [PubMed] [Google Scholar]
- 58.Möller B., Kukoc-Zivojnov N., Okamgba S., Kessler U., Puccetti E., Ottmann O.G., Kaltwasser J.P., Hoelzer D., Ruthardt M.. Folinic acid antagonizes methotrexate-induced differentiation of monocyte progenitors. Rheumatol, Int. 2002; 22:60–67. [DOI] [PubMed] [Google Scholar]
- 59.Freed E.F., Baserga S.J.. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res. 2010; 38:4798–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trainor P.A., Merrill A.E.. Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim. Biophys. Acta. 2014; 1842:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freed E.F., Prieto J.L., McCann K.L., McStay B., Baserga S.J.. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 2012; e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larsen D.H., Stucki M.. Nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 2016; 44:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum. Mol. Genet. 2007; 16:R21–R27. [DOI] [PubMed] [Google Scholar]
- 64.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 65.Sanij E., Hannan R.D.. The role of UBF in regulating the structure and dynamics of transcriptionally active rRNA genes chromatin. Epigenetics. 2009; 4:374–382. [DOI] [PubMed] [Google Scholar]
- 66.O'Sullivan A.C., Sullivan G.J., McStay B.. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 2002; 22:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Putnam C.D., Copenhaver G.P., Denton M.L., Pikaard C.S.. The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol. Cell. Biol. 1994; 14:6476–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mais C., Wright J.E., Prieto J.L., Raggett S.L., McStay B.. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005; 19:50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beaudin A.E., Stover P.J.. Insights into metabolic mechanisms underlying folate-responsive neural tube defects: a minireview. Birth. Defects. Res. A Clin. Mol. Teratol. 2009; 85:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berry R.J., Li Z., Erickson J.D., Li S., Moore C.A., Wang H., Mulinare J., Zhao P., Wong L.Y., Gindler J. et al. Prevention of neural-tube defects with folic acid in China. China-U.S. Collaborative Project for Neural TubeDefect Prevention. N. Engl. J. Med. 1999; 341:1485–1490. [DOI] [PubMed] [Google Scholar]
- 71.Czeizel A.E., Dudás I.. Prevention of the first occurrence of neural-tube defects by periconceptional vitamin supplementation. N. Engl. J. Med. 1992; 327:18321–18325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.