Abstract
Pseudomonas aeruginosa airway infections are a major cause of morbidity and mortality in patients with cystic fibrosis. Treatment of established infections is difficult, even with microbiologically active agents. Thus, prevention of infection is an important goal of management. Isolates from cystic fibrosis patients appear to originate from the environment but adapt to the milieu of the airway of the cystic fibrosis patient and evolve toward a common phenotype. Identification of the antigens expressed early in infection may lead to novel targets for vaccine development. Immunogenic peptides were identified in a J404 random nonapeptide phage display library with serum from cystic fibrosis patients obtained within the first year of P. aeruginosa infection. One hundred sixty-five reactive clones were verified by plaque lift assays, and their inserts were sequenced. The sequenced nonapeptides were compared with the published sequence of strain PAO1, identifying homologies to 76 genes encoding outer membrane and secreted proteins. The majority of these were proteins involved in small-molecule transport, membrane structural proteins, and secreted factors. An in silico analysis was performed that suggested that the occurrence of multiple matches to predominantly outer membrane and secreted proteins was not attributable to random chance. Finally, gene expression array data from early isolates of P. aeruginosa from cystic fibrosis patients was compared with the results from phage display analysis. Eleven outer membrane and secreted proteins were common between the two data sets. These included genes involved in iron acquisition, antibiotic efflux, fimbrial biogenesis, and pyocin synthesis. These results demonstrate the feasibility and validity of this novel approach and suggest potential targets for future development.
Pseudomonas aeruginosa is the most significant pulmonary pathogen infecting patients with cystic fibrosis. It has long been known that P. aeruginosa infects the majority of adults with cystic fibrosis, being reported in more than 80% of individuals with cystic fibrosis over the age of 18 years (11). However, serologic studies have recently suggested that a significant proportion of children with cystic fibrosis acquire the organism in the first few years of life (3, 42). In fact, both of these studies have demonstrated that initial serology is positive on average 6 to 12 months before P. aeruginosa can be cultured from oropharyngeal and bronchoalveolar lavage specimens. This suggests that intermittent infection occurs much earlier than previously thought and may be related to the early inflammation seen in cystic fibrosis.
This early acquisition of P. aeruginosa appears to be from the environment, with each individual having a genotypically unique strain (3, 24). As the disease progresses, the organism undergoes adaptation and evolves toward the “classic” P. aeruginosa phenotype associated with cystic fibrosis: mucoid, antibiotic resistant, nonmotile, and nonserotypeable based on rough lipopolysaccharide.
Approaches to the management of P. aeruginosa airway infections in cystic fibrosis patients have included aggressive antimicrobial treatment, frequently resulting in a high level of multiple antibiotic resistance (7, 8); antibiotic prophylaxis or suppressive therapy (18, 20, 31, 23); and active immunization (12, 10). In patients who have recently acquired P. aeruginosa, the organism can be eradicated with antibiotics (31, 20, 18). However, once the infection has become chronic, eradication is virtually impossible, even with active antimicrobial therapy (37, 32, 30). Thus, a Pseudomonas vaccine aimed at young children with cystic fibrosis who are not yet infected with P. aeruginosa would appear to have merit. Nonetheless, no efficacious vaccine has yet been reported in cystic fibrosis. This may be related to host factors, such as an exaggerated inflammatory response or inability to recognize specific antigens, or it may be due to the genetic and phenotypic plasticity of the organism.
Identifying antigens that are immunogenic early in cystic fibrosis, when there is the opportunity for prevention of chronic airway infection, would be a useful first step in vaccine development. Our finding of antibodies directed against P. aeruginosa in young children with cystic fibrosis led to the hypothesis that the host response might be useful in identifying the genes that are turned on early in the course of disease (3). We used a random peptide phage display library panned with cystic fibrosis patient sera in an attempt to identify potential P. aeruginosa therapeutic targets. Validation of the candidate proteins obtained via this approach was done statistically, with an in silico simulation, and by confirmation with gene expression array data, with early isolates from a natural history study of young children with cystic fibrosis (3, 34).
MATERIALS AND METHODS
Phage display library.
The J404 nonapeptide phage display library (kindly provided by J. Burrit, Montana State University) was produced in the filamentous phage M13KBst, a derivative of M13mp18 (14). The library expresses nine random amino acid peptide sequences as an amino-terminal fusion with the minor capsid protein pIII and bears a gene for kanamycin resistance (4, 5). The library has a complexity of approximately 5 × 108 unique phages. Escherichia coli XL-1 Blue was used as a recipient for the phage display library and cultured in 2×YT medium supplemented with 20 μg of tetracycline per ml.
Patient and control sera.
Human serum samples were obtained from two cystic fibrosis patients infected with P. aeruginosa for less than a year (cystic fibrosis serum), from two non-cystic fibrosis patients with acute P. aeruginosa urinary tract infection (acute-phase serum), and 14 healthy individuals (control serum). Sera were pooled within each category and tested for the presence of antibodies to P. aeruginosa exotoxin A by enzyme-linked immunosorbent assay, with recombinant exotoxin A-coated enzyme-linked immunosorbent assay plates, and a goat anti-human immunoglobulin G (Fc-specific)-horseradish peroxidase conjugate (Sigma-Aldrich Corporation, St. Louis, Mo.) for detection.
Panning of the phage library.
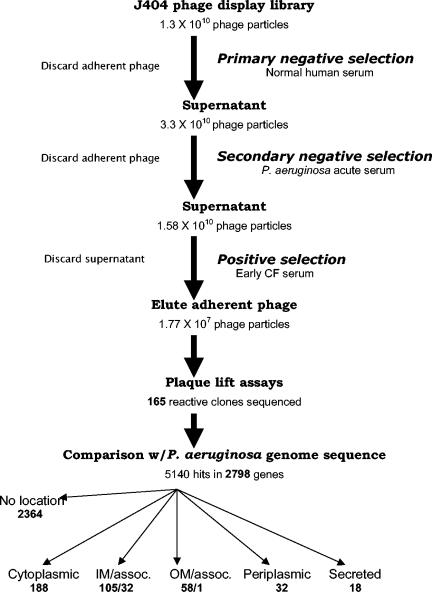
The wells of a 96-well microtiter plate were coated overnight at 4°C with anti-human immunoglobulin G (Fc specific) (Sigma) in 0.1 M NaHCO3, pH 9.6. Each well was blocked with 200 μl of 5% bovine serum albumin (Sigma) per well in phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM sodium phosphate, pH 7.2) for 1 h at room temperature and washed with PBS plus 0.05% Tween (PBS-T). Fifty microliters of the pooled control serum was added to each well, incubated for 1 h at room temperature, and washed (Fig. 1, primary negative selection). After another hour of blocking, 50 μl of the J404 nonapeptide phage display library, containing 1.3 × 1010 PFU, was added to each well and incubated for 3 h at room temperature. Unbound phage particles were used for further analysis. Phage titers in supernatants were determined by 10-fold dilutions in Luria-Bertani (LB) soft agar and subsequently amplified in E. coli XL-1 Blue. The supernatant of these expression cultures, containing the phage particles, was purified by polyethylene glycol precipitation and subjected to a second cycle of panning, this time against the acute-phase sera (Fig. 1, secondary negative selection). This panning was similar to the first except for the use of acute-phase serum in a dilution of 1:5 in 5% bovine serum albumin-PBS. Amplified phage particles from the second supernatant were used for affinity selection on cystic fibrosis serum (1:5 in 5% bovine serum albumin-PBS) (Fig. 1, positive selection). Unbound phage were removed, and the bound cystic fibrosis-specific phage were eluted with 0.1 M glycine, pH 2.2. The eluted phage particles were neutralized to pH 7 immediately.
FIG. 1.
Scheme for panning and evaluation of potential targets. Details are given in the text. The number of phage particles panned/recovered in each component is noted beneath the component. The total number of genes encoding proteins in each cytoplasmic location is noted beneath the location. IM, inner membrane; OM, outer membrane; assoc., associated.
Plaque-lift screening.
Plaques were screened for reactivity with cystic fibrosis patient sera by immunoblotting. Affinity-selected phage particles from the third cycle of panning were grown as isolated plaques overnight and blotted to a nylon membrane (Roche Diagnostics Corporation, Indianapolis, Ind.) for 4 h at room temperature, blocked with 5% nonfat dry milk-PBS for 1 h, and probed with the same cystic fibrosis sera (1:500) used for affinity selection (1 h at room temperature). The blot was washed in PBS-T, incubated with a goat anti-human immunoglobulin G-horseradish peroxidase (Sigma), washed three times with PBS-T, and developed by adding substrate solution (Super Signal West Pico Chemiluminescent Substrate; Pierce Biotechnology, Rockford, Ill.) according to the instructions of the manufacturer. Plaques that produced a signal on the blot were chosen for sequencing of the displayed inserts.
Sequencing of displayed inserts.
Phage particles with the desired reactivity in immunoblots of plaques were picked and used for infecting E. coli, and the supernatant after overnight growth was precipitated with polyethylene glycol. The pellets were lysed by boiling in 10 mM Tris-EDTA, pH 7.5. Insert DNA from individual clones was sequenced with the Big Dye terminator sequencing kit (PE Biosystems, Foster City, Calif.) with primer J534,3 (5′-GTTTTGTCGTCTTTCCAGACG-3′). Analysis of the DNA and protein sequences was done by comparison to the published sequence of P. aeruginosa PAO1 (40). Additional information was obtained from the PseudoCAP annotation project (www.pseudomonas.com).
Source of P. aeruginosa clinical isolates.
Four clinical isolates from cystic fibrosis patients in a natural history study were used to evaluate gene expression early in the course of airway infection (3). All were obtained within the first 3 years of life, and two were the first isolates from individual patients (CF471 at 24 months and CF1188 at 18 months). Two were from bronchoalveolar lavage (CF471 and CF725), and two were from the upper airway (CF1188 and CF1488).
Isolation of total bacterial RNA and chromosomal DNA.
Total bacterial RNA was isolated from mid-logarithmic-phase P. aeruginosa with Tri-Reagent according to the manufacturer's protocol (Molecular Research Center, Inc., Cincinnati, Ohio). Chromosomal DNA was isolated with the DNeasy tissue kit (Qiagen, Valencia, Calif.). For each microarray experiment, independent RNA and DNA preparations were generated for use as templates.
Microarray construction.
Microarrays consisting of 5,600 predicted open reading frames, representing 98% of the PAO1 genome, were constructed. PCR products (200 to 500 bp) were spotted with a Molecular Dynamics (Sunnyvale, Calif.) Gene III arrayer on mirrored glass slides (R. Bumgarner, University of Washington). P. aeruginosa open reading frame numbers and gene names were taken from the Pseudomonas Genome Project (www.pseudomonas.com).
Probe preparation, hybridization, and data analysis.
For labeling of experimental (cystic fibrosis clinical isolates) and control (PAO1) RNAs, 20 μg of total bacterial RNA, 1 μg of GC-rich random hexamer (NSNSNSNSNS), and 1 ng of in vitro-transcribed green fluorescent protein (GFP) mRNA in a final volume of 9.5 μl were heated to 70°C for 10 min and then chilled on ice. To this mixture, 5 μl of 10x first-strand buffer (Gibco-BRL, Rockville, Md.), 2 μl of 0.1 M dithiothreitol, 1 μl of deoxynucleoside triphosphate mix (5 mM each dATP, dGTP, and dTTP and 1 mM dCTP) (Gibco-BRL, Rockville, Md.), 2 μl (2 nmol) of indocarbocyanine- or indodicarbocyanine-labeled dCTP (Amersham-Pharmacia, Piscataway, N.J.), 0.5 μl of RNasin (Promega, Madison, Wis.), and 1 μl of Superscript II RNA-dependent DNA polymerase (Gibco-BRL) were added. Following the addition of Superscript, the labeling reactions were incubated at room temperature for 10 min and then incubated at 42°C for 2 h. The RNA template was hydrolyzed by the addition of 1 μl of 5 M NaOH and incubated at 37°C for 15 min; 5 μl of 2 M MOPS (morpholinepropanesulfonic acid, pH 7.0) was added to neutralize the NaOH, and the volume was adjusted to 50 μl with H2O.
Labeled cDNA was purified, and probes were prepared and hybridized as previously described (16). Slides were scanned with a Molecular Dynamics confocal dual-laser scanner at 532 and 633 nm. Data for each independent experiment were analyzed with custom array analysis software developed by R. Bumgarner (Spot-On Software package). Image processing, data normalization, and error analysis were performed as previously described (16). For each cystic fibrosis clinical isolate analyzed, a minimum of two independent microarray experiments (two arrays and eight individual DNA spots per assay) were performed and analyzed independently to produce expression ratios and error estimates.
Statistical analysis.
BLAST was used to align the phage clone sequences with the published sequence of P. aeruginosa PAO1 (1). The expected value of 3,000 allowed matches with as few as three of nine amino acids in common, allowing the search for homologues to be broad. The identified epitopes were characterized in terms of cellular location and functional class in order to determine the accessibility of the protein to immune recognition and the likelihood of expression during early cystic fibrosis airway infection. To determine whether the observed epitopes were as likely to have been identified as any other set of random epitopes, computer simulation was employed to generate 100 random models of the PAO1 genome. The “proteins” in these models had peptide lengths identical to that of the PAO1 genome, since the number of times sequence alignments above the threshold (hits) occurred for each gene is sensitive to gene length. The amino acids for each “protein” were randomly selected with the same frequency of occurrence as in the PAO1 genome.
The phage nonapeptides were aligned with the set of each model genome to construct a distribution of the number of BLAST hits per “gene.” This provided a mechanism for estimating how many hits could be expected by chance alone as well as the error associated with these estimates. The standard deviation over the model genomes for each of the multiple hits per gene in the distribution was calculated as a measure of variability expected for random sequence. The probability of multiple BLAST hits of the nonapeptides for PAO1 was then compared with the probability of hits within “genes” from the computer-generated genomes. Confirmation of the accuracy of the model came from the finding that, within error, the distribution for PAO1 is identical to that of the simulated genomes. Similar distributions were calculated for two classes of proteins: known or predicted outer membrane and secreted proteins and a random subset of cytosolic proteins.
RESULTS
Affinity selection for clones with binding activity to antibodies in patient serum.
To identify immunogenic pseudomonal proteins, the J404 random nonapeptide phage display library was screened for peptides recognized by antibodies in serum from cystic fibrosis patients recently infected with P. aeruginosa. The affinity selection process (panning) used both primary and secondary negative selection of the phage display library, followed by positive selection with pooled cystic fibrosis patient sera (Fig. 1). At each step in panning, 4 × 1010 phage particles were incubated with the serum antibodies. The number of nonbinding phages was quantified after the first two affinity selections and in the eluate of the third panning. The phage titer in the supernatant decreased approximately twofold after two cycles of negative selection (normal human sera and acute-phase P. aeruginosa infection sera, respectively) and by more than 1,000-fold in the eluate of the third panning (positive selection with cystic fibrosis sera). This is indicative of specific enrichment, since nonspecific clones were removed by the preceding negative selections.
Identification of potential PAO1 genome targets.
To identify those nonapeptides resembling P. aeruginosa antigens, clones from the eluate of the final panning were used in plaque lift assays, and reactive clones were sequenced. A total of 165 immunoreactive phage clones were sequenced. BLAST was used to translate the sequences and compare them to the published sequence of P. aeruginosa PAO1 (www.pseudomonas.com). The BLAST search of the 165 sequences with an expected value of 3,000 resulted in 5,140 hits in 2,798 P. aeruginosa proteins (Fig. 1). Between three and nine amino acids of a nonapeptide were identical to a nine-amino-acid section of a query protein. For a single peptide, PA4100, all nine amino acids were identical. However, in more than 50% of the displayed peptides, no more than five amino acids matched the respective pseudomonal sequence. This analysis identified proteins that had multiple nonapeptide matches and included many known or predicted outer membrane and secreted proteins (http://www.cmdr.ubc.ca/bobh/omptotal.html).
Single P. aeruginosa proteins were hit between 1 and 18 times by independent clones. One thousand two hundred sixty-seven proteins were hit two or more times in different positions. The entire list of genes that were hit is available from J.L.B. Genes that were identified six or more times (total, 51 genes) are shown in Table 1. Four hundred twenty-two genes (1,021 hits) were identical, overlapping (at most six amino acids apart), or in close proximity (up to 25 amino acids apart). The greatest number of hits was in gene PA4982. This gene, encoding a probable two-component sensor, was recognized 18 times, including two identical and nine overlapping hits. Further multiple hits (greater than 10) were in PA0690 (14 hits), a hypothetical protein with 38% identity to a 100-kDa heme:hemopexin-binding protein of Haemophilus influenzae (9), and in PA3206 (11 hits), another probable two-component sensor. Each of these two proteins showed a high number of overlapping or proximate hits. The high frequency of recognition of these three proteins and the large number of overlapping hits within them suggests that they are antigenic and that the actual immunogenic epitope is larger than the respective nonapeptide. In addition, it implies that they are valid candidates that should be investigated further as possible therapeutic or vaccine targets.
TABLE 1.
Frequency of hits comparing results of phage display library with the PAO1 genome
| Gene | Product | Functional classa | No. of hits |
|---|---|---|---|
| PA4982 | Probable two-component sensor | Two-component regulatory systems | 18 |
| PA0690 | Hypothetical protein | HUU | 14 |
| PA3206 | Probable two-component sensor | Two-component regulatory systems | 11 |
| PA3327 | Probable nonribosomal peptide synthetase | Adaptation/protection | 10 |
| PA1895 | Hypothetical protein | HUU/membrane protein | 9 |
| PA2402 | Probable nonribosomal peptide synthetase | Putative enzyme | 9 |
| PA2424 | Probable nonribosomal peptide synthetase | Adaptation/protection | 9 |
| PA2727 | Hypothetical protein | HUU | 9 |
| PA3272 | Probable ATP-dependent DNA helicase | DNA RRMR | 9 |
| PA0413 | Probable component of chemotactic signal transduction system | Two-component system/chemotaxis/motility/attachment | 8 |
| PA1406 | Hypothetical protein | HUU | 8 |
| pscP | Translocation protein in type III secretion | Protein secretion/export | 8 |
| PA3840 | Conserved hypothetical protein | HUU | 8 |
| PA3907 | Hypothetical protein | HUU | 8 |
| PA3969 | Conserved hypothetical protein | HUU | 8 |
| PA4112 | Probable sensor/response regulator hybrid | Two-component regulatory system | 8 |
| PA4489 | Conserved hypothetical protein | HUU | 8 |
| PA0449 | Hypothetical protein | HUU | 7 |
| PA0470 | Probable hydroxamate-type ferrisiderophore receptor | Small-molecule transport | 7 |
| PA0845 | Conserved hypothetical protein | HUU | 7 |
| PA1433 | Conserved hypothetical protein | HUU/membrane protein | 7 |
| sucA | 2-Oxoglutarate dehydrogenase (E1 subunit) | Energy metabolism/amino acid biosynthesis | 7 |
| PA1874 | Hypothetical protein | HUU | 7 |
| PA2302 | Probable nonribosomal peptide synthetase | Putative enzyme | 7 |
| czcA | RND divalent metal cation efflux transporter CzcA | Transport/membrane protein | 7 |
| ftsK | Cell division protein FtsK | Cell division | 7 |
| PA3340 | Hypothetical protein | HUU/membrane protein | 7 |
| PA3986 | Hypothetical protein | HUU | 7 |
| rpoB | DNA-directed RNA polymerase beta chain | Transcription/RNA processing | 7 |
| priA | Primosomal protein N′ | DNA RRMR | 7 |
| PA5114 | Hypothetical protein | HUU/membrane protein | 7 |
| ppkA | Serine/threonine protein kinase PpkA | Adaptation/protection/TPMD | 6 |
| PA0669 | Probable DNA polymerase alpha chain | DNA RRMR/putative enzyme | 6 |
| PA0695 | Hypothetical protein | HUU | 6 |
| PA0719 | Hypothetical protein of bacteriophage Pf1 | HUU/phage, transposon, plasmid | 6 |
| PA0791 | Probable transcriptional regulator | Transcriptional regulator | 6 |
| PA1134 | Hypothetical protein | HUU | 6 |
| napB | Cytochrome c-type protein NapB precursor | Energy metabolism | 6 |
| napA | Periplasmic nitrate reductase protein NapA | Energy metabolism | 6 |
| PA1336 | Probable two-component sensor | Two-component regulatory system | 6 |
| PA1364 | Probable transmembrane sensor | Membrane protein/transcriptional regulator | 6 |
| PA1669 | Hypothetical protein | HUU/membrane protein | 6 |
| PA1782 | Probable serine/threonine-protein kinase | Adaptation/protection | 6 |
| cti | cis/trans isomerase | Fatty acid and phospholipid metabolism | 6 |
| PA2108 | Probable decarboxylase | Putative enzyme | 6 |
| PA2138 | Probable ATP-dependent DNA ligase | DNA RRMR | 6 |
| PA2305 | Probable nonribosomal peptide synthetase | Putative enzyme | 6 |
| PA2383 | Probable transcriptional regulator | Transcriptional regulator | 6 |
| PA2600 | Hypothetical protein | HUU | 6 |
| PA2819 | Hypothetical protein | HUU | 6 |
| PA2928 | Hypothetical protein | HUU | 6 |
| PA3075 | Hypothetical protein | HUU | 6 |
| PA3297 | Probable ATP-dependent helicase | Transcription, RNA processing | 6 |
| narG | Respiratory nitrate reductase alpha chain | Energy metabolism | 6 |
| PA4186 | Hypothetical protein | HUU | 6 |
| fptA | Fe(III)-pyochelin receptor precursor | Small-molecule transport | 6 |
| pchF | Pyochelin synthetase | Secreted factor/transport | 6 |
| PA4601 | Conserved hypothetical protein | HUU/membrane protein | 6 |
| tatB | Translocation protein TatB | Protein secretion/export | 6 |
| PA5384 | Probable lipolytic enzyme | Putative enzyme | 6 |
| PA5399 | Probable ferridoxin | Energy metabolism | 6 |
HUU, hypothetical, unclassified, unknown; DNA RRMR, DNA replication, recombination, modification, and repair; TPMD, translation, posttranslational modification, degradation.
Classification of phage clones by cellular location and function.
Cellular location impacts the ability of a given protein to elicit a host immune response. Thus, the cellular locations of the identified hits were examined. Two thousand three hundred sixty four of the 2,798 P. aeruginosa proteins hit had no cellular location identified (84.5%). This percentage is typical of the annotation of the entire Pseudomonas genome, where only 15% of proteins have an identified cellular location (www.pseudomonas.com). Of the remaining 434 hits, 188 different consensus sequences were identified from the phage display library that have homology to cytoplasmic proteins, 105 to inner membrane proteins, 32 to inner membrane associated proteins, 32 to periplasmic proteins, 58 to known and putative outer membrane proteins, 1 to an outer membrane-associated protein, and 18 to secreted proteins from the P. aeruginosa genome.
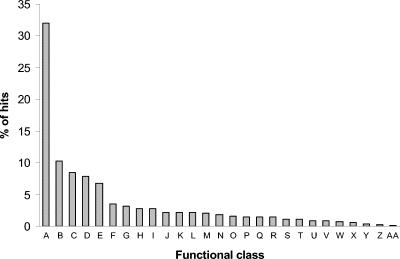
All proteins from the Pseudomonas PAO1 genome have also been classified into 27 functional classes according to their predicted or known function (www.pseudomonas.com). Thus, the distribution of functional classes of the 2,798 pseudomonal proteins that were identified with phage display was examined as well (Fig. 2). Most proteins that were identified belong to the hypothetical-unknown-unclassified class (32%), followed by membrane proteins (10.2%), proteins involved in the transport of small molecules (8.5%), and putative enzymes (7.9%). The percentage of gene products in different functional classes identified by phage display ranged from 41.4% for cell division proteins up to 85.7% (six of seven) for the proteins of the quinolone signal response (Pseudomonas quinolone signal [PQS]).
FIG. 2.
Distribution of functional classes of Pseudomonas genes that were sequenced from the J404 phage display library. Proteins of all 27 known functional classes were hit: (A) hypothetical, unknown, and unclassified, (B) membrane proteins, (C) small-molecule transport, (D) putative enzymes, (E) transcriptional regulators, (F) two-component regulatory systems, (G) amino acid biosynthesis, (H) energy metabolism, (I) adaptation and protection, (J) carbon compound catabolism, (K) translation, posttranslational modification, and degradation, (L) DNA replication, recombination, modification, and repair, (M) biosynthesis of cofactors and prosthetic groups, (N) cell wall, lipopolysaccharide, and capsule, (O) secreted factors, (P) protein secretion and export apparatus, (Q) motility and attachment, (R) central internal metabolism, (S) transcription and RNA processing, (T) nucleotide biosynthesis and metabolism, (U) fatty acid and phospholipid metabolism, (V) chemotaxis, (W) related to phages, transposons, and plasmids, (X) chaperones and heat shock proteins, (Y) cell division, (Z) antibiotic resistance/susceptibility, and (AA) quinolone signal response.
Characterization of hits in outer membrane and secreted proteins.
Since outer membrane and secreted proteins are generally more accessible and might stimulate an immune response, it seems likely that they would be more frequently recognized by cystic fibrosis patient sera. Sequence analysis and comparison of reactive phage peptides with the P. aeruginosa genome identified 58 outer membrane proteins (OMPs) (129 hits), corresponding to 48.3% of all known OMPs, and 18 secreted proteins (32 hits), corresponding to 69.2% of the known secreted proteins. Twelve functional classes were identified among the recognized OMPs, the most common being proteins involved in transport of small molecules (35.9%), followed by membrane proteins (27.1%), hypothetical-unknown-unclassified (15.3%), and proteins involved in secretion and export of proteins (11.8%). Several proteins were identified multiple times. PA0470, a probable hydroxamate-type ferrisiderophore receptor, was recognized seven times, and FptA (PA4221), an Fe(III)-pyochelin receptor precursor, was hit six times. Both proteins are involved in iron acquisition. Furthermore, PA1322, PA2911, and PA4837, three homologs of ferrichrome-iron receptors from other microorganisms, were each identified five times. PA1365 and PA0931, both siderophore receptor proteins, were recognized four and two times, respectively. These results suggest that iron acquisition proteins are expressed early in cystic fibrosis airway infections and are antigenic.
Seven different functional classes were identified among the secreted proteins. The majority of these belong to the category of secreted factors (60.5%). Other frequently identified classes included proteins associated with adaptation and protection (18.6%) and proteins involved in translation, posttranslational modification, and degradation (7%).
Pyocins (secreted factors) were also identified multiple times, including PA3866 (pyocin S4) was recognized four times, PA1150 (pyocin S2) three times, and PA0985 (pyocin S5). Pyocin S4 is lethal through tRNase activity (29). S-type pyocins (such as S2) are bacteriocins that have two components, a protein with DNase activity and an immunity protein that confers protection on the producing strain (36, 35, 13). Pyocin S5 kills bacterial cells through pore-forming activity. Although the production of pyocins in P. aeruginosa strains from cystic fibrosis patients can be lost over time (33), these data suggest that they are expressed in some strains early in infection and are antigenic.
Among the recognized secreted peptides were two clones with homology to ExoA (PA1148), the exotoxin A precursor. Cystic fibrosis patients infected with P. aeruginosa usually have high titers of anti-exotoxin A antibodies in their serum, and an enzyme-linked immunosorbent assay-based assay can be used for identification of infection (3, 42). Thus, the presence and identification of these clones help validate the method and experimental approach.
In silico analysis.
Identification of multiple hits within a specific protein was used to suggest the relative importance of that protein as a target. Because an expected value of 3,000 allowed matches with as few as three of nine amino acids in common, it is not reasonable to infer that any particular gene sequence found in the BLAST alignment is an epitope. However, an apparent predominance of multiple sequence alignment hits to outer membrane proteins led us to investigate whether hits to outer membrane or secreted proteins occurred more often than would be expected by chance.
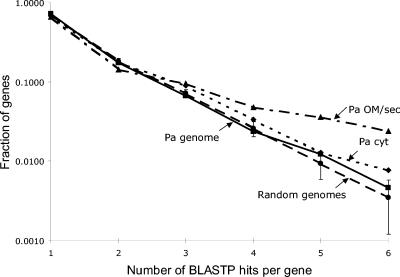
A simulation method was used to determine the distribution and its variability for multiple hits against 100 computer-generated pseudogenomes (random genomes in Fig. 3). The number of multiple hits to the P. aeruginosa genome (Pa genome in Fig. 3) is identical to the average of the random genomes within two standard deviations, indicating that the computer-generated genomes are a reasonable model for the P. aeruginosa genome. The number of BLAST hits per gene for the outer membrane and secreted proteins (Pa OM/sec in Fig. 3) is more than two standard deviations above the distribution for three or more hits, indicating that the outer membrane and secreted proteins are selected in the search more often than expected by random amino acid sequences. A set of 390 cytoplasmic proteins that would not be expected to be recognized unless a significant fraction of the bacteria were lysed in the experiment were also randomly chosen. The number of BLAST hits per gene for this set of cytoplasmic proteins (Pa cyt in Fig. 3) was not significantly different from the results for the entire P. aeruginosa genome.
FIG. 3.
Distribution of BLAST hits per gene for the P. aeruginosa (Pa) genome compared with the average over 100 random genomes. The variance of the random genome distribution for each number of BLAST hits per gene is indicated by ±2 standard deviation error bars. A selected set of 76 outer membrane and secreted proteins (Pa OM/sec) and 390 cytoplasmic proteins (Pa cyt) are also shown.
Comparison of phage display and gene array data.
A second method that was used to validate the importance of specific targets in the pathogenesis of cystic fibrosis airway disease was comparison of phage display results with transcriptional profiles of early cystic fibrosis clinical isolates. Microarray analysis was performed with four isolates from cystic fibrosis patients (CF471, CF725, CF1188, and CF1488) from early infection (3) and compared to the laboratory strain PAO1. The resulting transcriptional profiles indicated altered gene expression in isolates from cystic fibrosis patients compared to PAO1, both activation and repression of specific genes.
Transcriptional data on cystic fibrosis-activated genes was the target of comparison, because those genes encode proteins that are expressed during cystic fibrosis infection and therefore more likely to stimulate an immune response in cystic fibrosis patient serum. A total of 159 genes (277 hits) were identified by phage display among the 292 cystic fibrosis-activated genes (54.4%). A full listing of the 159 genes is available from J.L.B. Overlaps between the data sets were found with seven genes from isolate CF471 (70% of cystic fibrosis-activated genes in that strain), fourteen genes from isolate CF1488 (48.3%), 22 genes from isolate CF725 (57.9%), and 125 genes from isolate CF1188 (54.1%).
Among the four early isolates from cystic fibrosis patients, 10 genes were identified that were activated in at least two different strains (Table 2). Among those 10 were seven genes that were both activated in more than one strain and had multiple hits in the nonapeptide library. Several proteins were of particular interest because of a known or putative role in pathogenesis. PA0470 encodes a probable outer membrane hydroxamate-type ferrisiderophore receptor that was hit seven times in the phage display library. A second gene of interest is PA4175 (prpL), encoding a probable ArgC precursor that was activated in two early isolates from cystic fibrosis patients and was hit twice in the phage display library. PrpL, a secreted PvdS-regulated endoprotease, cleaves casein, lactoferrin, transferrin, elastin, and decorin and contributes to the ability of P. aeruginosa to persist in a rat chronic pulmonary infection model (43). Three of the identified proteins had homology to conserved hypothetical proteins (PA2854, PA3919, and PA4897). Also activated in multiple strains were two transcriptional regulators, algU, which encodes an alternative sigma factor, and PA3508, which encodes a probable transcriptional activator. Two genes that were hit twice and found to be activated in two strains were those encoding a probable asparagine synthetase (PA2084) and a probable permease from an ABC transporter (PA5095). A gene encoding a protein activator (Pra), which is a membrane-associated Ras-like protein that modulates GTP synthesis, was also identified (6).
TABLE 2.
Overlaps seen between cystic fibrosis-activated genes and those identified as immunogenic by phage display
| Gene | Gene product | Functional classa | No. of strains activated | No. of hits in nonapeptide library |
|---|---|---|---|---|
| PA0470 | Probable hydroxamate-type ferrisiderophore receptor | Small-molecule transport | 2 | 7 |
| algU | Sigma factor AlgU | Transcriptional regulator | 2 | 2 |
| PA2084 | Probable asparagine synthetase | Amino acid biosynthesis and metabolism | 2 | 2 |
| PA2854 | Conserved hypothetical protein | HUU | 2 | 5 |
| PA3508 | Probable transcriptional regulator | Transcriptional regulator | 2 | 1 |
| PA3919 | Conserved hypothetical protein | HUU | 2 | 1 |
| prpL | Probable endoproteinase ArgC precursor | Putative enzyme | 2 | 2 |
| pra | Protein activator | Carbon compound catabolism; small-molecule transport | 2 | 1 |
| PA4897 | Hypothetical protein | HUU | 2 | 2 |
| PA5095 | Probable permease of ABC transporter | Membrane protein; small-molecule transport | 2 | 2 |
HUU, hypothetical, unclassified, unknown.
Expression arrays from the early isolates from cystic fibrosis patients revealed cystic fibrosis-activated genes in all 27 functional classes with the single exception of the quinolone signal response class. Although the detection by phage display of six of the seven proteins in the PQS pathway suggests that these proteins are antigenic, transcriptional levels in none of the four isolates appeared different from that of the laboratory strain PAO1. PQS is required for the synthesis of secondary metabolites and extracellular enzymes, and mutations in PQS genes reduce the expression of pyocyanin and hydrogen cyanide biosynthesis (19).
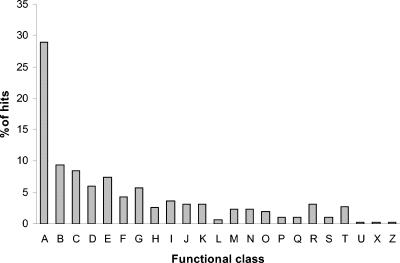
Proteins from 23 of the 26 functional classes were identified by both microarray and phage display data. No overlapping proteins were found in the categories of cell division, chemotaxis, or proteins related to phages. The distribution of functional classes is otherwise quite similar to that identified by phage display alone (Fig. 4). The majority of proteins belonged to the hypothetical, unclassified, unknown class (29%), followed by the membrane proteins (9.4%), proteins involved in the transport of small molecules (8.5%), and transcriptional regulators (7.4%).
FIG. 4.
Distribution of functional classes overlapping the microarray data. The transcriptional profiles of four isolates from cystic fibrosis patients were compared to that of PAO1. All cystic fibrosis-activated genes were used for comparison to the phage display data. Overlaps were found in 23 of 26 functional classes: (A) hypothetical, unknown, and unclassified, (B) membrane proteins, (C) small-molecule transport, (D) putative enzymes, (E) transcriptional regulators, (F) two-component regulatory systems, (G) amino acid biosynthesis, (H) energy metabolism, (I) adaptation and protection, (J) carbon compound catabolism, (K) translation, posttranslational modification, and degradation, (L) DNA replication, recombination, modification, and repair, (M) biosynthesis of cofactors and prosthetic groups, (N) cell wall, lipopolysaccharide, and capsule, (O) secreted factors, (P) protein secretion and export apparatus, (Q) motility and attachment, (R) central internal metabolism, (S) transcription and RNA processing, (T) nucleotide biosynthesis and metabolism, (U) fatty acid and phospholipid metabolism, (X) chaperones and heat shock proteins, and (Z) antibiotic resistance and susceptibility.
Among the 159 overlapping proteins were eight outer membrane proteins (16 hits) and three secreted proteins (nine hits) that were both detected by phage display and upregulated in the early isolates from cystic fibrosis patients. Outer membrane proteins that were detected by both methods include the probable hydroxamate-type ferrisiderophore receptor (PA0470) mentioned above, four hypothetical proteins (PA1613, PA1875, PA4540, and PA4897), and a probable secretion protein (PA4974). In addition, the OprM precursor (PA0427), which is part of the mexAB-oprM multiple antibiotic resistance efflux operon (25), and the probable fimbrial biogenesis usher protein CupB3 (PA4084) were identified in both analyses. The three known cup clusters encode different types of fimbrial structures and have a key role in the attachment of microorganisms to surfaces, a mechanism used to colonize tissues as well as abiotic surfaces (41). Among the secreted proteins in addition to PrpL were pyocins S2 and S4. Both were recognized in the nonapeptide library multiple times.
DISCUSSION
Development of an efficacious vaccine against P. aeruginosa that could be administered early in cystic fibrosis is an important goal, because antibiotic treatment of chronic infections almost never results in clearance of the organism (30, 32, 37). This is in contrast to the treatment of early P. aeruginosa infections in cystic fibrosis, where bacterial eradication has been reported (18, 20, 31). Chronic infections may be more difficult to treat because of the adaptive changes the organism undergoes in the airway of cystic fibrosis patients. Thus, prevention of colonization and early infection is imperative. Immunization or other therapy aimed at targets expressed early in cystic fibrosis lung infection is an attractive approach. The goal of this project was to identify accessible and immunogenic epitopes to target for vaccine development.
We describe the use of a random peptide phage display library to identify immunogenic proteins from P. aeruginosa. The use of phage display for the identification of immunogenic proteins has been demonstrated to be useful for development of vaccines against human diseases such as cancer (17, 38, 39) and many diverse human pathogens, including Neisseria meningitides (28, 22, 27), Echinococcus granulosus (21), and Trypanosoma cruzi (2). However, the use of a random peptide phage display library rather than a recombinant library made with DNA from the pathogen of interest is a novel technique that has been used in N. meningitidis to identify mimotopes of nonpeptide antigens (28). In the current study, only protein antigens have been examined thus far.
Two thousand seven hundred ninety-eight potentially immunogenic proteins were identified within the P. aeruginosa genome in the current study. Because such a large number of potential genome targets was identified, it was necessary to parse the data in order to designate specific proteins as valid candidates to explore. Thus, three criteria were established to designate proteins for further investigation: accessibility, immunogenicity, and expression in early isolates from cystic fibrosis patients.
Because accessibility was an important criterion for identification of candidate antigens, outer membrane and secreted proteins were a focus of this investigation. Of the 2,798 protein sequences hit, 76 were outer membrane or secreted. The most common outer membrane proteins identified were those involved in the transport of small molecules, particularly siderophores and proteins associated with iron acquisition. Among the secreted proteins, those designated secreted factors were most frequently identified, including three pyocins.
The number of times that a specific protein was hit was hypothesized to be a good measure of immunogenicity, especially if the same gene was hit in contiguous or overlapping sequences by independent clones. One thousand two hundred sixty seven genes were hit multiple times, and 422 were contiguous or overlapping. Three proteins particularly stood out with these criteria, including two two-component sensors and a heme:hemopexin binding protein (involved in iron acquisition). The high frequency of recognition of these three proteins and the large number of overlapping hits within them suggest that they are antigenic and likely to be expressed.
With accessibility and immunogenicity as criteria for further investigation, the number of candidate antigens was significantly decreased. This approach was validated by an in silico analysis of hits within specific protein genes. A Monte Carlo estimation of the frequency of variability for random alignments with BLAST was used to determine whether the multiple hits seen in outer membrane and secreted proteins were attributable to random chance (26, 15). The analysis performed here, with computer-generated genomes, demonstrated a clear difference between the number of hits within the entire PAO1 genome and the number within genes encoding outer membrane or secreted proteins.
The third criterion used to prioritize proteins for further investigation was expression in early cystic fibrosis airway isolates. Gene expression arrays were used to identify the proteins expressed in vitro by isolates obtained from cystic fibrosis patients early during infection. Comparing the data obtained from expression arrays with that generated by the phage display library, there was a significant overlap. Nearly 55% of the products of cystic fibrosis-activated genes were also identified by the phage display analysis. In the group of 159 proteins identified in both analyses were 11 outer membrane and secreted proteins, including those associated with iron uptake, antibiotic efflux, and fimbrial biogenesis. There were two proteins that were recognized in more than one clinical isolate, were hit multiple times in the phage display analysis, and are likely to be associated with virulence: these were a probable siderophore receptor (PA0470) and a secreted endoprotease that contributes to persistence in an animal model of chronic P. aeruginosa infection (prpL).
In addition to proteins specifically expressed in early isolates from cystic fibrosis patients, proteins that are always expressed in P. aeruginosa, whether from cystic fibrosis patients or not, were examined. Key among these were proteins from the PQS pathway. Since PQS is neither activated nor repressed in early or late isolates from cystic fibrosis patients (unpublished data) compared to PAO1, the proteins are probably present in all stages of the infection. Six of the seven proteins in the pathway were identified in the phage display analysis, suggesting that these are accessible and would potentially be good vaccine targets.
Taken together, the remarkable overlap between the transcriptome analysis and the phage screen for immunogenic proteins validates this experimental strategy. The data from this study demonstrate that an unbiased phage affinity approach can be used to identify novel putative vaccine candidates. The quality of these potential targets is reflected in multiple overlapping hits with high homology. The results presented here also suggest the importance of specific proteins during the natural course of infection with P. aeruginosa. Those candidates that have been suggested to be associated with virulence in animal models will be the first to be explored in future studies.
Acknowledgments
We thank James Burritt for providing the phage display library, Larry Gallagher for ongoing collaboration and advice, Jessica Foster for technical assistance, and Madonna Gordon and Alan Genatossio for obtaining patient samples.
This work was supported by a program project grant from the Cystic Fibrosis Foundation (program director, Samuel Miller; principal investigator, Jane Burns).
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, P., M. S. Leguizamon, C. A. Buscaglia, T. A. Pitcovsky, and O. Campetella. 2001. Multiple overlapping epitopes in the repetitive unit of the shed acute-phase antigen from Trypanosoma cruzi enhance its immunogenic properties. Infect. Immun. 69:7946-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444-452. [DOI] [PubMed] [Google Scholar]
- 4.Burritt, J. B., C. W. Bond, K. W. Doss, and A. J. Jesaitis. 1996. Filamentous phage display of oligopeptide libraries. Anal. Biochem. 238:1-13. [DOI] [PubMed] [Google Scholar]
- 5.Burritt, J. B., M. T. Quinn, M. A. Jutila, C. W. Bond, and A. J. Jesaitis. 1995. Topological mapping of neutrophil cytochrome b epitopes with phage-display libraries. J. Biol. Chem. 270:16974-16980. [DOI] [PubMed] [Google Scholar]
- 6.Chopade, B. A., S. Shankar, G. W. Sundin, S. Mukhopadhyay, and A. M. Chakrabarty. 1997. Characterization of membrane-associated Pseudomonas aeruginosa Ras-like protein Pra, a GTP-binding protein that forms complexes with truncated nucleoside diphosphate kinase and pyruvate kinase to modulate GTP synthesis. J. Bacteriol. 179:2181-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciofu, O., B. Giwercman, S. S. Pedersen, and N. Hoiby. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS 102:674-680. [PubMed] [Google Scholar]
- 8.Conway, S. P., K. G. Brownlee, M. Denton, and D. G. Peckham. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321-332. [DOI] [PubMed] [Google Scholar]
- 9.Cope, L. D., S. E. Thomas, J. L. Latimer, C. A. Slaughter, U. Muller-Eberhard, and E. J. Hansen. 1994. The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13:863-873. [DOI] [PubMed] [Google Scholar]
- 10.Cryz, S. J., Jr., A. Lang, A. Rudeberg, J. Wedgwood, J. U. Que, E. Furer, and U. Schaad. 1997. Immunization of cystic fibrosis patients with a Pseudomonas aeruginosa O-polysaccharide-toxin A conjugate vaccine. Behring Inst. Mitt. 98:345-349. [PubMed] [Google Scholar]
- 11.Cystic Fibrosis Foundation. 2003. 2002 CF data registry. Cystic Fibrosis Foundation, Bethesda, Md.
- 12.Doring, G., and F. Dorner. 1997. A multicenter vaccine trial using the Pseudomonas aeruginosa flagella vaccine IMMUNO in patients with cystic fibrosis. Behring Inst. Mitt. 98:338-344. [PubMed] [Google Scholar]
- 13.Duport, C., C. Baysse, and Y. Michel-Briand. 1995. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J. Biol. Chem. 270:8920-8927. [DOI] [PubMed] [Google Scholar]
- 14.Ebright, R., Q. Dong, and J. Messing. 1992. Corrected nucleotide sequence of M13mp18 gene III. Gene 114:81-83. [DOI] [PubMed] [Google Scholar]
- 15.Efron, B., and R. J. Tibshirani. 1993. An introduction to the bootstrap. Chapman and Hall, New York, N.Y.
- 16.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5:1341-1349. [DOI] [PubMed] [Google Scholar]
- 17.Fossa, A., L. Alsoe, R. Crameri, S. Funderud, G. Gaudernack, and E. B. Smeland. 2004. Serological cloning of cancer/testis antigens expressed in prostate cancer using cDNA phage surface display. Cancer Immunol. Immunother. 53:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frederiksen, B., C. Koch, and N. Hoiby. 1999. Changing epidemiology of Pseudomonas aeruginosa infection in Danish cystic fibrosis patients (1974-1995). Pediatr. Pulmonol. 28:159-166. [DOI] [PubMed] [Google Scholar]
- 19.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, R. L., J. Emerson, S. McNamara, J. L. Burns, M. Rosenfeld, A. Yunker, N. Hamblett, F. Accurso, M. Dovey, P. Hiatt, M. W. Konstan, R. Moss, G. Retsch-Bogart, J. Wagener, D. Waltz, R. Wilmott, P. L. Zeitlin, and B. Ramsey. 2003. Significant microbiological effect of inhaled tobramycin in young children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:841-849. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Sapienza, G., and R. E. Cachau. 2003. Identification of critical residues of an immunodominant region of Echinococcus granulosus antigen B. J. Biol. Chem. 278:20179-20184. [DOI] [PubMed] [Google Scholar]
- 22.Grothaus, M. C., N. Srivastava, S. L. Smithson, T. Kieber-Emmons, D. B. Williams, G. M. Carlone, and M. A. Westerink. 2000. Selection of an immunogenic peptide mimic of the capsular polysaccharide of Neisseria meningitidis serogroup A using a peptide display library. Vaccine 18:1253-1263. [DOI] [PubMed] [Google Scholar]
- 23.Hoiby, N., and C. Koch. 2000. Maintenance treatment of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Thorax 55:349-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung, A., I. Kleinau, G. Schonian, A. Bauernfeind, C. Chen, M. Griese, G. Doring, U. Gobel, U. Wahn, and K. Paul. 2002. Sequential genotyping of Pseudomonas aeruginosa from upper and lower airways of cystic fibrosis patients. Eur. Respir. J. 20:1457-1463. [DOI] [PubMed] [Google Scholar]
- 25.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manly, B. 1997. Randomization, bootstrap, and Monte Carlo methods in biology, 2nd ed. Chapman and Hall, London, United Kingdom.
- 27.Moe, G. R., and D. M. Granoff. 2001. Molecular mimetics of Neisseria meningitidis serogroup B polysaccharide. Int. Rev. Immunol. 20:201-220. [DOI] [PubMed] [Google Scholar]
- 28.Moe, G. R., S. Tan, and D. M. Granoff. 1999. Molecular mimetics of polysaccharide epitopes as vaccine candidates for prevention of Neisseria meningitidis serogroup B disease. FEMS Immunol. Med. Microbiol. 26:209-226. [DOI] [PubMed] [Google Scholar]
- 29.Parret, A. H., and R. De Mot. 2002. Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other gamma-proteobacteria. Trends Microbiol. 10:107-112. [DOI] [PubMed] [Google Scholar]
- 30.Ramsey, B. W., M. S. Pepe, J. M. Quan, K. L. Otto, A. B. Montgomery, J. Williams-Warren, K. M. Vasiljev, D. Borowitz, C. M. Bowman, B. C. Marshall, S. Marshall, and A. L. Smith. 1999. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. Cystic Fibrosis Inhaled Tobramycin Study Group. N. Engl. J. Med. 340:23-30. [DOI] [PubMed] [Google Scholar]
- 31.Ratjen, F., G. Doring, and W. H. Nikolaizik. 2001. Effect of inhaled tobramycin on early Pseudomonas aeruginosa colonisation in patients with cystic fibrosis. Lancet 358:983-984. [DOI] [PubMed] [Google Scholar]
- 32.Regelmann, W. E., G. R. Elliott, W. J. Warwick, and C. C. Clawson. 1990. Reduction of sputum Pseudomonas aeruginosa density by antibiotics improves lung function in cystic fibrosis more than do bronchodilators and chest physiotherapy alone. Am. Rev. Respir. Dis. 141:914-921. [DOI] [PubMed] [Google Scholar]
- 33.Romling, U., B. Fiedler, J. Bosshammer, D. Grothues, J. Greipel, H. von der Hardt, and B. Tummler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 34.Rosenfeld, M., R. L. Gibson, S. McNamara, J. Emerson, J. L. Burns, R. Castile, P. Hiatt, K. McCoy, C. B. Wilson, A. Inglis, A. Smith, T. R. Martin, and B. W. Ramsey. 2001. Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr. Pulmonol. 32:356-366. [DOI] [PubMed] [Google Scholar]
- 35.Sano, Y., and M. Kageyama. 1993. A novel transposon-like structure carries the genes for pyocin AP41, a Pseudomonas aeruginosa bacteriocin with a DNase domain homology to E2 group colicins. Mol. Gen. Genet. 237:161-170. [DOI] [PubMed] [Google Scholar]
- 36.Sano, Y., H. Matsui, M. Kobayashi, and M. Kageyama. 1993. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 175:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, A. L., G. Redding, C. Doershuk, D. Goldmann, E. Gore, B. Hilman, M. Marks, R. Moss, B. Ramsey, T. Rubio, et al. 1988. Sputum changes associated with therapy for endobronchial exacerbation in cystic fibrosis. J. Pediatr. 112:547-554. [DOI] [PubMed] [Google Scholar]
- 38.Somasundaram, R., K. Satyamoorthy, L. Caputo, H. Yssel, and D. Herlyn. 2004. Detection of HLA class II-dependent T helper antigen using antigen phage display. Clin. Exp. Immunol. 135:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somers, V. A., R. J. Brandwijk, B. Joosten, P. T. Moerkerk, J. W. Arends, P. Menheere, W. O. Pieterse, A. Claessen, R. J. Scheper, H. R. Hoogenboom, and S. E. Hufton. 2002. A panel of candidate tumor antigens in colorectal cancer revealed by the serological selection of a phage displayed cDNA expression library. J. Immunol. 169:2772-2780. [DOI] [PubMed] [Google Scholar]
- 40.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 41.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West, S. E., L. Zeng, B. L. Lee, M. R. Kosorok, A. Laxova, M. J. Rock, M. J. Splaingard, and P. M. Farrell. 2002. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA 287:2958-2967. [DOI] [PubMed] [Google Scholar]
- 43.Wilderman, P. J., A. I. Vasil, Z. Johnson, M. J. Wilson, H. E. Cunliffe, I. L. Lamont, and M. L. Vasil. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]