Abstract
Metagenomics (also referred to as environmental and community genomics) is the genomic analysis of microorganisms by direct extraction and cloning of DNA from an assemblage of microorganisms. The development of metagenomics stemmed from the ineluctable evidence that as-yet-uncultured microorganisms represent the vast majority of organisms in most environments on earth. This evidence was derived from analyses of 16S rRNA gene sequences amplified directly from the environment, an approach that avoided the bias imposed by culturing and led to the discovery of vast new lineages of microbial life. Although the portrait of the microbial world was revolutionized by analysis of 16S rRNA genes, such studies yielded only a phylogenetic description of community membership, providing little insight into the genetics, physiology, and biochemistry of the members. Metagenomics provides a second tier of technical innovation that facilitates study of the physiology and ecology of environmental microorganisms. Novel genes and gene products discovered through metagenomics include the first bacteriorhodopsin of bacterial origin; novel small molecules with antimicrobial activity; and new members of families of known proteins, such as an Na+(Li+)/H+ antiporter, RecA, DNA polymerase, and antibiotic resistance determinants. Reassembly of multiple genomes has provided insight into energy and nutrient cycling within the community, genome structure, gene function, population genetics and microheterogeneity, and lateral gene transfer among members of an uncultured community. The application of metagenomic sequence information will facilitate the design of better culturing strategies to link genomic analysis with pure culture studies.
INTRODUCTION
Microbiology has experienced a transformation during the last 25 years that has altered microbiologists' view of microorganisms and how to study them. The realization that most microorganisms cannot be grown readily in pure culture forced microbiologists to question their belief that the microbial world had been conquered. We were forced to replace this belief with an acknowledgment of the extent of our ignorance about the range of metabolic and organismal diversity.
This change fomented a revolution in microbiological thought. At the heart of this revolution was the convincing demonstration that the uncultured microbial world far outsized the cultured world and that this unseen world could be studied (105-108). This change in thinking was prompted by another, equally important realization: microorganisms underpin most of the geochemical cycles and many human health conditions that were previously thought to be driven by inorganic processes and stress, respectively. The glimmers of insight into the influence that microorganisms exert on the world propelled microbiologists to pursue the uncultured world. In 1931, Waksman optimistically believed that “a large body of information has accumulated that enables us to construct a clear picture…of…the microscopic population of the soil” (145), and in 1923 Bergey's Manual stated categorically that no organism could be classified without being cultured (133).
By the mid-1980s, however, microbiologists had lost this confidence, and the language and practice of microbiology changed to accommodate the vast unknown of uncultured life. Concepts, assumptions, images, and words needed to be replaced when it became evident that they were based upon the premise that microorganisms did not exist unless they could be cultured. Pace and colleagues highlighted the need for nontraditional techniques to understand the microbial world: “The simple morphology of most microbes provides few clues for their identification; physiological traits are often ambiguous. The microbial ecologist is particularly impeded by these constraints, since so many organisms resist cultivation, which is an essential prelude to characterization in the laboratory” (107).
In the ensuing years, microbiologists dedicated intense effort to describing the phylogenetic diversity of exotic and ordinary environments—ocean surfaces, deep sea vents, hot springs, soil, animal rumen and gut, human oral cavity and intestine. Many new lineages were classified based on their molecular signatures alone. The next challenge was to elucidate the functions of these new phylotypes and determine whether they represented new species, genera, or phyla of prokaryotic life. This challenge spawned various techniques, including metagenomics, the genomic analysis of assemblages of organisms. In a few years, the study of uncultured microorganisms has expanded beyond asking “Who is there?” to include the difficult question “What are they doing?”
The outcomes of the recognition of uncultured microorganisms are worthy of examination. One of these outcomes, metagenomics, is further shaping microbiology. Metagenomics has already opened new avenues of research by enabling unprecedented analyses of genome heterogeneity and evolution in environmental contexts and providing access to far more microbial diversity than has been viewed in the petri dish. This review will explore the origins of metagenomics and examine its recent application to microbial ecology and biotechnology.
HISTORY OF THE CULTURE DIVIDE
The current excitement about the uncultured world may make students of modern microbiology wonder why this aspect of microbiology was largely ignored for so long. It is worth tracing the origins of microbiology, which did not rely on culturing, and examine the reasons for the shift to culturing and the subsequent discoveries that rekindled interest in the uncultured world. This review will use the term uncultured microorganisms to capture the entire spectrum of organisms that are not cultured in a specific experiment. These may include microorganisms that we have not attempted to culture and those that have been resistant to culturing efforts but may submit to culturing in the future.
Early Microbiology and the Microscope
The roots of microbiology are firmly associated with the microscope. The first record of a human being's seeing a bacterial cell is in 1663. Antonie van Leeuwenhoek watched bacteria that he recovered from his own teeth through his homemade microscope. He was a keen observer and an outstanding maker of microscopes, and his observations and detailed illustrations of microbial life prompted many other observers (both scientists and nonscientists) to take an interest in the microscopic world. His colorful descriptions of bacteria made their study compelling; in his descriptions of the many shapes of the bacteria he sampled from his teeth, he marveled that one “shot through the water like a pike does through water” (30), firmly establishing that these tiny objects were, indeed, alive. For 200 years, microscopy enabled microbiologists to view heterotrophs, autotrophs, and obligate parasites alike.
Among the advances during this period of microbiology was the work of botanist Ferdinand Cohn, who classified many bacteria and described the life cycle of Bacillus subtilis based on his microscopic observations (60). Although mycologists such as Franz Unger had understood the concept of pure culture as early as the 1850s, it was in large part the emphasis on disease causality that solidified pure culture as the standard bacteriological technique for laboratory microbiology (49, 96). Robert Koch's postulates and his own innovation in developing culture media were instrumental in this shift, and from the 1880s forward, the microbiological world was divided into the cultured and the uncultured. Microbiologists were attracted to the power and precision of studies of bacteria in pure culture, and as a result, most of the knowledge that fills modern microbiology textbooks is derived from organisms maintained in pure culture.
Modern Microbiology—a Pure Culture Is Not Enough
Because culturing provided the platform for building the depth and detail of modern microbiological knowledge, for a long time microbiologists ignored the challenge to identify and characterize uncultured organisms. They focused instead on the rich source of discovery found in the readily cultured model organisms, and this contributed to the explosion of knowledge in microbial physiology and genetics in the 1960s to mid-1980s. Meanwhile, the study of uncultured microorganisms remained in the hands of a few persistent scientists who began to accumulate hints that flitted at the edge of the microbiological consciousness, suggesting that culturing did not capture the full spectrum of microbial diversity.
One of the indicators that cultured microorganisms did not represent much of the microbial world was the oft-observed “great plate count anomaly” (135)—the discrepancy between the sizes of populations estimated by dilution plating and by microscopy. This discrepancy is particularly dramatic in some aquatic environments, in which plate counts and viable cells estimated by acridine orange staining can differ by four to six orders of magnitude (66), and in soil, in which 0.1 to 1% of bacteria are readily culturable on common media under standard conditions (138, 139).
Brock and colleagues encountered microorganisms in Yellowstone hot springs that could not be cultured and others whose behavior in culture did not reflect their activities in situ. Many of the organisms could not be cultured on agar medium because their temperature requirements exceed the melting point of the agar. Therefore, elucidating the physiological function of microorganisms without culturing them required ingenuity. Brock's central technique involved the immersion of microscope slides in the spring for 1 to 7 days, followed by microscopic examination and often staining with fluorescent antibodies raised against cultured members of the taxonomic groups suspected to inhabit the environment (17, 21). This approach estimated in situ population sizes and growth rates, which indicated, for example, that certain strains of Sulfolobus grew in the hot springs at temperatures well below the optima in pure culture (103). The expanding body of evidence indicating that it was imperative to study physiology in the environment led Brock's group to determine which organisms in the hot spring were responsible for photosynthesis. To do so, they placed an opaque cover over the spring for a week. The spring lost its pink color, leading them to infer that the genus Synechococcus, typically pink in culture, was a major contributor to photosynthesis (25, 26).
Further evidence that drew attention to the uncultured world accumulated during the 1970s and 1980s. A study of oligotrophs indicated that incubation times longer than 25 days enhanced the recovery of certain organisms in culture (147). The food industry generated intense interest in “injured bacteria” in food—live organisms that cannot be cultured following stressful treatments such as heat, chilling, or desiccation but represent a significant risk to human health (41). The concept of organisms that were viable but not culturable emerged from the work of Colwell and colleagues, who showed that strains of Vibrio cholerae were indeed alive and virulent when isolated from aquatic environments (8) but did not grow in culture until after passage through a mouse or human intestine (35-37).
The confluence of these and many other scientific and technical advances steadily drew attention to the unculturable microbial world, but two discoveries figured significantly in the sharpened focus. The first was work on the diversity of soil bacteria, which demonstrated with DNA-DNA reassociation techniques that the complexity of the bacterial DNA in the soil was at least 100-fold greater than could be accounted for by culturing. This work suggested that the diversity of the uncultured world exceeded previous estimates (138). The second discovery was the demonstration that Helicobacter pylori causes gastric ulcers and cancer. Although spiral bacteria had been observed in the gastric mucosa of dogs in 1893 and in humans in 1906 (29), and correlations between the appearance of the bacteria and peptic ulcers were noted in 1938 (48), it was not until H. pylori was cultured that its role in disease was accepted (94, 95). Culturing was accompanied by the satisfaction of Koch's postulates on a human volunteer (94), providing definitive evidence for the causal relationship between the bacterium and ulcers.
Ironically, culturing was not that difficult. Plates accidentally incubated for 5 days instead of 3 revealed colonies later shown to be H. pylori (29). The fact that strong microscopic evidence for the role of H. pylori long preceded culturing and might have served as the basis for successful treatment decades earlier, perhaps reducing human suffering and mortality due to ulcers and cancer, did not escape the notice of microbiologists, medical practitioners, and the public. Whereas the studies of the complexity of the soil DNA demonstrated the diversity of the unknown world, the connection of uncultured bacteria and ulcers provided a striking example of the power of the undetected organisms. These discoveries provided compelling evidence that drew microbiologists to wrestle with the daunting challenge of devising strategies to access these organisms.
THE PARADIGM SHIFT
In 1985, an experimental advance radically changed the way we visualize the microbial world. Building upon the pioneering work of Carl Woese, which showed that rRNA genes provide evolutionary chronometers (148), Pace and colleagues created a new branch of microbial ecology (83, 134). They used direct analysis of 5S and 16S rRNA gene sequences in the environment to describe the diversity of microorganisms in an environmental sample without culturing (107, 134). The early studies were technically challenging, relying on direct sequencing of RNA or sequencing of reverse transcription-generated DNA copies. The next technical breakthrough arrived with the development of PCR technology and the design of primers that can be used to amplify almost the entire gene (62). This accelerated the discovery of diverse taxa as habitats across the earth were surveyed by the new technique (6, 50, 62, 126).
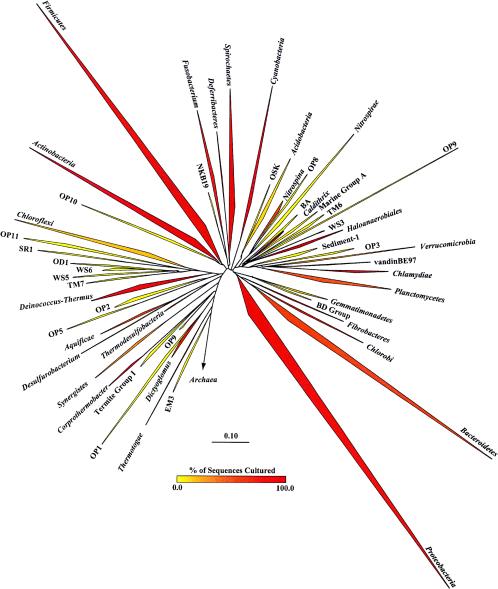
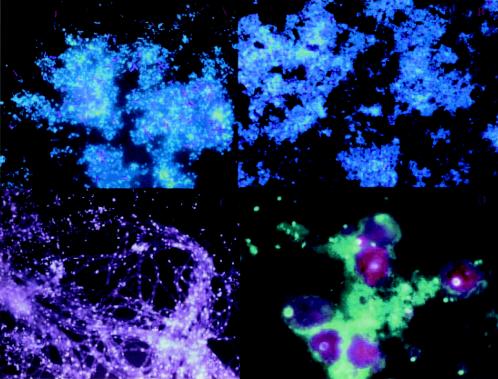
The application of PCR technology provided a view of microbial diversity that was not distorted by the culturing bias and revealed that the uncultured majority is highly diverse and contains members that diverge deeply from the readily culturable minority. Today, 52 phyla have been delineated, and most are dominated by uncultured organisms (Fig. 1) (114). The application of phylogenetic stains (Fig. 2) —nucleic acid probes with fluorescent labels that facilitate visualization of single cells in situ—led to a recrudescence of microscopy as a central tool of microbiology and microbial phylogeny (43, 63). Whereas traditional microscopy provides little phylogenetic information and fluorescent antibody studies require prior knowledge and culturing of an organism or one closely related to it to raise antibodies (17, 18), phylogenetic stains require only an rRNA sequence, which can be derived from an environmental sample without culturing. Phylogenetic stains corroborated evidence from PCR-based studies but provided quantitative information as well, because the findings are based on direct observation that is not subject to the skewing of organism abundance potentially observed with PCR (137).
FIG. 1.
Phylogenetic tree of Bacteria showing established phyla (italicized Latin names) and candidate phyla described previously (70, 74, 114) with the November 2003 ARB database (http://arb-home.de) (90) with 16,964 sequences that are >1,000 bp. The vertex angle of each wedge indicates the relative abundance of sequences in each phylum; the length of each side of the wedge indicates the range of branching depth found in that phylum; the redness of each wedge corresponds to the proportion of sequences in that phylum obtained from cultured representatives. Candidate phyla do not contain any cultured members.
FIG. 2.
Phylogenetic stains. Fluorescent in situ hybridization biofilm samples from Iron Mountain Mine, Calif. Nucleic acid probes were labeled with indodicarbocyanine, and DNA was stained nonspecifically with 4′,6′-diamidino-2-phenylindole. The nucleic acid probes are specific for (top left) Sulfobacillus spp., (top right) Archaea, (bottom left) Archaea on fungal filaments, and (bottom right) Eukarya. Reproduced from reference 5 with permission of the publisher.
rRNA Analysis and Culturing
In addition to providing a universal culture-independent means to assess diversity, 16S rRNA sequences also provided an aid to culturing efforts. Bacteria may be recalcitrant to culturing for diverse reasons—lack of necessary symbionts, nutrients, or surfaces, excess inhibitory compounds, incorrect combinations of temperature, pressure, or atmospheric gas composition, accumulation of toxic waste products from their own metabolism, and intrinsically slow growth rate or rapid dispersion from colonies (131). Testing myriad conditions requires focus on the critical variables, is challenging and laborious, and can only succeed if there is a sufficiently quantitative assay available to determine whether the organism of interest is enriched under a specific set of conditions.
Nucleic acid probes labeled with fluorescent tags provide such an assay, facilitating quantitative assessment of enrichment and growth. As a result, culturing efforts have intensified recently, and successes have included pure cultures of members of the SAR11 clade, now termed the genus Pelagibacter (34, 38, 113), which represents more than one-third of the prokaryotic cells in the surface of the ocean but was known only by its 16S rRNA signature until 2002 (38, 102, 113). The corollary to SAR11 in terrestrial environments is the Acidobacteria phylum (76, 121). Acidobacteria are abundant in soil, typically representing 20 to 30% of the 16S rRNA sequences amplified by PCR from soil DNA, but until recently only three members had been cultured (7, 56, 79, 81, 89, 97, 129, 132). Once again, the culture-independent indications that it was prevalent in the environment led to intensive efforts to culture members of the Acidobacteria phylum. The current efforts to culture new microorganisms will be advanced by the information that metagenomics can reveal about uncultured organisms (87).
Given that many organisms will not be coaxed readily into pure culture, a critical advance is to extend the understanding of the uncultured world beyond cataloging 16S rRNA gene sequences, and microbiologists have striven to devise methods to analyze the physiology and ecology of these diverse, uncultured organisms.
METAGENOMICS—CULTURE-INDEPENDENT INSIGHT
Among the methods designed to gain access to the physiology and genetics of uncultured organisms, metagenomics, the genomic analysis of a population of microorganisms, has emerged as a powerful centerpiece. Direct isolation of genomic DNA from an environment circumvents culturing the organisms under study, and cloning of it into a cultured organism captures it for study and preservation. Advances have derived from sequence-based and functional analysis in samples from water and soil and associated with eukaryotic hosts.
The word metagenomics was coined (69) to capture the notion of analysis of a collection of similar but not identical items, as in a meta-analysis, which is an analysis of analyses (64). (Community genomics, environmental genomics, and population genomics are synonyms for the same approach.) The idea of cloning DNA directly from environmental samples was first proposed by Pace (108), and in 1991, the first such cloning in a phage vector was reported (126). The next advance was the construction of a metagenomic library with DNA derived from a mixture of organisms enriched on dried grasses in the laboratory (71). Clones expressing cellulolytic activity were found in these libraries, which were referred to as zoolibraries, a term that has not been used widely in the field (71). The work of DeLong's group defined the field when they reported libraries constructed from prokaryotes in seawater (136). They identified a 40-kb clone that contained a 16S rRNA gene indicating that the clone was derived from an archaeon that had never been cultured. Construction of libraries with DNA extracted from soil lagged due to difficulties associated with maintaining the integrity of DNA during its extraction and purification from a soil matrix (14, 69, 80, 118) but eventually produced analyses analogous to those from seawater (39, 72, 118).
APPROACHES TO METAGENOMIC ANALYSIS
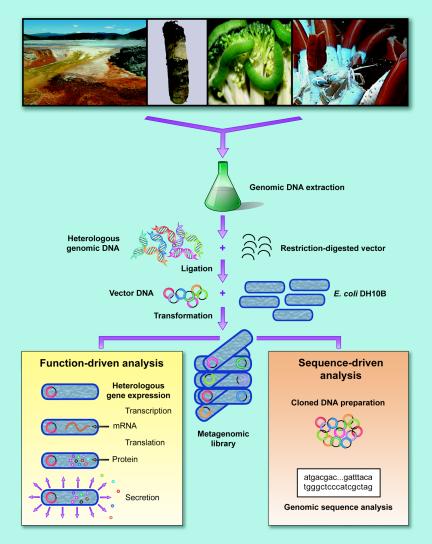
Metagenomic analysis involves isolating DNA from an environmental sample, cloning the DNA into a suitable vector, transforming the clones into a host bacterium, and screening the resulting transformants (Fig. 3). The clones can be screened for phylogenetic markers or “anchors,” such as 16S rRNA and recA, or for other conserved genes by hybridization or multiplex PCR (136) or for expression of specific traits, such as enzyme activity or antibiotic production (39, 44, 61, 78, 87, 88, 91, 93, 118, 125), or they can be sequenced randomly (141, 142). Each approach has strengths and limitations; together these approaches have enriched our understanding of the uncultured world, providing insight into groups of prokaryotes that are otherwise entirely unknown.
FIG. 3.
Construction and screening of metagenomic libraries. Schematic representation of construction of libraries from environmental samples. The images at the top from left to right show bacterial mats at Yellowstone, soil from a boreal forest in Alaska, cabbage white butterfly larvae, and a tube worm.
Sequence-Based Analysis
Sequenced-based analysis can involve complete sequencing of clones containing phylogenetic anchors that indicate the taxonomic group that is the probable source of the DNA fragment. Alternatively, random sequencing can be conducted, and once a gene of interest is identified, phylogenetic anchors can be sought in the flanking DNA to provide a link of phylogeny with the functional gene. Sequence analysis guided by the identification of phylogenetic markers is a powerful approach first proposed by the DeLong group, which produced the first genomic sequence linked to a 16S rRNA gene of an uncultured archaeon (136). Subsequently, they identified an insert from seawater bacteria containing a 16S rRNA gene that affiliated with the γ-Proteobacteria. The sequence of flanking DNA revealed a bacteriorhodopsin-like gene. Its gene product was shown to be an authentic photoreceptor, leading to the insight that bacteriorhodopsin genes are not limited to Archaea but are in fact abundant among the Proteobacteria of the ocean (11, 12).
A promising application of phylogenetic anchor-guided sequencing is to collect and sequence many genomic fragments from one taxon. In more complex environments and taxa, reassembly of a genome may not be feasible, but inference about the physiology and ecology of the members of the groups can be gleaned from sequence data. This approach has been initiated with clones from diverse soils carrying 16S rRNA genes that affiliate with the Acidobacteria phylum, which is abundant in soil and highly diverse (7, 28, 58, 76, 121) and about which little is known (85, 112). Complete sequencing of the estimated ≈500 kb of Acidobacterium DNA in metagenomic libraries may provide insight into the subgroups of bacteria in this phylum that have never been cultured.
The alternative to a phylogenetic marker-driven approach is to sequence random clones, which has produced dramatic insights, especially when conducted on a massive scale. The distribution and redundancy of functions in a community, linkage of traits, genomic organization, and horizontal gene transfer can all be inferred from sequence-based analysis. The recent monumental sequencing efforts, which include reconstruction of the genomes of uncultured organisms in a community in acid mine drainage (141) and the Sargasso Sea (142), illustrate the power of large-scale sequencing efforts to enrich our understanding of uncultured communities. These studies have made new linkages between phylogeny and function, indicated the surprising abundance of certain types of genes, and reconstructed the genomes of organisms that have not been cultured. These studies will be discussed in detail in the section entitled Biogeochemical Cycles.
The use of phylogenetic markers either as the initial identifiers of DNA fragments to study or as indicators of taxonomic affiliation for DNA fragments carrying genes of interest because of their function is limited by the small number of available markers that provide reliable placement in the Tree of Life. If a fragment of DNA that is of interest for other reasons does not carry a dependable marker, its organism of origin remains unknown. The collection of phylogenetic markers is growing, and as the diversity of markers increases, the power of this approach will also increase, making it possible to assign more fragments of anonymous DNA to the organisms from which they were isolated. Moreover, as more genomes are reconstructed, more genes will be linked to phylogenetic markers even though they were not cloned initially on the same fragment (141, 142).
Functional Metagenomics
Heterologous expression.
A powerful yet challenging approach to metagenomic analysis is to identify clones that express a function. Success requires faithful transcription and translation of the gene or genes of interest and secretion of the gene product, if the screen or assay requires it to be extracellular. Functional analysis has identified novel antibiotics (39, 61, 91, 142, 146), antibiotic resistance genes (44, 115), Na+(Li+)/H+ transporters (93), and degradative enzymes (71-73) The power of the approach is that it does not require that the genes of interest be recognizable by sequence analysis, making it the only approach to metagenomics that has the potential to identify entirely new classes of genes for new or known functions. The significant limitation is that many genes, perhaps most, will not be expressed in any particular host bacterium selected for cloning. In fact, there is an inherent contradiction in this approach—genes are cloned from exotic organisms to discover new motifs in biology, and yet these genes are required to be expressed in Escherichia coli or another domesticated bacterium in order to be detected. The diversity of the organisms whose DNA has been successfully expressed in E. coli is surprising (16, 33, 45, 57, 118-120), but heterologous expression remains a barrier to extracting the maximum information from functional metagenomics analyses.
Identifying active clones—screens, selections, and functional anchors.
The frequency of metagenomic clones that express any given activity is low. For example, in a search for lipolytic clones derived from German soil, only 1 in 730,000 clones showed activity (73). In a library of DNA from North American soil, 29 of a total of 25,000 clones expressed hemolytic activity (118). The scarcity of active clones therefore necessitates development of efficient screens and selections for discovery of new activities or molecules. Just as bacterial genetics relies on selections to detect low-frequency events, metagenomics will be advanced by seeking selectable phenotypes to increase the collection of active clones that can be compared, analyzed, and used to build a conceptual framework for functional analysis.
Several selections have proved to be fruitful. For example, the Daniel group designed a clever selection for Na+(Li+)/H+ antiporters that requires complementation of an E. coli mutant deficient in the three Na+/H+ antiporters (nhaA, nhaB, and chaA) enabling growth on medium containing 7.5 mM LiCl (93). This powerful selection facilitated the discovery of two novel antiporter proteins in a library of 1,480,000 clones containing DNA isolated from soil. Another selection strategy involved complementation of an E. coli mutant deficient in biotin production, which led to the isolation of seven new operons for biotin synthesis from enrichment cultures derived from samples of soil or horse excrement (52).
Selection for antibiotic resistance led to the isolation of a tetracycline resistance determinant from samples of the microbiota from the human mouth (44) and aminoglycoside resistance determinants from soil (115). The selection for aminoglycoside resistance identified nine clones, six of which encoded 6′-acetyltransferases that formed a new cluster based on sequence analysis. These genes were discovered in libraries containing a total of 4 Gb of DNA, or approximately 1 million genes, and thus their infrequent representation would have made it prohibitively laborious to discover them by a screen without a selection. This example illustrates the power of functional metagenomics—genes that are expressed in an ordinary host such as E. coli may be extraordinary and novel.
High-throughput screens can substitute when the functions of interest do not provide the basis for selection. For example, on certain indicator media, active clones display a characteristic and easily distinguishable appearance even when plated at high density. With the indicator dye tetrazolium chloride, Henne et al. (72) detected clones that utilize 4-hydroxybutyrate in libraries of DNA from agricultural or river valley soil. Very rare lipolytic clones in the same libraries were detected by production of clear halos on media containing rhodamine and either triolein or tributryin (73).
The discovery of new biological motifs will depend in part on functional analysis of metagenomic clones. Functional screens of metagenomic libraries have led to the assignment of functions to numerous “hypothetical proteins” in the databases. Innovation will be required to identify and overcome the barriers to heterologous gene expression and to detect rare clones efficiently in the immense libraries that are needed to represent all of the genomes in complex environments, such as soil. An emerging and powerful direction for metagenomic analysis is the use of functional anchors, which are the functional analogs of phylogenetic anchors. Functional anchors are functions that can be assessed rapidly in all of the clones in a library. When a collection of clones with a common function is assembled, they can be sequenced to find phylogenetic anchors and genomic structure in the flanking DNA. Such an analysis can provide a slice of the metagenome that cuts across clones with a different selective tool, determining the diversity of genomes that contain a particular function that can be expressed in the host carrying the library. Technological developments that promote functional expression and screening will advance this new frontier of functional genomics.
ECOLOGICAL INFERENCE FROM METAGENOMICS
The exigent questions in microbial ecology focus on how microorganisms form symbioses with eukaryotes, compete and communicate with other microorganisms, and acquire nutrients and produce energy. Thus far, metagenomics has provided insights into each of these areas, but in each instance, the challenge is to link the genomic information with the organism or ecosystem from which the DNA was isolated. Expression of a gene in a cultured host can establish gene function, but without the appropriate biological context, circumspection is required in drawing ecological inferences. Future technical innovations are needed to extend insights from metagenomics from inference to mechanistic analyses.
Symbiosis
Many bacterial symbionts that have highly specialized and ancient relationships with their hosts do not grow readily in culture. Many of them live in specialized structures, often in pure or highly enriched culture, in host tissues, making them ideal candidates for metagenomic analysis because the bacteria can be separated readily from host tissue and other microorganisms. This type of analysis has been conducted with Cenarchaeum symbiosum, a symbiont of a marine sponge (111), a Pseudomonas-like bacterium that is a symbiont of Paederus beetles (110), Buchnera aphidicola, an obligate symbiont of aphids (1), the Actinobacterium Tropheryma whipplei, the causal agent of the rare chronic infection of the intestinal wall (13, 31), and the Proteobacterium symbiont of the deep sea tube worm Riftia pachyptila (75). These systems provide good models for metagenomic analysis of more complex communities and thus warrant further attention in this review, although the term metagenomics typically connotes the study of multispecies communities. Therefore, the following section focuses on two of these obligate symbionts and the insight into their lifestyles offered by metagenomic analysis.
Buchnera-aphid symbiosis.
The first genome reconstruction of an uncultured organism was that of Buchnera aphidicola, the endosymbiont of aphids. The relationship between the bacterium and the insect is ancient, leaving each partner unable to function independently of the other, as is reflected in the genomic analysis. Moran's group isolated bacterial DNA from the insect and sequenced and reassembled the bacterial genome. The genus Buchnera contains a “reduced” genome of 564 open reading frames. Upon comparison with a reconstructed ancestral genome, 1,906 genes appear to have been lost. Most of the functions are associated with biosynthetic pathways contributed by the host, suggesting that the genome shrinkage is the result of the symbiotic lifestyle, which has become obligate because of gene loss (1, 42, 101).
The reconstruction of B. aphidicola's genome provided insights into the evolution of the symbiosis between the insect and bacterium, the biochemical mutual dependence that they have developed, and the mechanisms of genome shrinkage and rearrangement. The success of genome reconstruction with a single uncultured species provided part of the impetus needed to propose sequencing and reconstructing genomes in more complex assemblages.
Proteobacterium-tube worm symbiosis.
Riftia pachyptila, the deep sea tube worm, lives 2,600 m below the ocean surface, near the thermal vents that are rich in sulfide and reach temperatures near 400°C. The tube worm does not have a mouth or digestive tract, and therefore it is entirely dependent on its symbiotic bacteria, which provide the worm with food. The bacteria live in the trophosome, a specialized feeding sac inside the worm (32). The bacteria and trophosome constitute more than half of the animal's body mass. The bacteria oxidize hydrogen sulfide, thereby producing the energy required to fix carbon from CO2, providing sugars and amino acids (predominantly as glutamate) that nourish the worm (55, 84). The worm contributes to the symbiosis by collecting hydrogen sulfide, oxygen, and carbon dioxide and transporting them to the bacteria on hemoglobin-like molecules (3, 46, 149-153).
The bacterium is a member of the γ-Proteobacteria, as identified by 16S rRNA gene sequence (47). The bacteria have not been grown in pure culture in laboratory media, but they provide an excellent substrate for metagenomics because they reach high population density in the trophosome and exist there as a single species. Hughes et al. (75) isolated DNA from the bacterial symbiont and constructed fosmid libraries from it that were used to understand the physiology of the bacteria. Robinson et al. (116) identified a gene with similarity to ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) from the same fosmid library. All of the residues associated with the active site are conserved in the protein sequence deduced from the DNA sequence, and it has highest similarity with the RubisCO from Rhodospirillum rubrum. The characterization of this gene lends further support to the premise that the chemoautotrophic bacterial symbiont in R. pachyptila fixes carbon for its host.
The libraries were also screened for two-component regulators with a labeled histidine kinase gene as a probe. They identified a two-component system whose components complemented an envZ and a phoR creC double mutant, respectively. The discovery of a functional envZ homologue indicates that the symbiont carries a response regulator that is typical of γ-Proteobacteria, although the signals eliciting responses from these proteins have not yet been identified.
Genomic analysis of the symbiont also led to the identification of a gene encoding flagellin, which was expressed in E. coli and shown to direct the synthesis of flagella that are immunologically cross-reactive with Salmonella flagella. The presence of genes for flagella suggested to the authors that the endosymbiont has a free-living stage in its life cycle and may infect each generation of tube worms rather than being passed maternally (99).
Competition and Communication
What can metagenomics tell us about microbial competition and communication?
Competition for resources among community members selects for diverse survival mechanisms, including antagonism and mutualism among the members. Understanding these mechanisms is central to advancing the definition of principles that govern microbial community structure, function, and robustness. Historically, genetics has provided the most convincing evidence for traits contributing to microbial fitness. Classic mutant analysis has revealed genes required for microbial competition (9, 15, 20, 27, 82, 98, 100, 140, 143), antagonism (40, 51, 77, 117, 130), and mutualism (10, 54, 86, 92). Mutant analyses have provided the greatest advances in knowledge because screening mutants containing random mutations for effects on fitness has led to the identification of genes that would not have been predicted to play a role in microbial competition or mutualism.
Genes for competition and cooperation are hard to recognize based on sequence alone because the utility of their functions is entirely dependent on ecosystem context and the nature of the resources that are limiting. Therefore, genomics by itself does not provide a means to test ecological hypotheses or identify genes that confer fitness, but it can provide the basis for forming hypotheses. Ecological hypotheses are difficult to test in microorganisms that cannot be cultured or for which there are no genetic tools; however, functional genomics coupled with chemical ecology can yield informative answers. Chemical ecology involves the identification of small molecules with biological activity and proposed ecological function. These compounds can be identified through a variety of methods, including metagenomics. The addition of these molecules to communities can provide the basis for postulating their ecological roles in the community by measuring perturbations of community function. The following sections explore the discovery of small molecules in metagenomic libraries and postulate the ecological functions of these molecules in the organisms producing them.
Role of small molecules.
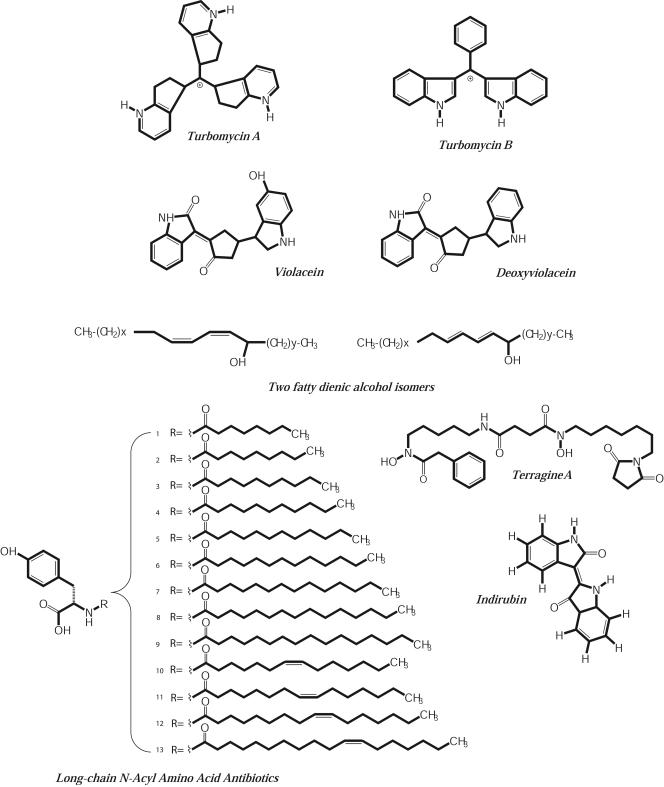
Small-molecule discovery by functional metagenomics has concentrated on antibiotics, which are of interest for their pharmaceutical applications as well as for their roles in ecosystem function. Traditional antibiotic screens for molecules that inhibit bacterial growth have led to the discovery of antibiotics in metagenomic libraries (Fig. 4). They have not been a rich source of novel antibiotics, likely because of the experimental limitations associated with the search. In studies that report frequencies, antibiotic-producing clones are detected at a frequency of approximately 1 producer per 104 clones (23, 24, 61). This low frequency hinders discovery because space and labor are required to conduct typical antimicrobial screens.
FIG. 4.
Antibiotics discovered in metagenomic libraries.
With standard inhibition assays, a Mycobacterium-inhibiting antibiotic, terragine, was discovered from a soil metagenomic clone maintained in Streptomyces lividans (146) and acyltyrosines from a clone maintained in E. coli (22, 24). Colored antibiotics represent a disproportionate share of those discovered because they can be identified visually. For instance, a clone noticed for its brown pigment was found to produce melanin, which masked orange and red pigments, two novel antibiotics, turbomycin A and turbomycin B (61). A purple-pigmented clone (23) produced violacein, previously shown to be an antibiotic made by the soil bacterium Chromobacterium violaceum. The sequence of the genes on the metagenomic clone diverged substantially from the C. violaceum violacein biosynthetic operon despite similar genetic organization (4, 23, 109), suggesting that the pathway on the metagenomic clone was derived from an organism other than C. violaceum. Osburne's group identified structurally related compounds, indirubin and indigo blue, in a soil metagenomic DNA library based on their blue color (91).
Sequence-based screening for small molecules.
The first polyketide synthases, enzymes involved in synthesis of polyketides, the broad class of antibiotics that includes erythromycin, epithilone, and rifamycin, were first cloned from soil with a PCR-based approach. Seow et al. (128) designed primers that hybridize with the highly conserved region of polyketide synthase genes and amplified novel polyketide synthase homologues directly from soil. This approach was adapted for screening metagenomic libraries by Osburne's group, who screened a 5,000-member metagenomic library for conserved regions of genes encoding type I polyketide synthase. Primers directed toward a conserved region of polyketide synthase I genes that flanks the active site of the ketoacyl synthetase domain were used to screen pools of 96 clones. The screen yielded 11 new polyketide synthase homologues that contained significant sequence similarity to polyketide synthase genes from cultured organisms. In addition, screening clones in both E. coli and Streptomyces lividans by chemical means revealed two novel compounds, fatty dienic alcohol isomers (Fig. 4).
Antibiotics as signal molecules.
If antibiotics evolved as mediators of functions other than warfare (42a), such as communication, antibiotic discovery will be expedited by screening metagenomic clones for signaling compounds as well as inhibitory compounds. The challenge is to develop assays that detect signaling by many compounds. A surprising result from the Davies group indicated that subinhibitory concentrations of many antibiotics induce quorum sensing despite no resemblance in structure to the acylated homoserine lactones that appear to be the natural inducers (65). This result presents a propitious opportunity—a single screen might capture molecules that are quorum-sensing inducers as well as antibiotics.
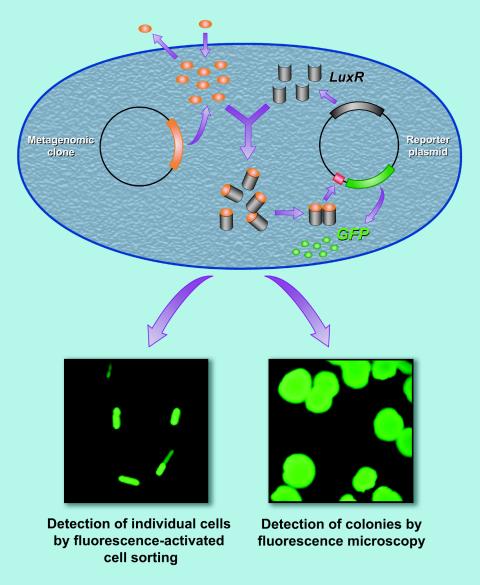
This opportunity was investigated by designing a high-throughput screen to identify compounds that induce the expression of genes under the control of a quorum-sensing promoter. The screen is intracellular, meaning the metagenomic DNA is in the same cell as the sensor for quorum-sensing induction (Fig. 5). The sensor is comprised of the luxR promoter, which is induced by acylated homoserine lactones, linked to gfp, and resides on a plasmid in an E. coli strain that did not induce quorum sensing itself (2). If an inducer of the luxR-mediated transcription of gfp is expressed from the metagenomic DNA, the cell fluoresces and can be captured by fluorescence-activated cell sorting or as a colony observed by fluorescence microscopy. Conversely, this sensor system can detect inhibitors of quorum sensing if acylated homoserine lactone is added to the medium and fluorescence-activated cell sorting is set to collect the nonfluorescent cells (Fig. 5). Metagenomic libraries from microbiota of the soil and from the midgut of the gypsy moth have been subjected to this screen, and an array of genes have been identified. Their products are under analysis, and some appear to differ from previously described quorum sensing inducers (L. Williamson, C. Guan, B. Borlee, and J. Handelsman, unpublished data).
FIG. 5.
Intracellular screen for quorum-sensing inducers. The biosensor, which detects molecules that induce luxR-regulated genes, resides inside the same cell as the metagenomic clone. An active clone fluoresces due to accumulation of green fluorescent protein and can be detected by fluorescence microscopy or captured by fluorescence-activated cell sorting.
Biogeochemical Cycles
Acid mine drainage.
An exciting potential of metagenomics is to provide community-wide assessment of metabolic and biogeochemical function. Analysis of specific functions across all members of a community can generate integrated models about how organisms share the workload of maintaining the nutrient and energy budgets of the community. The models can then be tested with genetic and chemical approaches. The best example of such an analysis is the nearly complete sequencing of the metagenome of a community in acid drainage of the Richmond mine, which represents one of the most extreme environments on Earth. The microbial community forms a pink biofilm that floats on the surface of the mine water. The drainage water below the biofilm has a pH of between 0 and 1 and high levels of Fe, Zn, Cu, and As (317, 14, 4, and 2 mM, respectively). The solution around the biofilm water is 42°C and microaerophilic. There is no source of carbon or nitrogen other than the gaseous forms in the air. The community is dominated by a few bacterial genera, Leptospirillum, Sulfobacillus, and sometimes Acidomicrobium, and one archaeal species, Ferroplasma acidarmanus, and other members of its group, the Thermoplasmatales. The mine is rich in sulfide minerals, including pyrite (FeS2), which is dissolved as a result of oxidation, which is catalyzed by microbial activity (5, 19).
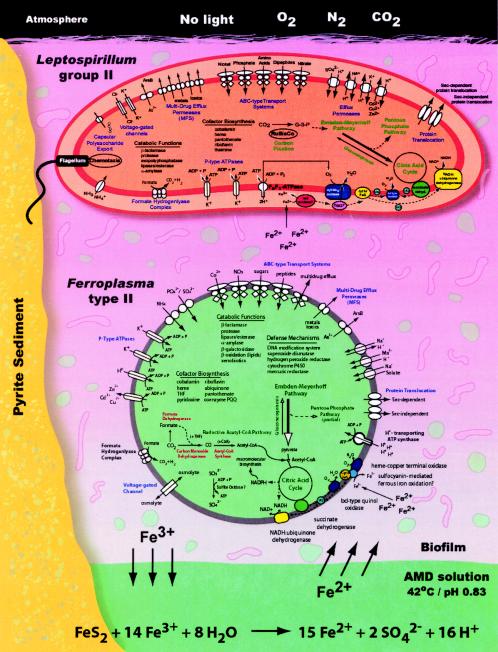
The simple community structure made it possible for Tyson et al. (141) to clone total DNA and sequence most of the community with high coverage. The G+C content of each clone provided a good indicator of its source because the G+C content of the genomes of the dominant taxa in the mine differ substantially (19) (Fig. 2). Sequence alignment of 16S rRNA and tRNA synthetase genes confirmed the organismal origins of the clones. Nearly complete genomes of Leptospirillum group II and Ferroplasma type II were reconstructed, and substantial sequence information for the other community members was reported.
The metagenomic sequence substantiated a number of significant hypotheses (Fig. 6). First, it appears that Leptospirillum group III contains genes with similarity to those known to be involved in nitrogen fixation, suggesting that it provides the community with fixed nitrogen. This was a surprise because the previous supposition was that a numerically dominant member of the community, such as Leptospirillum group II, would be responsible for nitrogen fixation. However, no genes for nitrogen fixation were found in the Leptospirillum group II genome, leading the authors to suggest that the group III organism is a keystone species that has a low numerical representation but provides a service that is essential to community function. Ferroplasma type I and II genomes contain no genes associated with nitrogen fixation but contain many transporters that indicate that they likely import amino acids and other nitrogenous compounds from the environment.
FIG. 6.
Metagenomics-based model of biogeochemical cycles mediated by prokaryotes in acid mine drainage. Cell metabolic cartoons constructed from the annotation of 2,180 open reading frames identified in the Leptospirillum group II genome and 1,931 open reading frames in the Ferroplasma type II genome. The cells are shown within a biofilm that is attached to the surface of an acid mine drainage stream. Reproduced from reference 141 with permission of the publisher.
Energy appears to be generated from iron oxidation by both Ferroplasma and Leptospirillum spp. The genomes of both groups contain electron transport chains, but they differ significantly. The genomes of Leptospirillum group II and III contain putative cytochromes that typically have a high affinity for oxygen. The cytochromes may play a role in energy transduction as well as in maintaining low oxygen tension, thereby protecting the oxygen-sensitive nitrogenase complex.
All of the genomes in the acid mine drainage are rich in genes associated with removing potentially toxic elements from the cell. Proton efflux systems are likely responsible for maintaining the nearly neutral intracellular pH, and metal resistance determinants pump metals out of the cells, maintaining nontoxic levels in the interior of the cells.
The acid mine drainage community provides a model for the analysis of other communities. Determining the origin of DNA fragments and assigning functions may be more difficult for communities that are phylogenetically or physiologically more complex (59), but the approach will be useful for all communities.
Sargasso Sea.
The Sargasso Sea is a complex and physically sprawling ecosystem compared with the contained acid mine drainage system. The inputs and outputs are more difficult to quantify, and the phylogeny of the community members has not been exhaustively surveyed. Venter et al. (142) embarked on the largest metagenomics project to date (affectionately dubbed megagenomics), in which they sequenced over 1 billion bp and claim to have discovered 1.2 million new genes. Intriguing inferences could be drawn because of the sheer size of the data set. They placed 794,061 genes in a conserved hypothetical protein group, which contains genes to which functions could not be confidently assigned. The next most abundant group contained 69,718 genes apparently involved in energy transduction. Among these were 782 rhodopsin-like photoreceptors, increasing the number of sequenced proteorhodopsin genes by almost 10-fold. Linkage of the rhodopsin genes to genes that provide phylogenetic affiliations, such as genes encoding subunits of RNA polymerase, indicated that the proteorhodopsins were distributed among taxa that were not previously known to contain light-harvesting functions, including the Bacteroides phylum (142).
The Sargasso Sea data set is a gold mine for further analysis. Intriguing hints about many aspects of ecosystem function abound and await further exploration. For example, an intriguing initial observation is that many of the genomes in the Sargasso Sea contain genes with similarity to those involved in phosphonate uptake or utilization of polyphosphates and pyrophosphates, which are present in this extremely phosphate-limited ecosystem. The phosphorus cycle is not well understood, and this collection of genomes provides a new route for discovery of the mechanisms of phosphorus acquisition and transformation. The understanding of nutrient cycling will be advanced by reconstruction of the genomes and the type of function-species analysis that Tyson et al. applied to the acid mine drainage community. The sequence data set from the Sargasso Sea provided the means to reassemble a number of genomes with criteria that include the depth of sequencing coverage, oligonucleotide frequencies, and similarity to previously sequenced genomes. The structures of these genomes individually and collectively will no doubt inform the development of models for nutrient cycling.
Population Genetics and Microheterogeneity
Metagenomic analysis has revealed that even apparently uniform populations contain substantial microheterogeneity. Within the population of Cenarchaeum symbiosum associated with the marine sponge, the rRNA genes are highly conserved, showing >99.2% identity, which indicates that the population comprises a single species. In the genomic regions flanking the rRNA genes, however, the DNA sequence identity drops to 87.8% (123). Similarly, Tyson et al. (141) found that the genomes of the species or groups in acid mine drainage varied in their uniformity. They found a high frequency of single-nucleotide polymorphisms among strains of the same species. The Ferroplasma type II group appears to contain a composite genome, with segments derived from three sources. In contrast, the Leptospirillum group II genome contained very few single-nucleotide or large-scale genome polymorphisms. These studies point to the importance of conducting genomic analysis on mixtures of strains to obtain a portrait of the heterogeneity within the species. In fact, metagenomics may provide insight into genome variation of organisms that can be readily cultured. If genetic variation in the environmental population is of interest, it may be more productive to clone the genome from the natural population than analyze the genomes of individuals cultured from it.
CONCLUSIONS AND FUTURE DIRECTIONS
Metagenomics has changed the way microbiologists approach many problems, redefined the concept of a genome, and accelerated the rate of gene discovery. The potential for application of metagenomics to biotechnology seems endless. Functional screens have identified new enzymes (39, 52, 53, 67, 68, 72, 73, 88, 93, 104, 118, 122, 124, 144) and antibiotics (22, 23, 39, 61, 69, 91, 110, 118, 146) and other reagents in libraries from diverse environments. A number of barriers have limited the discovery of new genes that provide insight into microbial community structure and function or that can be used to solve medical, agricultural, or industrial problems.
Realizing the potential for discovery from metagenomics is dependent on the advancement of methods that are central to library construction and analysis. For sequence-based approaches, the speed and cost of nucleotide sequencing will be a barrier of rapidly diminishing significance as sequencing technology continues to improve. Sequence-based assignment of function will also benefit from advances in detection of homology, which will increasingly rely on the tertiary structures of predicted proteins rather than simply on primary sequence. Advances that will facilitate the management and analysis of large libraries include bioinformatics tools to analyze vast sequence databases and reassemble multiple genomes rapidly and affordable gene chips for library profiling (127) or that readily distinguish clones that are expressing genes from those clones that are silent. Functional analysis will require more innovation in method development. Most important among these are strategies to improve heterologous gene expression and approaches for efficient screening of large libraries.
Microbiology has long relied on diverse methods for analysis, and metagenomics can provide the tools to balance the abundance of knowledge attained from culturing with an understanding of the uncultured majority of microbial life. Myriad environments on Earth have not been studied with culture-independent methods other than PCR-based 16S rRNA gene analysis, and they invite further analysis. Metagenomics may further our understanding of many of the exotic and familiar habitats that are attracting the attention of microbial ecologists, including deep sea thermal vents; acidic hot springs; permafrost, temperate, desert, and cold soils; Antarctic frozen lakes; and eukaryotic host organs—the human mouth and gut, termite and caterpillar guts, plant rhizospheres and phyllospheres, and fungi in lichen symbioses. With improved methods for analysis, funding stimulated by recent triumphs in the field, and attraction of diverse scientists to identify new problems and solve old ones, metagenomics will expand and continue to enrich our understanding of microorganisms.
REFERENCES
- 1.Abbot, P., J. H. Withgott, and N. A. Moran. 2001. Genetic conflict and conditional altruism in social aphid colonies. Proc. Natl. Acad. Sci. USA 98:12068-12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. Gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arp, A. J., M. L. Doyle, E. Di Cera, and S. J. Gill. 1990. Oxygenation properties of the two co-occurring hemoglobins of the tube worm Riftia pachyptila. Respir. Physiol. 80:323-334. [DOI] [PubMed] [Google Scholar]
- 4.August, P. R., T. H. Grossman, C. Minor, M. P. Draper, I. A. MacNeil, J. M. Pemberton, K. M. Call, D. Holt, and M. S. Osburne. 2000. Sequence analysis and functional characterization of the violacein biosynthetic pathway from Chromobacterium violaceum. J. Mol. Microbiol. Biotechnol. 2:513-519. [PubMed] [Google Scholar]
- 5.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 6.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baya, A. M., P. R. Brayton, V. L. Brown, D. J. Grimes, E. Russek-Cohen, and R. R. Colwell. 1986. Coincident plasmids and antimicrobial resistance in marine bacteria isolated from polluted and unpolluted Atlantic Ocean samples. Appl. Environ. Microbiol. 51:1285-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beattie, G. A., and J. Handelsman. 1993. Evaluation of a strategy for identifying nodulation competitiveness genes in Rhizobium leguminosarum biovar phaseoli. J. Gen. Microbiol. 139:529-538. [DOI] [PubMed] [Google Scholar]
- 10.Begum, A. A., S. Leibovitch, P. Migner, and F. Zhang. 2001. Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J. Exp. Bot. 52:1537-1543. [DOI] [PubMed] [Google Scholar]
- 11.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 12.Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 13.Bentley, S. D., M. Maiwald, L. D. Murphy, M. J. Pallen, C. A. Yeats, L. G. Dover, H. T. Norbertczak, G. S. Besra, M. A. Quail, and D. E. Harris. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 14.Berry, A. E., C. Chiocchini, T. Selby, M. Sosio, and E. M. H. Wellington. 2003. Isolation of high molecular weight DNA from soil for cloning into BAC vectors. FEMS Microbiol. Lett. 223:15-20. [DOI] [PubMed] [Google Scholar]
- 15.Bittinger, M. A., J. L. Milner, B. J. Saville, and J. Handelsman. 1997. rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol. Plant-Microbe Interact. 10:180-186. [DOI] [PubMed] [Google Scholar]
- 16.Black, C., J. A. M. Fyfe, and J. K. Davies. 1995. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 177:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohlool, B. B., and T. D. Brock. 1974. Immunofluorescence approach to the study of the ecology of Thermoplasma acidophilum in coal refuse material. Appl. Microbiol. 28:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohlool, B. B., and T. D. Brock. 1974. Population ecology of Sulfolobus acidocaldarius. II. Immunoecolgical studies. Arch. Microbiol. 97:181-194. [DOI] [PubMed] [Google Scholar]
- 19.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borthakur, D., and X. Gao. 1996. A 150-megadalton plasmid in Rhizobium etli strain TAL182 contains genes for nodulation competitiveness on Phaseolus vulgaris L. Can. J. Microbiol. 42:903-910. [DOI] [PubMed] [Google Scholar]
- 21.Bott, T. L., and T. D. Brock. 1969. Bacterial growth rates above 90 degrees C in Yellowstone hot springs. Science 164:1411-1412. [DOI] [PubMed] [Google Scholar]
- 22.Brady, S. F., C. J. Chao, and J. Clardy. 2002. New natural product families from an environmental DNA (eDNA) gene cluster. Am. Chem. Soc. 124:9968-9969. [DOI] [PubMed] [Google Scholar]
- 23.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 24.Brady, S. F., and J. Clardy. 2000. Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J. Am. Chem. Soc. 122:12903-12904. [Google Scholar]
- 25.Brock, T. D. 1967. Life at high temperatures. Science 158:1012-1019. [DOI] [PubMed] [Google Scholar]
- 26.Brock, T. D., and M. L. Brock. 1968. Measurement of steady-state growth rates of a thermophilic alga directly in nature. J. Bacteriol. 95:811-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromfield, E. S., D. M. Lewis, and L. R. Barran. 1985. Cryptic plasmid and rifampin resistance in Rhizobium meliloti influencing nodulation competitiveness. J. Bacteriol. 164:410-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 29.Buckley, M. J. M., and C. A. O'Morain. 1998. Helicobacter biology—discovery. Br. Med. Bull. 54:7-16. [DOI] [PubMed] [Google Scholar]
- 30.Bulloch, W. 1938. The history of bacteriology. Oxford University Press, New York, N.Y.
- 31.Cannon, W. R. 2003. Whipple's disease, genomics, and drug therapy. Lancet 361:1916. [DOI] [PubMed] [Google Scholar]
- 32.Cary, S. C., W. Warren, E. Anderson, and S. J. Giovannoni. 1993. Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol. Mar. Biol. Biotechnol. 2:51-62. [PubMed] [Google Scholar]
- 33.Chávez, S., J. C. Reyes, F. Cahuvat, F. J. Florencio, and P. Candau. 1995. The NADP-glutamate dehydrogenase of the cyanobacterium Synechocystis 6803: cloning, transcriptional analysis and disruption of the gdhA gene. Plant Mol. Biol. 28:173-188. [DOI] [PubMed] [Google Scholar]
- 34.Cho, J.-C., and S. J. Giovannoni. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine gammaproteobacteria. Appl. Environ. Microbiol. 70:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colwell, R. R., P. R. Brayton, D. Harrington, B. D. Tall, A. Huq, and M. M. Levine. 1996. Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 12:28-31. [DOI] [PubMed] [Google Scholar]
- 36.Colwell, R. R., and D. J. Grimes (ed.). 2000. Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 37.Colwell, R. R., M. L. Tamplin, P. R. Brayton, A. L. Gauzens, B. D. Tall, D. Harrington, M. M. Levine, S. Hall, A. Huq, and D. A. Sack. 1990. Environmental aspects of V. cholerae in transmission of cholera, p. 327-343. In R. B. Sack and Y. Zinnaka (ed.), Advances in research on cholera and related diarrhoeas, 7th ed. KTK Scientific Publications, Tokyo, Japan.
- 38.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courtois, S., C. M. Cappellano, M. Ball, F. X. Francou, P. Normand, G. Helynck, A. Martinez, S. J. Kolvek, J. Hopke, M. S. Osburne, P. R. August, R. Nalin, M. Guerineau, P. Jeannin, P. Simonet, and J. L. Pernodet. 2003. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Currie, C. R. 2001. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu. Rev. Microbiol. 55:357-380. [DOI] [PubMed] [Google Scholar]
- 41.Dahl Sawyer, C. A., and J. J. Pestka. 1985. Foodservice systems: presence of injured bacteria in foods during food product flow. Annu. Rev. Microbiol. 39:51-67. [DOI] [PubMed] [Google Scholar]
- 42.Daubin, V., N. A. Moran, and H. Ochman. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301:829-832. [DOI] [PubMed] [Google Scholar]
- 42a.Davies, J. 1990. What are antibiotics? Archaic functions for modern activities. Mol. Microbiol. 4:1227-1232. [DOI] [PubMed] [Google Scholar]
- 43.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 44.Diaz-Torres, M. L., R. McNab, D. A. Spratt, A. Villedieu, N. Hunt, M. Wilson, and P. Mullany. 2003. Novel tetracycline resistance determinant from the oral metagenome. Antimicrob. Agents Chemother. 47:1430-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding, M. a. D. B. Y. 1993. Cloning and analysis of the leuB gene of Leptospira interrogans serovar pomona. J. Gen. Microbiol. 139:1093-1103. [DOI] [PubMed] [Google Scholar]
- 46.Distel, D. L., E. F. DeLong, and J. B. Waterbury. 1991. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl. Environ. Microbiol. 57:2376-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Distel, D. L., D. J. Lane, G. J. Olsen, S. J. Giovannoni, B. Pace, N. R. Pace, D. A. Stahl, and H. Felbeck. 1988. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J. Bacteriol. 170:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doenges, J. L. 1938. Spirochaetes in the gastric glands of Macacus rhesus and humans without definite history of related disease. Proc. Soc. Exp. Biol. Med. 38:536-538. [Google Scholar]
- 49.Drews, G. 1999. Ferdinand Cohn: a promoter of modern microbiology. Nova Acta Leopoldina: Abhandlungen der Deutschen Akademie der Naturforscher Leopoldiana, Neue Folge, vol. 80.
- 50.Eden, P. A., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 51.Emmert, E. A., and J. Handelsman. 1999. Biocontrol of plant disease: a (gram-) positive perspective. FEMS Microbiol. Lett. 171:1-9. [DOI] [PubMed] [Google Scholar]
- 52.Entcheva, P., W. Liebl, A. Johann, T. Hartsch, and W. R. Streit. 2001. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Appl. Environ. Microbiol. 67:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eschenfeldt, W. H., L. Stols, H. Rosenbaum, Z. S. Khambatta, E. Quaite-Randall, S. Wu, D. C. Kilgore, J. D. Trent, and M. I. Donnelly. 2001. DNA from uncultured organisms as a source of 2,5-diketo-d-gluconic acid reductases. Appl. Environ. Microbiol. 67:4206-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faucher, C., F. Maillet, J. Vasse, C. Rosenberg, A. A. van Brussel, G. Truchet, and J. Denarie. 1988. Rhizobium meliloti host range nodH gene determines production of an alfalfa-specific extracellular signal. J. Bacteriol. 170:5489-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felbeck, H., and J. Jarchow. 1998. Carbon release from purified chemoautotrophic bacterial symbionts of the hydrothermal vent tubeworm Riftia pachyptila. Physiol. Zool. 71:294-302. [DOI] [PubMed] [Google Scholar]
- 56.Felske, A., A. Wolterink, R. Van Lis, W. M. De Vos, and A. D. Akkermans. 2000. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 66:3998-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreyra, R. G., F. C. Soncini, and A. M. Viale. 1993. Cloning, characterization, and functional expression in Escherichia coli of chaperonin (groESL) genes from the prototrophic sulfur bacterium Chromatium vinosum. J. Bacteriol. 175:1514-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galperin, M. Y. 2004. Metagenomics: from acid mine to shining sea. Environ. Microbiol. 6:543-545. [DOI] [PubMed] [Google Scholar]
- 60.Geison, G. L. 1981. Cohn, Ferdinand Julius. In C. C. Gillispie (ed.), Dictionary of scientific biography, vol. 3. Scribner, New York, N.Y.
- 61.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R. Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 63.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glass, G. V. 1976. Primary, secondary, and meta-analysis of research. Educ. Res. 5:3-8. [Google Scholar]
- 65.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grimes, D. J., R. W. Atwell, P. R. Brayton, L. M. Palmer, D. M. Rollins, D. B. Roszak, F. L. Singleton, M. L. Tamplin, and R. R. Colwell. 1986. The fate of enteric pathogenic bacteria in estuarine and marine environments. Microbiol. Sci. 3:324-329. [PubMed] [Google Scholar]
- 67.Gupta, R., Q. K. Beg, and P. Lorenz. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59:15-32. [DOI] [PubMed] [Google Scholar]
- 68.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 70.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Healy, F. G., R. M. Ray, H. C. Aldrich, A. C. Wilkie, I. L. O., and K. T. Shanmugam. 1995. Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose. Appl. Microbiol. Biotechnol. 43:667-674. [DOI] [PubMed] [Google Scholar]
- 72.Henne, A., R. Daniel, R. A. Schmitz, and G. Gottschalk. 1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henne, A., R. A. Schmitz, M. Bomeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hughes, D. S., H. Felbeck, and J. L. Stein. 1997. A histidine protein kinase homolog from the endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. Appl. Environ. Microbiol. 63:3494-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions acidobacteria, actinobacteria, proteobacteria, and verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keel, C., Z. Ucurum, P. Michaux, M. Adrian, and D. Haas. 2002. Deleterious impact of a virulent bacteriophage on survival and biocontrol activity of Pseudomonas fluorescens strain CHA0 in natural soil. Mol. Plant-Microbe Interact. 15:567-576. [DOI] [PubMed] [Google Scholar]
- 78.Knietsch, A., S. Bowien, G. Whited, G. Gottschalk, and R. Daniel. 2003. Identification and characterization of coenzyme B12-dependent glycerol dehydratase- and diol dehydratase-encoding genes from metagenomic DNA libraries derived from enrichment cultures. Appl. Environ. Microbiol. 69:3048-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozdroj, J., and J. D. van Elsas. 2001. Structural diversity of microorganisms in chemically perturbed soil assessed by molecular and cytochemical approaches. J. Microbiol. Methods 43:197-212. [DOI] [PubMed] [Google Scholar]
- 80.Krsek, M., and E. M. H. Wellington. 1999. Comparison of different methods for the isolation and purification of total community DNA from soil. J. Microbiol. Methods 39:1-16. [DOI] [PubMed] [Google Scholar]
- 81.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lagares, A., G. Caetano-Anolles, K. Niehaus, J. Lorenzen, H. D. Ljunggren, A. Puhler, and G. Favelukes. 1992. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J. Bacteriol. 174:5941-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laue, B. E., and D. C. Nelson. 1994. Characterization of the gene encoding the autotrophic ATP sulfurylase from the bacterial endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 176:3723-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Long, S. R. 2001. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 125:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lorenz, P., K. Liebeton, F. Niehaus, and J. Eck. 2002. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13:572-577. [DOI] [PubMed] [Google Scholar]
- 88.Lorenz, P., and C. Schleper. 2002. Metagenome—a challenging source of enzyme discovery. J. Mol. Catal. B Enzym. 19:13-19. [Google Scholar]
- 89.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 90.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MacNeil, I. A., C. L. Tiong, C. Minor, P. R. August, T. H. Grossman, K. A. Loiacono, B. A. Lynch, T. Phillips, S. Narula, R. Sundaramoorthi, A. Tyler, T. Aldredge, H. Long, M. Gilman, D. Holt, and M. S. Osburne. 2001. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 3:301-308. [PubMed] [Google Scholar]
- 92.Madinabeitia, N., R. A. Bellogin, A. M. Buendia-Claveria, M. Camacho, T. Cubo, M. R. Espuny, A. M. Gil-Serrano, M. C. Lyra, A. Moussaid, F. J. Ollero, M. E. Soria-Diaz, J. M. Vinardell, J. Zeng, and J. E. Ruiz-Sainz. 2002. Sinorhizobium fredii HH103 has a truncated nolO gene due to a −1 frameshift mutation that is conserved among other geographically distant S. fredii strains. Mol. Plant-Microbe Interact. 15:150-159. [DOI] [PubMed] [Google Scholar]
- 93.Majernik, A., G. Gottschalk, and R. Daniel. 2001. Screening of environmental DNA libraries for the presence of genes conferring Na+(Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. 183:6645-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marshall, B. J., J. A. Armstrong, D. B. McGechie, and R. J. Glancy. 1985. Attempt to fulfil Koch's postulates for pyloric Campylobacter. Med. J. Aust. 142:436-439. [DOI] [PubMed] [Google Scholar]
- 95.Marshall, B. J., D. B. McGechie, P. A. Rogers, and R. J. Glancy. 1985. Pyloric campylobacter infection and gastroduodenal disease. Med. J. Aust. 142:439-444. [DOI] [PubMed] [Google Scholar]
- 96.Mazumdar, P. M. H. 1995. Species and specificity: an interpretation of the history of immunology. Cambridge University Press, New York, N.Y.
- 97.McCaig, A. E., S. J. Grayston, J. I. Prosser, and L. A. Glover. 2001. Impact of cultivation on characterisation of species composition of soil bacterial communities. FEMS Microbiol. Ecol. 35:37-48. [DOI] [PubMed] [Google Scholar]
- 98.Michiels, J., H. Pelemans, K. Vlassak, C. Verreth, and J. Vanderleyden. 1995. Identification and characterization of a Rhizobium leguminosarum bv. phaseoli gene that is important for nodulation competitiveness and shows structural homology to a Rhizobium fredii host-inducible gene. Mol. Plant-Microbe Interact. 8:468-472. [DOI] [PubMed] [Google Scholar]
- 99.Millikan, D. S., H. Felbeck, and J. L. Stein. 1999. Identification and characterization of a flagellin gene from the endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. Appl. Environ. Microbiol. 65:3129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Milner, J. L., R. S. Araujo, and J. Handelsman. 1992. Molecular and symbiotic characterization of exopolysaccharide-deficient mutants of Rhizobium tropici strain CIAT899. Mol. Microbiol. 6:3137-3147. [DOI] [PubMed] [Google Scholar]
- 101.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 102.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 103.Mosser, J. L., B. B. Bohlool, and T. D. Brock. 1974. Growth rates of Sulfolobus acidocaldarius in nature. J. Bacteriol. 118:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oh, H. J., K. W. Cho, I. S. Jung, W. H. Kim, B. K. Hur, and G. J. Kim. 2003. Expanding functional spaces of enzymes by utilizing whole genome treasure for library construction. J. Mol. Catal. B Enzym. 26:241-250. [Google Scholar]
- 105.Olsen, G. J., N. R. Pace, M. Nuell, B. P. Kaine, R. Gupta, and C. R. Woese. 1985. Sequence of the 16S rRNA gene from the thermoacidophilic archaebacterium Sulfolobus solfataricus and its evolutionary implications. J. Mol. Evol. 22:301-307. [DOI] [PubMed] [Google Scholar]
- 106.Pace, N. R. 1995. Opening the door onto the natural microbial world: molecular microbial ecology. Harvey Lect. 91:59-78. [PubMed] [Google Scholar]
- 107.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 108.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1985. Analyzing natural microbial populations by rRNA sequences. ASM News 51:4-12. [Google Scholar]
- 109.Pemberton, J. M. 1991. Cloning and heterologous expression of the violacein biosynthesis gene cluster from Chromobacterium violaceum. Curr. Microbiol. 22:355-358. [Google Scholar]
- 110.Piel, J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Pro. Natl. Acad. Sci. USA 93:6241-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quaiser, A., T. Ochsenreiter, C. Lanz, S. C. Schuster, A. H. Treusch, J. Eck, and C. Schleper. 2003. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol. Microbiol. 50:563-575. [DOI] [PubMed] [Google Scholar]
- 113.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 114.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 115.Riesenfeld, C. S., R. M. Goodman, and J. Handelsman. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6:981-989. [DOI] [PubMed]
- 116.Robinson, J. J., J. L. Stein, and C. M. Cavanaugh. 1998. Cloning and sequencing of a form II ribulose-1,5-biphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 180:1596-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robleto, E. A., K. Kmiecik, E. S. Oplinger, J. Nienhuis, and E. W. Triplett. 1998. Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl. Environ. Microbiol. 64:2630-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rondon, M. R., R. M. Goodman, and J. Handelsman. 1999. The Earth's bounty: assessing and accessing soil microbial diversity. Trends Biotechnol. 17:403-409. [DOI] [PubMed] [Google Scholar]
- 120.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. USA 96:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 122.Sandler, S. J., P. Hugenholtz, C. Schleper, E. F. DeLong, N. R. Pace, and A. J. Clark. 1999. Diversity of radA genes from cultured and uncultured Archaea: comparative analysis of putative RadA proteins and their use as a phylogenetic marker. J. Bacteriol. 181:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schleper, C., E. F. DeLong, C. M. Preston, R. A. Feldman, K. Y. Wu, and R. V. Swanson. 1998. Genomic analysis reveals chromosomal variation in natural populations of the uncultured psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 180:5003-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schleper, C., R. V. Swanson, E. J. Mathur, and E. F. DeLong. 1997. Characterization of a DNA polymerase from the uncultivated psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 179:7803-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 126.Schmidt, T. M., E. F. DeLong, and N. R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sebat, J. L., F. S. Colwell, and R. L. Crawford. 2003. Metagenomic profiling: Microarray analysis of an environmental genomic library. Appl. Environ. Microbiol. 69:4927-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Seow, K. T., G. Meurer, M. Gerlitz, E. Wendt-Pienkowski, C. R. Hutchinson, and J. Davies. 1997. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J. Bacteriol. 179:7360-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Silo-Suh, L. A., B. J. Lethbridge, S. J. Raffel, H. He, J. Clardy, and J. Handelsman. 1994. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol. 60:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simu, K., and A. Hagstrom. 2004. Oligotrophic bacterioplankton with a novel single-cell life strategy. Appl. Environ. Microbiol. 70:2445-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Society of American Bacteriologists. 1923. Bergey's manual of determinative bacteriology. Williams and Wilkins, Baltimore, Md.
- 134.Stahl, D. A., D. J. Lane, G. J. Olsen, and N. R. Pace. 1985. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl. Environ. Microbiol. 49:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 136.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. DeLong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment front a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suzuki, M., M. S. Rappé, and S. J. Giovannonij. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 140.Triplett, E. W., and M. J. Sadowsky. 1992. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46:399-428. [DOI] [PubMed] [Google Scholar]
- 141.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 142.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 143.Vinuesa, P., F. Neumann-Silkow, C. Pacios-Bras, H. P. Spaink, E. Martinez-Romero, and D. Werner. 2003. Genetic analysis of a pH-regulated operon from Rhizobium tropici CIAT899 involved in acid tolerance and nodulation competitiveness. Mol. Plant-Microbe Interact. 16:159-168. [DOI] [PubMed] [Google Scholar]
- 144.Voget, S., C. Leggewie, A. Uesbeck, C. Raasch, K. E. Jaeger, and W. R. Streit. 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 69:6235-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Waksman, S. A., and R. L. Starkey. 1931. The soil and the microbe. John Wiley, New York, N.Y.
- 146.Wang, G. Y., E. Graziani, B. Waters, W. Pan, X. Li, J. McDermott, G. Meurer, G. Saxena, R. J. Andersen, and J. Davies. 2000. Novel natural products from soil DNA libraries in a streptomycete host. Org. Lett. 2:2401-2404. [DOI] [PubMed] [Google Scholar]
- 147.Whang, K., and T. Hattori. 1988. Oligotrophic bacteria from rendzina forest soil. Antonie van Leeuwenhoek 54:19-36. [DOI] [PubMed] [Google Scholar]
- 148.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zal, F., B. Kuster, B. N. Green, D. J. Harvey, and F. H. Lallier. 1998. Partially glucose-capped oligosaccharides are found on the hemoglobins of the deep-sea tube worm Riftia pachyptila. Glycobiology 8:663-673. [DOI] [PubMed] [Google Scholar]
- 150.Zal, F., F. H. Lallier, B. N. Green, S. N. Vinogradov, and A. Toulmond. 1996. The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila. II. Complete polypeptide chain composition investigated by maximum entropy analysis of mass spectra. J. Biol. Chem. 271:8875-8881. [DOI] [PubMed] [Google Scholar]
- 151.Zal, F., F. H. Lallier, J. S. Wall, S. N. Vinogradov, and A. Toulmond. 1996. The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila. I. Reexamination of the number and masses of its constituents. J. Biol. Chem. 271:8869-8874. [DOI] [PubMed] [Google Scholar]
- 152.Zal, F., E. Leize, F. H. Lallier, A. Toulmond, A. Van Dorsselaer, and J. J. Childress. 1998. S-Sulfohemoglobin and disulfide exchange: the mechanisms of sulfide binding by Riftia pachyptila hemoglobins. Proc. Natl. Acad. Sci. USA 95:8997-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zal, F., T. Suzuki, Y. Kawasaki, J. J. Childress, F. H. Lallier, and A. Toulmond. 1997. Primary structure of the common polypeptide chain b from the multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila: an insight on the sulfide binding-site. Proteins 29:562-574. [PubMed] [Google Scholar]