Abstract
The ability of fungi to produce both meiospores and mitospores has provided adaptive advantages in survival and dispersal of these organisms. Here we provide evidence of an endogenous mechanism that balances meiospore and mitospore production in the model filamentous fungus Aspergillus nidulans. We have discovered a putative dioxygenase, PpoC, that functions in association with a previously characterized dioxygenase, PpoA, to integrate fatty acid derived oxylipin and spore production. In contrast to PpoA, deletion of ppoC significantly increased meiospore production and decreased mitospore development. Examination of the PpoA and PpoC mutants indicate that this ratio control is associated with two apparent feedback loops. The first loop shows ppoC and ppoA expression is dependent upon, and regulates the expression of, nsdD and brlA, genes encoding transcription factors required for meiospore or mitospore production, respectively. The second loop suggests Ppo oxylipin products antagonistically signal the generation of Ppo substrates. These data support a case for a fungal “oxylipin signature-profile” indicative of relative sexual and asexual spore differentiation.
The adaptive success of any organism depends in large part on its ability to sense and respond appropriately to environmental stimuli. Development and survival are highly dependent on proper responses to extracellular signals. In both prokaryotes and eukaryotes a number of developmental signals are derived from common lipogenic origins, suggesting the possibility of a conserved cross kingdom cell-to-cell communication network (11, 20, 42, 56). In the prokaryotic kingdom, several species of lipogenic diffusible molecules regulate a variety of responses (bioluminescence, virulence, biofilm formation, etc.) in a density-dependent manner through the quorum-sensing mechanism (50). In mammals, fatty acid-derived oxylipins (e.g., prostaglandins and leukotrienes) regulate inflammation and other homeostatic responses through an autocrine-paracrine sensing system (19). In plants, similarly structured oxylipins regulate the expression of host defense genes against pathogen and pests and play a major role in the formation of phytohormones and tissue development (5, 17, 34). However, remarkably little is known of lipid signaling in the development and survival of the highly successful kingdom, the Fungi.
Fungi are ubiquitous eukaryotes that are estimated to comprise a quarter of the entire biomass on earth and consist of nearly 1.5 million species, with only 5% identified thus far (25). They are the primary degraders of cellulose and lignin and devastating pathogens of plants and animals. Their success is attributed to their multilateral reproductive strategies, which are uniquely represented by the development of specialized reproductive cells, the meiospore and mitospore. These two spores provide the sexual and asexual modes of fungal reproduction that occur in distinct reproductive organs (3, 4). Sexual reproduction is characterized by the fusion of two nuclei, followed by meiosis and the production of meiospores, and results in a high incidence of genetic recombination and the generation of new genotypes upon which natural selection acts to adapt readily to a multitude of environmental conditions. In many fungi, sexual reproduction usually occurs only once a year and lends adaptive benefits, such as dormancy (overwintering) and drought resistance, to the organism (3, 46, 53). In general, asexual or somatic reproduction is repeated several times during the fungal life cycle, contributing to dispersal by the production of large number of individual mitospores (3, 46, 53). Numerous species in all fungal phyla are able to reproduce both sexually and asexually, and phylogenetic studies indicate this to be the ancestral condition of most taxa (37, 53).
The genus Aspergillus comprises a diverse group of species with many members capable of producing only mitospores, a few that produce only meiospores, and several that can produce both spores (49). The homothallic genetic model Aspergillus nidulans is a classic example of the latter, producing both meiospores (e.g., ascospores) and mitospores (e.g., conidia) (Fig. 1). Studies examining both modes of reproduction in A. nidulans describe multiplex tissue development regulated by myriad nutritional and environmental factors, including components of an intrinsic signal transduction pathway that balance vegetative growth with spore development and control the onset of ascosporogenesis and conidiation (2, 6, 11). Ascospore formation in A. nidulans requires the GATA-type transcription factor NsdD, necessary for cleistothecia (sexual fruiting bodies bearing the ascospores) and Hülle cell production (21). Conidia formation in A. nidulans requires the function of BrlA, a zinc finger transcription factor essential for conidiophore development (45). Deletion of either gene blocks formation of the respective meiotic or mitotic fruiting body, resulting in a strict asexual morph (ΔnsdD) or a strict sexual morph (ΔbrlA).
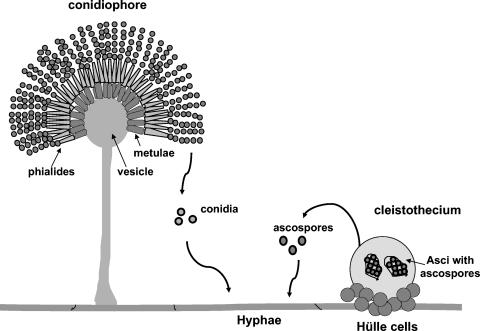
FIG. 1.
Life cycle of A. nidulans. Both asexual mitotic spores (conidia) and sexual meiotic spores (ascospores) can germinate, produce hyphae, and create a mature colony. The asexual fruiting body called conidiophore bears the conidia on the top of conidiogenous cells called phialides that arise on metulae, a layer of cells over the vesicle surface. The sexual fruiting body called the cleistothecium contains asci with eight ascospores each and is covered with the thick-walled Hülle cells.
In contrast to NsdD and BrlA, which are solely involved in the regulation of the sexual or the asexual cycle, respectively, physiological studies of Champe and el-Zayat (12) led to the identification of secreted lipogenic signal molecules, collectively named “psi factor” (for precocious sexual inducer), that govern the timing and balance of meiotic to mitotic spore development. Biochemical analysis showed that A. nidulans psi factor is an endogenous mixture of hormone-like oxylipins composed of hydroxylated oleic (18:1), linoleic (18:2), and linolenic (18:3) acid molecules termed psiβ, psiα, and psiγ, respectively (9, 39). The position and number of hydroxylations of the fatty acid backbone further identifies the psi compounds as psiB, psiC, and psiA (40). Feeding studies carried out for linoleic acid-derived psiα molecules reported that psiBα and psiCα stimulated ascospore and inhibited conidial development, whereas psiAα had the opposite effect (12, 13). Studies were not performed for purified oleic or linolenic derivatives. A potential role for oxylipins in cross kingdom communication as well as in balancing the ascospore/conidia ratio in A. nidulans was described by Calvo et al. (10) characterizing the effects of plant oxylipins on spore development in seed infecting aspergilli. Furthermore, numerous biochemical and physiological studies of oomycetes (protists resembling fungi in lifestyle), yeasts, and filamentous fungi have associated oxylipin synthesis with either meiospore or mitospore development (26, 32, 42). Fatty acids (e.g., farnesoic acid) also regulate morphological transitions in the human pathogen Candida albicans (43), and recently a bacterial virulence factor structurally similar to farnesoic acid was shown to inhibit the dimorphic transition in C. albicans (56).
The first genetic evidence to support a role for oxylipins in directing the meiospore-mitospore balance emerged from studies by Tsitsigiannis et al. (55), which identified an A. nidulans dioxygenase (enzymes that catalyze oxygenation of unsaturated fatty acids), PpoA, required for biosynthesis of the linoleic psi factor component, psiBα. PpoA localizes in lipid bodies of conidiophores, Hülle cells, and cleistothecia (Fig. 1). Deletion of ppoA significantly reduced the level of psiBα and increased the ratio of conidia to ascospores fourfold. In contrast, forced expression of ppoA resulted in elevated levels of psiBα and decreased the ratio of conidia to ascospores sixfold. These results correlated with previous studies from Champe's research group (12, 13).
Here we describe the characterization of another A. nidulans putative fatty acid dioxygenase, PpoC, responsible for formation of the oleic acid-derived psi factor component, psiBβ. PpoC and PpoA exhibit distinct antagonistic regulation of meiospore and mitospore development. In contrast to the ΔppoA mutant, deletion of ppoC significantly increased ascospore production and decreased conidial development. ppoC and ppoA regulation of spore development appeared to be mediated by brlA and nsdD. Biochemical and transcriptional examination of the PpoA and PpoC mutants also indicated that their products may serve as antagonistic molecular signals of lipogenic genes through regulatory feedback loops in the cellular machinery of the fungus. We hypothesize the existence of a fungal “oxylipin signature-profile” that plays a role in modifying fatty acid biosynthesis and provides a fitness mechanism (46) to the organism by temporally balancing meiospore (dormancy) to mitospore (dispersal) development.
MATERIALS AND METHODS
Fungal strains, growth conditions, and genetic manipulations.
The A. nidulans strains used in the present study (Table 1) were grown on glucose minimal medium (GMM) (9) with appropriate supplements as needed at 37°C in continuous dark or white light. Sexual crosses and protoplast transformation of A. nidulans strains were conducted according to standard techniques (44, 58). Developmental cultures were grown on GMM, and asexual and sexual induction was performed as previously described (21, 55). The cultures of RNA shown in Fig. 6 to 8 were grown by inoculating 30 ml of liquid GMM with 106 spores of the appropriate strain/ml before incubation for 14, 24, 48, and 72 h (stationary conditions) prior to harvesting. Radial growth tests were performed in triplicate with ∼1,000 conidia centered on 30-ml GMM plates, and growth rates were recorded as the colony diameter over time. For germination tests, A. nidulans wild-type and ΔppoA, ΔppoC, and ΔppoA ΔppoC mutant strains were inoculated into minimal medium at 106 spores/ml and then shaken for 24 h at 300 rpm at 37°C. Samples were examined at 2-h intervals and germination rate was determined by counting 100 conidia. For microscopic observations of the different strains in liquid cultures, the same experimental procedures were used and samples were examined every 24 h for asexual and sexual related structures over a period of 7 days. Mycelial weight of lyophilized tissue was assessed after 5 days cultures in liquid GMM. Stereoscopic analysis of the different developmental stages of the ΔppoC single and double mutants was performed with cultures grown on solid GMM under dark or light conditions, recording the observations on a daily basis. Cells were visualized by using an Olympus BX60F-3 microscope and an Olympus SZ-60 stereoscope, and images were captured by an Olympus digital camera (Olympus America, Inc.).
TABLE 1.
A. nidulans strains used in this study
| Fungal straina | Genotype | Source or reference |
|---|---|---|
| RDIT12.3 | biA1 argB2 metG1 ΔppoA::metG veA | 55 |
| RDIT44.10 | pabaA1 biA1 pyroA4 metG1 veA1 trpc801 | 55 |
| TTMK1.97 | argB2 metG1 ΔppoC::trpC veA1 trpC801 | This study |
| RDIT30.92 | argB2 metG1 veA1 trpC801 | 55 |
| RTMK22.13 | pabaA1 biA1 pyroA4 metG1 ΔppoA::metG veA trpC801 | This study |
| RDIT58.21 | argB2 ΔppoA::trpC pyroA4 metG1 veA trpC801 | This study |
| RDIT55.7 | pyroA4 veA trpC801 | This study |
| TDIT11.12 | argB2 ΔppoC::trpC ppoC::pyroA methG1 veA trpC801 | This study |
| RDIT58.3 | ΔppoC::trpC pyroA4 veA trpC801 | This study |
| TU85 | pabaA1 biA1 argB2 pyroA4 ΔbrlA::argB veA | L. Yager |
| RDIT2.1 | metG1 veA | This study |
| RDIT86.7 | argB2 ΔbrlA::argB veA | This study |
| KHH52 | pabaA1 yA2 ΔargB::trpC ΔnsdD::argB trpC801 | 21 |
| KHH62 | pabaA1 yA2 ΔargB::trpC niiA(p)::nsdD trpC801 | 21 |
| Prototrophic isogenic strains | ||
| RDIT9.32 | veA | 55 |
| RDIT12.9 | metG1 ΔppoA::metG veA | 55 |
| RDIT58.12 | ΔppoC::trpC veA trpC801 | This study |
| RDIT54.7 | ΔppoC::trpC metG1 ΔppoA::metG veA trpC801 | This study |
| RDIT92.6 | ΔppoC::trpC ppoC::pyroA veA trpC801 | This study |
| RDIT81.10 | metG1 ΔppoA::metG veA gpdA(p)::ppoA::trpC | 55 |
| RRAW5.2 | argB2 ΔodeA::argB veA | R.A. Wilson |
| RDIT87.8 | niiA(p)::nsdD veA | This study |
| RDIT88.13 | ΔnsdD::argB veA | This study |
Strains starting with a “T” are original transformants, and strains beginning with an “R” are recombinants after a sexual cross. Some of the strains are not described in the text but were used for sexual crosses to create the final prototrophic strains.
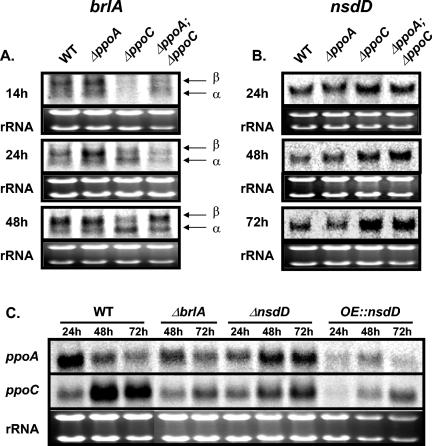
FIG. 6.
BrlA and NsdD are involved in the regulation of meiospore/mitospore ratio in A. nidulans. (A and B) Gene expression analysis of the spore specific transcriptional regulators brlA (asexual) (A) and nsdD (sexual) (B) in wild-type and ΔppoA, ΔppoC, and ΔppoA ΔppoC mutant strains. (A) Temporal delay of brlA transcripts (α and β) in the conidium-deficient ΔppoC and ΔppoA ΔppoC mutants. (B) nsdD gene is upregulated in the ascospore-overproducing ΔppoC and ΔppoA ΔppoC mutant strains. (C) ppoA and ppoC expression is altered in ΔbrlA, ΔnsdD, and overexpression nsdD (OE::nsdD) strains. Strains were grown in stationary liquid GMM at 37°C, and mycelia were harvested at the indicated time points. Equal loading of total RNA (20 μg) is depicted by ethidium bromide staining of the rRNA.
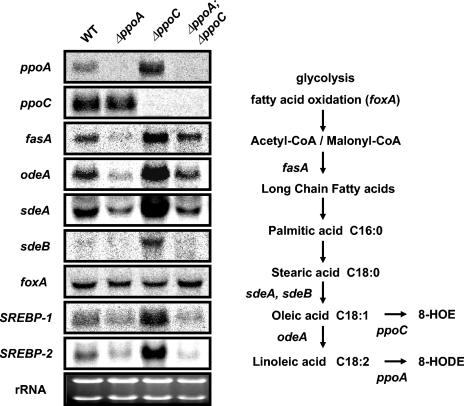
FIG. 8.
Antagonistic transcriptional regulation of fatty acid biosynthetic genes by PpoA and PpoC. Cultures of A. nidulans wild-type, ΔppoA, ΔppoC, and ΔppoA ΔppoC strains were grown at 37°C in stationary liquid GMM for 72 h and analyzed for the expression profile of fatty acid anabolic genes. The biosynthetic role of each gene is depicted in the flow chart of fatty acid biosynthesis in A. nidulans. Equal loading of total RNA (20 μg) is depicted by ethidium bromide staining of the rRNA.
Nucleic acid manipulations.
Construction, maintenance, and isolation of recombinant plasmids were performed by using standard techniques (48). Fungal chromosomal DNA was extracted from lyophilized mycelia by using previously described techniques (36). Total RNA was extracted from lyophilized mycelia by using TRIzol reagent (Invitrogen Co.) according to the manufacturer's recommendations. Approximately 20 μg of total RNA were used for Northern analysis with a 1.2% agarose-1.5% formaldehyde gel transferred to Hybond-XL membrane (Amersham Pharmacia Biotech). The PCR product obtained with primers ppoC-F16 (5′-TTTGCTTTTCCCGTCGCCGTCTC-3′) and ppoC-R18 (5′-CATTAGATCAGAGAACAACGA-GAC-3′) with the cosmid pLCJ14 as a template was used as a ppoC DNA probe for Southern and Northern hybridizations. Expression studies for the different genes were performed with appropriate probes: a 1.7-kb SalI-NdeI odeA fragment from plasmid pAMC30.4 (9), a 1.4-kb EcoRI-XhoI sdeA fragment from pRAW10 (57), a 1-kb HindIII-SphI sdeB fragment from pRAW18 (57), a 4.5-kb SalI brlA fragment from pTA111 (1), a 1-kb fasA PCR product obtained with the primers fasABF1 5′-GGATGGCCGGTAAGGTATTT-CTGG-3′ and fasABR1 (5′-GGTACACATGCCCTCCG-3′) (8), a 2.5-kb fox2 PCR product (amplified by the primers 5′-CCGTATCATCAACACCGCCT-3′ and 5′-GCGTTACGATGAAAA-AATTG-3′) (38), a 3-kb SREBP-1 PCR product obtained with the primers SREBP1-5′ (5′-GATTTCCAACTCTTCTCTCCCGC-3′) and SREBP1-3′ (5′-GGACTCTCCCACAAGCGCAGGTT-3′), a 0.8-kb SREBP-2 PCR product obtained with the primers SREBP2-5′ (5′-ATGTCTCCTGATCCCCTGTCGGC-3′) and SREBP2-3′ (5′-AACAGTATGCATTTGCTC-TTCTCTC-3′), a 4-kb ppoA PCR product obtained with primers ppoA-F2 and ppoA-R2 (55), and a 1.3-kb nsdD PCR product obtained with nsdD-5′ (5′-CCACATCTCCTGCTCCTCTCGTT-3′) and nsdD-3′ (5′-AGTGTCTTGGGTTTGAGGTTCGA-3′) (21). Detection and quantification of signals were carried out with a PhosphorImager-SI and ImageQuant software (Molecular Dynamics). Nucleotide sequences were analyzed and compared by using the Sequencher (Gene Codes Co.) and CLUSTAL W (http://www.ebi.ac.uk/clustalw/) software programs.
Molecular cloning, disruption, and cDNA isolation of the A. nidulans ppoC gene.
The ppoC gene was identified by tblastn search of the CEREON Genomics A. nidulans database (Monsanto Microbial Sequence Database), based on the amino acid sequence of linoleate diol synthase (Lds) cloned from Gaeumannomyces graminis (28) that was used as the query sequence. Oligonucleotides ppoC-F1 (5′-ACTACAACCCCCGCAACCTG-3′) and ppoC-R1 (5′-TGGTCGTAGTGGCGTGTAGG-3′) were designed based on the obtained contig ANI61C9558 predicting a fragment with high identity to Lds and PpoA. These primers were used to amplify a 1.2-kb fragment by PCR, with A. nidulans genomic DNA as a template. This PCR product was used as a probe to screen the A. nidulans pLORIST genomic cosmid library (Fungal Genetics Stock Center, Kansas City, Kans.). Two strongly hybridized overlapping cosmids, pLDF08 and pLCJ14, were identified and were further used as templates to sequence the entire open reading frame (ORF) of the ppoC gene, as well as ∼2,000 bp of the 5′- and the 3′-untranslated flanking regions in both DNA strands. A 9.1-kb SacII-SpeI fragment from the cosmid pLCJ14 containing the ppoC gene was subcloned into pBluescript, generating the plasmid pTMK2.2. The ppoC gene has been assigned accession no. AY613780 in the GenBank database.
The λZAP 24-h developmental cDNA library from A. nidulans (Fungal Genetics Stock Center) was used to isolate the ppoC cDNA according to the supplier's protocols. 5′ and 3′ ends were further confirmed by using RACE (rapid amplification of cDNA ends) technology and the Gene Racer kit (Invitrogen Co.). Pfu polymerase (Invitrogen) was used to amplify the corresponding fragments. Sequencing analysis of the resulting clones was performed to determine the positions of the introns and the amino acid sequence of the ppoC.
The ppoC deletion construct pTMK6.15, which included the trpC marker gene and ppoC flanking sequences, was constructed in the following manner. First, the modified primer pairs ppoC-5DF1-SacII (5′-CCCTCCCCGCGGGTGACTATAAT-3′) and ppoC-5DR1-XhoI (5′-GCAACATTGTGGGTCTCGAGAAGC-3′) were used to PCR amplify a 1.2-kb flanking region at the 5′-untranslated region (5′UTR) of the ppoC ORF with the plasmid pTMK2.2 as a template. The resulting amplified SacII-XhoI PCR fragment was subcloned into pBluescript, yielding the vector pTMK3.12. Next, the modified primers ppoC-3DF1-XhoI (5′-TACTGTAATGACTCGAGGACGAGG-3′) and ppoC-3DR2-KpnI (5′-TGCTTGAAGGGTACCTTATATGCCT-3′) were used to amplify the 1,000-bp flanking region at the 3′UTR of the ppoA ORF with the cosmid pTMK2.2 as a template. The amplified XhoI-KpnI 3′ flanking region was further ligated into XhoI-KpnI-digested pTMK3.12, generating the plasmid pTMK5.16 Finally, the 4,164-bp XhoI fragment from the pTA11 plasmid (7) containing the A. nidulans trpC cassette was inserted into the XhoI site of the pTMK5.16. The resulting disruption vector pTMK6.15 was used to transform the A. nidulans strain RDIT30.92 to tryptophan prototrophy, creating the transformant TTMK1.97, where the entire ppoC ORF was deleted. Gene replacement and ectopic integration were confirmed by PCR and Southern analysis. The ΔppoC allele was introduced in a veA background by sexual recombination of TTMK1.97 with RDIT55.7 to give the prototrophic strain RDIT58.12. The double mutant ΔppoA ΔppoC was created by a sexual cross between TTMK1.97 and RTMK22.13 (Table 1).
Complementation of the ΔppoC strain RDIT58.21 was achieved by using the vector pBJK2.6. The plasmid pBJK2.6 was created by inserting the 9.1-kb SacII-SpeI fragment from the plasmid pTMK2.2 containing the promoter, the coding sequence, and the termination cassette of ppoC into pJW53 (J.-W. Bok and N. Keller, unpublished data). pJW53 harbors a 1.8-kb fragment of the 5′ portion of the A. nidulans pyridoxine gene, which can complement pyroA4 mutation by single crossing over. TDIT11.12 was one of the pyridoxine prototrophs containing the ppoC::pyroA allele. TDIT 11.12 was further crossed with RDIT58.3 to give the complemented ΔppoC prototroph RDIT92.6.
Fatty acid analysis.
The strains were grown on 15 ml of liquid GMM in petri dishes under stationary conditions at 37°C under a dark regime. Mycelial mats were collected after 72 h, lyophilized, weighted, and homogenized mechanically by using an Ultra-Turax T25 dispenser (Ika Werke GmbH & Co. KG). Fatty acid methyl esters (FAMEs) were prepared by incubation of the mycelial homogenate in 2% H2SO4 in methanol at 80°C for 90 min. FAMEs were extracted into an equal volume of a n-hexane-chloroform (4:1 [vol/vol]) mixture and subsequently washed with distilled water. This step was repeated three times. Collected hexane fractions were combined, concentrated under a nitrogen stream, and dissolved in a small volume of hexane. To convert hydroxylated FAMEs (OH-FAMEs) into corresponding trimethylsilyl ether (TMSi) derivatives, the methanol phase was removed in vacuo, and the remaining residue was dissolved in 80 μl of a mixture of N′,O-bis(trimethylsilyl)trifluoroacetamide and trimethylchlorosilane (99:1 [vol/vol]; Sylon BFT kit; Supelco). The reaction was incubated at 90°C for 30 min, and OTMSi-FAMEs were recovered with in a small volume of hexane. Both FAMEs and OH-FAMEs were separated by gas chromatography (Thermoquest Trace GC) on an RTX-5MS 0.25-μm fused silica column (Restek Corp., Bellafonte, Pa.) and identified by mass spectrometry on an inline Finningan Polaris mass spectrometer. One microliter of the sample was injected into gas chromatograph equipment programmed as follows: 80°C (held for 2 min), increased at 20°C min−1 up to 220°C, followed by 30°C min−1 to 300°C, and then held at 300°C for 2 min. The injector temperature was 300°C. Helium (1 ml min−1, constant flow) was used as a carrier gas. For mass spectrometry electron impact mode was used, and the ion source was 280°C. The electron energy was 70 eV, the ionization current was 100 μmA, and the scan speed was 0.6 s per decade. Scans were recorded in a range from 35 to 600 atomic mass units (amu). Fatty acids were identified by comparison of retention times with a set of authentic fatty acids standards, whereas hydroxylated derivatives of fatty acids were identified by mass spectrometry on the basis of their fragmentation patterns reported elsewhere (9, 18, 55).
Physiological and morphological studies.
All strains used for physiological studies were prototrophic. RDIT9.32 (55) strain was used as the wild type. Asexual and sexual spore production studies were performed on plates containing 30 ml of solid 1.5% GMM. For each plate, 5 ml of top layer with cool melted 0.7% agar-GMM containing 106 conidia of the appropriate strain was added. Cultures were incubated in continuous dark or light at 37°C. Illumination was carried out in an incubator equipped with General Electric 15-W broad-spectrum fluorescent light bulbs (F15T12CW) placed 50 cm below the plates. A core 12.5 mm in diameter was removed from each plate at the appropriate time interval and homogenized for 1 min in 3 ml of sterile water supplemented with 0.01% Tween 80 to facilitate the release of the hydrophobic spores. Asexual and sexual spores were counted by using a hemocytometer. The experiments were performed with four replicates. Spore data were statistically compared by analysis of variance and Fisher least significant difference by using the Statistical Analysis System (SAS Institute, Cary, N.C.).
RESULTS
PpoC encodes a putative fatty acid dioxygenase.
Disruption of A. nidulans ppoA led to a strain defective in producing the linoleic acid- derived oxylipin psiBα (55). To further characterize genes involved in psi factor biosynthesis, we compared the A. nidulans publicly available genome database (Cereon Genomics; Monsanto) with the oxylipin-producing linoleate diol synthase (lds) gene (a ferric hemeprotein with fatty acid dioxygenase and hydroperoxide isomerase activities) from the filamentous fungus G. graminis var. graminis (28). A DNA fragment likely to encode a fatty acid dioxygenase was identified and used to characterize the gene, termed ppoC (for psi-producing oxygenase; GenBank accession number AY613780). Genomic and cDNA analysis showed that PpoC encodes an 1,117-amino-acid polypeptide containing 11 introns (Fig. 2). Comparative sequence analysis by using CLUSTAL W indicated that there is 44% identity between PpoC and PpoA and 40% identity between PpoC and G. graminis Lds. PpoC shares also high similarity with the Magnaporthe grisea linoleate diol synthase (15) and the Ssp1 protein from Ustilago maydis (29), as well as with various predicted proteins from existing filamentous fungal databases. PpoC orthologs are absent in the yeasts Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Candida albicans but present in the human dimorphic pathogen Histoplasma capsulatum.
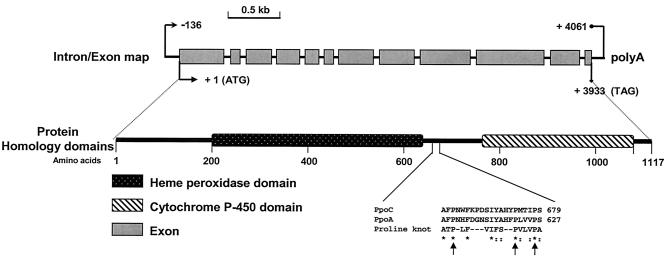
FIG. 2.
Molecular characterization of the A. nidulans PpoC locus. The ppoC genomic DNA sequence contains 11 introns and 12 exons. The major transcription initiation site was found 136 bp upstream of the first ATG codon, and the polyadenylation site (polyA) is located 128 bp downstream from the ORF stop codon. The deduced translated 1,117-amino-acid polypeptide contains predicted heme peroxidase and cytochrome P-450 domains and a sequence that aligns with the proline knot motif, an essential motif for targeting proteins to lipid bodies. Arrows indicate the conserved proline residues.
Analysis of the PpoC protein by using the PFAM database (http://pfam.wustl.edu) indicated that the PpoC residues 181 to 650 have domains similar to animal heme peroxidases and residues 765 to 1080 have domains similar to cytochrome P450 oxygenase (Fig. 2). The PpoC peptide contains six possible N glycosylation sites that might play a role in the maturation of PpoC, three cAMP-dependent protein kinase phosphorylation sites, and several protein kinase C phosphorylation sites. The PpoC amino acid sequence, like those of PpoA and Lds, shared high similarity with various mammalian cyclooxygenases or prostaglandin synthases, ranging from 25 to 26% identity and 38 to 42% similarity. PpoC also contains a putative hydrophobic subdomain known as a “proline knot” that is characteristic for targeting plant proteins to lipid bodies (Fig. 2). PpoA and Ssp1 also contain the proline knot motif and are localized to lipid bodies (29, 55). Taken together, these data suggest that PpoC is a paralog of PpoA and G. graminis Lds.
Creation of a ΔppoC strain.
A ppoC deletion mutant was generated by replacing the wild-type copy of ppoC with the trpC gene. Random screening of 100 transformants by PCR and Southern analysis revealed three transformants with identical phenotypes showing the DNA size fragments expected for a trpC replacement of ppoC (data not shown). One transformant was selected and crossed to produce a prototrophic ΔppoC strain, which was used for further physiological and molecular analyses. The ΔppoA ΔppoC double mutant was also obtained by a sexual cross. Complementation of the ΔppoC strain with a functional copy of ppoC restored the wild-type phenotype, thus confirming that the effects on sexual and asexual sporulation described below were solely due to the deletion of ppoC (data not shown).
Oxylipin and total fatty acid composition are altered in the ΔppoC strain.
The role of PpoC as a putative fatty acid dioxygenase was explored by analyzing both oxylipin and fatty acid composition of the ΔppoC mutants. Assessment of the two most abundant psi factor components, the oleic acid-derived psiBβ [8-HOE or 8-hydroxy-9(Z)-octadecanoic acid] and the linoleic acid-derived psiBα [8-HODE or 8-hydroxy-9(Z),12(Z)-octadecadienoic acid] revealed that deletion of the ppoC allele resulted in almost complete elimination of psiBβ molecules (in Table 2, hydroxylation of the fatty acid backbone designates the psi compounds as psiB [8′ hydroxy-], psiC [5′,8′ dihydroxy-], and psiA [the lactone ring of psiC at 5′ position]) (40). Previous studies showed that deletion of ppoA resulted in a strain deficient in producing psiBα. The double mutant was deficient in the production of both oleic and linoleic acid-derived psiB factors (Table 2). The presence of linoleic or oleic acid-derived psiA or psiC was not detected in any samples in accordance with previous studies (9, 55; data not shown). These data demonstrated that PpoC is involved in the production of psiB oxylipins and suggested that oleic acid is a preferable substrate for PpoC.
TABLE 2.
Psi factor composition of mycelia of A. nidulans oxylipin mutants
| Strain | Mean amt (μg) of psi-FAME/g of mycelium (dry wt)a ± SE
|
|
|---|---|---|
| psiBβ (8-HOE) | psiBα (8-HODE) | |
| Wild typeb | 5.87 ± 0.70 | 2.19 ± 0.87 |
| ΔppoAb mutant | 7.70 ± 0.69* | 0.22 ± 0.19** |
| ΔppoC mutant | 0.98 ± 0.81** | 2.37 ± 0.86 |
| ΔppoA ΔppoC mutant | 1.31 ± 0.69** | 0.53 ± 0.27** |
psi-FAME, psi fatty acid methyl esters. The analysis was carried out with 72-h-old mycelia grown in liquid GMM under stationary conditions at 37°C in the dark. Values are the means of three replications. Statistical analysis was performed by using the Student t test, and significance to wild-type oxylipin composition is indicated as follows: *, P < 0.05; **, P < 0.001. psiBβ, 8-HOE (8-hydroxy oleic acid); psiBα, 8-HODE (8-hydroxy linoleic acid).
Wild-type and ΔppoA values are from reference 54. The psi analysis was performed at the same time for all the Δppo mutants.
Fatty acid composition was analyzed from mycelia grown under dark conditions at 37°C at the same conditions that psi analysis was carried out. Table 3 shows the fatty acid profile of the wild-type and ΔppoA, ΔppoC, and ΔppoA ΔppoC mutant strains. In each strain palmitic, stearic, oleic, and linoleic acids are the most prevalent fatty acids. The ΔppoA strain showed no statistical differences in the amount of the individual fatty acids compared to the wild type. However, ΔppoC showed an increase in palmitic acid and a decrease in stearic acid compared to the wild type. The same pattern was maintained in the double mutant. Deletion of ppoC also led to a twofold increase in the total fatty acid composition per gram of mycelium, whereas ΔppoA strain showed a small but significant decrease in total fatty acid composition. The double mutant did not show a statistically significant alteration in the amount of total fatty acids compared to the wild type.
TABLE 3.
Fatty acid composition of mycelia of A. nidulans oxylipin mutantsa
| Strain | Mean wt (%) of major FAMEb ± SE
|
Total fatty acid (%) ± SE | |||
|---|---|---|---|---|---|
| Palmitic acid (16:0) | Stearic acid (18:0) | Oleic acid (18:1) | Linoleic acid (18:2) | ||
| Wild type | 31.10 ± 0.85 | 14.80 ± 1.20 | 14.70 ± 3.30 | 37.20 ± 3.75 | 1.09 ± 0.32 |
| ΔppoA mutant | 30.00 ± 2.25 | 13.30 ± 0.15 | 17.70 ± 0.75 | 39.90 ± 1.60 | 0.68 ± 0.03 |
| ΔppoC mutant | 38.18 ± 1.70 | 10.36 ± 0.25 | 14.35 ± 2.15 | 34.15 ± 0.15 | 2.10 ± 0.06 |
| ΔppoA ΔppoC mutant | 34.42 ± 1.85 | 10.28 ± 0.05 | 14.96 ± 0.85 | 37.84 ± 1.10 | 0.83 ± 0.06 |
The analysis was carried out with 72-h-old mycelia grown in liquid GMM under stationary conditions at 37°C in the dark. Values are the means of three replications.
FAME, fatty acid methyl esters. The weight percent FAME is based on the lyophilized weight of mycelia.
PpoA and PpoC antagonistically regulate meiospore and mitospore development.
Detailed physiological studies of the effect of linoleic acid derived oxylipins on A. nidulans development suggested that some components decreased the conidia/ascospore ratio, whereas others had the opposite effect (10, 12, 13). In particular, psiBα was implicated in increasing ascospore numbers (12). This finding was genetically supported by examination of two ppoA mutants. Deletion of ppoA resulted in a strain devoid of psiBα with an increased conidia/ascospore ratio, whereas the overexpression of ppoA strain (OE::ppoA) overproduced psiBα and ascospores (55). Although oleic acid-derived psi factor components were chemically characterized in A. nidulans, bioassays with purified molecules have not been described (9, 39, 40). Here we found that, as with the ΔppoA strain, neither the ΔppoC mutant nor the ΔppoA ΔppoC double mutant had any obvious effects upon the conidia germination (data not shown), growth pattern (Fig. 4A), or morphology of vegetative hyphae (data not shown). However, the kinetics of mitospore and meiospore development was oppositely regulated in ΔppoC and ΔppoA ΔppoC mutants compared to the ΔppoA strain.
FIG. 4.
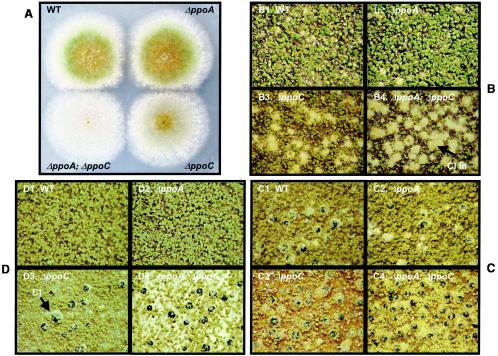
ppoA and ppoC genes are essential for balancing conidiophore/cleistothecia formation. Cultures of A. nidulans wild-type (B1, C1, and D1), ΔppoA (B2, C2, and D2), ΔppoC (B3, C3, and D3) and ΔppoA ΔppoC (B4, C4, and D4) were grown at 37°C on solid GMM. (A) Deletion of ppoC delays conidiophore formation. Five-day-old cultures of point-inoculated strains (inoculum, 103 conidia) under light conditions. (B to D) Induction of the sexual sporulation and suppression of the asexual fruiting bodies in ΔppoC and ΔppoA ΔppoC strains. The opposite is observed in the ΔppoA mutant. Each strain was inoculated with 106 conidia/plate, and cultures were grown for 2 days under dark (B), 8 days under dark (C), and 8 days under light (D) conditions. Black balls are cleistothecia (“Cl” in panel D3), fuzzy balls are cleistothecia initials (“Cl In” in B4), and smaller green spheres are conidiophore heads.
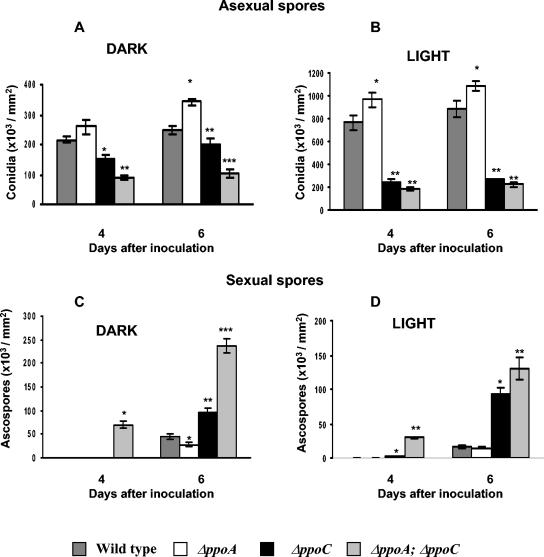
Conidia and ascospore production was assessed on GMM under light and dark conditions at 37°C (Fig. 3). Spore production was measured 4 and 6 days after inoculation. In contrast to the ΔppoA phenotype, the ΔppoC and ΔppoA ΔppoC mutants produced significantly fewer conidia but significantly more ascospores than the wild-type strain under both dark and light conditions (P < 0.001) (Fig. 3 and 4). These results were maintained over a time period of 10 days (data not shown). Overall, the ratio of conidia to ascospores decreased ∼3-fold in the ΔppoC mutant and 15-fold in the ΔppoA ΔppoC mutant after 6 days of cultivation in the dark. In contrast, ΔppoA led to a fourfold increase in the conidium/ascospore ratio (55).
FIG. 3.
ΔppoC and ΔppoA ΔppoC have decreased conidia and increased ascospore production compared to wild-type under both dark and light conditions (P < 0.001). Cultures of A. nidulans wild-type, ΔppoA, ΔppoC, and ΔppoA ΔppoC were grown at 37°C under dark and light conditions in GMM. Conidium production of 4- and 6-day-old cultures grown in the dark (A) or in light (B) and ascospore production of 4- and 6-day-old cultures grown in the dark (C) or in light (D). Values of ascospores in the dark (4 days) for wild-type, ΔppoA mutant, and ΔppoC mutant strains were low and cannot be represented in the graph (wild type, 390 ± 158; ΔppoA, 120 ± 69; ΔppoC, 330 ± 102). Values are the mean of four replicates, and error bars represent standard errors. Columns with asterisks represent values for the same day that differ significantly from the wild type (P < 0.001).
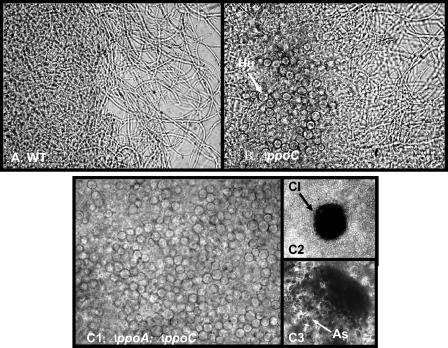
Not only were conidia/ascospore ratios decreased in the ΔppoC and ΔppoA ΔppoC strains but development proceeded in an aberrant fashion. Precocious development of Hülle cells, cleistothecia, and ascospores (Fig. 3 and 4) was visibly apparent in these strains, whereas conidiophore development was delayed 4 to 5 h for the ΔppoC and 8 to 10 h for the ΔppoA ΔppoC mutant (Fig. 4A). In radial growth experiments ΔppoC and ΔppoA ΔppoC strains showed approximately 1- and 2-mm retardation of the mature conidiophore zone, respectively. In addition, the ΔppoC and ΔppoA ΔppoC mutants were able to produce Hülle cells and cleistothecia in liquid shake cultures in GMM after 24 h in contrast to ΔppoA mutant and wild-type strains, which are unable to form these structures under these conditions (Fig. 5).
FIG. 5.
Deletion of ppoC induces sexual development in liquid cultures. Cultures of A. nidulans wild-type (A), ΔppoC (B), and ΔppoA ΔppoC (C) were grown at 37°C in liquid GMM. Abundant presence of Hülle cells after 2 days growth in liquid cultures of ΔppoC and ΔppoA ΔppoC strains (B and C). The double mutant was able to produce mature and fertile cleistothecia after 2 days of growth (C2 and C3). Hu, Hülle cells; Cl, cleistothecium; As, ascospores.
Changes in the meiotic/mitotic spore ratio are correlated with brlA and nsdD expression.
Transcriptional regulators specific for each spore stage have been described for A. nidulans. BrlA is a zinc finger transcription factor essential for conidiophore development (45), and NsdD is a GATA-type transcription factor required for cleistothecia development (21). Loss of either locus generates strains unable to produce either conidia (ΔbrlA) or ascospores (ΔnsdD), although the alternative spore type is produced normally in these mutants. We were interested in determining whether the changes we observe in ascospore and conidia production in the ppo mutants would be reflected in brlA and/or nsdD expression.
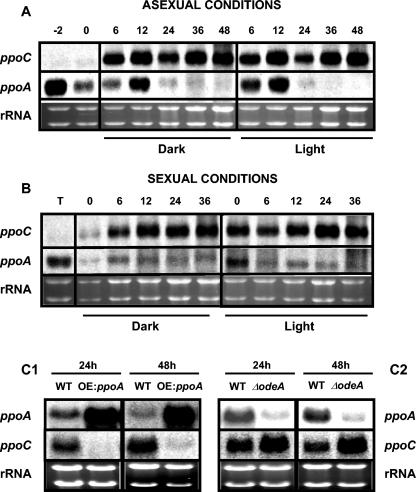
Mutations in brlA result in the “bristle” phenotype, characterized by the fact that conidiophores lack their normal components—vesicles, metulae, phialides, and conidia (Fig. 1). The brlA locus consists of overlapping transcription units, designated α and β, with α transcription initiating within β intronic sequences. As shown in Fig. 6A, the accumulation of brlA transcripts showed a temporal delay by ∼12 to 24 h in the ΔppoC and ΔppoA ΔppoC mutants. In addition, the two different brlA transcripts are aberrantly regulated in the ΔppoC mutant, where there is a more pronounced expression of brlAα than brlAβ at 48 h. In contrast, an increase of the brlA transcripts was observed in ΔppoA at 24 h. These transcriptional changes correlated positively with the relative decrease (ΔppoC and ΔppoA ΔppoC) and increase (ΔppoA) in conidial production.
Deletion of nsdD prevents Hülle cell, cleistothecia, and subsequently ascospore formation, whereas overexpression of nsdD leads to an increase in sexual development (21). Expression analysis of the nsdD gene demonstrated that is upregulated in the ascospore-overproducing strains ΔppoC and ΔppoA ΔppoC especially at 72 h (Fig. 6B), a time point that coincides with the initiation of sexual development in stationary liquid cultures. Loss of ppoA showed a slight decrease in nsdD expression at 72 h (Fig. 6B).
To further examine possible regulatory interactions between ppoA/ppoC and brlA/nsdD, expression studies were carried out in ΔbrlA and ΔnsdD strains and in strains overexpressing nsdD grown in stationary liquid GMM. ppoC expression was repressed in ΔbrlA, ΔnsdD, and OE::nsdD strains, whereas ppoA expression was not significantly affected in the ΔbrlA background at the examined time points but was induced in the ΔnsdD background and repressed at 24 h in the overexpression nsdD background (Fig. 6C). These results indicated that there is a reciprocal regulation between the ppo genes/gene products and these developmental transcription factors.
Interactive regulatory loops connect ppoA and ppoC expression.
We considered that the balance of ascospore and conidia production by PpoA and PpoC could implicate a regulatory relationship between these two factors and/or their products. Experiments to test this hypothesis were carried out at the transcriptional level. Previous data demonstrated that ppoA expression is correlated with the initiation of asexual and sexual fruiting body formation in A. nidulans (55). ppoC is not expressed under vegetative conditions (liquid cultures; Fig. 7A, time points −2 and 0), but mRNA studies performed throughout the asexual and sexual life cycle of A. nidulans showed that ppoC is expressed after induction over a longer time and at significantly higher levels than ppoA (Fig. 7A and B). The OE::ppoA strain, which accumulates high levels of psiBα (55), led to a significant suppression of ppoC transcript (Fig. 7C1). In addition, mRNA analysis showed that ppoC was upregulated and that ppoA was significantly downregulated in the ΔodeA mutant (OdeA is a Δ12-oleic acid desaturase converting oleic acid to the polyunsaturated linoleic acid) (Fig. 7C2), a strain that accumulates high levels of psiBβ and no psiBα (9).
FIG. 7.
ppoC and ppoA are differentially regulated under asexually (A) and sexually (B) induced cultures. Mycelia of the wild-type strain were synchronized by 18 h of vegetative growth in liquid shaken GMM (time zero) and developmentally induced on solid GMM to obtain asexual (A) and sexual (B) tissue types for RNA isolation at appropriate time intervals under dark or light conditions. Time points represents hours after asexual or sexual induction, respectively. Induction of asexual sporulation was performed under normal aeration conditions. The time point “−2” corresponds to 16 h of vegetative growth culture, 2 h before the transfer to solid medium for asexual induction. For the induction of sexual sporulation 18-h-liquid-grown mycelia were transferred onto solid medium, and the plates were sealed with parafilm for 20 h. After 20 h, the plates were unsealed, and at different time points samples were collected for RNA analysis (time represents hours after induction: 0 to 36 h). The time point “T” corresponds to the sample that was collected at the time of transfer to the solid media and before the initiation of sexual induction. (C) Differential regulation of ppoA and ppoC expression as was demonstrated by their transcript analysis in OE::ppoA (C1) and ΔodeA (C2) strains. Equal loading of total RNA (20 μg) is depicted by ethidium bromide staining of the rRNA. The time points of mycelium harvest are indicated above the lanes.
Antagonistic regulation of fatty acid anabolic genes by ppoA and ppoC.
The fatty acid composition of Δppo mutants demonstrated that the total percentage of the fatty acids was decreased in the ΔppoA strain and increased in the ΔppoC strain (Table 3). To see whether the biochemical difference was reflected at the transcript level, we examined the expression of several fatty acid biosynthetic genes in the mutant strains grown under the same conditions that fatty acid extraction was performed. Fatty acid synthase α-subunit (fasA) encodes the central enzyme in de novo lipogenesis (8), catalyzing the condensation of acetyl coenzyme A (acetyl-CoA) and malonyl-CoA into long-chain fatty acids. fasA expression was significantly upregulated in the ΔppoC mutant and downregulated in the ΔppoA strain (Fig. 8). This correlated with the total fatty acid content shown in Table 3. Similar expression patterns were obtained for the desaturase genes sdeA and sdeB (both Δ9-stearic acid desaturases converting stearic acid to the monounsaturated oleic acid) (57), odeA (9), and the oxylipin biosynthetic gene PpoA (55) (Fig. 8). In contrast, gene expression was closer to that of wild-type for the ΔppoA ΔppoC mutant, which did not differ in total fatty acid percentage from the wild type. It appeared that only the anabolic pathways were regulated by ppoA and ppoC since expression of foxA, encoding the catabolic d-bifunctional enzyme (enoyl-CoA hydratase and hydroxyacyl-CoA dehydrogenase) required for β-oxidation (38) was not altered in these mutants (Fig. 8).
These results were reminiscent of the feedback regulation of fatty acid biosynthesis already described in A. nidulans sdeA and odeA mutants (9, 57) and other eukaryotes, including yeast (54) and mammals (16). This coordination of lipid homeostasis is governed by end product feedback regulation of transcription. In animals this occurs through the proteolytic release of transcriptionally active sterol regulatory element binding proteins (SREBPs) from intracellular membranes (47). Mammalian genomes include two SREBP genes (basic helix-loop-helix leucine zipper transcription factors) that possess considerable selectivity difference in their target genes and bind to the sterol regulatory element (SRE) DNA motif in the promoters of lipogenic genes (46). Polyunsaturated fatty acids, including oxylipins such as prostaglandins and leukotrienes, appear to coordinately inhibit lipogenic gene transcription by rapidly reducing the nuclear content of SREBP-1 proteins. Search of the A. nidulans databases (Whitehead) led to the discovery of two putative transcription factors that showed similarity with the mammalian SREBP genes named SREBP-1 (locus AN7661.2, 38 to 40% identity in conserved areas) and SREBP-2 (locus AN7170.2, 28 to 38% identity in conserved areas). Expression analysis of these two putative lipogenic transcription factors in Δppo mutants led to the conclusion that SREBP-1 and SREBP-2 are significantly upregulated in the ΔppoC strain and downregulated in the ΔppoA and ΔppoA ΔppoC mutants, thus likely mediating the regulation of the anabolic genes involved in the fatty acid metabolism described above (Fig. 8). These results indicated that oxylipins may play a regulatory role in A. nidulans fatty acid metabolism as they act in mammals.
DISCUSSION
A central issue in fungal biology lies in elucidating the exogenous and endogenous factors required for meiospore and mitospore reproduction. Aside from their pivotal roles in fungal dissemination and survival, fungal spores comprise mainly the primary and secondary infection particles of plant pathogenic fungi (3, 4). Usually, the asexual spore serves as both primary and secondary inoculum of infection, whereas in some ascomycetes the overwintering sexual spore is the source of primary inoculum (3, 4). The experiments presented here show two genes that can influence the process of meiospore and mitospore development relative to each other. In the present study, we characterized ppoC, encoding a putative fatty acid dioxygenase in the model organism A. nidulans. PpoC is involved in the production of oleic acid-derived psi factor and has an opposing function to the previously characterized PpoA. Both enzymes serve as essential signaling regulators of mitotic-meiotic spore balance. To our knowledge, this is the first genetic study identifying a mechanism integrating an antagonistic orchestration of asexual and sexual reproduction.
PpoC is involved in the production of oleic acid oxylipins.
Chemical analysis of the ΔppoA, ΔppoC, and ΔppoA ΔppoC mutants demonstrated that PpoC, in contrast to the previously characterized PpoA, is probably involved in the production of psiBβ. The double mutant is crippled in its ability to produce both psiBα and psiBβ (Table 2). However, we cannot exclude the possibility that PpoA and PpoC are involved in the production of other oxylipin species: either downstream products of psiBα and psiBβ and/or derivatives of alternative fatty acids. Oxylipin-generating enzymes (dioxygenases, lipoxygenases, cyclooxygenases, etc.) frequently exhibit activity toward more than one substrate. For example, the G. graminis dioxygenase Lds can oxygenate oleic, α-linolenic, and ricinoleic acid, as well as linoleic acid (52). Since the double mutant still produces some psiB molecules, our results also suggest the presence of another enzyme capable of generating these products. This is similar to the situation in plants in which several lipoxygenases are involved in producing the same oxylipins (reference 17 and references therein).
Regulatory links between ppoA and ppoC: mechanism for an oxylipin signature.
Champe's physiological and biochemical characterization of psi factor in the 1980s was among the first studies to uncover a mechanism that shifted the meiospore-mitospore balance in fungi and was certainly the first to implicate the role of oxylipins in this phenomenon (12, 13). The phenotype of the ppoA mutant seemingly supports his findings. However, since Champe and coworkers did not assay the effects of purified oleic acid oxylipins on A. nidulans, we are unable to directly compare the ΔppoC phenotype to any published work. We also note that the application of a purified metabolite differs in many respects to the absence of a gene. Because deletion of ppoA and ppoC showed pleiotrophic effects, including changes in oxylipin make-up, fatty acid composition, and gene expression, we hesitate to attribute a particular role to a single oxylipin species. Rather, we suggest that oxylipin molecules as a whole are important in generating differentiation processes in A. nidulans in a manner similar to that described in plants.
In-depth analyses of plant oxylipin pathways have uncovered complex and tight control of oxylipin production, presumably required for appropriate development in changing environmental milieus (5). The temporal and spatial activity of different oxylipin biosynthetic enzymes appears to be of fundamental importance for normal growth; this is especially true for lipoxygenase isoforms (17). Activities and compartmentalization of the biosynthetic oxylipin enzymes is of paramount importance in determining the oxylipin profiles that will lead to the appropriate developmental pathway. Recent analyses indicate that the phyto-oxylipin pool of a given organelle, tissue, plant, or species confers an “oxylipin signature” to that respective entity (24, 35). It is proposed that the oxylipin signature is predictive of the execution of specific developmental pathways for the organism (5, 17).
Our findings suggest a similar conserved complex control exists in fungi and that a fungal oxylipin signature could be predictive of meiospore and mitospore development in any given fungal isolate. The expression profiles of ppoA and ppoC suggested that ppoA transcripts are important for the initiation of conidiophores and cleistothecia but ppoC needs to be at significantly higher transcriptional levels after asexual induction for normal sporulation (Fig. 7A and B). Inhibition of ppoC expression in a ppoA overexpression strain suggests the possibility of feedback regulation between ppoA and ppoC, ostensibly via oxylipin production (Fig. 7C and Fig. 8). This latter point is supported from results showing suppression of ppoA and induction of ppoC in a ΔodeA strain (Fig. 7C). Inactivation of OdeA (9), a Δ-12 desaturase required for linoleic acid biosynthesis, results in a strain that produces a sixfold increase in the oleic acid-derived psiBβ but no psiBα. We speculate that fungal oxylipin production is dependent on stimuli that can lead to alterations of the developmental schedule to withstand adverse or favorable environmental conditions.
BrlA and NsdD: mediators of oxylipin signaling?
The abnormal sporulation patterns of the Δppo mutants led us to investigate the expression profiles of the major developmental transcription factors BrlA and NsdD. The proper expression of brlA during asexual spore formation in A. nidulans is critical for the development of the conidiophores and for the activation of other developmentally specific genes (2). Our experiments indicated that the delay in conidiation in ΔppoC and ΔppoA ΔppoC mutants was at least partially mediated by the delay and alteration in the expression of brlAα and brlAβ that are individually essential for the formation of morphologically normal conidiophores (22, 23). brlAβ contains two ORFs: a small ORF, μORF, that is located upstream of the brlA initiation codon, and a large ORF that encodes brlAβ. The translation of μORF inhibits the translation of brlAβ ORF (BrlA) that in turn is required for brlAα transcription, leading to the activation of a series of conidiation genes. In our studies, brlAα was more abundant in ppoC mutants (Fig. 6A), indicating a role for PpoC and/or its enzymatic products in regulating the transcriptional ratio of brlAα: brlAβ or translation of the μORF that restricts the synthesis of brlAβ. Examination of the promoter regions of ppoA and ppoC revealed the presence of several putative BrlA response elements (data not shown) (14), further supporting our results that the transcriptional regulation of ppoA and ppoC is under the control of BrlA. In contrast to brlA expression, expression of the sexual stage transcription factor nsdD was upregulated in the ascospore overproducing the ΔppoC and ΔppoA ΔppoC strains and downregulated in the ΔppoA mutant, a strain that produces fewer ascospores than does the wild type (Fig. 6B). These results suggest that Ppo regulation of ascospore and conidial development is at least partially mediated through the nsdD and brlA transcription factors as summarized in our proposed model in Fig. 9.
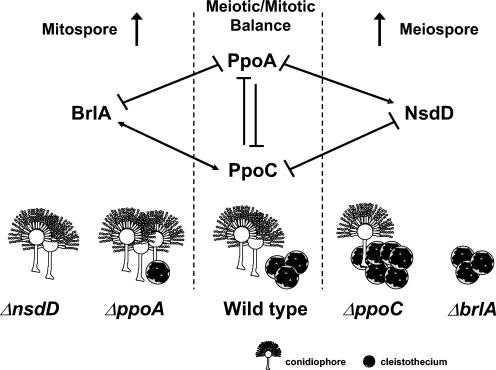
FIG. 9.
Proposed model depicting the genetic relationship between PpoC, PpoA, BrlA, and NsdD to establish the meiotic/mitotic ratio in A. nidulans. PpoA and PpoC are involved in linoleic and oleic acid oxylipin production, respectively. BrlA and NsdD are transcription factors regulating mitotic and meiotic development, respectively. The numbers of individual conidiophores and cleistothecia are indicative of the asexual/sexual ratio in each mutant.
Our studies also support a case for reciprocal regulation of ppo expression by BrlA and NsdD. Figure 6C shows that BrlA acts as a positive regulator of ppoC and a negative regulator of ppoA and NsdD acts as a negative regulator for both ppoA and ppoC. Based on these results we speculate that ppoC is regulated by BrlA and NsdD through a feedback mechanism and serves as a positive regulator of asexual and negative regulator of sexual development (Fig. 9). The fact that both BrlA and NsdD act as negative regulators of ppoA and that brlA and nsdD expression was not greatly affected by the ppoA deletion may indicate that ppoA acts downstream or in different pathways at transcriptional or translational levels to regulate the asexual and sexual cycles.
This apparent feedback loop between ppoA-ppoC and nsdD-brlA indicates a mechanism by the organism to maintain tight control of the meiospore/mitospore ratio and eliminate the possibility of a failure in the developmental mechanism. We conjecture that this interaction occurs in the reproductive tissues of the fungus. Our previous study demonstrated that PpoA accumulates predominantly in lipid bodies found in Hülle cells, nascent cleistothecia, and mature conidiophores—the tissues containing the highest concentrations of lipid bodies (55). PpoC, like PpoA, contains a proline knot motif in the central domain of the polypeptide (Fig. 2) that is crucial for targeting and anchoring proteins to lipid bodies (41) and is also likely to localize to these same tissues. Although no studies are available showing BrlA or NsdD localization, it is logical to assume that they would also be present in these tissues, which are dependent on their function.
Transcriptional regulation of lipid homeostasis.
Deletion of ppoC led to a significant increase in the transcription of genes involved in fatty acid biosynthesis and a concomitant increase in the total amount of fatty acids in the fungal thallus. On the other hand, ΔppoA lowered the transcriptional level of the lipogenic genes. Studies in primary rat hepatocytes and cultured 3T3-L1 adipocytes showed that arachidonic acid-derived oxylipin metabolites (e.g., prostaglandin E2) suppress the expression of the fatty acid synthase (FAS) through a G-protein-coupled receptor prostanoid signal transduction cascade (30). Thus, in analogy to these studies, we hypothesize that PpoC and PpoA product(s) modulate SREBP expression indirectly, perhaps by instigating autocrine-paracrine antagonistic signaling cascades that couple meiospore and mitospore production to a host of other developmental programs in A. nidulans, including fatty acid anabolism.
Conclusions.
With the characterization of ppoA and ppoC, we provide evidence of an endogenous system balancing meiospore and mitospore production in A. nidulans. Orthologs of both of these genes are found in filamentous fungi and, coupled with the numerous studies linking oxylipin production with fungal sporulation (27, 31, 33, 42, 51), support a case for conservation of an oxylipin-driven mechanism controlling, among other cellular processes, sexual and asexual differentiation. We propose that fungal oxylipins serve as autocrine or paracrine signals generated in response to—and enabling the fungus to respond appropriately to—specific environmental parameters. Since previous studies showed that Aspergillus spp. respond to seed oxylipins in a manner similar to that of psiBα and psiBβ (10), it is reasonable to postulate that host oxylipins can mimic and/or interfere with endogenous fungal oxylipins on a cellular basis, thus affecting the outcome of the host-fungal interaction. Recent evidence suggests that endogenous unsaturated fatty acids regulate morphological transitions and virulence in C. albicans (43) and have structural and functional homologs in prokaryotes (56), suggesting that fatty acids or the downstream oxylipins act as signals of cross-kingdom cell-cell communications. It remains to be examined whether microbial oxylipins act as virulence factors in these same interactions. Noverr et al. (42) postulated that microbial oxylipins can modulate disease pathogenesis and host immunity responses. Certainly, a better understanding of the molecular mechanisms that govern fungal oxylipin metabolism could contribute to the design of novel chemicals or other strategies that can reduce the survival and spread of pathogenic fungi.
Acknowledgments
This study was funded by NRI 2001-35319-10996 (N.P.K.) and a Novartis (Syngenta) Crop Protection Graduate Fellowship (D.I.T.).
We are grateful to the Jan G. Jaworski (Donald Danforth Plant Science Center, St. Louis, Mo.) lab for assistance with the gas chromatography-mass spectrometry analysis. We thank Christina Hull (Department of Biomolecular Chemistry, University of Wisconsin-Madison) for critical review of the manuscript.
Footnotes
This study is dedicated to the memory of Sewall Champe.
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrios, G. N. 1997. Plant pathology, 4th ed. Academic Press, Inc., San Diego, Calif.
- 4.Alexopoulos, C. J., C. W. Mims, and M. Blackwell. 1996. Introductory mycology, 4th ed. John Wiley & Sons, Inc., New York, N.Y.
- 5.Blee, E. 2002. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7:315-322. [DOI] [PubMed] [Google Scholar]
- 6.Braus, G. H., S. Krappmann, and S. E. Eckert. 2002. Sexual development in ascomycetes: fruit body formation of Aspergillus nidulans, p. 215-244. In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, Inc., New York, N.Y.
- 7.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, D. W., T. H. Adams, and N. P. Keller. 1996. Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc. Natl. Acad. Sci. USA 93:14873-14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo, A. M., H. W. Gardner, and N. P. Keller. 2001. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276:25766-25774. [DOI] [PubMed] [Google Scholar]
- 10.Calvo, A. M., L. L. Hinze, H. W. Gardner, and N. P. Keller. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65:3668-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo, A. M., R. A. Wilson, J. W. Bok, and N. P. Keller. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66:447-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champe, S. P., and A. A. el-Zayat. 1989. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 171:3982-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Champe, S. P., P. Rao, and A. Chang. 1987. An endogenous inducer of sexual development in Aspergillus nidulans. J. Gen. Microbiol. 133:1383-1387. [DOI] [PubMed] [Google Scholar]
- 14.Chang, Y. C., and W. E. Timberlake. 1993. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristea, M., A. E. Osbourn, and E. H. Oliw. 2003. Linoleate diol synthase of the rice blast fungus Magnaporthe grisea. Lipids 38:1275-1280. [DOI] [PubMed] [Google Scholar]
- 16.Duplus, E., M. Glorian, and C. Forest. 2000. Fatty acid regulation of gene transcription. J. Biol. Chem. 275:30749-30752. [DOI] [PubMed] [Google Scholar]
- 17.Feussner, I., and C. Wasternack. 2002. The lipoxygenase pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 53:275-297. [DOI] [PubMed] [Google Scholar]
- 18.Fox, S. R., A. Akpinar, A. A. Prabhune, J. Friend, and C. Ratledge. 2000. The biosynthesis of oxylipins of linoleic and arachidonic acids by the sewage fungus Leptomitus lacteus, including the identification of 8R-hydroxy-9Z,12Z-octadecadienoic acid. Lipids 35:23-30. [DOI] [PubMed] [Google Scholar]
- 19.Funk, C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294:1871-1875. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 21.Han, K. H., K. Y. Han, J. H. Yu, K. S. Chae, K. Y. Jahng, and D. M. Han. 2001. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41:299-309. [DOI] [PubMed] [Google Scholar]
- 22.Han, S., and T. H. Adams. 2001. Complex control of the developmental regulatory locus brlA in Aspergillus nidulans. Mol. Genet. Genomics 266:260-270. [DOI] [PubMed] [Google Scholar]
- 23.Han, S., J. Navarro, R. A. Greve, and T. H. Adams. 1993. Translational repression of brlA expression prevents premature development in Aspergillus. EMBO J. 12:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hause, B., I. Stenzel, O. Miersch, H. Maucher, R. Kramell, J. Ziegler, and C. Wasternack. 2000. Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24:113-126. [DOI] [PubMed] [Google Scholar]
- 25.Hawksworth, D. L. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105:1422-1432. [Google Scholar]
- 26.Herman, R. P. 1998. Oxylipin production and action in fungi and related organisms, p. 115-130. In A. F. Rowley, H. Kuhn, and T. Schewe (ed.), Eicosanoids and related compounds in plants and animals. Princeton University Press, Princeton, N.J.
- 27.Herman, R. P., and C. A. Herman. 1985. Prostaglandins or prostaglandin-like substances are implicated in normal growth and development in oomycetes. Prostaglandins 29:819-830. [DOI] [PubMed] [Google Scholar]
- 28.Hornsten, L., C. Su, A. E. Osbourn, P. Garosi, U. Hellman, C. Wernstedt, and E. H. Oliw. 1999. Cloning of linoleate diol synthase reveals homology with prostaglandin H synthases. J. Biol. Chem. 274:28219-28224. [DOI] [PubMed] [Google Scholar]
- 29.Huber, S. M., F. Lottspeich, and J. Kamper. 2002. A gene that encodes a product with similarity to dioxygenases is highly expressed in teliospores of Ustilago maydis. Mol. Genet. Genomics 267:757-771. [DOI] [PubMed] [Google Scholar]
- 30.Jump, D. B. 2002. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr. Opin. Lipidol. 13:155-164. [DOI] [PubMed] [Google Scholar]
- 31.Kerwin, J. L., C. A. Simmons, and R. K. Washino. 1986. Eicosanoid regulation of oosporogenesis by Lagenidium giganteum. Prostaglandins Leukot. Med. 23:173-178. [DOI] [PubMed] [Google Scholar]
- 32.Kock, J. L., C. J. Strauss, C. H. Pohl, and S. Nigam. 2003. The distribution of 3-hydroxy oxylipins in fungi. Prostaglandins Other Lipid Mediat. 71:85-96. [DOI] [PubMed] [Google Scholar]
- 33.Kock, J. L., P. Venter, D. Linke, T. Schewe, and S. Nigam. 1998. Biological dynamics and distribution of 3-hydroxy fatty acids in the yeast Dipodascopsis uninucleata as investigated by immunofluorescence microscopy: evidence for a putative regulatory role in the sexual reproductive cycle. FEBS Lett. 427:345-348. [DOI] [PubMed] [Google Scholar]
- 34.Kolomiets, M. V., D. J. Hannapel, H. Chen, M. Tymeson, and R. J. Gladon. 2001. Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13:613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramell, R., O. Miersch, R. Atzorn, B. Parthier, and C. Wasternack. 2000. Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves. Implications for different signaling pathways. Plant Physiol. 123:177-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, B. S., and J. W. Taylor. 1990. Isolation of DNA from fungal mycelia and single spores, p. 282-287. In M. A. Innis, D. H. Gelfand, J. S. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 37.Leslie, J. F., and K. K. Klein. 1996. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics 144:557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maggio-Hall, L. A., and N. P. Keller. 6 October 2004. Mitochondrial β-oxidation in Aspergillus nidulans. Mol. Microbiol. 10.1111/j.1365-2958.2004.04340.x. [DOI] [PubMed]
- 39.Mazur, P., H. V. Meyers, and K. Nakanishi. 1990. Structural elucidation of sporogenic fatty acid metabolites from Aspergillus nidulans. Tetrahedron Lett. 31:3837-3840. [Google Scholar]
- 40.Mazur, P., K. Nakanishi, A. A. E. El-Zayat, and S. P. Champe. 1991. Structure and synthesis of sporogenic psi factors from Aspergillus nidulans. J. Chem. Soc. Chem. Commun. 20:1486-1487. [Google Scholar]
- 41.Murphy, D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325-438. [DOI] [PubMed] [Google Scholar]
- 42.Noverr, M. C., J. R. Erb-Downward, and G. B. Huffnagle. 2003. Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clin. Microbiol. Rev. 16:517-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh, K. B., H. Miyazawa, T. Naito, and H. Matsuoka. 2001. Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc. Natl. Acad. Sci. USA 98:4664-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-239. [DOI] [PubMed] [Google Scholar]
- 45.Prade, R. A., and W. E. Timberlake. 1993. The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 12:2439-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pringle, A., and J. W. Taylor. 2002. The fitness of filamentous fungi. Trends Microbiol. 10:474-481. [DOI] [PubMed] [Google Scholar]
- 47.Rawson, R. B. 2003. The SREBP pathway: insights from Insigs and insects. Nat. Rev. Mol. Cell. Biol. 4:631-640. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Samson, R. A. 1994. Taxonomy: current concepts of Aspergillus systematics, p. 1-22. In J. E. Smith (ed.), Aspergillus, vol. 7. Plenum Press, New York, N.Y.
- 50.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 51.Strauss, T., A. Botha, J. L. Kock, I. Paul, D. P. Smith, D. Linke, T. Schewe, and S. Nigam. 2000. Mapping the distribution of 3-hydroxylipins in the Mucorales using immunofluorescence microscopy. Antonie Leeuwenhoek 78:39-42. [DOI] [PubMed] [Google Scholar]
- 52.Su, C., and E. H. Oliw. 1996. Purification and characterization of linoleate 8-dioxygenase from the fungus Gaeumannomyces graminis as a novel hemoprotein. J. Biol. Chem. 271:14112-14118. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, J., D. Jacobson, and M. Fisher. 1999. The evolution of asexual fungi: reproduction, speciation, and classification. Annu. Rev. Phytopathol. 37:197-246. [DOI] [PubMed] [Google Scholar]
- 54.Trotter, P. J. 2001. The genetics of fatty acid metabolism in Saccharomyces cerevisiae. Annu. Rev. Nutr. 21:97-119. [DOI] [PubMed] [Google Scholar]
- 55.Tsitsigiannis, D. I., R. Zarnowski, and N. P. Keller. 2004. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 279:11344-11353. [DOI] [PubMed] [Google Scholar]
- 56.Wang, L. H., Y. He, Y. Gao, J. E. Wu, Y. H. Dong, C. He, S. X. Wang, L. X. Weng, J. L. Xu, L. Tay, R. X. Fang, and L. H. Zhang. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903-912. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, R. A., P.-K. Chang, A. Dobrzyn, J. M. Ntambi, R. Zarnowski, and N. P. Keller. 2004. Two Δ9-stearic acid desaturases are required for Aspergillus nidulans growth and development. Fung. Gen. Biol. 41:501-509. [DOI] [PubMed] [Google Scholar]
- 58.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]