Photosynthesis is dependent on light. Photosynthetic organisms struggle their entire lives to optimize photosynthetic function and minimize photooxidative damage in response to light quantity and quality. When the absorbed light energy exceeds the capacity of photosynthetic energy consumption, overreduction of electron transport carriers and accumulation of excitation energy in the light-harvesting-antennae may occur. The latter favors the production of excited triplet-state chlorophyll molecules (3Chl) that can interact with O2 to cause the formation of reactive singlet oxygen (1O2). The former favors the direct reduction of O2 by photosystem I (PSI) and the subsequent generation of reactive oxygen species, such as superoxide (O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (·OH) (5, 6, 75). These reactive oxygen species are able to cause photo-oxidative damage to photosystem II (PSII), a primary target for photoinhibition (2, 4, 9), but also to PSI (59, 60), in particular under weak light at chilled temperatures (111, 121). Thus, adaptation mechanisms that balance the energy input, through photochemistry via the photosynthetic machinery, with the energy output through CO2 assimilation and other metabolic pathways, are essential for plant survival.
The function of the light-harvesting complexes (LHCs) is, as the name implies, to increase the effective absorption cross-section of the photosystems and to supply them with excitation energy. The LHCs bind chlorophyll a and b but lack any photochemical activity of their own. Both PSI and PSII have their own core antenna pigments, but the addition of the LHCs increases the number of antenna pigments connected to each reaction center by a factor of 2 to 4, depending upon the conditions (47). Energy transfer operates in a time scale of femtoseconds to picoseconds. The transfer time of excitation energy between neighboring Chl molecules in the antenna has been estimated to be 100 to 300 fs (45). It had long been known that the pigment molecules must be closely packed to allow such fast and efficient transfer, a supposition beautifully confirmed by the emergence of the X-ray crystal structures of PSI (65), PSII (42, 66, 135), and LHCI-PSI (12) in the last several years (see discussion below). On average, excitation energy makes 100 to 1,000 such hops between antenna chlorophylls before it is trapped at a reaction center. Depending upon the arrangement of pigment molecules within LHCs and of LHCs vis-à-vis the photosystems, excitation energy can be directed to specific photosystems, temporarily trapped on low-energy chlorophylls, or even converted to heat by “nonphotochemical quenching.” It does not require a great stretch of imagination to see how evolution might have taken advantage of such possibilities for the development of regulatory mechanisms to deal with variable illumination.
The regulation of light energy input and distribution, by the dynamic regulation of the light-harvesting system, probably plays the most important role in balancing the light and dark reactions of photosynthesis. Light-harvesting systems and their functioning in algae and vascular plants have been addressed and discussed in numerous reviews (47, 58, 69, 101). Over the last decade the eukaryotic unicellular green alga Chlamydomonas reinhardtii has emerged as a potent model system for studying assembly, function, and regulation of the photosynthetic machinery in general (48, 51, 57, 98). Chlamydomonas has proven to be an excellent genetic model system for investigating different features of the regulation of light energy input and distribution, such as the role of the violaxanthin cycle in protection from photo-oxidative stress (89, 90) or the function of a specific PSII-associated major light-harvesting protein in nonradiative dissipation of excess excitation energy under high-light conditions (39) and other aspects that are addressed in more detail below. Further strengths of this model system evolve from an ongoing genomic project (>180,000 expressed sequence tag [EST] sequences have been obtained, and the second version of >9-fold whole-genome shotgun coverage was recently released by the U.S. Department of Energy Joint Genome Institute in February 2004), powerful genetics, and molecular techniques, as well as applicability to in-depth biochemical and structural analyses. In this review it is our aim to discuss recent developments in elucidating the composition, structural features, and dynamics of light-harvesting proteins in response to environment changes in C. reinhardtii.
COMPOSITION OF LIGHT-HARVESTING SYSTEMS IN C. REINHARDTII
PSI and PSII represent the two basic types of photosynthetic reaction centers present on this planet: type 1 (using FeS clusters as terminal electron acceptors) and type 2 (using quinones), respectively. It must be remembered that PSI and PSII each possess a core antenna system that has been remarkably well conserved through evolutionary time between cyanobacteria, green algae, and plants. The core antennae are tightly associated with their reaction center, made of a heterodimer of the D1 and D2 subunits in PSII and the last five α-helices of PsaA and PsaB in PSI. In PSII, the core antenna is mainly contributed by the CP43 and CP47 subunits. In PSI, the core antenna pigments are bound by the first six α-helices of the PsaA and PsaB subunits. Thus, the arrangements of reaction center and core antenna are very similar between PSI and PSII (135). What differs between the two photosystems, and even between the same photosystem from different organisms, is the arrangement of the peripheral antenna. In the process of trapping, excitation energy from photons absorbed by the peripheral antenna is first transferred to the core antenna and then to the reaction center, where it drives charge separation.
The lhca and lhcb gene families in green plants encode several LHC proteins that collect and transfer light energy to the reaction centers of PSI and PSII, respectively. In vascular plants the lhcb gene family is composed of numerous genes; 15 different lhcb genes have been described in the Arabidopsis genome (61). In C. reinhardtii, several lhcb genes have also been reported. Searching the Chlamydomonas EST database, Elrad et al. (38, 39) identified nine genes that potentially encode Lhcb polypeptides composing the major PSII antenna (Lhcbm). For the naming of the Lhcb proteins, we follow the nomenclature as given by Elrad and Grossman (38). The major PSII antenna is organized in trimers and connected to the PSII core via minor Lhcb proteins. Two gene products (Lhcb4 and Lhcb5) that correspond to the minor antenna proteins of vascular plants (CP29 and CP26) have also been described for C. reinhardtii (120). Separation of thylakoid membranes by two-dimensional gel electrophoresis (2-DGE) and subsequent mass spectrometric analysis has identified seven distinct Lhcb proteins (113). Although the major Lhcb proteins are highly homologous, peptides unique for specific lhcb gene products can be recognized. Tryptic digestion of isolated LHCII trimers, separation of the resulting peptides by reversed-phase chromatography, and mass spectrometric analysis confirmed the Lhcb proteins identified by 2-DGE and revealed the presence of an eighth Lhcb protein (113). It is of note that the mass spectrometric data indicated different N-terminally processed forms of Lhcbm3 and Lhcbm6. Gene-tagging experiments confirmed the presence of differentially N-terminal processed Lhcbm6 proteins. The mass spectrometric analysis also identified phosphorylation of a Thr residue in the N-terminal part of the Lhcbm3 protein. Interestingly, the N-terminal protein processing of Lhcbm3 leads to removal of the phosphorylation site and may therefore represent a novel regulatory mechanism (113). Another unexpected finding is that the transit peptide of Lhcb4 (CP29) from C. reinhardtii is not removed but undergoes acetylation and phosphorylation (123), demonstrating that processing of the transit peptide after protein import into the chloroplast is not mandatory in Chlamydomonas.
The lhca gene family in Arabidopsis is composed of the products of six nuclear genes (lhca1 to -6), of which genes lhca5 and lhca6 are only marginally expressed. It is expected that these latter two subunits are only present in substoichiometric amounts compared to subunits Lhca1 to -4 (61). In C. reinhardtii at least seven distinct LHCI subunits have been identified (11, 120). From seven Lhca proteins, N-terminal amino acid sequences have been obtained by Edman amino acid sequencing (120, 122). Separation of isolated PSI complexes from C. reinhardtii by 2-DGE revealed the presence of about 18 LHCI protein spots, suggesting an even higher variability of Lhca proteins (56). Mass spectrometric analysis of the 2-DGE-separated Lhca protein spots confirmed this large variability and identified nine different Lhca proteins from C. reinhardtii (113). Of these nine, Lhca2 and Lhca9 were not found by N-terminal Edman amino acid sequencing (122). For the naming of the Lhca proteins we follow the nomenclature of Stauber et al. (113) (Table 1). Structural data obtained by electron microscopy (EM) indicate that the LHCI-PSI complex from Chlamydomonas is larger and contains between 11 and 14 light-harvesting proteins per reaction center compared to eight Lhca (Lhca1 to -4) proteins per PSI complex in vascular plants (18, 22, 44, 67). Based upon phylogenetic analysis, Lhca proteins from C. reinhardtii can be separated into five distinct classes (38, 122). Each of these classes is related to a vascular plant homologue. In this fashion the Chlamydomonas Lhca1 and Lhca3 vascular plant homologues are represented by one gene product and are the most conserved in respect to similarity to the plant homologues among the nine polypeptides. Lhca2-like and Lhca5-like homologues are represented each by two gene products. A fifth class, which is represented by three gene products in C. reinhardtii, is related to Lhca2 and Lhca4 of vascular plants, as pointed out by Teramoto et al. (120). It was noted that the range of Lhca protein expression in Chlamydomonas can be rather large and may differ up to 50-fold between the highest and lowest expressed polypeptides (113). These data indicate that the variability of Lhca proteins, both in their sequence and expression level, is greater in Chlamydomonas compared to vascular plants. It is tempting to speculate that the consequent variability in the composition of the LHCs could be an important regulatory parameter in response to changing physiological conditions.
TABLE 1.
Comparison of Lhca nomenclature used by Stauber et al. (113), Elrad et al. (38), and Tokutsu et al. (122)a
| Database annotation
|
Protein name used by:
|
|||
|---|---|---|---|---|
| C. reinhardtii EST (cDNA) contigb | Genomic gene modelc | Stauber et al. (113) | Elrad et al. (38) | Tokutsu et al. (122) |
| AF104633 | C_1460019 | Lhca1 | Lhca4 | LhcI-6 |
| 20020630.8317.1 | C_490067 | Lhca2 | Lhca9 | Not listed |
| 20020630.1214.1 | C_1610027 | Lhca3 | Lhca6 | LhcI-2 |
| 20020630.608.1 | C_320083 | Lhca4 | Lhca5 | LhcI-1 |
| 20020630.154.1 | C_130138 | Lhca5 | Lhca1 | LhcI-3 |
| 20020630.6026.2 | C_430022 | Lhca6 | Lhca8 | LhcI-5 |
| 20020630.7235.1 | C_100004 | Lhca7 | Lhca2 | LhcI-7 |
| AY171231d | ||||
| 20011023.3781.1 | C_270001 | Lhca8 | Lhca3 | LhcI-4 |
| AF244524 | C_100008 | Lhca9 | Lhca2 | Not listed |
For naming of the Lhca polypeptides, we follow the nomenclature of Stauber et al. (113).
The reference indicates either the C. reinhardtii EST assembly available from the Chlamydomonas Genetics Center or the accession numbers of previously reported genes.
Genomic gene models differ partially from cDNA sequences; Lhca9 is only found as a C-terminal fragment in the genomic database.
Lacks amino acids in positions 16 to 32 of the immature protein sequence compared to the C. reinhardtii EST contig.
STRUCTURAL ASPECTS
In order to understand the molecular remodeling of LHCI or LHCII complexes in response to changes in nutrient status or ambient light, knowledge of the overall architecture of the photosystems is crucial. The attainment in the last 2 years of a 2.5- and a 4.4-Å resolution structure for cyanobacterial and plant PSI (12, 65), respectively, and the body of evidence accumulated for PSII (66, 135), currently at a 3.5-Å resolution (42), has greatly enhanced our knowledge. Four of the five high-resolution X-ray structures are from the cyanobacterium Thermosynechococcus elongatus, but it is generally accepted that the core composition and structure of the photosystems are fundamentally similar for all organisms using oxygenic photosynthesis. Comparisons possible thus far with a higher plant 10-Å resolved PSII dimer structure, by electron crystallography (50), have already given rise to discussion regarding observed structural differences, proposed to be mainly due to differences in the composition of minor, peripherally located, polypeptide components of the PSII core complex. In terms of function, however, these differences have brought a new focus to old questions, such as how the light-harvesting antennae bind to each photosystem core and how LHC remodeling operates to optimize photosynthetic function and minimize photo-oxidative stress under diverse environmental conditions.
Photosynthetic structures in the eukaryote Chlamydomonas have only recently been probed structurally by intermediate resolution electron microscopic techniques (44, 67, 86), including larger structures composed not only of the reaction centers of PSI or PSII but also the concomitant light-harvesting assemblies that service them.
EUKARYOTIC PSII LIGHT-HARVESTING SUPERCOMPLEXES
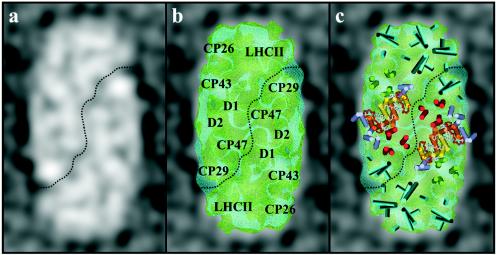
The first LHC-PS “supercomplex” observed, an LHCII-PSII complex from spinach, forms an excitonically coupled pigment network that captures light energy and transfers it to a dimeric PSII reaction center (16, 21, 50). This type of supercomplex has also been found in C. reinhardtii, and negatively stained particles have been characterized by single-particle analysis to a 30-Å resolution in three dimensions (86). It displays considerable similarity, given allowances for negative stain, to the nonstained cryo-EM spinach LHCII-PSII supercomplex (Fig. 1), with dimensions of ∼330 by 165 Å and 110 Å in depth. The structural resolution of the spinach supercomplex has been improved to 17 Å through the introduction of cryotechniques (86, 87), as shown in Fig. 1a. Comparisons can readily be made between the two structures, revealing that the PSII oxygen-evolving core dimer is centrally located and flanked by two clusters of the chlorophyll a/b-binding subunits Lhcb1, -2, -4, and -5. Each cluster is composed of a LHCII trimer (Lhcb1 and -2), which is coupled, both structurally and excitonically, to the central core dimer region via Lhcb4 and -5 monomers (49). It is anticipated that Lhcb4 and -5, together with an Lhcb6 component, can facilitate the binding of additional LHCII trimers, which probably contain Lhcb3, as well as Lhcb1 and Lhcb2, to the edge of the supercomplex (49), thus allowing for even higher-order structures, some of which have been recently observed (19-21). It should be noted that Lhcb6 (CP24) is missing in C. reinhardtii, indicating that although a similarity between supercomplexes of spinach and Chlamydomonas has been observed (see above) subtle difference do exist.
FIG. 1.
Structural comparisons between the internal distribution of LHCII and PSII subunits of the higher plant spinach and Chlamydomonas. (a) Two-dimensional membrane domain region of a nonstained cryo-EM-derived spinach LHCII-PSII supercomplex three-dimensional map at 17 Å in grayscale (85); (b) overlay of the three-dimensional membrane domain sections from spinach (green) and Chlamydomonas (blue) to emphasize the similarity in size and shape; (c) modeling of the transmembrane helical organization of the spinach core dimer derived from electron crystallography (49). These data have been overlaid with permission and include the Lhcb components (blue cylinders), modeled as helices based on the electron crystallographic structure of monomeric LHCII (70).
EUKARYOTIC PSI LIGHT-HARVESTING SUPERCOMPLEXES
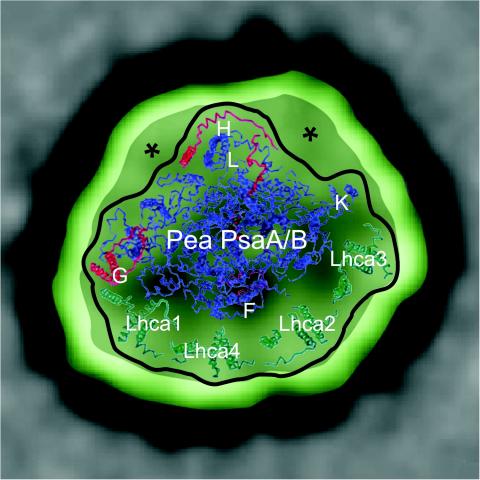
Visualization of a supercomplex composed of monomeric PSI, surrounded asymmetrically by a light-harvesting region composed of Lhca1 to -4 proteins, was made by using EM on spinach PSI-200 complexes (18). This LHCI-PSI supercomplex has now been observed in C. reinhardtii (44, 67), and a 30-Å three-dimensional model has been calculated (67). The typical LHCI-PSI supercomplex structure, resolved in two dimensions in the 20- to 30-Å range, has dimensions of ca. 220 by 180 Å and a height of 105 Å for the latter three-dimensional structure. A number of different sizes have been reported, which is likely to reflect the inherently labile nature of such complexes and the number of LHC components they are able to bind under various conditions. One cannot yet discount the existence of LHCI-PSI supercomplexes with a complete ring of LHCI components surrounding the central PSI core, but use of milder solubilization conditions for extraction of the complexes from membranes prior to imaging may be required for their visualization. Such a situation was true for the LHCII-PSII megacomplexes, which are composed of the more stable LHCII-PSII supercomplex core and additional LHCII components. It may be the case that if complexes are isolated “locked” into the state 2, LHCI-PSI supercomplexes will be observed with bound LHCII components. It should noted that the recently published 4.4-Å crystal structure of pea PSI revealed the presence of only four Lhca proteins per PSI reaction center (12), which contrasts with the finding that up to 14 Lhca subunits were proposed to surround the core of PSI, as assessed by EM (18, 22, 44, 67). In the modeling study presented by Kargul et al. (67), only one form (perhaps the most stable) of an LHCI-PSI supercomplex was observed in their C. reinhardtii preparations. This has been updated here to reflect the convergence with recent knowledge gleaned from crystallography (Fig. 2). In light of the new pea PSI crystal structure, it will be interesting to see whether the larger size of the LHCI antenna of C. reinhardtii (44, 67) and the larger number of Lhca polypeptides, compared to vascular plants, reflect a greater flexibility to optimize function in various environmental conditions. It is noteworthy that recent proteomic data revealed the presence of at least seven distinct Lhca polypeptides in PSI preparations from tomato (Lycopersicon esculentum), strongly indicating that PSI complexes with various Lhca compositions also exist in vascular plants (115, 134). The pea PSI structure reveals that the Lhca proteins are bound rather asymmetrically to the core of PSI. Lhca1 seems to be strongly bound by PsaG and loop 1 of PsaB. The latter structural motif is absent from the cyanobacterial PSI, whereas the other Lhca are more weakly attached to the core. A weak interaction between Lhca3 and PsaK can also be visualized. Therefore, remodeling of the antenna complex is more likely to occur at the PsaK/Lhca3 side than at the PsaG/Lhca1 side. This is consistent with the finding that the remodeling of LHCI induced by iron deficiency primarily affects PsaK and Lhca3 in C. reinhardtii (81).
FIG. 2.
A Chlamydomonas LHCI-PSI supercomplex observed by Kargul et al. (67), grayscale, overlaid to scale with the 4.4 Å resolved, membrane intrinsic, pea LHCI-PSI structure, obtained with permission (12). The pea structure, emphasized by the bold outline, has been positioned by placing its four copies of LHCI (labeled Lhca1 to -4; in green) into the four-domain asymmetric crescent of density observed by Kargul et al. (67). The remaining densities now visualized in the Chlamydomonas structure are labeled with an asterisk. Locations of other notable Psa subunits are labeled in white. The green outer band represents the putative detergent shell of ∼15 Å, which will also have been encompassed by negative stain.
Interestingly, the “open” side of the LHCI-PSI supercomplex harbors subunits PsaH, PsaL, and PsaO, which form a docking side for Lhcb polypeptides that bind to the PSI supercomplex under state 2 conditions (64, 73, 132).
Although the LHCI is closely associated with the PSI core complex, the LHCI subunits are normally synthesized in PSI-deficient mutants in C. reinhardtii (129). It should be noted that recent results with the Arabidopsis PSI assembly mutant HCF101 show that LHCI polypeptides are stable in vascular plants in the absence of PSI (72, 114). The LHCI complex was isolated from a C. reinhardtii ΔpsaB mutant and consisted of six Lhca polypeptides compared to nine in a LHCI-PSI supercomplex (117). Interestingly, Lhca3 and two minor polypeptides, Lhca2 and Lhca9, were lost during the purification procedure, although they were present in thylakoid membranes. This strongly indicates that Lhca2, Lhca3, and Lhca9 are not required for the stable oligomeric structure of the LHCI complex and that the association of these polypeptides with the LHCI complex is stabilized by the presence of the PSI core complex. Perhaps the Lhca2, Lhca3, and Lhca9 polypeptides function as linker proteins that allow the stable formation of the LHCI-PSI supercomplex. This would explain why Lhca3 is a prime target in the adaptation to iron deficiency (see above) and why the connection between LHCI and PSI becomes destabilized by proteolytic processing of this subunit (Fig. 1). The fact that an oligomeric LHCI complex can be isolated in the absence of PSI is a surprise in light of the new pea PSI crystal structure (12), which further underscores the differences in LHCI composition and structure between algae and vascular plants. From a functional point of view it is interesting that a functional association between LHCI and PSII has been observed in mutants of C. reinhardtii lacking PSI (32, 129), indicating that the presence of a LHCI complex in the absence of PSI might be of physiological importance under conditions in which PSI is more strongly compromised than PSII and free LHCI complexes are available to interact with PSII.
It should be noted that only the monomeric form of PSI, with or without LHCs attached, has been seen in eukaryotes. Trimers of PSI, a common oligomeric form in cyanobacterial systems, have only been observed in the case of artifactual association of monomers into larger oligomeric structures (18), and this has been attributed to the truncation in eukaryotes of PsaL (12), a polypeptide that is required for PSI trimerization in cyanobacteria (26). Indeed, higher-order light-harvesting structures composed of 18 copies of the IsiA polypeptide (which resembles CP43 of PSII) bound as a ring around trimeric PSI have been observed in iron-stressed cyanobacteria (13, 14, 17) or constitutively in green oxyphotobacteria (15), further emphasizing the diverse nature of the light-harvesting mechanisms used in oxygenic photosynthetic organisms. The inherent heterogeneity of higher plant and/or green algal photosynthetic structures, as demonstrated by the range of lhcb and lhca genes identified, greatly increases the difficulty of structural studies that rely on purity (e.g., crystallization for X-ray diffraction studies). To date, this has been overcome to a certain degree by “computer purification” single-particle analysis classification methods (87, 100). Further structural knowledge of the protein scaffold that holds and orients pigment molecules involved in light harvesting and light dissipation will be important for the understanding of the micromolecular dynamics of excitation energy transfer.
FUNCTION OF LHCS: EXCITATION ENERGY TRANSFER TO PSI AND PSII
The structures of the LHC proteins nicely explain their capabilities, which will be briefly reviewed here, before we address the ways in which these properties might be regulated. Energy transfer and equilibration in PSI occurs on the same time scale as primary charge separation, making it experimentally difficult to unravel these processes. Spectral equilibration (from higher- to lower-energy chromophores) and decay of excitations in the core PSI antenna take place with time constants of ∼5 and ∼25 ps, respectively. Primary charge separation occurs with a time constant of ∼4 ps (53), although the overall decay of excitations on the core antenna usually takes longer (25 to 30 ps) (52). Energy transfer within the antenna of PSI is strongly affected by a subset of Chls, the so-called “red Chls,” which have absorption maxima of >700 nm. Excitation energy tends to accumulate on these pigments, especially at low temperature. The location of these Chls has been the subject of much discussion, but it is clear that the number of red Chls and their excitation and emission maxima are very species dependent (45). In cyanobacteria, at least some of the red Chls are likely to be located at the interface between monomers in the PSI trimer because of the augmentation of red Chl emission (>720 nm) seen upon trimerization. In the core of eukaryotic PSI, there appears to be a small set of Chls that absorb at 705 nm and emit at 720 nm, but the major contributor to far-red fluorescence is LHCI, which contains Chls that absorb at 710 nm and emit at 730 nm in vascular plants (∼710 nm) in Chlamydomonas (11, 118, 129). These Chls appear to be localized to the Lhca1/Lhca4 heterodimer (68, 103). Interestingly, recombinant Lhca3 reconstituted with pigment resembles Lhca4 in its low-temperature fluorescence emission spectrum. Both Lhca proteins exhibit the most red-shifted fluorescence emission among the four Lhca proteins (103, 105), indicating that the excitation energy of the antenna may migrate via a low-energy chlorophyll of Lhca3 toward the PSI core chlorophylls. A recent ultrafast study (77, 78) found a 50-ps energy transfer component in the reconstituted Lhca1-Lhca4 heterodimer, which was absent in both monomers. This was ascribed to energy transfer from Lhca1 to Lhca4, which contains the lowest-energy (730-nm) pigment. The recent structure of pea LHCI-PSI (12) reveals three “linker Chls” at the interface between Lhca1 and Lhca4, which are the likely conduits of this efficient intersubunit energy transfer. In cyanobacteria the red Chls seem to be near P700, and they have been proposed to increase the efficiency of trapping by concentrating excitation energy near to the trap. However, they are also likely to slow trapping, given that in Chlamydomonas PSI, which has essentially no red Chls in its core antenna (76), the trapping rate is much faster (83). Thus, the situation is clearly different in eukaryotes, where the major red Chls are in the peripheral rather than the core antenna. The addition of LHCI to PSI has a significant effect upon the trapping rate, with the trapping time of 22 ps in PSI increasing to ca. 70 to 130 ps in LHCI-PSI. This is attributed to three main factors: the increase in the number of Chls, the higher proportion of lower-energy Chls, and the longer average distance between pigments in LHCI-PSI versus PSI.
In photosystem II, it had been assumed that trapping took place from a state in which excitation energy was thermally equilibrated (27, 102), but a recent theoretical study based upon the X-ray structure of PSII came to the opposite conclusion (126). Instead, it appears that there is a fast (<5 ps) phase of antenna energy redistribution within each complex of pigments followed by a much slower (100-ps) phase of redistribution between antenna and reaction center (RC) complexes. None of the Chls in the theoretical model were at their predicted Boltzmann level during the fluorescence decay. This is a consequence of the arrangement of pigments within the core of PSII. Unlike the situation in PSI, in which there is a complete ring of Chls surrounding the RC, the Chls bound by CP43 and CP47 are relatively isolated from each other (135). As trapping is taking place, there is constant redistribution of excitation energy to CP47, on the periphery of which is the red-most Chl of PSII (106). There are four antenna Chls located between the RC and the two core antenna complexes of PSII that seem to be especially important for transfer of excitation energy to the trap. Omission of either of the two most important led to a serious deviation from the fit to the experimental data obtained with Synechocystis cells lacking PSI (126).
DYNAMICS OF LIGHT-HARVESTING PROTEINS
The thylakoids of higher plants, and occasionally green algae, form flattened membranes that are segregated into stacked granal and unstacked stromal lamellae. The PSI and ATP synthase complexes are localized to the stromal lamellae, whereas the majority of active PSII resides in the grana (3, 8). Cytochrome b6f is present in both types of lamellae (1). In Chlamydomonas, the asymmetry in the distribution of the photosynthetic complexes has been shown in an immunocytochemical study with thylakoid membranes (125). Furthermore, the thylakoid membrane is a highly dynamic membrane system, where specific outer Lhcb proteins can shuttle between PSI in the stroma and PSII in the grana in order to optimize overall electron transport (69). In general, EM techniques of freeze-etch and freeze fracture, coupled with comparative studies of wild-type organisms and mutants deficient in PSI, PSII, or their LHC antennae and with antibody labeling (91), have enabled the localization of these protein complexes within the various lamellae of Chlamydomonas.
Regulation of this dynamic system in response to light can be divided into short- and long-term responses. Short-term responses include nonphotochemical thermal dissipation of excess energy (33, 58, 88), which will not be discussed here, and the process of state transitions. The latter process leads to a redistribution of excitation energy between PSII and PSI due to a reorganization of the antennae (23, 32, 84). In this process, LHCII polypeptides become phosphorylated under high-light conditions, when the plastoquinol pool is reduced (state II), which causes them to dissociate from PSII and migrate to the stromal lamellae, where they are functionally incorporated into the peripheral antenna of PSI (31, 32). Under oxidizing conditions (state I), the absorbance cross-sections of the two photosystems are nearly balanced in C. reinhardtii and change to 0.15 for PSII and 0.85 for PSI under reducing conditions (state II) (32). These changes in the distribution of excitation energy are accompanied by changes in the way the photosynthetic electron transfer operates. Cells perform mainly linear electron transfer in state I, whereas cyclic electron transfer dominates in state II (43). Cytochrome b6f has also been observed by immuno-EM to laterally redistribute along the thylakoids during state transitions (124). The identification of the thylakoid-associated serine-threonine kinase Stt7 confirmed the importance of LHCII phosphorylation in the process of state transitions (34). A C. reinhardtii stt7 mutant is deficient in LHCII phosphorylation and is also impaired in state transitions. It appears that a homolog of Stt7 exists in Arabidopsis. Interestingly, another thylakoid membrane protein kinase TAK1 was identified in Arabidopsis (109). TAK1 antisense plants displayed reduced TAK1 protein levels and exhibited reduced LHCP phosphorylation and impaired state transitions (108). Taken together, these results indicate that different kinases may control the process of state transitions. The activity of the kinase(s) responsible for LHCP phosphorylation appear to be controlled by the activity and/or occupation of the Qo site of the cytochrome b6f complex. By reverse genetics a Chlamydomonas mutant strain was constructed that showed no concerted oxidation of plastoquinol at the Qo (133). As a result, the mutant was locked in state I, since no phosphorylation of LHC proteins and no migration of LHCP to PSI occurred in this mutant under state II conditions, demonstrating that plastoquinol binding by the Qo pocket is required for LHCII kinase activation (133). Participation of the Qo site in the activation of the LHCP kinase was already suggested by Vener et al. (127, 128), since activation of the kinase correlated with the presence of plastoquinol at the Qo site.
Phosphorylation of PSII core subunits PsbD, PsbC, and PsbH has been reported for Chlamydomonas (29, 30, 35). In addition, a small protein of ∼5 kDa is also phosphorylated and has been identified as the product of psbI or psbF (35). Interestingly, a Chlamydomonas mutant BF4 that is strongly depleted in LHCII, as well as CP29 and CP26, does not display any PSII core phosphorylation (37). In the light of these results, it has been postulated that PSII core subunit phosphorylation is a prerequisite for stable LHCII-PSII supercomplex formation (36). However, as an alternative explanation, the absence of LHCII may have caused the impact in PSII core phosphorylation. Disruption of a small chloroplast-encoded open reading frame ycf9 resulted in reduction of CP26 in C. reinhardtii and tobacco (99, 116) and also CP29 to a lesser extent in tobacco (116). The disruption also changed considerably the phosphorylation of PSII core subunits in Chlamydomonas and tobacco (116). Ycf9 is shown to copurify with PSII core complexes from C. reinhardtii and tobacco and is absent from Fud7 and F34 mutants deficient in PSII but is present in the BF4 mutant deficient in LHCII (116). Therefore, Swiatek et al. concluded that the Ycf9 gene product is a bona fide PSII subunit and named it PsbZ. The fact that CP26 is depleted from the PsbZ-deficient mutant, but PsbZ is present in LHCII mutant BF4 strongly indicates that phosphorylation of the PSII core is at least partially dependent on the presence of CP26, whereas PsbZ is involved in binding of CP26 to PSII and therefore in the formation of the LHCII-PSII supercomplex. The formation of the supercomplex as a function of the phosphorylation status of PSII and LHC polypeptides and the functional connection to light-harvesting and/or light-dissipation remains to be investigated.
In algae the absorbance cross-sections of the photosynthetic reaction centers may also be modulated by long-term responses to ambient light intensity (10, 40, 71, 74, 95, 107, 119). The importance of balancing light-harvesting capacity and photosynthetic electron transfer to avoid overexcitation of the antennae is brought to light by the photosensitive phenotype of the PsaF-deficient strain (54). The psaF mutant was disrupted in its capacity for electron transfer between plastocyanin and PSI, showed an over-reduced QA by transient fluorescence induction and thermoluminescence measurements, and died at light intensities above 400 μE m−2 s−1 (41, 55). Screening for light-resistant suppressors yielded a strain in which the LHC polypeptides are not properly assembled within the thylakoid membrane and not functionally connected to the reaction centers (54). Thus, the inability of a large part of the excitation energy to reach the reaction center can provide protection against high light in this mutant. Interestingly, it has been shown that photoinactivation of PSII is slowed in C. reinhardtii strains unable to photo-oxidize the PQ pool and therefore unable to grow photoautotrophically (46, 131). This may indicate that under photoinhibitory conditions linear electron transfer from PSII to PSI may mediate the production of reactive oxygen species at PSI, which in turn cause a decline in PSII activity. This scenario is supported by the fact that high light causes an increase in lipid peroxidation and a concomitant decrease in PSII content in mutants affected either at the oxidizing or reducing sides of PSI (110). Therefore, downregulation of the antennae of both photosystems in order to decrease overall delivery of excitation energy and to balance the input between the photosystems is crucial to minimize photo-oxidative damage under high-light conditions.
The LHCs themselves may be responsible, at least in part, for the light sensitivity of some C. reinhardtii mutant strains. It has been noted that anaerobic (or microaerobic) conditions can alleviate the photosensitivity phenotype of mutants containing low levels of PSI (96, 97). Mutants with low antenna can even partially alleviate the extreme photosensitivity of mutants containing no PSI. This extreme light sensitivity is seen even in mutants completely lacking both PSI and PSII (24), in which all cellular chlorophyll is bound by LHC. Thus, at least some of the photo-oxidative reactions leading to photo-induced death must occur at the level of LHC pigments. These reactions are most likely singlet-triplet exchange reactions with molecular oxygen, leading to production of singlet oxygen (5, 6).
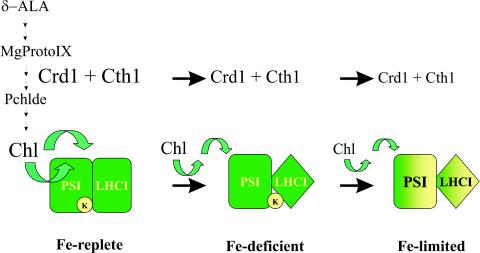
Other protective mechanisms operate when Chlamydomonas cells experience nutrient deprivation. In general, lack of sulfur, phosphorous, nitrogen, or iron, as well as low CO2 availability, lead to a downregulation of photosynthetic activity in C. reinhardtii (7, 81, 92, 94, 112, 130). In the case of nitrogen deficiency a specific loss of the cytochrome b6/f complex has been reported (25, 94). The importance of adjusting photosynthetic activity in response to nutrient deficiency is dramatized by the phenotype of the sac1 mutant (28), which is unable to reduce photosynthetic electron transfer capacity when sulfur levels fall and dies in the light within 2 days. A new response has been described that occurs during adaptation of C. reinhardtii to iron deficiency and leads to the alteration and remodeling of the LHC system even before downsizing of the photosynthetic machinery is induced (81). Changes in the physical coupling between the LHCI antenna system and PSI are already evident at iron levels (1 μM iron) at which a chlorosis phenotype is not obvious but expression of a marker gene for the iron nutritional status of the culture is maximally induced (81). Low-temperature fluorescence emission spectra, as well as biochemical data, indicate that functional coupling between LHCI and PSI is largely lost at this iron concentration. Based on the observation that the PSI subunit PsaK is more strongly affected at the onset of iron deficiency than other PSI subunits, Moseley et al. (81) proposed that this disconnection is mediated by a change in the physical properties of PsaK in response to a change in plastid iron content. This is in line with the fact that PsaK is a peripheral PSI chlorophyll-binding subunit (65) and functions in the connection of the Lhca2/Lhca3 heterodimer to PSI (62, 63). Interestingly, PsaK is largely depleted from the PSI complex in the crd1 mutant grown under normal conditions. The crd1 mutant was originally described as a copper response defect mutant (80) that has a strong chlorotic phenotype under conditions of copper deficiency, where it loses PSI and LHCI (80). The crd1 gene product is an iron-dependent aerobic oxidative cyclase. C. reinhardtii contains two isoforms of this protein, Crd1 and Cth1, which display a complementary pattern of expression dependent upon oxygen supply and copper nutritional status (82). These cyclases are supposed to function in the chlorophyll biosynthetic pathway, as shown for a bacterial homologue (93). Given the fact that PsaK is affected by a change in cyclase activity, Moseley et al. (81) proposed further that the chlorophyll-binding sites of PsaK are sensitive to flux through the chlorophyll biosynthetic pathway, which in turn is affected by the activity of the Cth1/Crd1 enzymes (Fig. 3). Consequently, a drop in the plastid iron levels would reduce cyclase activity and thereby result in a functional uncoupling between LHCI and PSI (Fig. 3). Adaptation to more severe iron-deprivation results in remodeling LHCI, leading to degradation of the Lhca polypeptides and possibly to the induction of novel LHC proteins. Mass spectrometric analyses demonstrated that one of these new LHCI subunits is actually an N-terminally processed Lhca3 (Chlamydomonas) Lhca3 is closely related to the vascular plant Lhca3 (113, 122; E. J. Stauber and M. Hippler, unpublished data). Interestingly, the N termini of the Lhca1 and Lhca4 proteins have been reported to be important in functional interactions with each other (104). Therefore, differential processing of Lhca3 may well alter the functional connections in the antenna network and contribute to the uncoupling of LHCI and PSI. Immunochemical analyses revealed that the abundance of LHCII subunits is also subject to change before chlorosis symptoms become visible (81), which indicates that the LHCII antenna may also be remodeled in response to iron deficiency. Furthermore, severe iron deficiency seems to induce the decoupling of LHCII from PSII, as indicated by 77-K fluorescence data, a phenomenon that has also be reported for land plants by using PSII-associated fluorescence (79). The protective function of these adaptations as a defense program to cope with photo-oxidative stress is illustrated by the enhanced growth of a PsaF-deficient strain either in iron-deficient conditions or in combination with the crd1 mutation (81), both of which lead to downregulation of LHCI. Therefore, adaptation to iron deficiency is another example where photosynthetic function is optimized by graduated modulation of light harvesting to minimize photo-oxidative damage.
FIG. 3.
Model of the progressive adaptation to iron depletion. Iron-deficient conditions induce remodeling of photosystem I antenna, degradation, replacement,and change in subunit associations. Further iron limitations result in a downregulation of abundance of the LHCI-photosystem I complex.
CONCLUSIONS
In light of recent and in expectation of future high-resolution X-ray structures of algal or higher plant photosystems, it will still be necessary to translate the structural information into mechanisms of how the light-harvesting systems couple functionally to their respective reaction center cores. All this will need to be understood in terms of the dynamic processes ranging from initial biogenesis, through to the state transitions, and any adaptation to different environmental conditions. One would hope that an organism, such as Chlamydomonas, which is suitable for reverse genetics, genomic, and proteomics, coupled to a structural classification technique such as single-particle analysis, will lead to rapid advances in our structural understanding of these dynamics.
Acknowledgments
We thank Sabeeha Merchant for help with Fig. 3.
M.H. acknowledges support from the Research Foundation of the University of Pennsylvania. J.N. currently holds a Royal Society University Research Fellowship and funding from the BBSRC UK research council. K.R. acknowledges support by the National Institutes of Health (NIGMS grant GM66345-01).
REFERENCES
- 1.Albertsson, P. A. 1995. The structure and function of the chloroplast photosynthetic membrane: a model for the domain organization. Photosynth. Res. 46:141-149. [DOI] [PubMed] [Google Scholar]
- 2.Allakhverdiev, S. I., V. V. Klimov, and R. Carpentier. 1997. Evidence for the involvement of cyclic electron transport in the protection of photosystem II against photoinhibition: influence of a new phenolic compound. Biochemistry 36:4149-4154. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, B., and J. M. Anderson. 1980. Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta 593:427-440. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, B., and S. Styring. 1991. Photosystem II: molecular-organization, function, and acclimation. Curr. Top. Bioenergetics 16:1-81. [Google Scholar]
- 5.Asada, K. 1994. Mechanisms for scavenging reactive molecules generated in chloroplasts under light stress. BIOS Scientific Publishers, Oxford, United Kingdom.
- 6.Asada, K. 1996. Radical production and scavenging in the chloroplast, p. 123-150. In Advances in photosynthesis: photosynthesis and the environment. Kluwer Academic Publishers, Amsterdam, The Netherlands.
- 7.Badger, M. R., A. Kaplan, and J. A. Berry. 1980. Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a carbon-dioxide concentrating mechanism. Plant Physiol. 66:407-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber, J. 1982. Influence of surface charges on thylakoid structure and function. Annu. Rev. Plant Physiol. 33:261-295. [Google Scholar]
- 9.Barber, J., and B. Andersson. 1992. Too much of a good thing: light can be bad for photosynthesis. Trends Biochem. Sci. 17:61-66. [DOI] [PubMed] [Google Scholar]
- 10.Baroli, I., and A. Melis. 1998. Photoinhibitory damage is modulated by the rate of photosynthesis and by the photosystem II light-harvesting chlorophyll antenna size. Planta 205:288-296. [DOI] [PubMed] [Google Scholar]
- 11.Bassi, R., S. Y. Soen, G. Frank, H. Zuber, and J. D. Rochaix. 1992. Characterization of chlorophyll a/b proteins of photosystem I from Chlamydomonas reinhardtii. J. Biol. Chem. 267:25714-25721. [PubMed] [Google Scholar]
- 12.Ben-Shem, A., F. Frolow, and N. Nelson. 2003. Crystal structure of plant photosystem I. Nature 426:630-635. [DOI] [PubMed] [Google Scholar]
- 13.Bibby, T. S., J. Nield, and J. Barber. 2001. Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412:743-745. [DOI] [PubMed] [Google Scholar]
- 14.Bibby, T. S., J. Nield, and J. Barber. 2001. Three-dimensional model and characterization of the iron-stress induced CP43′-photosystem I supercomplex isolated from the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 46:43246-43252. [DOI] [PubMed] [Google Scholar]
- 15.Bibby, T. S., J. Nield, F. Partensky, and J. Barber. 2001. Oxyphotobacteria antenna ring around photosystem I. Nature 413:590. [DOI] [PubMed] [Google Scholar]
- 16.Boekema, E. J., B. Hankamer, D. Bald, J. Kruip, J. Nield, A. F. Boonstra, J. Barber, and M. Rogner. 1995. Supramolecular structure of the photosystem II complex from green plants and cyanobacteria. Proc. Natl. Acad. Sci. USA 92:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boekema, E. J., A. Hifney, A. E. Yakushevska, M. Piotrowski, W. Keegstra, S. Berry, K. P. Michel, E. K. Pistorius, and J. Kruip. 2001. A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412:745-748. [DOI] [PubMed] [Google Scholar]
- 18.Boekema, E. J., P. E. Jensen, E. Schlodder, J. F. van Breemen, H. van Roon, H. V. Scheller, and J. P. Dekker. 2001. Green plant photosystem I binds light-harvesting complex I on one side of the complex. Biochemistry 40:1029-1036. [DOI] [PubMed] [Google Scholar]
- 19.Boekema, E. J., J. F. van Breemen, H. van Roon, and J. P. Dekker. 2000. Conformational changes in photosystem II supercomplexes upon removal of extrinsic subunits. Biochemistry 39:12907-12915. [DOI] [PubMed] [Google Scholar]
- 20.Boekema, E. J., H. van Roon, F. Calkoen, R. Bassi, and J. P. Dekker. 1999. Multiple types of association of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Biochemistry 38:2233-2239. [DOI] [PubMed] [Google Scholar]
- 21.Boekema, E. J., H. Van Roon, J. F. Van Breemen, and J. P. Dekker. 1999. Supramolecular organization of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Eur. J. Biochem. 266:444-452. [DOI] [PubMed] [Google Scholar]
- 22.Boekema, E. J., R. M. Wynn, and R. Malkin. 1990. The structure of spinach photosystem I studied by electron microscopy. Biochim. Biophys. Acta 1017:49-56. [Google Scholar]
- 23.Bonaventura, C., and J. Myers. 1969. Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189:366-383. [DOI] [PubMed] [Google Scholar]
- 24.Boudreaux, B., F. MacMillan, C. Teutloff, R. Agalarov, F. Gu, S. Grimaldi, R. Bittl, K. Brettel, and K. Redding. 2001. Mutations in both sides of the photosystem I reaction center identify the phylloquinone observed by electron paramagnetic resonance spectroscopy. J. Biol. Chem. 276:37299-37306. [DOI] [PubMed] [Google Scholar]
- 25.Bulte, L., and F. A. Wollman. 1992. Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur. J. Biochem. 204:327-336. [DOI] [PubMed] [Google Scholar]
- 26.Chitnis, V. P., and P. R. Chitnis. 1993. PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 336:330-334. [DOI] [PubMed] [Google Scholar]
- 27.Dau, H., and K. Sauer. 1996. Exciton equilibration and Photosystem II exciton dynamics: a fluorescence study on Photosystem II membrane particles of spinach. Biochim. Biophys. Acta Bioenergetics 1273:175-190. [Google Scholar]
- 28.Davies, J. P., F. H. Yildiz, and A. Grossman. 1996. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15:2150-2159. [PMC free article] [PubMed] [Google Scholar]
- 29.Dedner, N., H. E. Meyer, C. Ashton, and G. F. Wildner. 1988. N-terminal sequence analysis of the 8 kDa protein in Chlamydomonas reinhardtii: localization of the phosphothreonine. FEBS Lett. 236:77-82. [Google Scholar]
- 30.Delepelaire, P. 1984. Partial characterization of the biosynthesis and integration of the photosystem-Ii reaction centers in the thylakoid membrane of Chlamydomonas reinhardtii. EMBO J. 3:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delosme, R., D. Béal, and P. Joliot. 1994. Photoacoustic detection of flash-induced charge separation in photosynthetic systems: spectral dependence of the quantum yield. Biochim. Biophys. Acta 1185:56-64. [Google Scholar]
- 32.Delosme, R., J. Olive, and F.-A. Wollman. 1996. Changes in light energy distribution upon state transitions: an in vivo photoacoustic study of the wild type and photosynthesis mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1273:150-158. [Google Scholar]
- 33.Demming-Adams, B., A. M. Gilmore, and W. W. Adams III. 1996. Photoprotection and other responses of plants to highlight stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43:599-626. [Google Scholar]
- 34.Depege, N., S. Bellafiore, and J. D. Rochaix. 2003. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299:1572-1575. [DOI] [PubMed] [Google Scholar]
- 35.de Vitry, C., B. A. Diner, and J. L. Popo. 1991. Photosystem II particles from Chlamydomonas reinhardtii: purification, molecular weight, small subunit composition, and protein phosphorylation. J. Biol. Chem. 266:16614-16621. [PubMed] [Google Scholar]
- 36.de Vitry, C., J. Olive, D. Drapier, M. Recouvreur, and F. A. Wollman. 1989. Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109:991-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vitry, C., and F. A. Wollman. 1988. Changes in phosphorylation of thylakoid membrane-proteins in light-harvesting complex mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 933:444-449. [Google Scholar]
- 38.Elrad, D., and A. R. Grossman. 2004. A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45:61-75. [DOI] [PubMed] [Google Scholar]
- 39.Elrad, D., K. K. Niyogi, and A. R. Grossman. 2002. A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14:1801-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escoubas, J. M., M. Lomas, J. LaRoche, and P. G. Falkowski. 1995. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 92:10237-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farah, J., F. Rappaport, Y. Choquet, P. Joliot, and J. D. Rochaix. 1995. Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 14:4976-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira, K. N., T. M. Iverson, K. Maghlaoui, J. Barber, and S. Iwata. 2004. Architecture of the photosynthetic oxygen-evolving center. Science 303:1831-1838. [DOI] [PubMed] [Google Scholar]
- 43.Finazzi, G., A. Furia, R. P. Barbagallo, and G. Forti. 1999. State transitions, cyclic and linear electron transport and photophosphorylation in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1413:117-129. [DOI] [PubMed] [Google Scholar]
- 44.Germano, M., A. E. Yakushevska, W. Keegstra, H. J. van Gorkom, J. P. Dekker, and E. J. Boekema. 2002. Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525:121-125. [DOI] [PubMed] [Google Scholar]
- 45.Gobets, B., and R. van Grondelle. 2001. Energy transfer and trapping in photosystem I. Biochim. Biophys. Acta Bioenergetics 1507:80-99. [DOI] [PubMed] [Google Scholar]
- 46.Gong, H. S., and I. Ohad. 1991. The PQ/PQH2 ratio and occupancy of photosystem II-QB site by plastoquinone control the degradation of D1 protein during photoinhibition in vivo. J. Biol. Chem. 266:21293-21299. [PubMed] [Google Scholar]
- 47.Grossman, A. R., D. Bhaya, K. E. Apt, and D. M. Kehoe. 1995. Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu. Rev. Genet. 29:231-288. [DOI] [PubMed] [Google Scholar]
- 48.Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez, D. Rokhsar, J. Shrager, C. D. Silflow, D. Stern, O. Vallon, and Z. Zhang. 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2:1137-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hankamer, B., E. Morris, J. Nield, C. Gerle, and J. Barber. 2001. Three-dimensional structure of the Photosystem II core dimer of higher plants determined by electron microscopy. J. Struct. Biol. 135:262-269. [DOI] [PubMed] [Google Scholar]
- 50.Hankamer, B., J. Nield, D. Zheleva, E. Boekema, S. Jansson, and J. Barber. 1997. Isolation and biochemical characterisation of monomeric and dimeric photosystem II complexes from spinach and their relevance to the organisation of photosystem II in vivo. Eur. J. Biochem. 243:422-429. [DOI] [PubMed] [Google Scholar]
- 51.Harris, E. H. 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:363-406. [DOI] [PubMed] [Google Scholar]
- 52.Hastings, G., F. A. Kleinherenbrink, S. Lin, and R. E. Blankenship. 1994. Time-resolved fluorescence and absorption spectroscopy of photosystem I. Biochemistry 33:3185-3192. [DOI] [PubMed] [Google Scholar]
- 53.Hastings, G., F. A. Kleinherenbrink, S. Lin, T. J. McHugh, and R. E. Blankenship. 1994. Observation of the reduction and reoxidation of the primary electron acceptor in photosystem I. Biochemistry 33:3193-3200. [DOI] [PubMed] [Google Scholar]
- 54.Hippler, M., K. Biehler, A. Krieger-Liszkay, J. van Dillewjin, and J. D. Rochaix. 2000. Limitation in electron transfer in photosystem I donor side mutants of Chlamydomonas reinhardtii: lethal photo-oxidative damage in high light is overcome in a suppressor strain deficient in the assembly of the light harvesting complex. J. Biol. Chem. 275:5852-5859. [DOI] [PubMed] [Google Scholar]
- 55.Hippler, M., F. Drepper, J. Farah, and J. D. Rochaix. 1997. Fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii requires PsaF. Biochemistry 36:6343-6349. [DOI] [PubMed] [Google Scholar]
- 56.Hippler, M., J. Klein, T. Fink, T. Allinger, and P. Hoerth. 2001. Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 28:595-606. [DOI] [PubMed] [Google Scholar]
- 57.Hippler, M., B. Rimbault, and Y. Takahashi. 2002. Photosynthetic complex assembly in Chlamydomonas reinhardtii. Protist 153:197-220. [DOI] [PubMed] [Google Scholar]
- 58.Horton, P., A. V. Ruban, and R. G. Walters. 1994. Regulation of light harvesting in green plants (indication by nonphotochemical quenching of chlorophyll fluorescence). Plant Physiol. 106:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue, K., Y. Fuji, E. Yokoyama, K. Matsuura, K. Hiyama, and H. Sakurai. 1989. The photoinhibition site of photosystem I in isolated chloroplast under extremely reducing conditions. Plant Cell Physiol. 30:65-71. [Google Scholar]
- 60.Inoue, K., H. Sakurai, and T. Hiyama. 1986. Photoinactivation of photosystem I in isolated chloroplast. Plant Cell Physiol. 27:961-968. [Google Scholar]
- 61.Jansson, S. 1999. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4:236-240. [DOI] [PubMed] [Google Scholar]
- 62.Jansson, S., B. Andersen, and H. V. Scheller. 1996. Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 112:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jensen, P. E., M. Gilpin, J. Knoetzel, and H. V. Scheller. 2000. The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J. Biol. Chem. 275:24701-24708. [DOI] [PubMed] [Google Scholar]
- 64.Jensen, P. E., A. Haldrup, S. Zhang, and H. V. Scheller. 2004. The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J. Biol. Chem. 279:24212-24217. [DOI] [PubMed] [Google Scholar]
- 65.Jordan, P., P. Fromme, H. T. Witt, O. Klukas, W. Saenger, and N. Krauss. 2001. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411:909-917. [DOI] [PubMed] [Google Scholar]
- 66.Kamiya, N., and J. R. Shen. 2003. Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc. Natl. Acad. Sci. USA 100:98-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kargul, J., J. Nield, and J. Barber. 2003. Three-dimensional reconstruction of a light-harvesting complex I-Photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii: insights into light harvesting for PSI. J. Biol. Chem. 278:16135-16141. [DOI] [PubMed] [Google Scholar]
- 68.Knoetzel, J., I. Svendsen, and D. J. Simpson. 1992. Identification of the photosystem I antenna polypeptides in barley: isolation of three pigment-binding antenna complexes. Eur. J. Biochem. 206:209-215. [DOI] [PubMed] [Google Scholar]
- 69.Kruse, O. 2001. Light-induced short-term adaptation mechanisms under redox control in the PS II-LHCII supercomplex: LHC II state transitions and PS II repair cycle. Naturwissenschaften 88:284-292. [DOI] [PubMed] [Google Scholar]
- 70.Kühlbrandt, W., D. N. Wang, and Y. Fujiyoshi. 1994. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367:614-621. [DOI] [PubMed] [Google Scholar]
- 71.Ley, A. C., and D. C. Mauzerall. 1982. The reversible decline of oxygen flash yields at high flash energies evidence for total annihilation of excitations in Photosystem II. Biochim. Biophys. Acta 680:174-180. [Google Scholar]
- 72.Lezhneva, L., K. Amann, and J. Meurer. 2004. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37:174-185. [DOI] [PubMed] [Google Scholar]
- 73.Lunde, C., P. E. Jensen, A. Haldrup, J. Knoetzel, and H. V. Scheller. 2000. The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408:613-615. [DOI] [PubMed] [Google Scholar]
- 74.Maxwell, D. P., S. Falk, and N. P. A. Huner. 1995. Photosystem II excitation pressure and development of resistance to photoinhibition. 1. Light-harvesting complex-Ii abundance and zeaxanthin content in Chlorella vulgaris. Plant Physiol. 107:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mehler, A. H. 1951. Studies on reaction of illuminated chloroplasts. I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 33:65-77. [DOI] [PubMed] [Google Scholar]
- 76.Melkozernov, A. N. 2001. Excitation energy transfer in Photosystem I from oxygenic organisms. Photosynthesis Res. 70:129-153. [DOI] [PubMed] [Google Scholar]
- 77.Melkozernov, A. N., S. Lin, V. H. Schmid, H. Paulsen, G. W. Schmidt, and R. E. Blankenship. 2000. Ultrafast excitation dynamics of low energy pigments in reconstituted peripheral light-harvesting complexes of photosystem I. FEBS Lett. 471:89-92. [DOI] [PubMed] [Google Scholar]
- 78.Melkozernov, A. N., V. H. R. Schmid, G. W. Schmidt, and R. E. Blankenship. 1998. Energy redistribution in heterodimeric light-harvesting complex LHCI-730 of photosystem I. J. Phys. Chem. B 102:8183-8189. [Google Scholar]
- 79.Morales, F., N. Moise, R. Quilez, A. Abadia, J. Abadia, and I. Moya. 2001. Iron deficiency interrupts energy transfer from a disconnected part of the antenna to the rest of Photosystem II. Photosynthesis Res. 70:207-220. [DOI] [PubMed] [Google Scholar]
- 80.Moseley, J., J. Quinn, M. Eriksson, and S. Merchant. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moseley, J. L., T. Allinger, S. Herzog, P. Hoerth, E. Wehinger, S. Merchant, and M. Hippler. 2002. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21:6709-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moseley, J. L., M. D. Page, N. Pergam, M. Eriksson, J. Quinn, J. Soto, S. Theg, M. Hippler, and S. Merchant. 2002. Reciprocal expression of two di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14:673-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Müller, M. G., J. Niklas, W. Lubitz, and A. R. Holzwarth. 2003. Ultrafast transient absorption studies on photosystem I reaction centers from Chlamydomonas reinhardtii I: a new interpretation of the energy trapping and early electron transfer steps in photosystem I. Biophys. J. 85:3899-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murata, N. 1969. Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim. Biophys. Acta 172:242-251. [DOI] [PubMed] [Google Scholar]
- 85.Nield, J., C. Funk, and J. Barber. 2000. Supermolecular structure of photosystem II and location of the PsbS protein. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nield, J., O. Kruse, J. Ruprecht, P. da Fonseca, C. Buchel, and J. Barber. 2000. Three-dimensional structure of Chlamydomonas reinhardtii and Synechococcus elongatus photosystem II complexes allows for comparison of their oxygen-evolving complex organization. J. Biol. Chem. 275:27940-27946. [DOI] [PubMed] [Google Scholar]
- 87.Nield, J., E. V. Orlova, E. P. Morris, B. Gowen, M. van Heel, and J. Barber. 2000. 3D map of the plant photosystem II supercomplex obtained by cryoelectron microscopy and single particle analysis. Nat. Struct. Biol. 7:44-47. [DOI] [PubMed] [Google Scholar]
- 88.Niyogi, K. K. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed] [Google Scholar]
- 89.Niyogi, K. K., O. Bjorkman, and A. R. Grossman. 1997. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Niyogi, K. K., O. Bjorkman, and A. R. Grossman. 1997. The roles of specific xanthophylls in photoprotection. Proc. Natl. Acad. Sci. USA 94:14162-14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olive, J., and O. Vallon. 1991. Structural organization of the thylakoid membrane: freeze-fracture and immunocytochemical analysis. J. Electron Microsc. Tech. 18:360-374. [DOI] [PubMed] [Google Scholar]
- 92.Peltier, G., and G. W. Schmidt. 1991. Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 88:4791-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pinta, V., M. Picaud, F. Reiss-Husson, and C. Astier. 2002. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J. Bacteriol. 184:746-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Plumley, F. G., and G. W. Schmidt. 1989. Nitrogen-dependent regulation of photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 86:2678-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Polle, J. E., J. R. Benemann, A. Tanaka, and A. Melis. 2000. Photosynthetic apparatus organization and function in the wild type and a chlorophyll b-less mutant of Chlamydomonas reinhardtii: dependence on carbon source. Planta 211:335-344. [DOI] [PubMed] [Google Scholar]
- 96.Redding, K., F. MacMillan, W. Leibl, K. Brettel, J. Hanley, A. W. Rutherford, J. Breton, and J. D. Rochaix. 1998. A systematic survey of conserved histidines in the core subunits of Photosystem I by site-directed mutagenesis reveals the likely axial ligands of P700. EMBO J. 17:50-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rochaix, J., N. Fischer, and M. Hippler. 2000. Chloroplast site-directed mutagenesis of photosystem I in Chlamydomonas: electron transfer reactions and light sensitivity. Biochimie 82:635-645. [DOI] [PubMed] [Google Scholar]
- 98.Rochaix, J. D. 2002. Chlamydomonas, a model system for studying the assembly and dynamics of photosynthetic complexes. FEBS Lett. 529:34-38. [DOI] [PubMed] [Google Scholar]
- 99.Ruf, S., K. Biehler, and R. Bock. 2000. A small chloroplast-encoded protein as a novel architectural component of the light-harvesting antenna. J. Cell Biol. 149:369-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruprecht, J., and J. Nield. 2001. Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction. Prog. Biophys. Mol. Biol. 75:121-164. [DOI] [PubMed] [Google Scholar]
- 101.Sandona, D., R. Croce, A. Pagano, M. Crimi, and R. Bassi. 1998. Higher plants light harvesting proteins. Structure and function as revealed by mutation analysis of either protein or chromophore moieties. Biochim. Biophys. Acta 1365:207-214. [DOI] [PubMed] [Google Scholar]
- 102.Schatz, G. H., H. Brock, and A. R. Holzwarth. 1988. Kinetic and energetic model for the primary processes in photosystem Ii. Biophys. J. 54:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmid, V. H., K. V. Cammarata, B. U. Bruns, and G. W. Schmidt. 1997. In vitro reconstitution of the photosystem I light-harvesting complex LHCI-730: heterodimerization is required for antenna pigment organization. Proc. Natl. Acad. Sci. USA 94:7667-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmid, V. H., H. Paulsen, and J. Rupprecht. 2002. Identification of N- and C-terminal amino acids of Lhca1 and Lhca4 required for formation of the heterodimeric peripheral photosystem I antenna LHCI-730. Biochemistry 41:9126-9131. [DOI] [PubMed] [Google Scholar]
- 105.Schmid, V. H., S. Potthast, M. Wiener, V. Bergauer, H. Paulsen, and S. Storf. 2002. Pigment binding of photosystem I light-harvesting proteins. J. Biol. Chem. 277:37307-37314. [DOI] [PubMed] [Google Scholar]
- 106.Shen, G., J. J. Eaton-Rye, and W. F. Vermaas. 1993. Mutation of histidine residues in CP47 leads to destabilization of the photosystem II complex and to impairment of light energy transfer. Biochemistry 32:5109-5115. [DOI] [PubMed] [Google Scholar]
- 107.Smith, B. M., P. J. Morrissey, J. E. Guenther, J. A. Nemson, M. A. Harrison, J. F. Allen, and A. Melis. 1990. Response of the photosynthetic apparatus in Dunaliella salina (green-algae) to irradiance stress. Plant Physiol. 93:1433-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Snyders, S., and B. D. Kohorn. 2001. Disruption of thylakoid-associated kinase 1 leads to alteration of light harvesting in Arabidopsis. J. Biol. Chem. 276:32169-32176. [DOI] [PubMed] [Google Scholar]
- 109.Snyders, S., and B. D. Kohorn. 1999. TAKs, thylakoid membrane kinases associated with energy transduction. J. Biol. Chem. 274:9137-9140. [DOI] [PubMed] [Google Scholar]
- 110.Sommer, F., M. Hippler, K. Biehler, N. Fischer, and J. D. Rochaix. 2003. Comparative analysis of photosensitivity in photosystem I donor and acceptor side mutants of Chlamydomonas reinhardtii. Plant Cell Environment 26:1881-1892. [Google Scholar]
- 111.Sonoike, K., I. Terashima, M. Iwaki, and S. Itoh. 1995. Destruction of photosystem I iron-sulfur centers in leaves of Cucumis sativus L. by weak illumination at chilling temperatures. FEBS Lett. 362:235-238. [DOI] [PubMed] [Google Scholar]
- 112.Spalding, M. H., R. J. Spreitzer, and W. L. Ogren. 1983. Reduced inorganic carbon transport in a CO2-requiring mutant of Chlamydomonas reinhardtii. Plant Physiol. 73:273-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stauber, E. J., A. Fink, C. Markert, O. Kruse, U. Johanningmeier, and M. Hippler. 2003. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2:978-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stockel, J., and R. Oelmuller. 2004. A novel protein for photosystem I biogenesis. J. Biol. Chem. 279:10243-10251. [DOI] [PubMed] [Google Scholar]
- 115.Storf, S., E. J. Stauber, M. Hippler, and V. H. Schmid. 2004. Proteomic analysis of the photosystem I light-harvesting antenna in tomato (Lycopersicon esculentum). Biochemistry 43:9214-9224. [DOI] [PubMed] [Google Scholar]
- 116.Swiatek, M., R. Kuras, A. Sokolenko, D. Higgs, J. Olive, G. Cinque, B. Muller, L. A. Eichacker, D. B. Stern, R. Bassi, R. G. Herrmann, and F. A. Wollman. 2001. The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13:1347-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Takahashi, Y., T. Yasui, J. E. Stauber, and M. Hippler. 2003. Comparison of the subunit compositions of the PSI-LHCI supercomplex and the LHCI complex in the green alga Chlamydomonas reinhardtii. submitted. [DOI] [PubMed]
- 118.Takahashi, Y., T. Yasui, J. E. Stauber, and M. Hippler. 2004. Comparison of the subunit compositions of the PSI-LHCI supercomplex and the LHCI complex in the green alga Chlamydomonas reinhardtii. Biochemistry 43:7816-7823. [DOI] [PubMed] [Google Scholar]
- 119.Teramoto, H., A. Nakamori, J. Minagawa, and T. Ono. 2002. Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiol. 130:325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Teramoto, H., T. Ono, and J. Minagawa. 2001. Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 42:849-856. [DOI] [PubMed] [Google Scholar]
- 121.Terashima, I., S. Funayama, and K. Sonoike. 1994. The site of photoinhibition in leaves of Cucumis sativus L. at low temperature is photosystem I and not photosystem II. Planta 193:300-306. [Google Scholar]
- 122.Tokutsu, R., H. Teramoto, Y. Takahashi, T. A. Ono, and J. Minagawa. 2004. The light-harvesting complex of photosystem I in Chlamydomonas reinhardtii: protein composition, gene structures and phylogenic implications. Plant Cell Physiol. 45:138-145. [DOI] [PubMed] [Google Scholar]
- 123.Turkina, M. V., A. Villarejo, and A. V. Vener. 2004. The transit peptide of CP29 thylakoid protein in Chlamydomonas reinhardtii is not removed but undergoes acetylation and phosphorylation. FEBS Lett. 564:104-108. [DOI] [PubMed] [Google Scholar]
- 124.Vallon, O., L. Bulte, P. Dainese, J. Olive, R. Bassi, and F. A. Wollman. 1991. Lateral redistribution of cytochrome b6/f complexes along thylakoid membranes upon state transitions. Proc. Natl. Acad. Sci. USA 88:8262-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vallon, O., F. A. Wollman, and J. Olive. 1986. Lateral distribution of the main protein complexes of the photosynthetic apparatus in Chlamydomonas reinhardtii and in spinach: an immunocytochemical study using intact thylakoid membranes and a Ps-Ii enriched membrane preparation. Photobiochem. Photobiophys. 12:203-220. [Google Scholar]
- 126.Vasil'ev, S., P. Orth, A. Zouni, T. G. Owens, and D. Bruce. 2001. Excited-state dynamics in photosystem II: insights from the X-ray crystal structure. Proc. Natl. Acad. Sci. USA 98:8602-8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vener, A. V., P. J. Van Kan, A. Gal, B. Andersson, and I. Ohad. 1995. Activation/deactivation cycle of redox-controlled thylakoid protein phosphorylation: role of plastoquinol bound to the reduced cytochrome bf complex. J. Biol. Chem. 270:25225-25232. [DOI] [PubMed] [Google Scholar]
- 128.Vener, A. V., P. J. van Kan, P. R. Rich, I. I. Ohad, and B. Andersson. 1997. Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc. Natl. Acad. Sci. USA 94:1585-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wollman, F.-A., and P. Bennoun. 1982. A new chlorophyll-protein complex related to Photosystem I in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 680:352-360. [Google Scholar]
- 130.Wykoff, D. D., J. P. Davies, A. Melis, and A. R. Grossman. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zer, H., O. Prasil, and I. Ohad. 1994. Role of plastoquinol oxidoreduction in regulation of photochemical reaction center IID1 protein turnover in vivo. J. Biol. Chem. 269:17670-17676. [PubMed] [Google Scholar]
- 132.Zhang, S., and H. V. Scheller. 2004. Light-harvesting complex II binds to several small subunits of photosystem I. J. Biol. Chem. 279:3180-3187. [DOI] [PubMed] [Google Scholar]
- 133.Zito, F., G. Finazzi, R. Delosme, W. Nitschke, D. Picot, and F. A. Wollman. 1999. The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 18:2961-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zolla, L., S. Rinalducci, A. M. Timperio, and C. G. Huber. 2002. Proteomics of light-harvesting proteins in different plant species. Analysis and comparison by liquid chromatography-electrospray ionization mass spectrometry. Photosystem I. Plant Physiol. 130:1938-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zouni, A., H. T. Witt, J. Kern, P. Fromme, N. Krauss, W. Saenger, and P. Orth. 2001. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409:739-743. [DOI] [PubMed] [Google Scholar]