Abstract
Research in the macaque monkey suggests that cortical areas with similar microstructure are more likely to be connected. Here, we examine this link in the human cerebral cortex using 2 magnetic resonance imaging (MRI) measures: quantitative T1 maps, which are sensitive to intracortical myelin content and provide an in vivo proxy for cortical microstructure, and resting-state functional connectivity. Using ultrahigh-resolution MRI at 7 T and dedicated image processing tools, we demonstrate a systematic relationship between T1-based intracortical myelin content and functional connectivity. This effect is independent of the proximity of areas. We employ nonlinear dimensionality reduction to characterize connectivity components and identify specific aspects of functional connectivity that are linked to myelin content. Our results reveal a consistent spatial pattern throughout different analytic approaches. While functional connectivity and myelin content are closely linked in unimodal areas, the correspondence is lower in transmodal areas, especially in posteromedial cortex and the angular gyrus. Our findings are in agreement with comprehensive reports linking histologically assessed microstructure and connectivity in different mammalian species and extend them to the human cerebral cortex in vivo.
Keywords: cortical microstructure, functional connectivity, high-resolution MRI, myelin imaging
Introduction
Comprehensive research in macaque monkeys has revealed an intricate link between cortical microstructure and connectivity (see Pandya et al. 2015 for a review). In particular, long-range connections preferentially occur between areas with similar microstructural features (e.g. Pandya and Sanides 1973; Pandya and Yeterian 1985, 1990; Barbas and Pandya 1989). Microstructural similarity has therefore been suggested as a general connectivity principle—or wiring rule—of the mammalian cortex (Barbas 2015; Pandya et al. 2015). This claim is supported by quantitative analyses of the macaque (Beul et al. 2015), cat (Beul et al. 2014), and mouse (Goulas et al. 2016a) connectomes, but remains largely unexplored in the human brain.
The aforementioned animal studies rely on combined tract-tracing and postmortem histology, invasive approaches that are unsuitable to study the human brain. While magnetic resonance imaging (MRI)-based techniques to assess cortical connectivity have been widely adopted, the analysis of human cortical microstructure remains challenging. Some recent studies have related cytoarchitectonic data, available from the literature, to connectivity measures derived from MRI in a separate group of subjects. Thus, van den Heuvel et al. (2015, 2016) demonstrated that the degree of connectivity of an area is related to the size and complexity of its layer III pyramidal neurons, a finding that is in line with previous work in macaque monkeys (Scholtens et al. 2014). Moreover, Goulas et al. (2016b) showed that the existence of connections between 2 areas is partly predicted by their similarity in supragranular cell density. These findings lend strong support for a relationship between macroscale connectivity and microstructural features in the human cortex. Here, we investigate this link in vivo, using high-resolution MRI measures, sensitive to connectivity and microstructural features, both acquired from the same subjects.
With recent advances in MRI technology, it has become possible to derive measures related to cortical microstructure in the living human brain. Classic histological approaches to describe cortical microstructure focus on the distribution of cells (“cytoarchitectonics” e.g. Brodmann 1909) or myelinated fibers (“myeloarchitectonics” e.g. Vogt and Vogt 1919) within the cortical sheet. Because the longitudinal relaxation time (T1) in MRI is sensitive to gray-matter myelin content (Bock et al. 2009; Stüber et al. 2014), maps of intracortical T1 have been introduced as an in vivo proxy for cortical microstructure and revived interest in myeloarchitectonic approaches (Nieuwenhuys 2013). These maps exhibit a decrease in estimated myelin content from primary to transmodal areas (Glasser and Van Essen 2011; Tardif et al. 2015), which is in line with findings from histological studies (e.g., Hopf 1955, 1956; Hopf and Vitzthum 1957). The co-occurrence of local changes in intracortical T1 maps with certain architectonic (Geyer et al. 2011; Glasser and Van Essen 2011), functional (Bridge et al. 2005; Sigalovsky et al. 2006), and topographic (Dick et al. 2012; Sereno et al. 2013) boundaries, as well as rapid changes in functional connectivity patterns (Glasser et al. 2016), exemplifies their usefulness to investigate cortical organization.
Cortical connectivity patterns are now commonly investigated in vivo through resting-state functional connectivity (Biswal et al. 1995). While functional connectivity can reflect indirect links between areas (Adachi et al. 2012), it is largely constrained by anatomical connections (Skudlarski et al. 2008; Honey et al. 2009; Hermundstad et al. 2013).
With this study, we investigate the relationship between in vivo measures sensitive to intracortical myelin and functional connectivity in the human cerebral cortex. We employ ultrahigh field MRI and dedicated image processing tools to compare functional connectivity patterns with cortical differences in T1-based myelin estimates at a submillimeter resolution. Motivated by the coherent findings in other mammalian species, we hypothesize that similarity in intracortical myelin relates to functional connectivity, thereby constituting a wiring rule of human cerebral cortex.
Materials and Methods
Data
MRI data were acquired and published previously in 2 independent studies (Gorgolewski et al. 2015; Tardif et al. 2016). Here, we use data sets of 9 subjects who took part in both studies (5 females, mean age = 24.8 years, standard deviation = 1.2 years, see Supplementary Table 1 for subject identifiers). All subjects gave written informed consent and the studies were approved by the Ethics Committee of the University of Leipzig. Data were acquired on a 7T MR scanner (MAGNETOM, Siemens Healthcare) equipped with a 24-channel Nova head coil. In the first study (Tardif et al. 2016), a structural scan was acquired at an isotropic resolution of 0.5 mm using the MP2RAGE sequence (Marques et al. 2010, inversion time (TI)1/TI2 = 900/2750 ms, repetition time (TR) = 5 s, echo time (TE) = 2.45 ms, α1/α2 = 5°/3°, bandwidth = 250 Hz/px, echo spacing = 6.8 ms, partial Fourier = 6/8, scan time = 28 min per hemisphere) and an optimized radiofrequency pulse to minimize sensitivity to B1 inhomogeneity (Hurley et al. 2010). The resulting images were used to generate whole brain T1 maps. In the second study (Gorgolewski et al. 2015), a total of 4 whole brain resting-state scans were acquired in 2 sessions at an isotropic resolution of 1.5 mm, using a T2*-weighted echo-planar imaging sequence (field of view = 192 × 192 mm2, 70 slices, TR = 3 s, TE = 17 ms, α = 70°, bandwidth = 1116 Hz/Px, partial Fourier 6/8, GRAPPA acceleration with iPAT factor of 3, 300 volumes per scan). In the first of these 2 sessions, an additional short structural scan (11 min) was acquired using the MP2RAGE sequence with similar parameters as above but an isotropic resolution of 0.7 mm. The uniform T1-weighted (T1w) image generated from this scan was used as an intermediate registration target to project the resting-state data into the space of the high-resolution T1 map and will be referred to as the “low-resolution T1w image.”
System Environment and Software
All data processing and analyses were performed on Linux servers running Ubuntu 12.04.5 LTS. The following software tools were used for data processing: CBS Tools (v3.0, Bazin et al. 2014) as a plugin for MIPAV (v7.0.1, McAuliffe et al. 2001) and JIST (v2.0, Lucas et al. 2010), ANTs (v2.1.0, Avants et al. 2011), Nipype (v0.11.0, Gorgolewski et al. 2011), Nilearn (v0.2.3, Abraham et al. 2014), Meshlab (v1.3.0, meshlab.sourceforge.net), Freesurfer (v5.3.0, Dale et al. 1999; Fischl et al. 1999), and AFNI (v16.1.28, Cox 1996). Custom scripts were written in Python 2.7 using numerous libraries (Jones et al. 2001; McKinney 2010; Seabold and Perktold 2010; Pedregosa et al. 2011; van der Walt et al. 2011; Droettboom et al. 2016; Waskom et al. 2016). More details and all code can be found at https://github.com/juhuntenburg/myelinconnect.
Preprocessing of Structural Data
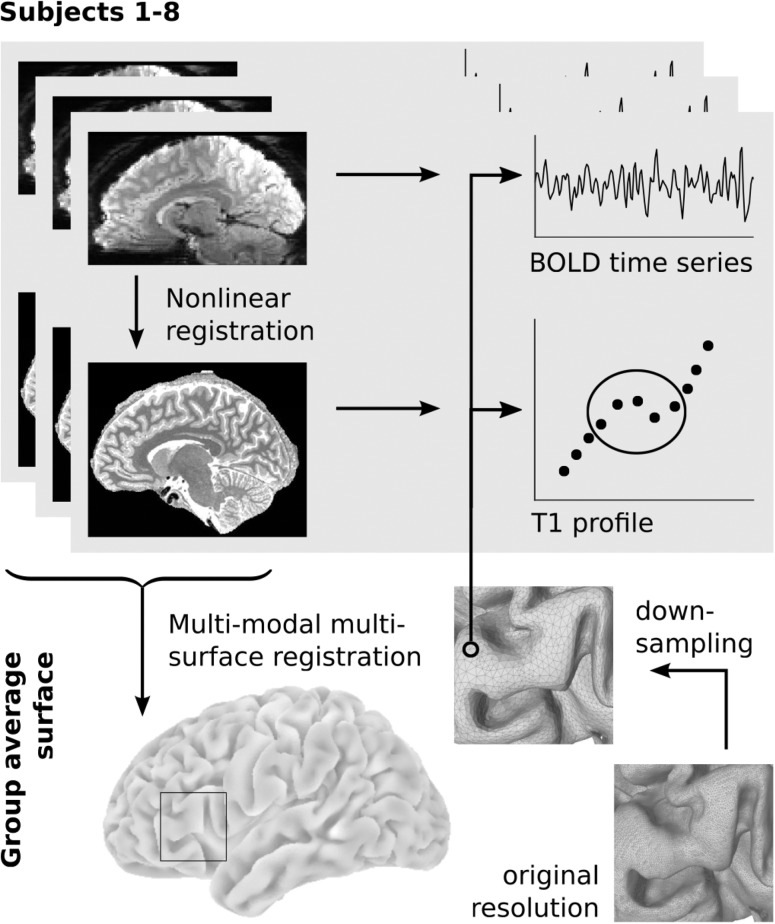
Preprocessing of the high-resolution T1 maps was carried out using CBS Tools (Fig. 1). Tissue segmentation and reconstruction of white matter (WM), midcortical, and pial surfaces was performed as described previously (Bazin et al. 2014). Cortical thickness between the WM and pial surface was calculated. For each subject, 9 intracortical surfaces were generated between the WM and pial surface. The radial position of each surface was determined using a volume-preserving approach (Waehnert et al. 2014), which accounts for the dependence of layer thickness on cortical folding described by Bok (1929). For each node of the subject's midcortical surface, T1 values were sampled in radial direction on the surfaces (WM, 9 intracortical, pial), yielding a vector of 11 values that was assigned to each node. We will refer to this vector as the “T1 profile” (cf. Waehnert et al. 2014, 2016; Dinse et al. 2015). We used the multimodal, multicontrast surface registration approach (MMSR), introduced by Tardif et al. (2015), to derive a study-specific surface template from the high-resolution T1 maps. The resulting group-average surface mesh was simplified from approximately 700k to 70k nodes using MeshLab's Quadric Edge Collapse Decimation filter (percent reduction = 0.11, quality threshold = 1.0, preserving mesh boundaries, normals, and topology, optimal placement, postsimplification clean). This template surface provides the common reference space for subsequent analyses. For visualization purposes, the group template surface was partly inflated using Meshlab's Taubin Smooth filter (lambda = 0.6, mu = −0.3, steps = 150). For coregistration of the resting-state data, the low-resolution T1w images were preprocessed using FreeSurfer's standard recon-all pipeline.
Figure 1.
Data extraction workflow. Resting-state images were nonlinearly coregistered to the structural space of the same subject. A group-specific surface template was created using midcortical surfaces and intracortical T1 contrasts of all subjects in a multimodal multisurface registration approach. The group-average surface was downsampled and projected into the space of each subject for sampling of BOLD time series and T1 profiles. Cortical depth profiles were sampled according to an equi-volumetric principle; only the central values were averaged to minimize partial volume effects.
Preprocessing of Resting-State Data
Preprocessing of the resting-state time series was streamlined in a reusable pipeline using Nipype. For each resting-state scan, the first 5 volumes were removed to ensure steady-state magnetization. After simultaneous slice time and motion correction, as implemented in Nipy (Roche 2011), a temporal median image was created and bias field corrected using N4 as implemented in ANTs (Tustison et al. 2010). This median image was coregistered to the high-resolution T1 map of the same individual in 2 steps: in a linear step, both the median image and the high-resolution T1 map were registered to the low-resolution T1w image using Freesurfer's boundary-based registration with 6 degrees of freedom (Greve and Fischl 2009). The 2 resulting transformations were concatenated and used to project the median image into the space of the high-resolution T1 map. After carefully masking both images, a second nonlinear registration step was performed using ANTs’ symmetric diffeomorphic transformation model (SyN) and fast cross-correlation similarity metric (Avants et al. 2008). This last step is intended to account for nonlinear distortions in the functional data. All transformations were combined and used to project data from the individual's functional to the high-resolution structural space in a single interpolation (Fig. 1). Voxel-wise temporal denoising of the slice time and motion corrected time series was performed using Nilearn. Confounds that were removed from the time series included linear trends, the 6 motion regressors and their first derivatives, intensity outliers (z > 3) and motion outliers (mean composite norm > 0.5 mm), as identified by Nipype's rapidart algorithm, as well as regressors reflecting the signal in the WM and cerebrospinal fluid (CSF), following the aCompCor approach (Behzadi et al. 2007). For the latter, WM and CSF masks were derived using the MGDM segmentation as implemented in CBS Tools (Bazin et al. 2014), and eroded by 4 (WM) or 2 (CSF) voxels, respectively, to avoid partial volume effects with the gray matter. Nilearn's high_variance_confounds method with default settings was used to extract 5 high variance temporal components from voxels within each of the 2 masks, resulting in a total of 10 components that were included in the regression. The denoised time series were temporally normalized and bandpass filtered to a frequency range of 0.01–0.1 Hz. One subject was excluded due to excessive motion (composite norm max > 3 mm, mean > 0.15 mm across the 4 resting-state scans) and insufficient coregistration quality as assessed by visual inspection (Supplementary Fig. 1). Data of the remaining 8 subjects passed these criteria and were used for subsequent analyses.
Masks
The medial wall was delineated by hand on both hemispheres of the inflated group template surface mesh using AFNI's Surface Mapper. Additionally, masks were created to exclude nodes with signal quality of at least 1.5 standard deviations below the mean in at least one session of one subject and one imaging modality. Signal quality in each subject's structural image was derived from the second inversion images of the MP2RAGE sequence: the probability that a data point represents foreground (data, modeled by a uniform distribution) rather than background (noise, modeled by an exponential distribution) was calculated based on a distribution mixture model. The derived measure represents a heuristic definition of available signal in the T1 maps. Signal quality in the resting-state images was assessed separately for each subject and session using the temporal signal-to-noise ratio (tSNR). After slice time and motion correction, but before nuissance regression, each voxel's tSNR was calculated by dividing its temporal mean BOLD signal by the temporal standard deviation of its BOLD signal. All masks were combined and used to exclude low fidelity regions from subsequent analyses.
Intracortical T1 and T1 Difference Matrix
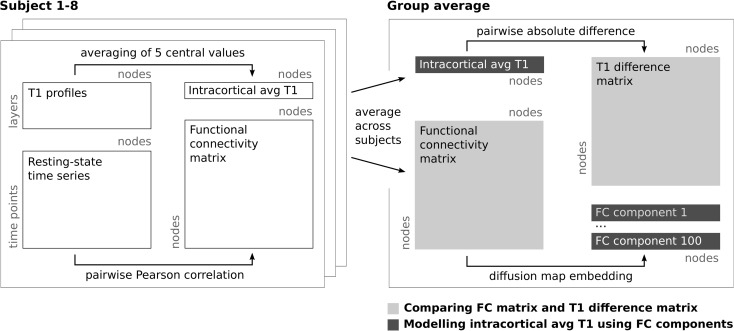
For each subject, the 5 central values (45%) of the T1 profiles were averaged to create a single intracortical average T1 value at each surface node, which is minimally biased by partial volume effects with the WM and CSF (Figs 1 and 2). Throughout the manuscript, we assume this intracortical average T1 value to largely reflect intracortical myelin content. Limitations of this assumption will be addressed in “Discussion” section. Intracortical average T1 values were slightly smoothed on the individual midcortical surface using a Gaussian kernel with full-width-half-maximum (FWHM) of 1.5 mm, to prepare them for sampling on the lower resolution template surface. The group template surface was projected into each individual's native space using the MMSR-derived transformations and the intracortical average T1 was sampled from the closest point on the subject's original high-resolution surface. Thus, each node on the group-level surface was associated with a single intracortical T1 value for each subject. These values were subsequently averaged across subjects to derive the group-level intracortical T1 map. A T1 difference matrix was generated from this map by calculating the absolute difference in group-level intracortical T1 for each pair of nodes on the surface template. High values in this matrix therefore indicate node pairs that differ substantially in their intracortical average T1 values while values close to zero indicate pairs with very similar values. (See Fig. 2 for a schematic of the processing steps..
Figure 2.
Data analysis workflow. Schematic describing the processing steps from the single subject T1 profiles and resting-state time series to the comparison of group-level intracortical average (avg) T1 and functional connectivity (FC). In the first analysis (light gray), the high-dimensional functional connectivity and T1 difference matrices are correlated. In the second analysis (dark gray), intracortical T1 maps and a linear combination of functional connectivity components are compared in a single dimension.
Functional Connectivity Matrix and Comparison to T1 Difference Matrix
The fully preprocessed resting-state time series were projected into the individual high-resolution structural space. Subsequently, the time series data of each subject were sampled onto the group template surface at midcortical depth and smoothed along the cortex using a Gaussian kernel (FWHM = 3 mm). Due to its lower spatial resolution, the resting-state data were not sampled at different intracortical depth levels. Whole brain functional connectivity matrices were derived for each subject and session by calculating the Pearson's product–moment correlation between the time series of each pair of nodes. All matrices were Fisher r-to-z-transformed and averaged across subjects and sessions. The resulting average functional connectivity matrix was back transformed to Pearson's r values. (See Fig. 2 for a schematic of the processing steps.)
Pearson's product–moment correlation coefficient was calculated between the upper triangles of the group-average functional connectivity and T1 difference matrices. Moreover, functional connectivity and T1 difference patterns were related to each other in a node-wise fashion: Pearson's product–moment correlation coefficient was calculated between each row of the functional connectivity matrix (representing functional connectivity of one node to all other cortical nodes) with the same row in the T1 difference matrix (representing the T1 difference of the same node to all other cortical nodes). This results in one value per node, indicating how well its functional connectivity to any other node can be predicted from its T1 difference to that node (and vice versa). The resulting map was compared with the group average of signal quality measures in both imaging modalities, assessed as described in “Masks” section (see Supplementary Fig. 2). To control for the effect of spatial distance, the above analyses were repeated after regressing Euclidean distances between nodes in 3D space against both functional connectivity and T1 difference. Thus, both the upper triangle correlation and node-wise correlations were recalculated for the residuals of the functional connectivity and T1 difference after distance regression. Euclidean distance was also correlated to T1 difference and functional connectivity separately (Supplementary Fig. 3). Importantly, Euclidean distance is a biased estimate of the actual length of fiber tracts through the WM, which systematically underestimates certain pathways. Inferences drawn using it as a measure of connection length must therefore be evaluated in the light of this serious limitation. To control for the relationship between cortical thickness (CT) and intracortical T1, CT was smoothed (FWHM = 1.5 mm), sampled in the subjects’ native space and averaged on the group template surface. A matrix of absolute difference in group-level CT between each pair of nodes was regressed against the T1 difference matrix, and vice versa, before correlation to functional connectivity.
Functional Connectivity Decomposition and Comparing Components to T1
To disentangle which particular functional connectivity features underlie the relationship to intracortical T1, we decomposed the functional connectivity matrix into a set of one-dimensional components, reflecting distinct aspects of cortical connectivity (see Fig. 2). For this, the group average functional connectivity matrix was transformed into a positive similarity matrix L through increasing each element by 1 and then dividing it by 2. Thus, nodes with highly correlated signals are considered similar, while nodes with highly anticorrelated signals are considered dissimilar. Nonlinear dimensionality reduction of L was performed using diffusion maps (Coifman and Lafon 2006, Implemented in https://github.com/satra/mapalign.) In short, diffusion maps embed the high-dimensional connectivity data in low-dimensional space by first transforming the similarity matrix L in a Markov chain with transition probability matrix M:
If the diffusion operator α is set to 1, M is equal to the classical normalized graph Laplacian. Here, we use α = 0.5 in which case transition in the Markov chain approximates Fokker–Planck diffusion and the method is less sensitive to nonuniform sampling of the data on the underlying manifold. The eigenvectors of M constitute the coordinate system of the embedded space. Here, we embed the 70k × 70k similarity matrix L in 100 dimensions, which we will refer to as functional connectivity components FC1–100. These components are naturally ordered by decreasing eigenvalues, that is, FC1 explains most of the variance, etc. To approximate the amount of variance explained by individual components, we make the simplifying assumption that FC1–100 together explain 100% of the variance (FC100 explains 0.29% of this variance). Note that these values refer to variance in the transition matrix M, not in the original functional connectivity matrix.
Diffusion maps have been used for the analysis of resting-state fMRI data before and in-depth discussions can be found in Langs et al. (2015a, 2015b) and Margulies et al. (2016). In contrast to linear decomposition approaches, such as principal component analysis, nonlinear methods retain both local and global relationships between data points in the embedded space without requiring kernel manipulations. Diffusion maps, specifically, are more robust to noise in the connectivity matrix than other nonlinear dimensionality reduction techniques, such as Isomap (Tenenbaum et al. 2000).
Pearson's product–moment correlation coefficient was calculated between the first component of the functional connectivity embedding (FC1) and intracortical T1. To account for heteroscedasticity, we assessed the significance of this correlation using the robust sandwich variance estimator (White 1980) as implemented in Statsmodels. Higher order functions were fitted as well, to assess whether they would provide a better description of the relationship between FC1 and T1. In a next step, intracortical T1 was modeled as a linear combination of one or multiple FC components using Ordinary Least Squares regression as implemented in Scikit-learn such that:
We compared all possible combinations of the first 20 FC components (FC1–20, each explaining >1%, together explaining 61.6% of the variance in M, 1 048 575 different models). Again, significance of the coefficients was tested using the robust sandwich variance estimator.
Model Comparison
To compare the performance of combinations of FC components to model intracortical T1, we derived the Bayesian information criterion (BIC) as:
where p is the number of parameters and n the number of data points used for fitting, R is the data range, is the residual variance and the sum of squared residuals. The derivation of the BIC closely follows that in Bazin and Vezien (2005) and can be found in the Supplementary Methods. We estimated from local variance in the group average T1 data using all surface nodes that were not excluded by the mask:
where is an estimate of the median absolute difference between neighboring T1 values. Assuming that these differences follow a half Gaussian distribution their median is given by and . The data range R is set to the range of group average T1 values not excluded by the mask.
Permutation Tests
Random data sets were simulated through the following steps: first, values for each node on the surface template were drawn from a normal distribution, creating 1000 data sets per hemisphere. Second, data were smoothed on the surface applying differently sized Gaussian smoothing kernels (FWHM = 1.5, 3, 6, 12, and 24 mm). Third, after masking, the values of each random data set were adjusted to those of the original intracortical T1 map using histogram matching. Each random data set was fitted as described for the T1 data in Functional Connectivity Decomposition and Comparing Components to T1, employing the models consisting of either FC1 alone or a linear combination of FC1, 5, and 6 (the best performing model in the previous analysis). The coefficient of determination was calculated for all models and used as null distribution to assess significance of the fit achieved for the real intracortical T1 map. Note that for this analysis, the left and right hemispheres were treated separately for both the random and the real T1 data. Using whole brain data would result in an unfair comparison because both the FC components and the original T1 map show high interhemispheric symmetry while the random data do not.
Results
Functional Connectivity and Intracortical T1 Are Systematically Related
We created 2 group average matrices: one containing the difference in intracortical T1 between each pair of nodes on the surface (“Intracortical T1 and T1 difference matrix”) and a second one with the functional connectivity between each pair of nodes (“Functional connectivity matrix and comparison to T1 difference matrix,” see also Fig. 2). We found both matrices to be anticorrelated (Pearson's r = −0.34, P < 0.0001, ). Thus, the more similar 2 nodes are in their T1 values, the higher their functional connectivity.
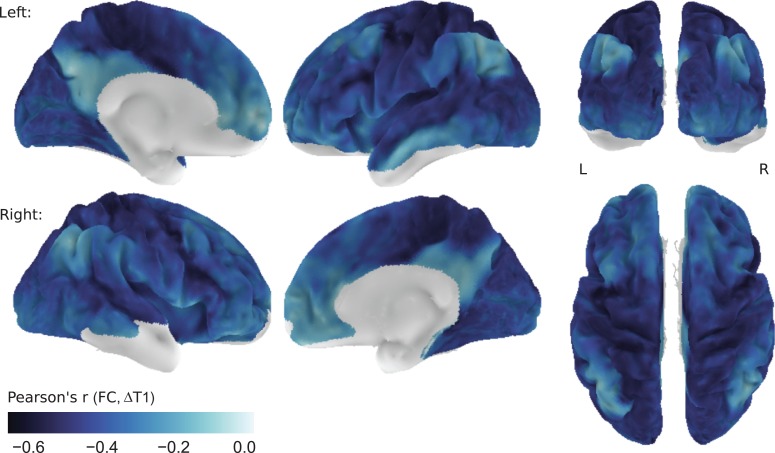
The Strength of this Link Varies Across the Cortex
To investigate whether this inverse relationship between functional connectivity and T1 difference is homogeneous across the cortex, we also calculated the correlation in a node-wise fashion. For each surface node with sufficient signal quality in both imaging modalities (cf. “Masks”), we compared its functional connectivity with all other nodes with the respective T1 differences to those nodes. This results in one value per surface node, indicating how well T1 difference predicts functional connectivity for this particular node. Figure 3 shows that the relationship between functional connectivity and T1 difference follows a distinct pattern across the cortex. Unimodal areas display a particularly strong link between the 2 measures. Conversely, regions that show little correlation between functional connectivity and T1 differences include posteromedial cortex, superior portions of the inferior parietal lobule, superior temporal sulcus and middle temporal gyrus, medial prefrontal regions, anterior insular cortex and posterior portions of the superior and middle frontal gyrus. The correspondence between intracortical T1 and connectivity strength is thus generally stronger in unimodal than in transmodal areas. This effect shows little dependence on the signal quality in the structural (Pearson's r = −0.18, P < 0.0001) and resting-state (Pearson's r = −0.09, P < 0.0001) data. In fact, particularly low correspondence of functional connectivity and T1 difference in posteromedial cortex and the inferior parietal lobule coincides with high signal quality in both imaging modalities (Supplementary Fig. 2).
Figure 3.
Node-wise correlation of functional connectivity and T1 difference. For each surface node, the correlation between its functional connectivity to all other nodes and its T1 difference to all other nodes is shown. Values closer to zero (white) indicate a weaker linear relationship between connectivity strengths and T1 differences. Correlation values are shown on the left (L) and right (R) hemisphere of the group average surface. Nodes with low signal quality in either imaging modality (predominantly in orbitofrontal and ventral temporal areas) were excluded from the analysis.
We observed the same general pattern of correspondence between functional connectivity and differences in a T1w/T2w-based estimate of intracortical myelin content in a large, independent data set (n = 820), made available through the Human Connectome Project (HCP, Glasser et al. 2013; van Essen et al. 2013, see Supplementary Methods and Supplementary Fig. 7 for details).
The Correspondence is Independent of Euclidean Distance
It is well known that connectivity decreases with spatial distance (e.g., Young 1992; Scannell et al. 1995) and reasonable to assume a similar relationship for microstructural similarity. We therefore investigated whether the observed relationship between functional connectivity and T1 difference is driven by a common dependence on spatial distance. To this end we created a third matrix, describing the Euclidean distance of each pair of nodes on the surface template. Of note, Euclidean distance is a biased estimate of connection length and the ensuing results must be interpreted in due consideration of this limitation. As expected, functional connectivity and Euclidean distance were highly correlated (Pearson's r = −0.41, P < 0.0001, , Supplementary Fig. 3a ). However, T1 difference was independent of Euclidean distance when calculated across the whole cortex (Pearson's r = 0.01, P < 0.0001, Supplementary Fig. 3b ). After regressing Euclidean distance against both T1 difference and functional connectivity, the anticorrelation between residual T1 difference and residual functional connectivity remained essentially unchanged (Pearson's r = −0.37, P < 0.0001, ).
We also repeated the node-wise analysis after regressing T1 difference and functional connectivity against Euclidean distance for each node separately. The overall pattern of the relationship between functional connectivity and T1 is not affected by distance regression (Supplementary Fig. 4). Only some areas, such as supplementary motor area, midcingulate cortex, and medial temporal gyrus, show increased anticorrelation. Thus, the relationship between functional connectivity and T1 is largely independent of a shared dependence on spatial distance.
The Relationship Persists When Controlling for Cortical Thickness
Beul et al. (2015) found cortical thickness (CT) to be related to microstructural differentiation and cortical connectivity. However, in their study, the association between CT and connectivity disappeared when controlling for microstructural similarity, while microstructure was still related to connectivity when controlling for thickness. We found a similar pattern in our data. Difference in CT between nodes showed a strong correlation to T1 difference (Pearson's r = 0.70, P < 0.0001) and a moderate correlation to functional connectivity (Pearson's r = −0.28, P < 0.0001). Regressing CT difference against T1 difference reduced, but did not remove, the correlation between T1 difference and functional connectivity (Pearson's r = −0.20, P < 0.0001 vs. r= −0.34, P < 0.0001 before CT regression). However, CT difference was no longer related to functional connectivity after T1 difference had been regressed from it (Pearson's r = −0.06 , P < 0.0001). Intracortical T1 thus seems to capture variance in functional connectivity patterns beyond what is accounted for by CT alone.
Specific Connectivity Components Account for the Relationship to T1
After establishing an association between functional connectivity and T1 difference in high-dimensional space, we sought to identify the low-dimensional features of functional connectivity that specifically underlie this link. To this end, we decomposed the group-level functional connectivity matrix using nonlinear dimensionality reduction. The result are 100 components (FC1–100), each representing a distinct aspect of functional connectivity in a single dimension. The value assigned to a given node in each of these dimensions indicates its position along a spectrum of connectivity patterns: nodes with similar values on a given component resemble each other in their connectivity pattern, or more precisely, in the aspect of connectivity that is captured by this particular component. Nodes with very different values on the same component have more distinct connectivity profiles. Unlike independent component analysis, the method applied here does not enforce spatial independence of different components, but results in a set of gradients spanning the entire cortex (see “Functional Connectivity Decomposition and Comparing Components to T1” section for more details). This approach enables us to describe the relationship between the distribution of intracortical T1 and the main modes of variation of connectivity patterns across the cortex.
The Principal Gradient of Functional Connectivity
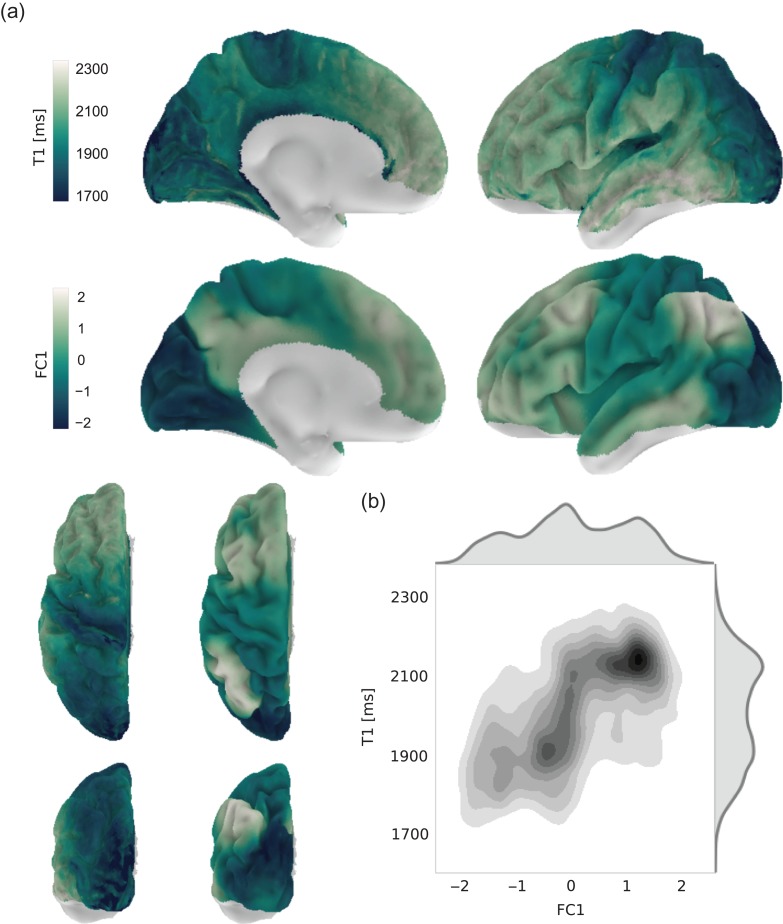
We initially focused on the principal gradient (FC1) that captures the main variation of functional connectivity patterns across the cortex (12% of the variance). As visible in Figure 4a, FC1 spans a gradient between unimodal visual, motor, somatosensory, and auditory areas at one end of the spectrum, and higher order association areas in frontal, parietal, and temporal cortex at the other (see also Supplementary Fig. 5).
Figure 4.
Relationship between intracortical T1 and the principal component of functional connectivity. (a) Intracortical T1 (top row and left column of bottom row) and the principal component of functional connectivity decomposition (FC1, middle row and right column of bottom row) are shown on the surface of the left hemisphere. Nodes with low signal quality in either imaging modality were excluded from the analysis. The surface plots for the right hemisphere are highly comparable and are shown in Supplementary Figure 5. (b) Bivariate distribution of FC1 and T1 values across both hemispheres.
The one-dimensional representation of functional connectivity in FC1 can directly be compared with the group-average intracortical T1 map (see Fig. 2). This map is used as an estimate of intracortical myelin content, with higher T1 values indicating less myelin (Waehnert et al. 2016). Limitations of this assumption will be discussed below. It reveals the highest estimated intracortical myelin content in primary visual, somatosensory, auditory, and motor regions. Intermediate values are found in posteromedial cortex, especially ventral portions adjacent to the corpus callosum, as well as superior parietal cortex, and relatively low myelin content prevails in prefrontal cortex, middle and inferior temporal gyrus as well as the inferior parietal lobule (Fig. 4a and Supplementary Fig. 5).
Node-wise comparison of the T1 and FC1 map reveals a substantial spatial correlation (Fig. 4b, Pearson's r = 0.53, P < 0.0001, ), exceeding the overall correlation in high-dimensional space. This implies that FC1 specifically captures an aspect of functional connectivity, which is related to intracortical T1. However, clear differences between the spatial layout of FC1 and T1 remain, most prominently in the superior part of the inferior parietal lobule and posteromedial cortex. Moreover, the bivariate distribution indicates a nonlinear relationship between FC1 and T1 (Fig. 4b). We therefore fitted higher order models to the data and compared them using an adapted version of the BIC (see “Model Comparison” section). Although more complex functions, such as polynomials or a sigmoid, explained the data better, their overall performance was worse, due to the higher number of parameters in these models (Supplementary Fig. 6).
Again, we found a similar relationship between FC1 and T1w/T2w-based intracortical myelin content in the HCP data set (n = 820, Pearson's r = 0.52 , P < 0.0001, see Supplementary Methods and Supplementary Fig. 8 for details).
Including Multiple Components Improves the Fit
The first component of the functional connectivity decomposition captures the main variance in the data. Still, a considerable amount of variance is explained by subsequent components (Fig. 5a), which might be essential to establish the link to intracortical T1. We therefore investigated whether a linear combination of multiple FC components can improve the fit to intracortical T1. We focused on components that each explains more than 1% of the variance (FC1–20). Every possible linear combination of these 20 components was fitted to the intracortical average T1 data. We then compared all models using the same implementation of BIC as in the previous section. A lower BIC value indicates a better balance of model fit and complexity.
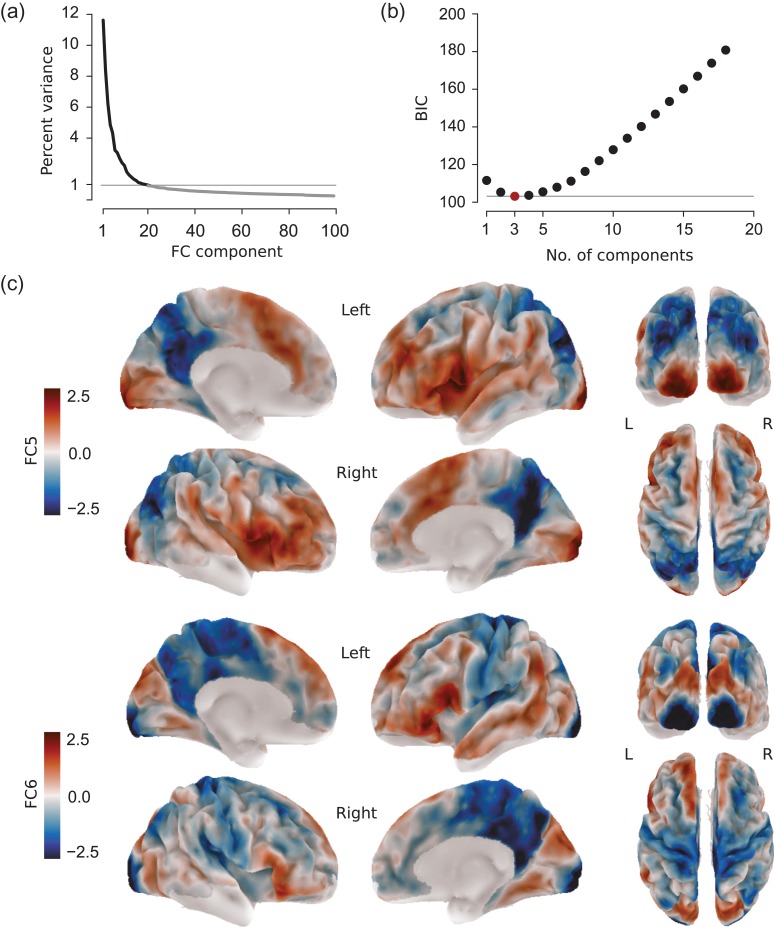
Figure 5.
Using multiple connectivity components to fit intracortical T1. (a) Variance in the connectivity transition matrix Mexplained by the different embedding components. (b) BIC values of the best performing model by number of components employed. The best model (FC1, 5, 6) is highlighted in red. (c) Connectivity components FC5 (top row) and FC6 (bottom row), together with FC1 constituting the best performing model, are shown on the left (L) and right (R) hemisphere of the group average surface. Values indicate the relative position of a given surface node along the respective component and have no unit. Nodes with low signal quality in either imaging modality were excluded from the analysis.
Figure 5b shows the best performing model for each group of models employing the same number of FC components. Importantly, the model selection procedure is stable in that the best model in each group is equal to the best model in the previous group, except for the addition of one further. Moreover, the results are resistant against modest variation in the empirically determined noise prior (Supplementary Table 2). The most parsimonious model to fit intracortical T1 is a linear combination of component FC1, 5, and 6:
FC5 (explaining ca. 4% variance) contrasts posteromedial cortex and the superior parietal lobule at one end of the spectrum with inferior frontal gyrus, anterior insula, superior temporal cortex, ventral parts of precentral and postcentral gyrus, inferior parietal lobule, and visual cortex at the other (Fig. 5c, top). The most remarkable feature of FC6, which explains approximately 3% of variance, is that it separates primary sensorimotor regions from secondary regions and unimodal association areas within a given modality (Fig. 5c, bottom).
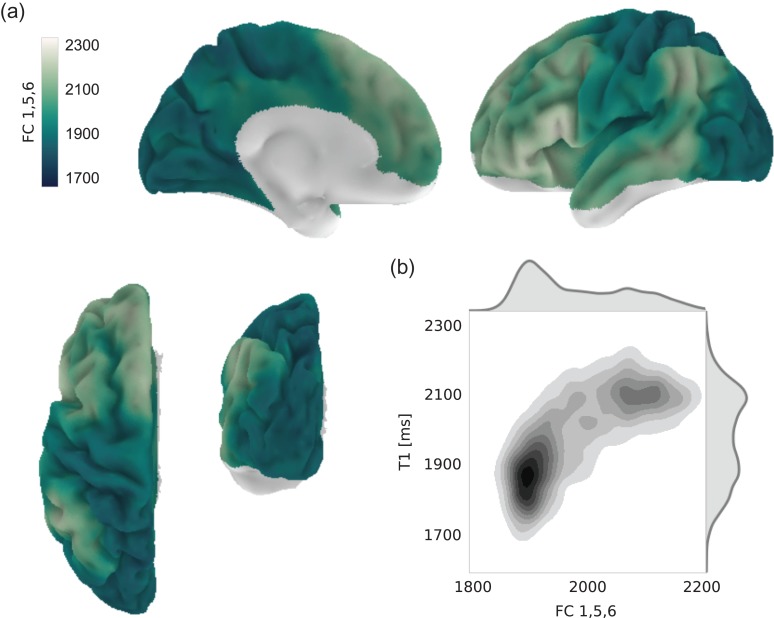
Combining FC1, 5, and 6 most notably improves the fit to intracortical T1 in posteromedial cortex (Fig. 6a). Moreover, visual regions become more similar in value to somatomotor and auditory regions than for FC1 alone. As shown in Figure 6b, including FC5 and FC6 also increases node-wise correlation between T1 and modeled T1 (Pearson's r = 0.67, P < 0.0001, , a quadratic model was fitted as well but without substantial gain, Supplementary Fig. 6). Differences to intracortical T1, however, remain. In particular, compared with the original T1 map, regions around the angular gyrus still display relatively high values in the combined FC1, 5, 6 map. The fitted T1 values also do not extend as far into the low range as original T1 values, resulting in residual differences especially in primary sensorimotor regions.
Figure 6.
Combination of connectivity components providing the best fit to intracortical T1. (a) Result of modeling T1 as a linear combination of connectivity components FC 1, 5, and 6, shown on the left hemisphere. Nodes with low signal quality in either imaging modality were excluded from the analysis. The surface plots for the right hemisphere are highly comparable and are shown in Supplementary Figure 5. (b) Bivariate distribution of modeled and original T1 values across both hemispheres.
The Correspondence Is Independent of Data Smoothness
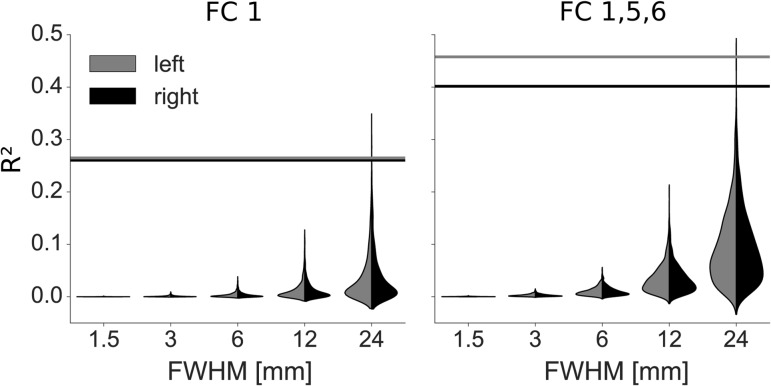
In order to ensure that the spatial correlation values obtained in the previous section do not result from spatial autocorrelation alone, we performed permutation testing. We simulated 1000 random data sets per hemisphere and smoothed them with different kernel sizes, ranging from the kernel applied to the original T1 maps (FWHM = 1.5 mm) to very large kernels (FWHM = 24 mm). We next fitted general linear models based on FC1 alone or FC1, 5, 6 to the random maps, as was done for the real data. Figure 7 shows that the model fit obtained for the real data far exceeds what is expected from data smoothness alone, even for large smoothing kernels (P < 0.005 in all cases). This is true for both hemispheres and both models, although the more complex model naturally fits the random data better than the simple model.
Figure 7.
Validation of model fit. Random, smoothed data sets were fitted using linear models containing only FC1 (left) or FC1, 5, and 6 (right). The distributions of resulting R2 values for the left (grey) and right (black) hemisphere are shown as split violin plots. Horizontal lines indicate the values obtained when fitting respective models to the actual map of myelin content (FWHM = 1.5 mm).
Discussion
We demonstrated a systematic relationship between functional connectivity and a T1-based estimate of intracortical myelin in the human cortex. Regions with similar intracortical T1 show higher functional connectivity than regions that differ in intracortical T1. We initially illustrated the link to T1 in the high-dimensional space of the full functional connectivity matrix. By embedding functional connectivity into a set of one-dimensional components, we then identified particular functional connectivity features that relate to intracortical T1. Across both approaches, posteromedial cortex and superior portions of the inferior parietal lobule stood out as regions of low correspondence between functional connectivity and intracortical T1. In the following, we detail how the neurobiological signature of these regions, as well as characteristics of the MRI-based measures, can help to understand this effect.
Assessing Cortical Myelin and Connectivity in vivo
In this study, we used intracortical T1 to estimate myelin content. Validation against histological data has confirmed that T1 contrast reflects myelin in cortical gray matter (Bock et al. 2009; Geyer et al. 2011; Stüber et al. 2014), displaying a gradient of decreasing myelin density from primary toward transmodal regions (cf. Vogt and Vogt 1919; Hopf 1955, 1956; Hopf and Vitzthum 1957; Sanides 1962). Iron concentration has also been shown to contribute to intracortical T1 contrast (Stüber et al. 2014) and an alternative interpretation of the current results is a systematic relationship between functional connectivity and iron levels. However, intracortical iron is strongly colocalized to myelin (Fukunaga et al. 2010), with ferritin particles embedded in the myelin sheath functioning as a storage for oligodendrocytes, that requires iron for the production and repair of myelin (Connor and Menzies 1996; Todorich et al. 2009). Therefore, independent of the exact contributions of iron and myelin to the T1 contrast, it appears justified to interpret T1 as to largely reflect the distribution of intracortical myelin.
We acquired quantitative T1 maps using the MP2RAGE sequence (Marques et al. 2010), like many recent works (Dick et al. 2012; Sereno et al. 2013; Tardif et al. 2015; Waehnert et al. 2016). This approach has the advantage of minimizing sensitivity to B1 inhomogeneities, allowing for quantitative intersubject and intersite comparison, and disentangling T1 from contribution other factors such as proton density and T2*, present in standard T1-weighted images (Turner 2015; Weiskopf et al. 2015). Alternative quantitative MRI techniques, such as magnetization transfer imaging (Dousset et al. 1992) and myelin water imaging (Mackay et al. 1994), are more specific to myelin than T1, but provide lower spatial resolution. Moreover, quantitative T1 has recently been characterized by the highest intrasubject and intersubject reliability in a comparison of several approaches to map intracortical myelin (Haast et al. 2016). Some studies have used a ratio of T1w over T2w images to assess intracortical myelin (Glasser and Van Essen 2011; Glasser et al. 2014, 2016). The relationship of T1w/T2w contrast to myelin content has not yet been validated and does not allow for quantitative comparison across studies. However, the spatial distribution of the T1w/T2w ratio generally appears similar to quantitative T1 maps and we were able to confirm the main results of the current study using T1w/T2w-based estimates of intracortical myelin. As myelin imaging becomes more abundantly used, it will be crucial to carefully compare different in vivo approaches with each other as well as with histological data.
For the in vivo assessment of cortical connectivity, we used resting-state functional connectivity, which has become a widely used tool in human neuroimaging. Large-scale functional connectivity patterns robustly reproduce across subjects (Damoiseaux et al. 2006), test–retest sessions (Shehzad et al. 2009; Zuo and Xing 2014), sites, and protocols (Biswal et al. 2010). Most importantly, resting-state functional connectivity is largely constrained by anatomical connections, as demonstrated in multiple studies in the human brain (Skudlarski et al. 2008; Honey et al. 2009; Hermundstad et al. 2013) and in comparison with tract-tracing data in monkeys (Vincent et al. 2007; Miranda-Dominguez et al. 2014). Since functional connectivity is based on temporal correlations, however, it does not necessarily reflect monosynaptic connections (Honey et al. 2009; Adachi et al. 2012). It is thus important to keep in mind that resting-state functional connectivity measures aspects of cortical organization that are closely related to, but not directly measuring, structural connections.
Microstructural Similarity as a Wiring Principle of Cerebral Cortex
Microstructural similarity has been suggested as a candidate principle underlying the organization of cortical connections (Barbas 2015; Pandya et al. 2015). Respective links between microstructure and connectivity were established in the cortex of different mammalian species, by combining invasive tract-tracing and postmortem histology (Beul et al. 2014, 2015; Scholtens et al. 2014; Pandya et al. 2015; Goulas et al. 2016a). Due to the invasive nature of this approach, it cannot directly be translated to the human brain. Two recent studies have addressed this challenge by comparing diffusion-weighted MRI data to cytoarchitectonic atlas information (von Economo and Koskinas 1925), demonstrating a relationship between connectivity and layer III neuron complexity (van den Heuvel et al. 2015, 2016), as well as supragranular cell density (Goulas et al. 2016b). Spatial accuracy, however, is inevitably limited when mapping atlas information in stereotactic space and the discrete parcellation scheme prohibits the analysis of gradual changes or variable boundaries. We pursued an alternative approach, directly comparing in vivo measures related to cortical connectivity and microstructure in the same group of individuals at high spatial resolution. While these data facilitate the extraction of intracortical T1 values with high spatial precision, it is important to note that the current resolution does not allow the investigation of cortical features at the microscale. Differences in intracortical average T1 are likely to reflect differences in the underlying microstructure, namely the density of myelinated fibers. However, multiple microscale configurations can result in the same average T1 value when measured at a relatively coarse resolution of 0.5 mm and averaged across different cortical depth. Despite these limitations, we found that intracortical average T1 is related to functional connectivity, indicating that functional connectivity is higher between areas with similar intracortical myelin levels. Our results thus extend the aforementioned line of research to the living human brain and support the recognition of microstructural similarity as a general wiring rule of cerebral cortex.
Clearly, we do not mean to imply that microstructure is the sole determinant of connectivity. A well-established wiring rule states that 2 areas are more likely to be connected if they are close to each other in space (Young 1992; Scannell et al. 1995). This distance rule, however, cannot account for the existence of long-range connections and it must be assumed that additional factors are at play. In the current study, we confirm that a relationship exists between Euclidean distance and functional connectivity (Supplementary Fig. 3a ), but ensure that the correlation between functional connectivity and T1 is not driven by spatial proximity (Supplementary Fig. 4). While Euclidean distance is a biased estimate of connection length, entailing significant limitations of inferences drawn on its basis, our findings can be interpreted as a provisional indication that cortical microstructure and spatial distance are independent factors shaping functional connectivity (cf. Goulas et al. 2016a).
Moreover, similarity in cortical thickness (CT) has previously been shown to be related to connectivity in the macaque monkey (Beul et al. 2015) and we find it to be associated with functional connectivity in our sample. Similarly to Beul et al. (2015), we could show that this relationship disappears when controlling for the shared variance of CT and intracortical T1. In contrast, intracortical T1 captures variance in functional connectivity beyond what is explained by CT. Our results suggest that CT can be viewed as a pragmatic surrogate marker for cortical microstructure, capturing some of the same aspects of cortical organization as intracortical T1 (cf. Wagstyl et al. 2015).
Deviations in Transmodal Cortex
While a general link between cortical microstructure and functional connectivity is supported by our data, we also find important deviations. Regions in which functional connectivity and intracortical T1 are least correlated consistently lie in transmodal cortex, especially in posteromedial cortex and the inferior parietal lobule. This pattern persists through different analytic approaches (Figs 3 and 4) and cannot be attributed to the proximity of areas or the distribution of noise in the data (Supplementary Figs 2 and 4).
One possible explanation for the observed deviations are methodological challenges of our MRI-based approach. When visually comparing our T1-based measures of intracortical myelin with histologically derived maps of overall myelin density from Adolf Hopf (recently reviewed in Nieuwenhuys and Broere 2016), the general spatial pattern appears similar. Noteworthy differences, however, exist in posteromedial cortex and the inferior parietal lobule. In Hopf's map, these regions have lower myelin densities than most frontal regions, while the opposite relationship holds for our data (Fig. 4a) as well as for previously published myelin maps (e.g., Glasser and Van Essen 2011; Tardif et al. 2015). Strikingly, the same regions show the weakest link between functional connectivity and intracortical T1 in our study. It is therefore, possible, that the lower correspondence in posteromedial and inferior parietal regions is an artefact of in vivo derived T1-based myelin measures. Moreover, our group-average functional connectivity estimates could be impaired by interindividual differences in functional connectivity patterns, which are relatively high in inferior parietal regions (Mueller et al. 2013), although this does not apply to posteromedial cortex.
In addition to methodological differences, it is important to keep in mind that previous studies were performed in nonhuman primates or lower mammals. Compared with those species, transmodal areas in particular are thought to have undergone massive expansion in the human lineage (Hill et al. 2010). It has been suggested that this rapid expansion, by way of increasing the distance to molecular patterning centers, relaxed developmental constraints and allowed for new properties to emerge in transmodal areas (Buckner and Krienen 2013). Indeed, transmodal, as compared with unimodal, areas show denser and more variable connectivity patterns (Pandya et al. 1988; Mueller et al. 2013). Cortical folds in these areas emerge later during development (Hill et al. 2010), are more variable across individuals (Zilles et al. 1997), and show a less straightforward relationship to architectonically defined boundaries (Fischl et al. 2008). Myelination in transmodal areas also occurs late during development (Flechsig 1920) and remains relatively light in the adult brain. According to Braitenberg (1962), a major function of myelination in cortical gray matter is to inhibit the formation of new connections. Low myelin content in transmodal regions might therefore entail increased synaptic plasticity and make these areas more suitable to support adaptive behavior and learning. Finally, gene expression profiles distinguish resting-state networks (Krienen et al. 2016) and hubs (Vértes et al. 2016) related to sensorimotor functions from those in transmodal association and paralimbic regions.
Taken together, transmodal cortex appears to differ from unimodal areas in displaying more complex and variable connectivity and morphological patterns, as well as molecular and cellular characteristics that point toward increased plasticity. It could therefore be hypothesized that wiring in these regions is governed by flexible mechanisms as well. More rigid wiring rules, such as spatial proximity or microstructural similarity, might consequently have less impact. However, not all of transmodal cortex shows equally low correspondence between T1 and functional connectivity in our analyses. The most pronounced deviations occur in posteromedial cortex and the inferior parietal lobule, in particular the angular gyrus. Interestingly, these regions share a very distinctive connectivity profile. They have been identified as central hubs of structural connectivity in the human cortex (Hagmann et al. 2008) and, unlike most other regions, exhibit both high distant and high local functional connectivity (Sepulcre et al. 2010). Below we discuss how these connectional features might influence the correspondence of functional connectivity to intracortical T1 in posteromedial and inferior parietal cortex.
Investigating Connectivity Patterns Through Spatial Gradients
As described in the previous section, one characteristic of those areas in which functional connectivity and intracortical T1 are least associated is that they have particularly dense and diverse connectivity patterns. For instance, posteromedial cortex has heterogeneous functional and structural connectivity to unimodal, transmodal, and paralimbic areas (Margulies et al. 2009). It is conceivable that only a portion of these connections are explained by microstructural similarity. Only certain features of the functional connectivity patterns would then relate to intracortical T1, but the link is obscured when considering all functional connections of an area at once. We addressed this possibility by decomposing the functional connectivity space into different components, each representing specific aspects of functional connectivity as one dimension. We could thus describe the relationship between the distribution of intracortical T1 and the main modes of variation of connectivity patterns across the cortex.
The first component of this decomposition (FC1), spanning a gradient between primary sensorimotor and transmodal association regions (Fig. 4a, cf. Margulies et al. 2016), showed a stronger relationship to intracortical T1 than the full functional connectivity matrix. Deviations in transmodal regions, especially in posteromedial cortex, were still present in this comparison but substantially alleviated when including 2 additional components of the functional connectivity decomposition (FC 5 and 6) in a simple linear model of intracortical T1 (Figs 5 and 6). Importantly, the added components were not the ones that explained most of the remaining variance, indicating that significant aspects of functional connectivity show no strong relationship to the distribution of intracortical T1. When scrutinizing those particular features which improve the fit to intracortical T1, it appears that FC6 captures fine differences between primary and higher order unimodal sensorimotor areas that are not present in FC1. The most important property of FC5 in the current context could be that it separates postcentral transmodal areas, which showed the strongest deviation from intracortical T1 when using FC1 alone, from transmodal areas in medial and ventrolateral (but not dorsolateral) frontal cortex. It should be noted that the foregoing interpretation of FC components remains tentative, as it is based on only a subset of salient features in the complex spatial patterns of these maps.
We consider decomposing functional connectivity patterns into overlapping gradients, rather than spatially discrete parcels, a promising new approach to investigate cortical organization. Recent work indicates that functional connectivity gradients closely relate to the functional specialization of the cortex (Haak et al. 2016; Margulies et al. 2016). The arrangement of cortical areas in spatial gradients has already been noted by Vogt and Vogt (1919) and Brockhaus (1940) and is fundamental to the evolutionary accounts of cortical organization by Sanides (1962) and Pandya et al. (2015). This concept does not conflict with the existence of areal boundaries, but rather attempts to situate individual areas within an overarching layout. Gradient-based approaches to describe cortical organization might therefore become a fruitful complement to discrete parcellations (e.g Glasser et al. 2016).
Limitations and Future Directions
In this study, we used publicly available high quality MRI data (Gorgolewski et al. 2015; Tardif et al. 2016) and dedicated tools to harness the high spatial resolution (Bazin et al. 2014; Waehnert et al. 2014; Tardif et al. 2015). Nevertheless, our analyses and conclusions are subject to several limitations, which might partly be overcome in future studies.
One caveat is that the high field strength exacerbates the problem of MRI susceptibility artefacts. In our data, both structural and functional images suffer from low signal-to-noise ratio in orbitofrontal and ventral temporal areas. The functional data additionally display strong distortions in these areas, which cannot be corrected completely by nonlinear coregistration to the anatomy. To prevent unreliable signal in respective areas from driving our results, we decided to exclude them from our analyses (ca. 1.5% of surface nodes, see “Masks” section) and all results and statements only pertain to the included regions.
Further, given the distribution of our data (Figs 4 and 6), it might be asked whether linear regression is the appropriate analytic approach. We modeled the data using higher order functions, which, however, did not entail substantial improvements when accounting for model complexity (Supplementary Fig. 6). To address the presence of heteroscedasticity, we used a robust sandwich variance estimator for significance testing (see “Functional Connectivity Decomposition and Comparing Components to T1” section). A more difficult question is, whether the multimodal distribution of the data reflects a principled division of the cortex in different classes which should be treated as discrete clusters. In our opinion, the considerable amount of data points falling between the denser data clouds indicate the existence of gradual changes in cortical features, which would be neglected by clustering approaches (see also “Investigating Connectivity Patterns Through Spatial Gradients” section). We thus favored a regression approach here, but a more in-depth treatment of this question may be subject to future studies.
We pursue a group-average approach using a study-specific template derived with a state-of-the art multimodal surface-based registration algorithm (Tardif et al. 2015). While such approaches have been shown to outperform volumetric and single modality registration techniques (Robinson et al. 2014; Tardif et al. 2015), intersubject averaging inevitably introduces inaccuracies in areas with high interindividual differences. Prospective work might circumvent this issue by investigating the relationship of functional connectivity and intracortical myelin within individual subjects (cf. Glasser et al. 2016).
Our estimate of intracortical myelin content takes into account the dependence of layer thickness on cortical morphology and minimizes partial volume effects. However, we use an intracortical average value, while classic myeloarchitectonic techniques are largely based on the differential distribution of myelinated fibers across the different cortical laminae (e.g., Vogt and Vogt 1919). Radial differences in myelin content can be comparable in size to differences between cortical areas (Lutti et al. 2014) and quantitative analyses of myelin or cell density profiles, running perpendicular to the cortical surface, have been used for observer-independent definition of areal boundaries in postmortem data (Schleicher et al. 1999; Annese et al. 2004; Eickhoff et al. 2005). Initial approaches to leverage the radial distribution of intracortical T1 in MR images in a similar way show promise that this information can be used to distinguish cortical areas (Dinse et al. 2015, Marques et al. 2016; Waehnert et al. 2016). Methods to extract and analyze such T1 profiles are still in their infancy, but more elaborate and robust approaches are likely to emerge from this line of research soon. Similarly, first studies demonstrating layer-resolved fMRI contrast (Goense et al. 2012; Hubert et al. 2015; Guidi et al. 2016) raise the possibility of differentiating functional connectivity between input and output layers. While the current study uncovers a general link between functional connectivity and intracortical T1, we expect that increasing spatial resolution in MRI data (Gallichan et al. 2015; Stucht et al. 2015) and improved image processing techniques will make it possible to refine this observation to the scale of individual laminar compartments in the near future.
Conclusion
The current resurgence of interest in human cortical anatomy is closely intertwined with methodological advances, providing the tools to investigate it in vivo. Some well-studied phenomena, emerging from decades of research in experimental animals, can now be explored noninvasively in the human brain. In the current study, we focus on a link between cortical microstructure and connectivity, which has consistently been described in animal studies, but until now had not been investigated in the living human brain. We demonstrate a systematic relationship between T1-based intracortical myelin content and resting-state functional connectivity. Our results support the view that microstructural similarity represents a general wiring rule of mammalian cortex. Although methodological difficulties persist and questions about underlying mechanisms remain unresolved, our findings line up with a growing body of work targeted at demystifying the complex connectivity patterns of human cerebral cortex.
Supplementary Material
Notes
Conflict of Interest: None declared.
Supplementary Material
Supplementary material are available at Cerebral Cortex online.
Funding
Data for the confirmatory analysis were provided by the HCP, and the Washington University, University of Minnesota, and Oxford University Consortium (Principal Investigators David Van Essen and Kamil Ugurbil; grant 1U54MH091657) funded by 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research, and the McDonnell Center for Systems Neuroscience at Washington University. S.S.G. was partially supported by NIH grants 1R01EB020740-01A1, 1P41EB019936-01A1, 3R01MH092380-04S2, and 1U01MH108168-01. A.G. was supported by a Humboldt research fellowship from the Alexander von Humboldt Foundation.
References
- Abraham A, Pedregosa F, Eickenberg M, Gervais P, Mueller A, Kossaifi J, Gramfort A, Thirion B, Varoquaux G. 2014. Machine learning for neuroimaging with scikit-learn. Front Neuroinform. 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Osada T, Sporns O, Watanabe T, Matsui T, Miyamoto K, Miyashita Y. 2012. Functional connectivity between anatomically unconnected areas is shaped by collective network-level effects in the macaque cortex. Cereb Cortex. 22(7):1586–1592. [DOI] [PubMed] [Google Scholar]
- Annese J, Pitiot A, Dinov ID, Toga AW. 2004. A myelo-architectonic method for the structural classification of cortical areas. Neuroimage. 21(1):15–26. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 12(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 54(3):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas H, Pandya DN. 1989. Architecture and intrinsic connections of the prefrontal cortex in the rhesus monkey. J Comp Neurol. 286:353–375. [DOI] [PubMed] [Google Scholar]
- Barbas H. 2015. General cortical and special prefrontal connections: principles from structure to function. Annu Rev Neurosci. 38(1):269–289. [DOI] [PubMed] [Google Scholar]
- Bazin PL, Vezien JM. 2005. Integration of geometric elements, Euclidean relations, and motion curves for parametric shape and motion estimation. IEEE Trans Pattern Anal. 27(12):1960–1976. [DOI] [PubMed] [Google Scholar]
- Bazin PL, Weiss M, Dinse J, Schäfer A, Trampel R, Turner R. 2014. A computational framework for ultra-high resolution cortical segmentation at 7Tesla. Neuroimage. 93(Pt 2):201–209. [DOI] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. 2007. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 37(1):90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beul SF, Grant S, Hilgetag CC. 2014. A predictive model of the cat cortical connectome based on cytoarchitecture and distance. Brain Struct Funct. 220(6):3167–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beul SF, Barbas H, Hilgetag CC. 2015. A predictive structural model of the primate connectome. arXiv:151107222. [DOI] [PMC free article] [PubMed]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 34(4):537–541. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, et al. 2010. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 107(10):4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock NA, Kocharyan A, Liu JV, Silva AC. 2009. Visualizing the entire cortical myelination pattern in marmosets with magnetic resonance imaging. J Neurosci Methods. 185(1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok S. 1929. Der Einfluß der in den Furchen und Windungen auftretenden Krümmungen der Großhirnrinde auf die Rindenarchitektur. Z Gesamte Neurol Psychiatr. 12:682–750. [Google Scholar]
- Braitenberg V. 1962. A note on myeloarchitectonics. J Comp Neurol. 118(2):141–156. [DOI] [PubMed] [Google Scholar]
- Bridge H, Clare S, Jenkinson M, Jezzard P, Parker AJ, Matthews PM. 2005. Independent anatomical and functional measures of the V1/V2 boundary in human visual cortex. J Vision. 5(2):93–102. 2.1. [DOI] [PubMed] [Google Scholar]
- Brockhaus H. 1940. Die Cyto-und Myeloarchitektonik des Cortex claustralis und des Claustrum beim Menschen. J Psychol Neurol. 49(4–6):249–348. [Google Scholar]
- Brodmann K. 1909. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Leipzig (Germany): Johann Ambrosius Barth. [Google Scholar]
- Buckner RL, Krienen FM. 2013. The evolution of distributed association networks in the human brain. Trends Cogn Sci. 17(12):1–18. [DOI] [PubMed] [Google Scholar]
- Coifman RR, Lafon S. 2006. Diffusion maps. Appl Comput Harmon Anal. 21(1):5–30. [Google Scholar]
- Connor JR, Menzies SL. 1996. Relationship of iron to oligodendrocytes and myelination. Glia. 17:83–93. [DOI] [PubMed] [Google Scholar]
- Cox R. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 29(29):162–173. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 9(2):179–194. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. 2006. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 103(37):13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick F, Tierney AT, Lutti A, Josephs O, Sereno MI, Weiskopf N. 2012. In vivo functional and myeloarchitectonic mapping of human primary auditory areas. J Neurosci. 32(46):16095–16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinse J, Haertwich N, Waehnert MD, Tardif CL, Schäfer A, Geyer S, Preim B, Turner R, Bazin PL. 2015. A cytoarchitecture-driven myelin model reveals area-specific signatures in human primary and secondary areas using ultra-high resolution in-vivo brain MRI. Neuroimage. 114:71–87. [DOI] [PubMed] [Google Scholar]
- Dousset V, Grossman RI, Ramer KN, Schnall MD, Young LH, Gonzalez-Scarano F, Lavi E, Cohen JA. 1992. Experimental allergic encephalomyelitis and multiple sclerosis: lesion characterization with magnetization transfer imaging. Radiology. 182(2):483–491. [DOI] [PubMed] [Google Scholar]
- Droettboom M, Caswell TA, Firing E, McDougall D, Ivanov P, Giuca M, Seppänen JK, Evans J, Cimarron, Silvester S, Nielsen JH, et al. 2016. matplotlib: matplotlib v1.5.1. 10.5281/zenodo.44579 [DOI]
- von Economo C, Koskinas G. 1925. Die Cytoarchitektonik der Hirnrinde des erwachsenen Menschen. Wien (Austria): Springer. [Google Scholar]
- Eickhoff S, Walters NB, Schleicher A, Kril J, Egan GF, Zilles K, Watson JDG, Amunts K. 2005. High-resolution MRI reflects myeloarchitecture and cytoarchitecture of human cerebral cortex. Hum Brain Mapp. 24(3):206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis: II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 9(2):195–207. [DOI] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, Augustinack J, Hinds O, Yeo BTT, Mohlberg H, Amunts K, Zilles K. 2008. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 18(8):1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechsig PE. 1920. Anatomie des menschlichen Gehirns und Rückenmarks auf myelogenetischer Grundlage. Leipzig (Germany): Thieme. [Google Scholar]
- Fukunaga M, Li TQ, van Gelderen P, de Zwart JA, Shmueli K, Yao B, Lee J, Maric D, Aronova MA, Zhang G, et al. 2010. Layer-specific variation of iron content in cerebral cortex as a source of MRI contrast. Proc Natl Acad Sci USA. 107(8):3834–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichan D, Marques JP, Gruetter R. 2015. Retrospective correction of involuntary microscopic head movement using highly accelerated fat image navigators (3D FatNavs) at 7T. Magn Reson Med. 1039:1030–1039. [DOI] [PubMed] [Google Scholar]
- Geyer S, Weiss M, Reimann K, Lohmann G, Turner R. 2011. Microstructural parcellation of the human cerebral cortex – from brodmann's post-mortem map to in vivo mapping with high-field magnetic resonance imaging. Front Hum Neurosci. 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. 2011. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 31(32):11597–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, et al. 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Goyal MS, Preuss TM, Raichle ME, Van Essen DC. 2014. Trends and properties of human cerebral cortex: Correlations with cortical myelin content. Neuroimage. 93:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. 2016. A multi-modal parcellation of human cerebral cortex. Nature. 536:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goense J, Merkle H, Logothetis NK. 2012. High-resolution fMRI reveals laminar differences in neurovascular coupling between positive and negative BOLD responses. Neuron. 76(3):629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO. 2011. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Front Neuroinform. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Mendes N, Wilfling D, Wladimirow E, Gauthier CJ, Bonnen T, Ruby FJ, Trampel R, Bazin PL, Cozatl R, et al. 2015. A high resolution 7-Tesla resting-state fMRI test-retest dataset with cognitive and physiological measures. Sci Data. 2:140054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A, Uylings HBM, Hilgetag CC. 2016. a. Principles of ipsilateral and contralateral cortico-cortical connectivity in the mouse. Brain Struct Funct. 10.1007/s00429-016-1277-y. [DOI] [PubMed] [Google Scholar]
- Goulas A, Werner R, Beul SF, Saering D, van den Heuvel M, Triarhou LC, Hilgetag CC. 2016. b. Cytoarchitectonic similarity is a wiring principle of the human connectome. bioRxiv. 10.1101/068254. [DOI] [Google Scholar]
- Greve DN, Fischl B. 2009. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 48(1):63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi M, Huber L, Lampe L, Gauthier CJ, Moeller HE. 2016. Lamina-dependent calibrated BOLD response in human primary motor cortex. Neuroimage. 141:250–261. [DOI] [PubMed] [Google Scholar]
- Haak KV, Marquand AF, Beckmann CF. 2016. Connectopic mapping with resting-state fMRI. arXiv:160207100. [DOI] [PubMed]
- Haast RAM, Ivanov D, Formisano E, Uludag K. 2016. Reproducibility and reliability of quantitative and weighted T1 and T2∗ mapping for myelin-based cortical parcellation at 7 Tesla. Front Neuroanat. 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Van Wedeen J, Sporns O. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6(7):1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermundstad AM, Bassett DS, Brown KS, Aminoff EM, Clewett D, Freeman S, Frithsen A, Johnson A, Tipper CM, Miller MB, et al. 2013. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc Natl Acad Sci USA. 110(15):6169–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, Feldman Barrett L, Hilgetag CC, de Reus MA. 2015. Bridging cytoarchitectonics and connectomics in human cerebral cortex. J Neurosci. 35(41):13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Scholtens LH, de Reus MA, Kahn RS. 2016. Associated microscale spine density and macroscale connectivity disruptions in schizophrenia. Biol Psychiatry. 80(4):293–301. [DOI] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D. 2010. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci USA. 107(29):13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci USA. 106(6):2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf A. 1955. Über die Verteilung myeloarchitektonischer Merkmale in der isokortikalen Schläfenlappenrinde beim Menschen. J Hirnforsch. 2(1):36–54. [Google Scholar]
- Hopf A. 1956. Über die Verteilung myeloarchitektonischer Merkmale in der Stirnhirnrinde beim Menschen. J Hirnforsch. 2(4):311–333. [PubMed] [Google Scholar]
- Hopf A, Vitzthum HG. 1957. Über die Verteilung myeloarchitektonischer Merkmale in der Scheitellappenrinde beim Menschen. J Hirnforsch. 3(2/3):83–104. [PubMed] [Google Scholar]
- Huber L, Goense J, Kennerley AJ, Trampel R, Guidi M, Reimer E, Ivanov D, Neef N, Gauthier CJ, Turner R, et al. 2015. Cortical lamina-dependent blood volume changes in human brain at 7T. Neuroimage. 107:23–33. [DOI] [PubMed] [Google Scholar]
- Hurley AC, Al-Radaideh A, Bai L, Aickelin U, Coxon R, Glover P, Gowland PA. 2010. Tailored RF pulse for magnetization inversion at ultrahigh field. Magn Reson Med. 63(1):51–58. [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P, et al. 2001. Scipy: Open source scientific tools for python. Online resource: http://www.scipy.org/ (19 July 2016, last date accessed).
- Krienen FM, Yeo BTT, Ge T, Buckner RL, Sherwood CC. 2016. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci USA. 113(4):E469–E478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langs G, Golland P, Ghosh SS. 2015. a. Predicting activation across individuals with resting-state functional connectivity based multi-atlas label fusion. Med Image Comput Comput Assist Interv. 9350:313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langs G, Wang D, Golland P, Mueller S, Pan R, Sabuncu MR, Sun W, Li K, Liu H. 2015. b. Identifying shared brain networks in individuals by decoupling functional and anatomical variability. Cereb Cortex. 26(10):4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas BC, Bogovic JA, Carass A, Bazin PL, Prince JL, Pham DL, Landman BA. 2010. The Java Image Science Toolkit (JIST) for rapid prototyping and publishing of neuroimaging software. Neuroinformatics. 8(1):5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutti A, Dick F, Sereno MI, Weiskopf N. 2014. Using high-resolution quantitative mapping of R1 as an index of cortical myelination. Neuroimage. 93:176–188. [DOI] [PubMed] [Google Scholar]
- Mackay A, Whittall K, Adler J, Li D, Paty D, Graeb D. 1994. In vivo visualization of myelin water in brain by magnetic resonance. Magn Reson Med. 31(10):673–677. [DOI] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M. 2009. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci USA. 106(47):20069–20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, Bezgin G, Petrides M, Jefferies E, Smallwood J. 2016. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 113(44):12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R. 2010. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage. 49(2):1271–1281. [DOI] [PubMed] [Google Scholar]
- Marques JP, Khabipova D, Gruetter R.. 2016. Studying cyto and myeloarchitecture of the human cortex at ultra-high field with quantitative imaging: R1, R2* and susceptibility. Neuroimage. 147:152–163. [DOI] [PubMed] [Google Scholar]
- McAuliffe MJ, Lalonde FM, McGarry D, Gandler W, Csaky K, Trus BL. 2001. Medical image processing, analysis & visualization in clinical research. Comp Med Sci. 14th IEEE:381–386.
- McKinney W. 2010. Data structures for statistical computing in python. In: van der Walt S, Millman J, editors, Proceedings of the 9th Python in Science Conference. p. 51–56.
- Miranda-Dominguez O, Mills BD, Grayson D, Woodall A, Grant KA, Kroenke CD, Fair DA. 2014. Bridging the gap between the human and macaque connectome: a quantitative comparison of global interspecies structure-function relationships and network topology. J Neurosci. 34(16):5552–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BTT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H. 2013. Individual variability in functional connectivity architecture of the human brain. Neuron. 77(3):586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuys R. 2013. The myeloarchitectonic studies on the human cerebral cortex of the Vogt-Vogt school, and their significance for the interpretation of functional neuroimaging data. Brain Struct Funct. 218(2):303–352. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R, Broere CAJ. 2016. A map of the human neocortex showing the estimated overall myelin content of the individual architectonic areas based on the studies of Adolf Hopf. Brain Struct Funct. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya DN, Sanides F. 1973. Architectonic parcellation of the temporal operculum in rhesus monkey and its projection pattern. Zeit Anat Entwicklungs. 161:127–161. [DOI] [PubMed] [Google Scholar]
- Pandya DN, Yeterian EH. 1985. Architecture and connections of cortical association areas In: Peters A, Jones EG, editors. Association and auditory cortices. Cerebral cortex, Vol 4 Boston (MA): Springer US. [Google Scholar]
- Pandya DN, Seltzer B, Barbas H. 1988. Input-output organization of the primate cerebral cortex In: Steklis D, Erwin J, editors. Comparative primate biology 4: Neurosciences. New York (NY): Alan R. Liss. [Google Scholar]
- Pandya DN, Yeterian EH. 1990. Prefrontal cortex in relation to other cortical areas in rhesus monkey: architecture and connections In: Uylings H, van Eden C, de Bruin JPC, Feenstra M, Pennartz C, editors. The prefrontal cortex: its structure, function and pathology. Progress in brain research, Vol. 85 Amsterdam (Netherlands): Elsevier. [DOI] [PubMed] [Google Scholar]
- Pandya DN, Seltzer B, Petrides M, Cipolloni PB. 2015. Cerebral cortex: architecture, connections, and the dual origin concept. Oxford (UK): Oxford University Press. [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res. 12:2825–2830. [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M. 2014. MSM: a new flexible framework for Multimodal Surface Matching. Neuroimage. 100:414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche A. 2011. A four-dimensional registration algorithm with application to joint correction of motion and slice timing in fMRI. IEEE Trans Med Imaging. 30(8):1546–1554. [DOI] [PubMed] [Google Scholar]
- Sanides F. 1962. Die Architektonik des menschlichen Stirnhirns. Berlin-Heidelberg (Germany): Springer. [Google Scholar]
- Scannell JW, Blakemore C, Young MP. 1995. Analysis of connectivity in the cat cerebral cortex. J Neurosci. 15(2):1463–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher A, Amunts K, Geyer S, Morosan P, Zilles K. 1999. Observer-independent method for microstructural parcellation of cerebral cortex: a quantitative approach to cytoarchitectonics. Neuroimage. 9(1):165–177. [DOI] [PubMed] [Google Scholar]
- Scholtens LH, Schmidt R, de Reus MA, van den Heuvel MP. 2014. Linking macroscale graph analytical organization to microscale neuroarchitectonics in the macaque connectome. J Neurosci. 34(36):12192–12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold S, Perktold J. 2010. Statsmodels: Econometric and statistical modeling with python. In: Proceedings of the 9th Python in Science Conference.
- Sepulcre J, Liu H, Talukdar T, Martincorena I, Yeo BTT, Buckner RL. 2010. The organization of local and distant functional connectivity in the human brain. Plos Comput Biol. 6(6):e1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereno MI, Lutti A, Weiskopf N, Dick F. 2013. Mapping the human cortical surface by combining quantitative T1 with retinotopy. Cereb Cortex. 23(9):2261–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AMC, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, et al. 2009. The resting brain: unconstrained yet reliable. Cereb Cortex. 19(10):2209–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalovsky IS, Fischl B, Melcher JR. 2006. Mapping an intrinsic MR property of gray matter in auditory cortex of living humans: a possible marker for primary cortex and hemispheric differences. Neuroimage. 32(4):1524–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skudlarski P, Jagannathan K, Calhoun VD, Hampson M, Skudlarska BA, Pearlson G. 2008. Measuring brain connectivity: diffusion tensor imaging validates resting state temporal correlations. Neuroimage. 43(3):554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber C, Morawski M, Schäfer A, Labadie C, Wähnert M, Leuze C, Streicher M, Barapatre N, Reimann K, Geyer S, et al. 2014. Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. Neuroimage. 93(Pt 1):95–106. [DOI] [PubMed] [Google Scholar]
- Stucht D, Danishad KA, Schulze P, Godenschweger F, Zaitsev M, Speck O. 2015. Highest resolution in vivo human brain MRI using prospective motion correction. Plos One. 10(7):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif CL, Schäfer A, Waehnert M, Dinse J, Turner R, Bazin PL. 2015. Multi-contrast multi-scale surface registration for improved alignment of cortical areas. Neuroimage. 111:107–122. [DOI] [PubMed] [Google Scholar]
- Tardif CL, Schäfer A, Trampel R, Villringer A, Turner R, Bazin PL. 2016. Open science CBS neuroimaging repository: sharing ultra-high-field MR images of the brain. Neuroimage. 124:1143–1148. [DOI] [PubMed] [Google Scholar]
- Tenenbaum JB, de Silva V, Langford JC. 2000. A global geometric framework for nonlinear dimensionality reduction. Science. 290:5500. [DOI] [PubMed] [Google Scholar]
- Todorich B, Pasquini JM, Garcia CI, Paez PM, Connor JR. 2009. Oligodendrocytes and myelination: The role of iron. Glia. 57:467–478. [DOI] [PubMed] [Google Scholar]
- Turner R. 2015. Myelin imaging In: Toga AW, editor. Brain mapping: an encyclopedic reference, Vol. 1 Amsterdam (Netherlands): Elsevier. Acquisition Methods, Methods and Modeling. [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. 2010. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 29(6):1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, WU-Minn HCP Consortium . 2013. The WU-Minn human connectome project: an overview. Neuroimage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vértes PE, Rittman T, Whitaker KJ, Romero-Garcia R, Váša F, Kitzbichler M, Fonagy P, Dolan RJ, Jones PB, Goodyer IM, NSPN Consortium, Bullmore ET . 2016. Gene transcription profiles associated with intra-modular and inter-modular hubs in human fMRI networks. Philos Trans R Soc Lond B Biol Sci. 371:735–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. 2007. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 447(7140):83–86. [DOI] [PubMed] [Google Scholar]
- Vogt C, Vogt O. 1919. Allgemeinere Ergebnisse unserer Hirnforschung. J Psychol Neurol. 25:279–468. [Google Scholar]
- Waehnert MD, Dinse J, Weiss M, Streicher MN, Waehnert P, Geyer S, Turner R, Bazin PL. 2014. Anatomically motivated modeling of cortical laminae. Neuroimage. 93(Pt 2):210–220. [DOI] [PubMed] [Google Scholar]
- Waehnert MD, Dinse J, Schäfer A, Geyer S, Bazin PL, Turner R, Tardif CL. 2016. A subject-specific framework for in vivo myeloarchitectonic analysis using high resolution quantitative MRI. Neuroimage. 125:94–107. [DOI] [PubMed] [Google Scholar]
- van der Walt S, Colbert SC, Varoquaux G. 2011. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng. 13(2):22–30. [Google Scholar]
- Wagstyl K, Ronan L, Goodyer IM, Fletcher PC. 2015. Cortical thickness gradients in structural hierarchies. Neuroimage. 111:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom M, St-Jean S, Evans C, Warmenhoven J, Meyer K, Martin M, Rocher L, Hobson P, Bachant P, Nagy T, et al. 2016. seaborn: v0.7.0 (January 2016). 10.5281/zenodo.45133 [DOI]
- Weiskopf N, Mohammadi S, Lutti A, Callaghan MF. 2015. Advances in MRI-based computational neuroanatomy: from morphometry to in-vivo histology. Curr Opin Neurol. 28(4):313–322. [DOI] [PubMed] [Google Scholar]
- White H. 1980. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica. 48(4):817–838. [Google Scholar]
- Young MP. 1992. Objective analysis of the topological organization of the primate cortical visual system. Nature. 358:152–155. [DOI] [PubMed] [Google Scholar]
- Zilles K, Schleicher A, Langemann C, Amunts K, Morosan P, Palomero-Gallagher N, Schormann T, Mohlberg H, Bürgel U, Steinmetz H, et al. 1997. Quantitative analysis of sulci in the human cerebral cortex: Development, regional heterogeneity, gender difference, asymmetry, intersubject variability and cortical architecture. Hum Brain Mapp. 5(4):218–221. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Xing XX. 2014. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci Biobehav Rev. 45:100–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.