Abstract
Agricultural productivity is severely hampered by drought in many parts of the globe. It is well-known that wild plant species can tolerate drought better when compared with their closely related cultivated plant species. Better drought adaptation of wild species over cultivated ones is accounted for their ability to differentially regulate gene expression. miRNAs, known to regulate gene expression at the post-transcriptional level, are admitted to play an important role in plant adaptation to stresses. This study aims at evaluating miRNA dynamics in a drought-tolerant wild Ipomoea campanulata L. and drought-sensitive cultivated Jacquemontia pentantha (Jacq.) of the family Convolvulaceae under ex situ drought. Sequencing profiles revealed that 34 conserved miRNA families were analogous between the two species. Drought altered expression levels of several of these miRNAs in both the species. Drought-tolerant I. campanulata showed upregulation of miR398, miR168, miR858, miR162 and miR408, while miR394 and miR171 were downregulated. Drought-sensitive J. pentantha showed upregulation of miR394, miR156, miR160, miR164, miR167, miR172, miR319, miR395, miR396, miR403 and downregulation of miR157. Basal miRNA levels and their drought mediated regulation were very different between the two species. Differential drought sensitivities of these two plant species can be attributed to these innate variations in miRNA levels and their expression.
Keywords: Convolvulaceae, Drought, MiRNA dynamics, Wild and cultivated species
Introduction
Drought severely reduces crop yield world-wide and with changing climate, its probability of occurrence is predicted to rise (Yinpeng et al. 2009). One of the strategies for improving drought tolerance of crop plants is to explore the drought tolerance mechanisms in tolerant plants (Tuberosa and Salvi 2006; Ashraf 2010). Cultivated plants are grown in benign environments for higher yields where they hardly confront water shortage, making them to become drought-sensitive over the years (Halpin 2005; Tuberosa and Salvi 2006). By contrast, wild plants, found naturally in arid and semi-arid regions, grow under highly fluctuating levels of water availability, which makes them to express innate tolerance mechanisms that are not found in cultivated plants (Akashi et al. 2008). Attempts were made to unravel such mechanisms using gene or protein expression profiles but such efforts have mostly utilized crop plants. Rarely a crop plant or a cultivated plant’s response is being compared to a related wild species. Moreover, post-transcriptional gene regulation is a less understood phenomenon compared to the transcriptional regulation in response to drought. Several recent studies have shown that miRNAs that act as post-transcriptional regulators are regulated by drought and other abiotic stresses revealing a new layer of gene regulation that is very important for adapting to stress conditions (Sunkar et al. 2007, 2012; Ghorecha et al. 2013). Furthermore, miRNA regulation differs between drought sensitive and tolerant genotypes under stress (Kulcheski et al. 2011).
Thus far, 8496 mature miRNAs are identified from 74 plant species (miRBase release-21) (http://www.mirbase.org/cgi-bin/browse.pl). Most of these miRNAs are identified from model and crop species. There is little information available for miRNAs from non-model wild species. Compared with the cultivated plant species, naturally occurring, closely related wild species exhibit tolerance to stress. Identifying such differential tolerance mechanisms could assist in improving the performance of cultivated species. We previously compared the differences in selected biochemical parameters along with the expression analysis of some of the conserved miRNAs using small RNA blot analysis in wild, drought-tolerant (Ipomoea campanulata L.) and cultivated but drought-sensitive Jacquemontia pentantha (Jacq.) growing under in situ conditions (Ghorecha et al. 2014). The present study has been carried out to evaluate miRNAs dynamics using sequencing-based strategy by identifying the entire spectrum of conserved miRNAs as well as their responsiveness to drought in ex situ grown wild and cultivated species (I. campanulata and J. pentantha respectively), belonging to the same family, Convolvulaceae.
Materials and methods
Experiments were carried out in greenhouse with no artificial day length control. Day and night temperatures were maintained at 35 and 25 °C respectively ±1 °C. Stem cuttings of similar length and girth were obtained from healthy and non-stressed natural populations of I. campanulata and J. pentantha (Ghorecha et al. 2014). Cuttings were trimmed to remove the leaves (except 2–3 at the tip). Lower ends of these cuttings were dipped in Rootone (containing IBA) to promote rooting. They were planted in standard pots (32 cm diameter top, 25 cm diameter base and 25 cm depth) filled with homogenous garden soil. Potted plants for each species were watered daily to the soil capacity and allowed to grow for 2 months so as to ensure that all of the clones have attained uniform size. At this stage, most of the plants of I. campanulata and J. pentantha showed 13–15 and 35–37 leaves respectively. Five pots each, for both control and drought stress, were arranged in a complete randomized block design. Drought stress was applied by withholding water supply for 2 days, as at this stage cultivated species showed leaf dropping. There was no change in the watering cycle of control plants. As an indicator of stress, leaf relative water content (RWC) was measured in mature leaves (3rd, 5th and 7th from tip) of both the species under control and drought stress, according to Catsky (1960). For measuring RWC, five leaves each from control and stressed plants were weighed immediately after sampling to measure the Fresh weight (FW). These leaves were immersed in distilled water for 5 h and Turgid weight (TW) was measured. The leaves were then oven-dried at 60 °C for 2 days and Dry weight (DW) was measured. RWC was calculated using the formula RWC = (FW − DW)/(TW − DW) × 100.
Total RNA was extracted from mature leaves (6th from tip) of control and stressed plants using TRIzol reagent (Invitrogen). The quality and concentration of RNA was measured with the help of 1% agarose gel and ND1000 spectrophotometer (Thermo Scientific). LC Sciences (http://www.lcsciences.com, Houston, TX, USA) has sequenced small RNAs using Illumina platform. Small RNAs were selected in accordance with standard quality control of LC Sciences and reads containing clear adaptor sequences were further processed. Using in-house written software, the adaptor sequences were removed to obtain 18–30 nt small RNAs for which both total and unique read numbers were established (Jagadeeswaran et al. 2012). Non-coding RNAs such as rRNAs, tRNAs, snRNAs and snoRNAs were removed from the unique reads and then using BLASTn, small RNA reads were searched against the known plant miRNAs in the database (miRBase version 21 available at http://microrna.sanger.ac.uk/sequences/). Small RNA sequences that aligned to the known plant miRNAs in miRBase were identified as conserved miRNAs. For comparing the differential expression of miRNAs in wild and cultivated species towards drought stress, the normalized miRNA expression levels (actual miRNA reads*1,000,000/total count of clean reads) were used. To further validate the sequencing profiles, small RNA blot analysis was performed as described (Jagadeeswaran et al. 2012).
Results and discussion
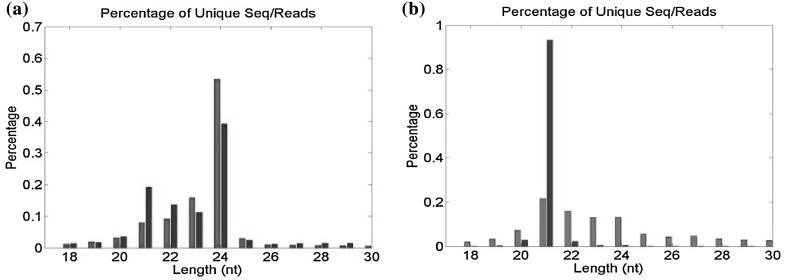
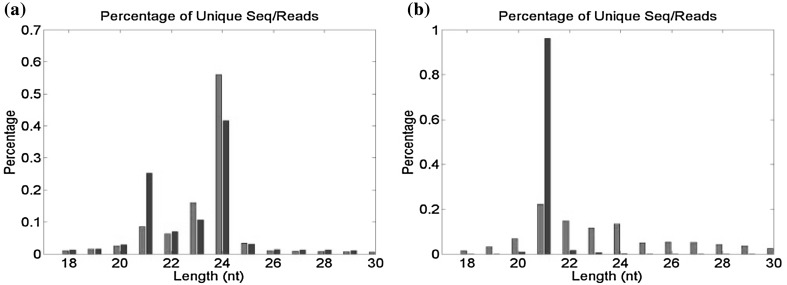
Upon exposure to drought stress, leaf wilting was observed in J. pentantha while no such symptoms were seen in I. campanulata. At this stage, leaf RWC of drought-stressed J. pentantha and I. campanulata were 55 and 40% lower than the control plants, respectively. Lesser decrease in RWC in I. campanulata is indicative of its ability to hold water during drought stress, making it as drought-tolerant compared to J. pentantha. Similarly, it was reported that drought tolerance is associated with the capacity to maintain high RWC levels (Stoyanov 2005; Keyvan 2010). Differential drought tolerance of wild and cultivated species of Helianthus annuus was correlated with the differences in gene expression profiles (Mayrose et al. 2011). Gene expression is orchestrated at both transcriptional and post-transcriptional level (Vaucheret and Fagard 2001). At the post-transcriptional level, the messenger RNAs can rapidly be regulated by selectively repressing the translation of some mRNAs while allowing the others to express (Holcik and Sonenberg 2005; Leung and Sharp 2007). As miRNAs are crucial post-transcriptional regulators of plant development and stress responses (Yang et al. 2007; Sunkar et al. 2012), their dynamics have been analyzed here in I. campanulata and J. pentantha grown under control and drought. The sequences obtained from small RNA libraries are presented in Table 1. The obtained total small RNA reads from control and drought-stressed libraries ranged between 4 and 8 million (Table 1). The abundances of unique 18–30 nt small RNA reads were almost similar in the control libraries of both the species. In the controls of both the species, highest reads of total unique small RNAs were observed for 24-nt size class followed by 23-nt size class (Figs. 1a, 2a). The abundance of small RNA reads in the control libraries of the two species was comparable to those reported in Lagenaria siceraria, Cucurbita moschata, Cucurbita pepo and watermelon (Jagadeeswaran et al. 2012). However, control and drought stress libraries of Medicago truncatula and Hordeum vulgare were reported to show the highest peak of small RNA reads at 24-nt and second highest reads for 21-nt size class (Wang et al. 2011; Hackenberg et al. 2015). Similar kind of size distribution of drought stressed small RNA libraries of the two species in this study have been observed with the highest peak of total small RNA reads at 24-nt followed by 21-nt size class (Figs. 1a, 2a). It indicated that drought stressed libraries of the two species showed the second highest abundance of unique 21-nt small RNAs, which was seen for 23-nt size class in the control libraries. The 21-nt class of small RNAs is typically composed of miRNAs and tasiRNAs (trans-acting short interfering RNAs) (Montes et al. 2014). Amongst the unique miRNA reads, the size distribution revealed the highest abundance for 21-nt size class in both control and drought libraries of both the species (Figs. 1b, 2b). The abundance of small RNAs in this class (21-nt) was prominently higher in drought libraries of both the species as compared to control libraries. Overall, results from both the species suggested altered miRNA levels under drought exposure.
Table 1.
Statistics of small RNA reads for I. campanulata control library (IcC); I. campanulata drought stressed library (IcD); J. pentantha control library (JpC) and J. pentantha drought stressed library (JpD)
| Category | IcC | IcD | JpC | JpD | ||||
|---|---|---|---|---|---|---|---|---|
| Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | Unique reads | Total reads | |
| miRNAs | 2981 | 628,066 | 2351 | 394,931 | 1755 | 682,937 | 2217 | 557,632 |
| pre-miRNAs | 5741 | 676,322 | 4414 | 420,180 | 4185 | 699,640 | 5072 | 566,222 |
| ncRNAs | 84,596 | 1,128,955 | 92,895 | 823,379 | 112,212 | 1,971,704 | 85,376 | 980,687 |
| repeats | 34,600 | 244,242 | 34,771 | 199,246 | 38,727 | 699,137 | 34,360 | 2,38,450 |
| Total | 3,482,433 | 7,814,660 | 2,010,241 | 4,610,207 | 1,343,665 | 4,973,757 | 2,340,667 | 5,333,042 |
Fig. 1.
Size distribution of total unique small RNA (a) and unique mature miRNA (b) reads identified from control (blue bars) and drought stressed (red bars) libraries of I. campanulata
Fig. 2.
Size distribution of total unique small RNA (a) and unique mature miRNA (b) reads identified from control (blue bars) and drought stressed (red bars) libraries of J. pentantha
Although identification of miRNAs is difficult for plants lacking genome sequence information, the miRNA homology search can identify miRNAs from such plants (Zhang et al. 2006). Deep sequencing of small RNAs was effectively used to identify conserved miRNAs from plants that lack genome information (Wang et al. 2012; Guzman et al. 2012; Lukasik et al. 2013). In the present study, small RNAs from leaves of control and drought-exposed I. campanulata and J. pentantha were sequenced. The sequence analysis revealed the identification of 41 conserved miRNA families in I. campanulata (represented by 213 and 177 miRNAs in control and drought-stressed libraries, respectively) (Table 2). In the case of J. pentantha, 35 conserved miRNA families (150 and 176 miRNAs from control and drought-stressed libraries, respectively) have been identified (Table 2). Overall, 34 miRNA families overlapped between these two plant species and the remaining miRNAs (miR169, miR447, miR473, miR477, miR4995, miR5141, miR530-5 and miR3630-3) were only identified in either of the species which had relatively lower expression (Table 2). In Eugenia uniflora and Vigna mungo (lacking genome sequence information), a total of 45 (consisting 204 miRNAs) and 19 (consisting 45 miRNAs) conserved miRNA families were identified respectively (Guzman et al. 2012; Paul et al. 2014). Number of miRNAs expressed in drought stress library was lower than control in I. campanulata and vice versa in J. pentantha. Differential expression of miRNAs and their profiles, in control and drought treated libraries of tolerant and sensitive species was reported previously by attributing the variation to species-specific expression (Candar-Cakir et al. 2016). Expression of miR477 was specifically identified in the wild species, I. campanulata. Similar to this observation, expression of miR477 was reported in leaves of wild progenitor of Cassava as compared to the cultivated ones (Chen et al. 2015). miR477 was identified from Populus trichocarpa, Vitis vinifere and Nicotiana tabaccum (Lu et al. 2005; Jaillon et al. 2007; Tang et al. 2012). In P trichocarpa, miR477 is predicted to target a member of the GRAS gene family and a NAC-domain mRNA that are largely involved in developmental patterning (Lu et al. 2005; Laufs et al. 2004; Mallory et al. 2004). Similar kind of inferences can be extended to the expression of miR477 observed in the wild species I. campanulata of this study.
Table 2.
List of miRNA families identified from control and drought stress libraries of I. campanulata (Ic) and J. pentantha (Jp)
| S.No. | miRNA families | Normalized miRNA reads (RPTM) of Ic | Normalized miRNA reads (RPTM) of Jp | ||
|---|---|---|---|---|---|
| Control | Drought | Control | Drought | ||
| 1 | miR156 | 19,712 | 18,030 | 7807 | 16,355 |
| 2 | miR157 | 961 | 603 | 1094 | 516 |
| 3 | miR159 | 4687 | 4306 | 8694 | 11,575 |
| 4 | miR160 | 413 | 347 | 141 | 300 |
| 5 | miR162 | 1760 | 3525 | 1060 | 1628 |
| 6 | miR164 | 394 | 217 | 54 | 377 |
| 7 | miR165 | 265 | 275 | 597 | 416 |
| 8 | miR166 | 987,328 | 1,063,453 | 2,393,340 | 1,560,620 |
| 9 | miR167 | 20,394 | 12,785 | 6215 | 17,180 |
| 10 | miR168 | 20,408 | 73,216 | 37,636 | 52,762 |
| 11 | miR171 | 242 | 106 | 12 | 34 |
| 12 | miR172 | 2905 | 1740 | 235 | 1693 |
| 13 | miR319 | 807 | 833 | 117 | 309 |
| 14 | miR390 | 14 | 17 | 432 | 654 |
| 15 | miR393 | 41 | 35 | 2 | 39 |
| 16 | miR394 | 586 | 234 | 229 | 653 |
| 17 | miR395 | 67 | 17 | 32 | 131 |
| 18 | miR396 | 227,384 | 152,399 | 77,038 | 192,734 |
| 19 | miR397 | 3 | 0 | 16 | 0 |
| 20 | miR398 | 19 | 121 | 826 | 793 |
| 21 | miR399 | 74 | 30 | 54 | 71 |
| 22 | miR403 | 58,562 | 63,151 | 97 | 371 |
| 23 | miR408 | 138 | 319 | 3673 | 1873 |
| 24 | miR1310 | 36 | 48 | 177 | 129 |
| 25 | miR2111 | 10 | 0 | 0 | 6 |
| 26 | miR2111-5 | 228 | 226 | 0 | 2 |
| 27 | miR2911 | 47 | 37 | 52 | 30 |
| 28 | miR482 | 4 | 7 | 0 | 4 |
| 29 | miR5139 | 6 | 9 | 0 | 2 |
| 30 | miR530 | 22 | 33 | 2 | 0 |
| 31 | miR6173 | 26 | 46 | 38 | 26 |
| 32 | miR6478 | 49 | 13 | 24 | 38 |
| 33 | miR858 | 1574 | 3258 | 0 | 2 |
| 34 | miR894 | 1 | 4 | 0 | 4 |
| 35 | miR169 | 22 | 0 | – | – |
| 36 | miR447 | 0 | 4 | – | – |
| 37 | miR473 | 0 | 4 | – | – |
| 38 | miR477 | 9 | 22 | – | – |
| 39 | miR4995 | 1 | 0 | – | – |
| 40 | miR5141 | 3 | 2 | – | – |
| 41 | miR530-5 | 17 | 15 | – | – |
| 42 | miR3630-3 | – | – | 0 | 2 |
“–” indicates lack of expression
Interestingly, normalized miRNA levels revealed differences in miRNA abundances in the untreated leaves of these two plant species. Overall, miR166, followed by miR396 are most abundantly expressed families in the control samples of both plant species (Table 2). Similar abundances for miR166 was reported in Glycine max, Sorghum bicolor and Cucurbit species (Li et al. 2011; Zhang et al. 2011; Jagadeeswaran et al. 2012). However, expression of miR166 is almost two fold greater in J. pentantha, compared to I. campanulata. miR166 is known to target HD-ZIP III transcription factor and regulate diverse facets of plant development such as leaf polarity, xylem differentiation in root, and modulation of lateral root growth under drought stress (Jung and Park 2007; Sakaguchi and Watanabe 2012; Bakhshi et al. 2016). Differential abundances of miR166, observed in this study, could imply differential regulation of its target/s in wild and cultivated species studied here, impacting some of the targeted responses. Similarly, several other miRNA families such as miR156, miR160, miR164, miR167, miR172, miR319 and miR403 were more abundantly expressed in I. campanulata, while miR159, miR168 and miR408 were more abundantly expressed in J. pentantha (Table 2). In general, evolutionarily conserved miRNAs are most abundantly expressed miRNAs in plants (Sunkar and Jagadeeswaran, 2008; Zhang et al. 2006). More strikingly, the differences in miRNA abundances were extremely high for miR403 in I. campanulata (almost 600 fold greater than J. pentantha) whereas miR858 and miR2111-5 were found only in I. campanulata (Table 2). Both the species showed only two miR403 isoforms and miR403 is mostly found restricted to dicot families like Malvaceae, Vitaceae, Salicaceae and Solanaceae (Jagtap and Shivaprasad 2014). miR403 regulates AGO2 expression in Arabidopsis (Allen et al. 2005). Its innate high levels in I. campanulata may be indicative of its differential role from J. pentantha. On the other hand, J. pentantha showed 30 and 40 fold greater levels of miR390 and miR398, respectively, than compared to I. campanulata (Table 2). miR390 regulates developmental timing and patterning in plants through the miR390/tasiRNA/ARF regulatory system while miR398 is crucial for plant stress responses as it regulates CuZnSOD levels (Sunkar et al. 2006). Association between miR398 and CuZnSOD levels was previously established in these two species (Ghorecha et al. 2014).
Several miRNA levels were altered in both the species under drought. miRNA families having considerable expression (>100 RPTM) were considered for analysis. Those miRNAs that differed by at the least one fold are classified as drought-responsive miRNAs in this study. Similar fold change criterion was previously used in identifying differentially regulated miRNAs (Wang et al. 2016). Analysis of this study revealed about 2 downregulated (miR171, miR394) and 5 (miR162, miR168, miR398, miR858, miR408) upregulated miRNAs in I. campanulata, and, one (miR157) downregulated and 10 (miR394, miR156, miR160, miR164, miR167, miR172, miR319, miR395, miR396, miR403) upregulated miRNA families in J. pentantha (Table 3). Of the upregulated miRNAs in I. campanulata, miR398 is the most upregulated that showed 5.37 fold increase under drought (Table 3). Most upregulated miRNA families in J. pentantha include miR172, miR164, miR395 and miR403 (Table 3).
Table 3.
Expression pattern of drought responsive miRNAs (“−” sign before a value indicates decrease and others indicate an increase in expression) in I. campanulata and J. pentantha
| Plant species | miRNA families | Fold change |
|---|---|---|
| I. campanulata | miR398 | 5.37 |
| miR168 | 2.59 | |
| miR408 | 1.31 | |
| miR858 | 1.07 | |
| miR162 | 1 | |
| miR171 | −1.28 | |
| miR394 | −1.50 | |
| J. pentantha | miR172 | 6.20 |
| miR164 | 5.98 | |
| miR395 | 3.09 | |
| miR403 | 2.82 | |
| miR394 | 1.85 | |
| miR167 | 1.76 | |
| miR319 | 1.64 | |
| miR396 | 1.5 | |
| miR160 | 1.13 | |
| miR156 | 1.09 | |
| miR157 | −1.12 |
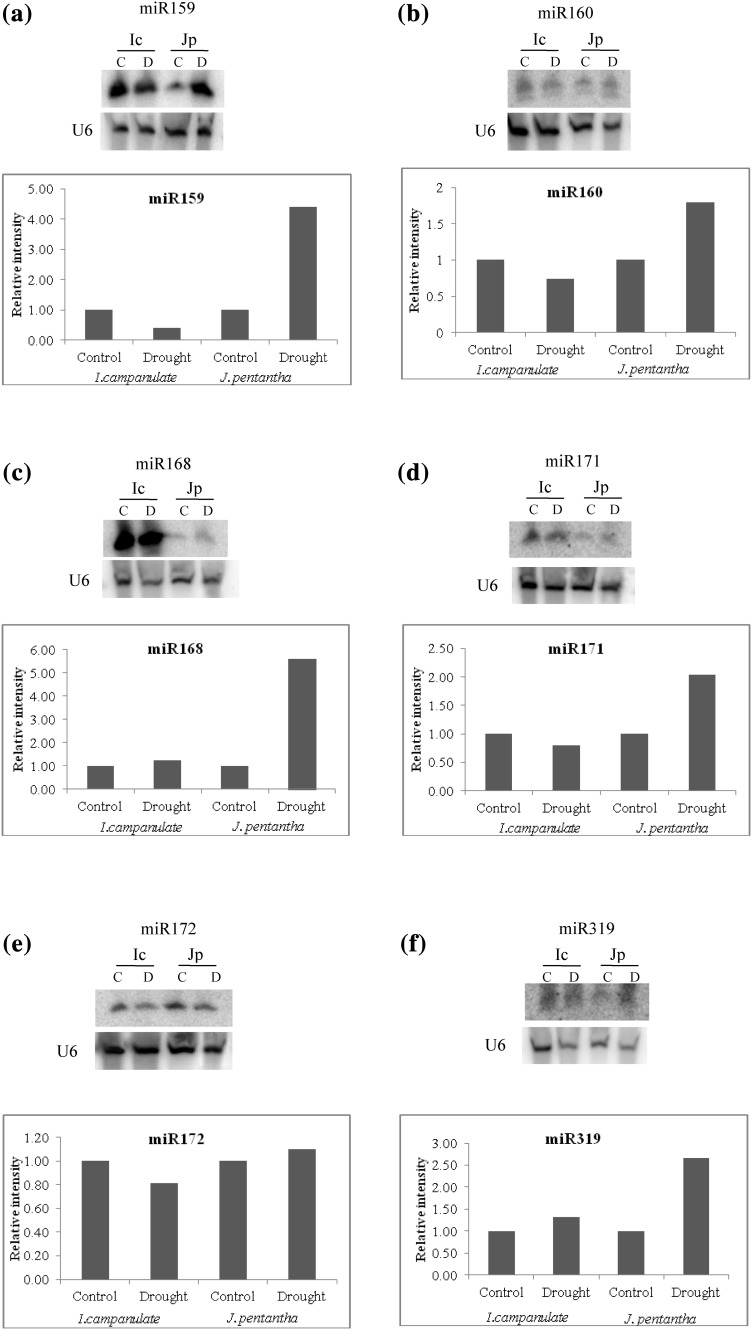
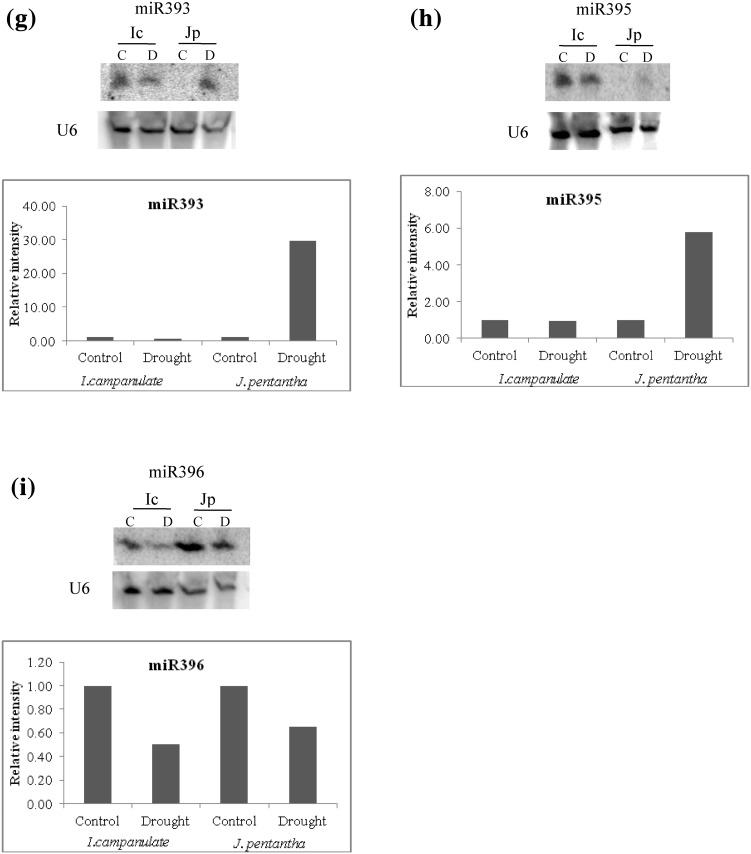
The expression pattern of few of these drought-responsive miRNAs was validated by small RNA blot analysis (Fig. 3). miR168 and miR319 were upregulated while miR396 was downregulated in both the species (Fig. 3c, f, i). Others 6 miRNAs showed downregulation in I. campanulata and were upregulated in J. pentantha (Fig. 3a, b, d, e, g, h). Except for miR396 in J. pentantha, the regulation pattern (i.e. up or down-regulation) of the miRNAs selected for validation were comparable to their respective sequencing profiles under stress. The lack of correlation between sequencing and northern blot results for miR396 expression in stressed J. pentantha could not be attributed to the source of RNA as same RNA samples were used for sequencing and Northern blot analysis. This lack of correlation between sequencing-based profiling and Northern profiling may largely be attributed to the biased-ligation with the adapters (Reddy et al. 2009) or sequencing problems. Although miR393 was not found as differentially regulated by drought as per our cut off criterion, the small RNA blot analysis revealed its induction by drought in J. pentantha but not in I. campanulata (Fig. 3g). In I. campanulata, its levels were higher (revealed by both sequencing and blot analysis) in control samples than in J. pentantha (Table 2; Fig. 3g). miR393 mediated regulation of auxin signalling is important for innate immunity of plants against pathogen attack (Navarro et al. 2006; Robert-Seilaniantz et al. 2011). It has been frequently reported that miR393 is upregulated by drought in several plant species (Sunkar et al. 2012) and miR393 dynamics in drought stressed J. pentantha is comparable to these reports.
Fig. 3.
Expression level of conserved miRNAs in I. campanulata (Ic) and J. pentantha (Jp) leaves growing under Control (C) and Drought (D) conditions as analysed through Northern blotting. U6 (small nuclear RNA) was used as loading control and relative accumulation of all miRNAs (to that of control) was quantified by normalizing their intensity values in accordance to that of U6
Interestingly, differential regulations were found for miR408 (upregulated by 1.31 fold in I. campanulata but downregulated by slightly less than one fold {not shown due to the cut off criterion} in J. pentantha) and miR394 (upregulated by 1.85 fold in J. pentantha but downregulated by 1.50 fold in I. campanulata) under drought in these two species (Table 3). miR408 was reported to accumulate in drought stressed M. truncatula (Trindade et al. 2010). Within the rice genotypes, miR408 levels were only elevated in drought tolerant ones (Nagina-22 and Vandana) but not in drought-sensitive genotypes under drought stress (Mutum et al. 2013). Corresponding to the levels of miR408 in rice, the target plantacyanin-like protein showed inverse expression profile indicating that miR408-plantacyanin regulation may play a role in drought tolerance (Mutum et al. 2013). Our findings on sequencing profile of miR408 revealed similar results as upregulation of miR408 in tolerant I. campanulata and downregulation in sensitive J. pentantha under drought stress. Similar to miR408, miR394 also regulates leaf development (i.e. by targeting LEAF CURLING RESPONSIVENESS {LCR} gene). It is known to be drought responsive in Arabidopsis and other species (Song et al. 2012; Liu et al. 2008; Kantar et al. 2011; Shuai et al. 2013). Upregulation of miR394 in J. pentantha is comparable to those reported in cotton (0.87 fold increase) exposed to drought stress (Xie et al. 2014). In another study, miR394 was shown to be downregulated in high-tolerant sugarcane cultivar but upregulated in the sensitive cultivars (Gentile et al. 2013). In these cultivars, miR394 was predicted to target gene encoding protein, NSP-interacting kinase (NIK) that is involved in plant development and its response to biotic stresses. Downregulation of miR394 in tolerant I. campanulata could play a similar role as reported in this plant species. This needs further investigation.
miR160 was known to regulate Auxin Response Factors (ARF) in plants (Liu et al. 2007; Ballen-Taborda et al. 2013; Turner et al. 2013). In Arabidopsis, miR160 appears to regulate auxin-ABA crosstalk, playing crucial role in seed germination (Liu et al. 2007). Drought mediated downregulation of miR160 was reported in Cassava, Populus trichocarpa and Sorghum bicolor (Ballen-Taborda et al. 2013; Shuai et al. 2013; Hamza et al. 2016). Contrary to this, miR160 was reported to accumulate in Prunus persica and Sugarcane under drought (Eldem et al. 2012; Gentile et al. 2015). miR160 showed downregulation in I. campanulata and upregulation in J. pentantha under drought. Downregulation of miR160 in I. campanulata is reminiscent of what was observed in relatively drought tolerant Sorghum (Hamza et al. 2016). Under in situ drought, miR160 was downregulated in both the species (Ghorecha et al. 2014) indicating that miR160 in I. campanulata showed unaltered regulation under in situ and ex situ drought while it differed in J. pentantha. Likewise, many other miRNAs including miR156, miR159, miR171, miR172, miR396, miR393 of I. campanulata and miR319, miR172 of J. pentantha showed unaltered regulation under in situ and ex situ drought. Contrary to this, several other miRNAs including miR168, miR319, miR398, miR408, miR395 of I. campanulata and miR156, miR159, miR160, miR168, miR171, miR398, miR408, miR396, miR393, miR395 of J. pentantha showed differential regulation under ex situ drought of this study and in situ drought study reported earlier (Ghorecha et al. 2014). Downregulation of miR398 (and corresponding increase in CuZnSODs) in I. campanulata was reported under in situ stress (Ghorecha et al. 2014), which was not seen in the present study. In our view, discrepancies in the regulation of these miRNAs, under in situ drought, could be attributed to possible influence of other uncontrolled environmental variables, which is normally not the case under ex situ drought. Similar kind of differential expression of miRNAs was observed in sugarcane that is grown in greenhouse versus field conditions (Gentile et al. 2015).
Overall, our study suggests that miRNAs in these two plant species are differentially regulated by drought. Moreover, the basal miRNA levels were very different between the two plant species. Further studies are required to analyze the effect of altered miRNAs on their target genes, which would help in understanding how miRNA regulation affects their targets genes and this, in turn, alters drought tolerance in these two plant species.
Acknowledgements
VG and NSRK are thankful to UGC-DRS program for financial assistance. VG is grateful to RS for providing lab facilities.
References
- Akashi K, Yoshimura K, Nanasato Y, Takahara K, Munekage Y, Yokota A. A wild plant resources for studying molecular mechanisms of drought/strong light stress tolerance. Plant Biotechnol. 2008;25:257–263. doi: 10.5511/plantbiotechnology.25.257. [DOI] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ashraf M. Inducing drought tolerance in plants: recent advances. Biotechnol Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Bakhshi B, Fard EM, Nikpay N, Ebrahimi MA, Bihamta MR, Mardi M, Salekdeh GH. MicroRNA signatures of drought signaling in rice root. PLoS ONE. 2016;11:e0156814. doi: 10.1371/journal.pone.0156814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballen-Taborda C, Plata G, Ayling S, Rodriguez-Zapata F, Lopez-Lavalle LAB, Duitama J, Tohme J. Identification of cassava MicroRNAs under abiotic stress. Int J Genom. 2013;1:857–986. doi: 10.1155/2013/857986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candar-Cakir B, Arican E, Zhang B. Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J. 2016;14:1727–1746. doi: 10.1111/pbi.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catsky J. Determination of water deficit in disks cut out from leaf blades. Biol Plant. 1960;2:76–78. doi: 10.1007/BF02920701. [DOI] [Google Scholar]
- Chen X, Xia J, Xia Z, Zhang H, Zeng C, Lu C, Zhang W, Wang W. Potential functions of microRNAs in starch metabolism and development revealed by miRNA transcriptome profiling of cassava cultivars and their wild progenitor. BMC Plant Biol. 2015;15:33. doi: 10.1186/s12870-014-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldem V, Akçay UC, Ozhuner E, Bakır Y, Uranbey S, Unver T. Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS ONE. 2012;7:e50298. doi: 10.1371/journal.pone.0050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Ferreira TH, Mattos RS, Dias LI, Hoshino AA, Carneiro MS, Souza GM, Calsa T, Jr, Nogueira RM, Endres L, Menossi M. Effects of drought on the microtranscriptome of field-grown sugarcane plants. Planta. 2013;237:783–798. doi: 10.1007/s00425-012-1795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Dias LI, Mattos RS, Ferreira TH, Menossi M. MicroRNAs and drought responses in sugarcane. Front Plant Sci. 2015 doi: 10.3389/fpls.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorecha V, Krishnayya NSR, Sunkar R. Impact of climate change on MicroRNA expression in plants. In: Tuteja N, Gill SS, editors. Climate change and plant abiotic stress tolerance. Hoboken: Wiley; 2013. pp. 507–520. [Google Scholar]
- Ghorecha V, Patel K, Ingle S, Sunkar R, Krishnayya NSR. Analysis of biochemical variations and microRNA expression in wild (Ipomoea campanulata) and cultivated (Jacquemontia pentantha) species exposed to in vivo water stress. Physiol Mol Biol Plants. 2014;20:57–67. doi: 10.1007/s12298-013-0207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman F, Almerao MP, Korbes AP, Loss-Morais G, Margis R. Identification of microRNAs from Eugenia uniflora by high-throughput sequencing and bioinformatics analysis. PLoS ONE. 2012;7:e49811. doi: 10.1371/journal.pone.0049811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Gustafson P, Langridge P, Shi BJ. Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol J. 2015;13:2–13. doi: 10.1111/pbi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C. Gene stacking in transgenic plants—the challenge for 21st century plant biotechnology. Plant Biotechol J. 2005;3:141–155. doi: 10.1111/j.1467-7652.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- Hamza NB, Sharma N, Tripathi A, Sanan-Mishra N. MicroRNA expression profiles in response to drought stress in Sorghum bicolor. Gene Expr Patterns. 2016 doi: 10.1016/j.gep.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Nimmakayala P, Zheng Y, Gowdu K, Reddy UK, Sunkar R. Characterization of the small RNA component of leaves and fruits from four different cucurbit species. BMC Genom. 2012;3:329. doi: 10.1186/1471-2164-13-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagtap S, Shivaprasad PV. Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants. BMC Genom. 2014;15:1. doi: 10.1186/1471-2164-15-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury J, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jung JH, Park CM. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta. 2007;225:1327–1338. doi: 10.1007/s00425-006-0439-1. [DOI] [PubMed] [Google Scholar]
- Kantar M, Lucas SJ, Budak H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta. 2011;233:471–484. doi: 10.1007/s00425-010-1309-4. [DOI] [PubMed] [Google Scholar]
- Keyvan S. The effects of drought stress on yield, relative water content, proline, soluble carbohydrates and chlorophyll of bread wheat cultivars. J Anim Plant Sci. 2010;8:1051–1060. [Google Scholar]
- Kulcheski FR, Oliveira LF, Molina LG, Almerao MP, Rodrigues FA, Marcolino J, Barbosa JF, Stolf-Moreira R, Nepomuceno AL, Marcelino-Guimaraes FC, Abdelnoor RV, Nascimento LC, Carazzolle MF, Pereira GA, Margis R. Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genom. 2011;12:307. doi: 10.1186/1471-2164-12-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Li H, Dong Y, Yin H, Wang N, Yang J, Liu X, Wang Y, Wu J, Li X. Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol. 2011;11:1–12. doi: 10.1186/1471-2229-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Tian X, Li Y, Wu C, Zheng C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress–responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik A, Pietrykowska H, Paczek L, Szweykowska-Kulinska Z, Zielenkiewicz P. High-throughput sequencing identification of novel and conserved miRNAs in the Brassica oleracea leaves. BMC Genom. 2013;14:801. doi: 10.1186/1471-2164-14-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Mayrose M, Kane NC, Mayrose I, Dlugosch KM, Rieseberg LH. Increased growth in sunflower correlates with reduced defences and altered gene expression in response to biotic and abiotic stress. Mol Ecol. 2011;20:4683–4694. doi: 10.1111/j.1365-294X.2011.05301.x. [DOI] [PubMed] [Google Scholar]
- Montes RAC, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martinez N, Meyers BC, Green PJ, de Folter S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun. 2014;5:1–15. doi: 10.1038/ncomms4722. [DOI] [PubMed] [Google Scholar]
- Mutum RD, Balyan SC, Kansal S, Agarwal P, Kumar S, Kumar M, Raghuvanshi S. Evolution of variety-specific regulatory schema for expression of osa-miR408 in indica rice varieties under drought stress. FEBS J. 2013;280:1717–1730. doi: 10.1111/febs.12186. [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Paul S, Kundu A, Pal A. Identification and expression profiling of Vigna mungo microRNAs from leaf small RNA transcriptome by deep sequencing. J Integr Plant Biol. 2014;56:15–23. doi: 10.1111/jipb.12115. [DOI] [PubMed] [Google Scholar]
- Reddy AM, Zheng Y, Jagadeeswaran G, Macmil SL, Graham WB, Roe BA, Desilva U, Zhang W, Sunkar R. Cloning, characterization and expression analysis of porcine microRNAs. BMC Genom. 2009;10:1. doi: 10.1186/1471-2164-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Sakaguchi J, Watanabe Y. miR165/166 and the development of land plants. Dev Growth Differ. 2012;54:93–99. doi: 10.1111/j.1440-169X.2011.01318.x. [DOI] [PubMed] [Google Scholar]
- Shuai P, Liang D, Zhang Z, Yin W, Xia X. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genom. 2013;14:233. doi: 10.1186/1471-2164-14-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JB, Huang SQ, Dalmay T, Yang ZM. Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 2012;53:1283–1294. doi: 10.1093/pcp/pcs080. [DOI] [PubMed] [Google Scholar]
- Stoyanov Z. Effects of water stress on leaf water relations of young bean plants. JCEA. 2005;6:5–14. [Google Scholar]
- Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Tang S, Wang Y, Li Z, Gui Y, Xiao B, Xie J, Zhu Q, Fan L. Identification of wounding and topping responsive small RNAs in tobacco (Nicotiana tabacum) BMC Plant Biol. 2012;12:28. doi: 10.1186/1471-2229-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade I, Capitao C, Dalmay T, Fevereiro MP, Santos DM. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta. 2010;231:705–716. doi: 10.1007/s00425-009-1078-0. [DOI] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S. Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci. 2006;11:405–412. doi: 10.1016/j.tplants.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013;162:2042–2055. doi: 10.1104/pp.113.220699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Fagard M. Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 2001;17:29–35. doi: 10.1016/S0168-9525(00)02166-1. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen L, Zhao M, Tian Q, Zhang WH. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011;12:1. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Gao F, Li S, Zhu Y, Yang P. Identification of conserved and novel microRNAs from Liriodendron chinense floral tissues. PLoS ONE. 2012;7:e44696. doi: 10.1371/journal.pone.0044696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Tang S, Liu J, Zhang H, Zhi H, Jia G, Diao X. Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genet. 2016;17:1. doi: 10.1186/s12881-015-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Wang Q, Sun R, Zhang B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J Exp Bot. 2014;66:789–804. doi: 10.1093/jxb/eru437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Xue L, An L. Functional diversity of miRNA in plants. Plant Sci. 2007;172:423–432. doi: 10.1016/j.plantsci.2006.10.009. [DOI] [Google Scholar]
- Yinpeng L, Ye W, Wang M, Yan X. Climate change and drought: a risk assessment of crop-yield impacts. Clim Res. 2009;39:31–46. doi: 10.3354/cr00797. [DOI] [Google Scholar]
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zheng Y, Jagadeeswaran G, Li Y, Gowdu K, Sunkar R. Identification and temporal expression analysis of conserved and novel microRNAs in Sorghum. Genomics. 2011;98:460–468. doi: 10.1016/j.ygeno.2011.08.005. [DOI] [PubMed] [Google Scholar]