Summary
During development of the central nervous system, there is a shift in the subunit composition of NMDA receptors (NMDARs) resulting in a dramatic acceleration of NMDAR-mediated synaptic currents. This shift coincides with upregulation of the GluN2A subunit and triheteromeric GluN1/2A/2B receptors with fast deactivation kinetics, whereas expression of diheteromeric GluN1/2B receptors with slower deactivation kinetics is decreased. Here, we show that allosteric interactions occur between the glutamate-binding GluN2 subunits in triheteromeric GluN1/2A/2B NMDARs. This allosterism is dominated by the GluN2A subunit and results in functional properties not predicted by those of diheteromeric GluN1/2A and GluN1/2B NMDARs. These findings suggest that GluN1/2A/2B NMDARs may maintain some signaling properties of the GluN2B subunit while having the kinetic properties of GluN1/2A NMDARs, and highlight the complexity in NMDAR signaling created by diversity in subunit composition.
Keywords: Triheteromeric NMDA receptors, allosteric interaction, amino-terminal domain, glutamate, excitatory synaptic transmission, open probability, crosslinking
eTOC Blurb
Sun et al. demonstrate asymmetric inter-GluN2 allosteric interactions within triheteromeric GluN1/2A/2B NMDARs that result in open probability and deactivation kinetics similar to diheteromeric GluN1/2A receptors. This finding highlights the complexity in NMDAR signaling endowed by diversity in subunit composition.
Introduction
During excitatory neurotransmission in the central nervous system, glutamate is released into the synaptic cleft where this neurotransmitter reaches a high peak concentration (~1 mM) for a brief duration (~1 ms) (Clements et al., 1992). In this short time, glutamate will bind ionotropic glutamate receptors and initiate receptor conformational changes that lead to channel gating. However, the component of this synaptic response that is mediated NMDA-type glutamate receptors continues for tens to hundreds of millisecond after synaptic glutamate is removed, during which time, the receptors transition between glutamate-bound open and closed conformational states until glutamate eventually unbinds and the synaptic response is terminated (Lester et al., 1990). For NMDA receptors, these functional properties are controlled by the subunit composition (Gielen et al., 2009; Monyer et al., 1994; Yuan et al., 2009). The subunit diversity among NMDA receptors and assembly of different receptor subtypes with distinct functional properties enable precise tuning of the synaptic response and allows variation in the physiological roles of NMDA receptors during neuronal development (Paoletti et al., 2013; Sanz-Clemente et al., 2013; Traynelis et al., 2010)
The developmental transition from diheteromeric GluN1/2B receptors to triheteromeric GluN1/2A/2B receptors (Akazawa et al., 1994; Gray et al., 2011; Monyer et al., 1994; Rauner and Kohr, 2011; Sheng et al., 1994; Watanabe et al., 1992) accompanies maturation of neuronal circuits and alterations in synaptic plasticity (Dumas, 2005; Paoletti et al., 2013). Our understanding of how GluN2 subunits combine to shape the functional properties of triheteromeric receptors is lacking considering the abundance of GluN1/2A/2B receptors in the adult brain and the dramatic, developmental shift in channel properties. Allosteric interactions may occur between the GluN2 glutamate-binding subunits, but the influence of such interactions on functional NMDAR properties has not been directly addressed. Our findings describe an unpredicted and asymmetric interaction between the GluN2 subunits of in triheteromeric GluN1/2A/2B receptors, which is the most abundant NMDAR in the adult forebrain.
Results
Asymmetric allosteric interaction occurs within GluN1/2A/2B triheteromeric NMDARs
We used an endoplasmic reticulum (ER) retention signal system to selectively express triheteromeric GluN1/2A/2B (A/B) receptors, which are assembled from two glycine-binding GluN1 and two different glutamate-binding GluN2 subunits (Hansen et al., 2014) (Fig. 1A, Fig. S1). The deactivation time constant of A/B receptors (50 ± 3 ms, n=18) is slightly slower than that of diheteromeric GluN1/2A (A/A) receptors (36 ± 1 ms, n=6) and much faster than that of diheteromeric GluN1/2B (B/B) receptors (333 ± 17 ms, n=8) (Fig. 1B). These results are consistent with previous reports on triheteromeric receptor function (Hansen et al., 2014; Stroebel et al., 2014; Tovar et al., 2013), and suggest that the function of GluN1/2A/2B is more similar to that of GluN1/2A compared to GluN1/2B. Furthermore, this observation raises the intriguing possibility that allosteric interactions exist between GluN2 subunits in GluN1/2A/2B NMDARs that endow triheteromeric receptors with unique functional properties compared to the diheteromeric GluN1/2A and GluN1/2B receptors. Since NMDAR activation requires agonist binding to all four subunits (Benveniste and Mayer, 1991), the receptor deactivation rate in the continuous presence of glycine may be defined largely by the GluN2 subunit with fastest glutamate unbinding. To address this assumption and to conclusively test whether interactions occur between GluN2A and GluN2B subunits, we introduced two cysteine mutations into GluN2, which spontaneously form a disulfide bond that locks the ligand binding domain (LBD) in a conformation similar to the glutamate-bound state (GluN2A: K487C + N687C; GluN2B: K488C + N688C; hereafter Acc and Bcc, respectively; Blanke and VanDongen, 2008; Dai and Zhou, 2016; Kussius and Popescu, 2010). Thus, this disulfide bond will generate “constitutively liganded” GluN2 subunits with high efficiency (Fig. S2). We used the ER retention signal method (Hansen et al., 2014) to generate receptors containing one crosslinked GluN2 and one wildtype GluN2. These receptors require glutamate binding only to the wildtype GluN2 subunit for activation (Fig. S3) and allows direct evaluation of deactivation from a single GluN2 subunit in the tetrameric receptor. Crosslinking one GluN2A subunit within A/A receptors (Acc/A) doubled the deactivation time constant from 36 ± 1 ms (n=6) to 73 ± 5 ms (n=5) (Fig. 1C), consistent with two equivalent, independent binding sites in the A/A receptor. Similarly, crosslinking one GluN2B subunit (Bcc/B) also resulted in a doubling of the B/B deactivation time constant from 333 ± 17 ms (n=8) to 680 ± 26 ms (n=9) (Fig. 1D).
Figure 1. Subunit interactions within GluN1/2A/2B triheteromeric NMDARs.
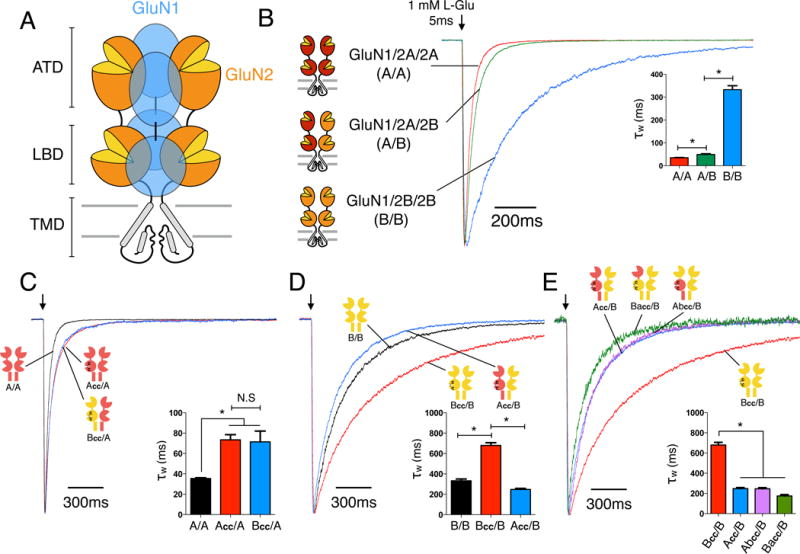
(A) Cartoon of the heterotetrameric NMDAR structure with three distinctive layers formed by the amino-terminal domains (ATDs), ligand binding domains (LBDs), and transmembrane domains (TMDs). The majority of NMDARs are assembled from two glycine-binding GluN1 and two glutamate-binding GluN2 subunits. Diheteromeric NMDARs are assembled from GluN1 and one type of GluN2 subunit, such as GluN1/2A (A/A) and GluN1/2B (B/B), whereas triheteromeric NMDARs are assembled from GluN1 and two different GluN2 subunits, such as GluN1/2A/2B (A/B). (B) Deactivation kinetics of diheteromeric A/A, B/B and triheteromeric A/B NMDARs in response to a 5ms pulse of 1mM L-glutamate pulse indicated by the arrow (* indicates significantly different, p<0.05, one-way ANOVA with Tukey posttest). (C) Deactivation kinetics of A/A receptors and NMDARs containing one wildtype GluN2A and one mutant subunit with crosslinked LBD (Acc/A or Bcc/A). Crosslinked LBDs are generated by introducing two cysteine mutations (GluN2A: K487C + N687C; GluN2B: K488C + N688C;) which are designed to mimic a “constitutively liganded” state. (D) Deactivation kinetics of B/B, Bcc/B and Acc/B receptors. (E) Deactivation kinetics of receptors with one wildtype GluN2B and one crosslinked chimeric subunit with the ATD interchanged between GluN2A and GluN2B (Abcc/B or Bacc/B), compared to those of Bcc/B and Acc/B. All current responses are normalized to their peaks. See also Figures S1–S4.
Next, we tested whether the deactivation rate from a single GluN2 subunit is dependent on the identity of the other GluN2 subunit in the receptor complex. First, we expressed one wildtype GluN2A subunit with one crosslinked GluN2B subunit (Bcc/A). These Bcc/A receptors deactivated with a time constant (71 ± 8 ms, n=9) that is not different from the Acc/A receptor (73 ± 5 ms, n=5) (Fig. 1C), indicating that deactivation from the GluN2A subunit is independent of co-expression with another GluN2A or GluN2B subunit. Surprisingly, receptors with one crosslinked GluN2A subunit and one wildtype GluN2B subunit (Acc/B) deactivate ~2.7 fold faster (248 ± 10 ms, n=10) than Bcc/B receptors (680 ± 26ms, n=9) (Fig. 1D). This is a marked acceleration in receptor kinetics compared to the deactivation time course that would be predicted if the GluN2 subunits function independently as suggested by the crosslinked Acc/A and Bcc/B receptors (Fig. 1C,D). This unexpected result therefore indicates that an asymmetric interaction exists between GluN2A and GluN2B subunits within triheteromeric GluN1/2A/2B receptors, in which the GluN2A subunit greatly accelerates glutamate unbinding from the GluN2B subunit (compare Bcc/B and Acc/B; Fig. 1D). In stark contrast, GluN2B has no effect on glutamate unbinding from the GluN2A subunit (compare Acc/A and Bcc/A; Fig. 1C). This unidirectional interaction shapes the deactivation time of triheteromeric GluN1/2A/2B receptors (50 ms), and brings the deactivation time closer to that of diheteromeric GluN1/2A receptors (36 ms). In theory, the deactivation time of GluN1/2A/2B receptors with independent deactivation from the two GluN2 subunits would be 66 ms (see Methods).
The extracellular amino-terminal domain (ATD) of GluN2 subunits has been shown to mediate much of the variation in functional properties among NMDAR subtypes (Gielen et al., 2009; Yuan et al., 2009). To gain insight to the structural determinants of the dominant effect of GluN2A on glutamate unbinding from GluN2B, we introduced the GluN2A ATD into the crosslinked GluN2B subunit (denoted Bacc) and the GluN2B ATD into the crosslinked GluN2A subunit (Abcc). For Abcc/B and Bacc/B receptors, the deactivation times were markedly accelerated compared to Bcc/B receptor (Abcc/B: 246 ± 11 ms, n=7, Bacc/B 188 ± 18 ms, n=5; compared to Bcc/B: 680 ± 26 ms, n=9) (Fig. 1E). These results show that including full length or parts of GluN2A always results in faster GluN2B deactivation, indicating the dominant role of GluN2A.
GluN2A ATD and LBD+TMD dominate channel deactivation
To determine whether the dominant effect of the GluN2A subunit is preserved in receptors with two functioning glutamate binding sites, we expressed the chimeric GluN2A and GluN2B subunits with swapped ATDs as both diheteromeric receptors, which contain two identical GluN2 subunits, and triheteromeric receptors, which contain two different GluN2 subunits. Introducing the GluN2B ATD into a single GluN2A subunit within A/A receptors (i.e., A/Ab receptors.) slightly slowed receptor deactivation from 36 ± 1 ms (n = 6) to 48 ± 3 ms (n = 5) (Fig. 2A). Introducing the GluN2B ATD into both GluN2A subunits (Ab/Ab) further slowed the deactivation to 80 ± 8 ms (n=6) (Fig. 2A), but was still much faster than the deactivation of B/B receptors (333 ± 17 ms, n=8). This result indicates that domains in addition to the GluN2 ATD determine deactivation, consistent with previous studies (Gielen et al., 2009; Hansen et al., 2013; Vance et al., 2011; Yuan et al., 2009). In stark contrast to domain swapping in A/A receptors, introducing the GluN2A ATD into a single GluN2B subunit within B/B receptors (B/Ba) markedly accelerated deactivation from 333 ± 17 ms (n=8) to 89 ± 8 ms (n=7) and replacing both ATDs with those from GluN2A (Ba/Ba) did not further change the time constant (81 ± 9 ms, n=7) (Fig. 2B). These results further support that the GluN2A ATD plays a dominant role in determining receptor deactivation, since incorporating a single copy of the GluN2A ATD into B/B receptors causes a substantial acceleration of the deactivation time course that is not further accelerated by incorporating two copies of GluN2A ATD, whereas the GluN2B ATD does not have comparable dominant effects in A/A receptors. Similar to the ATD, the GluN2A LBD+TMD also dominates receptor deactivation. The Ab/B receptors essentially correspond to B/B receptors with the GluN2A LBD+TMD introduced to a single GluN2B subunit, and the deactivation of these Ab/B receptors is reduced from 333 ± 17 ms (n=8) for B/B to 166 ± 7 ms (n=6) (Fig. 2C). In contrast, introducing a GluN2B LBD+TMD into A/A receptors (Ba/A) failed to change deactivation (A/A: 36 ± 1 ms, n=6, Ba/A: 36 ± 3 ms, n=4) (Fig. 2C).
Figure 2. Effects of domain swapping on NMDAR deactivation kinetics.
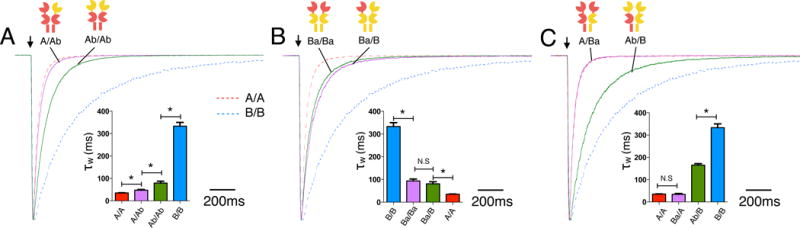
(A) Deactivation kinetics of A/A receptors with one (A/Ab) or both ATDs (Ab/Ab) swapped to the GluN2B ATD. The red and blue dotted lines indicate deactivation time course for wildtype A/A and B/B receptors, respectively (same in B and C). (B) Deactivation of B/B receptors with single ATD (Ba/B) or both ATDs (Ba/Ba; green trace) swapped to the GluN2A ATD. (C) Deactivation of A/A receptors with a single GluN2A LBD+TMD swapped to the GluN2B LBD+TMD (Ba/A) and B/B receptors with a single GluN2B LBD+TMD swapped to the GluN2A LBD+TMD (Ab/B).
GluN2A ATD and LBD+TMD dominate channel open probability
To determine if GluN2 subunit interactions affect other properties of NMDAR gating, we tested whether the same subunit manipulations that altered deactivation also affect NMDAR open probability. We used the open channel blocker (+)-MK-801 (1 μM) to block NMDA receptor-mediated steady-state currents and used the rate of block to estimate open probability. MK-801 blocked A/A and B/B currents with time constants of 92 ± 4 ms (n=6) and 292 ± 20 ms (n=6), respectively (Fig. 3A). Assuming a linear relationship between open probability and MK-801 block rate (Hansen et al., 2013), A/A steady-state open probability is ~3 times higher than that of B/B receptors (0.48 vs 0.15, Table 1) (Erreger et al., 2005; Yuan et al., 2009). MK-801 blocked A/B receptors with a slightly slower time constant (107 ± 3 ms, n=6, corresponding to an open probability of 0.41) than A/A receptors (92 ± 4 ms, n=6, open probability is 0.48) (Fig. 3A, E). This result demonstrates that GluN2A not only dominates the deactivation time course, but also open probability. To further pinpoint the domain that determines open probability, we used the chimeric GluN2 subunits to switch single or both copies of the ATD within A/A or B/B receptors. The open probabilities of these chimeric subunits mirror their deactivation times in that the GluN2A subunit also dominates open probability of triheteromeric GluN1/2A/2B receptors, and that this effect is mainly mediated by the GluN2A ATD (Fig. 3, Table 1). This dominant role of the GluN2A subunit is consistent with structural interactions between the ATD and LBD of triheteromeric GluN2A/GluN2B-containing receptors described in (Lü et al., 2017).
Figure 3. Effect of domain swapping on open probability.
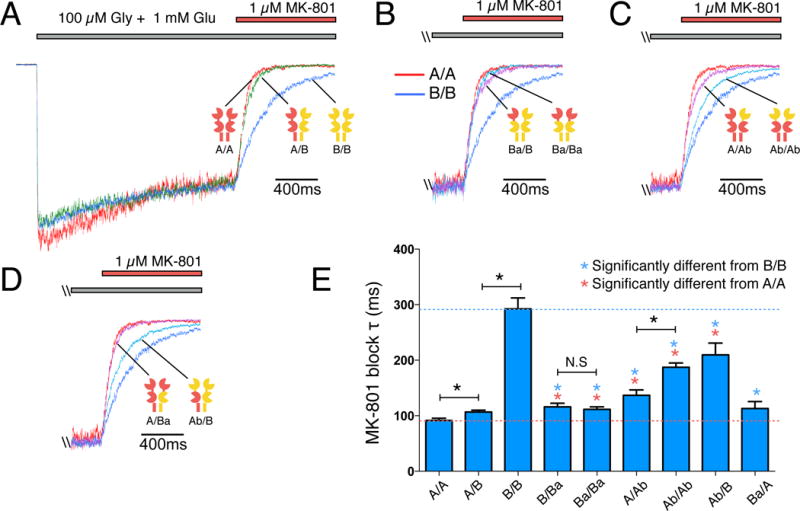
(A) The open channel blocker MK-801 (1 μM) inhibits steady state currents activated by 100 μM glycine and 1 mM glutamate from A/A, A/B and B/B receptors at different rates, consistent with distinct channel open probabilities. (B) MK-801 block of B/B receptors with a single ATD (Ba/B; magenta trace) or both ATDs (Ba/Ba; light blue trace) swapped to the GluN2A ATD. Red and blue traces indicate currents for A/A and B/B receptors, respectively (same in C and D). (C) MK-801 block of A/A receptors with a single ATD (A/Ab; magenta trace) or both ATDs (Ab/Ab; light blue trace) swapped to the GluN2B ATD. (D) MK-801 block of A/A receptors with a single LBD+TMD swapped to the GluN2B LBD+TMD (A/Ba; magenta trace) and B/B receptors with a single LBD+TMD swapped to the GluN2A LBD+TMD (Ab/B; light blue trace). (E) Bar graph with a summary of time constants for MK-801 block (τ). Data are mean ± SEM.
Table 1. Summary of deactivation time constants and MK-801 block time constant.
Deactivation time constants and time constants for MK-801 block were obtained by fitting current traces using double exponential and single exponential functions, respectively. For estimation of open probability (Po), 1 μM (+)-MK-801 was used to block currents activated by 100 μM glycine plus 1 mM glutamate at pH 8.0. MK-801 block rates (1/τ) for all receptors were divided by the block rate of diheteromeric GluN1/2A receptors (A/A) to obtain the scaling factors and Po was calculated by multiplying the previously determined Po of GluN1/2A receptors at pH 8.0 (Yuan et al., 2009) by the scaling factor determined for each receptor. Data are shown as mean ± SEM. n.d: not determined.
| Group | τfast (ms) | τslow (ms) | τfast % | τweighted (ms) | n | MK-801 (1 μM) block τ (ms) | Estimated Po | n |
|---|---|---|---|---|---|---|---|---|
| A/A | 31 ± 1 | 111 ± 14 | 92 ± 2 | 36 ± 1 | 6 | 92 ± 4 | 0.48 | 6 |
| A/B | 39 ± 3 | 139 ± 15 | 85 ± 3 | 50 ± 3 | 18 | 107 ± 3 | 0.41 | 6 |
| B/B | 174 ± 12 | 586 ± 35 | 60 ± 4 | 333 ± 17 | 8 | 292 ± 20 | 0.15 | 6 |
| Acc/A | 44 ± 4 | 171 ± 9 | 77 ± 3 | 73 ± 5 | 5 | n.d | ||
| Bcc/A | 43 ± 3 | 246 ± 40 | 86 ± 4 | 71 ± 8 | 9 | n.d | ||
| Bcc/B | 224 ± 27 | 1079 ± 58 | 46 ± 3 | 680 ± 26 | 9 | n.d | ||
| Acc/B | 119 ± 13 | 470 ± 51 | 59 ± 6 | 248 ± 10 | 10 | n.d | ||
| Abcc/B | 138 ± 12 | 434 ± 54 | 61 ± 8 | 246 ± 11 | 7 | n.d | ||
| Bacc/B | 91 ± 2 | 310 ± 29 | 55 ± 7 | 188 ± 18 | 5 | n.d | ||
| A/Ab | 38 ± 2 | 134 ± 23 | 89 ± 3 | 49 ± 3 | 6 | 137 ± 10 | 0.32 | 3 |
| Ab/Ab | 55 ± 7 | 178 ± 21 | 78 ± 6 | 80 ± 8 | 6 | 187 ± 8 | 0.24 | 6 |
| Ba/B | 58 ± 9 | 192 ± 37 | 68 ± 11 | 89 ± 8 | 6 | 116 ± 6 | 0.38 | 4 |
| Ba/Ba | 50 ± 4 | 160 ± 25 | 67 ± 8 | 81 ± 9 | 7 | 111 ± 5 | 0.40 | 3 |
| Ba/A | 30 ± 2 | 87 ± 7 | 89 ± 4 | 36 ± 3 | 4 | 113 ± 13 | 0.39 | 4 |
| Ab/B | 109 ± 4 | 434 ± 57 | 81 ± 3 | 166 ± 7 | 6 | 209 ± 21 | 0.21 | 5 |
Discussion
Subunit allosteric interactions within triheteromeric NMDARs
Recent cryo-EM data suggest that structure of triheteromeric GluN1/2A/2B NMDARs is unique because it breaks the two-fold symmetry of diheteromeric GluN1/2B receptors (Lü et al., 2017). Our study shows a direct functional consequence of losing this structural symmetry: GluN2A and GluN2B interact asymmetrically, and GluN2A dominates this interaction resulting in a high open probability and fast deactivating receptor. We therefore identify functional consequences of asymmetry in GluN1/2A/2B NMDARs that were not detected in previous reports describing triheteromeric receptor function (Hansen et al., 2014; Stroebel et al., 2014; Tovar et al., 2013) and cannot be determined from static cryo-EM protein structures (Lü et al., 2017). Since the cloning of NMDA receptor subunits 25 years ago, these allosteric interactions between glutamate binding GluN2 subunits have not been revealed, and because of this, the GluN2 subunits have been assumed to operate independently (Clements and Westbrook, 1991).
Significance of GluN2A-dominant allosteric interaction during development
The developmental inclusion of GluN2A in triheteromeric NMDARs substantially speeds up channel deactivation (Carmignoto and Vicini, 1992; Flint et al., 1997; Gray et al., 2011; Roberts and Ramoa, 1999; Stocca and Vicini, 1998) and therefore shortens the window for coincidence detection (Erreger et al., 2005). How GluN2A accomplishes this feat is not entirely clear. Our results indicate that GluN2A dictates not only the fast deactivation, but also increases open probability, in part, through allosteric interactions that override the kinetic properties of the GluN2B subunit. Previous studies suggest that the fast deactivation time course of triheteromeric GluN1/2A/2B receptors is governed by the GluN2A subunit, but the mechanistic basis for the fast deactivation time course has not been explicitly investigated (Hansen et al., 2014; Stroebel et al., 2014; Tovar et al., 2013). Here, we demonstrate that a GluN2A-dominant allosteric interaction speeds up deactivation from the neighboring GluN2B subunit (Fig. 1D) and results in a 26% faster deactivation time constant (50ms) for triheteromeric GluN1/2A/2B receptors than expected if subunits were assumed to be independent (66ms) (Fig. 1B, also see Methods). The high open probability of triheteromeric receptors suggests the GluN2A subunit also dominates receptor gating (Fig. 3A). Thus, the GluN2A-dominant allosteric interaction forces the triheteromeric GluN1/2A/2B receptor into a “GluN1/2A-like” state, and significantly shapes the amplitude and duration of current flux, and presumably Ca2+ dynamics. Since the diheteromeric GluN1/2B to triheteromeric GluN1/2A/2B receptor transition is ubiquitous in the forebrain during development and coincides with learning, the fast deactivating triheteromeric NMDAR is likely crucial for reliable adult brain function. Consistent with this, GluN2A conditional KO mice show altered synaptic plasticity and disrupted learning (Andreescu et al., 2011; Sakimura et al., 1995; Zhao and Constantine-Paton, 2007). The GluN2A-dominant allosteric interaction is necessary for producing NMDARs with faster kinetics and higher Po that enables a balance optimal for information processing and storage (Dumas, 2005).
In addition, the kinetic dominance of the GluN2A subunit in triheteromeric NMDARs results in a GluN1/2A-like receptor while maintaining the presence of a GluN2B subunit. GluN2B subunits are involved in critical functions such as receptor trafficking, localization and intracellular signaling (Barria and Malinow, 2005; Delaney et al., 2013; Foster et al., 2010; Lau and Zukin, 2007; Tang et al., 2010). Whether triheteromeric GluN1/2A/2B NMDARs maintain the intracellular signaling properties of GluN1/2A and GluN1/2B diheteromeric receptors remains an open question. Tang et al., (2010) suggested a dominant role for the GluN2B subunit in controlling trafficking of triheteromeric GluN1/2A/2B receptors to recycling endosomes and showed that recycling of GluN2B-containing NMDARs in wildtype neurons is not significantly different from GluN2A-deficient neurons. Such dominant properties of GluN2B in regulating certain trafficking properties of triheteromeric GluN1/2A/2B NMDARs is in contrast to our demonstration of the dominant role of GluN2A in receptor function and highlight the complexity created by subunit diversity in NMDARs. The regulation and localization of GluN2A and GluN2B by MAGUK proteins (e.g. PSD-95 and SAP102) remains elusive (Sanz-Clemente et al., 2013). This could be due, at least in part, to unexpected properties of triheteromeric GluN1/2A/2B NMDARs and the absence of these receptors in working models of NMDAR trafficking. Previous studies on the trafficking properties of GluN2A and GluN2B subunits may have overlooked the possibility that triheteromeric GluN1/2A/2B NMDARs have distinct regulatory and signaling properties resulting from the dominant behavior of one intracellular GluN2 C-terminal domain over the other. Future studies are needed to address the possibility that GluN1/2A/2B triheteromeric receptors retain the interactions with intracellular proteins that preferentially bind to the intracellular C-terminal domain of the GluN2B subunit, such as RAS-guanine-nucleotide-releasing factor 1 (RASGRF1) (Krapivinsky et al., 2003), PSD-95-neuronal nitric oxide synthase complex (Martel et al., 2012) and Ca2+/calmodulin-dependent protein kinase II (CaMKII) (Barria and Malinow, 2005). It is possible that triheteromeric GluN1/2A/2B NMDARs have the hybrid properties of being kinetically dominated by the GluN2A subunit while maintaining the intracellular signaling or trafficking properties of the GluN2B subunit, thereby allowing these receptors to play unique physiological roles.
In this study, we provide key insights on how GluN2A and GluN2B cooperatively determine the function of triheteromeric GluN1/2A/2B NMDARs. The asymmetric allosteric interactions between GluN2 subunits highlight an important feature of NMDARs. That is, the function of triheteromeric receptors cannot be accurately predicted solely from the function of the corresponding diheteromeric receptors. As demonstrated in this study, unexpected properties can arise from combining different subunits into the NMDAR complex. Furthermore, our findings support the possibility that unique intra- and inter-subunit interfaces exist in triheteromeric GluN1/2A/2B NMDARs, which are not present in diheteromeric GluN1/2A and GluN1/2B NMDARs (Lü et al., 2017). The distinct structure and function of GluN1/2A/2B NMDARs create opportunities for the development of novel modulators, which are selective for triheteromeric over diheteromeric receptors and could provide a path forward for selectively targeting GluN1/2A/2B NMDARs in the treatment of neurological and psychiatric disorders.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Craig Jahr (jahr@ohsu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Xenopus laevis oocytes
Xenopus laevis oocytes (Xenopus 1, http://www.xenopus1.com) were used for receptor expression and two-electrode voltage clamp (TEVC) recordings. Group sample sizes were chosen based on previous studies (Hansen et al., 2013, 2014) and no statistical methods were used to predetermine sample size. Recordings were obtained 2–5 days following cRNA injection.
Cell culture
HEK293 Tet-On Advanced cells were used for heterologous expression and patch-clamp recordings. Cells were obtained from Clontech and were not further authenticated. Cells were grown and maintained using standard protocols at 37 °C with 5% CO2. Group sample sizes were chosen based on previous studies (Hansen et al., 2013, 2014) and no statistical methods were used to predetermine sample size. Recordings were obtained 24–48 hr following transfection.
METHOD DETAILS
DNA constructs
Rat cDNAs for GluN1-1a (GenBank accession number U08261; hereafter, GluN1), GluN2A (D13211), and GluN2B (U11419) were provided by Drs. S. Heinemann (Salk Institute) and S. Nakanishi (Osaka Bioscience Institute). The GluN2B cDNA was modified to remove a T7 RNA polymerase termination site without changing the amino acid sequence as previously described (Hansen et al., 2014). GluN2 cDNAs containing ER retention signal sequences at the intracellular C-terminus (C1 and C2) are identical to constructs used in (Hansen et al., 2014). Briefly, the cDNAs encoding C1 and C2 were inserted in place of the stop codon in the open reading frame of GluN2A to generate 2AC1 and 2AC2. The C-terminal domain of GluN2B was then replaced by the C-terminal domain of the GluN2A subunits 2AC1 and 2AC2. Residues 1–844 of resulting chimeric subunits denoted 2BAC1 and 2BAC2 were identical to GluN2B, and the remaining residues were identical to residues 844–1541 and 844–1533 from 2AC1 and 2AC2, respectively. In this study, receptors assembled from G1uN1, 2AC1, and 2AC2 are denoted A/A, receptors assembled from G1uN1, 2BAC1, and 2BAC2 are denoted B/B, and receptors assembled from G1uN1, 2AC1, and 2BAC2 are denoted A/B. The GluN2A with the GluN2B ATD (denoted Ab) was generated by swapping the first 404 amino acids from GluN2B with the first 403 amino acids in GluN2A. The GluN2B with the GluN2A ATD (denoted Ba) was generated by swapping the first 403 amino acids from GluN2A with the first 404 amino acids in GluN2B. For constructs encoding subunits with LBDs stabilized in the glutamate-bound conformation (i.e. crosslinked subunits), double cysteine mutations were introduced to GluN2A (K487C and N687C) and GluN2B (K488C and N688C). Sample size range: 3–9. Recordings were not performed blind.
Two-Electrode Voltage-Clamp Recordings
Preparation and injection of Xenopus oocytes were performed essentially as previously described (Hansen et al., 2013). For experiments using C1- and C2-tagged GluN2 subunits, the cRNAs encoding GluN1, as well as C1- and C2-tagged GluN2, were injected at a 1:6:6 ratio at a total volume of 50 nl (10ng total cRNA). Recordings were performed 2–5 days after cRNA injection at room temperature (23°C) using an Axoclamp 900A amplifier (Molecular Devices, Sunnyvale, CA). Oocytes were placed in a custom-made chamber and continuously perfused (approximately 5 ml/min) with oocyte recording solution containing (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, 0.01 EDTA (pH 7.4 with NaOH). Recording electrodes were filled with 3.0 M KCl, and current responses were recorded at a holding potential of −60 mV. Data acquisition, voltage control, and application of solutions were controlled using pClamp software (Molecular Devices, Sunnyvale, CA). The reliability of the ER retention system to selectively express triheteromeric NMDARs in Xenopus oocytes was evaluated as previously described (Hansen et al., 2014) (see Fig. S1). Sample size range: 3–9. Recordings were not performed blind.
Whole cell patch clamp recordings
HEK293 Tet-On Advanced cells (Clontech) were transfected with plasmid cDNAs encoding GluN1 (together with EGFP in the pTRE-tight vector) and C1- and C2-tagged GluN2 subunits at a ratio of 1:2 using the calcium phosphate precipitation method as previously described (Hansen et al., 2014). Whole-cell voltage-clamp recordings were performed using an Axopatch 1D amplifier (Molecular Devices, Union City, CA) at room temperature. The holding potential was −60 mV. The electrodes were filled with internal solution containing (in mM) 110 D-gluconate, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, and 0.3 NaGTP (pH 7.35 with CsOH), and the extracellular recording solution was composed of (in mM) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, 0.01 EDTA, 20 mM D-mannitol (pH 7.4 with NaOH). Rapid solution exchange (open tip solution exchange had 10–90% rise times below 1 ms) was achieved using a two-barrel theta-glass pipette controlled by a piezobimorph. Data were acquired at 20kHz, filtered at 5–10kHz, and analyzed with Axograph software (axographx.com). NMDAR deactivation kinetics were obtained by fitting currents activated by 5 ms pulses of 1 mM glutamate in the presence of 100 μM glycine using a double exponential function: I = X*e (−t/taufast) + Y*exp(−t/tauslow), where X and Y are the current amplitudes of the fast and slow components, respectively. Tauweighted values were calculated using equation: Tauweighted = (X*taufast + Y*tauslow)/(X+Y). For triheteromeric GluN1/2A/2B receptors (A/B), the weighted deactivation was 50 ms, which is faster than the deactivation rate assuming independent GluN2A and GluN2B subunits. From the receptors with a single crosslinked GluN2 subunit, we can estimate glutamate deactivation from single GluN2 subunits using Acc/A (73ms) and Bcc/B (680ms) receptors (Fig. 1C, D). If GluN2A and GluN2B subunits deactivate independently in A/B receptors the overall deactivation time constant should be: 1/(1/73 ms + 1/680 ms) = 66 ms, which is slower than the measured time constant 50 ms, and closer to the deactivation time constant calculated using the kinetics of Bcc/A and Acc/B (Fig. 1C, D, blue traces): 1/(1/71 ms + 1/248 ms) = 55 ms. This is consistent with GluN2A-dominant allosteric interactions enabling faster deactivation of A/B. To estimate whether the leak from A/A could account for the observed difference between measured and predicted time constants of the triheteromeric receptors (50 ms vs 66 ms), we determined the magnitude of A/A escape currents needed to change the deactivation time constant from the predicted 66 ms to the observed 50 ms. We assume that the A/A escape currents will be detected as a contribution to the fast component of the A/B deactivation course (see Table 1). We find that ~60% of amplitude of the measured current would have to be mediated by escaped A/A to cause the A/B deactivation time constant to be shifted from 66 ms to 50 ms (Fig. S4). This level of A/A escape currents is inconsistent with comparisons of ifenprodil inhibition between HEK293 cells and Xenopus oocytes using the same expression system (Hansen et al., 2014). Hansen et al. (2014) found almost identical levels of ifenprodil inhibition for A/B in HEK293 cells and Xenopus oocytes, which is inconsistent with ~60% of the current response in HEK293 cells being mediated by escaped A/A. The experiments were not randomized, but oocytes and HEK cells expressing a particular construct were randomly selected for recordings. Outliers were not excluded from analysis.
Evaluation of LBD crosslinking efficiency
When expressed in HEK cells, Acc/Bcc receptors produced maximum current in responses to 100 μM glycine and were insensitive to glutamate (Fig. S2), indicating the crosslink efficiency is nearly 100%. When co-expressing GluN1 with crosslinked GluN2-C1 and wildtype GluN2-C2 in Xenopus oocytes, the resulting receptors show enhanced glutamate potency and a decreased Hill coefficient, consistent of receptors with only one glutamate binding site (Fig. S3). Concentration-response data were fitted by equation: I = Imax/(1 + 10^((logEC50 − log[A]) * nH)), where Imax is the maximum current in response to the agonist, nH is the Hill slope, [A] is the agonist concentration, and EC50 is the agonist concentration that produces half-maximum response.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± standard error of the mean (S.E.M). Sample number (n) values are indicated in the results section and figures and refers to the total number of recorded HEK293 cells or oocytes. Statistical differences between groups were examined by One Way ANOVA with Tukey’s post hoc test using Prism 6 software (GraphPad) and all datasets are assumed to follow normal distribution based on Prism evaluation; P values less than 0.05 were considered statistically significant.
Supplementary Material
Highlights.
Allosterism occurs between GluN2 subunits in triheteromeric GluN1/2A/2B NMDARs.
These allosteric interactions are asymmetric and dominated by the GluN2A subunit.
GluN2A-dominant interactions span multiple domains in the NMDAR.
The allosteric interactions endow GluN1/2A/2B with the function of GluN1/2A NMDARs.
Acknowledgments
We thank Brett Carter and Delia Chiu from the Jahr laboratory and Gina Bullard, Feng Yi, and Genevieve Lind from the Hansen laboratory for their helpful discussions and technical assistance. This work was supported by the National Institutes of Health (R21NS091337, P20GM103546, and R01NS097536).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
W.S, K.B.H and C.E.J designed the experiments and wrote the paper, W.S conducted the experiments.
References and Notes
- Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol. 1994;347:150–160. doi: 10.1002/cne.903470112. [DOI] [PubMed] [Google Scholar]
- Andreescu CE, Prestori F, Brandalise F, D’Errico A, De Jeu MT, Rossi P, Botta L, Kohr G, Perin P, D’Angelo E, et al. NR2A subunit of the N-methyl D-aspartate receptors are required for potentiation at the mossy fiber to granule cell synapse and vestibulo-cerebellar motor learning. Neuroscience. 2011;176:274–283. doi: 10.1016/j.neuroscience.2010.12.024. [DOI] [PubMed] [Google Scholar]
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Benveniste M, Mayer ML. Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys J. 1991;59:560–573. doi: 10.1016/S0006-3495(91)82272-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AM. Constitutive activation of the N-methyl-D-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem. 2008;283:21519–21529. doi: 10.1074/jbc.M709190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmignoto G, Vicini S. Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science. 1992;258:1007–1011. doi: 10.1126/science.1279803. [DOI] [PubMed] [Google Scholar]
- Clements JD, Lester RA, Tong G, Jahr CE, Westbrook GL. The time course of glutamate in the synaptic cleft. Science. 1992;258:1498–1501. doi: 10.1126/science.1359647. [DOI] [PubMed] [Google Scholar]
- Clements JD, Westbrook GL. Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron. 1991;7:605–613. doi: 10.1016/0896-6273(91)90373-8. [DOI] [PubMed] [Google Scholar]
- Dai J, Zhou HX. Semiclosed Conformations of the Ligand-Binding Domains of NMDA Receptors during Stationary Gating. Biophys J. 2016;111:1418–1428. doi: 10.1016/j.bpj.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney AJ, Sedlak PL, Autuori E, Power JM, Sah P. Synaptic NMDA receptors in basolateral amygdala principal neurons are triheteromeric proteins: physiological role of GluN2B subunits. J Neurophysiol. 2013;109:1391–1402. doi: 10.1152/jn.00176.2012. [DOI] [PubMed] [Google Scholar]
- Dumas TC. Developmental regulation of cognitive abilities: modified composition of a molecular switch turns on associative learning. Prog Neurobiol. 2005;76:189–211. doi: 10.1016/j.pneurobio.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AC, Maisch US, Weishaupt JH, Kriegstein AR, Monyer H. NR2A subunit expression shortens NMDA receptor synaptic currents in developing neocortex. J Neurosci. 1997;17:2469–2476. doi: 10.1523/JNEUROSCI.17-07-02469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KA, McLaughlin N, Edbauer D, Phillips M, Bolton A, Constantine-Paton M, Sheng M. Distinct roles of NR2A and NR2B cytoplasmic tails in long-term potentiation. J Neurosci. 2010;30:2676–2685. doi: 10.1523/JNEUROSCI.4022-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011;71:1085–1101. doi: 10.1016/j.neuron.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jorgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol. 2013;84:114–127. doi: 10.1124/mol.113.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Manasian Y, Ivanov A, Tyzio R, Pellegrino C, Ben-Ari Y, Clapham DE, Medina I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Kussius CL, Popescu GK. NMDA receptors with locked glutamate-binding clefts open with high efficacy. J Neurosci. 2010;30:12474–12479. doi: 10.1523/JNEUROSCI.3337-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Lester RA, Clements JD, Westbrook GL, Jahr CE. Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature. 1990;346:565–567. doi: 10.1038/346565a0. [DOI] [PubMed] [Google Scholar]
- Lü W, Du J, Goehring A, Gouaux E. Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science. 2017 doi: 10.1126/science.aal3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel MA, Ryan TJ, Bell KF, Fowler JH, McMahon A, Al-Mubarak B, Komiyama NH, Horsburgh K, Kind PC, Grant SG, et al. The subtype of GluN2 C-terminal domain determines the response to excitotoxic insults. Neuron. 2012;74:543–556. doi: 10.1016/j.neuron.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Rauner C, Kohr G. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem. 2011;286:7558–7566. doi: 10.1074/jbc.M110.182600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EB, Ramoa AS. Enhanced NR2A subunit expression and decreased NMDA receptor decay time at the onset of ocular dominance plasticity in the ferret. J Neurophysiol. 1999;81:2587–2591. doi: 10.1152/jn.1999.81.5.2587. [DOI] [PubMed] [Google Scholar]
- Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, et al. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature. 1995;373:151–155. doi: 10.1038/373151a0. [DOI] [PubMed] [Google Scholar]
- Sanz-Clemente A, Gray JA, Ogilvie KA, Nicoll RA, Roche KW. Activated CaMKII couples GluN2B and casein kinase 2 to control synaptic NMDA receptors. Cell reports. 2013;3:607–614. doi: 10.1016/j.celrep.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- Stocca G, Vicini S. Increased contribution of NR2A subunit to synaptic NMDA receptors in developing rat cortical neurons. J Physiol. 1998;507(Pt 1):13–24. doi: 10.1111/j.1469-7793.1998.013bu.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Carvalho S, Grand T, Zhu S, Paoletti P. Controlling NMDA receptor subunit composition using ectopic retention signals. J Neurosci. 2014;34:16630–16636. doi: 10.1523/JNEUROSCI.2736-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TT, Badger JD, 2nd, Roche PA, Roche KW. Novel approach to probe subunit-specific contributions to N-methyl-D-aspartate (NMDA) receptor trafficking reveals a dominant role for NR2B in receptor recycling. J Biol Chem. 2010;285:20975–20981. doi: 10.1074/jbc.M110.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, McGinley MJ, Westbrook GL. Triheteromeric NMDA receptors at hippocampal synapses. J Neurosci. 2013;33:9150–9160. doi: 10.1523/JNEUROSCI.0829-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Simorowski N, Traynelis SF, Furukawa H. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun. 2011;2:294. doi: 10.1038/ncomms1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann RA, Fanger CM, Anderson DR, Sirivolu VR, Paschetto K, Gordon E, Virginio C, Gleyzes M, Buisson B, Steidl E, et al. MPX-004 and MPX-007: New Pharmacological Tools to Study the Physiology of NMDA Receptors Containing the GluN2A Subunit. PLoS ONE. 2016;11:e0148129. doi: 10.1371/journal.pone.0148129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- Yi F, Mou TC, Dorsett KN, Volkmann RA, Menniti FS, Sprang SR, Hansen KB. Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors. Neuron. 2016;91:1316–1329. doi: 10.1016/j.neuron.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JP, Constantine-Paton M. NR2A−/− mice lack long-term potentiation but retain NMDA receptor and L-type Ca2+ channel-dependent long-term depression in the juvenile superior colliculus. J Neurosci. 2007;27:13649–13654. doi: 10.1523/JNEUROSCI.3153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


