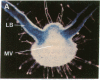
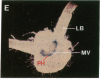
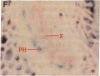
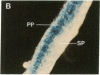
Abstract
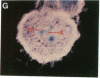
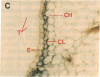
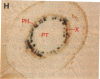
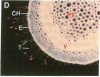
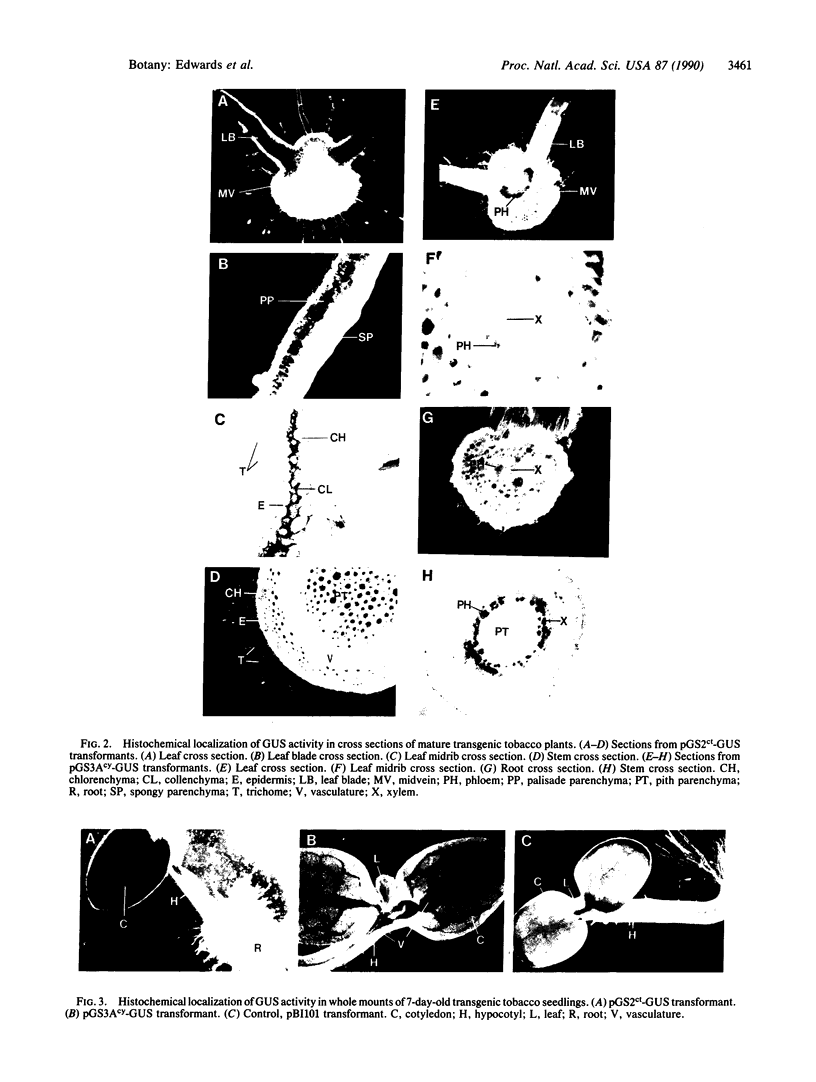
Chloroplast and cytosolic isoforms of glutamine synthetase (GS; EC 6.3.1.2) are encoded by separate nuclear genes in plants. Here we report that the promoters for chloroplast GS2 and cytosolic GS3A of Pisum sativum confer nonoverlapping, cell-specific expression patterns on the beta-glucuronidase (GUS) reporter gene in transgenic tobacco. The promoter for chloroplast GS2 directs GUS expression within photosynthetic cell types (e.g., palisade parenchymal cells of the leaf blade, chlorenchymal cells of the midrib and stem, and photosynthetic cells of tobacco cotyledons). The promoter for chloroplast GS2 retains the ability to confer light-regulated gene expression in the heterologous transgenic tobacco system in a manner analogous to the light-regulated expression of the cognate gene for chloroplast GS2 in pea. These expression patterns reflect the physiological role of the chloroplast GS2 isoform in the assimilation of ammonia generated by nitrite reduction and photorespiration. In contrast, the promoter for cytosolic GS3A directs expression of GUS specifically within the phloem elements in all organs of mature plants. This phloem-specific expression pattern suggests that the cytosolic GS3A isoenzyme functions to generate glutamine for intercellular nitrogen transport. In germinating seedlings, the intense expression of the cytosolic GS3A-GUS transgene in the vasculature of cotyledons reveals a role for cytosolic GS in the mobilization of seed storage reserves. The distinct, cell-specific patterns of expression conferred by the promoters for chloroplast GS2 and cytosolic GS3A indicate that the corresponding GS isoforms perform separate metabolic functions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- A simple and general method for transferring genes into plants. Science. 1985 Mar 8;227(4691):1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Benfey P. N., Ren L., Chua N. H. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989 Aug;8(8):2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984 Nov 26;12(22):8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Darnell J. E., Jr Transcriptional control of the mouse prealbumin (transthyretin) gene: both promoter sequences and a distinct enhancer are cell specific. Mol Cell Biol. 1986 Dec;6(12):4697–4708. doi: 10.1128/mcb.6.12.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke S. H., Schrader L. E., Miller M. G. Low Temperature Effects on Soybean (Glycine max [L.] Merr. cv. Wells) Free Amino Acid Pools during Germination. Plant Physiol. 1978 Oct;62(4):642–647. doi: 10.1104/pp.62.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. W., Coruzzi G. M. Photorespiration and light act in concert to regulate the expression of the nuclear gene for chloroplast glutamine synthetase. Plant Cell. 1989 Feb;1(2):241–248. doi: 10.1105/tpc.1.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B. G., Day H. M., Turton J. F., Shen W. J., Cullimore J. V., Oliver J. E. Two glutamine synthetase genes from Phaseolus vulgaris L. display contrasting developmental and spatial patterns of expression in transgenic Lotus corniculatus plants. Plant Cell. 1989 Apr;1(4):391–401. doi: 10.1105/tpc.1.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C., Oliver J. E., Forde B. G., Saarelainen R., Miflin B. J. Primary structure and differential expression of glutamine synthetase genes in nodules, roots and leaves of Phaseolus vulgaris. EMBO J. 1986 Jul;5(7):1429–1435. doi: 10.1002/j.1460-2075.1986.tb04379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B., Bouet C., King B., Layzell D., Jacobs F., Verma D. P. Glutamine synthetase genes are regulated by ammonia provided externally or by symbiotic nitrogen fixation. EMBO J. 1987 May;6(5):1167–1171. doi: 10.1002/j.1460-2075.1987.tb02350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B., Gadal P. Glutamine Synthetase in Rice: A COMPARATIVE STUDY OF THE ENZYMES FROM ROOTS AND LEAVES. Plant Physiol. 1980 Oct;66(4):619–623. doi: 10.1104/pp.66.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R. B., Klee H. J. Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: Role of T-DNA borders in the transfer process. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4428–4432. doi: 10.1073/pnas.83.12.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Burgess S. M., Hirsh D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore G. M., Shah D. M. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- McNally S. F., Hirel B., Gadal P., Mann A. F., Stewart G. R. Glutamine Synthetases of Higher Plants : Evidence for a Specific Isoform Content Related to Their Possible Physiological Role and Their Compartmentation within the Leaf. Plant Physiol. 1983 May;72(1):22–25. doi: 10.1104/pp.72.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin B. J. The location of nitrite reductase and other enzymes related to amino Acid biosynthesis in the plastids of root and leaves. Plant Physiol. 1974 Oct;54(4):550–555. doi: 10.1104/pp.54.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Sosa M., Sonnewald U., Frommer W., Stratmann M., Schell J., Willmitzer L. Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J. 1989 Jan;8(1):23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. R. Introduction, translocation, and distribution of viruses in plants. Adv Virus Res. 1965;11:163–221. doi: 10.1016/s0065-3527(08)60546-1. [DOI] [PubMed] [Google Scholar]
- Tingey S. V., Coruzzi G. M. Glutamine Synthetase of Nicotiana plumbaginifolia: Cloning and in Vivo Expression. Plant Physiol. 1987 Jun;84(2):366–373. doi: 10.1104/pp.84.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingey S. V., Tsai F. Y., Edwards J. W., Walker E. L., Coruzzi G. M. Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J Biol Chem. 1988 Jul 15;263(20):9651–9657. [PubMed] [Google Scholar]
- Tingey S. V., Walker E. L., Coruzzi G. M. Glutamine synthetase genes of pea encode distinct polypeptides which are differentially expressed in leaves, roots and nodules. EMBO J. 1987 Jan;6(1):1–9. doi: 10.1002/j.1460-2075.1987.tb04710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E. L., Coruzzi G. M. Developmentally Regulated Expression of the Gene Family for Cytosolic Glutamine Synthetase in Pisum sativum. Plant Physiol. 1989 Oct;91(2):702–708. doi: 10.1104/pp.91.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallsgrove R. M., Turner J. C., Hall N. P., Kendall A. C., Bright S. W. Barley mutants lacking chloroplast glutamine synthetase-biochemical and genetic analysis. Plant Physiol. 1987 Jan;83(1):155–158. doi: 10.1104/pp.83.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H. C., Powell G. K., Dekker E. E. Glutamine Synthetase of Germinating Peanuts : PROPERTIES OF TWO CHROMATOGRAPHICALLY DISTINCT FORMS AND THEIR ACTIVITY TOWARD 4-METHYLENEGLUTAMIC ACID. Plant Physiol. 1982 Jan;69(1):41–47. doi: 10.1104/pp.69.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]